Abstract
The small molecules thalidomide, lenalidomide, and pomalidomide induce the ubiquitination and proteasomal degradation of the transcription factors Ikaros (IKZF1) and Aiolos (IKZF3) by recruiting a Cys2-His2 (C2H2) zinc finger domain to Cereblon (CRBN), the substrate receptor of the CRL4CRBN E3 ubiquitin ligase. Here we screened the human C2H2 zinc finger proteome for degradation in the presence of thalidomide analogs, identifying 11 zinc finger degrons. Structural and functional characterization of the C2H2 zinc finger degrons demonstrates how diverse zinc finger domains bind the permissive drug-CRBN interface. Computational zinc finger docking and biochemical analysis predict that more than 150 zinc fingers bind the drug-CRBN complex in vitro, a larger number than previously anticipated, and we show that selective zinc finger degradation can be achieved through compound modifications. Our results provide a rationale for therapeutically targeting transcription factors that were previously considered undruggable.
Introduction
Thalidomide and its derivatives, lenalidomide and pomalidomide, are effective therapies for the hematologic malignancies multiple myeloma, del(5q) myelodysplastic syndrome, and mantle cell lymphoma (1). Thalidomide analogs bind Cereblon (CRBN), the substrate receptor of the CUL4-RBX1-DDB1-CRBN (CRL4CRBN) E3 ubiquitin ligase and alter its substrate selectivity to recruit, ubiquitinate and degrade seemingly unrelated proteins including Ikaros (IKZF1), Aiolos (IKZF3), Casein kinase 1 alpha (CK1α) and GSPT1 (2–7). Degradation of these targets in part explains the therapeutic effects of thalidomide analogs. IKZF1 and IKZF3 are lymphocyte lineage transcription factors (8, 9) that are essential for the survival of the malignant plasma cells in multiple myeloma. CK1α is essential for the survival of hematopoietic stem cells, and heterozygous deletion of its gene in del(5q) myelodysplastic syndrome provides a therapeutic window for eliminating the malignant stem cell clone (10).
IKZF1 and IKZF3 belong to the family of Cys2-His2 (C2H2) zinc finger (ZF) proteins (11). These ZF proteins share a conserved C2H2 ZF fold composed of a β-hairpin and an α-helix held together by pairs of zinc-coordinating cysteine and histidine residues. Because amino acids in the α-helical portion of some ZF domains recognize DNA base-pairs in a sequence-specific manner (12), the approximately 800 C2H2 ZF proteins are predicted to comprise the largest group of transcription factors in the human genome (13). Transcription factors have remained challenging drug targets due to the absence of druggable active sites (14). Thalidomide analogs, however, induce degradation of IKZF1 and IKZF3, raising the possibility that other C2H2 ZF-containing transcription factors are similarly destabilized.
E3 ubiquitin ligases recognize their substrates through degrons, short stretches of primary sequence that are necessary and sufficient for the interaction with substrate receptors of ubiquitin ligases (15). Previous work has implicated the second C2H2 ZF domain in IKZF1 and IKZF3 (5, 7), and ZF4 of ZFP91 as the drug-inducible degrons (5, 7, 16). Unexpectedly, the known thalidomide analog targets, IKZF1/3, ZFP91, CK1a and GSPT1 do not share obvious primary sequence similarity with the exception of a glycine residue located in a β-hairpin, which diverges from the canonical destruction motif paradigm. Although the majority of C2H2 ZFs are structurally similar to the ZF degrons of IKZF1/3 and ZFP91, with 4,661 out of 6,572 carrying a glycine residue at an equivalent position, proteome-wide mass-spectrometry demonstrated selective degradation of endogenous IKZF1/3, and ZFP91 in multiple cell lines (3, 4, 16). To identify determinants of drug-induced substrate specificity, we set out to characterize the human ZF ‘degrome’ amenable to degradation in the presence of CRL4CRBN and thalidomide analogs and examine whether compound modifications change ZF selectivity.
A C2H2 ZF library screen identifies novel degrons and protein targets
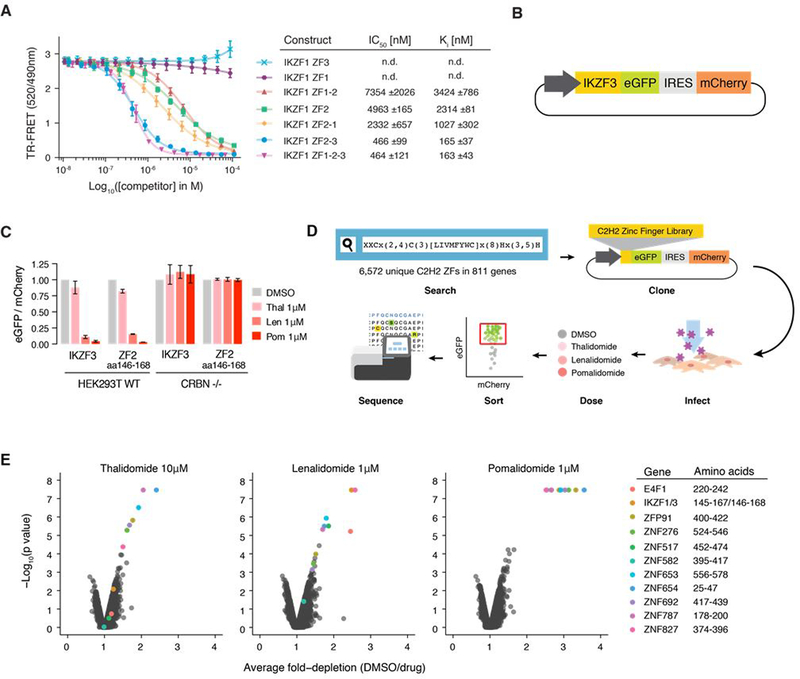
To characterize the minimal C2H2 ZF degron of IKZF1/IKZF3, we first generated different IKZF1 deletion constructs and measured their affinity for the drug-CRBN complex in vitro using time-resolved fluorescence resonance energy transfer (TR-FRET) (Fig. 1A and fig. S1, A and B). IKZF1 ZF2 (amino acid residues (aa) 141–174) was the shortest construct that showed binding to CRBN-pomalidomide, with a Ki of 2314 ±81 nM. Higher affinity binding to CRBN-pomalidomide in vitro was observed with an IKZF1 construct spanning ZF2 and ZF3 (aa 143–243(Δ197–238); Ki 165±37 nM). Replacing ZF3 with ZF1 in the context of the ZF2-ZF3 construct (ZF2-ZF1) decreased the binding affinity by a factor of 6 (Ki 1027±302 nM). IKZF1 binding to CRBN-pomalidomide is thus driven by ZF2 in vitro, with minor but specific contributions from the C-terminal ZF3.
Fig. 1. C2H2 ZF library screen identifies 11 C2H2 ZFs degraded in the presence of thalidomide, lenalidomide, or pomalidomide.
(A) TR-FRET counter titration. Unlabeled IKZF1 constructs (0.01–100 μM) titrated to preassembled Alexa488DDB1ΔB-CRBN-pomalidomide-biotinIKZF1ZF1−2−3 (200 nM Alexa488DDB1ΔB-CRBN, 100nM biotinIKZF1ZF1−2−3, 5μM pomalidomide). (B) Schematic of the protein degradation reporter vector (IRES: Internal ribosome entry site). (C) HEK293T WT and CRBN−/− cells expressing IKZF3 constructs in the degradation reporter were treated for 20 hours with DMSO or drug then analyzed by flow cytometry to quantify the DMSO-normalized ratio of eGFP/mCherry fluorescence (exp. rep. = 3, tech. rep. = 3, bar heights indicate mean of experimental replicates, error bars indicate 95% CI). (D) Schematic of the human C2H2 ZF library screen. (E) Average fold-depletion of sequencing read counts (DMSO/drug) and corresponding p values (empirical rank-sum test-statistic) for the 5,611 C2H2 ZFs with raw read count >200 in all three control replicates (exp. rep. = 3, labeled data points possess FDR<0.01 in at least one of the three drugs).
To identify the minimal construct required for IKZF1/IKZF3 degradation in cells, we created a lentiviral reporter vector that enabled us to use flow cytometry to compare the fluorescence of degron-tagged enhanced green fluorescent protein (eGFP) to untagged mCherry (Fig. 1B) (17). We transduced WT and CRBN−/− human embryonic kidney cells (HEK293T) with the degradation reporter containing IKZF3 deletion constructs and treated the cells with thalidomide, lenalidomide or pomalidomide. Deletion of ZF2 (aa 146–168) in full–length IKZF3 rendered the reporter resistant to drug treatment, whereas deletion of the other ZFs had little or no effect (fig. S1C). Accordingly, IKZF3 ZF2 (aa 146–168), which is identical to IKZF1 ZF2 (aa 145–167; fig. S1D), was sufficient to confer CRBN-dependent degradation of the reporter (Fig. 1C). Together, these results establish IKZF1/IKZF3 ZF2 as the minimal unit required for thalidomide analog-induced CRBN binding in vitro and for CRL4CRBN-dependent degradation in cells.
Having demonstrated that a single ZF is sufficient to induce degradation of the eGFP/mCherry reporter, we sought to screen the entire human C2H2 ZF proteome for degradation in the presence of thalidomide analogs. To analyze the human ZF ‘degrome’ independent of cell-type specific expression levels (fig. S1, E and F) and accessibility of ZFs in the context of full-length proteins engaged in macromolecular assemblies, cDNAs for 6,572 unique C2H2 ZFs from the human proteome matching the PROSITE (18) ZF motif x(2)-C-x(2,4)-C-x(3)-[LIVMFYWC]-x(7)-H-x(3,5)-H were synthesized and inserted into the degradation reporter vector. HEK293T cells were transduced with this C2H2 ZF library and then treated with dimethyl sulfoxide (DMSO), thalidomide, lenalidomide or pomalidomide. Fluorescence activated cell sorting (FACS) was used to isolate eGFP+/mCherry+ cells, and the relative number of read counts of each ZF was quantified by next-generation sequencing (Fig. 1D). A ZF was considered degraded if read counts were significantly under-represented in drug-treated eGFP+/mCherry+ cells relative to DMSO-treated control cells.
