Abstract
Various stresses increase disease susceptibility and accelerate aging, and increasing evidence suggests that these effects can be transmitted over generation. Epidemiological studies suggest that stressors experienced by parents affect the longevity of their offspring, possibly by regulating telomere dynamics. Telomeres are elongated by telomerase and shortened by certain stresses as well as telomere repeat-containing RNA (TERRA), a telomere transcript. However, the mechanism underlying the transgenerational effects is poorly understood. Here, we show that TNF-α, which is induced by various psychological stresses, induces the p38-dependent phosphorylation of ATF7, a stress-responsive chromatin regulator, in mouse testicular germ cells. This caused a release of ATF7 from the TERRA gene promoter in the subtelomeric region, which disrupted heterochromatin and induced TERRA. TERRA was transgenerationally transmitted to zygotes via sperm and caused telomere shortening. These results suggest that ATF7 and TERRA play key roles in paternal stress-induced telomere shortening in the offspring.
INTRODUCTION
There has been a long-standing interest in whether environmental factors such as stress modulate the epigenetic modifications, and could thereby have long-lasting effects on health, sometimes even in subsequent generations. Various stresses increase disease susceptibility and accelerate aging (1), and increasing evidence suggests that these effects can be transmitted over generation (2). Epidemiological studies suggests that stressors experienced by parents affect the longevity of their offspring (3), possibly by regulating telomere dynamics (4). Telomeres, which consist of tandem TTAGGG repeats and are associated shelterin multi-protein complex, maintain the integrity of chromosome ends during cell division (5,6). In most somatic cells, telomere length shortens with each cell division and is therefore an important marker of aging (7). Some epidemiological studies suggested telomere length is maternally and paternally inherited (8–10), but the mechanism remains unknown.
Telomeres can be elongated enzymatically by telomerase, a complex consisting of a catalytic subunit (TERT) and an RNA subunit, which counterbalances the effects of cell division (5,6,11). Several types of stress decrease telomere length that may, in part, affect aging: exposure to psychosocial stress is associated with telomere shortening (12); prenatal stress exposure causes shorter telomere length later in life (13). In addition, in vitro study demonstrated that telomere length is shortened by oxidative stress (14), which was also supported by some empirical studies using in model organisms (15). Recently we demonstrated that the stress-responsive chromatin regulator ATF7 mediates telomere shortening induced by TNF-α (16), which is induced by various stresses including psychological stress (17). Telomerase is mainly recruited to telomeres through TPP1 (18–21), while the Ku complex was also shown to recruit telomerase to telomere (22). ATF7 and telomerase are localized on telomeres via interactions with the Ku complex. In response to TNF-α, ATF7 was phosphorylated by p38, leading to the release of ATF7 and telomerase from telomeres, which causes telomere shortening (17).
Telomere regions are transcribed from the adjacent subtelomeric heterochromatin regions by RNA polymerase II in a conserved manner from yeast to humans which generates TERRA (telomeric repeat-containing RNAs) (23,24). Thus, TERRA contains the telomere repeat sequence in addition to sequences unique to the subtelomeric region of each chromosome, and ranges in size from 100 bases to more than 100 kb. TERRA expression is regulated developmentally and also in the cell-cycle dependent manner (24,25). Early observations indicated that TERRA is localized to the ends of mammalian chromosomes with the foci associated with inactive X chromosome (24,26). More recent studies showed that TERRA is localized to the pseudoautosomal region of sex chromosomes and specific genes on the autosomes, as well as the telomeric regions (27). Multiple in vitro and in vivo studies indicated that TERRA inhibits the telomerase activity and shortens telomere length (24,27–32). TERRA was also shown to form RNA-DNA hybrids at telomeres and to stimulate homologous recombination between telomeric ends by recruiting Rad51 (25,33–35). Furthermore, TERRA contributes to form telomeric heterochromatin by interacting with HP1, histone H3K9 methyltransferases, and shelterins (36).
ATF7 is a vertebrate member of the ATF2 subfamily of transcription factors, which belong to the ATF/CREB superfamily (37–39). ATF2 proteins are phosphorylated by p38 in response to various stresses, including inflammatory cytokines, psychological stress, and pathogen infection (40,41). In the absence of stress, yeast Atf1, vertebrate ATF7 and Drosophila ATF2 (dATF2) contributes to the formation of heterochromatin and heterochromatin-like structure by recruiting histone H3K9 tri- and di-methyltransferases independently from the RNAi-dependent mechanism, thus silencing the transcription of target genes (42–45,16). In response to various stresses, ATF7 and dATF2 are phosphorylated, which causes the release of ATF7, dATF2 and histone H3K9 tri- and di-methyltransferases from chromatin; this leads to transcriptional activation that can be maintained for long periods (43,45,16), and in some cases it is transmitted to the next generation (44). Thus, ATF7 functions as the stress-responsive chromatin regulator.
In the present study, we have asked whether treatment of male mice with TNF-α, which is induced by various stresses including psychological stress, affects the telomere length of offspring. Paternal TNF-α treatment induces TERRA in testicular germ cells via the p38-dependent phosphorylation of ATF7 and a disruption of subtelomeric heterochromatin. TERRA is transmitted to zygotes via sperm, resulting to telomere shortening in the male offspring.
MATERIALS AND METHODS
Mice
Congenic Atf7+/– mice in the C57BL/6J genetic background were described previously (43). The mice used in this study were maintained under specific pathogen-free conditions on a 12 h light–dark cycle and fed a normal diet (CE-2 from CLEA Inc., composed of 12 kcal% fat, 29 kcal% protein and 59 kcal% carbohydrates). Experiments were performed in accordance with the guidelines of the Animal Care and Use Committee of the RIKEN Institute.
Paternal TNF-α injection and preparation of MEFs and PMBCs
Male wild-type (WT) and Atf7+/– mice aged 7–8 weeks were used for TNF-α injection. Typically, TNF-α (10 μg/kg weight) was intraperitoneally administered to mice daily for 6 weeks. Then, 8–9 week-old female mice were mated within 3 days. MEFs were prepared from E14.5 embryos. PMBCs were prepared from blood collected from 3- or 8-week-old pups.
Measurement of telomere length by Q-FISH
Telomere length in MEFs was analyzed by Q-FISH as reported previously (46) using three MEF lines from independent pregnant mice. MEFs were cultured for 72 h in DMEM containing 10% FBS, treated with colcemid (0.1 μg/ml, Wako, Japan) for 4 h, and then subjected to hypotonic shock and methanol/acetic acid fixation. Cell suspensions were dropped onto clean glass slides and used for hybridization experiments. The metaphase chromosomes were hybridized using the peptide nucleic acid–FISH method (47). A Cy3-labeled (CCCTAA)3 peptide nucleic acid (PNA) probe (Telo C, catalogue number F1002; Fasmac, Japan) was used to label the telomeres. The chromosomes were counterstained with TO-PRO-3 Iodide (Thermofisher, T3605). Q-FISH and image analysis were performed as described previously (46,47). The fluorescence intensity of telomeres was analyzed with the TFL-Telo V2 software package (Terry Fox Laboratory, BC Cancer Research Center, Canada). Telomere signals were analyzed in each group of samples, and the telomere intensity of individual arms in metaphase spreads was measured. Abnormal telomere structures were quantified using captured images.
Measurement of telomere length by real-time PCR
DNA was purified using the DNeasy Blood & Tissue Kit (Qiagen). Average telomere length was measured by real-time quantitative PCR as previously described. In this assay, the average telomere length ratio is determined by quantifying telomeric DNA using a specifically designed primer sequence, and then dividing that value by the level of a single-copy gene measured in the same sample (36B4). The primer sequences used are shown in Supplementary Table S1. Real-time PCR was performed using 1.5 ng of DNA as a template in an ABI 7500 real-time PCR instrument with SYBR Green PCR Master Mix (Applied Biosystems) as previously described (48,49). The PCR conditions for telomeres were as follows: 95°C for 10 min, and 25 cycles of 95°C for 15 s and 54°C for 2 min. The PCR conditions for 36B4 were as follows: 95°C for 10 min, and 35 cycles of 95°C for 15 s, 52°C for 20 s, and 72°C for 32 s. For each PCR reaction, a standard curve was generated using serial dilutions of known amounts of DNA. The telomere signal (T) was normalized to the signal from the single-copy gene 36B4 (S) to generate a T/S ratio, indicative of relative telomere length.
Measurement of telomere length by TRF (telomere restriction fragment) assay
MEFs (5 × 106) were embedded in agarose using a CHEF Genomic DNA Plug kit (Bio-Rad) and digested overnight with excess amounts of restriction enzymes MboI. Cells in agarose were loaded on 1% agarose gels and separated by electrophoresis in 0.5 × TBE, Resolved DNA was transferred to Biodyne B membrane (Pall Biosupport) by capillary transfer. The membrane was hybridized overnight in hybridization buffer (7% sodium dodecyl sulfate, 1% BSA, 0.5 M phosphate [pH 7.2], and 1 mM EDTA) at 42°C with 32P-labeled oligonucleotide telomeric probes (Supplementary Table S1). Signals were visualized and quantitated on an Image Analyzer 2500 (Fuji).
TERRA FISH
TERRA FISH was performed using MEFs or testicular germ cells essentially as described previously (23). Cells were permeabilized with CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES [pH 7.0], 0.5% Triton X-100 and 10 mM vanadyl ribonucleoside complex [VRC]) and fixed in 4% paraformaldehyde. After dehydration, cells were hybridized in hybridization buffer (2× SSC, 2 mg/ml BSA, 10% dextran sulphate sodium, 20% formamide, 10 mM VRC, and 0.3 ug/ml cy3-TelC) overnight at 37°C. Then, cells were washed three times in 2× SSC and 50% formamide and three times in 2× SSC at 39°C, each 5 min. Finally, the cells were washed once at room temperature in 2× SSC and couterstained by TO-PRO-3 Iodide (Thermofisher, T3605). Images were acquired using the LMS510 confocal microscope. TERRA foci were quantified using ImageJ software (US National Institutes of Health).
