Abstract
8-Oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) is one of the major DNA modifications and a potent pre-mutagenic lesion prone to mispair with 2′-deoxyadenosine (dA). Several thousand residues of 8-oxodG are constitutively generated in the genome of mammalian cells, but their genomic distribution has not yet been fully characterized. Here, by using OxiDIP-Seq, a highly sensitive methodology that uses immuno-precipitation with efficient anti–8-oxodG antibodies combined with high-throughput sequencing, we report the genome-wide distribution of 8-oxodG in human non-tumorigenic epithelial breast cells (MCF10A), and mouse embryonic fibroblasts (MEFs). OxiDIP-Seq revealed sites of 8-oxodG accumulation overlapping with γH2AX ChIP-Seq signals within the gene body of transcribed long genes, particularly at the DNA replication origins contained therein. We propose that the presence of persistent single-stranded DNA, as a consequence of transcription-replication clashes at these sites, determines local vulnerability to DNA oxidation and/or its slow repair. This oxidatively-generated damage, likely in combination with other kinds of lesion, might contribute to the formation of DNA double strand breaks and activation of DNA damage response.
INTRODUCTION
One of the most common processes that causes genomic lesions is DNA oxidation, due to pro-oxidant species generated during endogenous metabolism. Indeed, cellular processes such as energy production by mitochondria and/or enzymatic activities lead to the production of Reactive Oxygen Species (ROS), that preferentially oxidize 2′-deoxyguanosine in the DNA double-helix, forming 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG) (1,2). 8-oxodG is considered a potent premutagenic lesion, due to its ability to pair with both cytosine and adenine residues, thus causing G:C to T:A transversions during DNA replication (3,4). 8-oxodG is an effective biomarker of oxidative stress, and its accumulation in the genome has been associated with cancer, aging, and cardiovascular diseases (1,5,6). Moreover, 8-oxodG has been proposed as a new potential independent prognostic factor in breast cancer (7).
8-OxodG is constantly repaired in unperturbed conditions to ensure genome stability. Indeed, 8-oxodG:dC pairs are repaired by the Base Excision Repair (BER) pathway (8–10). Intriguingly, components of BER machinery have also been involved in transcription, suggesting an inherent physiologically intertwined relationship between transcription and DNA repair (11–16). This is not surprising since over the last decade it has become increasingly evident that transcription, replication and DNA repair are closely integrated and constantly threatened by multiple intrinsic processes such as endogenous oxidative stress (17–24). It has been estimated that a typical human cell undergoes ∼70 000 lesions per day, the majority of which are single strand breaks arising from oxidatively-generated damage during metabolism, or base hydrolysis (4). Thus, oxidation of guanine residues represents a major threat to genome integrity and identification of preferential sites of 8-oxodG within the genome is crucial to our understanding of the pathways contributing to ROS-induced genome instability and the repair mechanisms involved. To date, genomic distribution of 8-oxodG remains poorly characterized. Fluorescence in-situ detection of 8-oxodG on metaphase chromosomes from human peripheral lymphocytes showed that 8-oxodG immunoreactivities are often located in boundary regions of R and/or G bands, known as transition zones of DNA replication timing. In the same study, as many as 10,000 8-oxodGs per nucleus were found (25). Combination of immuno-precipitation assay and microarray hybridization on the genome of normal rat kidney cells, revealed that 8-oxodG is preferentially located at gene deserts (26). Interestingly, no differences in 8-oxodG levels were found when comparing poorly- and highly-expressed genes, while a strong correlation with lamina-associated domains (LADs) suggested that the spatial location of genomic DNA in the nucleus determines its susceptibility to oxidation (26). Chromatin immuno-precipitation followed by high-throughput sequencing (ChIP-seq) analysis of 8-oxodG distribution in normal and hypotoxic rat pulmunary artery endothelial cells showed association between 8-oxodG and hypoxia-induced transcription changes (27). More recently, following chemical labeling of 8-oxodG with biotin, Ding and colleagues reported the genome-wide distribution of 8-oxodG in MEFs (28).
Old reports showed that single-stranded DNA (ssDNA), a hallmark of stress, is more sensitive to oxidation than double-stranded DNA (dsDNA) (27,29,30). These observations suggest that genomic sites showing persistent ssDNA (e.g. in the presence of stable R-loops, or alternative (non-B) DNA structures, such as Z-DNA, cruciforms, intramolecular triplexes and quadruplexes, etc.) might be hotspots of oxidatively-generated damage (27). Persistent ssDNA can also form as a consequence of transcription/replication clashes, when transcription and replication machineries pause, because of head-on collisions that can be direct, or indirect (i.e. because of positive supercoils that accumulate ahead of both machineries) (31,32). In particular, since very long genes found at Common Fragile Sites (CFSs) need more than one cell cycle to be entirely transcribed, it has been proposed that the frequent transcription/replication clashes occurring at those sites might lead to replication fork stalling that, in turn, might favor chromosomal fragility (31,32). However, instability might also result from secondary DNA structures and/or specific chromatin features, as suggested by the observation that not all the active long genes are prone to breakage.
We report here the genome-wide distribution of 8-oxodGs in MCF10A and in MEFs. We developed the OxiDIP-Seq that combines immuno-precipitation of single-stranded DNA with high-throughput sequencing to map the 8-oxodG both in human and mouse genomes. Furthermore, when OxiDIP-Seq was compared to γH2AX ChIP-Seq data, a distinctive co-enrichment of 8-oxodG and γH2AX was found within the gene body of transcribed long genes in both genomes. Moreover, we found a significant enrichment of 8-oxodG at DNA replication origins (ORIs), suggesting that accumulation of 8-oxodG at ORIs of active long genes significantly contributes to the inherent instability of these genomic regions.
MATERIALS AND METHODS
Cell culture and treatments
MCF10A cells were cultured in 1:1 mixture DMEM-F12 supplemented with 5% horse serum, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera enterotoxin and 20 ng/ml epidermal growth factor, and incubated at 37°C in humidified atmosphere with 5% CO2 (33). Mouse embryonic fibroblasts, MEFs (3T9-MycER), were grown in DMEM medium supplemented with 10% serum, penicillin/streptomycin and 2 mM l-Gln. For UV treatment, exponentially growing cells were irradiated with 254-nm UV light at 40 J/m2. For NAC treatment, 1 mM N-acetyl cysteine (A7250, Sigma-Aldrich) was added to the medium for 2 h before being collected as previously described.
8-oxodG enrichment from ssDNA and G4-containing oligomers
The IP was performed as described (28) with the following changes: oligomers with 8-oxodG were designed with flanking Renilla primers for qPCR quantification (ssDNA = 5′-GGAATTATAATGCTTATCTACGTGCGACGGCCAGTGTAGTTGGAGCTC/8oxodG/TGGCGTAGGCAAGAGTGTCATAGCTGGTAAAAGGTCTTCATTTTTCGCAAG and G-quadruplex DNA = 5- GGAATTATAATGCTTATCTACGTGCCCCGCCCCCCGGGGCGGGCC/8oxodG/GGGGCGGGGTCCCGGCGGGGCGGAGCCATGTAAAAGGTCTTCATTTTTCGCAAG-3′). Each IP reaction was performed with 3 fmol 8-oxodG-containing oligomer and a large excess (104 fold) of random 8-oxodG – free oligomers, with the following antibodies: 4 μl of polyclonal antibody against anti-8-oxodG (AB5830 Millipore); 2 μg monoclonal antibody anti-8-oxodG (Trevigen, 0.5 mg/ml) and 4 μl of IgG. The IP efficiency was calculated by qPCR as % of immuno-precipitated DNA over input. The following primers were used in qPCR: Oligo-Renilla-Fwd GGA ATT ATA ATG CTT ATC TAC GTG C and Oligo-Renilla-Rev CTT GCG AAA AAT GAA GAC CTT TTA C.