Of the 6,572 ZFs, 5,611 had sufficient representation in the sequencing data to be assayed (>200 read counts). At a false discovery rate (FDR) of <0.01, pomalidomide depleted 11 C2H2 ZFs, each from different proteins, including IKZF1/IKZF3 ZF2 (Fig. 1E). Lenalidomide and thalidomide targeted a variable subset of these 11 ZFs. When cloned into the degradation reporter vector and tested in isolation, the 11 ZFs exhibited degradation in the presence of all three compounds (Fig. 2A and fig. S2A).
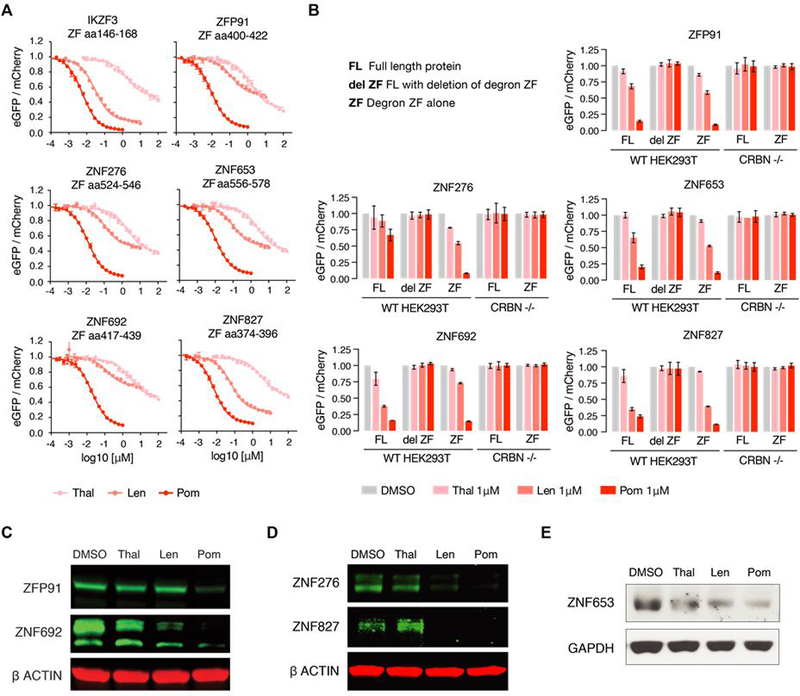
Fig. 2. Six proteins are degraded by thalidomide, lenalidomide, or pomalidomide conditional on the presence of a C2H2 ZF degron.
(A) HEK239T WT and CRBN−/− cells expressing the 11 ZFs identified in the screen in the degradation reporter were treated for 20 hours with drug then analyzed by flow cytometry to measure the DMSO-normalized ratio of eGFP/mCherry fluorescence (exp. rep. = 3, tech. rep. =3, dots represent average of experimental replicates, error bars indicate range). ZFs not shown are in fig S2A. (B) HEK293T WT or CRBN−/− cells expressing full-length (FL), FL with ZF deleted (del ZF), or ZF alone cDNA constructs were treated for 20 hours with DMSO or 1μM drug then analyzed using flow cytometry to measure the DMSO-normalized ratio of eGFP/mCherry fluorescence (exp. rep. = 3, tech. rep. = 3, bar heights indicate mean of exp. rep., error bars indicate 95% CI). Proteins not shown are in fig. S2B. (C) KG1 (human acute myeloid leukemia), (D) WM266–4 (human melanoma), and (E) MOLM-16 cells (human acute myeloid leukemia) were treated with DMSO or 1μM drug for 20 or 24 hours after which protein lysates were harvested, run on a polyacrylamide gel, and immunoblotted for the specified targets (images representative of 3 experimental replicates).
We next used the degradation reporter to determine whether the 11 ZFs destabilized their respective full-length protein and found that 6 of the 11 full-length proteins were degraded in the presence of drug (IKZF1/IKZF3, ZNF692, ZFP91, ZNF276, ZNF653 and ZNF827) (Fig. 2B and fig. S2B). All 6 ZFs that mediated degradation of their full-length protein carry an additional ZF C-terminal to the one identified in the library screen, whereas 5 of 6 ZFs that did not mediate degradation of their corresponding full-length protein do not posses a proximal C-terminal ZF. These results are consistent with our findings that while IKZF1/IKZF3 ZF2 constitutes the minimal ZF degron using the degradation reporter, an IKZF1 construct spanning ZF2 and ZF3 conferred higher affinity binding to CRBN-pomalidomide than ZF2 alone in vitro (Fig. 1A). We confirmed degradation of endogenous ZNF692, ZFP91, ZNF276, ZNF653 and ZNF827 in the presence of pomalidomide using western blotting (Fig. 2, C–E). Additionally, we demonstrated binding of hemagglutinin (HA)-tagged, full-length ZFP91 and ZNF692 to endogenous CRBN in the presence of all three compounds by immunoprecipitation (fig. S2C).
In summary, a screen of 6,572 C2H2 ZFs identified 11 C2H2 ZFs degraded in the presence of thalidomide, lenalidomide or pomalidomide, 6 of which mediate degradation of their respective full-length protein. Four of these full-length proteins were previously unknown thalidomide analog targets (fig. S2D).
Identification of amino acid loci critical to degradation
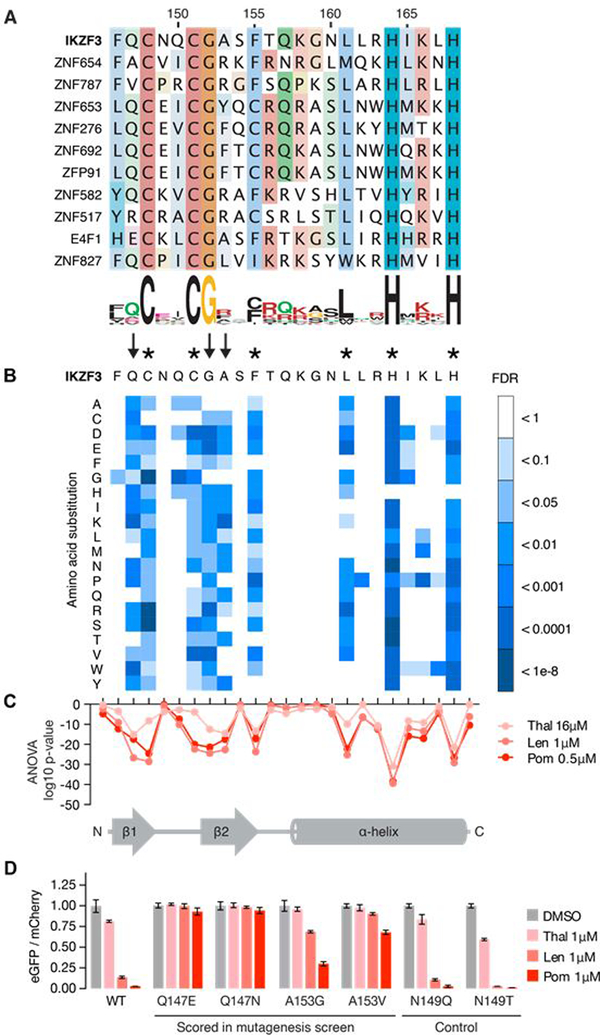
Sequence alignment of the C2H2 ZF hits (Fig. 3A) highlighted shared residues that are part of the PROSITE C2H2 ZF search motif (IKZF3 C148, C151, H164, H168, F155), residues that stabilize the ternary fold of the ZF domain (IKZF3 L161), and residues that frequently appear within C2H2 ZF domains (IKZF3 G152). The PROSITE ZF search motif covered sequences in which the β-hairpin zinc-coordinating cysteines are separated by 2, 3 or 4 residues (C-x(2,4)-C), but only C-x(2)-C ZFs were found destabilized in the screen (Fig. 3A). Insertion of a glycine residue between the β-hairpin residues N148 and Q149 of the IKZF1 ZF2-ZF3 construct (IKZF3 N149 and Q150; fig. S1D) compromised binding in vitro (fig. S3A). This suggests that the ZF ‘degrome’ is restricted to an inter-cysteine spacing of two residues. Besides the inter-cysteine spacing, residues that stabilize the ternary ZF fold, or residues that frequently appear in C2H2 ZF domains, no discernable sequence consensus could be detected among the 11 ZFs (Fig. 3A). The identified ZF degrons thus revealed structural features common to C2H2 ZF domains that alone cannot explain selective degradation of only 11 ZFs, while 4,661 structurally similar ZFs were present in the library.
Fig. 3. Identification of amino acids required for drug-induced degradation of a C2H2 ZF degron.
(A) Sequence alignment of the 11 C2H2 ZFs with FDR<0.01 in at least one drug condition (amino acids colored by property). (B) Saturation mutagenesis screen of IKZF3 aa 130–189 in presence of lenalidomide displayed as heat map of the FDR for mutant amino acids (unpaired, one-sided t-test, FDR correction performed within each column, tech. rep. = 3). Asterisks indicate amino acids required for the ZF fold and arrows indicate non-structural IKZF3 residues required for degradation. Complete results for all 60 amino acids are located in fig. S3C (C) ANOVA p values for difference in frequency of mutant amino acids at each position in IKZF3 aa 130–189 (DMSO vs drug). Complete results for all 60 aa are located in fig. S3D. (D) HEK293T cells expressing IKZF3 ZF2 constructs in the degradation reporter were treated for 20 hours with DMSO or 1μM drug after which flow cytometry was used to measure the DMSO-normalized ratio of eGFP/mCherry fluorescence (exp. rep. = 1, tech. rep. = 3, bar height is the average of tech. rep, error bars denote 95% CI).
To test whether the divergent, non-structural residues contribute to a functional ZF degron, we synthesized a mutagenesis library of IKZF3 spanning aa 130–189 such that at each of the 60 loci all 19 possible amino acid substitutions were represented. The pooled library was inserted into the degradation reporter vector and transduced into HEK293T cells. We treated the cells with DMSO, thalidomide, lenalidomide or pomalidomide, and used FACS to isolate drug-treated eGFP-/mCherry+ cells or DMSO-treated eGFP+/mCherry+ control cells. Next-generation sequencing was used to quantify the relative number of read counts for each substitution (fig. S3B).