In vitro fertilization (IVF)-generated zygotes and two-cell embryos were used for TERRA FISH as described previously (50). Cells were seeded in Denhardt's solution coated coverslips and dried down. Then, cells were fixed in 3% paraformaldehyde for 10 min and permeabilized in PBS with 0.5% Triton X-100 and 2 mM VRC for 2 min. The cells were then hybridized and washed as described above.
qRT-PCR
RNA was purified from various tissues and cells using the Trizol reagent (Invitrogen) with DNase I treatment. TERRA and ATF7 mRNA were examined by qRT-PCR using SYBR green real-time PCR Master Mix (TOYOBO). The primers used are described in Supplementary Table S1.
Quantification of TERRA from mature spermatozoa
Caput epididymis was incubated in M2 medium (Sigma) for 1 h at 37°C, and whole medium containing sperm cells was collected through a 70 μm nylon mesh. Pelleted cells were incubated in somatic cell lysis buffer (PBS containing 0.1% SDS and 0.5% Triton X-100) for 10 min to remove somatic cells. The sperm cell suspension was mildly sonicated using a Handy Sonic UR-20P sonicator (Tomy, Japan) to separate the sperm head from the tail, and the high-density sperm head fraction was collected by centrifugation in 80% Percoll. RNA was purified using the Trizol reagent (Invitrogen) with DNase I treatment. To measure the TERRA level by poly(A)-tailed qRT-PCR, poly(A) tailing and first-strand cDNA synthesis were performed using a miRNA cDNA Synthesis Kit (Invitrogen), and cDNA levels were measured using the SYBR green qPCR Mix (TOYOBO). The primers used are described in Supplementary Table S1. To analyse the size of TERRA by northern blotting, RNA was prepared from spermatozoa, loaded onto a 6% polyacrylamide gel, ran in TBE buffer, and then trans-blotted to a Biodyne B membrane. The membrane was hybridized with telomere probe C24 as described above.
Western blotting of ATF7 and pATF7
Testicular germ cells were lysed in SDS sample buffer with mild sonication. After denaturation by boiling, the samples were subjected to 7.5% SDS-PAGE. Anti-ATF7 (1A7) and ant-p71 ATF2 (CST, 9221) were used to detect ATF7 and pATF7/pATF2.
qChIP and ChIP/slot-blot hybridization
ChIP assays were performed as described previously (22). Germ cells or MEFs were crosslinked in 0.5% formaldehyde for 8 min at room temperature, and glycine was added to a final concentration of 0.125 M to quench the crosslinking reaction. For the ATF7 ChIP assay, chromatin was digested in 200 U MNase on ice for 1 h, and then solubilized and sonicated in lysis buffer (50 mM Tris–HCl [pH 8.0], 10 mM EDTA, 1% SDS and protease inhibitor cocktail). For other assays, the chromatin was directly solubilized and sonicated in lysis buffer. Immunoprecipitation was performed overnight at 4°C with anti-ATF7 (2F10), anti-histone H3 (ab1791, Abcam), anti-histone H3K9me3 (ab8898, Abcam) and anti- histone H3K9me2 (ab1220, Abcam) antibodies. Normal anti-mouse antibody or anti-rabbit antibody was used as the control. Immunocomplexes were recovered using Dynabeads Protein G beads (10009D, Life Technologies), washed three times with wash buffer (50 mM HEPES–KOH [7.0], 0.5 M LiCl, 1 mM EDTA, 0.7% deoxycholate, and 1% NP40), and incubated at 65°C in elution buffer [50 mM Tris–HCl (pH 8.0), 10 mM EDTA, 1% SDS and 200 mg/ml proteinase K] to release proteins. The DNA was further purified using a QIAquick PCR Purification Kit (Qiagen) and eluted in water. Eluted DNA samples were blotted onto a Biodyne B membrane for hybridization by telomere probes or amplified using SYBR green qPCR Mix (TOYOBO). The primers and probes used are described in Supplementary Table S1. Hybridization was performed overnight in hybridization buffer containing 7% Sodium dodecyl sulphate, 1% BSA, 0.5 M phosphate buffer (pH 7.2), and 1 mM EDTA at 42°C with 32P-labeled oligonucleotide telomeric probes. The signals were visualized and quantified using Image Analyzer 2500 (Fuji).
Co-immunoprecipitation of ATF7 and Suv39h1
Testicular germ cells were lysed by mild sonication in NETN buffer [420 mM NaCl, 0.5% NP40, 20 mM Tris–HCl (pH 7.5) and 1 mM EDTA]. Lysates were diluted 3-fold in TE buffer and incubated in anti-ATF7 (2F10) antibody. The immunocomplexes were collected using Dynabeads Protein G beads (10009D, Life Technologies), washed three times in wash buffer [150 mM NaCl, 20 mM NaCl, 0.5% NP40, 20 mM Tris–HCl (pH 7.5) and 1 mM EDTA], and subjected to western blotting with anti-Suv39h1 antibodies (05–615, Millipore).
Preparation of TERRA and antisense oligonucleotide for injection experiments
DNA fragments containing 50 tandem repeats of TTAGGG (wild-type telomere repeat) or ATACCG (mutant telomere repeat) with the flanking sequences containing PstI sites were synthesized by Takara and cloned into the pGEM vector. The sequences are shown in Supplementary Table S1. The 300-nucleotide WT and mutant TERRA were synthesized from the BamH1- or BstX1-digested vector using a MEGAscript T7 Transcription Kit (Thermo Fisher).
Recently, a locked nucleic acid (LNA)-gapmer against TERRA was successfully used to knock-down TERRA, and the LNA was used at both 5′ and 3′ ends, whereas the middle portion of the DNA-RNA hybrid was targeted by RNase H (16). However, RNase H expression was not detected in the mouse zygote, cleaved embryo, and morula (See Unigene site, https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Mm.182470). Therefore, a 16-nucleotide TERRA antisense LNA-oligonucleotide was used, in which LNA was used for all the nucleotides, to simply mask TERRA. TERRA antisense and control oligonucleotides were synthesized by Exiqon (www.exiqon.com) with modified LNA bases and phosphothiolated backbone modifications. The sequences are shown in Supplementary Table S1.
Zygotes
Spermatozoa were prepared from untreated, TNF-α-, or saline-injected C57BL/6J male mice, and used for IVF with oocytes prepared from C57BL/6J female mice to generate zygotes. Zygotes were used for injection of TERRA or antisense oligonucleotides. Zygotes and embryos that had reached the two-cell stage were used for RNA-FISH or transferred into the oviducts of pseudopregnant ICR female mice for the development of E14.5 embryos, which were used for preparation of MEFs.
Freezing and thawing of spermatozoa
Spermatozoa from TNF-α- or saline-injected C57BL/6J male mice were frozen before IVF to generate zygotes under the same experimental conditions. The spermatozoa were frozen according to the method developed by Nakagata and Takeshima (51) with slight modifications. In brief, the epididymides were incised with fine scissors, and the spermatozoa were allowed to disperse in 100 μl of the sperm cryopreservation solution (R18S3) comprising 18% raffinose (Difco, Becton Dickinson, Franklin Lakes, NJ, USA) and 3% dehydrated skim milk (Difco, Becton Dickinson). The sperm suspension was divided into eight 10-μl aliquots. Each aliquot was placed in a 0.25 ml plastic straw and cooled in liquid nitrogen (LN2) vapor for 10–20 min, and then immersed directly in LN2. For thawing, the straws containing frozen spermatozoa were removed from the LN2 and immersed in a water bath at 37°C for 15 min.
IVF
Superovulation was induced in female C57BL/6J mice by injection of 7.5 IU equine chorionic gonadotropin (eCG; Peamex, Sankyo Co., Tokyo, Japan), followed 48–50 h later by 7.5 IU human chorionic gonadotropin (hCG; Gonatropin, ASKA Pharmaceutical, Tokyo, Japan). Cumulus-oocyte complexes were collected from the oviducts and transferred into human tubal fluid (HTF) medium at 37°C under 5% CO2 in humidified air. Epididymides collected from a 10-week-old male were incised with fine scissors, and spermatozoa were allowed to disperse in 200 μl HTF medium droplets and preincubated for 1–1.5 h. Insemination was performed by adding 2 μl of preincubated spermatozoa to the droplets containing cumulus–oocyte complexes.
IVF using frozen-thawed epididymal spermatozoa was performed as described previously (52,53) with slight modifications. Cumulus-oocyte complexes were collected from the oviducts and preincubated for 1–1.5 h in 80 μl droplets of HTF medium (54) supplemented with 1.25 mM reduced glutathione (MilliporeSigma Canada Co.) (55,56). Five microliters of frozen-thawed sperm suspensions were diluted in 200 μl of sperm preincubation medium [HTF containing 0.4 mM methyl-β-cyclodextrin (57,58) and 0.1 mg/ml polyvinyl alcohol (PVA), but without bovine serum albumin] and incubated at 37°C under 5% CO2 in humidified air for 45–60 min. Insemination was performed by adding preincubated spermatozoa to the droplets containing oocytes at concentrations of 400–500 spermatozoa/μl.
At 3–4 h after insemination, oocytes were separated from spermatozoa and cumulus cells using a fine glass pipette, and transferred into 10-μl droplets of CZB medium (59) containing 5.6 mM glucose, 0.1 mg/ml PVA, and 3.0 mg/ml bovine serum albumin. The zygotes obtained were used for TERRA FISH and injection of TERRA or antisense oligonucleotides.
TERRA mRNA and antisense oligonucleotide injection
TERRA mRNA or antisense LNA-oligonucleotides were diluted with nuclease-free water to 1 or 5 μg/ml and injected into the cytoplasm of pronuclear stage embryos at 5–6 h after IVF using a Piezo-driven micromanipulator (Primetech, Ibaraki, Japan). Injected embryos were cultured in CZB medium at 37°C under 5% CO2 in humidified air. Embryos that had reached the two-cell stage after 24 h in culture were transferred into the oviducts of pseudopregnant ICR female mice on day 0.5 (the day following sterile mating with a vasectomized male mouse) and then developed to E14.5 embryos to prepare MEFs.
Statistics
Results are presented as the mean ± standard deviation (SD). Differences between groups were examined for statistical significance using Student's t-test. Fisher's Z test was used for comparisons of relationships in the Q-FISH assay of telomere length.
Data availability
All data are available from the corresponding author upon reasonable request.
