Abstract
RNA splicing is a highly regulated process dependent on sequences near splice sites. Insertions of Alu retrotransposons can disrupt splice sites or bind splicing regulators. We hypothesized that some common inherited polymorphic Alu insertions are responsible for splicing QTLs (sQTL). We focused on intronic Alu variants mapping within 100 bp of an alternatively used exon and screened for those that alter splicing. We identify five loci, 21.7% of those assayed, where the polymorphic Alu alters splicing. While in most cases the Alu promotes exon skipping, at one locus the Alu increases exon inclusion. Of particular interest is an Alu polymorphism in the CD58 gene. Reduced CD58 expression is associated with risk for developing multiple sclerosis. We show that the Alu insertion promotes skipping of CD58 exon 3 and results in a frameshifted transcript, indicating that the Alu may be the causative variant for increased MS risk at this locus. Using RT-PCR analysis at the endogenous locus, we confirm that the Alu variant is a sQTL for CD58. In summary, altered splicing efficiency is a common functional consequence of Alu polymorphisms including at least one instance where the variant is implicated in disease risk. This work broadens our understanding of splicing regulatory sequences around exons.
INTRODUCTION
Alternative splicing of transcripts is pervasive in the genome, with >95% of genes generating more than one mRNA by alternative splicing (1). This provides flexible substrates for gene and protein evolution (e.g. (2–4)) and enhances the cellular and functional complexity in eukaryotes (reviewed in (5,6)). In addition to enhancing mRNA and protein diversity, alternative splicing can also regulate the levels of protein produced by incorporating or excluding a stop codon in the processed transcript or shifting the protein open reading frame. When splicing is skewed beyond normal alternative splicing, the aberrant resultant isoform can result in a disease phenotype. In between these two extremes, it is possible that common variants subtly alter mRNA splicing with modest phenotypic outcomes, such as increasing common disease risk (7). Here, we investigated whether commonly occurring polymorphic Alu elements can alter splicing and affect mRNA isoform prevalence. In particular, these structural variants exist in the population with both the Alu-containing allele and the pre-insertion (no Alu) allele present (e.g. (8)).
Alu elements are short interspersed elements (SINE) that have proliferated in primate genomes through a ‘copy-paste’ mechanism of retrotransposition. There are over 1.1 million Alu elements in the human genome (9). Alu elements are ∼300 bp in length (approximately 280 bp plus a poly A tail) and are derived from 7SL RNA (reviewed in (10)). Alu elements can profoundly affect mRNA splicing. De novo Alu insertions at splice acceptor and splice donor sites can disrupt splicing and result in disease-causing alleles (11–14). A subset of Alu elements that have become fixed in human populations (i.e. homozygous present insertions in all individuals) can contribute to natural mRNA isoform diversity. Exonization of all or part of an Alu sequence is well documented at many loci and frequently occurs because Alu elements anti-sense to a gene can provide splice acceptor sites (e.g. (15,16)). Alu elements can also affect mRNA splicing isoforms by creating circular RNAs (17,18) or in ectopic assays by altering recognition of splice sites (19,20).
As new Alu insertions caused by retrotransposition occur at essentially random locations with respect to exons, it is notable that there is a depletion of Alu elements within 100 or 150 bp of an exon (20,21). This suggested to us that purifying selection eliminates those Alu insertions positioned to affect mRNA splicing. We hypothesized that polymorphic Alu insertions close to exons would be likely to affect mRNA splicing, potentially with phenotypic consequences. In this study, we identify reported polymorphic Alu elements mapping to this underrepresented zone near exons and test their impact on mRNA splicing. Using a splicing reporter assay to evaluate 23 loci, we find five where the Alu affects mRNA isoform representation by promoting either exon skipping or inclusion.
MATERIALS AND METHODS
Intronic Alu distribution
Polymorphic Alu elements with insertion sites mapped to single base pair resolution (8,22) were narrowed to those that map within annotated RefSeq genes (GRCh37/hg19, UCSC genome browser). All AluY elements were obtained from UCSC genome browser RepeatMasker (23) track (GRCh37/hg19, UCSC genome browser). Genome-wide exon coordinates and type were obtained from HEXevent (24). Exons were defined as alternatively spliced if the exon is skipped in any reported transcript isoform, or alternative 5′ and/or 3′ ends were reported for the exon (24). The distance between each Alu element and the nearest exon was determined using bedtools closest tool (25). Distribution of either AluY or polymorphic Alu elements was graphed as the total number of elements mapping within 25 bp bins over the total number of intronic elements.
Ectopic minigene reporter assays
The pSpliceExpress vector ((26), Addgene) contains two constitutive rat insulin exons flanking a an intron modified for Gateway cloning (Invitrogen). For each tested locus, the alternatively used exon and ∼2000 bp of flanking intronic sequence was amplified using primers listed in Supplementary Table S1. The region was amplified from DNA of Centre d'Étude du Polymorphisme Humain (CEPH) Utah Residents with Northern and Western European Ancestry (CEU) (Coriell Institute for Medical Research, Camden, NJ, USA) using individuals who were either homozygous for the Alu insertion or for the empty (non-Alu containing) allele. These fragments were cloned into the minigene reporter intron using Gateway cloning (Invitrogen) and Sanger sequence verified to ensure no other sequence difference existed outside of the Alu genotype. Two independent clones with the polymorphic Alu present and two constructs without the Alu present were generated and sequence verified. Each vector was transfected into 293T cells using Fugene HD (Promega) with manufacturer's suggested protocol. After 24 h, RNA was isolated using Quick RNA MicroPrep Kit (Zymo Research) per manufacturer's suggested protocols. cDNA was synthesized with iScript cDNA Synthesis Kit (BioRad) per manufacturer's protocol using 1 μg of RNA and random primers. PCR was performed with primers binding within the rat insulin exons (5′-CAGCACCTTTGTGGTTCTCA-3′, 5′-AGAGCAGATGCTGGTGCAG-3′). The relative quantification of alternatively spliced RNA isoforms was performed on ethidium bromide stained agarose gels with band intensities normalized for DNA fragment length. A representative gel image with the independent clones is shown for each locus. Two separate transfections were performed for each independent clone. For each locus, we performed ANOVA analysis and the only variable with significant differences was the Alu genotype. Therefore, quantifications from the two independent clones and their replicates were combined resulting in four data points for each type of construct (i.e. with or without Alu) for each locus tested. The quantification of each clone from each experiment is available in Supplemental Figure S1. Quantification is graphed as percent of transcripts that skip the alternative exon. Unpaired t-tests were used to compare the percent exon skipping for each construct type for a given locus and adjusted for multiple comparisons when necessary as described below.
To define further the mechanism by which the Alu polymorphism alters splicing, additional constructs were generated for the CD58 and SLC2A9 loci. These sequences included one of the following being cloned in place of the polymorphic Alu sequence: AluYa5 consensus sequence with an average length polyA tail (Supplementary Table S2), randomized AluYa5 consensus sequence (with the polyA tail sequence scrambled with the rest of the Alu sequence), or randomized sequence with GC content matching the pre-insertion integration site (the same length as the polymorphic Alu including the polyA tail) (Supplementary Table S2). For all randomized sequences, two different sequences were evaluated at each locus to decrease the potential to confound results by introducing cryptic regulators. When results differed for the scrambled Alu sequence at CD58, a third scrambled sequence was also evaluated (Supplementary Table S2). These sequences were synthesized (Invitrogen) and cloned in place of the polymorphic Alu element in the vector using Gibson Assembly (New England BioLabs). Final vectors were sequence verified to ensure seamless replacement of the polymorphic Alu sequence with the desired synthesized fragments. Because seven or eight constructs were evaluated at SLC2A9 and CD58 respectively, the P-value of 0.05 was adjusted to 0.0024 or 0.0019 to account for the 21 or 26 unpaired t-tests that were performed.
