Abstract
N 6-methyladenosine (m6A) constitutes one of the most abundant internal RNA modifications and is critical for RNA metabolism and function. It has been previously reported that viral RNA contains internal m6A modifications; however, only recently the function of m6A modification in viral RNAs has been elucidated during infections of HIV, hepatitis C virus and Zika virus. In the present study, we found that enterovirus 71 (EV71) RNA undergoes m6A modification during viral infection, which alters the expression and localization of the methyltransferase and demethylase of m6A, and its binding proteins. Moreover, knockdown of m6A methyltransferase resulted in decreased EV71 replication, whereas knockdown of the demethylase had the opposite effect. Further study showed that the m6A binding proteins also participate in the regulation of viral replication. In particular, two m6A modification sites were identified in the viral genome, of which mutations resulted in decreased virus replication, suggesting that m6A modification plays an important role in EV71 replication. Notably, we found that METTL3 interacted with viral RNA-dependent RNA polymerase 3D and induced enhanced sumoylation and ubiquitination of the 3D polymerase that boosted viral replication. Taken together, our findings demonstrated that the host m6A modification complex interacts with viral proteins to modulate EV71 replication.
INTRODUCTION
Chemical modifications of RNA are critical for RNA metabolism, function, and localization. One of the most abundant internal RNA modifications is N6-methyladenosine (m6A), which is catalyzed by a methyltransferase complex consisting of METTL3, METTL14, WTAP and other proteins such as KIAA1429, RBM15 and RBM15B (1–11). In turn, FTO and ALKBH5 (10–13) constitute m6A demethylases that remove the methyl groups from RNA. YTH proteins bind to m6A sites and play critical roles in various biological processes, such as mRNA stability (14–16), RNA structure (17), mRNA nuclear export (13), mRNA splicing (16) and translation (18,19). Overall, the internal m6A modification of mRNA is mainly distributed in translation start sites, stop codons, and 3′ untranslated regions (3′ UTRs) (2,5,20).
The internal m6A modification of viral RNA was identified 40 years ago during infections of viruses that replicate in nucleus, such as influenza virus, simian virus 40 (SV40), and Rous sarcoma virus (RSV) (21–26). However, only recently the function of m6A began to be unraveled during virus infection. During infections of human immunodeficiency virus (HIV), SV40, Kaposi's sarcoma-associated herpesvirus (KSHV) and influenza virus, it has been shown that these viral RNAs contain m6A modifications, which affect viral replication and gene expression (27–34). Viruses that whose genomes replicate in the cytoplasm, such as VSV, vaccinia virus and reovirus, have also been reported to contain m6A-modified 5′ caps (35–38). Other cytoplasm replicating viruses, such as hepatitis C virus (HCV), Zika virus (ZIKV), dengue virus, yellow fever virus and West Nile virus, have been shown to contain internal m6A modifications that are involved in virus replication (39). All together, these reports indicate that cellular m6A methyltransferases may be active in the cytoplasm and that viral RNA is perhaps m6A-methylated during infection.
As an important human pathogen, enterovirus type 71 (EV71) is a non-enveloped single-stranded RNA virus, belonging to the family Picornaviridae (40) that has three genotypes (A, B and C) and several sub-genotypes (41). In recent years, EV71 has been the major pathogenic enterovirus responsible for hand-foot-and-mouth disease epidemics in Asia. Notably, since 2008, large-scale outbreaks of hand-foot-and-mouth disease have been reported yearly in mainland China, which resulted in hundreds of deaths and were caused mainly by the c4a clade of the C4 EV71 subtype (42).
In this study, we demonstrated that EV71 RNA contains m6A modification and investigated its function during EV71 C4 subtype infection. We found that the expression and localization of m6A methyltransferases, demethylases, and binding proteins were affected upon virus infection. Moreover, perturbation of the expression of m6A-related proteins or mutation of the m6A modification sites altered viral replication, suggesting that the host m6A machinery is involved in viral replication. Notably, we showed that the m6A methyltransferase METTL3 not only interacted with viral RNA-dependent RNA polymerase (RdRp) 3D, but also induced sumoylation and ubiquitination of the polymerase, which have been reported to facilitate its stability and boost viral replication (43). Taken together, our findings implied that m6A modification of EV71 RNA constitutes an important process in the regulation of viral replication.
MATERIALS AND METHODS
Cell culture
Vero (American Type Culture Collection (ATCC), Manassas, VA, USA; CCL-81), HEK293T (ATCC, CRL-11268) and RD (ATCC, CCL-136) cells were cultured in Dulbecco's modified Eagle's medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (Gibco) with 5% CO2 at 37°C.
Viruses
EV71 (strain XF; Microorganisms & Viruses Culture Collection Center (MVCCC)) was obtained from the MVCCC, Wuhan Institute of Virology (WIV), Chinese Academy of Sciences (CAS). Viruses were amplified and titrated by 50% tissue culture infectious dose (TCID50) in Vero cells using the Reed–Muench formula (44).
m6A-Methylated RNA immunoprecipitation (MeRIP) and Northern blotting
Total RNA was extracted from Vero cells infected with strain EV71-XF at a multiplicity of infection (MOI) of 0.1 using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). In vitro EV71 RNA was transcribed from a cDNA plasmid (45) linearized by HindIII using the MEGAscript® T7 Kit (Ambion, Austin, TX, USA) according to the manufacturer's protocols. For MeRIP, 300 μg of total RNA or 10 μg in vitro transcribed EV71 RNA were incubated with an anti-m6A antibody (Synaptic Systems, Goettingen, Germany) or a IgG antibody in 300 μl of immunoprecipitation (IP) Buffer (150 mM NaCl, 0.1% NP-40, 10 mM Tris–HCl, pH 7.4) for 2 h at 4°C. The mixture was then incubated with 20 μl of anti-rabbit antibody conjugated magnetic beads (NEB, Ipswich, MA, USA; S1432S), which were then washed three times with 500 μl of IP buffer, followed by rotating for 2 h at 4°C. Beads were washed six times with 500 μl of IP buffer and then incubated with 300 μl of elution buffer (5 mM Tris–HCl, pH 7.5, 1 mM EDTA, pH 8.0, 0.05% sodium dodecyl sulfate (SDS), and 4.2 μl of 20 mg/ml proteinase K) for 1.5 h at 50°C. The eluted RNA was extracted with phenol/chloroform and precipitated with ethanol.
All the RNAs collected from MeRIP were separated on 1% agarose/2.2 M formaldehyde gels in running buffer (20 mM MOPS, 5 mM sodium acetate, 1 mM EDTA, pH 7.0) for 13 h at 28 V. The RNAs were transferred to Hybond-N+ membranes in 20× SSC buffer (3.0 M NaCl, 0.3 M sodium citrate) overnight. UV-crosslinked to a membrane, and hybridized with a DIG-labelled EV71 probe (nt 1–7405). Probe detection was performed using the DIG Luminescent Detection Kit II (Roche, Madison, WI, USA) according to the manufacturer's instructions. Signals were developed on a ChemiDoc™ MP imaging system (Bio-Rad Laboratories, Berkeley, CA, USA).
MeRIP-Seq
MeRIP-Seq of the EV71 methylome was carried out according to a previously published protocol (46). In brief, total cellular RNA extracted from EV71-infected Vero cells was fragmented by ZnCl2 followed by ethanol precipitation. Fragmented RNA was incubated with an anti-m6A antibody (Synaptic Systems, 1:300). MeRIP was conducted as previously described (46). The eluted RNA and input were subjected to high-throughput sequencing using standard protocols (Illumina, San Diego, CA, USA). The MeRIP-Seq data were analyzed as described previously (32).
Ultra-high performance liquid chromatography-tandem mass spectrometry (UHPLC-MS/MS)
EV71 stock (1 L at 2 × 108 TCID50/ml) was concentrated by ultracentrifugation at 26 000 rpm in a SW28Ti rotor (Beckman Coulter, Brea, CA, USA) for 2 h at 4°C. Viral RNA was extracted using an RNeasy mini kit (QIAGEN, Venlo, The Netherlands). UHPLC-MS/MS analysis was performed on an Agilent 1290 UHPLC system coupled with an ESI-triple quadrupole mass spectrometer (G6410B or G6495, Agilent Technologies, Santa Clara, CA, USA) according to previously published instructions (47).