The screen highlighted 9 loci in IKZF3 ZF2 whose amino acid identities were critical for degradation (Fig. 3, B and C, and fig. S3, C and D). These loci again included residues that maintain the tertiary ZF fold (IKZF3 C148, C151, F155, L161, H164, H168) and the β-hairpin glycine residue (IKZF3 G152). In addition, the screen highlighted two non-structural residues (IKZF3 Q147 and A153) that varied among the other ZF degrons (Fig. 3A), but were important for degradation of the IKZF3 ZF2 reporter (Fig. 3, B and C). Mutation of these two residues impaired degradation in validation experiments, whereas N149, a residue that was not highlighted in the mutagenesis screen, tolerated mutation (Fig. 3D). These data show that in addition to the residues maintaining the ZF fold, non-structural amino acids (IKZF3 Q147, G152 and A153) contribute to degron specificity.
Structures of CRBN bound to pomalidomide and two different ZF degrons
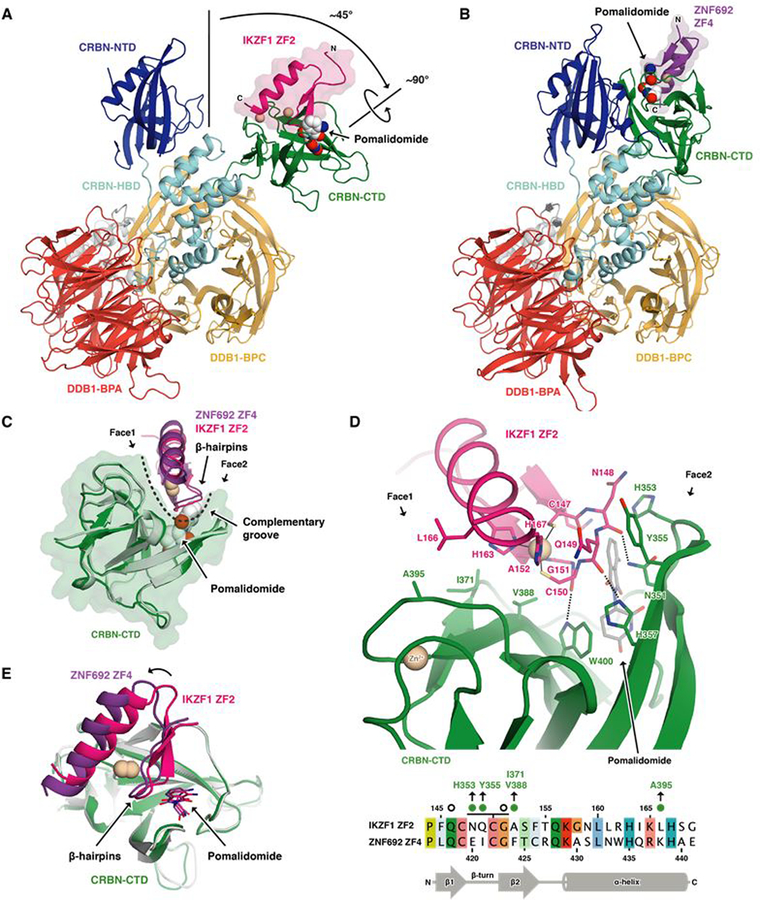
To examine how the varying, non-structural residues within ZF degrons contribute to selective CRBN binding, we determined crystal structures of DDB1ΔBPB-CRBN bound to pomalidomide and IKZF1 ZF2 (aa 141–174) or ZNF692 ZF4 (aa 416–442) (Fig. 4, A and B). Unlike previous CRBN structures, CRBN bound IKZF1 ZF2 in an ‘open’ conformation, with its N- and C-terminal domains separated and stabilized by crystal contacts (Fig. 4A and fig. S4A). As supported by the ZNF692 structure, release of the C-terminal domain is not required for ZF binding (Fig. 4B).
Fig. 4. The pomalidomide-CRBN interface accommodates C2H2 ZF degrons with distinct amino acid sequences and properties.
(A) Crystal structure of DDB1ΔB-CRBNΔN40 bound to pomalidomide and IKZF1ZF2. Zinc ions are shown as beige spheres. (B) Crystal structure of DDB1ΔB-CRBNΔN40 bound to pomalidomide and ZNF692ZF4. CTD, carboxy-terminal domain; HBD, helical bundle domain; NTD, amino-terminal domain. BPA, BPC: β-propeller A and B. (C) Superposition of the CRBN-CTDs bound to IKZF1ZF2 and ZNF692ZF4. A dashed line indicates the complementary groove. (D) Side chain interactions between IKZF1ZF2, CRBN and pomalidomide (dashed lines indicate hydrogen bonds) and sequence alignment of IKZF1ZF2 and ZNF692ZF4 (amino acids colored by property). Green dots or open circles indicate side chain interactions with CRBN or pomalidomide, respectively. A black line indicates ZF residues involved in backbone interactions with CRBN and/or pomalidomide. (E) Superposition of both CRBN-CTDs shows differences in IKZF1/ZNF692 ZF orientation.
Attempts to crystalize the IKZF1 ZF2-ZF3 construct resulted in poorly diffracting crystals. To dissect the contribution of the C-terminal ZF to binding affinity, we employed an unbiased docking approach that placed ZF3 on the neighboring N-terminal domain of ‘closed’ CRBN (fig. S4B). This model positions the C-terminus of ZF2 adjacent to the N-terminus of ZF3 consistent with the 5 amino acid linker between the two ZFs. The model highlights three consecutive arginine residues of ZF3 (IKZF1 R183-R185) at the CRBN-ZF2 interface that are not present in the ZF2-ZF1 construct (Fig. 1A and fig. S4C). Mutation of R185 to alanine, or replacement of all three arginines with the corresponding amino acids in ZF1 (IGP) reduced the affinity of the ZF2-ZF3 construct similar to that of ZF2-ZF1 (Fig. 1A) or ZF2 alone as predicted by our model (fig. S4D). These results show that IKZF1 ZF2 is the primary determinant for drug binding, and identify features of ZF3 that contribute to high affinity binding to the drug-CRBN complex (Fig. 1A and fig. S4B).
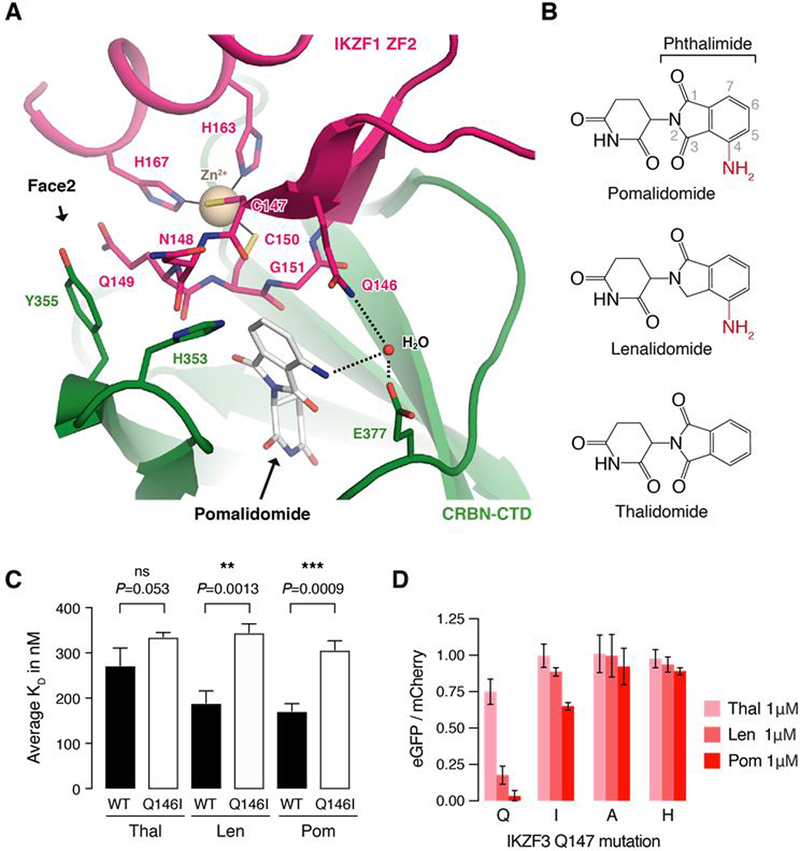
Crystal structures of CRBN-pomalidomide bound to IKZF1 ZF2 and ZNF692 ZF4 reveal the detailed side chain interactions between CRBN, pomalidomide and the ZFs. Both IKZF1 ZF2 and ZNF692 ZF4 bind a complementary groove on the CRBN C-terminal domain (Fig. 4C), interact with the compound through their β-hairpin loops, and match the overall backbone conformation of previous models of the ZF-drug-CRBN complex (fig. S4E). In each structure, the β-hairpin glycine of the ZF packs against the phthalimide ring of pomalidomide similar to the β-hairpins of CK1α (6) and GSPT1 (5), affirming the structural importance of a glycine residue at this position (Fig. 4D and fig. S4F). As the CRBN groove is narrow (Fig. 4C), ZF-CRBN interactions at both sides of the groove together determine binding. IKZF1 ZF2 N148 and Q149 (IKZF3 N149 and Q150; fig. S1D) form hydrophobic interactions through their amphipathic side chains with CRBN residues H353 and Y355 (Fig. 4D), explaining CRBN compatibility with diverse amino acids at these ZF positions. IKZF1 residues A152 and L166 (IKZF3 A153 and L167) face CRBN residues V388, I371 and A395 located on the opposite side of the groove (Fig. 4, C and D). While ZNF692 ZF4 and IKZF1 ZF2 show similar binding modes (Fig. 4C), the position of ZNF692 ZF4 is offset with respect to IKZF1 ZF2 (Fig. 4E and fig. S4, G and H), without significant differences in binding affinity (fig. S4I). Thus, the two structures demonstrate how CRBN accommodates ZF degrons with varying side chain properties at the ZF-CRBN interface (Fig. 3A; IKZF3 N148: E, N, K, R, V, P; Q149: I, V, R, L, A; A153: Y, F, L, R and L167: V, I, K, N).