Additional analysis at CD58 locus
Genotyping and linkage disequilibrium analysis using a 30 trio reference panel of Centre d'Étude du Polymorphisme Humain (CEPH) Utah Residents with Northern and Western European Ancestry (CEU) (Coriell Institute for Medical Research, Camden, NJ) was previously reported (22). LD plots were generated using Haploview (27) as previously described (22). Lymphoblastoid cell lines were grown in RPMI with 15% fetal bovine serum at 37°C in 5% carbon dioxide. RNA was isolated with Trizol (Invitrogen) and cDNA was prepared as above. To evaluate the endogenous CD58 locus, RT-PCR was performed with primers (5-TGGTTCTGTCTGGTTTTCTGTC-3′, 5′-TGGTGTTGTGTATGGGAATGT-3′) binding in constitutive exons flanking the alternative exon and intronic polymorphism. Amplicons were run on an agarose gel and quantified as above. To quantify these results using a second method, the experiment was repeated and samples were also run on the Fragment Analyzer Automated CE System (Advanced Analytical) and quantified with the accompanying PROsize software. Similar results were obtained for quantification with peak height or peak area; data shown uses peak height. For either method, samples were stratified based on genotype and unpaired t-test was performed with the p-value of 0.05. We also quantified the splice variants at the endogenous locus using qRT-PCR. Primers were designed to differentiate the alternative isoforms (Supplementary Figure S2). To measure the skip of exon 3, we used a primer spanning exons 2 and 4, resulting in a product only when exon 3 was skipped. To measure the exon 3 inclusion, we used a primer that binds within exon 3. In both cases, the isoform-specific primer was paried with a shared primer in exon 2. Overall gene expression level was detected with downstream primers that would amplify the mRNA regardless of exon 3 inclusion/skip. GAPDH expression was another tested control; using either overall CD58 or GAPDH yielded comparable results. RNA was isolated and cDNA prepared as above for seven samples with varying Alu genotypes (Supplementary Figure S2). qRT-PCR was performed in 20 μl reactions with 1 μl of cDNA with SsoAdvanced universal SYBR green supermix (BioRad). Reactions were initially denatured at 98°C for 30 s, and then 45 cycles of 98°C for 10 s and 60°C for 30s. Thermal dissociation curves resulted in a single clean peak for each primer pair and reaction. Ct values were determined using the standard parameters of the program. All –RT reactions had a Ct value that was not determined as no amplification was detected. For each primer pair, each sample was tested in triplicate. Relative expression of each the skip isoform and the isoform including exon 3 was calculated as 2−ΔCt where ΔCt = CtCD58 primer pair – CtGAPDH within each sample. We combined results for Individuals with the same genotype and graphed them together. Depicted results are from one experiment (Supplementary Figure S2). A second experiment gave similar results.
RESULTS
Candidate polymorphic Alu elements for splicing effects
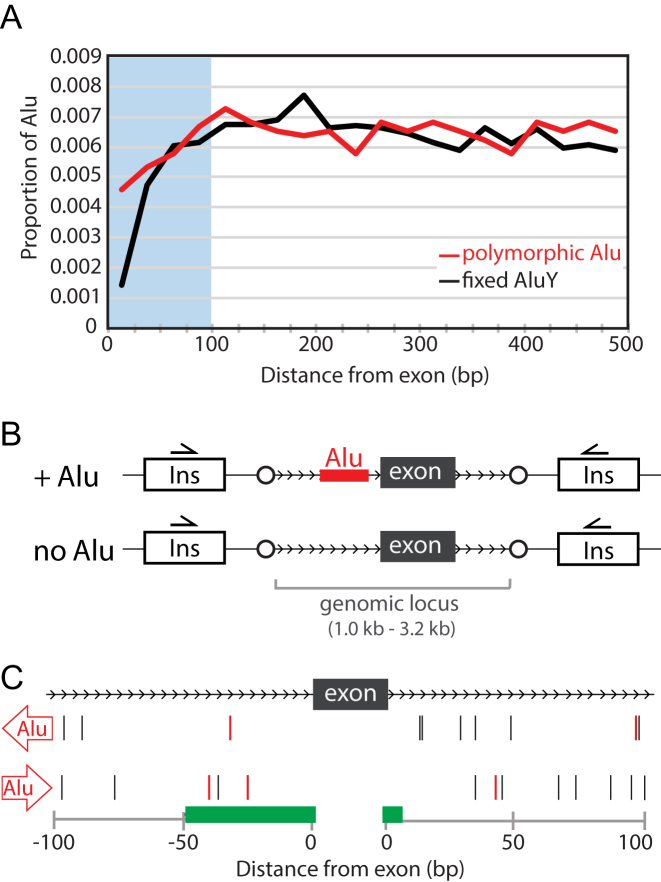
To identify Alu elements that alter splicing, we focused on those that occur within a short distance to the nearest exon. Most sequence determinants of splicing occur proximal to exon boundaries (e.g. (28,29)), and we expected that the Alu elements altering splicing would most likely fall within the previously reported ‘underrepresentation zone’ that spans 100 bp 5′ and 3′ of exons (21). To confirm the depletion of Alu elements near exons reported by Lev-Maor et al. and Zhang et al. (20,21), we assessed the distribution of fixed-present AluY insertion sites in 25 bp bins based on distance to exons. We recapitulate the underrepresentation of Alu elements near exons. This depletion is evident in the regions both immediately upstream and downstream of the exon. Moreover, Zhang et al. also showed a depletion of the relatively recently acquired, polymorphic Alu insertions near exons, suggesting that this effect results from rapid selection against these insertions (21). We evaluated this using a recently updated, more inclusive catalog of polymorphic Alu elements (8), and we also confirmed the depletion of polymorphic Alu insertion variants within this 100 bp underrepresentation zone (Figure 1A). This depletion is not absolute, however; in all, we identified 168 polymorphic Alu elements that map within 100 bp of an exon (Supplementary Table S3).
Figure 1.
Evaluation of polymorphic Alu elements mapping near exons. (A) Alu elements are depleted near exons. Proportions of intronic polymorphic Alu elements (red) or reference AluY (black) are shown in 25 bp bins from the nearest exon. (B) Two constructs for each locus were evaluated using ectopic mini-gene splicing assays. The genomic locus encompassing the alternatively used exon (black) and flanking sequence was cloned into the vector containing rat insulin (Ins) exons using attP sequences (circles) and Gateway cloning (Invitrogen). (C) Assayed polymorphic Alu elements are representative of all combinations of distance and position relative to the exon and orientation relative to gene transcription. Those with effects in the ectopic assay are in red. Green bars indicate regions associated with known splicing regulatory sequence. Upstream of the exon these regulatory sequences (from left to right) include the branch site and polypyrimidine tract (the exact distance from the exon can vary, so the most common range is depicted). At the upstream exon/intron boundary is the splice acceptor site. At the downstream exon/intron boundary is the splice donor site.