RESULTS
Telomere shortening in male offspring by paternal TNF-α exposure
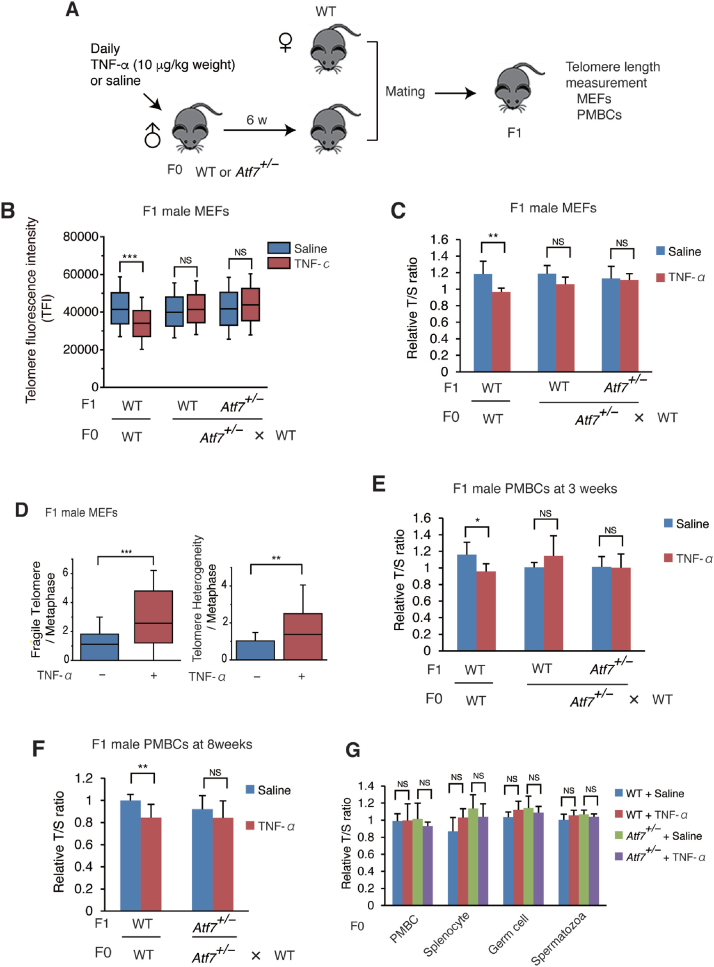
To examine whether paternal stress exposure induces telomere shortening in the offspring, various doses of TNF-α were injected daily into 8-week-old male mice (F0 mice) for 4, 6 or 11 weeks, and telomere length in the peripheral blood mononuclear cells (PMBCs) of 3-week-old offspring (mixture of male and female F1 mice) was measured by Q-PCR. Paternal injection of TNF-α (10 or 20 μg/kg weight) for 6 or 11 weeks induced telomere shortening in the offspring (Supplementary Figure S1A and B). Therefore, injection of TNF-α at 10 μg/kg weight for 6 weeks was used as the experimental condition in the present study (Figure 1A). Quantitative fluorescence in situ hybridization (Q-FISH) and Q-PCR indicated that paternal TNF-α exposure induced telomere shortening in F1 male mouse embryonic fibroblasts (MEFs) (Figure 1B and C and Supplementary Figure S1C). Paternal TNF-α treatment increased the frequency of telomere fragility and telomere length heterogeneity, which are hallmarks of telomere dysfunction (60) (Figure 1D and Supplementary Figure S1D). Telomere shortening was also observed in PMBCs of 3- and 8-week-old F1 male mice (Figure 1F).
Figure 1.
Paternal TNF-α exposure induces telomere shortening in the male offspring via ATF7, but not in paternal germ cells. (A) Scheme of experimental procedures. Eight-week-old male mice (F0) were daily administered with TNF-α (10 μg/kg weight) or saline for 6 weeks, and then mated with WT female mice. Telomere length of the offspring (F1) was measured. (B, C) Paternal TNF-α treatment induces telomere shortening in MEFs from male offspring. Wide-type (WT) or Atf7+/– 8-week-old male mice (F0) (n = 3) were treated daily with TNF-α (10 μg/kg weight) or saline for 6 weeks and then mated with WT female mice (n = 3). MEFs (n = 3, three independent MEFs from three independent pregnant mice) were prepared from WT or Atf7+/– offspring (F1) for measurement of telomere length by Q-FISH (B). Raw data are shown in Supplementary Figure S1C. Middle lines in the colored boxes indicate medians; top and bottom edges, 25th and 75th percentiles; and whiskers, 10th and 90th percentiles. ***P < 0.001; NS, not significant. Telomere length in MEFs (n = 7, 6; 4, 9; 3, 5 for each group from three three independent pregnant mice) from the offspring (F1) were measured by Q-PCR (C). Relative telomere length, expressed as the T/S ratio, is shown ± SD. **P < 0.01; ***P < 0.001; NS, not significant. (D) Paternal TNF-α exposure increases the frequency of telomere fragility and telomere length heterogeneity in MEFs of male offspring. WT male offspring MEFs (n = 3, three independent MEFs from three independent pregnant mice) were prepared as described above, and the frequency of telomere fragility and telomere length heterogeneity were examined. Box plots show the abnormal telomere number per methaphase (n = 26, 31; for each group from three independent male F0 mice). Middle lines in the coloured boxes indicate medians; top and bottom edges, 25th and 75th percentiles; and whiskers, 10th and 90th percentiles. **P < 0.01; ***P < 0.001. Raw images are shown in Supplementary Figure S1D. (E, F) Paternal TNF-α exposure induces telomere shortening in peripheral blood mononuclear cells (PMBCs) from male offspring. Paternal mice were treated as above. PMBCs (n = 6, 6; 6, 5; 6, 5; n = 6, 4; 5, 5 for each group from mice generated from three independent pregnant mice, in e and f, respectively) were prepared from the 3- (E) or 8-week-old (F) male pups (F1), and telomere length was determined by Q-PCR as above. *P < 0.05; **P < 0.01; NS, not significant. (G) TNF-α exposure condition used did not induce telomere shortening in various tissues of F0 male mice. WT and Atf7+/– (n = 3) male F0 mice were treated with TNF-α or saline for 6 weeks as described above. Telomere length of PMBC, splenocytes, testicular germ cells and spermatozoa from F0 mice (n = 3) were determined by Q-PCR as above.
In these experiments, mating was finished within a short period (3 days), since TNF-α was not injected during mating. To test the role of ATF7, Atf7 homozygous (Atf7–/–) male F0 mice could not be used because they exhibited anxiety-related behaviours including low sexual behaviour, as reported previously (43), and mating could not be finished within 3 days. Therefore, we used Atf7 heterozygous (Atf7+/–) mice, which have normal sexual behaviour. In Atf7+/– male F0 mice, paternal TNF-α injection did not induce telomere shortening in MEFs and PMBCs of wild-type (WT) and Atf7+/– male offspring (Figure 1B, C, E, and F and Supplementary Figure S1C), suggesting a critical role for ATF7.
We initially speculated that TNF-α induces telomere shortening via ATF7 in testicular germ cells, as recently reported (16), and the shortened telomeres are transmitted to the next generation. However, the TNF-α injection condition used in the present study did not cause telomere shortening in the blood cells, spleen, testicular germ cells, or spermatozoa of F0 mice (Figure 1G). We recently reported that daily injection of TNF-α (20 μg/kg weight) into pregnant mice from embryonic day (E) 2.5 to E14.5 induces telomere shortening in embryos in an ATF7- and telomerase-dependent manner (16). This indicates that the absence of TNF-α-induced telomere shortening in F0 mice may be due to differences in the TNF-α injection conditions. These results indicate that telomere shortening induced by paternal TNF-α treatment is not caused by the transmission of shortened telomeres to the offspring (Supplementary Table S2).
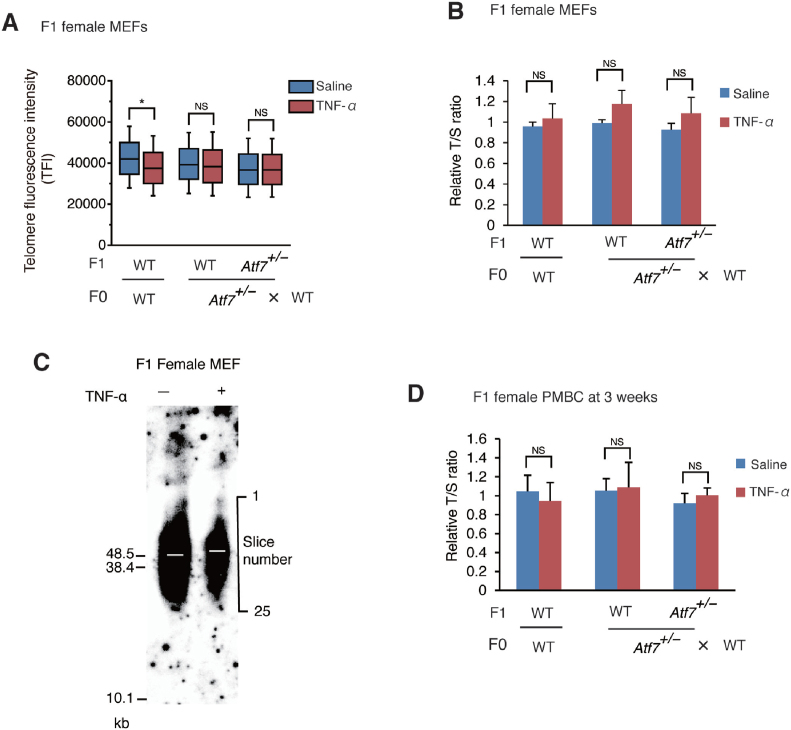
Unlike the response in male offspring, the degree of telomere shortening induced by paternal TNF-α injection was lower in female offspring MEFs than in male MEFs, as shown by Q-FISH, and no telomere shortening was detected by Q-PCR and TRF assays (Figure 2A, B, and C and Supplementary Figure S2). Telomere shortening in PMBCs of 3-week-old F1 female offspring was not induced by paternal TNF-α exposure (Figure 2C). Thus, paternal TNF-α treatment induced telomere shortening only in the male offspring.
Figure 2.