IKZF1 Q146 forms the only side chain interaction with the compound (Fig. 5A), and mutation of the equivalent residue in IKZF3 (Q147) stabilized the reporter in the saturation mutagenesis experiment (Fig. 3, B and D). The IKZF1 Q146 side chain (IKZF3 Q147) packs against the phthalimide group of pomalidomide and forms a water-mediated hydrogen bond with the C4 amino group of the compound (Fig. 5, A and B and fig. S5A). Thalidomide lacks the C4 amino group (Fig. 5B) and mutation of IKZF1 Q146 (IKZF3 Q147) to isoleucine, which removes the ability to form a hydrogen bond with the drug, equalizes the binding affinity of IKZF1 ZF2-ZF3 to CRBN across the three compounds accordingly (Fig. 5C and fig. S5, B and C). Equivalent mutations in IKZF3 ZF2 (Q147I/A/H) stabilized the degradation reporter in cells (Fig. 5D, fig. S5D), explaining preferential binding of IKZF1/IKZF3 to CRBN engaged with pomalidomide or lenalidomide over thalidomide in vitro (6), and the contribution of this residue to compound selectivity in vivo (fig. S5E). Strikingly, 7 of the 11 ZF degrons carry an equivalent glutamine residue at this position (Fig. 3A).
Fig. 5. IKZF3 Q147 interacts with the amino group on pomalidomide.
(A) Side chain interactions between IKZF1 ZF2, CRBN, and pomalidomide. Dashed lines indicate hydrogen bonds. (B) Chemical structures of pomalidomide, lenalidomide, and thalidomide with differences highlighted in red. (C) Inhibitory constants (Ki) for wild-type (WT) and Q146I mutant IKZF1ZF2-ZF3 in the presence of thalidomide, lenalidomide, and pomalidomide in TR-FRET counter titration experiments. (D) HEK293T cells expressing IKZF3 ZF2 Q147 mutants in the degradation reporter were treated for 20 hours with DMSO or drug and then analyzed by flow cytometry to measure the DMSO-normalized ratio of eGFP/mCherry fluorescence (exp. rep. = 3, tech. rep. = 3, bar height is average of exp. rep, error bars indicate 95% CI). The complete set of Q147 amino acid mutants is located in fig. S5D.
ZF degrons show epistatic properties during CRL4CRBN engagement
The crystal structures demonstrate that ZFs with diverse amino acid sequences are compatible with the drug-CRBN interface. To test whether the variable residues are interchangeable between ZF degrons, we swapped the β-hairpin and α-helix of IKZF3 ZF2 with that of ZFP91 ZF4. Replacing the IKZF3 ZF2 β-hairpin with the ZFP91 ZF4 β-hairpin resulted in a construct that was degraded more efficiently than either of the parent molecules, however, replacing the IKZF3 α-helix with the ZFP91 α-helix resulted in a ZF domain that was resistant to degradation (fig. S5F). Furthermore, introducing a single residue mutation at the drug-ZF interface of IKZF3, Q147 to a glutamic acid residue, stabilized the IKZF3 reporter (IKZF3 ZF2 Q147E; Fig. 3D), whereas an equivalent glutamic acid allows robust reporter degradation in the sequence context of E4F1 (Fig. 3A and fig. S2A). Despite the low degree of sequence similarity across the 11 ZF degrons (Fig. 3A), only distinct amino acid combinations at the drug-CRBN interface result in degradation. ZF degrons are thus defined by the sequence context of their amino acid side chains that contact the drug-CRBN interface (Fig. 4D), which suggests an epistatic relationship of these variable ZF residues during CRL4CRBN engagement and explains the absence of a primary sequence consensus.
In silico identification of novel ZFs that interact with the drug-CRBN interface
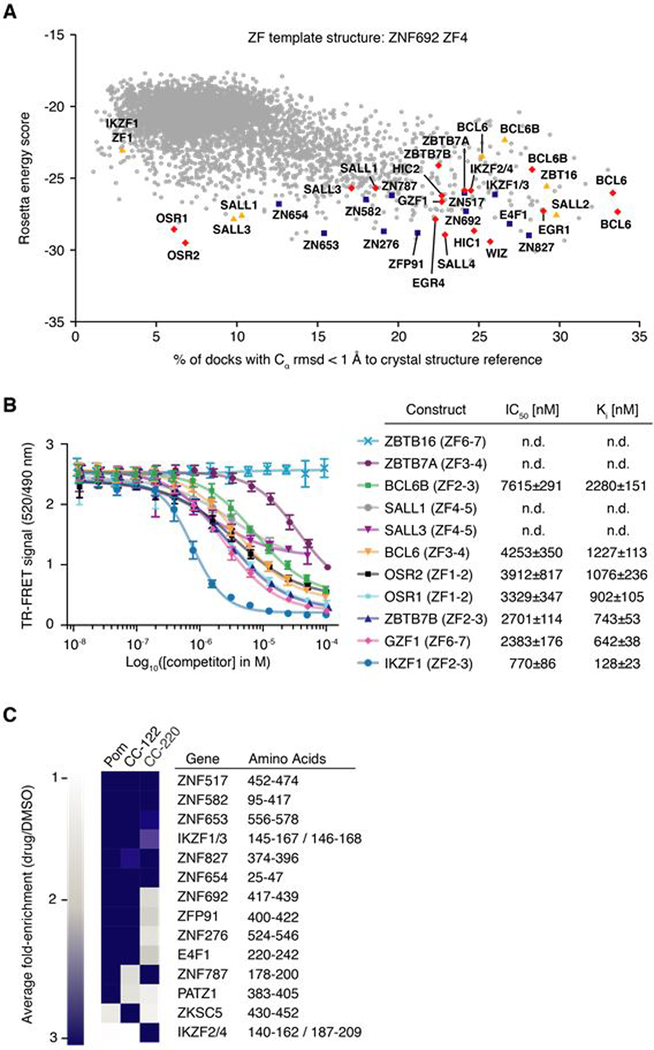
Only 11 of the 4,661 structurally similar ZFs were degraded in the library screen, yet the ZF degron appears to be complex. To create a semi-quantitative model of the observed degradation pattern with respect to our crystal structures, we employed computational docking to account for multiple, potentially epistatic ZF interactions at the drug-CRBN interface. The Rosetta software package (19) was used to computationally dock all human C2H2 ZFs into the complementary binding groove of CRBN. The Rosetta interface energy scores and the structural similarity to our ZF-pomalidomide-CRBN crystal structures were calculated to assess the docking trials (Fig. 6A and fig. S6, A and B). Using the ZNF692 template structure, we accurately predicted all 11 ZFs identified in the library screen (Fig. 6A), found 40 ZFs with lower interface energy scores than the ZNF692 ZF4 template, and identified 108 ZFs scoring better than the lowest ranking ZF found degraded in the library screen. Overall, different docking trials revealed an overlapping set of ~50–150 ZFs with interface scores similar to, or better than those of ZFs found degraded in the library screen (Fig. 6A and fig. S6, A and B), suggesting that an unexpectedly large number of ZFs are capable of binding the drug-CRBN interface.
Fig. 6. Thalidomide analogs with chemical modifications at the ZF-interface target different sets of C2H2 ZF degrons.
(A) ZF library docking results using RosettaDock with the CRBN C-terminal domain bound to pomalidomide and ZNF692ZF4 as a crystal structure reference. Blue squares indicate degraded ZFs from the library screen, red diamonds highlight ZFs that bind pomalidomide-CRBN in vitro, and yellow triangles mark ZFs for which CRBN-binding could not be confirmed in vitro or in vivo (B) TR-FRET counter titration. Unlabeled dual ZF candidate constructs (0.01–100 μM) titrated to preassembled Alexa488DDB1ΔB-CRBN-pomalidomide-biotinIKZF1ZF1−2−3 (500 nM Alexa488DDB1ΔB-CRBN, 50 nM biotinIKZF1ZF1−2−3, 5 μM pomalidomide). (C) Average fold-enrichment of sequencing read counts (drug/DMSO) for the 11 previously identified ZF degrons and 3 novel ZF hits.
Based on biological relevance, 21 of these ZFs were selected and tested for CRBN engagement in TR-FRET experiments, either as individual ZFs (fig. S6C) or fused to their respective C-terminal ZF (i.e. similar to the IKZF1 ZF2-ZF3 construct) (Fig. 6B). 16 ZFs bound CRBN-pomalidomide in vitro (BCL6, BCL6B, EGR1, EGR4, GZF1, HIC1, HIC2, SALL1, SALL3, SALL4, OSR1, OSR2, WIZ, ZBTB7A, ZBTB7B), with similar (WIZ ZF7) or lower binding affinities (~5–20 fold) than the respective IKZF1 ZF2-ZF3 or ZF2 constructs (Fig. 6B and fig. S6, C, D, and E). Analogously, IKZF1 ZF1, a ZF that was not predicted to bind CRBN in silico (Fig. 6A), did not bind CRBN in vitro (fig. S6C). Thus, of the 33 ZFs individually tested in this study, 28 are recruited to pomalidomide-engaged CRBN. Our in silico ZF docking approach is thus capable of predicting ZFs that interact with the drug-CRBN complex with over 80% accuracy (fig. S6F).
Those ZFs that bound CRBN-pomalidomide in the biochemical assay were subsequently tested for degradation in cells using the degradation reporter. Of the 16 ZFs, WIZ ZF7 showed partial degradation in the presence of pomalidomide, the best in vitro binder in the set (fig. S6E and fig. S7, A and B). Examination of published proteome-wide mass spectrometry data confirmed degradation of endogenous, full-length WIZ in WSU-DLCL2 (human diffuse large B cell lymphoma) and TMD8 (human diffuse large B cell lymphoma) cells in the presence of lenalidomide and the thalidomide analog CC-122 (fig. S7C) (20). As the majority of these ZFs are recruited to pomalidomide-engaged CRBN in vitro, but not destabilized by the same compound in cells, our results suggest that small changes in ZF-binding affinity disproportionally influence ZF degradation in cells.