To identify polymorphic Alu sequences that affect exon usage, we focused on insertion variants near alternatively spliced exons. We considered both exons that are skipped and those with altered splice sites. We found 73 polymorphic Alu elements within 100 bp of an alternatively spliced exon, which is not significantly different from the expected rate (P = 0.3125, chi square test). However, these were of particular interest to us because we hypothesized that the polymorphic Alu element may be a determinant of splice-site choice. Further, as many of these exons are of lengths not divisible by three, isoforms including or excluding these exons would likely result in a frameshift (n = 41).
Polymorphic Alu elements near exons share features typical of polymorphic Alu elements genome-wide. Most of these Alu elements are full length or nearly full length (270 bp average plus the polyA tail, range 72–284 bp), with only two of these Alu variants having significantly shorter sequence (i.e. the Alu variants at DRAM1 and P2RX7). Also as expected, the majority of these elements are from the youngest, most commonly polymorphic Alu subfamilies (57.5%), including AluYa5, AluYb8 and AluYb9. Although more of these elements are anti-sense with respect to the gene, 42 compared to 31 that are sense with respect to the gene, this is not significantly different from expected (P = 0.1979, chi square test). Similarly, these polymorphic Alu elements map equally upstream (n = 40) and downstream (n = 33) of alternatively used exons (P = 0.671, chi-square test).
Polymorphic Alu elements can affect exon usage
To determine if presence of each Alu variant alters incorporation of its nearby exon, we evaluated exon splicing in an ectopic minigene-splicing assay (e.g. (26,30)). We used the pSpliceExpress vector, which enables cloning of genomic loci into the minigene reporter by recombination for higher throughput (26). This vector contains two constitutive rat insulin exons flanking an intron into which we cloned the sequences to be tested, the exon(s) and surrounding intronic sequences with and without corresponding Alu variant (Figure 1B). We evaluated transcript isoforms after expression of the minigene reporter in 293T cells, using RT-PCR primers specific to the constitutive rat insulin exons. In all, we tested 23 loci where a polymorphic Alu element mapped within 100 bp of an exon (Supplementary Table S1). In selecting loci, we captured examples of every combination of Alu orientation and position relative to the gene and nearby exon (Figure 1C) and a variety of common Alu subfamilies (Supplementary Table S3). We identified significant effects of the Alu insertion on exon usage at five of these loci (data from those without significant effects are in Supplementary Figure S3).
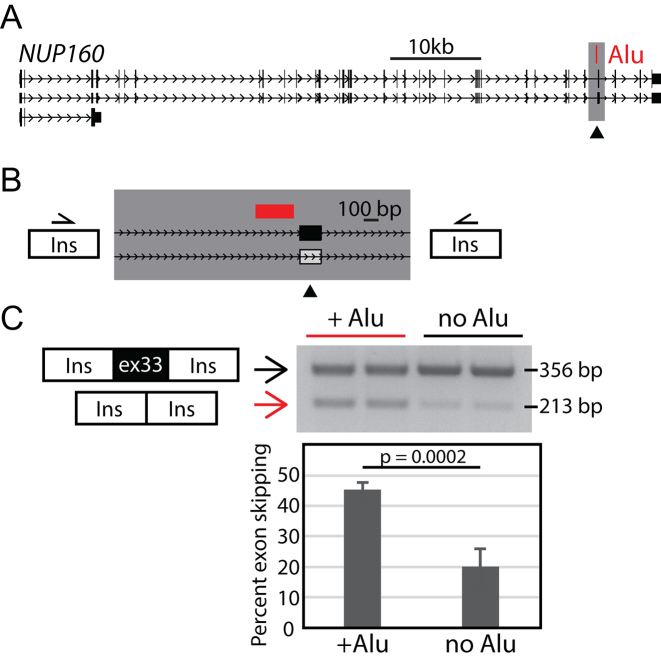
At one locus where we detected an effect, a polymorphic Alu element maps 41 bp upstream of exon 33 of the NUP160 gene (Figure 2A). NUP160 encodes Nucleoporin 160, a member of the 120-MD nuclear pore complex that mediates nucleoplasmic transport. Exon 33 of this gene is a near constitutive exon, but EST data (JD448821) suggest that it is skipped in a minor transcript isoform; skipping of the 143 bp exon 33 would result in a frameshift in the mRNA open reading frame. The 262 bp AluYh3a3 element at NUP160 is oriented antisense with respect to the gene. To determine its effect on exon usage, we tested a 1743 bp fragment of this locus, both with and without the Alu element present (Figure 2B), in the minigene reporter assay. We detect two different splice events with both constructs. Sanger sequencing of the RT-PCR products confirmed that one event includes the NUP160 exon 33 and the other skips the NUP160 exon. Both spliced products are detected with and without the Alu insertion; however, when the Alu is present, the exon is skipped significantly more often, 45.2%, compared to only 20% when the Alu is not present (P < 0.001) (Figure 2C, Supplemental Figure S1). This indicates that at least in the reporter assay this Alu polymorphism has an effect on exon usage; the presence of the Alu promotes exon skipping.
Figure 2.
Polymorphic Alu results in increased exon skipping at NUP160. (A) NUP160 locus. A polymorphic AluYh3a3 (red) is sense with respect to NUP160 transcription and maps 41 bp upstream of alternatively used exon 33 (arrowhead). The region included in the splicing reporter is highlighted in gray. (B) Spicing reporter assay of Alu insertion effects at NUP160 locus. Minigene-splicing reporters contain the genomic segment (gray box, also from A) with and without the Alu present (red). White box indicates exon is skipped in rare isoforms according to EST data. (C) A representative gel showing splicing assay amplicons from two independent clones for each construct assayed. Two bands of indicated sizes were quantified on an agarose gel after PCR with primers in the rat insulin (Ins) exons. The larger product incorporates NUP160 exon 33. Data were combined with a second replicate and graphed. Error bars are the standard deviation of the four values for each construct. Unpaired t-test results are shown.