Spike-in experiment with 8-oxodG-containing oligomers
The spike-in experiment to test the specificity of the antibodies was performed as described (34) with the following changes: 64 pg of both 8-oxodG- (described above) and dG-containing oligonucleotides (ssDNA = GTGGTGTGCAGCGAGAATAGAAACGACGGCCAGTGTAGTTGGAGCTCGTGGCGTAGGCAAGAGTGTCATAGCTGTTTCCTAACGACATCTACAACGAGCG) were mixed together with 1 μg of genomic DNA with undetectable endogenous 8-oxodG levels from MCF10 treated cell with 1 mM NAC for 2 h. IP was performed as indicated for OxiDIP (see next paragraph). The relative enrichment of 8-oxodG was calculated by qPCR as % of immuno-precipitated DNA over input using the following primers: Oligo-Renilla (described above) for 8-oxodG-containing oligonucleotide and Oligo-FireFly for dG-containing oligonucleotide (Fwd-CGCTCGTTGTAGATGTCGTTAG and Rev- GTGGTGTGCAGCGAGAATAG).
The spike-in experiment to test the sensitivity of the antibodies was performed as described (35) with the following changes: 1 μg of genomic DNA with undetectable endogenous 8-oxodG levels, from NAC-treated MCF10 cells, was added to increasing amounts (from 0.5 to 64 pg) of 8-oxodG–containing oligonucleotides (above described). IP was performed as indicated for OxiDIP. The IP efficiency was calculated by qPCR as % of immuno-precipitated DNA over input.
LC–MS/MS
Stock solutions (8-oxodG, dG and (15N5) 8-hydroxy-2-deoxyguanosine) were prepared in methanol at a concentration of 50 mg/l of each analyte. Final 1 mg/L individual analyte standard solutions were prepared by serial dilutions from stock solutions at 0.5, 1, 5, 25, 50 pg/μl and used for calibration curves. All standards were kept in the dark, under nitrogen, at −20°C before LC–MS/MS analysis. Genomic DNA from growing MCF10A cells was extracted by using Dneasy Blood&Tissue kit (Cat. no. 69504, QIAGEN). Furthermore, 50 μM N-tert-Butyl-α-phenylnitrone (B7263, Sigma) was added to Dneasy Blood&Tissue to preserve the oxidized state of DNA (36). DNA samples were hydrolyzed in HCl 0.1 M for 30 min at 30°C until a clear solution was obtained. Samples were then dried in a SpeedVac and 10 μl of methanol was added into an LC vial for analysis. 4 μl of heavy 8-oxodG were added before any sample treatment. Samples 1 μl were analysed by using an Agilent 6400 Series Triple Quadrupole LC/MS system with a HPLC 1100 series binary pump (Agilent. Waldbronn, Germany). The mobile phase was generated by mixing eluent A (0.1% Formic Acid and eluent B (methanol) at a flow rate of 0.2 ml/min. The elution gradient was from 5% A to 95% B in 6 min. The tandem mass spectrometry analysis was performed in positive MRM mode. A standard solution of 500 pg/μl of dG, 8-oxodG, and (15N5) 8-oxodG were individually infused to establish the optimal instrument settings for each compound. Experimental automatic tuning using MassHunter Optimizer was employed to define ionization polarity, to select the best product ion (Q3 ion) and to optimize both the collision energy (CE) and the declustering potential (DP). Extracted mass chromatogram peaks of the analytes were integrated using Agilent MassHunter Quantitative Analysis software (B.05.00). Peak areas of the corresponding analytes were then used as quantitative measurements for assay performance assessments such as variation, linearity etc. Linearity was determined using standard solutions and matrix matched calibrations. Standard calibration curves were constructed by plotting peak areas against concentration (pg/μl) and linear functions were applied to the calibration curves. Data were integrated by Mass Hunter quantitative software showing a linear trend in the calibration range and a coefficients of determination (R2) greater than 0.99 for all analytes. The limits of detection (LODs) for each species were determined by making 10 replicate measurements of blank samples spiked with low concentrations of each analyte and calculated as LOD = 3*SD. LOQ was determined as the concentration when the S/N ratio was 10. The MRM transitions and all the instrumental and analytical parameters are summarised in Supplementary Table S1. The possible effect of acid hydrolysis on the extent of oxidation was tested by submitting an aliquot of dG to acidic hydrolysis followed by LC–MRM mass spectrometry analysis. Supplementary Figure S1 shows that no evidence of 8-oxodG was recorded in the TIC chromatograms, only exhibiting the peaks corresponding to the dG transitions. For quantitative analyses, samples were spiked with a known amount of (15N5) 8-oxodG, submitted to acidic hydrolysis as previously described and directly analysed by tandem mass spectrometry in MRM scan mode. The analytes concentrations were calculated in pg/μl and then expressed in ppm (i.e. number of 8-oxodGs per million of dGs). As an example, Supplementary Figure S1 shows the MRM transitions recorded for 8-oxodG before and after UV treatment, indicating an increase of 8-oxodG following UV exposure. Notably, direct comparison between HCl treatment and enzymatic degradation (data not shown) of the genomic DNA for LC–MS/MS quantification of 8-oxodG showed very similar results.
OxiDIP-sequencing and quantitative 8-oxodG immuno-precipitation assays
Genomic DNA from growing MCF10A cells or from growing MEFs was extracted by using Dneasy Blood&Tissue kit (Cat. no. 69504, QIAGEN). 10 μg of genomic DNA per immuno-precipitation were sonicated in 100 μl TE buffer (100 mM Tris–HCl pH 8.0, 0.5 M EDTA pH 8.0) to generate random fragments ranging in size between 200 and 800 bp using Bioruptor Plus UCD-300. 4 μg of fragmented DNA in 500 μl TE Buffer were denatured for 5 min at 95°C and immuno-precipitated over night at 4°C with 4 μl of polyclonal antibodies against 8-Hydroxydeoxyguanosine (AB5830 Millipore) in a final volume of 500 μl IP buffer (110 mM NaH2PO4, 110 mM Na2HPO4 ph 7.4, 0.15 M NaCl, 0.05% Triton X-100, 100 mM Tris–HCl pH 8.0, 0.5 M EDTA pH 8.0) under constant rotation. The immuno-precipitated complex was incubated with 50 μl Dynabeads Protein G (Cat. No. 10003D, ThermoFisher Scientific, previously saturated with 0.5% bovine serum albumine diluted in PBS) for 3 h at 4 °C, under constant rotation, and washed three times with 1 ml Washing buffer (110 mM NaH2PO4, 110mM Na2HPO4 pH 7.4, 0.15 M NaCl, 0.05% Triton X-100). The beads–antibody–DNA complexes were then disrupted by incubation in 200 μl Lysis buffer (50 mM Tris–HCl pH 8, 10 mM EDTA pH 8, 1% SDS, 0.5 mg/ml Proteinase K) for 4 h at 37°C, and 1 h at 52°C following addition of 100 μl Lysis buffer. The immuno-precipitated DNA was purified by using MinElute PCR Purification kit (Cat. No. 28004, QIAGEN) in a final volume of 72 μl EB buffer (provided in the kit). All the steps of OxiDIP-Seq protocol, including the washes of the immunocomplexes, were carried out in low-light conditions. Furthermore, 50 μM N-tert-butyl-α-phenylnitrone (stock solution: 28 mM in H2O; B7263, Sigma) was added to each Dneasy Blood&Tissue buffer, IP and washing buffers, to preserve the oxidized DNA (36).