Different thalidomide analogs target distinct sets of ZFs for degradation
We next examined whether thalidomide derivatives with chemical alterations at the drug-ZF interface induce degradation of the computationally predicted and biochemically validated ZF hits that are not destabilized by pomalidomide in cells. We therefore treated cells expressing these ZFs in the eGFP/mCherry degradation reporter with the previously reported thalidomide derivatives CC-122 (20), CC-220 (21) and CC-885 (5) (fig. S7, A and B). The modified thalidomide analogs induced mild but significant degradation for some of these ZFs, including 2 ZFs destabilized by CC-122 (WIZ ZF6, BCL6 ZF3), 2 by CC-220 (BCL6B ZF2, HIC2 ZF5), and 8 by CC-885 (BCL6 ZF3, BCL6B ZF2, OSR1 ZF1, ZBTB7B ZF2, SALL3 ZF4, SALL4 ZF2, ERG1 ZF3, SALL1 ZF4). While the effects are small, our findings suggest that different thalidomide analogs target different ZFs for degradation, including ZFs identified by computational docking, rendering them suitable candidates for drug development.
To comprehensively determine if the chemically distinct thalidomide analogs induce degradation of different sets of ZFs, we repeated the C2H2 ZF library screen in the presence of pomalidomide, CC-122, and CC-220 (Fig. 6C, fig. S8, A and B). The 11 previously identified ZF hits again scored in the screen with pomalidomide (fig. S8B). Consistent with our earlier finding that CC-122 and CC-220 were capable of degrading ZFs not affected by pomalidomide, all three thalidomide analogs exhibited distinct ZF degradation patterns across the ZF library (Fig. 6C). Notably, IKZF2 ZF2 was selectively degraded by CC-220 (Fig. 6C). IKZF2 ZF2 differs from IKZF3 ZF2 by a single amino acid substitution at the drug-ZF interface, Q147H (Fig. 5A and fig. S1D). This amino acid change decreases binding to CRBN-pomalidomide in vitro by a factor 2–3 (fig. S5B), and stabilizes the respective ZF reporter in cells (Fig. 5D). In validation experiments, the IKZF2 ZF2 reporter was degraded by more than 40% in the presence of CC-220 (fig. S8, C and D). These results demonstrate that thalidomide analogs with chemical alterations at the drug-ZF interface promote degradation of distinct sets of ZFs and are capable of converting ZFs that bind CRBN-pomalidomide weakly in vitro into degraded ZFs in cells.
Discussion
We utilized a combination of functional and structural approaches to define the molecular basis of C2H2 ZF recruitment to the drug-engaged CRL4CRBN ubiquitin ligase. Thalidomide, lenalidomide and pomalidomide mediate CRL4CRBN-dependent degradation of a larger number of proteins than previously appreciated through a C2H2 ZF degron (3, 4, 16). We identified 15 individual ZFs and 7 full-length ZF-containing proteins that are degraded by thalidomide derivatives in functional or computational screens. Crystal structures, in vitro binding, and cellular degradation assays illustrate that 28 ZFs (including IKZF2) with diverse amino acid sequences bind the same drug-CRBN interface (fig. S6E).
The majority of E3 ligase degrons are characterized by conserved primary sequence motifs that lie in unstructured regions of the protein substrate (15). In contrast, the more than 28 C2H2 ZFs accommodated by the drug-CRBN interface show surprisingly little sequence conservation, apart from residues that stabilize the ternary ZF fold. ZF degrons bind a complementary groove on the CRBN surface that fits the overall shape of the C2H2 ZF domain, bringing different amino acids of different ZF degrons in contact with the same drug-CRBN interface. Substitution of a ZF residue at one site of the CRBN interface can be compensated by amino acid changes at another site of the ZF, explaining the observed epistatic properties of the ZF degrons. Recruitment and degradation are thus influenced by the complementarity of the substrate to the drug-CRBN binding groove and the sequence context of the contacting ZF residues, making the overall “shape” of the ZF the important binding determinant, rather than its primary sequence. In addition to the complex ZF-CRBN side chain interaction, direct contacts between thalidomide analogs and the ZF provide additional layers of specificity (fig. S5D). Unbiased functional and computational approaches are thus required to identify the full complement of ZF degrons capable of drug-dependent binding and degradation.
The discovery that multiple C2H2 ZFs are drug targets raises the possibility that a large number of ZFs could be subject to thalidomide analog-based degradation. Supporting this notion, 28 of the ~50–150 additional C2H2 ZFs nominated by in silico docking were shown to bind the drug-CRBN complex, demonstrating that the CRBN surface is more permissive than suggested by the number of targets identified in the ZF library screens. Small differences in affinity between the ZF and the drug-CRBN complex in vitro translate into large differences in degradation in cells, which are likely due to competition for CRBN occupancy by multiple ZFs as well as other substrates that utilize the same binding surface (fig. S5, Fig. 6, C and D and fig. S7). If ZF degradation depends on the ZF occupancy of CRL4CRBN, it is possible that high local concentration of the ZF or CRL4CRBN, or low concentrations of competing ZFs, could compensate for low binding affinity leading to degradation under such conditions. Given the CRL4CRBN protein architecture and its ~100 Å ubiquitination radius (6, 7, 22), we expect degradation of full-length ZF proteins to be less dependent on lysine accessibility, and instead primarily determined by the protein synthesis rate and the affinity/binding kinetics of the internal ZF degron to CRBN, which can be modulated by the drug.
We observed that thalidomide analogs with chemical modifications at the drug-ZF interface are capable of converting ZFs with weak affinity for the CRBN-pomalidomide complex into degraded targets. Most strikingly, IKZF2 ZF2 is not degraded by pomalidomide, lenalidomide, or CC-122, but is efficiently degraded by CC-220. The crystal structures suggest that ZF side chains, particularly those proximal to the drug at IKZF1 position 146, 148 and 153 (IKZF3 147, 149 and 154), interact with thalidomide analogs, and that chemical alterations can modulate ZF specificity (fig. S5D). ZFs that bind weakly but are not degraded in response to one compound could therefore serve as starting points for the development of new thalidomide analogs that selectively and efficiently degrade such ZFs. Several of the ZFs identified as degraded in functional experiments, or in vitro binders that could be targeted with further drug development, have been implicated in human disease. For example, BCL6 is an oncoprotein in lymphomas, ZFP91 is implicated in NF-κB signaling (23), and ZNF827 is reported to be an essential scaffolding protein for alternative lengthening of telomeres (ALT) (24). HIC1, HIC2, GZF1, OSR1, OSR2, and SALL4 have all been implicated in development and could contribute to developmental abnormalities in fetuses exposed to thalidomide (25–30).
The ~800 C2H2 ZF-containing proteins are the largest class of putative transcription factors in the human proteome (13). Our results suggest that degradation of C2H2 ZF-containing proteins through novel thalidomide derivatives may be a generalizable paradigm for targeting “undruggable” transcription factors and provides a structural and functional starting point to explore the extent to which CRBN-binding small molecules may be used to target ZF proteins for therapeutic intervention.
Materials and Methods
Cell Lines
| # | Cell Line | Tissue/Cell Type | Source | Medium | Temp. | Sex | Identity Verified? |
|---|---|---|---|---|---|---|---|
| 1 | HEK293T | Human embryonic kidney cell | Genetic perturbations platform, Broad Institute | DMEM + 10% fetal bovine serum | 37°C | Female | No |
| 2 | KG1 | Acute myeloid leukemia (AML) | Broad Institute Cell Line Repository | RPMI + 10% Fetal bovine serum + penicillin and streptomycin | 37°C | Male | Yes, by Broad cell line repository |
| 3 | WM226.4 | Melanoma | Broad Institute Cell Line Repository | DMEM + 10% Fetal Bovine Serum | 37°C | Unknown | Yes, by Broad cell line repository |
| 4 | MOLM16 | Acute myeloid leukemia (AML) | ATCC | RPMI + 20% fetal bovine serum + 10 ng/mL hGM-CS | 37°C | Female | No |
Compounds
| # | Compound | Company | Order # |
|---|---|---|---|
| 1 | Thalidomide | SelleckChem | S1193 |
| 2 | Lenalidomide | SelleckChem | No longer in stock |
| 3 | Pomalidomide | SelleckChem | S1567 |
| 4 | CC-122 | MedChem Express | HY-100507 |
| 5 | CC-885 | Axon MedChem | 2645 |
| 6 | CC-220 | Medkoo | 528831 |
Degradation Reporter Vectors
| # | Name | Contents | Resistance Marker |
Backbone | Addgene | Used for |
|---|---|---|---|---|---|---|
| 1 | Artichoke | SFFV.BsmBICloneSite-17aaRigidLinker-EGFP.IRES.mCherry.cppt.EF1α.PuroR | AmpicillinR |
Lentiviral (pLKO, taken from lentiGuide-Puro, Addgene #52963) |
#73320 | Full-length proteins, IKZF3 aa130–189, saturation mutagenesis screen |
| 2 | Cilantro 2 | PGK.BsmBICloneSite-10aaFlexibleLinker-EGFP.IRES.mCherry. cppt.EF1α.PuroR | #74450 | C2H2 zinc finger library screen, C2H2 zinc fingers in isolation |
Antibodies
| # | Target | Species | Mono/Poly | Company | Cat. # |
|---|---|---|---|---|---|
| 1 | human ZFP91 | rabbit | Polyclonal | Bethyl Laboratories | A303–245A |
| 2 | human ZNF692 | rabbit | Polyclonal | Abcam | ab204595 |
| 3 | human CRBN | rabbit | Polyclonal | Novus Biologicals | NBP1–91810 |
| 4 | HA-affinity gel | mouse | Monoclonal | Sigma | E6779 |
| 5 | human ZNF276 | rabbit | Polyclonal | Novus | NBP1–03342 |
| 6 | human ZNF827 | rabbit | Polyclonal | Sigma | HPA021166 |
| 7 | human ZNF653 | mouse | Polyclonal | Novus Biologicals | H00115950-B01P |
| 8 | human β-Actin | Mouse | Monoclonal | Cell Signaling Technology | 3700 |
| 9 | human GAPDH | Rabbit | Monoclonal | Cell Signaling Technology | 2118 |
Protein degradation reporter analysis
Two versions of the protein degradation reporter were used for the assays described in the main text: “Artichoke”, which possesses a 17aa rigid linker at the N-terminus of eGFP, was used for full-length proteins or large fragments (Addgene #73320, Benchling link to vector map and sequence: https://benchling.com/s/dRHlqBaZ). “Cilantro 2”, which possesses a 10aa flexible linker at the N-terminus of eGFP, was used for individual C2H2 zinc fingers (Addgene #74450, Benchling link to vector map and sequence: https://benchling.com/s/k2PYU86Q).