Polymorphic Alu elements can promote exon skipping or exon inclusion
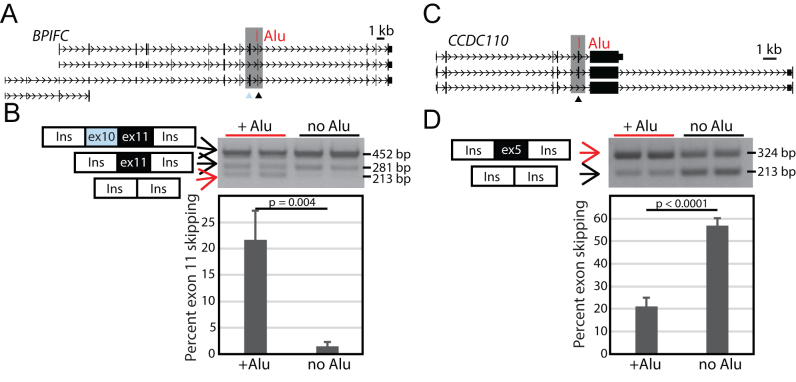
We identified 3 other polymorphic Alu elements that increase skipping of nearby exons similar to that observed at NUP160. One example is a polymorphic Alu element at 22q12.3 in BPIFC (Figure 3A). This gene encodes a BPI Fold Containing Family C protein that binds lipids and lipopolysaccharides. There are 8 alternatively used exons within this gene. One of these, exon 11 of the transcript, which is rarely skipped, maps 25 bp from a 261 bp polymorphic AluYa5 element (Figure 3A). The polymorphic AluYa5 is oriented in sense with respect to BPIFC. To test the effect of this Alu on mRNA splicing, we cloned a 2252 bp fragment encompassing the interval of BPIFC exons 10 and 11, both of which are alternatively incorporated exons, and flanking intronic sequences with and without the Alu element (Figure 3A). We detected three bands in the ectopic assay, and Sanger sequencing confirmed these bands correspond to inclusion of both exons, only exon 11, and neither exon (Figure 3B); inclusion of only exon 10 in the absence of exon 11 was not detected, consistent with annotated isoforms. We detected a notable increase in the skipping of both exons 10 and 11 in the presence of the Alu element. With the Alu, 21.6% of transcripts skip both exons 10 and 11 compared to only 1.6% of transcripts which skip these exons when the Alu is absent (P = 0.004) (Figure 3B, Supplemental Figure S1). Therefore, in the ectopic assay, this Alu polymorphism affects exon usage similarly to that at the NUP160 locus; the Alu promotes exon skipping. Similarly, we identified polymorphic Alu elements that promote skipping of a nearby exon at two other loci, SLC2A9 and CD58, which we will detail in subsequent sections.
Figure 3.
Polymorphic Alu elements can increase exon skipping or inclusion. (A) BP1FC locus. A polymorphic AluYa5 (red) is sense to the BP1FC gene and maps 25 bp upstream from alternatively used exon 11 (black arrowhead). There is also an alternatively used exon 10 (blue arrowhead) within the region that was evaluated in the splicing reporter (gray box). (B) A representative gel showing splicing assay amplicons from two independent clones for each construct assayed. Three spliced products were detected. Data were combined with a second replicate and the percent exon skipping of exon 11 is graphed with error bars indicating the standard deviation of the 4 values for each construct; the Alu results in increased exon skipping. (C) CCDC110 locus. A polymorphic AluY (red) is sense to the CCDC110 gene and maps 42 bp downstream of alternatively used exon 33 (black arrowhead). (D) Splicing assays were performed and are shown as in (B). At this locus, the polymorphic Alu element increases exon inclusion. Unpaired t-test results are shown.
We found that polymorphic Alu elements can alter exon usage in the opposite manner as well. While previous examples presented here indicate that the Alu elements promote exon skipping, at 4q35.1, we identified a polymorphic Alu element that promotes the inclusion of a nearby, annotated alternatively used exon in CCDC110 (NM_001145411.1). This alternatively used exon 5 is 111 bp in length. An Alu polymorphism maps 42 bp downstream of this exon (Figure 3C). The 257 bp AluY is in the same orientation as the CCDC110 gene, which encodes Coiled-coil Domain Containing 110 protein. We tested a 1077 bp region encompassing CCDC110 exon 5 and flanking intronic sequence in the minigene assay with and without the Alu present. When the Alu is present, the percent inclusion for exon 5 increases from 43.2% without the Alu to 79% when the Alu is present (Figure 3D, Supplemental Figure S1) (P < 0.0001). Therefore, in the ectopic assay, this polymorphic Alu element promotes the inclusion of the nearby alternatively used exon. Together with the other examples, this indicates that polymorphic Alu elements can alter inclusion rates for an exon in either direction: promoting exon inclusion or exon skipping.
A polymorphic Alu at SLC2A9 promotes exon skipping in a sequence independent manner
To test the hypothesis that Alu polymorphisms alter mRNA splicing resulting in an effect on gene function and disease risk, we evaluated two instances where an Alu variant near an alternatively spliced exon occurred in proximity to a genome-wide association study (GWAS) signal (22).
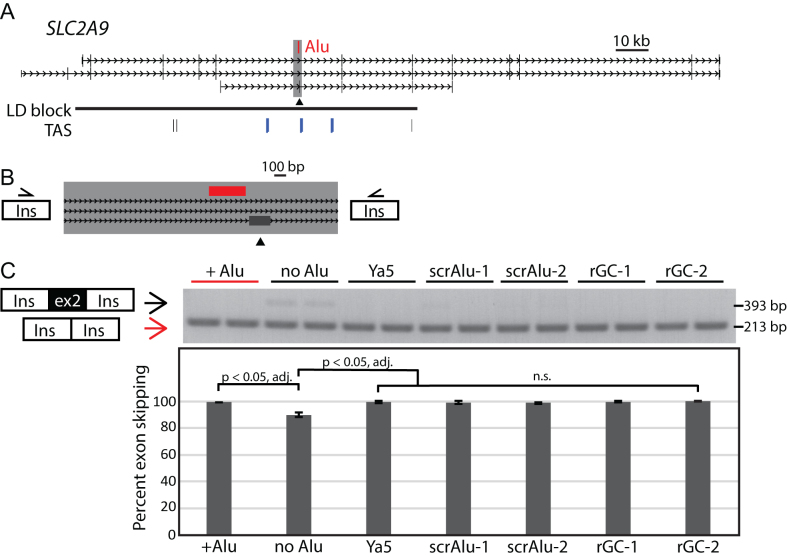
At the first of these, a 284 bp AluYi6 maps 31 bp upstream of an alternatively used exon in the SLC2A9 gene that encodes Solute Carrier Family 2 Member 9 protein that is involved in transmembrane transport of urate and fructose (Figure 4A). This locus has previously been associated with serum uric acid levels and risk for developing gout. Seven (7) trait-associated SNPs were reported, including rs4481233 and rs7442295 (P = 6 × 10−34 and 2 × 10−15) (e.g. (31,32)) which define a 105 kb linkage disequilibrium (LD) (r2 > 0.8) block. We previously reported that an Alu variant mapping to this interval is in moderate LD with two of these trait-associated SNPs and specifically associated with the protective haplotype (22). This moderate level of LD (r2 = 0.56, D’ = 1 with confidence intervals of 0.79–1) indicates that the Alu may cooperate with other variants on the same haplotype to affect SLC2A9 function. To evaluate effects of this Alu polymorphism on splicing, we cloned a 2312 bp fragment that encompassed the alternatively used exon and Alu polymorphism into our ectopic splicing vector (Figure 4B). We tested the locus with and without the Alu present in the construct. In contrast to other loci examined, the alternative exon is part of the minor isoform and is skipped in the majority of transcripts; this is exon 2 of this isoform (DA631706) in which it is included, but it is skipped in all other isoforms. This splice pattern was recapitulated in the ectopic assay, which also showed more frequent skipping when the Alu is present (Figure 4C, Supplemental Figure S1). Skipping is near complete when the Alu is present, 99%, compared to when the Alu is absent, 89.9% (P < 0.01).
Figure 4.