Formaldehyde-crosslinked RNA-immunoprecipitation (RIP)
Two 10-cm plates of 95% confluent RD cells were used for each sample. Cells were crosslinked by adding phosphate buffered saline (PBS) containing 1% methanol-free formaldehyde and incubated for 10 min at 37°C. Cross-linking was terminated by the addition of 2.5 M glycine to a final concentration of 0.125 M. Cells were washed three times with ice-cold PBS and scraped off the plates, followed by centrifugation at 800 × g for 3 min at 4°C. Cell pellets were resuspended in 400 μl of RIP buffer (150 mM KCl, 25 mM Tris-HCl pH 7.4, 5 mM EDTA, 0.5 mM dithiothreitol (DTT), 0.5% NP40, 100 U/ml RNase inhibitor, 100 μM phenylmethylsulfonyl fluoride (PMSF) and 1 μg/ml proteinase Inhibitors). The lysates were centrifugated at 16 000 × g for 10 min, and the supernatant containing the protein-RNA complexes was subjected to IP overnight with an anti-Flag (Sigma-Aldrich, St. Louis, MO, USA) or mouse IgG (control) antibody. On the following day, pre-blocked protein-G agarose beads (30 μl) were added to each sample for 1 h at 4°C. The beads were then washed three times each with the washing buffer (300 mM KCl, 25 mM Tris–HCl pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, 100 U/ml RNase inhibitor, 100 μM PMSF and 1 μg/ml proteinase Inhibitors), followed by three washes with the RIP buffer. After proteinase K digestion, the RNA samples were extracted using TRIzol.
Primer extension analysis
A primer extension analysis using Tth DNA polymerase and AMV reverse transcriptase was performed as previously described (48). Briefly, DNA primers for 28S RNA (A4190: 5′-GAG CTC GCC TTA GGA CAC CTG CG-3′) or EV71 RNA (A3055: 5′–TTG TGT TCC CCG AAT GTG GGA TAT CCG-3′, A4555: 5′-TGT TGC TTA TAA CCG TCA AAA TGA TCC GGG-3′) were 5′ end-labeled with 32P using T4 polynucleotide kinase (NEB, Ipswich, MA, USA) and [γ-32P] ATP (PerkinElmer, Waltham, MA, USA) according to a standard method. Radiolabeled primer (2 μl) and 10 μg of total RNA extracted from normal HEK293T cells or 20 μg poly(A) RNA from EV71-infected Vero cells were mixed in annealing solution followed by denaturation at 95°C for 10 min. After addition of the enzymes (2 μl; final enzyme concentration of 0.05 U/μl for Tth pol and 0.3 U/μl for AMV RT), the mixtures were heated at 55°C (Tth) or 37°C (AMV) for 2 min. A dTTP solution was then added to a final concentration of 100 μM, and the mixtures were incubated at 55°C (Tth) or 37°C (AMV) for 30 min. The final products were resolved on a 20% denaturing polyacrylamide gel. The signal was developed on a ChemiDoc™ MP imaging system (Bio-Rad).
Western blot analysis
Cells were seeded 1 day before infection at 80% confluence and infected with virus (MOI = 1). The cells were then harvested and lysed at the indicated times. The cell lysates were centrifuged and quantified, then denatured by boiling in a loading buffer for 10 min, and were subjected to SDS polyacrylamide gel electrophoresis (SDS-PAGE). Western blot analysis was performed using a standard protocol (49).
Antibodies and reagents used
Primary antibodies used in our study are as follows: mouse monoclonal antibody against GAPDH (60004-1-lg, Proteintech, Rosemont, IL, USA), rabbit polyclonal antibody against GAPDH (10494-1-AP, Proteintech), mouse monoclonal antibody against beta-actin (sc47778, Santa Cruz Biotechnology, Dallas, TX, USA), rabbit monoclonal antibody against METTL3 (15073-1-AP, Proteintech), anti-METTL14 (SAB1104405, Sigma-Aldrich), anti-WTAP (ab155655, Abcam, Cambridge, UK), anti-ALKBH5 (ab69325, Abcam), anti-FTO (ab124892, Abcam), anti-YTHDF1 (17479-1-AP, Proteintech), anti-YTHDF2 (24744-1-AP, Proteintech), anti-YTHDF3 (25537-1-AP, Proteintech), anti-YTHDC1 (14392-1-AP, Proteintech), anti-Histone 3 (GTX122148, GeneTex), anti-Flag (F1804-1 MG, Sigma-Aldrich), anti-HA (66006-1-Ig, Proteintech) and a mouse polyclonal antibody against EV71 VP1 generated in house. The secondary antibodies used in the study are goat anti-mouse IgG and goat anti-rabbit IgG were supplied by AntiGene Biotech GmbH, Stuttgart, Germany. Alexa Fluor 488, Alexa Fluor 568-conjugated secondary antibodies, and Hoechst 33258 were purchased from Invitrogen.
Immunofluorescence confocal microscopy
Vero cells were seeded in six-well plates 1 day before infection at ∼50% confluence, then the cells were infected with EV71 (MOI = 1) and incubated for the indicated times. Indirect immunofluorescence assay was performed as described previously (50). In brief, cells were washed three times with PBS and fixed in 3.7% paraformaldehyde in PBS for 15 min, then permeabilized in 0.5% Triton X-100 for 3 min and blocked in 3% bovine serum albumin for 1 h at room temperature. Cells were incubated with primary antibodies at a dilution as manufactures suggested overnight at 4°C, then washed three times with PBS and stained with a respective secondary antibody for 1 h at room temperature. Nuclei were stained with Hoechst. The slides were observed under a PerkinElmer VoX confocal microscope.
Short hairpin (sh) RNA-mediated gene silencing
shRNAs specific to each gene used in the study are as follows: METTL3 (shMETTL3-1: 5′-GCC AAG GAA CAA TCC ATT GTT-3′, shMETTL3-2: 5′-CGT CAG TAT ATT GGG CAA GTT-3′), FTO (shFTO-1: 5′-TCA CCA AGG AGA CTG CTA TTT-3′, shFTO-2: 5′-GAT CCA AGG CAA AGA TTT ACT-3′), YTHDF1 (shYTHDF1-1: 5′-CCC GAA AGA GTT TGA GTG GAA-3′, shYTHDF1-2: 5′-CCC TAC CTG TCC AGC TAT TAC-3′), YTHDF2 (shYTHDF2-1: 5′-CCA CAG GCA AGG CCC AAT AAT-3′, shYTHDF2-2: 5′-AAG GAC GTT CCC AAT AGC CAA-3′) and YTHDF3 (shYTHDF3-1: 5′-GAT AAG TGG AAG GGC AAA TTT-3′, shYTHDF3-2: 5′-TAA GTC AAA GAA GAC GTA TTA-3′), and YTHDC1 (shYTHDC1-1: 5′-TGG ATT TGC AGG CGT GAA TTA-3′, shYTHDC1-2: 5′-CAC CAG AGA CCA GGG TAT TTA-3′). They were cloned into the pLKO.1-TRC vector (Addgene plasmid 10878, Cambridge, MA, USA) and packaged into lentiviruses according to the manufacturer's instructions. Stable knockdown cell lines were generated by lentiviral infection followed with puromycin selection. Vero cells were selected under puromycin at 10 μg/ml, while HEK293T and RD cells were at 2 μg/ml.