HEK293T cells were lentivirally transduced with the respective construct in the protein degradation reporter vector and selected with 2μg/mL puromycin for at least 2 days to select for cells stably expressing the reporter. Following a 20 hour drug treatment, flow cytometry was used to quantify eGFP and mCherry fluorescence. In FLOWJO (flow cytometry analysis software), a parameter was derived which calculated the eGFP/mCherry fluorescence ratio on a single-cell basis. The geometric mean of the ratio for a given drug-treated sample was normalized to the average of three DMSO-treated controls. For each experiment, three technical replicates and three experimental replicates were performed unless otherwise indicated.
C2H2 zinc finger library screen
To curate a list of C2H2 zinc fingers in the human proteome we used PROSITE (18) (http://prosite.expasy.org/scanprosite/) to search the proteome for occurrences of the C2H2 motif X-X-C-x(2,4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3,5)-H (parameters: UniprotKB, SwissProt Database, no alternative splice variants). A total of 6,819 C2H2 ZFs were found (6,572 unique) in 811 genes. The zinc finger amino acid sequences were reverse translated into codon-optimized DNA sequences in silico and appended with BsmBI restriction enzyme sites. The ssDNA oligonucleotide library was then synthesized in a pooled format using array-based synthesis (CustomArray, http://customarrayinc.com/). The library was converted to dsDNA by low cycle PCR amplification with NEBNext polymerase (NEB M0541L), digested with BsmBI (NEB R0580), purified using the Qiagen MinElute PCR purification kit (Qiagen 28004), ligated into the “Cilantro 2” degradation reporter vector (Addgene #74450), and purified using the QIAquick PCR purification kit (Qiagen 28104). ElectroMAX Stbl4 Competent bacteria (Thermo Fischer Scientific #11635018) were transformed with the library and plated onto large-format format LB plates with carbepencillin as chemical selection and grown at 30°C overnight. Resulting colonies were scraped, pooled, and maxiprepped using the Qiagen Plasmid Plus Maxi Kit (Qiagen #12963). Lentivirus was generated from the C2H2 zinc finger library using HEK293T cells.
To execute the screen, we performed three experimental replicates of the following: on day 0 HEK293T cells were lentivirally infected with the library, and on day 1 were selected with 2 μg/mL of puromycin. On day 3 the cells were plated in puromycin-free media, then on day 4 the cells were treated for 20 hours with either DMSO, thalidomide (10 μM), lenalidomide (1 μM), or pomalidomide (1 μM). A baseline control of unsorted DMSO-treated cells were harvested and pelleted, and the DMSO and drug-treated cells then underwent fluorescence-activated cell sorting for eGFP+/mCherry+ and EGFP-/mCherry+ cells (SONY Cell Sorter). Genomic DNA was harvested from the baseline control and sorted cells using the QiaAmp DNA Blood Mini and Midi kits (Qiagen #51104 and #51183). The zinc finger sequences were then PCR amplified from the genomic DNA with primers appended with Illumina primers and barcodes. As a control, the original plasmid DNA library was also amplified. The resulting amplicons were then pooled and sequenced on the Illumina NextSeq.
To reduce noise and false positives caused by zinc fingers with low numbers of reads, zinc fingers with less than 200 raw reads in the DMSO-treated unsorted control were removed from the library. This threshold reduced the library size from 6819 to 5856. Reads were aligned to the library using bowtie and read counts for each experimental condition were normalized as reads per million. To identify C2H2 zinc fingers degraded in the presence of drug, we calculated the DMSO/drug fold-depletion in eGFP+/mCherry+ read counts for each experimental replicate. Zinc fingers were then ranked on basis of their fold-depletion, and the three experimental replicates were integrated by calculating a rank-sum empirical p-value and FDR for each zinc finger.
The library screens with CC-122 and CC-220 were performed as described above with small modifications. HEK293T were infected in three experimental replicates and selected as above. On day 4 post-infection, HEK293T cells were treated for 20 hours with either DMSO, pomalidomide (1 μM), CC-122 (1 μM) or CC-220 (1 μM). and 5 % low eGFP cells were sorted either on H800S Cell Sorter or ASTRIOS EQ Sorter. Sorted cells were lysed (1 mM CaCl2, 3 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 10 mM Tris pH 7.5, 0.2 mg/ml Proteinase K) and subjected to two-step PCR amplification with primers appended with Illumina adaptors, barcodes, and “staggers” to increase library sequence diversity. The resulting amplicons were pooled and sequenced on Illumina NovaSeq S2 platform.
Read count tables were generated with ShortRead and dplyr package (R version 3.4.2). To reduce noise, three filtering criteria were applied independently to remove the least reliable zinc fingers a) zinc fingers with less than 500 reads in any of three DMSO replicate b) zinc fingers with coefficient of variance in top 0.3 quantile between three DMSO replicates c) zinc fingers with coefficient of variance in top 0.3 quantile between three drug replicates. These thresholds reduced the library size from 6,819 to 3,206 ZFs. ZFs were ranked based on the fold-enrichment in normalized read counts in the eGFP-/mCherry+ sorting gate (drug/DMSO) and p-values and FDRs were calculated as above.
Immunoblots
KG1, WM266–4, or MOLM-16 cells were treated with either DMSO or 1 μM of thalidomide, lenalidomide, or pomalidomide, after which protein lysates were harvested and run on a polyacrylamide gel, transferred to nitrocellulose membrane, and blotted for ZFP91, ZNF692, ZNF276, ZNF827, or ZNF653. Immunoprecipitation was achieved by overexpressing HA-tagged ZFP91 and ZNF692 in KG1 cells. Protein lysates were collected and then incubated with anti-HA sepharose beads (Sigma #E6779) in the presence of 1 μM of thalidomide, lenalidomide, or pomalidomide overnight at 4°C. Proteins were eluted using HA-peptide and incubating at 30°C for 5 minutes. Eluates and whole cell lysate were run on a polyacrylamide gel, transferred to a nitrocellulose membrane, and immunoblotted for HA and CRBN.
Saturation Mutagenesis
The saturation mutagenesis library for IKZF3 130–189 aa was designed such that each of the possible 19 mutant amino acids were represented at every locus in the 60 aa construct, yielding a library size of ~1,200 elements (20 possible aa × 60 loci). The ssDNA oligonucleotide library was synthesized on an Agilent array, PCR amplified to form dsDNA, and purified of frame-shifting mutations in bacterial vector as described in Melnikov et. al. Nucleic Acids Research 2014. The resulting plasmid library was digested with BsmBI and cloned into the lentiviral “Artichoke” degradation reporter vector (Addgene #73320). The pooled plasmid library was used to make a lentiviral library from HEK293T cells.
To execute the screen we performed three technical replicates of the following: on day 0 HEK293T cells were transduced with the saturation mutagenesis lentiviral library, and on day 1 were selected in 2 μg/mL puromycin. On day 4 the cells were passaged into puromycin-free media and the infection efficiency was calculated to be 50%. On day 5 the cells were plated for dosing with drug, and then on day 6 the cells were dosed for 20 hours with DMSO, 16 μM thalidomide, 1 μM lenalidomide, or 0.5 μM pomalidomide. On day 7 the cells underwent fluorescence activated cell sorting of the eGFP+/mCherry+ (DMSO) or eGFP-/mCherry+ (thalidomide, lenalidomide, pomalidomide) cell population on a BD FACS Aria.
Genomic DNA was harvested from the sorted cell populations using the QIAamp DNA blood micro kit (Qiagen 56304). A two-step PCR was used to amplify the IKZF3 mutagenesis library from the genomic DNA and add Illumina primers, adaptors, and sample barcodes. The resulting amplicons were then pooled and sequenced on the Illumina MiSeq.
Reads were aligned to the library to obtain read counts for each mutation and wild-type sequences. Only reads which contained a single mutation were analyzed. Read counts within each experimental condition were normalized and log2 transformed. To identify amino acid mutations which impaired degradation we compared the frequency of each mutation in the DMSO-treated eGFP+/mCherry+ population and drug-treated eGFP-/mCherry+ populations using an unpaired, one-sided t-test with FDR correction performed within each of the 60 loci. To more generally identify loci in which mutations impaired degradation we used an ANOVA test statistic to compare the relative frequency of reads containing mutations in the DMSO and drug-treated conditions.