Conversion of ssDNA to dsDNA was obtained by Random Primers DNA Labeling System (Cat. No. 18187-013, ThermoFisher Scientific). Library preparation was performed as described (37) using 2 ng of DIP or Input DNA. Prior to sequencing, libraries were quantified using Qubit (Invitrogen) and quality-controlled using Agilent Bioanalyzer. 50 bp single-end sequencing was performed using Illumina HiSeq 2000 platform according to standard operating procedures. Reads were quality checked and filtered with NGS-QC Toolkit (38). Alignments were performed with Bowtie (39) and BWA (40) to hg18 or mm9 using default parameters. SAMtools (41) and bedtools (42) were used for filtering steps and file formats conversion. The peaks were identified from uniquely mapped reads without duplicates using MACS (43) (P < 1e–5 and fold enrichment >7). DNA Input was used as control. UCSC genome browser was used for data visualization. For qPCR analysis, 3 μl of 8-oxodG immuno-precipitated DNA (antibody AB5830, Millipore) was analysed in duplicate by quantitative PCR, using SYBR Green 2X PCR Master Mix (Applied Biosystems). The primer sets used in OxiDIP-qPCR from two biological replicates are indicated in Supplementary Table S2.
γH2AX ChIP-sequencing
Chromatin extracts from MCF10A cells were performed as described (33). 10 ng of ChIP (or Input) DNA were used to prepare ChIP-Seq libraries with TruSeq ChIP Sample Prep Kit (Illumina) according to the manufacturer's instructions. 50 bp single-end sequencing was performed using Illumina HiSeq 2000 platform. Reads were quality checked and filtered with ngsqctoolkit. Alignments were performed with Bowtie and BWA to hg18 using default parameters. SAMtools and bedtools were used for filtering steps and file formats conversion. The peaks were identified from uniquely mapped reads without duplicates using SICER (44) and FDR 0.01 was used as cutoff. DNA Input was used as control. UCSC genome browser was used for data visualization. Two biological independent experiments were performed and tested for reproducibility with Pearson correlation coefficient analysis (0.91), P < 2.2 × 10−15. γH2AX ChIP-Seq in MEFs were from GSE63861. Fastq data were filtered as described above and aligned with BWA to the mm9 using default parameters. HOMER (45) was used for peak detection and Input DNA was used as control.
Bioinformatic and statistical analyses
ChIP-Seq data were subjected to unbiased clustering using the SeqMINER 1.3.2 platform (46). The clustering was performed using a list of unique genes (hg18 or mm9) and the most expressed transcript (deriving from analysis of the GRO-Seq data) for each known gene symbol. All the gene loci, regardless of their length, were divided in 200 bins (20 from the 5 kb upstream the TSS, 160 from the gene body, and 20 from the 5 kb downstream the TTS), thus allowing direct comparisons. The length for all the bins upstream and downstream the gene body was constant (i.e. 250 bp), while it changed for the 160 bins from the gene body, as it depends on gene size; the signal from each bin was expressed as the highest number of overlapping reads within the bin. The 200 density values measured at each gene locus resulted in one vector, representing the relative distribution of the ChIP signal over the whole region (i.e. 5 kb upstream the TSS to 5 kb downstream the TTS). SeqMINER k-means unbiased clustering was performed using distances computed from the sets of vectors defined above (one vector per gene) to identify genes showing similar read densities within the specified genomic window. Thus, each of the four clusters obtained by this procedure represents a group of genes having similar distribution of 8-oxodG read densities over the gene locus. k = 4 was the lowest number of clusters providing the best separation of the 8-oxodGs signals from the analysed genes (n ∼ 20 000). Results did not change when the gene body was divided in <160 bins (down to 16), to accommodate the bins of short genes to the length of the sequenced reads.
Statistical significance of the observed differences in expression levels and gene lengths among the gene clusters was evaluated by one-way ANOVA test followed by pairwise comparison of means (Bonferroni post hoc analysis). Statistical significance of the overlap between human and mouse Cluster #3 genes was evaluated by means of hypergeometric test followed by post hoc analysis.
ChIP-Seq peaks were annotated using PAVIS (47). The hg18 genomic coordinates of peaks identified in MCF10A cells were converted to hg38 coordinates before annotation by using the UCSC tool liftover, whereas the mm9 coordinates of peaks identified in MEF cells were used for the annotation. Relative peak enrichment was determined with Fisher test of bedtools suite. Linear correlations between γH2AX and 8-oxodG signals were tested by means of Pearson's correlation test on the list of unique genes.
RNA-Seq were analysed with RAP pipeline (48) with default parameters, transcript assembly and abundance estimation were performed with Cufflink and the relative abundance measured in FPKM. Differential expression analyses were performed with HTSeq and DESeq, respectively. Fastq data for MCF10A and MEF RNA-Seq were retrieved as reported in Supplementary Table S11.
Gene set enrichment analyses were performed using GSEA/MSigDB tool on the 1609 genes of the Cluster #3 that showed the highest 8-oxodG signals both in human and mouse cells.
GRO-Seq and Pol-II-Ser2 in MCF10A were from ArrayExpress (E-MTAB-742) and GEO data NCBI (GSE45715), respectively. FASTQ files were aligned using Bowtie algorithm for identifying uniquely mapping region allowing for a maximum of two mismatches. GRO-Seq read quantifications were performed using HTSeq (49); reads mapping −2.5 kb upstream the TSS to the end of the corresponding gene were considered, and transcription levels were expressed as RPKM. GRO-Seq in MEFs was from GEO data NCBI (GSE27037) and analysed as above.
ORI density of each gene was expressed as the number of replication origins (i.e., ORC1 binding sites or Short Nascent Strands peaks identified in human and mouse cells, respectively) found within the body of the gene (from TSS to TTS), per kb. Statistical significance of the observed differences was evaluated by one-way ANOVA test followed by pairwise comparison of means (Bonferroni post hoc analysis).
The numbers of: genes, ORI-containing genes, ORIs within the gene body, ORIs overlapping with 8-oxodG peaks, and genes with ORIs overlapping with 8-oxodG peaks, reported in Supplementary Table S9, were computed with bedtools suite. Montecarlo approach was devised to test the enrichment of the overlap between 8-oxodG peaks and ORIs within the body of Cluster #3 genes. Montecarlo procedure was built in order to compute empirical P values associated with the number of observed Cluster #3 genes containing at least one ORI overlapping with 8-oxodG peaks. In each realization, the number of ORIs overlapping random 8-oxodG peaks and the number of distinct genes containing these ORIs were drown out under the null hypothesis that 8-oxodG peaks were randomly distributed over the genome. In particular, the following resampling procedure was implemented: (i) select a random permutation of the genomic coordinates of 8-oxodG sites over the corresponding reference genome; (ii) take the subset of ORC binding sites containing at least one random 8-oxodG peak from step (i); (iii) count the number of genes from Cluster #3 containing ORC binding sites from step (ii). We repeated these steps 1000 times. We then compared the observed number of genes containing ORIs overlapping with 8-oxodG peaks with the corresponding series of 1000 random realizations from the Montecarlo simulation, and considered the observed value as statistically significant if it was greater than all the simulated values.
Bedtools was used to analyse the overlap between genes in each cluster from MCF10A cells and CFSs mapped at the molecular level (50), or cancer deletions (Ref (51) and CosmicStructExport v80.tsv).
Density of 8-oxodG peaks previously identified in MEF (28) was determined using bedtools suite and measured for each gene as the number of OG-peaks/100 kb. Statistical significance of enrichment of ORI density in Cluster #3 genes was evaluated by means of one-way ANOVA test followed by pairwise comparison of means (Bonferroni post hoc analysis).
G4 analysis within the 8-oxodG peaks has been carried out by applying the Quadron tool (52), a machine learning algorithm, using default options. Ten random permutations of the 52 298 8-oxodG peaks were obtained with bedtools suite and analysed in Quadron.