Quantitative reverse-transcription PCR (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen). Reverse transcription was performed with 3 μg of total RNA using M-MLV reverse transcriptase (Invitrogen). qRT-PCR was performed using the SYBR Green Realtime PCR Master Mix (Toyobo, Osaka, Japan) on a CFX Connect Real-Time system (Bio-Rad). Relative gene expression levels were obtained by normalizing the quantification Cycle (Cq) values to those of GAPDH to yield 2−△△Cq. For Formaldehyde RIP-qPCR and MeRIP-qPCR, relative enrichment was normalized to inputs. The primers used for gene expression are as follows: EV71 (forward: 5′-CGA ATG CTA GTG ATG AGA GTA T-3′, reverse: 5′-GAG GAA GAT CTA TCT CCC CAA CT-3′) and GAPDH (forward: 5′-GAA GGT GAA GGT CGG AGT C-3′, reverse: 5′-GAA GAT GGT GAT GGG ATT TC-3′).
Sumoylation and ubiquitination assays
Sumoylation and ubiquitination assays were performed as described (43). Briefly, HEK293T cells were co-transfected with the indicated plasmids (Figure 7) using calcium phosphate reagent. Cells were cultured in Dulbecco's modified Eagle's medium (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (Gibco) with 5% CO2 at 37°C for 30 h and then lysed for western blotting and IP. The lysates were centrifuged at 16 000 × g at 4°C for 10 min. 50 μl of protein G Dynabeads was incubated with 10 μg of indicated antibody for 20 min, followed by incubation with the cell lysate for 25 min. The complexes were washed several times with PBST (PBS with 0.02% Tween 20) and were subjected to western blotting.
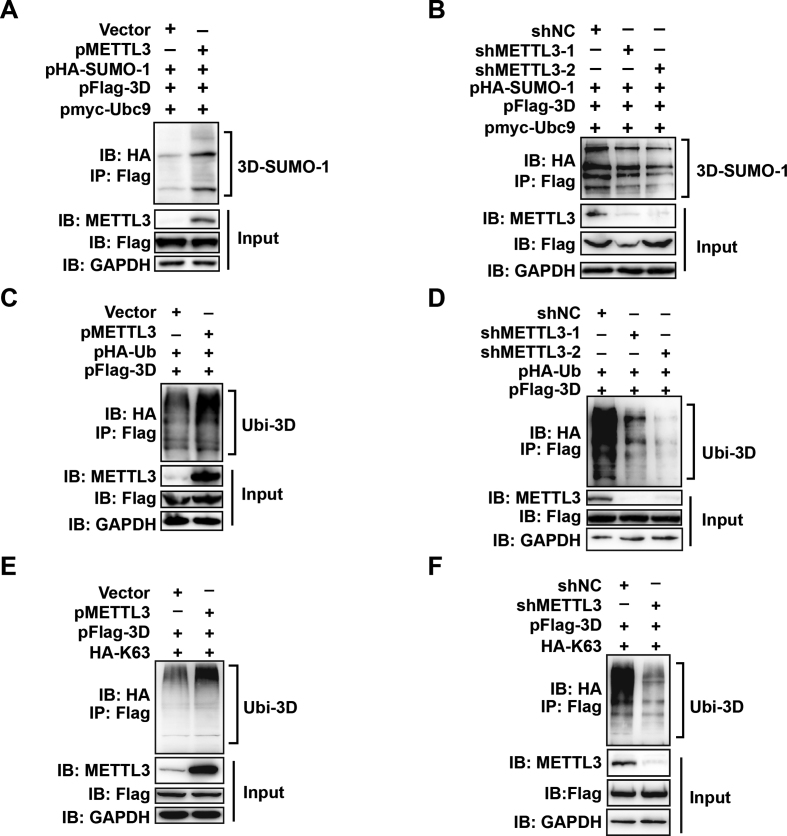
Figure 7.
The expression of METTL3 regulates the sumoylation and ubiquitination levels of 3D. (A &and B) Sumoylation assay. METTL3 was overexpressed or knocked down in HEK293T cells by transfection with pMETTL3 or shRNA, and then the cells were transfected with pFlag-3D, pHA-SUMO-1, and pMyc-Ubc9. IP and immunoblot analysis were performed using the indicated antibodies for the sumoylation assay. (C–F) Ubiquitination assay. HEK293T cells were transfected by pFlag-3D and pHA-Ub or pHA-K63 after METTL3 overexpression or knockdown. IP and immunoblot analyses were performed using the indicated antibodies.
Statistical analysis
Statistical analysis of qRT-PCR was performed using a two-tailed unpaired t-test in GraphPad Prism Software (La Jolla, CA, USA). Data are presented as the means ± standard error of the mean (SEM) (n = 3). All experiments were repeated at least three times.
RESULTS
EV71 RNA contains m6A residues
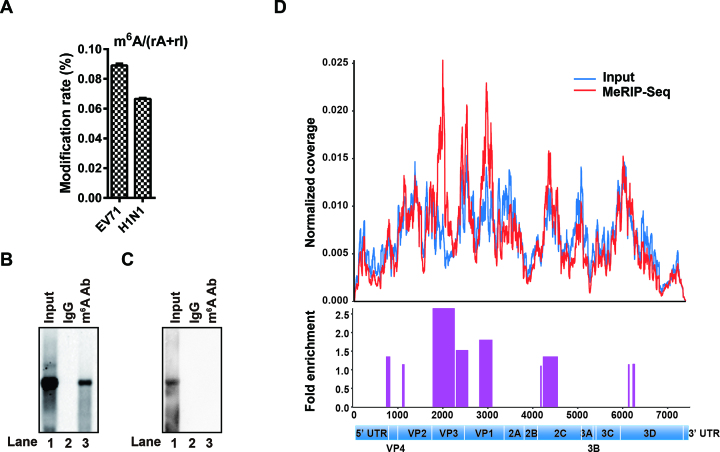
To investigate whether EV71 RNA was m6A-modified, RNAs of EV71 and Influenza A virus H1N1 were purified from large-scale infected culture. The presence of m6A residues was quantified by UHPLC-MS/MS. Notably, m6A in EV71 RNA accounted for 0.089% of adenosines, higher than that in Influenza A virus H1N1 RNA (0.066%), which had been reported to contain m6A (23,51) and served as a positive control (Figure 1A). To confirm our results, we isolated total RNA from EV71-infected Vero cells or from in vitro T7 transcribed EV71 genome. IP using an m6A-specific antibody was performed, followed by northern blot analysis. The results showed that in vivo EV71 RNA was pulled down by the anti-m6A antibody (Figure 1B, lane 3) whereas the in vitro transcribed EV71 RNA was not (Figure 1C, lane 3), suggesting that EV71 RNA contains m6A residues.
Figure 1.
EV71 genomic RNA contains m6A modifications. (A) UHPLC-MS/MS. EV71 RNA was harvested from large-scale culture and subjected to UHPLC-MS/MS analysis. The percentage of m6A among A residues is presented on the y axis. (B and C) MeRIP-Northern blotting. RNAs from virus infected Vero cells (B) or from in vitro T7 transcription (C) were incubated with IgG or m6A-specific antibody, followed by IP. RNAs were resolved on 1% agarose gels containing 2.2 M formaldehyde and transferred to Hybond-N+ membranes. The RNA signal was detected using an EV71 probe spanning from nt 1to nt 7405. (D) MeRIP-Seq. Total RNA was extracted from EV71-infected Vero cells and fragmented by ZnCl2. The fragmented RNA was subjected to IP by using an m6A-specific antibody followed by next-generation sequencing. Methylation coverage on the full-length input EV71 RNA and MeRIP-Seq are presented in blue and red, respectively. Representative of n = 3 determinations.
To map the m6A modification status in the EV71 RNA genome, MeRIP-Seq was performed (5,46). Several m6A peaks were identified across the EV71 RNA genome (Figure 1D), which were located in the VP, 3D and 2C coding region, suggesting that EV71 RNA is marked by m6A during infection. Collectively, our results demonstrated that EV71 RNA is modified at m6A during infection.