Protein purification
Human wild-type and mutant versions of DDB1 (Uniprot entry Q16531), CRBN (Q96SW2), IKZF1 (Q13422), ZNF692 (Q9BU19), BCL6 (P41182), BCL6B (Q8N143), ZBT7A (O95365), ZBT7B (O15156), GZF1 (Q9H116), ZBT16 (Q05516), HIC1 (Q14526), HIC2 (Q96JB3), WIZ (O95785), SALL1 (Q9NSC2), SALL2 (Q9Y467), SALL3 (Q9BXA9), SALL4 (Q9UJQ4), EGR1 (P18146), EGR4 (Q05215), OSR1 (Q8TAX0) and OSR2 (Q8N2R0) were subcloned into pAC-derived vectors (31) and recombinant proteins expressed as N-terminal His6, His-Spy, StrepII or StrepII-Avi fusions in Trichoplusia ni High Five insect cells using the baculovirus expression system (Invitrogen). His-DDB1ΔBPB-StrepII-CRBN and Spy-His-DDB1ΔBPB-His-CRBN were purified as described previously (6). High Five insect cells expressing truncated versions of wild-type or mutant StrepII-Avi-IKZF1 (ZF1–2–3: amino acids (aa) 83–255(Δ197–238), ZF1–2: aa 109–174, ZF2–3: aa 141–243(Δ197–238), ZF1: aa 114–142, ZF2: aa 141–174, ZF3: aa 169–243(Δ197–238)), ZNF692 (ZF4–5: aa 413–476, ZF4: aa 416–442), BCL6 (ZF3–4: aa 570–625), BCL6B (ZF2–3: aa 353–409, ZF3–4: aa 380–438), ZBT7A (ZF3–4: aa 434–492), ZBT7B (ZF2–3: aa 370–426), GZF1 (ZF6–7: aa 459–515), ZBT16 (ZF6–7: aa 542–598), HIC1 (ZF5: aa 587–615), HIC2 (ZF5: aa 585–613), WIZ (ZF6: aa 765–793, ZF7: aa 867–894), SALL1 (ZF4–5: aa 729–790, ZF9: aa 1158–1185), SALL2 (ZF9: aa 397–425), SALL3 (ZF4–5: aa 703–763, ZF9: aa 1137–1164), SALL4 (ZF2: aa 406–432), EGR1 (ZF3: aa 392–420), EGR4 (ZF3: aa 537–565), OSR1 (ZF1–2: aa 172–227) or OSR2 (ZF1–2: aa 169–224) were lysed by sonication in 50 mM Tris-HCl pH 8.0, 500 mM NaCl, 0.25 mM tris(2-carboxyethyl)phosphine (TCEP), 1 mM phenylmethylsulfonylfluoride (PMSF) and 1× protease inhibitor cocktail (Sigma-Aldrich). Following ultracentrifugation, the soluble fraction was passed over Strep-Tactin Sepharose affinity resin (IBA) and eluted in 50 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.25 mM TCEP, 2.5 mM D-desthiobiotin. Affinity tags were removed by overnight TEV protease treatment. For ion exchange chromatography, the protein was diluted 1:1 with buffer A (50 mM Tris-HCl pH 6.8, 10 mM NaCl, 0.25 mM TCEP) and passed over an 8 mL Poros 50HQ column. The flow through was passed over an 8 mL Poros 50HS column and eluted over a salt gradient mixing buffer A and buffer B (50 mM Tris-HCl pH 6.8, 1 M NaCl, 0.25 mM TCEP) over 15 column volumes to a final ratio of 80% buffer B. Poros 50HS peak fractions were concentrated and subjected to size exclusion chromatography in 50 mM HEPES pH 7.4, 200 mM NaCl and 0.25 mM TCEP. The concentrated proteins were flash cooled in liquid nitrogen and stored at −80°C.
Crystallization and data collection
The IKZF1 protein solution for crystallization contained 70 μM His-DDB1ΔBPB-StrepII-CRBNΔN40 with 80 μM pomalidomide and 350 μM cleaved IKZF1-ZF2 in size exclusion buffer. The DDB1ΔBPB-CRBNΔN40-pomalidomide-IKZF1ZF2 complex crystallized in an optimized Morpheus HT condition (32), containing 100 mM Morpheus buffer system 1 pH 6.5, 10 % (v/v) Morpheus NPS solution, 15 % (v/v) ethylene glycol and 9.5 % (w/v) poly(ethylene glycol) 5000 monomethyl ether (PEG 5K MME) in MRC 2 well format vapor diffusion crystallization plates (Swissci). Plates were incubated at 19°C and crystals appeared after 1–2 days. Crystals were flash cooled in liquid nitrogen in reservoir solution supplemented with 5 % (v/v) ethylene glycol for cryo-protection. Diffraction data were collected at the Swiss Light Source (beamline PXIII) with a Pilatus 2M detector (Dectris) at a wavelength of 1 Å and from crystals cooled to 100 K. Data were indexed and integrated with DIALS (33) and scaled with AIMLESS supported by other programs of the CCP4 suite (34), and converted to structure factor amplitudes with STARANISO (35).
ZNF692 protein solution for crystallization contained 70 μM His-DDB1ΔBPB-StrepII-CRBNΔN40 with 80 μM pomalidomide and 350 μM cleaved ZNF692-ZF4 in size exclusion buffer. Initial crystals grew after mixing the protein solution equally with a reservoir containing 70 mM Tris-HCl pH 8.5 and 14% (w/v) PEG 6000. These crystals were used for iterative streak seeding and seeds were added one day after pre-mixing reservoirs containing 70 mM Tris-HCl pH 7.0–8.5 and 13.7–14.5 % (w/v) PEG 5K MME equally with the protein solution using EasyXtal 15-well hanging drop vapor diffusion crystallization plates (Qiagen #132006). The final crystals grew over several days at 19°C with a reservoir containing 14.1% (w/v) PEG 5K MME and 70 mM Tris-HCl pH 7.5. Crystals were cryo-protected in a buffer containing 70 mM HEPES pH 7.5, 14.1% (w/v) PEG 5K MME and 20% (v/v) glycerol. Diffraction data were collected at the Swiss Light Source (beamline PXI) with an Eiger X 16M detector (Dectris) at a wavelength of 1 Å from crystals cooled to 100 K. Data were indexed and integrated by XDS (36), scaled with XSCALE, and converted to structure factor amplitudes with XDSCONV.
Structure determination and model building
The DDB1ΔBPB-CRBNΔN40-pomalidomide-IKZF1ZF2 complex crystallized in space group P212121 with four complexes in the crystallographic asymmetric unit (ASU). PHASER (37) was used to determine the structure by molecular replacement (MR) using a crystallographic model of DDB1ΔBPB-CRBNΔN40 (PDB entry 5FQD) omitting the CRBN-CTD, and our IKZF1ZF2 homology model (6) as search models. The DDB1ΔBPB-CRBNΔN40-pomalidomide-ZNF692ZF4 complex crystallized in space group P21 with two complexes in the ASU. PHASER was used to determine the structure by MR using a crystallographic model of DDB1ΔBPB-CRBNΔN40 (PDB entry 5FQD) as the search model. The initial models were improved by iterative cycles of building with COOT, and refinement using phenix.refine (38) and autoBUSTER (39), with ligand restraints generated with Grade (Global Phasing). Structural figures were generated with PyMOL (Schrödinger), and model quality was assessed with MOLPROBITY (40). Data processing statistics, refinement statistics and model quality parameters are summarised in Table S1.
Labeling of SpyCatcher with Alexa488-maleimide
Purified SpyCatcher (S50C) was incubated with DTT (8 mM) at 4°C for 1 h. DTT was removed using a S200 16/60 gel filtration column (GE healthcare) in a buffer containing 50 mM Tris-HCl pH 7.3 and 150 mM NaCl. Alexa488-C5-maleimide (Invitrogen) was dissolved in 100 % DMSO and mixed with SpyCatcher to achieve fourfold molar excess of Alexa488-C5-maleimide. SpyCatcher labelling was carried out at room temperature (RT) for 3 h in a vacuum desiccator and stored overnight at 4 °C. Labeled SpyCatcher was purified on a S200 16/60 gel filtration column in 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.25 mM TCEP and 10 % (v/v) glycerol, concentrated by ultrafiltration (Millipore), flash frozen (~50 μM) in liquid nitrogen and stored at −80 °C. Purified Spy-His-DDB1ΔBPB-His-CRBN was mixed with an equimolar concentration of Alexa488-SpyCatcher, incubated for 1 h at room temperature (RT), flash frozen in liquid nitrogen and stored at −80 °C (~35–45 μM).
Biotinylation of IKZF1
Purified StrepII-Avi-tagged IKZF1ZF1−2−3 was biotinylated in vitro at a concentration of 25–50 μM by incubation with final concentrations of 2.5 μM BirA enzyme and 0.2 mM D-biotin in 50 mM HEPES pH 7.4, 200 mM NaCl, 10 mM MgCl2, 0.25 mM TCEP and 20 mM ATP. The reaction was incubated for 1 h at RT and stored at 4°C for 14–16 h. Biotinylated IKZF1ZF1−2−3 was purified by gel filtration chromatography and stored at −80°C (~25 μM).
Time-resolved fluorescence resonance energy transfer (TR-FRET)
Increasing concentrations of Alexa488-SpyCatcher-labeled Spy-His-DDB1ΔBPB-His-CRBN (Alexa488DDB1ΔBPB-CRBN) were added to a mixture of biotinylated IKZF1ZF1−2−3 (biotinIKZF1ZF1−2–3) at 90 nM, terbium-coupled streptavidin at 4 nM (Invitrogen) and thalidomide analogs at 5 μM (final concentrations) in 384-well microplates (Greiner #784076) in a buffer containing 50 mM Tris pH 7.5, 100 mM NaCl, 0.1 % pluronic acid and 1 % DMSO (see also figure legends). Before TR-FRET measurements, reactions were incubated for 5 min at RT. After excitation of terbium (Tb) fluorescence at 337 nm, emission at 490 nm (Tb) and 520 nm (Alexa488) were measured with a 70 μs delay to reduce background fluorescence and the reactions were followed by recording 60 data points of each well over 1 h using a PHERAstar FS microplate reader (BMG Labtech). The TR-FRET signal of each data point was extracted by calculating the 520:490 nm ratio. Data were analysed with Prism 7 (GraphPad) assuming equimolar binding of biotinIKZF1ZF1–2–3 to Alexa488DDB1ΔBPB-CRBN using the equations described previously (6).
Counter titrations with unlabeled proteins were carried out by mixing 0.4–1 μM Alexa488DDB1ΔBPB-CRBN with 200 nM biotinIKZF1ZF1−2−3 in the presence of 8 nM terbium-coupled streptavidin and 10 μM thalidomide analog. After incubation for 15 min on ice, increasing amounts of unlabeled ZF constructs (0.02–200 μM) were added to the preassembled Alexa488DDB1ΔBPB-CRBN/biotinIKZF1ZF1−2−3 complexes in a 1:1 volume ratio and incubated for 5 min at RT. TR-FRET data were acquired as described above. The 520:490 nm ratios were plotted to calculate the half maximal inhibitory concentrations (IC50) assuming a single binding event using Prism 7 (GraphPad). IC50 values were converted to the respective Ki as described previously (41). Three technical replicates were carried out per experiment. The standard deviation was derived from the sum of the mean absolute error of 10 measurements of each data point and the standard deviation of the technical replicates.