This study was conducted using 0.05 as significance threshold; all statistical analyses, except seqMINER, were performed with R (R Development Core Team, 2016).
RESULTS
OxiDIP-Seq allows genome-wide mapping of 8-oxodG
In order to obtain the genome-wide mapping of 8-oxodG in human cells, we used the immortalized non-tumorigenic human breast epithelial MCF10A cells. First, to measure the amount of 8-oxodG in exponentially growing cells, genomic DNA was analysed by established and highly sensitive ultra-performance liquid chromatography tandem mass spectrometry (LC-MS/MS), which uses stable isotope-labelled (15N5) 8-oxodG as an internal standard for sample quantification. This approach led to an estimation of 2.5 8-oxodG/106 dG in MCF10 cells (Figure 1A and Supplementary Figure S1A and B). To further test the specificity of our LC–MS/MS approach, we used: (i) UV irradiation, which is known to induce, by intracellular photoreactions, the formation of reactive oxygen species (ROS) that, in turn, oxidize DNA (53–56) and (ii) N-acetylcysteine (NAC), which is an effective ROS scavenger (57). Consistently, while UV-irradiated MCF10A cells showed increased 8-oxodG levels (23 8-oxodG/106 dG), decrease in ROS levels led to undetectable 8-oxodG signals in NAC-treated cells (Figure 1A). Next, we compared two commercially available anti-8-oxodG antibodies in immuno-precipitation (IP) assays of 8-oxodG (OxiDIP) contained within synthetic sequences, or secondary structures commonly found in the genomic DNA: ssDNA and G-quadruplex DNA (G4-DNA). qPCR analyses with primer pairs specific for both synthetic ssDNA and G-quadruplex, following OxiDIP performed with the polyclonal antibodies, showed at least a nine-fold increase in the amount of immuno-precipitated DNA, compared to the monoclonal antibody (Ab M and Ab T, respectively, in Figure 1B). Therefore, only the polyclonal antibodies were used for the following analyses.
Figure 1.
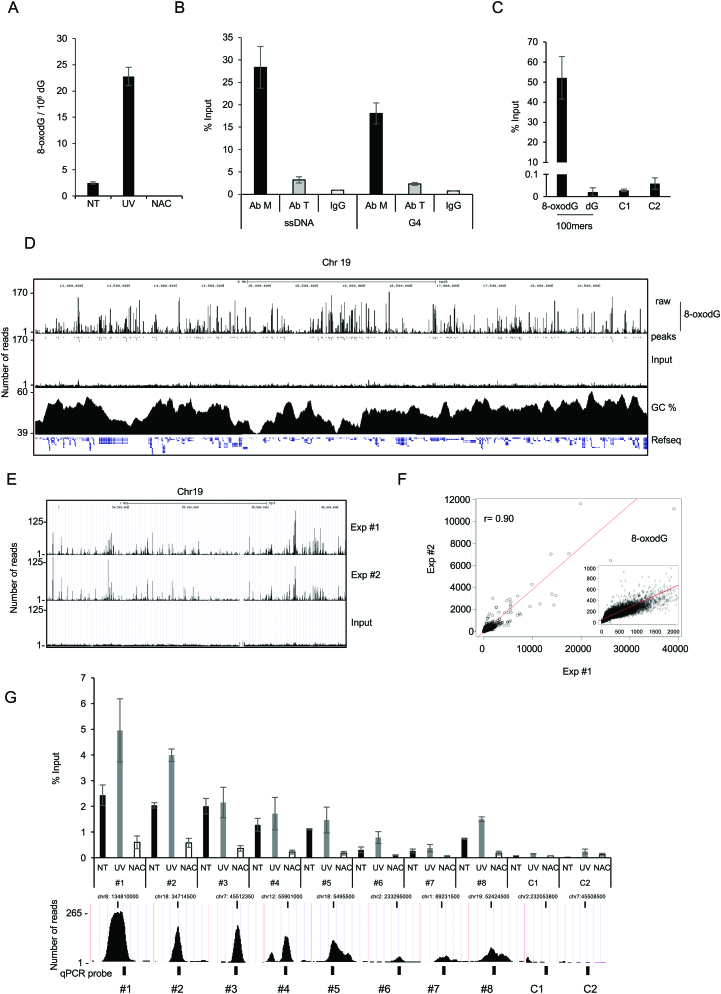
(A) Number of 8-oxodGs per million of dGs (8-oxodg/106 dG) measured by LC-MS/MS in untreated (NT), UV-irradiated (UV) and NAC-treated (NAC) MCF10A cells, as indicated. (B) Efficiency of polyclonal anti-8-oxodG from Millipore (Ab M), or monoclonal anti-8-oxodG from Trevigen (Ab T), and of anti-IgG antibodies in immuno-precipitation assays of 8-oxodG-containing synthetic ssDNA or G4 structures, as indicated. (C) Anti-8oxodG immuno-precipitation assay (% of input DNA, measured by qPCR; y axis) with equal amount (64 pg) of both synthetic oligonucleotides (8-oxodG-100mer and dG-100mer) added to 1 μg of NAC-treated genomic DNA. C1 and C2 indicate the same genomic negative control regions as in panel G. (D) Screenshot from the UCSC genome browser of 3.7 Mb from human chromosome 19 showing (top to bottom): OxiDIP-Seq signal profile, 8-oxodG peaks (dots), Input DNA, CG%, and RefSeq genes. (E) Screenshot from the UCSC genome browser of 2.1 Mb from human chromosome 19 showing OxiDIP-Seq signal profiles of two independent experiments (Exp #1 and #2) and Input DNA. (F) Scatter plot showing the correlation of the OxiDIP-Seq signals obtained in two independent experiments (Exp #1 and #2). Pearson's correlation coefficient (r), as indicated. Figure inset shows magnification of the high-density region. (G) OxiDIP-qPCR showing 8-oxodG enrichments (% of Input DNA) at eight different positive (#1–8) and two negative (C1, C2) regions, in untreated (black bar), UV- (gray), or NAC-treated (white) MCF10A cells. Screenshots from the UCSC genome browser show genomic position, 8-oxodG signal intensity, and qPCR probes (black box) of the selected regions. Data from two independent OxiDIP-qPCR assays are shown (±S.D.; P< 0.05 for all comparisons except for NT versus UV of peaks #3–#5; Student's t-test).
Aiming at a more precise estimation of the background of OxiDIP using input DNA from cells growing in unperturbed conditions, immuno-precipitated DNA (expressed as % of Input) was first evaluated for linearity by addition of increasing amounts of 8-oxodG-containing oligonucleotides (i.e. 0.5–64 pg of oligomers) to 1 μg of genomic DNA from NAC-treated MCF10A cells (Supplementary Figure S1C). We then performed OxiDIP experiments using equimolar amounts of both 8-oxodG- and dG-containing oligonucleotides (i.e. 64 pg) mixed with 1 μg of genomic DNA from NAC-treated cells. qPCR amplifications performed prior and after IP, with primer pairs specific for the 8-oxodG- or dG-containing oligonucleotides, showed that the 8-oxodG-containing oligonucleotide was specifically immuno-precipitated (>1000-fold more than the control; Figure 1C). Given the high specificity of the anti-8-oxodG antibodies, we carried out OxiDIP-Seq in MCF10A cells; genomic DNA from asynchronous MCF10A cells was extracted, fragmented by sonication, denatured, and immuno-precipitated using the specific anti-8-oxodG antibodies. The immuno-precipitated and input DNA were sequenced and the obtained sequence tags were aligned to the human genome (Figure 1D). MACS (Model-based Analysis for ChIP-Seq) analysis identified 52 298 genomic regions enriched in 8-oxodG (or high-confidence peaks: P < 1e–5 and Fold Enrichment>7; see Methods and Supplementary Table S2). Strikingly, an independent biological replicate showed 95% overlap with the first dataset of 8-oxodG peaks (Figure 1E), consistent with the very high correlation between the signals of the two OxiDIP-Seq experiments at the 8-oxodG peaks (r = 0.90; Figure 1F). OxiDIP-Seq data were validated by qPCR using eight 8-oxodG-positive regions with increasing tag densities, and two control regions (regions #1–8 and C1-C2, respectively; Figure 1G) in a third biological replicate in MCF10A untreated cells. Interestingly, the recovery of the two control regions was similar to the control oligonucleotide in the OxiDIP performed with control and 8-oxodG-containing oligonucleotides together (compare Figure 1C and Figure 1G), while the amount of recovered DNA from the 8-oxodG-positive regions was consistent with peak amplitude or proximity of probes to the peak summit (Figure 1G, black bars). Moreover, upon UV-irradiation of MCF10A cells, almost all the 8-oxodG – positive regions showed an increase in 8-oxodG levels (strong for regions #1, 2, 6 and 8, or mild for regions #4, 5 and 7) (Figure 1G, gray bars), while the intensity of 8-oxodG signals was drastically reduced in NAC-treated cells (Figure 1G, white bars), thus further confirming the specificity of our antibodies (compare Figure 1A and Figure 1G).