EV71 infection alters the expression patterns of m6A methyltransferases, demethylases and binding proteins
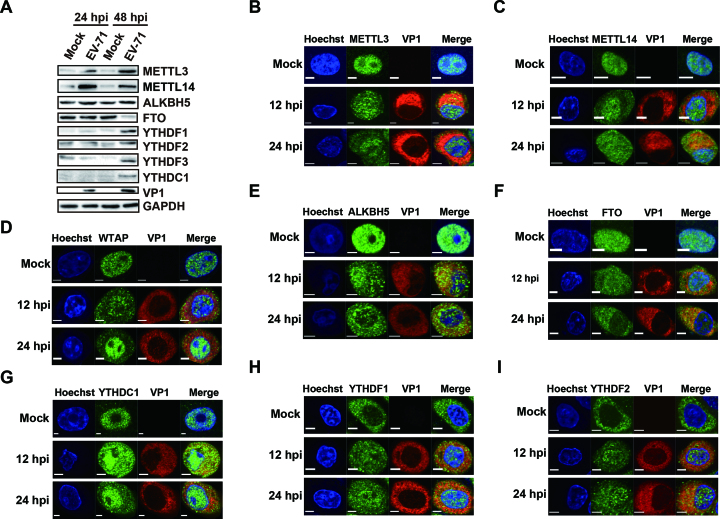
As EV71 replicates in the cytoplasm (52) and undergoes m6A modification, we next checked whether EV71 infection affected the expression of the host proteins related to m6A modification. Toward this end, EV71-infected Vero cells were analyzed by Western blotting using antibodies against m6A-related proteins. Virus infection was monitored by VP1 expression. We found that whereas expression of the methyltransferases, METTL3 and METTL14, was low under normal conditions, their expression increased at 24 and 48 h post-infection (hpi). In comparison, the expression of the demethylase ALKBH5 was not changed after infection; whereas that of FTO decreased at 48 hpi. Expression of the m6A binding proteins YTHDF1–3 and YTHDC1 increased at 48 hpi (Figure 2A). All these results suggested that the expression pattern of m6A proteins was altered during EV71 infection.
Figure 2.
EV71 infection influences the expression patterns of m6A methyltransferases, demethylases, and YTH proteins. (A) Western blotting. Vero cells infected with EV71 (MOI = 1) were harvested at 24 and 48 hpi. Western blotting was performed with antibodies as indicated. GAPDH was used as a loading control. (B–I) Confocal microscopy images of EV71- or mock-infected Vero cells. The nucleus (blue) and virus protein (red) were labeled with Hoechst and VP1-specific antibody, respectively. The methyltransferases, demethylases, and YTH proteins (green) were stained with antibodies as indicated. Scale bars, 5 μm.
Previous studies have indicated that METTL3, METTL14 and WTAP as well as the m6A demethylases FTO and ALKBH5 co-localize with nuclear speckle markers (7,12,13,53). Thus, we next determined whether EV71 infection affected the subcellular localization of these m6A methyltransferases, demethylases, along with YTH domain-containing proteins by staining Vero cells with the respective antibodies at 12 and 24 hpi. Methyltransferases, demethylases, and YTHDC1 were detected mostly in the nucleus under normal conditions. YTHDF1 and YTHDF2 were located in the cytoplasm without virus infection (Figure 2B–I). However, these proteins were all present in both the nucleus and cytoplasm after EV71 infection (Figure 2B–I). Accordingly, the ratio of methyltransferases, demethylases, and YTHDC1 in the cytoplasm versus nucleus was increased (Supplementary Figure S1, D-I); whereas that of YTHDF1 and YTHDF2 was decreased (Supplementary Figure S1J and K). Notably, we found that the localization of YTHDF3 was not affected by EV71 infection (Supplementary Figure S1A and L), which is similar to the nuclear protein histone 3 and cytoplasmic protein GAPDH (Supplementary Figure S1B, C, M and N). Taken together, these results indicated that the methyltransferases were localized to the cytoplasm during virus infection. The co-localization of methyltransferases and demethylases with viral protein VP1 (Figure 2B–F) implied that these proteins might interact with EV71 RNA in the cytoplasm.
m6A methyltransferases and demethylases regulate m6A of EV71 RNA and virus replication
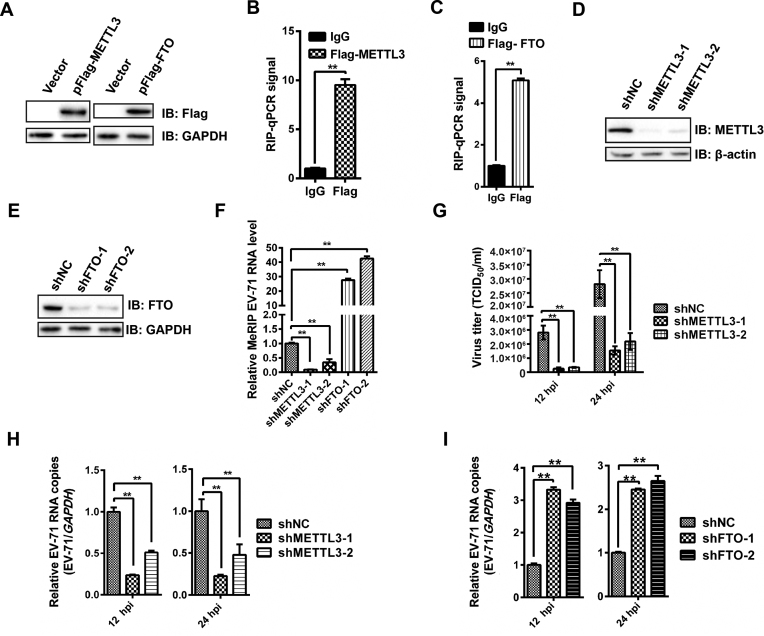
We next investigated the possibility that EV71 RNA m6A residues are modified by host methyltransferases and demethylases as EV71 itself does not encode any enzymes with internal m6A methyltransferase activity. Flag tagged METTL3 or FTO gene was expressed in RD cells by transfection (Figure 3A), and then qRT-PCR was performed following formaldehyde-crosslinked RIP using an anti-flag antibody to pull down METTL3- or FTO-bound RNA. Notably, EV71 RNA was pulled down by METTL3 and FTO (Figure 3B and C), which indicated that EV71 RNA could interact with METTL3 and FTO. We next knocked down endogenous METTL3 and FTO in Vero cells using shRNA (Figure 3D and E). m6A abundance in EV71 RNA was detected using qRT-PCR after MeRIP. In particular, the abundance of m6A in EV71 RNA was decreased by silencing METTL3 gene and increased by FTO depletion (Figure 3F). To further confirm this result, the expressions of METTL3 and FTO were restored by transfecting an shRNA-resistant cDNA (Supplementary Figure S2A and B). Interestingly, the level of m6A in EV71 RNA increased when METTL3 expression was restored and decreased when FTO expression was complemented (Supplementary Figure S2C). Taken together, our results indicated that m6A in EV71 RNA was regulated by METTL3 and FTO.
Figure 3.
Expression of methyltransferases and demethylases regulate m6A and the replication of EV71. (A) Western blotting. Flag-METTL3 and Flag-FTO were overexpressed in RD cells. The expression of METTL3 and FTO was checked using anti-Flag antibody. Vector-transfected cells were used as a control. (B & C) Formaldehyde-RIP-qPCR. Cell lysates from formaldehyde-crosslinking were subjected to IP with an anti-Flag antibody or IgG and quantified by qRT-PCR. IgG was used as a negative control. Unpaired Student's t-test was performed and data are presented as the means ± SEM (n = 3). **P ≤ 0.01. (D and E) Western blotting. METTL3 and FTO were knocked down in Vero cells by shRNA. Western blotting was carried out to check the expression of METTL3 and FTO. shNC was used as a control (F) MeRIP-qPCR. RNA was extracted from EV71-infected Vero cells in which METTL3 or FTO was knocked down, isolated by Me-RIP, and quantified by qRT-PCR. Total intracellular EV71 RNA was used as a control. Unpaired Student's t-test was performed and data are presented as the means ± SEM (n = 3). **P ≤ 0.01. (G) Viral titers (TCID50/ml) at 12 and 24 hpi. Vero cells in which METTL3 was knocked down or not were infected by EV71, and the supernatants were collected at indicated times post infection to measure virus titers as TCID50. The data presented are the mean viral titers and SDs from three independent experiments. Significant differences were determined using the Student t test (**P ≤ 0.01). (H and I) qRT-PCR. Total RNA was extracted at the indicated times from EV71-infected Vero cells in which METTL3 or FTO was knocked down or not. Quantification of EV71 RNA by qRT-PCR, with GAPDH used as a control. Unpaired Student's t-test was performed and data are presented as the means ± SEM (n = 3). *P ≤ 0.05, **P ≤ 0.01.