A polymorphic Alu increases exon skipping at SLC2A9 in a non-sequence specific manner. (A) SLC2A9 locus. A polymorphic AluYi6 (red) is antisense to SLC2A9 transcription and maps 34 bp upstream of alternatively used exon 2 (arrowhead). An LD block (horizontal black line) spans from chr4:9529704–9634418 (hg19) is defined as a region that is in strong LD (r2 > 0.8) encompassing six trait associated SNPs (TAS, vertical lines) associated with gout and serum urate levels; three are in moderate LD with the Alu (blue) r2 > 0.46 D’ = 1 (confidence intervals 0.8–1). SNPs from left to right are rs16890979, rs4475146, rs4481233, rs7442295, rs6449213 and rs775948. (B) Minigene-splicing vectors contain the region encompassing exon 2 and flanking sequence with or without the Alu (red). (C) Gel of representative splicing assay results showing two independent clones for each construct assayed. Two bands of indicated size were quantified from the agarose gel. Data were combined with a second replicate and graphed with error bars indicating the standard deviation of the 4 values for each construct. Constructs with the AluYi6, AluYa5 consensus sequence, scrambled Alu sequence (scrAlu), and spacer sequence with GC content matching the intron (randomized sequence with controlled GC content, rGC) are not statistically different from one another (n.s.). The difference between the no Alu (pre-insertion) construct and all other constructs is statistically significant (unpaired t-tests, *P-adjusted = 0.0002).
Given the clinical significance of this locus, we wanted to explore whether this suppressive effect on exon incorporation was an intrinsic feature of the Alu sequence (i.e., reflecting a sequence-specific effect of the Alu) or was caused by disruption to non-Alu regulatory sequences that function in the absence of the Alu (i.e. sequences that are intact only on the ‘empty’ allele). We hypothesized that the Alu could alter splicing by providing binding sites for splicing regulators (e.g. (33)) or that Alu RNA secondary structure (34,35) or overall GC content could regulate splice-site recognition (36). To test these models, we developed a series of reporter constructs. We replaced the AluYi6 with Alu sequence of a different, commonly polymorphic Alu subfamily (AluYa5 consensus sequence with an average size polyA tail, 5% divergent from AluYi6) or two different random scrambles of Alu sequence (the entire Alu sequence including the polyA tail was scrambled) to disrupt any protein binding sites or pre-mRNA secondary structure. We also tested the effect of two different randomized sequences of equal length to the Alu (including polyA tail) but with a GC content matching the integration site (46.6% GC compared to 61.6% GC for the Alu) (Supplementary Table S2 and methods). We found that all of these constructs resulted in >98% of transcripts skipping the SLC2A9 exon, similar to each other and to the AluYi6 containing construct (Figure 4C, Supplemental Figure S1). Specifically, the inclusion rate of SLC2A9 exon 2 is not statistically different for any of these constructs or the AluYi6-containing construct, but it is different relative to the construct containing no additional sequence (P < 0.05, adjusted for multiple testing). Thus, we conclude that the Alu polymorphism reduces inclusion of the alternatively used exon in SLC2A9, but does so independently of the specific Alu sequence. Since this is not a sequence-specific effect, we conclude that this Alu insertion variant disrupts splicing regulators that function when the Alu is not present.
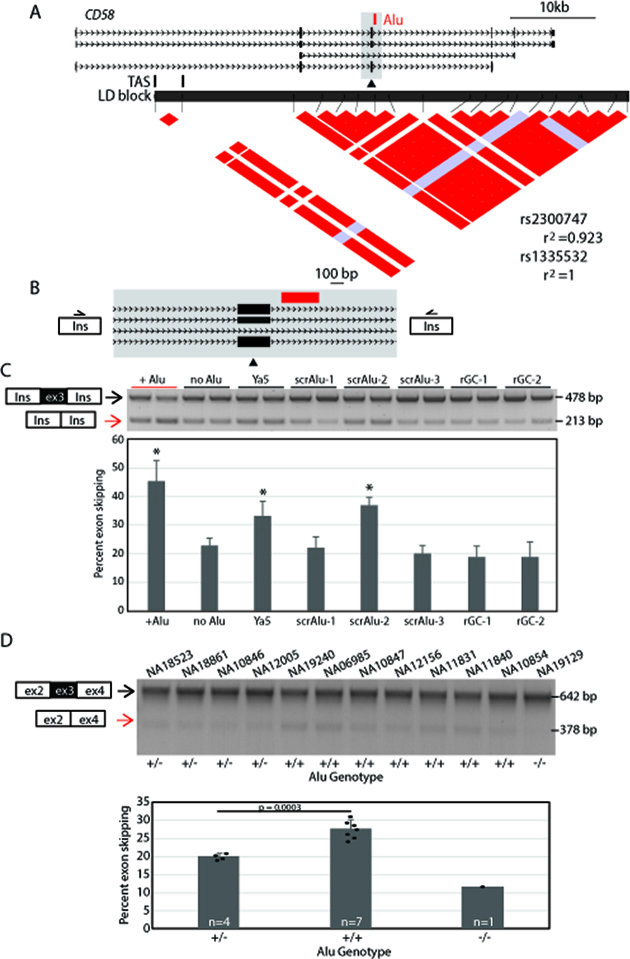
A polymorphic Alu responsible for CD58 exon skipping may affect multiple sclerosis risk
We previously reported an intronic Alu at CD58 mapping near a GWAS signal for multiple sclerosis (MS) risk (P = 3 × 10−16) (e.g. (22,37,38)). The risk allele is associated with lower expression (39) of CD58, a cell surface protein also known as lymphocyte function-associated antigen 3 (LFA-3), that is widely expressed in hematopoietic and non-hematopoietic cells. Reduced CD58 mRNA level is associated with both risk of developing MS and relapse of symptoms for those with MS (39). Fine mapping studies have narrowed the region to a 76 kb interval of CD58 (39) (Figure 5A). However, the causative variant leading to this disease risk has yet to been identified. A polymorphic 281 bp AluY element maps within this region, on the risk haplotype, and is in strong LD (r2 > 0.9) with the trait-associated SNPs identified by GWAS (Figure 5A).
Figure 5.
An Alu variant associated with MS risk is an sQTL at CD58. A) CD58 locus. Trait associated SNPs (TAS, vertical black lines) identify this as a multiple sclerosis risk locus by GWAS. The LD structure from the GWAS identified LD block (black horizontal line) was generated by pair-wise comparisons of variants. Two variants in near perfect LD (r2 > 0.9) are depicted with red coloring at the intersection of the LD plot. Gray-blue boxes indicate less LD between evaluated variants. The polymorphic Alu element is in LD with the 2 TASs, rs2300747 and rs1335532. LD values between the TAS and the polymorphic Alu are shown to the right of the plot. (B) Minigene-splicing assay of CD58 locus. The CD58 genomic locus (gray box, also from A) with and without the Alu present (red) encompasses alternatively used exon 3. (C) Representative gel of splicing assay results showing 2 independent clones for each construct assayed. Two bands of indicated size were quantified from agarose gel. Data were combined with a second replicate and graphed with error bars indicating the standard deviation of the four values for each construct. The construct containing the polymorphic AluY was compared to the allele without an Alu present and constructs where the polymorphic AluY was replaced with the AluYa5 consensus sequence, scrambled Alu sequence (scrAlu), and spacer sequence with GC content matching the intron (rGC). The polymorphic AluY has the greatest effect but is not statistically different from the AluYa5 consensus or scrAlu-2. These 3 constructs (+Alu, Ya5, and scrAlu-2) are denoted with a * because they are all statistically different than the construct with the empty naturally occurring allele (no Alu). This indicates an effect of these sequences on splicing relative to the no Alu allele. They are also statistically different from all constructs without a * (unpaired t-tests, P< 0.05, adjusted). (D) Analysis of CD58 splicing at the endogenous locus. With primers that bind in CD58 exons 2 and 4, two spliced products, of indicated sizes, were detected at the endogenous locus. Individuals (n = 12) with noted Alu genotypes were evaluated. Graph shows quantification based on genotype with each individual indicated by a black circle. Error bars are the standard deviation within the group. An unpaired t-test indicates a dose dependent effect between genotypes and percent exon skipping was detected.