Visual inspection of: (i) the 8-oxodG signal distribution (tag densities) along the genome, (ii) input DNA and (iii) the GC content profile, allowed the identification of 8-oxodG enrichments both in GC-rich and GC-poor regions, and of GC-rich regions almost devoid of 8-oxodGs (Figure 1D, and Supplementary Figure S2).
The G4-DNA was previously found to be enriched in 8-oxodG (28,58). In order to measure the occurrence of G4 within the 8-oxodG peaks, we used Quadron, a sequence-based computational model that was developed using large-scale machine learning from an extensive experimental G4 dataset obtained by G4-seq methodology (52). This model allows the identification of putative quadruplex sequences that do not actually form stable G4 structures. Strikingly, 19 235 8-oxodG peaks (37% of the total) contained potential G4 structures, the vast majority of which showed high folding potential (Supplementary Figure S3A, B). Furthermore, the model identified only 5350 to 5620 potential G4 structures when 52 298 regions were randomly positioned (n = 10 times) in the human genome (Supplementary Figure S3C). Thus, OxiDIP-Seq immuno-precipitate 8-oxodG within G4 structures formed in the human genome, as suggested by our analysis of synthetic oligonucleotides (Figure 1B).
Together, these results show the specificity of the antibodies used for 8-oxodG within ssDNA and G4s, and the reproducibility of our OxiDIP assay.
8-oxodG and γH2AX co-localize at transcribed regions
Next, we asked whether 8-oxodG peaks were enriched at specific regions of the human genome. At this purpose, we analysed the genomic distribution of 8-oxodG peaks, and found that: (i) 42% mapped within gene loci (i.e., promoter and gene body; Figure 2A); (ii) they were enriched within both gene body and promoter regions (P < 2.2e–16; Supplementary Table S4); (iii) 54% and 30% of the 8-oxodG peaks mapped within protein-coding and long non-coding genes, respectively (P < 2.2e–16; Figure 2B and Supplementary Table S4).
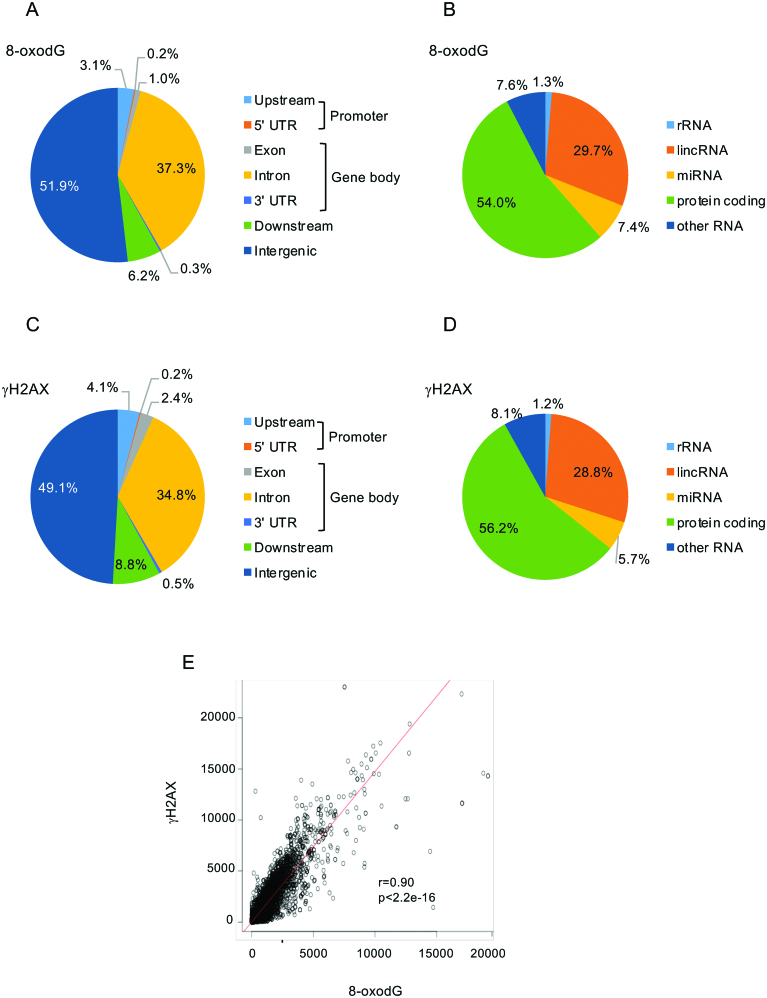
Figure 2.
Pie charts showing the annotation of 8-oxodG (panels A, B) and γH2AX (panels C, D) peaks. (E) Scatter plot showing the correlation between 8-oxodG and γH2AX signals within the RefSeq genes. Pearson's correlation coefficient (r), and P-value (p), as indicated.
In order to investigate the association between the presence of 8-oxodG peaks within genes and the activation of a DNA damage response (DDR) induced by DSB formation, we performed anti-γH2AX ChIP-Seq in MCF10A cells, and found 20 440 γH2AX-enriched regions. Interestingly, 42% mapped within gene loci (Figure 2C; P < 2.2e–16). Thus, genomic distributions of γH2AX and 8-oxodG peaks were almost identical (compare Figure 2A and C, and Figure 2B and D); consistently, similar to 8-oxodG peaks, 56% and 29% of γH2AX-enriched regions were found within protein-coding and long non-coding genes (P < 2.2e–16; Figure 2D and Supplementary Table S4).
Thus, we asked whether 8-oxodG signals at gene loci were associated with γH2AX. Strikingly, when we compared the tag densities of 8-oxodG and γH2AX within the gene body of the RefSeq genes, we found a very strong correlation (Pearson correlation test r = 0.9, P < 2.2e–16) (Figure 2E). Collectively, these data indicate that oxidation of guanosines in cells grown in unperturbed conditions strongly correlate with H2AX phosphorylation within the gene bodies, intriguingly suggesting that DNA oxidation at these sites is a potential source of constitutive endogenous double strand breaks (DSBs).
8-oxodG and γH2AX accumulate within the gene body of long genes with poor-to-moderate transcription levels
To investigate the role of transcription in the observed enrichment of 8-oxodG within gene bodies (Supplementary Table S4), we first measured the association between 8-oxodG and transcription levels. Pearson correlation between the tag densities of 8-oxodG and GRO-Seq within the gene body of the RefSeq genes was very poor (r = 0.05, P = 5.59e-14; Supplementary Figure S4A).