Paternal TNF-α treatment does not induce telomere shortening in the female offspring. (A, B, C) Paternal TNF-α treatment did not induce telomere shortening in MEFs from female offspring. WT or Atf7+/– 8-week-old male mice (F0) (n = 3) were daily administered with TNF-α as described in Figure 1A, and then mated with WT female mice (n = 3). MEFs (n = 3 from three independent pregnant mice) were prepared from female offspring (F1 mice) for measurement of telomere length by Q-FISH (A). *, P < 0.05; NS, not significant. Raw data of Q-FISH are shown in Supplementary Figure S2. Telomere length of MEFs (n = 3, 5; 3, 4; 5, 3 for each group from three independent pregnant mice) were measured by Q-PCR (B). Relative telomere length, expressed as the T/S ratio, is shown ± SD. NS, not significant. Telomere length was also measured by telomere restriction fragment (TRF) assay using the G24 probe (C). Three primary MEFs from independent pregnant mice were used. The radioactivity of each slice in each lane (slice number is shown on the right) was quantitated using Image Analyzer, and the position of mean radioactivity was calculated. White bars indicate the position (slice number) of mean radioactivity in various samples. (D) Paternal TNF-α treatment did not induce telomere shortening in PMBCs from female offspring. Paternal mice were treated as above. PMBCs (n = 6, 3; 6, 6; 6, 5 for each group from three independent pregnant mice) were prepared from the 3-week-old female pups (F1), and telomere length was determined by Q-PCR as above. NS, not significant.
Increase of TERRA level in F0 testicular germ cells and F1 male MEFs by paternal TNF-α exposure
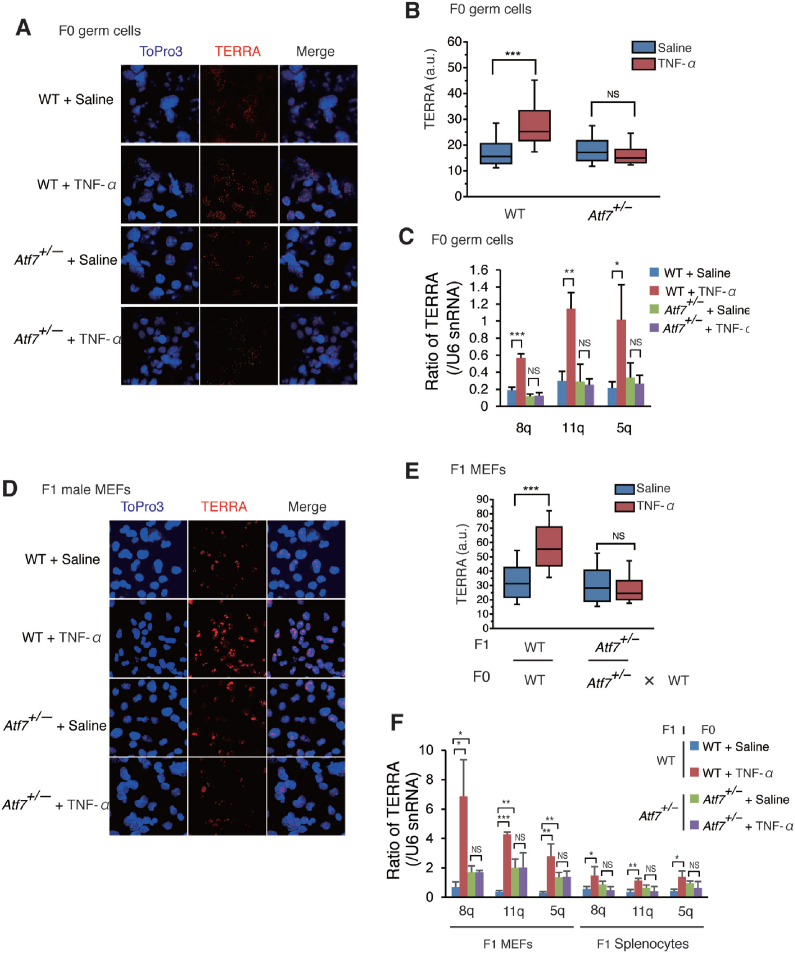
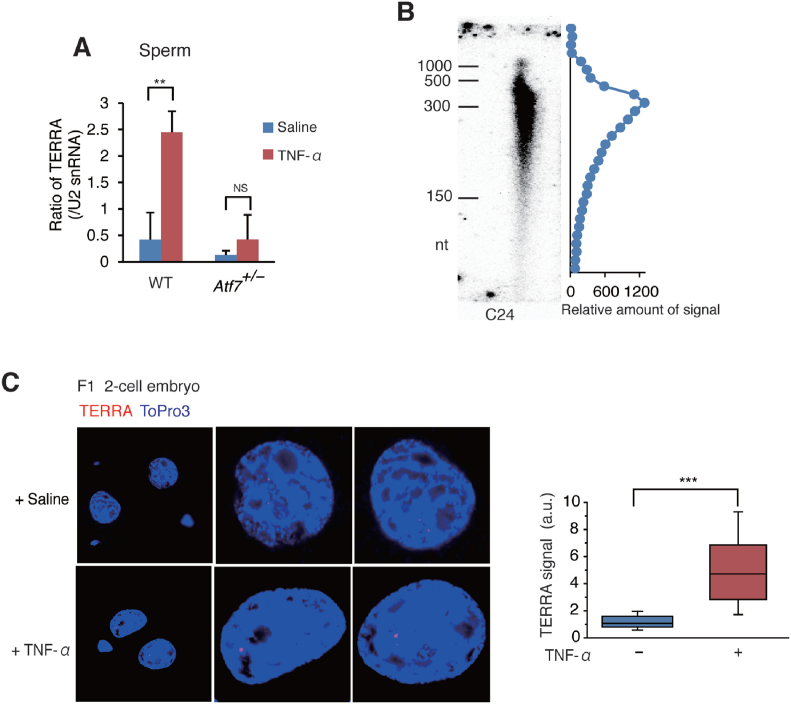
Data obtained by RNA-FISH indicated that paternal TNF-α treatment increased the level of TERRA in the testicular germ cells of F0 WT mice, but not in Atf7+/– mice (Figure 3A and B and Supplementary Figure S3A). The results of qRT-PCR showed that the levels of TERRA transcribed from chromosomes 8q, 11q, and 5q were increased by TNF-α in the testicular germ cells of F0 WT mice, but not in Atf7+/– mice (Figure 3C). Although TERRA induces telomere shortening (24,27–32), TNF-α-induced TERRA did not shorten telomere length in F0 testicular germ cells (Figure 1G). This could be attributed to the relatively high level of telomerase in testicular germ cells (61). Paternal TNF-α injection also increased TERRA levels in WT male offspring MEFs, but not in Atf7+/– male offspring MEFs or splenocytes from TNF-α-injected Atf7+/– F0 mice (Figure 3D–F and Supplementary Figure S3B and C). These results raise the possibility that TERRA was induced by TNF-α in testicular germ cells in an ATF7-dependent manner, and then transmitted to the offspring via sperm and reduced telomere length (Supplementary Table S2).
Figure 3.
TNF-α increases TERRA level in testicular germ cells and male offspring MEFs in an ATF7-dependent manner. (A–C) TNF-α increases TERRA level in an ATF7-dependent manner in F0 testicular germ cells. WT and Atf7+/– male F0 mice (n = 3) were treated with TNF-α or saline as described in Figure 1A. TERRA expression in F0 testicular germ cells was determined by RNA FISH. Typical data from RNA FISH is shown (A). A Tel-Cy3 RNA probe was used for measurement of TERRA expression. Topro3 was used to detect the nuclei. Box plots show the fluorescence intensity (arbitrary units, a.u.) of individual TERRA foci from nuclei (n = 98, 93; 92, 93 for each group from three independent male F0 mice) (B). Middle lines in the colored boxes indicate medians; top and bottom edges, 25th and 75th percentiles; and whiskers, 10th and 90th percentiles. ***P < 0.001; NS, not significant. TERRA expression in F0 testicular germ cells was determined by qRT-PCR (C). The average value relative to U6 snRNA ± SD is shown (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. (D–F) Paternal TNF-α exposure increases TERRA level in an ATF7-dependent manner in F1 male MEFs. F0 mice were injected with TNF-α as described in Figure 1A. TERRA level in MEFs from male F1 embryos was determined by RNA FISH. Typical data from RNA FISH is shown (D). Box plots representing the fluorescence intensity (a.u.) of individual TERRA foci from nuclei (n = 101; for each group, three independent MEFs from three independent pregnant mice are shown) (E). ***P < 0.001; NS, not significant. TERRA expression in F1 male MEFs or splenocytes from 3-week-old male F1 pups was measured by qRT-PCR as described above (n = 3 for each type of sample from three independent pregnant mice) (F). *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant.
ATF7-dependent and TNF-α-induced disruption of heterochromatin structure on TERRA gene promoter in testicular germ cells
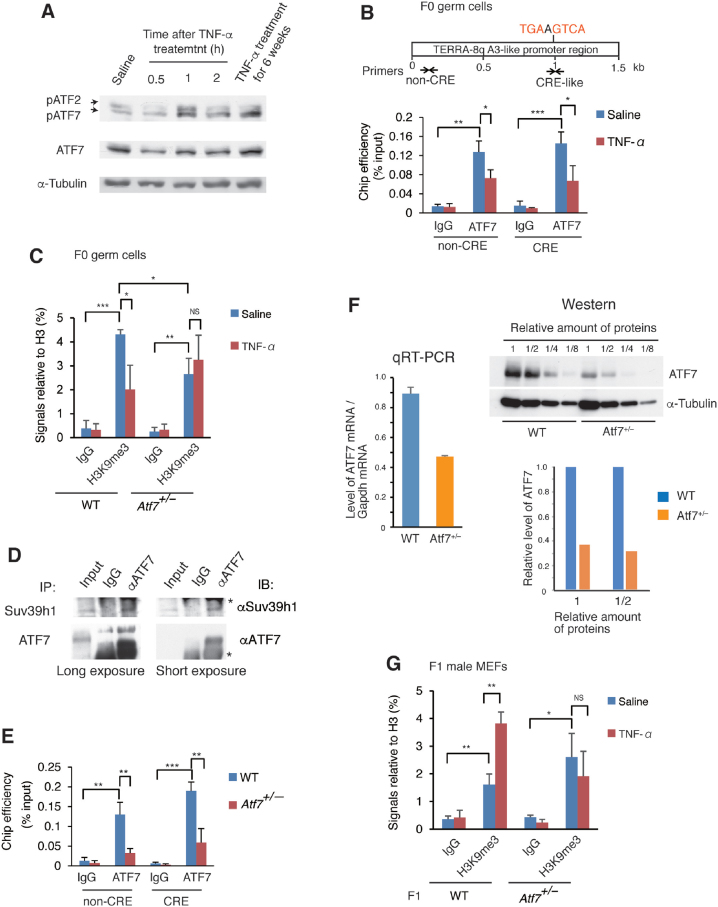
We investigated the role of ATF7 in the TNF-α-induced increase of TERRA in testicular germ cells. Either single injection or 6-week daily injections of TNF-α induced ATF7 phosphorylation in testicular germ cells (Figure 4A). The results of qChIP assays showed that ATF7 localized to both regions containing or lacking a cAMP response element (CRE)-like sequence, to which ATF7 binds directly, in the subtelomeric region on chromosome 8q (Figure 4B), suggesting that ATF7 binds directly and indirectly to this region via interactions with other factor(s). TNF-α-induced ATF7 phosphorylation caused a release of ATF7 from the TERRA gene promoter (Figure 4B). Consistent with the release of ATF7, TNF-α treatment decreased the histone H3K9 trimethylation (H3K9me3) level on the TERRA gene promoter (Figure 4C). Anti-ATF7 antibody co-immunoprecipitated the H3K9 tri-methyltransferase Suv39h1 with ATF7 in lysates of testicular germ cells (Figure 4D), suggesting that ATF7 recruits Suv39h1 to the TERRA gene promoter to establish the H3K9m3 mark.