To test the hypothesis that the polymorphic Alu element is the causative variant leading to increased disease risk by altering CD58 mRNA splicing, we investigated effects of the AluY on a nearby alternatively used exon; the AluY maps 97 bp downstream (3′) of the exon. The 265 bp exon 3 is skipped in the shortest isoform of CD58 (uc001ego.1, FLJ77504), altering the protein reading frame. We cloned a 2340 bp fragment containing the alternatively used exon and polymorphic Alu insertion site into the minigene reporter and measured exon inclusion (Figure 5B). The presence of the Alu results in a significant decrease in incorporation of the exon, 54.7% of the transcripts include the exon when the Alu is present as compared to 77.2% for the pre-insertion allele (Figure 5C, Supplemental Figure S1) (P < 0.05, adjusted). Therefore, presence of the Alu results in more exon skipping.
To further dissect the mechanism by which the Alu alters exon splicing, we replaced the Alu in the CD58-containing minigene reporter with AluYa5 subfamily consensus sequence (CD58 Alu is 2% divergent from AluYa5, Supplementary Figure S4), scrambled Alu sequences (62.6% GC), and randomized sequences of the same length matching the GC content of the pre-insertion intron (35.6% GC) (Supplementary Table S2). The polymorphic AluY sequence had the greatest effect on exon skipping of all constructs tested, with 45.3% of transcripts skipping exon 3 in the presence of the Alu (Figure 5C). The AluYa5 consensus sequence also resulted in a significant increase in exon 3 skipping relative to the empty allele, containing no Alu sequence (P<0.05, adjusted). The polymorphic variant at CD58 differs from the AluYa5 consensus sequence by 7 nucleotides distributed over the length of the elements (Supplementary Figure S4). The presence of the polymorphism results in relatively more exon skipping as compared to the AluYa5 consensus sequence (45.3% versus 33.2%), although this difference is not significant when accounting for the multiple comparisons made (Figure 5C, Supplemental Figure S1).
We next performed minigene reporter assays replacing the polymorphic AluY sequence with randomized sequences with varying nucleotide compositions. First, we scrambled the Alu sequence to retain the length and GC content of the naturally occurring polymorphism, but disrupt specific sequence motifs and RNA secondary structure. We initially evaluated two scrambled Alu sequences but these gave significantly different results (P < 0.05, adjusted) with 22% (scrAlu-1) or 37% (scrAlu-2) of transcripts skipping CD58 exon 3 when the scrambled sequences were present (Figure 5C, Supplemental Figure S1). Given the discrepant results, we tested a third version of scrambled Alu (scrAlu-3) and it behaved more similarly to scrAlu-1 with 20.1% of transcripts lacking exon 3. Next, we used randomized sequence that matched the GC content of the ‘empty’ allele intron (i.e. the integration site without the Alu insertion). Both of these random sequences (rGC-1, rGC-2) behaved similar to the construct with no Alu present resulting in 19% of transcripts lacking CD58 exon 3.
Altogether, these results indicate that the polymorphic AluY insertion variant at CD58 increases exon 3 skipping, and suggest that this effect is sequence dependent. While the naturally occurring AluY variant had the greatest effect of sequences we tested, the AluYa5 consensus sequence, and one of three scrambled Alu sequences also increase CD58 exon 3 skipping.
A polymorphic Alu element is a splicing QTL at CD58
We next wanted to evaluate haplotype effects on exon 3 splicing at the endogenous CD58 locus. CD58 is widely expressed in hematolymphoid cells, which is relevant to the MS disease phenotype and makes it ideal for splicing quantitative trait loci (sQTL) analysis as a large number of well-characterized, EBV-transformed lymphoblastoid cell lines (LCLs) expressing CD58 are readily accessible. Given this, we evaluated splicing patterns of endogenous CD58 in individuals from the Centre d’Etude du Polymorphism Humain (CEPH)/Utah Collection [CEU] HAPMAP population with differing CD58 haplotypes. We selected LCLs from 12 individuals; consistent with the reported minor allele frequency of 0.285 (8), seven of the samples were homozygous for the Alu insertion, four were heterozygous for the Alu, and one was homozygous for the empty (pre-insertion) allele (22). We isolated RNA from the LCLs, performed RT-PCR using primers in exons flanking the alternatively used exon, and used two different methods (agarose gel and fragment analyzer) to detect and quantify CD58 splice variants (Figure 5D, Supplementary Figure S5, methods). We identified a genotype-dependent effect on exon skipping. The presence of the Alu results in relatively more of the isoform lacking exon 3 (P = 7.96e–10, ANOVA). Further, homozygotes with two copies of the Alu-containing allele show greater exon skipping than heterozygotes having only one copy of the Alu-containing allele (P = 0.0003, t-test). Within one genotype, no significant differences were seen in percent exon skipping (P > 0.05, not significant, adjusted). To confirm this result, we used qRT-PCR to measure transcript isoforms independent of each other in a subset of the LCLs. We designed one primer pair to amplify only the exon 3-containing transcript (one primer binds within exon 3) and a second primer pair to selectively amplify the isoform without exon 3 (one primer spans the exon 2- exon 4 junction) (Supplemental Figure S2). We measured the ratio of the two isoforms and found there is more exon skipping in the presence of the Alu (P < 0.05, unpaired t-test) (Supplemental Figure S2). All of these results are in agreement with the ectopic minigene-splicing assay. Altogether, our findings demonstrate genetic evidence that the Alu insertion promotes skipping of CD58 exon 3.
DISCUSSION
Splicing and processing mRNA transcripts are key aspects of gene function, and genetic variants affecting these can be critical determinants of phenotypes (40,41). Sequences that regulate splicing are complex and not fully understood (e.g. (29)) and Alu insertions can alter their activities (11,13,19,20). In this report, we sought to delineate the impact of polymorphic Alu elements on alternative splicing. To this end, we evaluated 23 polymorphic Alu elements near exons for effects on exon splicing. All mapped within 100 bp of an exon, a zone where Alu elements are underrepresented genome-wide. Using a minigene reporter assay, we identified five of these loci where presence of the Alu variant altered splicing patterns, ∼22% of loci tested. These data indicate that common Alu variants can have effects on exon usage. While previous reports have identified effects of Alu sequences on splicing in minigene-splicing assays (19,20), our report is the first systematic study of common insertion polymorphisms for this effect. We now identify commonly occurring polymorphic Alu elements associated with altered exon incorporation rates.
That polymorphic Alu elements alter splice site choice may have physiological implications including as part of the common variant, common disease paradigm. Previously, we reported that a subset of polymorphic Alu elements may be functional variants contributing to disease risk detected by GWAS (22). Now, we demonstrate a mechanism by which this may occur. The polymorphic Alu element at the CD58 locus is in near-perfect LD with trait-associated SNPs identified for MS risk. Here, we demonstrate that the risk allele is a sQTL at CD58, and that the Alu promotes CD58 exon 3 skipping. This is predicted to result in a frame shifted protein, likely related to the reduced CD58 expression reported in MS patients (39).