We then asked whether 8-oxodG and γH2AX enrichments occur at the same genes. At this purpose, we compared 8-oxodG and γH2AX profiles across all human RefSeq genes; publicly available MCF10A datasets of RNA Polymerase II phosphorylated at the CTD serine 2 residue (Pol2-S2P) and GRO-Seq were included in the analysis. We used seqMINER k-means unbiased clustering that allows: (i) the analysis of the signal enrichment status in multiple tracks, (ii) an easy visualization of signal distribution over multiple loci, and iii) the identification of general patterns over the analysed dataset (i.e. ∼20 000 genes). Thus, all the gene loci, including the 5 kb both upstream the Transcription Start Site (TSS) and downstream the Transcription Termination Site (TTS), were binned in order to compare genes with different lengths, and 8-oxodG, γH2AX, Pol2-S2P and GRO-Seq signals were analysed (see Methods).
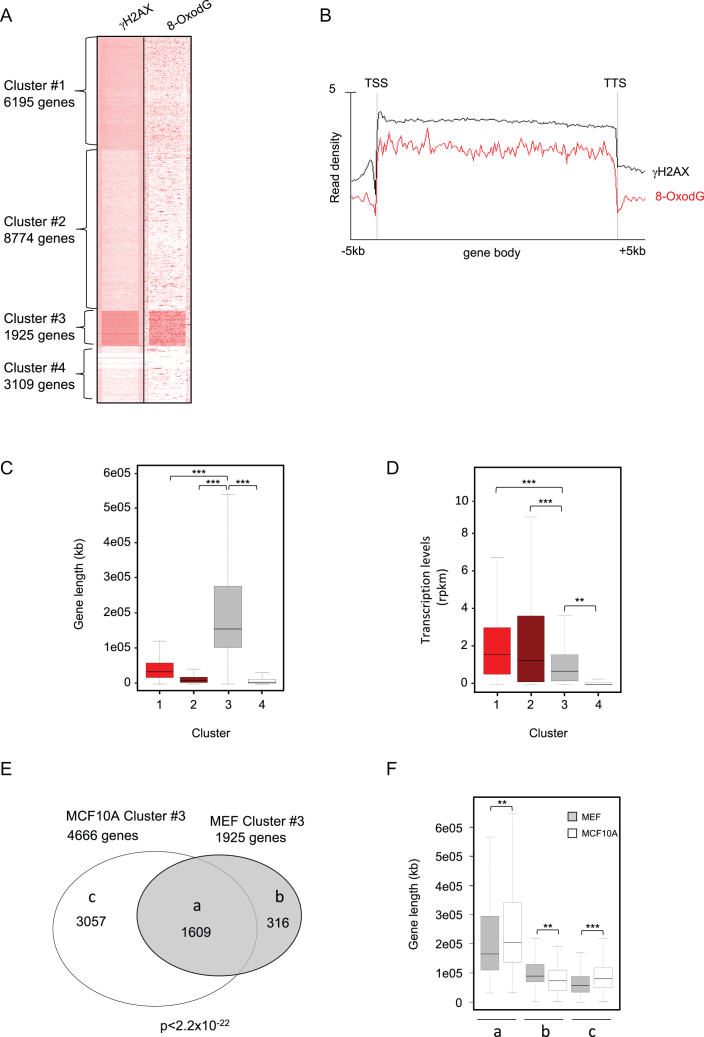
Visualization of the whole dataset of genes was achieved through heatmaps, which revealed four different clusters (#1–#4), with Cluster #3, containing 4666 genes, showing the strongest 8-oxodG and γH2AX signals (Figure 3A, Supplementary Figure S4B and Supplementary Table S5). Analysis of the average profiles of 8-oxodG and γH2AX signals showed that they were much stronger in the gene body of Cluster #3 genes than in the other clusters, with a sharp decrease at both the TSS and TTS, while they were similar in all the four clusters both upstream the TSS and downstream the TTS (Figure 3B).
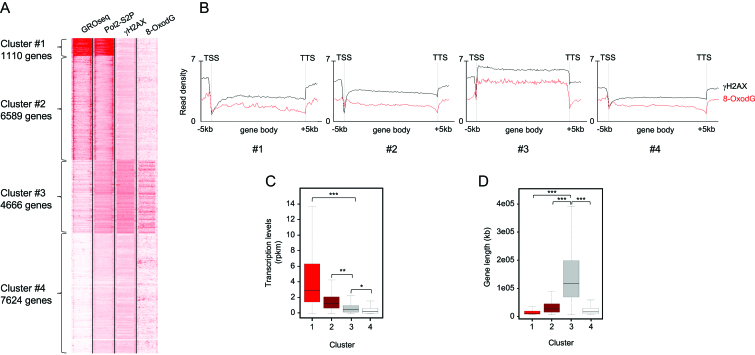
Figure 3.
(A) Heatmap showing GRO-Seq, Pol2-S2P, γH2AX ChIP-Seq, and 8-oxodG OxiDIP-Seq signals at the gene body, and within the 5 kb both upstream the TSS and downstream the TTS of the RefSeq genes. Gene clusters identified by SeqMINER unbiased k-means clustering (Cluster #1–#4), and the number of genes contained within each cluster, as indicated. (B) Read density profiles of 8-oxodG OxiDIP-Seq (red) and γH2AX ChIP-Seq (black) in Cluster #1–#4, as indicated. (C) Box plot showing the distribution of transcription levels (RPKM, measured by GRO-seq) of human genes within each cluster in MCF10A cells. (D) Box plot showing the length distribution of human genes within each cluster in MCF10A cells (P< 2.2e–16, ANOVA test; ***P< 2.2e–16, **P = 2.1e–3, *P = 2.4e–2, Bonferroni post hoc analysis for pairwise comparison).
Interestingly, Cluster #3 genes showed low-to-moderate transcription levels, as revealed by both GRO-Seq signals (Figure 3A and C) and RNA-Seq data (Supplementary Figure S4C). Cluster #1 genes were instead characterized by the highest transcription levels, as shown by GRO-Seq signals (Figure 3A and C) and RNA-Seq data (Supplementary Figure S4C). However, they showed very low γH2AX and 8-oxodG levels, comparable to those of Clusters #2 and #4, which contain genes with high-to-moderate or extremely low transcription levels, respectively (Figure 3A–C). Thus, gene body accumulation of γH2AX and 8-oxodG is not associated with high transcription levels.
We then asked whether the Cluster #3 genes showed specific genetic features, and found that they were much longer (median length of 111 kb) than the genes from all the other clusters (P < 2.2e–16; Figure 3D). Together, these data show that 8-oxodG and γH2AX enrichments preferentially occur within long genes with poor-to-moderate transcription levels.
In mouse embryo fibroblasts 8-oxodG and γH2AX showed same distribution as in MCF10A cells
8-oxodG-Seq (OG-Seq) data were recently obtained in MEFs by affinity purification of chemically biotin-labeled 8-oxodG) (28). In order to compare: i) OxiDIP-Seq to the published OG-Seq data, and ii) human OxiDIP-Seq data to the mouse ones, we performed OxiDIP-Seq in mouse embryo fibroblasts (MEFs). Using the same criteria as in MCF10A cells, 15 218 high-confidence 8-oxodG peaks were identified (Supplementary Table S6). Two biological replicates showed great overlap, as shown both locally (Figure 4A) and in the whole genome (by Pearson correlation test; r = 0.9). Similar to what we observed in human cells, 37% of 8-oxodG peaks mapped within gene loci, defined as above, and they were enriched within both gene body and promoter regions (P < 2.2e–16 and P = 1.3e–14; Figure 4B and Supplementary Table S4). Furthermore, 74% of the 8-oxodG peaks mapped within protein-coding genes (P < 2.2e–16; Figure 4C and Supplementary Table S4).