Figure 4.
TNF-α decreases H3K9me3 level on the TERRA gene promoter by releasing ATF7. (A) TNF-α induces ATF7 phosphorylation in testicular germ cells. Testicular germ cells were prepared from WT mice at the indicated times after TNF-α (10 μg/kg weight) or saline single injection. Testicular germ cells were also prepared from mice treated daily with TNF-α (10 μg/kg weight) for 6 weeks. Western blotting was performed using whole cell lysates of testicular germ cells to detect the indicated proteins. (B) TNF-α induces the release of ATF7 from the TERRA gene promoter on chromosome 8q (TEERA-8q) in the testicular germ cells of F0 mice. WT and Atf7+/– male F0 mice (n = 3) were treated with TNF-α or saline as described in Figure 1A. Modified chromatin immunoprecipitation (ChIP) was performed using MNase, testicular germ cells, and anti-ATF7 antibody. Primers to amplify the region in the TERRA-8q promoter, which contains the CRE-like site or lacks the CRE site, were used for Q-PCR. Average values relative to the input ± SD are shown (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (C) TNF-α decreases H3K9me3 levels on the TERRA gene promoter in WT but not in Atf7+/– testicular germ cells of F0 mice. WT and Atf7+/– male F0 mice (n = 3) were treated with TNF-α or saline as described above, and ChIP was performed using testicular germ cells, anti-H3K9me3, and anti-H3 antibody. Primers in the non-CRE region were used to amplify the TERRA-8q promoter region, and the average value of the H3K9me3 signal relative to H3 ± SD is shown (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001; NS, not significant. (D) Co-immunoprecipitation of ATF7 and Suv39h1. Cell lysates of testicular germ cells were immunoprecipitated with anti-ATF7 antibody or control IgG, and the immuno-complexes were subjected to western blotting using anti-Suv39h1. Asterisk indicates IgG. (E) Amount of ATF7 on the TERRA gene promoter in Atf7+/– testicular germ cells is less than half of that in WT cells. Modified qChIP assays were used to measure the ATF7 amount on the TERRA-8q gene promoter in WT and Atf7+/– F0 testicular germ cells (n = 3) as described above.*P < 0.05; **P < 0.01; ***P < 0.001. (F) Levels of ATF7 mRNA and protein in Atf7+/– testicular germ cells. The level of ATF7 mRNA in WT and Atf7+/– testicular germ cells was examined by qRT-PCR (left). To exaine the ATF7 protein level, Western blotting was performed using serial dilution of whole cell lysates (right upper), and quantification of ATF7 band is shown as a bar graph (right lower). (G) Paternal TNF-α treatment increases the level of H3K9me3 on the TERRA gene promoter in WT but not in Atf7+/– F1 male MEFs. WT and Atf7+/– male F0 mice (n = 3) were treated with TNF-α or saline as described above, and mated with female mice. MEFs were prepared from E14.5 F1 male embryos and used for ChIP with anti-H3K9me3 and anti-H3 antibodies. Primers in the non-CRE region were used to amplify the TERRA-8q promoter region, and the average value of the H3K9me3 signal relative to H3 ± SD is shown (n = 3 from three independent pregnant mice). *P < 0.05; **P < 0.01; NS, not significant.
The results of the modified qChIP assay, in which MNase was used to increase the sensitivity for the detection of the heterochromatin region, showed that the ATF7 amount on the TERRA gene promoter in Atf7+/– testicular germ cells was ∼30% of that in WT cells (Figure 4E). The results of qRT-PCR and Western blotting indicated that level of ATF7 mRNA and ATF7 protein in Atf7+/– testicular germ cells was about half (52.5%) and one-third of that in WT cells, respectively (Figure 4F). Thus, lower amount of ATF7 on the TERRA gene promoter than we had expected is due to decreased level of total ATF7 protein. Since we recently found the level of phosphorylated p38 is increased by the changing metabolism in Atf7+/– testicular germ cells (data not shown), enhanced ATF7 phosphorylation may induce its release from the target genes and ATF7 protein degradation. The H3K9me3 levels on the TERRA gene promoter in Atf7+/– testicular germ cells was lower than that in WT cells, and was not decreased by TNF-α (Figure 4C), which may be due to the presence of small amounts of ATF7 on the TERRA gene promoter.
In contrast to the findings in F0 testicular germ cells, paternal TNF-α injection increased the H3K9me3 level on the TERRA gene promoter in the MEFs of male WT offspring (Figure 4G). These results indicate that the decrease in H3K9me3 levels on the TERRA gene promoter in F0 testicular germ cells was not inherited by the MEFs of offspring (Supplementary Table S2). As in the TERRA gene promoter, TNF-α injection induced a release of ATF7 from telomeres and decreased the H3K9me3 level on the telomere in WT testicular germ cells (Supplementary Figure S4A and B); however, the decreased H3K9me3 level was not transmitted to the MEFs of male offspring (Supplementary Figure S4C and Supplementary Table S2). These results indicate that telomere shortening induced by paternal TNF-α injection was not caused by the inheritance of decreased H3K9me3 levels on the TERRA gene promoter and telomere. TERRA enhances heterochromatin formation by interacting with TRF2 (36), suggesting the possibility that TERRA transmitted to offspring increased the H3K9me3 level.
Transmission of TERRA to zygote via spermatozoa
Results of qRT-PCR indicated that the level of TERRA was ∼6-fold higher in sperm from TNF-α-injected mice than in that from control mice (Figure 5A and Supplementary Figure S5) Such an increase of TERRA level was not observed in Atf7+/— spermatozoa. These results indicate that TNF-α-induced TERRA in the testicular germ cells is transmitted to spermatozoa. Consistent with recent reports that the RNA size in spermatozoa is relatively small (62), TERRA in mature spermatozoa also showed a small size with a peak of ∼300 nucleotides (Figure 5B).
Figure 5.
Transgenerational transmission of TNF-α-induced TERRA into zygotes via spermatozoa. (A) TNF-α increases TERRA level in spermatozoa in an ATF7-dependent manner. WT and Atf7+/– male F0 mice (n = 3) were treated with TNF-α or saline as described in Figure 1A. TERRA expression in sperm was determined by poly(A)-tailed qRT-PCR. The average value relative to U2 snRNA ± SD is shown (n = 3). **P < 0.01; ***P < 0.001; NS, not significant. Control data for poly(A)-tailed qRT-PCR are shown in Supplementary Figure S5. WT and Atf7+/– male F0 mice (n = 3) were administered with TNF-α or saline as described in Figure 1A. TERRA expression in mature spermatozoa was determined by poly(A)-tailed qRT-PCR. Average value relative to U2 snRNA ± SD is shown (n = 3). **P < 0.01; NS, not significant. (B) Total RNA from spermatozoa was used for Northern blotting with 32P-labeled C24. The distribution of signal amounts is drawn on the right. (C) Transgenerational transmission of TNF-α-induced TERRA into zygotes. Spermatozoa was prepared from the TNF-α or saline-injected mice, and used for in vitro fertilization with the oocytes from female mice. The level of TERRA in two-cell embryos was determined by RNA FISH. Box plots representing the fluorescence intensity (arbitrary units, a.u.) of individual TERRA foci from nuclei (n = 40) are shown. ***P < 0.001. Typical RNA FISH data are shown.
The results of RNA-FISH experiments showed that the level of TERRA was 4-fold higher in the two-cell embryos generated using the sperm from the TNF-α-injected mice than in the control (Figure 5C), although TERRA was not detected in one-cell embryos. A detection of strong single dot of TERRA in two-cell embryos might be consistent with the previous observations that TERRA is concentrated on the inactive X chromosome (24–27). These results indicate that TERRA levels in one-cell embryos may be lower than the detectable level using FISH. The increased level of TERRA in one-cell embryos may enhance the transcription of the telomere region, where TERRA and ATRX are located, in two-cell embryos during zygotic gene activation, as TERRA enhances transcription by antagonizing ATRX (27).