Our results indicate that the mechanisms by which Alu elements alter mRNA isoform abundance are complex and locus-specific. We identified polymorphic Alu elements affecting splicing for elements both upstream and downstream of the alternative exon, sense and antisense to the gene, and at varying distances from the intron–exon junction (Figure 1C). Given that no Alu characteristic(s) was common to only Alu elements that effect splicing, we considered some genic features that may sensitize a locus to Alu-dependent alterations in splicing. We considered the lengths of both the alternatively spliced exon and the adjacent, Alu-containing intron. In both cases, these intervals did not differ between loci with and without an Alu effect (data not shown). We also considered that the Alu might only have an effect on exons with weak splice sites. However, we detect effects at loci with strong (e.g. CD58) or weak splice sites (e.g. SLC2A9). It is likely that some combination of Alu and genic sequences determine the impact (if any) of an Alu on splicing rates for a nearby exon. Because of the limited number of loci evaluated, we are not able to decipher the key regulatory code. Further, even among the loci where an Alu effected splicing, the direction and degree of that effect also varied; while the typical exon-proximal Alu element promotes exon skipping (Figures 2, 4 and 5), at one locus, the presence of the Alu element increases exon inclusion (Figure 3C and D).
Whether a polymorphic Alu element alters exon incorporation by disrupting or delivering key regulatory sequences also appears locus-dependent. We built a series of reporter constructs at two loci to address this question. At one, SLC2A9, randomized sequences had a similar effect on exon skipping indicating the Alu insertion likely has a disruptive effect (Figure 4). Consistent with this, the polymorphic Alu is located within a region that is replete in splicing regulatory sequences (i.e., within 40 bp upstream of an exon) (19). We suspect that many polymorphic Alu elements that fall within this region would have an effect on splicing by disrupting the relative positioning of key splicing sequences. Notably, however, an Alu insertion location relative to an exon is not a consistent predictor of the effect of the Alu on splicing patterns. While 3 of the Alu elements that induce exon skipping are a similar distance upstream of the exon, another locus with a polymorphic Alu element mapping to the same region did not have altered exon usage (Figure 1C).
In other cases, the Alu sequence itself may play a more sequence-specific role, such as at CD58 where the AluY insertion has an effect on exon usage not recapitulated by all randomized sequences tested (Figure 5C). As the CD58 polymorphic AluY maps almost 100 bp downstream of the alternatively used exon, it emphasize the longer-range effects of these structural variants on exon incorporation. Here, it appears that this Alu introduces sequences that promote exon skipping (Figure 5C). Native Alu sequences at this site may recruit splicing suppressor(s) or create secondary RNA structures that sterically hinder the splicing reaction. It is possible that at CD58, these mechanisms combine with others since scrambled Alu sequences inconsistently and partially recapitulate the effect (scrAlu-2, Figure 5C). These scrambled sequences likely remove specific protein binding sites and relieve significant secondary structures associated with Alu elements. Here, this Alu introduces sequences that suppress splicing and increases exon skipping (Figure 5C). Because Alu sequences contain cryptic splice sites, they can become exonized and part of the mature transcript (e.g. (4,15,16)). To suppress this exonization and maintain transcript integrity, hnRNP C binds to Alu elements preventing their exonization by competing with the splicing factor U2AF65 (33). While we do not detect exonization of the Alu at any of the loci evaluated here, this mechanism may explain the increased rate of exon skipping of exons nearby an Alu element. At loci with an Alu sequence dependent effect, such as CD58, the suppression of the Alu exonization may ‘spread’ beyond the limits of the Alu sequence, limiting the access of splicing factors to the nearby exon, resulting in additional exon skipping.
The minigene-splicing assay appears to reproduce genotype-dependent splicing patterns at the endogenous CD58 locus. The relatively high expression level of this gene in lymphoblastoid cell lines allowed us to evaluate splicing isoforms of the endogenous CD58 gene in individuals with various Alu variant genotypes. Our results demonstrated an Alu-allele dose-dependent increase in exon skipping. We were particularly interested in this locus because the Alu-containing CD58 haplotype is associated with MS risk, and decreased expression of CD58 has been associated with both disease risk and recurrence (e.g. (31,32)). Our results show that presence of the Alu on the risk allele results in increased exon 3 skipping, and that this effect is specific to this Alu sequence. Interestingly, increased skipping of an alternative exon of another gene, IL7R, has also been implicated in MS risk (e.g. (42,43)). The mechanism is different in these two cases. The IL7R alternative splice event reflects an interaction between cis-acting sequence variants and the trans-acting DDX39B regulator (43); DDX39B does not regulate CD58 splicing. Further studies will be necessary to fully elucidate how altered IL7R and CD58 gene functions contribute to MS risk.
Altogether, this work reveals a prevalent regulatory effect of polymorphic Alu elements mapping near alternative exons. At 22% of evaluated loci, we identified altered exon incorporation rates dependent on the Alu genotype. Further, it is likely that this is only part of the story since our experimental approach is not designed to capture tissue-specific or long-distance effects. It is well documented that mRNA isoform abundance can vary significantly between tissue types and developmental time points (e.g. (44–47)). While studies relating genotype and phenotype often encompass exonic variants, our work highlights the importance of also considering transcript structure and characterizing sequence variants adjacent to exons.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Multiple Sclerosis Society [PP-1704-27379]; National Institutes of Health [R01GM124531 to K.H.B.]. Funding for open access charge: Grant money.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J.. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008; 40:1413–1415. [DOI] [PubMed] [Google Scholar]
- 2. Xing Y., Lee C.. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:13526–13531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Modrek B., Lee C.J.. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 2003; 34:177–180. [DOI] [PubMed] [Google Scholar]
- 4. Sorek R., Ast G., Graur D.. Alu-containing exons are alternatively spliced. Genome Res. 2002; 12:1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matlin A.J., Clark F., Smith C.W.. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005; 6:386–398. [DOI] [PubMed] [Google Scholar]
- 6. Blencowe B.J. Alternative splicing: new insights from global analyses. Cell. 2006; 126:37–47. [DOI] [PubMed] [Google Scholar]
- 7. Manning K.S., Cooper T.A.. The roles of RNA processing in translating genotype to phenotype. Nat. Rev. Mol. Cell Biol. 2017; 18:102–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H. et al. . An integrated map of structural variation in 2,504 human genomes. Nature. 2015; 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. et al. . Initial sequencing and analysis of the human genome. Nature. 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 10. Deininger P.L., Moran J.V., Batzer M.A., Kazazian H.H. Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003; 13:651–658. [DOI] [PubMed] [Google Scholar]