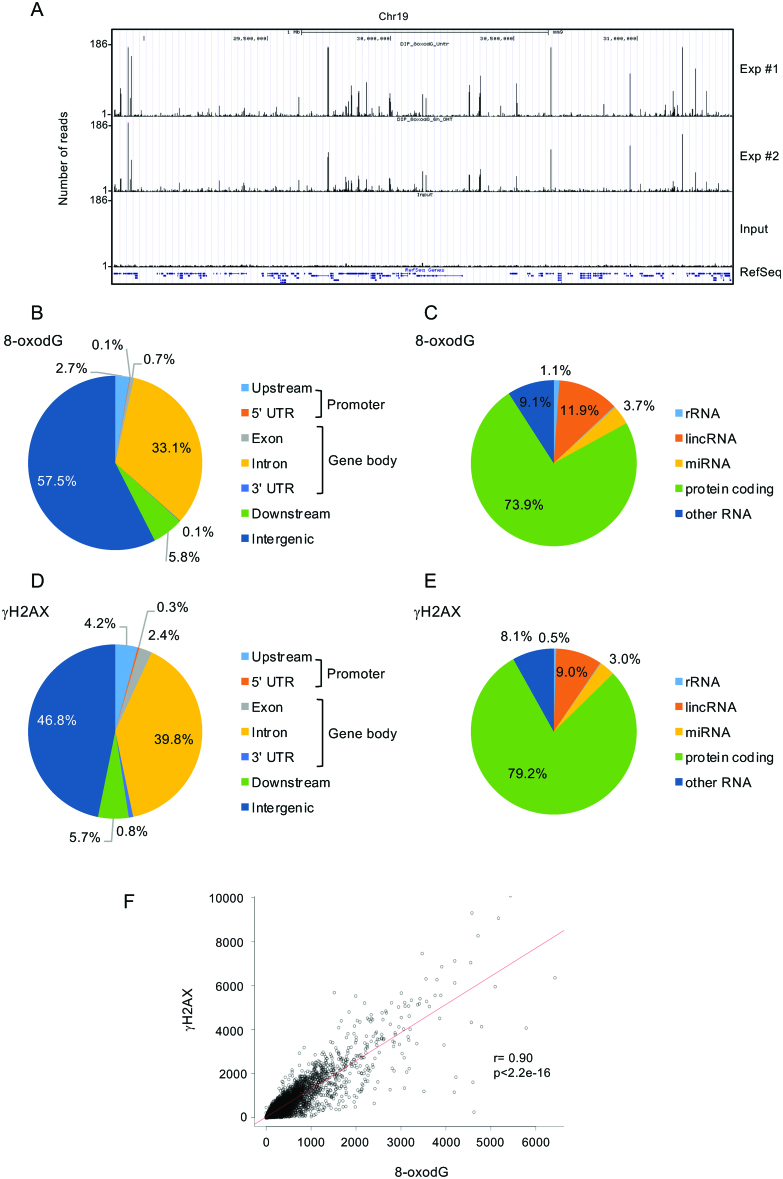
Figure 4.
(A) Screenshot from the UCSC genome browser of 2.6 Mb from mouse chromosome 19 showing (top to bottom): OxiDIP-Seq signal profiles of two independents OxiDIP-Seq experiments (Exp #1 and #2), Input DNA, and RefSeq genes. (D-E) Pie charts showing the annotation of 8-oxodG (panels B, C) and γH2AX (panels D, E) peaks. (F) Scatter plot showing the correlation of 8-oxodG and γH2AX signals within the RefSeq genes. Pearson's correlation coefficient (r), and P-value (p), as indicated.
Analysis of a publicly available dataset of γH2AX ChIP-Seq in MEFs showed 48% of γH2AX peaks within gene loci (P<2.2e-16; Figure 4D, and Supplementary Table S4). Furthermore, 79% of γH2AX peaks mapped within protein-coding genes (P < 2.2e–16; Figure 4E and Supplementary Table S4). Strikingly, as in human cells, we found a highly significant correlation between 8-oxodG and γH2AX signals within the mouse genes (Pearson correlation test, r = 0.9; P < 2.2e–16) (Figure 4F).
Unbiased clustering of 8-oxodG and γH2AX profiles across mouse RefSeq genes identified four different clusters. Cluster #3 genes (n = 1925) showed the strongest signals of both 8-oxodG and γH2AX (Figure 5A and Supplementary Table S7), which spread along the gene body, with a sharp decrease at TSS and TTS (Figure 5B and Supplementary Figure S5). Interestingly, as observed in MCF10A cells, these genes were much longer than those from all the other clusters (median length of 156 kb; Figure 5C), and they were among the least transcribed in MEFs, as shown by both GRO-Seq (Figure 5D) and RNA-Seq data (Supplementary Figure S4D). Comparison between OxiDIP-Seq and OG-Seq showed extremely poor overlap (<1%). Strikingly, however, analysis of 8-oxodG peak density from OG-Seq in the same gene clusters identified by OxiDIP-Seq, showed that Cluster #3 genes where by far the ones with the highest oxidatively-generated damage (Supplementary Figure S6).
Figure 5.
(A) Heatmap showing γH2AX ChIP-Seq and 8-oxodG OxiDIP-Seq signals within the gene body, and the 5 kb both upstream the TSS and downstream the TTS of the RefSeq genes. Gene clusters identified by SeqMINER unbiased k-means clustering (Cluster #1–#4), and the number of genes contained within each cluster, as indicated. (B) Read density profiles of 8-oxodG OxiDIP-Seq (red) an γH2AX ChIP-Seq (black) in Cluster #1–#4, as indicated. (C) Box plot showing the length distribution of mouse genes within each cluster in MEFs (P < 2.2e–16, ANOVA test; ***P < 2.2e–16, **P = 2.9e–4, Bonferroni post hoc analysis for pairwise comparison). (D) Box plot showing the distribution of transcription levels (RPKM, measured by GRO-seq) of mouse genes within each cluster in MEFs (P< 2.2e–16, ANOVA test; ***P < 2.2e–16, **P = 2.9e–4, Bonferroni post hoc analysis for pairwise comparison). (E) Venn diagram showing the number of common (group a) and mouse- (MEFs, group b) or human- (MCF10A, group c) specific Cluster #3 genes; P< 2.2e–16 (Hypergeometric test). (F) Gene length distribution of the common (a), mouse-specific (b) or human-specific (c) genes in human (white bars) and mouse (grey bars), as indicated (P< 2.2e–16, ANOVA test; ***P< 2.2e–16, **P = 1.0e–5, Bonferroni post hoc analysis for pairwise comparison).
Furthermore, the vast majority of the 1925 Cluster #3 genes (84%; n = 1609/1925) showed the highest 8-oxodG signals also in human cells (P < 2.2e–16; Figure 5E and Supplementary Table S8). Consistently, the remaining Cluster #3 mouse- and human-specific genes (n = 316 and n = 3057, respectively) were significantly longer than their orthologs (Figure 5F). Collectively, these findings demonstrate that both in human and mouse cells, the transcribed large genes are particularly prone to DNA oxidation.