Telomere shortening in male offspring by transmitted TERRA
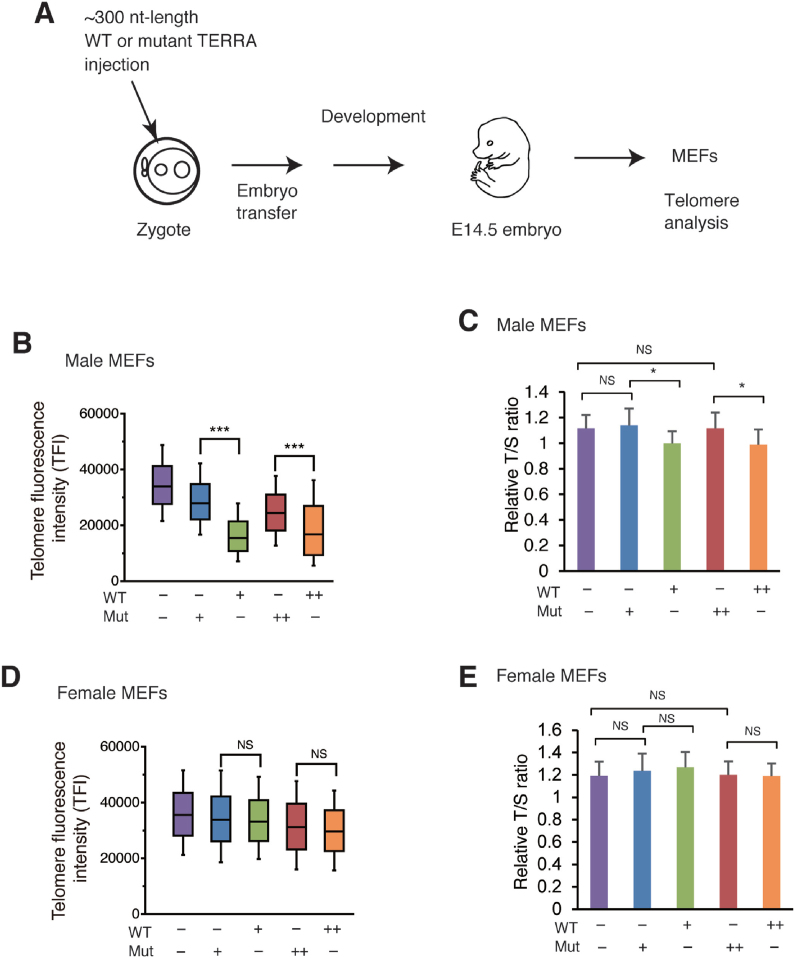
To test whether TERRA in zygotes induces telomere shortening in the developed embryos, WT and mutated TERRA molecules of ∼300 nucleotides in length were in vitro synthesized and injected into zygotes (Figure 6A). The results of Q-FISH and Q-PCR indicated that injection of WT TERRA (1 or 5 μg/ml) into zygotes reduced telomere length in male MEFs, but not in female MEFs, from developed embryos (Figure 6B–E and Supplementary Figure S6). Mutated TERRA also induced mild telomere shortening, although to a considerably lower degree than WT TERRA, suggesting that telomere shortening may be non-specifically induced.
Figure 6.
Injection of TERRA into zygotes induces telomere shortening in male but not female MEFs. (A) Scheme of experimental procedures. The 300-nucleotide TERRA containing WT (UUAGGG) and mutant (AUACCG) telomere repeat sequence was made by in vitro transcription, and injected into zygotes, which were generated by IVF using non-treated WT spermatozoa and oocytes. Zygotes were then transferred into the oviducts of pseudopregnant females, and MEFs were prepared from E14.5 male or female embryos. (B–E) MEFs (n = 3) were prepared from the male (B, C) or female (D, E) embryo and were used to analyze the telomere length by Q-FISH (B, D) or Q-PCR (C, E). Raw data of Q-FISH are shown in Supplementary Figure S6. The number of MEFs used for Q-PCR was 7, 6, 7, 13, 12 for each group (C) and 5, 11, 8, 7, 13 (E), respectively. *P < 0.05; ***P < 0.001; NS, not significant.
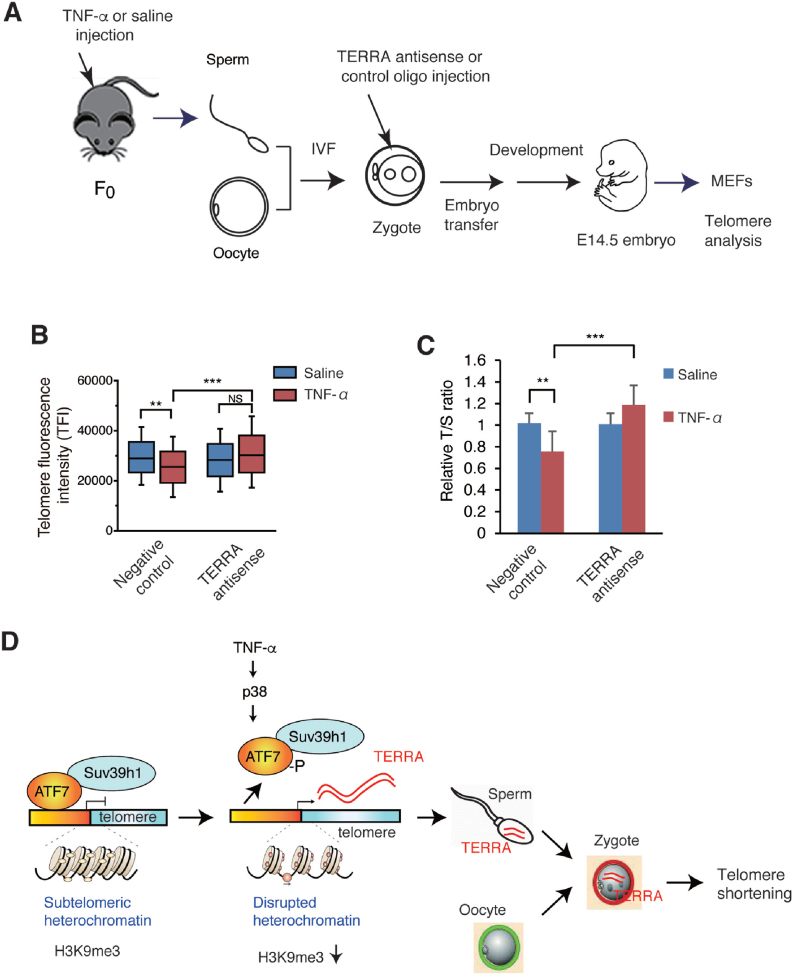
To further test whether TERRA in zygotes, which was derived from the sperm of TNF-α-injected mice, caused telomere shortening in the developed embryos, a TERRA antisense oligonucleotide was used (Figure 7A). Recently, a locked nucleic acid (LNA)-gapmer against TERRA was successfully used to knock-down TERRA, and the LNA was used at both 5′ and 3′ ends, whereas the middle portion of the DNA-RNA hybrid was targeted by RNase H (27). However, RNase H expression was not detected in the mouse zygote, cleaved embryo, and morula (see Unigene site, https://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Mm.182470). Therefore, a 16-nucleotide TERRA antisense LNA-oligonucleotide was used, in which LNA was used for all the nucleotides, to simply mask TERRA. Transfection of HeLa cells with the TERRA antisense oligonucleotide decreased the TERRA FISH signals, whereas control RNA had no effect (Supplementary Figure S7A and B). The results of slot-blot hybridization indicated that transfection of HeLa cells with TERRA antisense oligonucleotide did not decrease the level of TERRA (Supplementary Figure S7C), suggesting that this TERRA antisense oligonucleotide masks TERRA, whereas it does not downregulate TERRA. Injection of TERRA antisense oligonucleotide into zygotes generated from the sperm of TNF-α- or saline-injected mice cancelled the TNF-α-induced telomere shortening in MEFs, whereas control oligonucleotides had no effect (Figure 7B and C and Supplementary Figures S7D). These results support the notion that TNF-α-induced TERRA is transmitted to zygotes via sperm and induces telomere shortening in the offspring (Figure 7D).
Figure 7.
Injection of TERRA antisense LNA-oligonucleotide into zygotes blocked paternal TNF-α-induced telomere shortening. (A) Scheme of experimental procedures. WT male F0 mice (n = 2) were administered with TNF-α or saline as described in Figure 1A. Spermatozoa were collected for IVF, and the zygote was injected with TERRA antisense or control LNA-oligonucleotides. Zygotes were then transferred into the oviducts of pseudopregnant females, and MEFs were prepared from E14.5 male embryos. (B, C) Telomere length in MEFs was measured by Q-FISH (B) or Q-PCR (C). Three independent male MEFs from two independent pregnant mice were used for Q-FISH. Raw data of Q-FISH are shown in Supplementary Figure S7D. 7, 5, 13 and 13 for each type of MEFs from two independent pregnant mice were used for Q-PCR. **P < 0.01; ***P < 0.001; NS, not significant. (D) Model for paternal TNF-α-induced telomere shortening in offspring. In testicular germ cells, ATF7 silences TERRA gene transcription by forming heterochromatin structure on the TERRA gene promoter in the subtelomeric region via recruiting histone H3K9 trimethyltransferase, Suv39h1. TNF-α exposure induces ATF7 phosphorylation by p38, which causes a release of ATF7 from the TERRA gene promoter, resulting to disruption of heterochromatin structure and induction of TERRA gene transcription. TERRA is transgenerationally transmitted to zygotes via sperm, and induces telomere shortening in the male offspring.
DISCUSSION
In this study, we have demonstrated that paternal TNF-α treatment induces TERRA in the testicular germ cells, which is transmitted to the zygotes and shortens telomere length in the male offspring (Figure 7D). ATF7 has a key role to induce TERRA by TNF-α treatment. Previously, we showed that TNF-α induces ATF7 phosphorylation by p38, which then causes a release of ATF7 and the histone H3K9 trimethyltransferases, ESET or Suv39h1, from the heterochromatin-like region in the euchromatin, the pericentromeric heterochromatin, and telomere (43,16). Similar to these previous studies, TNF-α-induced ATF7 phosphorylation causes a release of ATF7 from the TERRA gene promoter in the subtelomeric region and telomeres in the testicular germ cells, resulting to a decrease of histone H3K9me3 level and TERRA induction (Figure 7D). This is consistent with the study that the histone H3K9me3 level and HP1α regulates TERRA gene transcription (63). Three histone H3K9 trimethyltransferases, Suv39h1, Suv39h2, and ESET, are expressed in testicular germ cells, and the interaction of ATF7 with Suv39h1 was shown by co-immunoprecipitation in the present study. However, we could not test the role of Suv39h2 due to a lack of appropriate antibody. Thus, it remains unknown which histone H3K9 trimethyltransferase most strongly contributes to heterochromatin formation on the subtelomeric region containing the TERRA gene promoter, because the availability and quality of appropriate antibodies for each enzyme.