Because the expression of endogenous methyltransferases or demethylases is known to affect HIV and HCV protein expression and virus production (27–29,39), we knocked down endogenous METTL3 (Figure 3D) or FTO (Figure 3E) in Vero cells, followed by EV71 infection to check whether methyltransferases or demethylases affect EV71 replication. Viral titer was measured as TCID50, and viral RNA copy numbers were quantified by qRT-PCR. Efficient knockdown of METTL3 resulted in significantly decrease in virus titer (Figure 3G) and copy numbers of EV71 RNA at both 12 and 24 hpi (Figure 3H). However, the genomic copy numbers of EV71 RNA were significantly increased when FTO was knocked down (Figure 3I). The expression of VP1 was significantly decreased when METTL3 was knocked down (Supplementary Figure S3). Complementation of the expression of METTL3 or FTO by shRNA-resistant cDNAs increased and decreased EV71 replication (Supplementary Figure S2D–F), respectively. Collectively, these results suggested that the m6A methyltransferase METTL3 and demethylase FTO have impacts on efficient EV71 replication.
YTH proteins regulate the replication of EV71
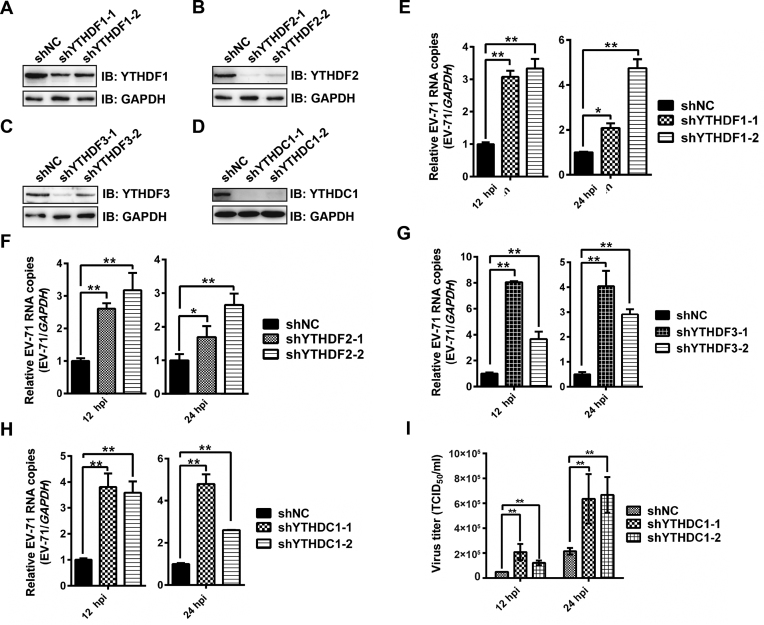
YTH proteins bind to m6A modifications on single-stranded RNA via a conserved domain located at the C terminus (27,39). In particular, YTH proteins bind to HIV and HCV viral RNA and play important roles in viral protein expression and virus release (27–29,39,54). Moreover, the cytoplasmic distribution and co-localization with EV71 VP1 protein (Figure 2, G-I & Supplementary Figure S1A) indicated that YTH proteins may also contribute to EV71 infection. To test this hypothesis, we separately knocked down YTH proteins in RD cells by using shRNA prior to EV71 infection (Figure 4, A-D). The viral genomic copies were quantified by qRT-PCR. Notably, knockdown of YTH proteins resulted in a significant increase in viral genomic copies at both 12 and 24 hpi (Figure 4E–H). Viral replication was decreased when the expression of YTH proteins was restored by shRNA-resistant cDNAs in knockdown cells (Supplementary Figure S4, A-H). We also checked virus titer when YTHDC1 was knocked down. Interestingly, knockdown of YTHDC1 in RD cells led to an increased viral titer (Figure 4I). However, knockdown and overexpression of YTHDF2 and YTHDF3 in Vero cells resulted in a decrease and an increase of viral replication, respectively (Supplementary Figure S5A–H). All together, these results suggested that the YTH proteins regulate EV71 infection.
Figure 4.
Replication of EV71 is regulated by YTH proteins in RD cells. (A–D) Western blotting. YTHDF1-3 and YTHDC1 were knocked down in RD cells by shRNA. The expression of YTH proteins was detected by western blotting with the respective antibodies. (E–H) qRT-PCR. Total RNA was extracted at indicated times from EV71-infected RD cells in which YTHDF1-3 or YTHDC1 were knocked down or not. Quantification of EV71 RNA by qRT-PCR, with GAPDH used as a control. Unpaired Student's t-test was performed. Data are presented as the means ± SEM (n = 3). *P ≤ 0.05, **P ≤ 0.01. (I) Viral titers (TCID50/ml) at 12 and 24 hpi. RD cells in which YTHDC1 was knocked down or not were infected by EV71, and the supernatants were collected at indicated times post infection to measure virus titers as TCID50. The data presented are the mean viral titers and SDs from three independent experiments. Significant differences were determined using the Student's t test (**P ≤ 0.01).
m6A site mutations in the EV71 RNA influence viral replication
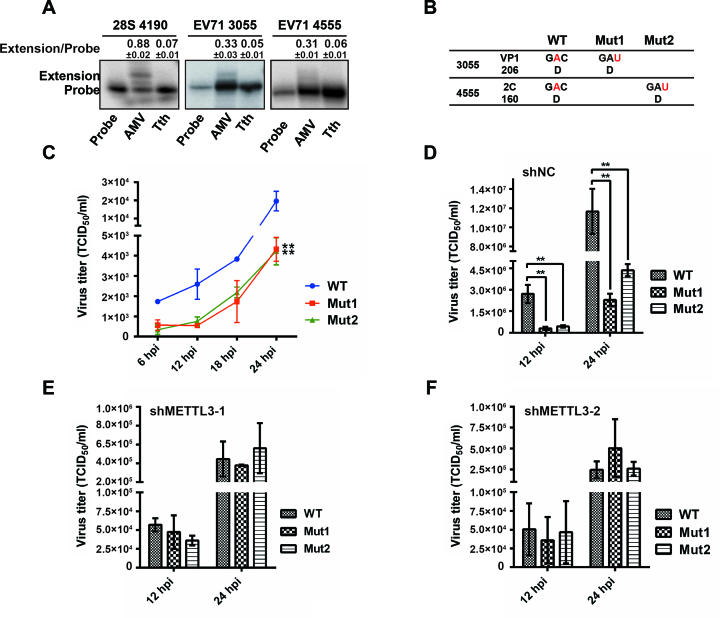
To test the function of specific m6A sites in the EV71 genome, we first identified potential m6A modification sites by primer extension analysis. The Tth polymerase enzyme discriminates m6A from A by significantly reducing the extension ability of DNA oligos, whereas AMV reverse transcriptase extends both m6A and A efficiently (28,48). To identify the m6A residues, we designed primers to test the m6A peaks identified by MeRIP-Seq. Tth polymerase failed to extend m6A sites at nt 3055 and nt 4555; whereas AMV polymerase extended these sites efficiently (Figure 5A), indicating that these A residues are methylated in the EV71 genome.
Figure 5.