Docking simulations
Docking simulations were carried out with the RosettaDock pipeline from the Rosetta molecular suite (version 3.8 release 2017.29.59598) (19). A non-redundant list of human ZF motifs was extracted from PROSITE with the search pattern (x(2)-C-x(2,4)-C-x(3)-[LIVMFYWC]-x(8)-H-x(3,5)-H), corresponding to the ZF constructs used for the cellular library screen. For the final docking simulation screens, the ZF boundaries were extended to correspond to the crystallization constructs, and the simulations were restricted to 4,645 of these ZFs matching the pattern (x(6)-C-x(2)-C-G-x(2)-[LIVMFYWC]-x(8)-H-x(3)-H-x(2)), constituting the major of class of C2H2-type ZFs, which are 21 residues long and contain a Gly residue following the second zinc-coordinating Cys residue. Docking was carried out between the CRBNCTD (320–425 aa) bound to pomalidomide and its structural zinc ion, and the ZF library. The CRBN-pomalidomide-ZF copies best described by electron density in their crystal structures (chains B and C in both crystal structures) were used as templates for ZF model generation, and as reference for root-mean-square derivation (RMSD) scoring of the docking simulations. Alternative ZF crystal structure templates were also selected from the Protein Databank (PDB) based on their scoring in initial docking simulations with either of the crystallized ZF sequences. ZF models were generated with the ‘align-and-mutate’ tools in COOT (42). Models with no further modification and versions optimized with Rosetta ‘relax’ were used in docking simulations.
After ‘prepacking’ optimizing of the models with Rosetta, CRBN-pomalidomide and the ZF were separated by 10 Å. One thousand docking simulations were then carried for each ZF, with the starting position randomly perturbed by up to 3° and 8 Å. Constraints were included to preserve the coordination geometry of the structural zinc ions. The docking simulations were evaluated with the beta_nov16 score function, which readily discriminated IKZF1ZF2 from IKZF2ZF2 whose domains differ only by a single amino acid (IKZF1 Q146H). For analysis of the various docking screens, scattering plots were constructed of the mean top five interface scores against the percentage of docked models with a C⍺ RMSD < 1 Å with respect to a crystallographic model of the complex. ZFs mapping to the funnel region of the scatter plot were scrutinized further.
To identify potential binding modes, IKZF1ZF3 was docked onto the CRBN-pomalidomide-IKZF1ZF2 complex with ClusPro (43) and the global docking procedure in Rosetta. The CRBN-pomalidomide-IKZF1 ZF2 model was extracted from the Ikaros crystal structure (chains B and C) and CRBN N-terminal domain was reoriented to match its usual domain-closed conformation as found in the ZNF692 crystal structure. Docking was carried out with IKZF1ZF3 homology models constructed using IKZF1ZF2 from the Ikaros crystal structure or PDB entry 4M9E. For Rosetta, the docking simulations were carried out as before except the starting positions of the models were rotated randomly, and docking was repeated 200,000 times for each run.
Supplementary Material
ACKNOWLEDGMENTS
We thank Donna Neuberg and Robert Redd for statistical analysis of the saturation mutagenesis screen, Cong Zhu for helpful discussions on how to clone the saturation mutagenesis library, Daniel Haldar for biological characterization of the novel targets, Jenny Chen for her assistance with analyzing the C2H2 ZF library screen, Patricia Rogers for help with FACS, and Shourya S. Roy Burman for advice with Rosetta docking. The Broad, Brigham and Women’s Hospital, Harvard, and MGH have filed patent applications directed to the work described in this paper.
Funding: B.L.E. received funding from the NIH (R01HL082945 and P01CA108631), the Howard Hughes Medical Institute, the Edward P. Evans Foundation, and the Leukemia and Lymphoma Society. N.H.T received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No [666068]). Q.L.S. was supported by award Number T32GM007753 from the National Institute of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences or the National Institutes of Health. G.P. was supported by the Human Frontier Science Program (HFSP Long-Term Fellowship LT000210/2014) and the European Molecular Biology Organization (EMBO Advanced Fellowship aALTF 761–2016). M.S. received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 702642. B.J.L. was supported by the National Health and Medical Research Council (Early Career Fellowships Grant APP1124979).
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: All data will be made publically available and materials will be available upon request.
References
- 1.Fink EC, Ebert BL, The novel mechanism of lenalidomide activity. Blood. 126, 2366–2369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu G et al. , The Myeloma Drug Lenalidomide Promotes the Cereblon-Dependent Destruction of Ikaros Proteins. Science. 343, 305–309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krönke J et al. , Lenalidomide Causes Selective Degradation of IKZF1 and IKZF3 in Multiple Myeloma Cells. Science. 343, 301–305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krönke J et al. , Lenalidomide induces ubiquitination and degradation of CK1α in del(5q) MDS. Nature. 523, 183–188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matyskiela ME et al. , A novel cereblon modulator recruits GSPT1 to the CRL4CRBN ubiquitin ligase. Nature, 1–24 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Petzold G, Fischer ES, Thomä NH, Structural basis of lenalidomide-induced CK1α degradation by the CRL4CRBN ubiquitin ligase. Nature, 1–16 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Fischer ES et al. , Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature, 1–16 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgopoulos K et al. , The Ikaros gene is required for the development of all lymphoid lineages. Cell. 79, 143–156 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Wang JH et al. , Aiolos regulates B cell activation and maturation to effector state. Immunity. 9, 543–553 (1998). [DOI] [PubMed] [Google Scholar]
- 10.Schneider RK et al. , Role of Casein Kinase 1A1 in the Biology and Targeted Therapy of del(5q) MDS. Cancer Cell. 26, 509–520 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobb BS et al. , Targeting of Ikaros to pericentromeric heterochromatin by direct DNA binding. Genes & Development. 14, 2146–2160 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavletich NP, Pabo CO, Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 261, 1701–1707 (1993). [DOI] [PubMed] [Google Scholar]
- 13.Najafabadi HS et al. , C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat. Biotechnol 33, 555–562 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Dang CV, Reddy EP, Shokat KM, Soucek L, Drugging the “undruggable” cancer targets. Nat Rev Cancer. 207, 73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guharoy M, Bhowmick P, Sallam M, Tompa P, Tripartite degrons confer diversity and specificity on regulated protein degradation in the ubiquitin-proteasome system. Nature Communications. 7, 10239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.An J et al. , pSILAC mass spectrometry reveals ZFP91 as IMiD-dependent substrate of the CRL4(CRBN) ubiquitin ligase. Nature Communications. 8, 15398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Global protein stability profiling in mammalian cells. Science. 322, 918–923 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Sigrist CJA et al. , New and continuing developments at PROSITE. Nucleic Acids Res. 41, D344–7 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaudhury S et al. , Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS ONE. 6, e22477 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagner PR et al. , CC-122, a pleiotropic pathway modifier, mimics an interferon response and has antitumor activity in DLBCL. Blood. 126, 779–789 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matyskiela ME et al. , A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos. J. Med. Chem., acs.jmedchem.6b01921 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Angers S et al. , Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature. 443, 590–593 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Jin X et al. , An atypical E3 ligase zinc finger protein 91 stabilizes and activates NF-kappaB-inducing kinase via Lys63-linked ubiquitination. Journal of Biological Chemistry. 285, 30539–30547 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Conomos D, Reddel RR, Pickett HA, NuRD-ZNF827 recruitment to telomeres creates a molecular scaffold for homologous recombination. Nat. Struct. Mol. Biol 21, 760–770 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Valenta T, Lukas J, Doubravska L, Fafilek B, Korinek V, HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. EMBO J. 25, 2326–2337 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dykes IM et al. , HIC2 is a novel dosage-dependent regulator of cardiac development located within the distal 22q11 deletion syndrome region. Circ. Res 115, 23–31 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Vallecillo-García P et al. , Odd skipped-related 1 identifies a population of embryonic fibro-adipogenic progenitors regulating myogenesis during limb development. Nature Communications. 8, 1218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mechanisms of Development. 107, 175–179 (2001). [DOI] [PubMed] [Google Scholar]
- 29.Morinaga T et al. , GDNF-inducible zinc finger protein 1 is a sequence-specific transcriptional repressor that binds to the HOXA10 gene regulatory region. Nucleic Acids Res. 33, 4191–4201 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohlhase J et al. , Mutations at the SALL4 locus on chromosome 20 result in a range of clinically overlapping phenotypes, including Okihiro syndrome, Holt-Oram syndrome, acro-renal-ocular syndrome, and patients previously reported to represent thalidomide embryopathy. J. Med. Genet 40, 473–478 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abdulrahman W et al. , A set of baculovirus transfer vectors for screening of affinity tags and parallel expression strategies. Anal. Biochem 385, 383–385 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Gorrec F, The MORPHEUS protein crystallization screen. J Appl Crystallogr. 42, 1035–1042 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Waterman DG et al. , The DIALS framework for integration software. CP Newslett. Protein Crystallography. 49, 13–15 (4AD). [Google Scholar]
- 34.Winn MD et al. , Overview of the CCP4 suite and current developments. Acta Crystallogr Sect D Biol Crystallogr. 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tickle IJ et al. , STARANISO. Cambridge, United Kingdom Global Phasing Ltd; (2017). [Google Scholar]
- 36.Kabsch W, IUCr, Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr Sect D Biol Crystallogr. 66, 133–144 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy AJ et al. , Phaser crystallographic software. J Appl Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Afonine PV et al. , Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr Sect D Biol Crystallogr. 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bricogne GBE, BUSTER version 2.11.5. Global Phasing Ltd. (2011). [Google Scholar]
- 40.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cer RZ, Mudunuri U, Stephens R, Lebeda FJ, IC50-to-Ki: a web-based tool for converting IC50 to Ki values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res. 37, W441–W445 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Comeau SR, Gatchell DW, Vajda S, Camacho CJ, ClusPro: an automated docking and discrimination method for the prediction of protein complexes. Bioinformatics. 20, 45–50 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.