We had originally speculated that inheritance of shortened telomere or heterochromatin disruption of telomere/subtelomeric region may be the mechanism of paternal TNF-α exposure-induced telomere shortening (Supplementary Table S2). However, TNF-α injection under the condition used in the present study did not cause telomere shortening in testicular germ cells, although different TNF-α injection condition, for instance by using more amount of TNF-α, might be able to induce telomere shortening. TERRA induces telomere shortening in multiple cell types (24,27–32), but TNF-α injection did not induce telomere shortening in testicular germ cells in spite of that it increased the TERRA level. This might be due to the relatively high level of telomerase in the testicular germ cells (61). Abundant telomerase in testicular germ cells is also supported by the observation that telomere length increases with age in human sperm in contrast to somatic tissue (64). Furthermore, a disruption of heterochromatin on subtelomeric region and telomeres in testicular germ cells, which is shown by decreased H3K9me3 level, was not inherited to the offspring. It was shown that TERRA contributes to form telomeric heterochromatin by interacting with HP1, histone H3K9 methyltransferases, and shelterins (36).
Both paternal TNF-α and TERRA injection into zygotes induced telomere shortening in the male offspring, but not in the female offspring. This indicates that increased level of TERRA shortens telomere length only in male cells. It was reported that TERRA signal is concentrated on the inactive X chromosome (24,26), suggesting that TERRA could be adsorbed by inactive X chromosome and may not induce telomere shortening in female cells. A recent study showed that TERRA binds not only to telomeres, but also to many genes on autosomes and the pseudoautosomal region of sex chromosomes, regulating the expression of many genes (27). These TERRA-binding genes are involved in chromatin and transcription regulation as well as telomere regulation. Therefore, the transmission of TNF-α-induced TERRA via sperm may affect the expression of these genes in addition to inducing telomere shortening.
In the testicular germ cells of Atf7+/– mice, TNF-α injection did not induce TERRA gene expression. The amount of ATF7 on the TERRA gene promoter in Atf7+/— testicular germ cells was only about 30% of that in WT cells. These results suggest that in Atf7+/– testicular germ cells, TNF-α injection did not disrupt heterochromatin on the TERRA gene promoter due to small amount of ATF7, which mediate the TNF-α-induced disruption of heterochromatin, and did not induce TERRA gene expression. We have recently found that this is due to increased level of ATF7 phosphorylation caused by changing metabolism (data not shown). In spite of small amount of ATF7 on the subtelomeric heterochromatin, the H3K9me3 level on the subtelomeric heterochromatin in Atf7+/— testicular germ cells was ∼60% of that in WT cells. Heterochromatin is established at early stage of development and maintained during later stage (65–67). The two mechanisms, involving the RNA interference machinery and the stress-responsive ATF2 family of transcription factors, independently contribute to the establishment and maintenance of heterochromatin (16,42–46,65,66,68). Therefore, a decrease of H3K9me3 level on the subtelomeric region, which is caused by localization of only small amount by ATF7, might be partly complemented by the RNA interference-dependent mechanism. In contrast to the case of subtelomeric region, the level of H3K9me3 level on telomeres was similar between WT and Atf7+/– testicular germ cells. This may suggest that effect of decreased amount of ATF7 on the heterochromatin structure may be different depending on the loci. Interaction of ATF7 with other chromatin factors, which are required for formation of heterochromatin, might be different depending on the loci.
The levels of H3K9me3 in Atf7+/– F0 germ cells and in WT TNF-α-treated F0 germ cells were comparable (Figure 4C), but expression of TERRA in Atf7+/– F0 germ cells were much lower than that in WT TNF-α treated germ cells (Figure 3B, C). We speculate that this might be due to negative feedback regulation of H3K9me3 level by TERRA. TERRA has two opposite functions for transcriptional regulation; one is to activate transcription through antagonizing ATRX, which is the SNF2-like protein with ATPase and helicase domains (27), and another is to enhance heterochromatin formation via interaction with telomere repeat factors 1/2 (TRF1/2), subunits of the origin recognition complex (ORC), heterochromatin protein 1 (HP1), and histone H3K9me3 (36). When TERRA is induced in TNF-α-treated WT testicular germ cells via decreasing the H3K9me3 level on the TERRA gene promoter, it enhances TERRA gene transcription. Accumulated TERRA then enhances the H3K9me3 level and heterochromatin formation. Due to this negative feed back regulation, the H3K9me3 level on the TERRA gene promoter in the TNF-α-treated WT testicular germ cells might not be significantly lower than that in Atf7+/– F0 germ cells.
F1 male MEFs that treated with TNF-α in the previous generation showed increased level of H3K9me3 at TERRA gene promoter (Figure 4G). Nevertheless they harbored elevated levels of TERRA (Figure 3D, E, F). We speculate that this also might be due to negative feedback regulation of H3K9me3 level by TERRA. As described above, accumulated TEERA enhances heterochromatin formation and increases the H3K9me3 (36). Since this function is coupled with DNA replication and cellular proliferation through interaction with ORC, an increase of H3K9me3 level by TERRA may be more evident in MEFs than testicular germ cells, because proliferation of germ cells is limited. TERRA in zygotes, which is transgenerationally transmitted from TNF-α-treated testicular germ cells, enhances TERRA gene transcription in 2-cell embryo and later stage of early embryo. As heterochromatin formation is evident after blastocyst stage during development, increase of H3K9me3 on the TERRA gene promoter by TERRA might be dominant at later stage of blastocyst, such as in MEFs.
Some epidemiological studies suggests that stressors experienced by parents affect the longevity and/or disease onset of their offspring by regulating telomere dynamics (3,4), but until now no empirical evidence was shown for the telomere shortening by parental stress and its molecular mechanism. To our knowledge, the present study is the first evidence to show the mechanism of telomere shortening by paternal stress. Inflammatory cytokines, including TNF-α, which are induced by psychological stress (17), and other stresses, including environmental stress, oxidative stress, and pathogen infection, induce ATF7 phosphorylation by p38 (16,41,43,45). The present study raises the possibility that various paternal stressors may cause telomere shortening in the offspring by the same mechanism. Although daily injection of TNF-α (10 μg/kg weight) for 6-weeks did not cause telomere shortening in F0 mice in this study, different stress conditions may induce telomere shortening in F0 mice. If stressors induce telomere shortening in F0 mice, such phenomena may appear to be the inheritance of paternal stress-induced telomere shortening. Thus, the present study may shed light on the mechanism of paternal inheritance of telomere length suggested by epidemiological studies (9).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Keiji Mochida for his advice for IVF, and the members of Ishii Lab for useful discussion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Japan Agency for Medical Research and Development (AMED) [16gm0510015h0004]; Ministry of Education, Culture, Sports, Science and Technology (MEXT) [16H01413]. Funding for open access charge: Japan Agency for Medical Research andDevelopment (AMED).
Conflict of interest statement. None declared.
REFERENCES
- 1. Feil R., Fraga M.F.. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2012; 13:97–109. [DOI] [PubMed] [Google Scholar]
- 2. Heard E., Martienssen R.A.. Transgenerational epigenetic inheritance: myths and mechanisms. Cell. 2014; 157:95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaati G., Bygren L.O., Pembrey M., Sjöström M.. Transgenerational response to nutrition, early life circumstances and longevity. Eur. J. Hum. Genet. 2007; 15:784–790. [DOI] [PubMed] [Google Scholar]
- 4. Haussmann M.F., Heidinger B.J.. Telomere dynamics may link stress exposure and ageing across generations. Biol. Lett. 2015; 11:20150396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blackburn E.H. Switching and signaling at the telomere. Cell. 2001; 106:661–673. [DOI] [PubMed] [Google Scholar]
- 6. Palm W., de Lange T.. How shelterin protects mammalian telomeres. Ann. Rev. Genet. 2008; 42:301–334. [DOI] [PubMed] [Google Scholar]
- 7. Harley C.B., Futcher A.B., Greider C.W.. Telomeres shorten during ageing of human fibroblasts. Nature. 1990; 345:458–460. [DOI] [PubMed] [Google Scholar]
- 8. Nordfjäll K., Larefalk Å., Lindgren P., Holmberg D., Roos G.. Telomere length and heredity: Indications of paternal inheritance. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:16374–16378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Njajou O.T., Cawthon R.M., Damcott C.M., Wu S.H., Ott S., Garant M.J., Blackburn E.H., Mitchell B.D., Shuldiner A.R., Hsueh W.C.. Telomere length is paternally inherited and is associated with parental lifespan. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:12135–12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eisenberg D.T., Hayes M.G., Kuzawa C.W.. Delayed paternal age of reproduction in humans is associated with longer telomeres across two generations of descendants. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:10251–10256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smogorzewska A., de Lange T.. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004; 73:177–208. [DOI] [PubMed] [Google Scholar]
- 12. Epel E.S., Blackburn E.H., Lin J., Dhabhar F.S., Adler N.E., Morrow J.D., Cawthon R.M.. Accelerated telomere shortening in response to life stress. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:17312–17315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Entringer S., Epel E.S., Kumsta R., Lin J., Hellhammer D.H., Blackburn E.H., Wüst S., Wadhwa P.D.. Stress exposure in intrauterine life is associated with shorter telomere length in young adulthood. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:E513–E518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richter T., von Zglinicki T.. A continuous correlation between oxidative stress and telomere shortening in fibroblasts. Exp. Gerontol. 2007; 42:1039–1042. [DOI] [PubMed] [Google Scholar]
- 15. Cattan V., Mercier N., Gardner J.P., Regnault V., Labat C., Mäki-Jouppila J., Nzietchueng R., Benetos A., Kimura M., Aviv A. et al. . Chronic oxidative stress induces a tissue-specific reduction in telomere length in CAST/Ei mice. Free Radic. Biol. Med. 2008; 44:1592–1598. [DOI] [PubMed] [Google Scholar]