Mutation of m6A sites influences the replication of EV71. (A) AMV reverse transcriptase and Tth DNA polymerase primer extension analysis of m6A sites in EV71. Total RNA was extracted from EV71-infected Vero cells. Primers were designed to probe for m6A modification at EV71 A3055 and A4555. AMV and Tth primer extensions for these two sites were performed using 10 μg poly(A)-enriched RNA from EV71-infected Vero cells. Extension of 28S RNA at A4190 served as a positive control. Relative gray intensity of extended bands in extension versus probe was quantified using ImageJ program. Data are presented as the means ± SD (n = 3). (B) Diagram of EV71 wild-type (WT) and m6A mutants. Sequences of WT and mutants are presented. (C) Growth curves of EV71 WT and m6A mutant in Vero cells. The cultures of Vero cells infected with EV71 WT or m6A mutants were harvested at the indicated times post infection to plot the growth curves. The data are shown as the means ± SD (n = 3). Significant differences were determined using Unpaired Student's t-test (*P ≤ 0.05, **P ≤ 0.01). (D–F) Viral titers (TCID50/ml) at 12 and 24 hpi. Vero cells in which METTL3 was knocked down or not were infected by EV71 WT, Mut1 or Mut2. At the indicated times post infection, the supernatants were collected to determine virus titers as TCID50. The data presented represent the mean viral titers and SDs from three independent experiments. Significant differences were determined using the Student's t test (**P ≤ 0.01).
To determine the role of the m6A sites on EV71 replication, C residues at nt 3056 and 4556 were mutated to inactivate the m6A modification in the EV71 infectious clone (Figure 5B), then Vero cells were infected by wild-type (EV71 WT) and mutant viruses (Mut1 and Mut2, respectively). Notably, virus titer was significantly decreased when m6A sites were mutated (Figure 5C and D), suggesting that m6A regulates viral replication, which is similar to the result in METTL3 depletion. The virus titer was not significantly altered when WT or mutant virus was infected in METTL3 knockdown Vero cells (Figure 5E and F), However, restored METTL3 expression resulted in a titer of WT virus higher than that of mutant viruses (Supplementary Figure S6B and C). Thus, these results supported that m6A sites in the EV71 genome play a role in viral replication.
METTL3 interacts with EV71 polymerase 3D and influences sumoylation and ubiquitination of the polymerase
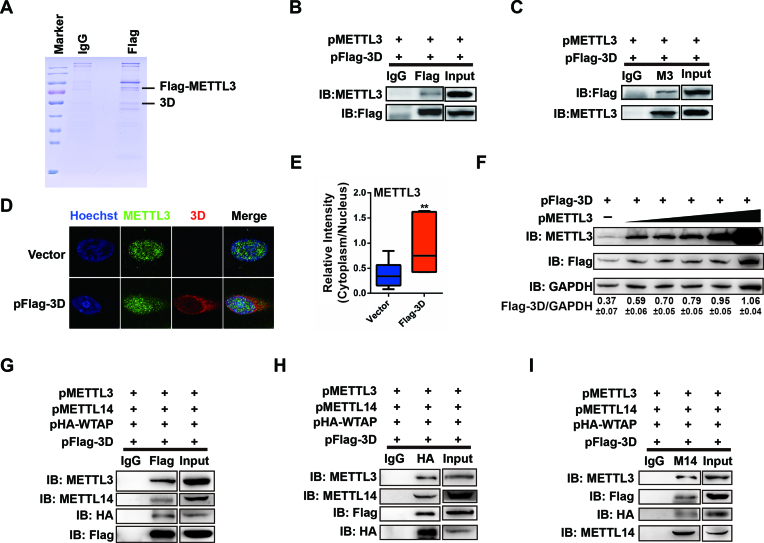
To investigate how the host m6A modification machinery affects EV71 replication, a Flag-tagged METTL3 cDNA was expressed in RD cells followed by EV71 infection. The cell lysate was subjected to IP with an anti-Flag antibody and analyzed by mass spectrometry (MS) (Figure 6A). In addition to the host proteins interacting with METTL3, the EV71 RdRp 3D protein was detected. To further confirm our results, pMETTL3 and pFLAG-3D were co-transfected into HEK293T cells. The IP experiment with an anti-METTL3 antibody followed by staining with anti-Flag or vice versa showed that METTL3 indeed interacted with 3D protein (Figure 6B and C) in the absence or presence of RNase A (Supplementary Figure S7A and B). We also checked the localization of METTL3 when Flag-tagged 3D was expressed in Vero cells. Although METTL3 was distributed in the nucleus without Flag-3D expression, it was present in both the nucleus and cytoplasm when Flag-tagged 3D was expressed (Figure 6D and E). Such co-localization of METTL3 and 3D supported the interaction between METTL3 and 3D. In addition, 3D expression was increased as more METTL3 was expressed by transfection (Figure 6F), suggesting that 3D expression was affected by the abundance of METTL3.
Figure 6.
METTL3 interacts with EV71 polymerase 3D. (A) Coomassie blue staining. pFlag-METTL3 was transfected into RD cells followed by EV71 infection. Cell lysates were subjected to IP with an anti-Flag antibody and then to Coomassie blue staining. (B and C) Western blotting. HEK293T cells were transfected with pMETTL3 and pFlag-3D. Co-IP was performed with anti-Flag or IgG (B) or anti-METTL3 or IgG (C) antibodies. The immunoblots were probed with the anti-Flag or anti-METTL3 antibodies. (D) Confocal microscopy images of Vero cells transfected by Vector or 3D. Costaining was performed using an anti-METTL3 antibody (green) and anti-Flag antibody (red), together with Hoechst to stain the nucleus (blue). (E) Relative fluorescence intensity of METTL3 in the cytoplasm versus the nucleus was quantified using ImageJ and graphed in box-and-whisker plots, representing the minimum, first quartile, median, third quartile, and maximum. Unpaired Student's t-test was performed (n ≥ 10). **P ≤ 0.01. (F) Western blotting. HEK293T cells were transfected with 1 μg of pFlag-3D and pMETTL3 (0, 0.5, 0.75, 1, 2 and 4 μg, respectively) in 30-mm dishes. The expression of METTL3 and 3D was detected by western blotting. Relative intensity of 3D versus GAPDH was quantified using ImageJ program. The data are shown as the means ± SD (n = 3). (G–I) Western blotting. HEK293T cells were transfected with pMETTL3, pMETTL14, pHA-WTAP, and pFlag-3D. Co-IP was performed with anti-Flag or IgG (G), or anti-HA or IgG (H), or anti-METTL14 or IgG (I) antibodies. The immunoblots were probed with indicated antibodies.
The methyltransferase complex is consisted of METTL3, METTL14, WTAP and other component proteins. To investigate whether METTL14 and WTAP interact with 3D, METTL14, HA-WTAP, METTL3 and Flag-3D were co-expressed in HEK293T cells, followed by IP with indicated antibodies (Figure 6G–I). Notably, METTL14 and WTAP were pulled down by 3D protein or vice versa (Figure 6G–I), indicating that METTL14 and WTAP interacted with 3D. Though 3D interacts with the main components of methyltransferase, the catalytic active site (DPPW) of METTL3 was not involved in the interaction between METTL3 and 3D (Supplementary Figure S7C and D). Furthermore, we found that the 3D mutant genomic RNA was pulled down by an anti-m6A antibody (Supplementary Figure S8D), suggesting that the m6A modification of viral RNA was not dependent on the interaction between 3D and METTL3.
As sumoylation and ubiquitination levels are known to enhance the stability of 3D and facilitate EV71 replication (43), and METTL3 expression is also linked to EV71 replication, we next investigated whether METTL3 affected the modification of 3D. To this end, METTL3 or shMETTL3, Flag-3D, HA-SUMO-1, myc-Ubc-9 or HA-Ub were expressed in HEK293T cells by transfection. We found that overexpression of METTL3 induced enhanced sumoylation and ubiquitination of 3D (Figure 7A and C) whereas METTL3 knockdown led to a decrease in 3D sumoylation or ubiquitination (Figure 7B and D). Futhermore, restored METTL3 expression in knockdown cells resulted in elevated 3D modification (Supplementary Figure S9A and B). Thus, these results suggested that the expression of METTL3 is involved in the regulation of viral 3D protein modification, the status of which affects viral replication.