- 11. Wallace M.R., Andersen L.B., Saulino A.M., Gregory P.E., Glover T.W., Collins F.S.. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991; 353:864–866. [DOI] [PubMed] [Google Scholar]
- 12. Gallus G.N., Cardaioli E., Rufa A., Da Pozzo P., Bianchi S., D’Eramo C., Collura M., Tumino M., Pavone L., Federico A.. Alu-element insertion in an OPA1 intron sequence associated with autosomal dominant optic atrophy. Mol. Vis. 2010; 16:178–183. [PMC free article] [PubMed] [Google Scholar]
- 13. Ganguly A., Dunbar T., Chen P., Godmilow L., Ganguly T.. Exon skipping caused by an intronic insertion of a young Alu Yb9 element leads to severe hemophilia A. Hum. Genet. 2003; 113:348–352. [DOI] [PubMed] [Google Scholar]
- 14. Tighe P.J., Stevens S.E., Dempsey S., Le Deist F., Rieux-Laucat F., Edgar J.D.. Inactivation of the Fas gene by Alu insertion: retrotransposition in an intron causing splicing variation and autoimmune lymphoproliferative syndrome. Genes Immun. 2002; 3:S66–S70. [DOI] [PubMed] [Google Scholar]
- 15. Sorek R., Lev-Maor G., Reznik M., Dagan T., Belinky F., Graur D., Ast G.. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in alu exons. Mol. Cell. 2004; 14:221–231. [DOI] [PubMed] [Google Scholar]
- 16. Krull M., Brosius J., Schmitz J.. Alu-SINE exonization: en route to protein-coding function. Mol. Biol. Evol. 2005; 22:1702–1711. [DOI] [PubMed] [Google Scholar]
- 17. Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L.. Complementary sequence-mediated exon circularization. Cell. 2014; 159:134–147. [DOI] [PubMed] [Google Scholar]
- 18. Kelly S., Greenman C., Cook P.R., Papantonis A.. Exon skipping is correlated with exon circularization. J. Mol. Biol. 2015; 427:2414–2417. [DOI] [PubMed] [Google Scholar]
- 19. Nakama M., Otsuka H., Ago Y., Sasai H., Abdelkreem E., Aoyama Y., Fukao T.. Intronic antisense Alu elements have a negative splicing effect on the inclusion of adjacent downstream exons. Gene. 2018; 664:84–89. [DOI] [PubMed] [Google Scholar]
- 20. Lev-Maor G., Ram O., Kim E., Sela N., Goren A., Levanon E.Y., Ast G.. Intronic Alus influence alternative splicing. PLoS Genet. 2008; 4:e1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y., Romanish M.T., Mager D.L.. Distributions of transposable elements reveal hazardous zones in mammalian introns. PLoS Comput. Biol. 2011; 7:e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Payer L.M., Steranka J.P., Yang W.R., Kryatova M., Medabalimi S., Ardeljan D., Liu C., Boeke J.D., Avramopoulos D., Burns K.H.. Structural variants caused by Alu insertions are associated with risks for many human diseases. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E3984–E3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000; 16:418–420. [DOI] [PubMed] [Google Scholar]
- 24. Busch A., Hertel K.J.. HEXEvent: a database of human EXon splicing events. Nucleic Acids Res. 2013; 41:D118–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kishore S., Khanna A., Stamm S.. Rapid generation of splicing reporters with pSpliceExpress. Gene. 2008; 427:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barrett J.C., Fry B., Maller J., Daly M.J.. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005; 21:263–265. [DOI] [PubMed] [Google Scholar]
- 28. Rosenberg A.B., Patwardhan R.P., Shendure J., Seelig G.. Learning the sequence determinants of alternative splicing from millions of random sequences. Cell. 2015; 163:698–711. [DOI] [PubMed] [Google Scholar]
- 29. Barash Y., Calarco J.A., Gao W., Pan Q., Wang X., Shai O., Blencowe B.J., Frey B.J.. Deciphering the splicing code. Nature. 2010; 465:53–59. [DOI] [PubMed] [Google Scholar]
- 30. Cooper T.A. Use of minigene systems to dissect alternative splicing elements. Methods. 2005; 37:331–340. [DOI] [PubMed] [Google Scholar]
- 31. Suhre K., Shin S.Y., Petersen A.K., Mohney R.P., Meredith D., Wagele B., Altmaier E. CardioGram . CardioGram Deloukas P., Erdmann J. et al. . Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011; 477:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wallace C., Newhouse S.J., Braund P., Zhang F., Tobin M., Falchi M., Ahmadi K., Dobson R.J., Marcano A.C., Hajat C. et al. . Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 2008; 82:139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zarnack K., Konig J., Tajnik M., Martincorena I., Eustermann S., Stevant I., Reyes A., Anders S., Luscombe N.M., Ule J.. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013; 152:453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sinnett D., Richer C., Deragon J.M., Labuda D.. Alu RNA secondary structure consists of two independent 7 SL RNA-like folding units. J. Biol. Chem. 1991; 266:8675–8678. [PubMed] [Google Scholar]
- 35. Shepard P.J., Hertel K.J.. Conserved RNA secondary structures promote alternative splicing. RNA. 2008; 14:1463–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amit M., Donyo M., Hollander D., Goren A., Kim E., Gelfman S., Lev-Maor G., Burstein D., Schwartz S., Postolsky B. et al. . Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012; 1:543–556. [DOI] [PubMed] [Google Scholar]
- 37. De Jager P.L., Jia X., Wang J., de Bakker P.I., Ottoboni L., Aggarwal N.T., Piccio L., Raychaudhuri S., Tran D., Aubin C. et al. . Meta-analysis of genome scans and replication identify CD6, IRF8 and TNFRSF1A as new multiple sclerosis susceptibility loci. Nat. Genet. 2009; 41:776–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. International Multiple Sclerosis Genetics Consortium Wellcome Trust Case Control, Consortium Sawcer S., Hellenthal G., Pirinen M., Spencer C.C., Patsopoulos N.A., Moutsianas L., Dilthey A., Su Z. et al. . Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011; 476:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Jager P.L., Baecher-Allan C., Maier L.M., Arthur A.T., Ottoboni L., Barcellos L., McCauley J.L., Sawcer S., Goris A., Saarela J. et al. . The role of the CD58 locus in multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5264–5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lappalainen T., Sammeth M., Friedlander M.R., t Hoen P.A., Monlong J., Rivas M.A., Gonzalez-Porta M., Kurbatova N., Griebel T., Ferreira P.G. et al. . Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013; 501:506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwan T., Benovoy D., Dias C., Gurd S., Provencher C., Beaulieu P., Hudson T.J., Sladek R., Majewski J.. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008; 40:225–231. [DOI] [PubMed] [Google Scholar]
- 42. Gregory S.G., Schmidt S., Seth P., Oksenberg J.R., Hart J., Prokop A., Caillier S.J., Ban M., Goris A., Barcellos L.F. et al. . Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat. Genet. 2007; 39:1083–1091. [DOI] [PubMed] [Google Scholar]
- 43. Galarza-Munoz G., Briggs F.B.S., Evsyukova I., Schott-Lerner G., Kennedy E.M., Nyanhete T., Wang L., Bergamaschi L., Widen S.G., Tomaras G.D. et al. . Human epistatic interaction controls IL7R splicing and increases multiple sclerosis risk. Cell. 2017; 169:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A.. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. U.S.A. 2008; 105:20333–20338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Venables J.P., Lapasset L., Gadea G., Fort P., Klinck R., Irimia M., Vignal E., Thibault P., Prinos P., Chabot B. et al. . MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat. Commun. 2013; 4:2480. [DOI] [PubMed] [Google Scholar]
- 46. Ghigna C., Giordano S., Shen H., Benvenuto F., Castiglioni F., Comoglio P.M., Green M.R., Riva S., Biamonti G.. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell. 2005; 20:881–890. [DOI] [PubMed] [Google Scholar]
- 47. Thiery J.P., Acloque H., Huang R.Y., Nieto M.A.. Epithelial-mesenchymal transitions in development and disease. Cell. 2009; 139:871–890. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.