- 16. Maekawa T., Liu B., Nakai D., Yoshida K., Nakamura K., Yasukawa M., Koike M., Takubo K., Chatton B., Ishikawa F. et al. . ATF7 mediates TNF-α–induced telomere shortening. Nucleic Acids Res. 2018; 46:4487–4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maes M., Song C., Lin A., De Jongh R., Van Gastel A., Kenis G., Bosmans E., De Meester I., Benoy I., Neels H. et al. . The effects of psychological stress on humans: increased production of pro-inflammatory cytokines and a Th1-like response in stress-induced anxiety. Cytokine. 1998; 10:313–318. [DOI] [PubMed] [Google Scholar]
- 18. Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O’Connor M.S., Songyang Z.. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature. 2007; 445:559–562. [DOI] [PubMed] [Google Scholar]
- 19. Wang F., Podell E.R., Zaug A.J., Yang Y., Baciu P., Cech T.R., Lei M.. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007; 445:506–510. [DOI] [PubMed] [Google Scholar]
- 20. Zhong F.L., Batista L.F., Freund A., Pech M.F., Venteicher A.S., Artandi S.E.. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell. 2012; 150:481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt J.C., Zaug A.J., Cech T.R.. Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell. 2016; 166:1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfingsten J.S., Goodrich K.J., Taabazuing C., Ouenzar F., Chartrand P., Cech T.R.. Mutually exclusive binding of telomerase RNA and DNA by Ku alters telomerase recruitment model. Cell. 2012; 148:922–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Azzalin C.M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J.. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007; 318:798–801. [DOI] [PubMed] [Google Scholar]
- 24. Schoeftner S., Blasco M.A.. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008; 10:228–236. [DOI] [PubMed] [Google Scholar]
- 25. Graf M., Bonetti D., Lockhart A., Serhal K., Kellner V., Maicher A., Jolivet P., Teixeira M.T., Luke B.. Telomere length determines TERRA and R-Loop regulation through the cell cycle. Cell. 2017; 170:72–85. [DOI] [PubMed] [Google Scholar]
- 26. Zhang L.-F., Ogawa Y., Ahn J.Y., Namekawa S.H., Silva S.S., Lee J.T.. Telomeric RNAs mark sex chromosomes in stem cells. Genetics. 2009; 182:685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chu H.P., Cifuentes-Rojas C., Kesner B., Aeby E., Lee H.G., Wei C., Oh H.J., Boukhali M., Haas W., Lee J.T.. TERRA RNA antagonizes ATRX and protects telomeres. Cell. 2017; 170:86–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Luke B., Panza A., Redon S., Iglesias N., Li Z., Lingner J.. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol. Cell. 2008; 32:465–477. [DOI] [PubMed] [Google Scholar]
- 29. Redon S., Reichenbach P., Lingner J.. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic. Acids. Res. 2010; 38:5797–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maicher A., Kastner L., Dees M., Luke B.. Deregulated telomere transcription causes replication-dependent telomere shortening and promotes cellular senescence. Nucleic. Acids. Res. 2012; 40:6649–6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pfeiffer V., Lingner J. TERRA promotes telomere shortening through exonuclease 1-mediated resection of chromosome ends. PLos Genet. 2012; 8:e1002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfeiffer V., Crittin J., Grolimund L., Lingner J.. The THO complex component Thp2 counteracts telomeric R-loops and telomere shortening. EMBO J. 2013; 32:2861–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Balk B., Maicher A., Dees M., Klermund J., Luke-Glaser S., Bender K., Luke B.. Telomeric RNA-DNA hybrids affect telomere-length dynamics and senescence. Nat. Struct. Mol. Biol. 2013; 20:1199–1205. [DOI] [PubMed] [Google Scholar]
- 34. López de Silanes I., Graña O., De Bonis M.L., Dominguez O., Pisano D.G., Blasco M.A.. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 2014; 5:4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yu T.Y., Kao Y.W., Lin J.J.. Telomeric transcripts stimulate telomere recombination to suppress senescence in cells lacking telomerase. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:3377–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P.M.. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell. 2009; 35:403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gaire M., Chatton B., Kedinger C.. Isolation and characterization of two novel, closely related ATF cDNA clones from HeLa cells. Nucleic Acids Res. 1990; 18:3467–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Maekawa T., Sakura H., Kanei-Ishii C., Sudo T., Yoshimura T., Fujisawa J., Yoshida M., Ishii S.. Leucine zipper structure of the protein CRE-BP1 binding to the cyclic AMP response element in brain. EMBO J. 1989; 8:2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hai T.W., Liu F., Coukos W.J., Green M.R.. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989; 3:2083–2090. [DOI] [PubMed] [Google Scholar]
- 40. Gupta S., Campbell D., Dérijard B., Davis R.J.. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science. 1995; 267:389–393. [DOI] [PubMed] [Google Scholar]
- 41. Seong K.H., Maekawa T., Ishii S.. Inheritance and memory of stress-induced epigenome change: roles played by the ATF-2 family of transcription factors. Genes Cells. 2012; 17:249–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jia S., Noma K., Grewal S.I.. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science. 2004; 304:1971–1976. [DOI] [PubMed] [Google Scholar]
- 43. Maekawa T., Kim S., Nakai D., Makino C., Takagi T., Ogura H., Yamada K., Chatton B., Ishii S.. Social isolation stress induces ATF-7 phosphorylation and impairs silencing of the 5-HT 5B receptor gene. EMBO J. 2010; 29:196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Seong K.H., Li D., Shimizu H., Nakamura R., Ishii S.. Inheritance of stress-induced, ATF-2-dependent epigenetic change. Cell. 2011; 145:1049–1061. [DOI] [PubMed] [Google Scholar]
- 45. Yoshida K., Maekawa T., Zhu Y., Renard-Guillet C., Chatton B., Inoue K., Uchiyama T., Ishibashi K., Yamada T., Ohno N. et al. . The transcription factor ATF7 mediates lipopolysaccharide-induced epigenetic changes in macrophages involved in innate immune memory. Nat. Immunol. 2015; 16:1034–1043. [DOI] [PubMed] [Google Scholar]
- 46. Takubo K., Aida J., Izumiyama N., Ishikawa N., Fujiwara M., Poon S.S., Kondo H., Kammori M., Matsuura M., Sawabe M. et al. . Chromosomal instability and telomere lengths of each chromosomal arm measured by Q-FISH in human fibroblast strains prior to replicative senescence. Mech. Ageing Dev. 2010; 131:614–624. [DOI] [PubMed] [Google Scholar]
- 47. Poon S.S., Lansdorp P.M.. Quantitative fluorescence in situ hybridization (Q-FISH). Curr. Protoc. Cell Biol. 2001; doi:10.1002/0471143030.cb1804s12. [DOI] [PubMed] [Google Scholar]
- 48. Cawthon R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002; 30:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Callicott R.J., Womack J.E.. Real-time PCR assay for measurement of mouse telomeres. Comp. Med. 2006; 56:17–22. [PubMed] [Google Scholar]
- 50. Okamoto I., Otte A.P., Allis C.D., Reinberg D., Heard E.. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004; 303:644–649. [DOI] [PubMed] [Google Scholar]
- 51. Nakagata N., Takeshima T.. Cryopreservation of mouse spermatozoa from inbred and F1 hybrid strains. Exp. Anim. 1993; 42:317–320. [DOI] [PubMed] [Google Scholar]
- 52. Mochida K., Hasegawa A., Otaka N., Hama D., Furuya T., Yamaguchi M., Ichikawa E., Ijuin M., Taguma K., Hashimoto M. et al. . Devising assisted reproductive technologies for wild-derived strains of mice: 37 strains from five subspecies of Mus musculus. PLoS One. 2014; 9:e114305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hasegawa A., Mochida K., Tomishima T., Ogura A.. Microdroplet in vitro fertilization can reduce the number of spermatozoa necessary for fertilizing oocytes. J. Reprod. Dev. 2014; 60:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Quinn P., Kerin J.F., Warnes G.M.. Improved pregnancy rate in human in vitro fertilization with the use of a medium based on the composition of human tubal fluid. Fertil. Steril. 1985; 44:493–498. [DOI] [PubMed] [Google Scholar]
- 55. Bath M.L. Inhibition of in vitro fertilizing capacity of cryopreserved mouse sperm by factors released by damaged sperm, and stimulation by glutathione. PLoS One. 2010; 5:e9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hasegawa A., Yonezawa K., Ohta A., Mochida K., Ogura A.. Optimization of a protocol for cryopreservation of mouse spermatozoa using cryotubes. J. Reprod. Dev. 2012; 58:156–161. [DOI] [PubMed] [Google Scholar]
- 57. Choi Y.H., Toyoda Y.. Cyclodextrin removes cholesterol from mouse sperm and induces capacitation in a protein-free medium. Biol. Reprod. 1998; 59:1328–1333. [DOI] [PubMed] [Google Scholar]
- 58. Takeo T., Nakagata N.. Reduced glutathione enhances fertility of frozen/thawed C57BL/6 mouse sperm after exposure to methyl-β-cyclodextrin. Biol. Reprod. 2011; 85:1066–1072. [DOI] [PubMed] [Google Scholar]
- 59. Chatot C.L., Ziomek C.A., Bavister B.D., Lewis J.L., Torres I.. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil. 1989; 86:679–688. [DOI] [PubMed] [Google Scholar]
- 60. Sfeir A., Kosiyatrakul S.T., Hockemeyer D., MacRae S.L., Karlseder J., Schildkraut C.L., de Lange T.. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009; 138:90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martín-Rivera L., Herrera E., Albar J.P., Blasco M.A.. Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:10471–10476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J., Farinelli L., Miska E., Mansuy I.M.. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014; 17:667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Arnoult N., Van Beneden A., Decottignies A.. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1α. Nat. Struct. Mol. Biol. 2012; 19:948–956. [DOI] [PubMed] [Google Scholar]
- 64. Kimura M., Cherkas L.F., Kato B.S., Demissie S., Hjelmborg J.B., Brimacombe M., Cupples A., Hunkin J.L., Gardner J.P., Lu X. et al. . Offspring's leukocyte telomere length, paternal age, and telomere elongation in sperm. PLoS Genet. 2008; 4:e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hall I.M, Shankaranarayana G.D., Noma K., Ayoub N., Cohen A., Grewal S.I.. Establishment and maintenance of a heterochromatin domain. Science. 2002; 297:2232–2237. [DOI] [PubMed] [Google Scholar]
- 66. Richards E.J., Elgin S.C.. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002; 108:489–500. [DOI] [PubMed] [Google Scholar]
- 67. Maison C., Almouzni G.. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell. Biol. 2004; 5:296–304. [DOI] [PubMed] [Google Scholar]
- 68. Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I., Martienssen R.A.. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002; 297:1833–1837. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available from the corresponding author upon reasonable request.