Finally, because K63-linked ubiquitination is a docking site for mediating protein-protein interactions or conformational changes (55), we examined whether METTL3 could regulate this ubiquitin-linked chain of 3D. HEK293T cells, in which METTL3 was overexpressed or knocked down, were expressed with Flag-3D and HA-K63, followed by IP and immunobloting. The result showed that K63-linked ubiquitination was increased or decreased significantly by METTL3 overexpression or knockdown (Figure 7E and F). This result was confirmed by the increase in K63-linked ubiquitination when METTL3 restored in knockdown cells (Supplementary Figure S9C).
DISCUSSION
In this study, we showed that EV71 RNA contains m6A modifications that play a critical role in viral replication. Knockdown of m6A methyltransferases, demethylases, and binding proteins in host cells affected viral replication. In addition, mutation of the m6A modification sites in the infectious clone decreased EV71 progeny virus production and protein expression, which is similar to the effect of METTL3 knockdown. Notably, we found that METTL3 interacted with viral RdRp 3D protein and regulated its modification to modulate viral replication. Thus, our findings demonstrated that m6A modification represents an important component in the regulation of EV71 replication.
m6A has been identified in infection of both RNA and DNA viruses. Assessment of the internal m6A modification status of EV71 RNA using MeRIP followed by Northern blotting revealed that an m6A antibody binds to EV71 RNA. UHPLC-MS/MS analysis of viral RNA confirmed that EV71 RNA contains m6A with a ratio of m6A/A of ∼0.09%. Moreover, m6A peaks spanning the full length of EV71 RNA were identified by MeRIP-Seq. Thus, these results revealed that EV71 RNA was m6A-methylated. However, as EV71 does not encode any known methyltransferase, host methyltransferases appear to play a critical role in m6A methylation.
We also found that the expression of m6A-related proteins was altered by virus infection. METTL3 and METTL14, the key components of methyltransferases, together with YTH proteins, were up-regulated. Among demethylases, however, the expression of FTO was decreased; whereas ALKBH5 was not affected by EV71 infection. These data implied that EV71 infection altered host proteins expression to enhance RNA m6A modifications, which may promote viral infection. The expression of ALKBH5 was not affected during EV71 infection. ALKBH5 preferentially binds to the CDS shortly after the start codon and only targets a small subset of m6A sites installed by METTL3 (56). Our results showed that the expression of FTO was linked to the replication of EV71, indicating that the demethylation of EV71 RNA was mainly catalyzed by FTO. Methyltransferases and demethylases localize in the nucleus of Vero cells while YTH proteins are expressed in the cytoplasm (Figure 2). However, METTL3, METTL14, FTO and YTH proteins were redistributed in both the nucleus and cytoplasm upon EV71 infection (Figure 2). A previous study showed that during heat shock, YTHDF2 relocalizes to the nucleus in order to prevent FTO-mediated demethylation of the 5′-UTR of stress-induced transcripts (57). EV71 infection may induce a cellular stress that is similar to the stress of heat shock, which may be the reason that m6A-related proteins relocated under EV71 infection. We also found that these m6A-related proteins co-localized with the viral capsid protein VP1, supporting that the m6A modification machinery can modify cytoplasmic EV71 RNA during EV71 infection.
m6A modification has been linked to viral replication and gene expression. Knockdown of methyltransferases or demethylases decrease or increase HIV replication (27–29). However, silencing of METTL3 or FTO has the opposite effect on the replication of HCV (39). Due to the different replication locations of HIV and HCV, we tested the effect of methyltransferases or demethylases on EV71 replication in Vero cells. Our results showed that m6A methyltransferases and demethylases regulate EV71 replication, which is in an agreement with the recent reports that the m6A machinery regulates HCV, HIV and influenza virus infection (27–30,39). In particular, knockdown of METTL3 down-regulated EV71 replication, which was similar to HIV. Moreover, mutation of the m6A modification sites in the infectious clone of EV71 followed by rescuing and titration of the mutant viruses decreased replication of the mutants viruses, compared to the wild-type virus, which was consistent with the result in METTL3 knockdown.
Knockdown of YTHDF2 and YTHDF3 in Vero cells led to significantly decreased viral replication at 12- and 24-hpi(Supplementary Figure S5E and F). Considering that the expression of YTHDF1-3 was low in Vero cells (Figure 2A), we knocked down YTH proteins in EV71-permissive RD cells. We found that knockdown of YTHDF1-3 proteins in RD cells led to increased EV71 replication, which was similar to HCV and ZIKV (39,54). Our results showed that YTH proteins have different roles on EV71 replication in different cell lines. However, the mechanism by which YTH proteins affect viral replication remains unknown. As Vero cells do not express interferon (IFN), we are currently studying whether the IFN signaling pathway plays a role in the m6A-regulated viral replication. We also tested whether the nuclear m6A binding protein YTHDC1 affects EV71 replication. The results showed that YTHDC1 positively regulates EV71 replication in RD cells, which is similar to YTHDF1, 2 and 3. Our data thus showed that both the nuclear and cytoplasmic m6A readers regulate viral replication in RD cells.
To address how EV71 hijacks the m6A modification machinery to facilitate virus replication, we performed METTL3 IP to check viral protein interactions. The results showed that METTL3 interacted with 3D. Notably, METTl14 and 3D are the components of 3D-METTL3 complex, indicating that the methyltransferase interact with viral protein. Moreover, in the presence of 3D expression, METTL3 was distributed in both the nucleus and cytoplasm, which confirms the interaction between METTL3 and 3D and may explain the presence of METTL3 in the cytoplasm during EV71 infection. However, the catalytic site of METTL3 was not involved in the interaction between METTL3 and 3D protein. Importantly, METTL3 promoted sumoylation and ubiquitination of 3D to facilitate viral replication (43), which indicates that the m6A modification machinery plays an important role in EV71 infection.
Collectively, we present evidence supporting the importance of m6A modification for the replication of EV71. The expression and localization of the m6A methylation machinery were affected by EV71 virus infection. The expression of proteins in the m6A methylation machinery in turn regulated viral replication. Mutation of the m6A sites decreased viral replication. Nevertheless, further studies are necessary to elucidate the detailed mechanisms, such as how the methyltransferases, demethylases and YTH proteins alter their localization and whether viral non-structural proteins play a role in the methylation process. In addition, our study suggests m6A modification may be a novel target for antivirals of EV71.
DATA AVAILABILITY
Complete CDS sequence of EV71 (strain XF) used in this work has been deposited with the National Center for Biotechnology Information under accession number JQ804832.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Hailin Wang for help with UHPLC-MS/MS, and all of the members of the laboratory of WXG for discussions and critical reading. We are grateful to Lei Zhang and Ding Gao of the Core Facility and Technical Support in the Wuhan Institute of Virology, CAS for their help in the next generation sequencing and confocal microscopy. We also thank Haizhou Liu and Di Liu for their assistance with MeRIP-Seq data analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Technology of China [2016YFC1200400]; Chinese Academy of Sciences [ZDRW-ZS-2016-4]; Open Research Fund Program of CAS Key Laboratory of Special Pathogens and Biosafety, Chinese Academy of Sciences [2015SPCAS002 to W.G.]. The funders had no role in the design, interpretation, or submission of this work for publication. Funding for open access charge: Chinese Academy of Sciences [ZDRW-ZS-2016-4].
Conflict of interest statement. None declared.
REFERENCES
- 1. Agarwala S.D., Blitzblau H.G., Hochwagen A., Fink G.R.. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012; 8:e1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 3. Horiuchi K., Kawamura T., Iwanari H., Ohashi R., Naito M., Kodama T., Hamakubo T.. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013; 288:33292–33302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patil D.P., Chen C.K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R.. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016; 537:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ping X.L., Sun B.F., Wang L., Xiao W., Yang X., Wang W.J., Adhikari S., Shi Y., Lv Y., Chen Y.S. et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D. et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014; 8:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhong S., Li H., Bodi Z., Button J., Vespa L., Herzog M., Fray R.G.. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008; 20:1278–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lewis C.J., Pan T., Kalsotra A.. RNA modifications and structures cooperate to guide RNA-protein interactions. Nat. Rev.. Mol. Cell Biol. 2017; 18:202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao B.S., Roundtree I.A., He C.. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017; 18:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G. et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011; 7:885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.M., Li C.J., Vagbo C.B., Shi Y., Wang W.L., Song S.H. et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013; 49:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S. et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015; 347:1002–1006. [DOI] [PubMed] [Google Scholar]
- 15. Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014; 505:117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J.. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014; 10:927–929. [DOI] [PubMed] [Google Scholar]
- 17. Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T.. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015; 518:560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.B., Jaffrey S.R.. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell. 2015; 163:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C.. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015; 161:1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meyer K.D., Jaffrey S.R.. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol Cell Biol. 2014; 15:313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finkel D., Groner Y.. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology. 1983; 131:409–425. [DOI] [PubMed] [Google Scholar]
- 22. Kane S.E., Beemon K.. Precise localization of m6A in Rous sarcoma virus RNA reveals clustering of methylation sites: implications for RNA processing. Mol. Cell. Biol. 1985; 5:2298–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krug R.M., Morgan M.A., Shatkin A.J.. Influenza viral mRNA contains internal N6-methyladenosine and 5′-terminal 7-methylguanosine in cap structures. J. Virol. 1976; 20:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gokhale N.S., Horner S.M.. RNA modifications go viral. PLoS Pathog. 2017; 13:e1006188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kennedy E.M., Courtney D.G., Tsai K., Cullen B.R.. Viral Epitranscriptomics. J. Virol. 2017; 91:e02263-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pereira-Montecinos C., Valiente-Echeverria F., Soto-Rifo R.. Epitranscriptomic regulation of viral replication. Biochim. Biophys. Acta. 2017; 1860:460–471. [DOI] [PubMed] [Google Scholar]
- 27. Kennedy E.M., Bogerd H.P., Kornepati A.V., Kang D., Ghoshal D., Marshall J.B., Poling B.C., Tsai K., Gokhale N.S., Horner S.M. et al. Posttranscriptional m(6)A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016; 19:675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lichinchi G., Gao S., Saletore Y., Gonzalez G.M., Bansal V., Wang Y., Mason C.E., Rana T.M.. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016; 1:16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tirumuru N., Zhao B.S., Lu W., Lu Z., He C., Wu L.. N(6)-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife. 2016; 5:e15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Courtney D.G., Kennedy E.M., Dumm R.E., Bogerd H.P., Tsai K., Heaton N.S., Cullen B.R.. Epitranscriptomic Enhancement of Influenza A Virus Gene Expression and Replication. Cell Host Microbe. 2017; 22:377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsai K., Courtney D.G., Cullen B.R.. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog. 2018; 14:e1006919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tan B., Liu H., Zhang S., da Silva S.R., Zhang L., Meng J., Cui X., Yuan H., Sorel O., Zhang S.W. et al. Viral and cellular N(6)-methyladenosine and N(6),2′-O-dimethyladenosine epitranscriptomes in the KSHV life cycle. Nat. Microbiol. 2018; 3:108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hesser C.R., Karijolich J., Dominissini D., He C., Glaunsinger B.A.. N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma-associated herpesvirus infection. PLoS Pathog. 2018; 14:e1006995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ye F., Chen E.R., Nilsen T.W.. Kaposi's Sarcoma-Associated herpesvirus utilizes and manipulates RNA N(6)-Adenosine methylation to promote lytic replication. J Virol. 2017; 91:e00466-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Banerjee A.K. 5′-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol. Rev. 1980; 44:175–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Boone R.F., Moss B.. Methylated 5′-terminal sequences of vaccinia virus mRNA species made in vivo at early and late times after infection. Virology. 1977; 79:67–80. [DOI] [PubMed] [Google Scholar]
- 37. Both G.W., Banerjee A.K., Shatkin A.J.. Methylation-dependent translation of viral messenger RNAs in vitro. PNAS. 1975; 72:1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moyer S.A., Banerjee A.K.. In vivo methylation of vesicular stomatitis virus and its host-cell messenger RNA species. Virology. 1976; 70:339–351. [DOI] [PubMed] [Google Scholar]
- 39. Gokhale N.S., McIntyre A.B., McFadden M.J., Roder A.E., Kennedy E.M., Gandara J.A., Hopcraft S.E., Quicke K.M., Vazquez C., Willer J. et al. N6-Methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe. 2016; 20:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brown B.A., Pallansch M.A.. Complete nucleotide sequence of enterovirus 71 is distinct from poliovirus. Virus Res. 1995; 39:195–205. [DOI] [PubMed] [Google Scholar]
- 41. McMinn P.C. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 2002; 26:91–107. [DOI] [PubMed] [Google Scholar]
- 42. Tan X., Huang X., Zhu S., Chen H., Yu Q., Wang H., Huo X., Zhou J., Wu Y., Yan D. et al. The persistent circulation of enterovirus 71 in People's Republic of China: causing emerging nationwide epidemics since 2008. PLoS One. 2011; 6:e25662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Y., Zheng Z., Shu B., Meng J., Zhang Y., Zheng C., Ke X., Gong P., Hu Q., Wang H.. SUMO modification stabilizes enterovirus 71 polymerase 3D to facilitate viral replication. J. Virol. 2016; 90:10472–10485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pizzi M. Sampling variation of the fifty percent end-point, determined by the Reed-Muench (Behrens) method. Hum. Biol. 1950; 22:151–190. [PubMed] [Google Scholar]
- 45. Shang B., Deng C., Ye H., Xu W., Yuan Z., Shi P.Y., Zhang B.. Development and characterization of a stable eGFP enterovirus 71 for antiviral screening. Antiviral Res. 2013; 97:198–205. [DOI] [PubMed] [Google Scholar]
- 46. Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G.. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013; 8:176–189. [DOI] [PubMed] [Google Scholar]
- 47. Basanta-Sanchez M., Temple S., Ansari S.A., D’Amico A., Agris P.F.. Attomole quantification and global profile of RNA modifications: Epitranscriptome of human neural stem cells. Nucleic Acids Res. 2016; 44:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harcourt E.M., Ehrenschwender T., Batista P.J., Chang H.Y., Kool E.T.. Identification of a selective polymerase enables detection of N(6)-methyladenosine in RNA. J. Am. Chem. Soc. 2013; 135:19079–19082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hao S., Zhang J., Chen Z., Xu H., Wang H., Guan W.. Alternative polyadenylation of human bocavirus at its 3′ end is regulated by multiple elements and affects capsid expression. J. Virol. 2017; 91:e02026-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen H., Pei R., Zhu W., Zeng R., Wang Y., Wang Y., Lu M., Chen X.. An alternative splicing isoform of MITA antagonizes MITA-mediated induction of type I IFNs. J. Immunol. 2014; 192:1162–1170. [DOI] [PubMed] [Google Scholar]
- 51. Narayan P., Ayers D.F., Rottman F.M., Maroney P.A., Nilsen T.W.. Unequal distribution of N6-methyladenosine in influenza virus mRNAs. Mol. Cell. Biol. 1987; 7:1572–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Solomon T., Lewthwaite P., Perera D., Cardosa M.J., McMinn P., Ooi M.H.. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet. Infect. Dis. 2010; 10:778–790. [DOI] [PubMed] [Google Scholar]
- 53. Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M.. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997; 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 54. Lichinchi G., Zhao B.S., Wu Y., Lu Z., Qin Y., He C., Rana T.M.. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016; 20:666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mallette F.A., Richard S.. K48-linked ubiquitination and protein degradation regulate 53BP1 recruitment at DNA damage sites. Cell Res. 2012; 22:1221–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou J., Wan J., Shu X.E., Mao Y., Liu X.M., Yuan X., Zhang X., Hess M.E., Bruning J.C., Qian S.B.. N(6)-Methyladenosine guides mRNA alternative translation during integrated stress response. Mol. Cell. 2018; 69:636–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.B.. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015; 526:591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete CDS sequence of EV71 (strain XF) used in this work has been deposited with the National Center for Biotechnology Information under accession number JQ804832.