Genes within CFSs, or frequently deleted in cancer, show high levels of 8-oxodG
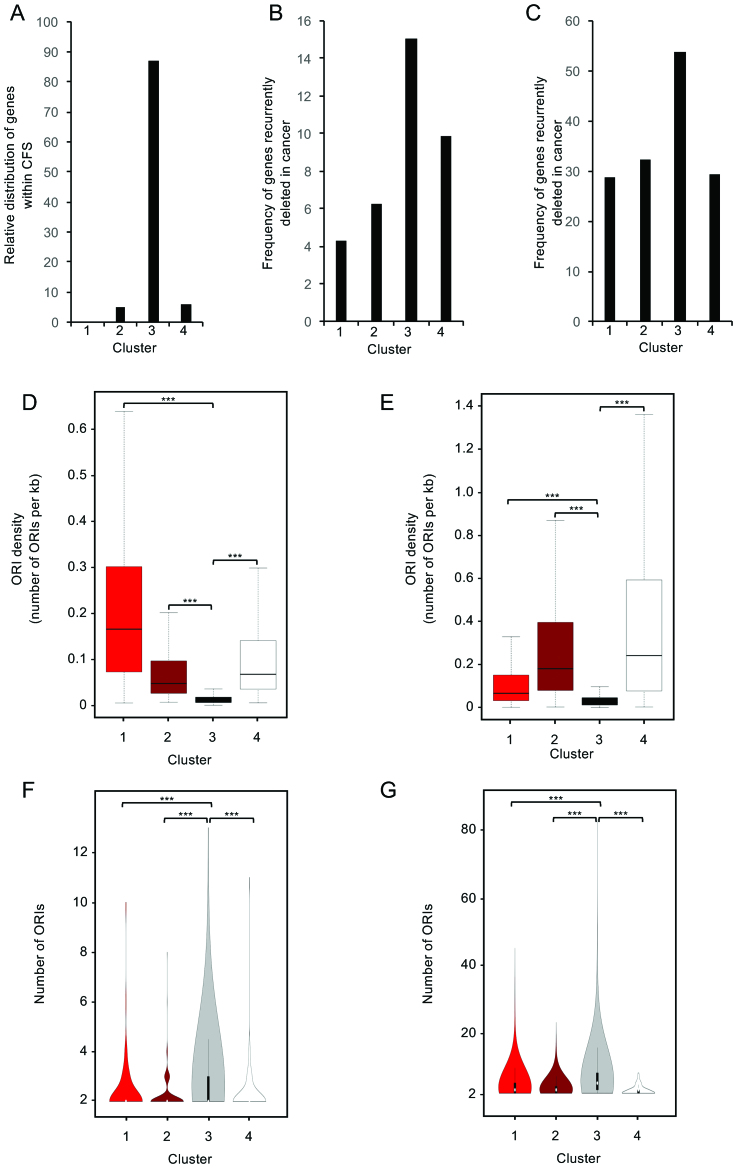
Transcribed long genes (>300 kb in length) have been associated with Common Fragile Sites (CFSs) (50,59). Thus, we investigated the distribution, within the four clusters, of the 85 genes which have been mapped at the molecular level within CFSs in human cells (59), and found 88% of them in Cluster #3 (n = 75/85; Figure 6A). Next, we investigated the frequency of genes recurrently deleted in cancer within the four clusters, using two different databases (i.e. Ref (51), and COSMIC, containing 1877 and 6785 genes, respectively). Strikingly, in both cases, the highest frequency of recurrently deleted genes in cancer was found in Cluster #3 (Figure 6B, C). Thus, genes showing co-enrichment of 8-oxodG and γH2AX are the most frequently associated with CFSs or recurrent deletions in cancers. We then compared the expression of the Cluster #3 genes in T47D breast cancer cells and MCF10A, and found that the majority of differentially expressed genes (56%; n = 1542) showed higher expression levels in the cancer cells (Supplementary Table S9). Cancer cells might thus experience higher frequency of transcription-replication conflicts at these genes.
Figure 6.
(A) Bar plot showing the distribution within the four cluster of the genes contained within molecularly mapped CFSs (P< 2.2e–16; X2-test). (B and C) Bar plots showing the frequency of the genes recurrently deleted in cancer within each cluster (Ref (51), panel B; COSMIC database, panel C). (D) Boxplots showing the ORI density (human ORC1 binding sites) within the four identified clusters, as indicated. (E) Boxplots showing the ORI density (mouse Short Nascent Strands sites) within the four identified clusters, as indicated (P< 2.2e–16, ANOVA test; ***P< 2.2e–16, Bonferroni post hoc analysis for pairwise comparison). (F, G) Violin plot showing the relative abundance of genes with the indicated number of ORIs within each gene cluster (only genes with ≥2 ORIs are shown) in MCF10A (panel F) and MEFs (panel G).
Finally, GSEA/MSigDB analysis of Cluster#3 genes common to MCF10A cells and MEFs (n = 1609) showed that they were enriched in: genome maintenance (UV response), oestrogen response (late and early response), epithelial to mesenchyme transition and cell signalling (TGFbeta, IL2-STAT5 and PI3K-AKT-mTOR) (Supplementary Table S9).
8-oxodG preferentially co-localizes with DNA replication origins within the body of long genes
Collisions between replication and transcription machineries have been proposed to be unavoidable for very large genes, since their transcription extends into the S phase of a subsequent cell cycle (50,60). Furthermore, it has been demonstrated that persistent RNA:DNA hybrids and ssDNA found at collision sites are particularly sensitive to damage (31,32). Thus, we hypothesized the presence of persistent DNA oxidation at sites of transcription-replication clashes within the gene body of Cluster #3 genes. To test this hypothesis, we first determined the density of ORIs within the four gene clusters using the available datasets of human ORC1 binding sites (61) and active mouse ORIs deriving from isolation of Short Nascent DNA Strands (62). Interestingly, Cluster #3 genes were extremely poor in ORIs both in human and mouse genomes (Figure 6D, E), consistent with the notion that ORI-deficient regions are strongly associated with CFSs and deletions in cancer (50,60). However, because of the length distribution of the genes contained within Cluster #3, it showed the highest proportion of genes with at least two ORIs within their gene body (Figure 6F, G and Supplementary Figure S7A, B). We then investigated the physical relationship between DNA oxidation and ORIs within the gene bodies, and found that in Cluster #3 the frequency of genes with 8-oxodG peaks mapping in the proximity (±2.5 kb) of ORIs was higher than in all the other clusters (Supplementary Figure S7C, D). The same was observed when considering the frequency of 8-oxodG peaks co-localizing with ORIs (Supplementary Figure S7E, F). In particular, 8-oxodG peaks significantly co-localized with ORIs within Cluster #3 genes (P<0.001; Supplementary Figure S8A, B and Supplementary Table S10). Strikingly, Cluster #3 genes that showed 8-oxodG peaks co-localizing with ORIs were significantly enriched both in MCF10A cells and MEFs (P < 0.001; Supplementary Figure S8C, D and Supplementary Table S10).
Together, these data show that though Cluster #3 genes are characterized by the lowest ORI density in the human and mouse genomes, they generally contain multiple ORIs, which are prone to guanosine oxidation. Increased oxidation levels, together with other kinds of lesion occurring at the ORIs within these genes, might thus contribute to the observed accumulation of γH2AX as a consequence of DSB formation.
DISCUSSION
In the present study, we describe a sensitive method (OxiDIP-Seq) for the genome-wide mapping of DNA oxidation. In diploid human mammary-epithelial cells (MCF10A) and mouse embryonic fibroblasts (MEFs) grown in unperturbed conditions, we found ∼52 000 and 15 000 8-oxodeoxyguanosine – enriched regions (or peaks), respectively. 37% of the 8-oxodG peaks identified in the human cells contained G4 structures, the vast majority of which possessed high folding potential, thus showing the efficiency of OxiDIP in the identification of 8-oxodGs within non-B DNA structures, and extending previous knowledge about the sensitivity of G4s to oxidation.
Unbiased clustering based on GROseq, OxiDIP-Seq and γH2AX ChIPseq profiles, allowed the identification of a significant fraction of coding and non-coding genes (in the order of thousands in both human and mouse cells) that show co-occurrence of high levels of DNA oxidation and γH2AX within their gene body, compared to the flanking regions (both upstream the TSS and downstream the TTS) and all the remaining genes. Strikingly, these genes are transcribed and generally long (median of 111 kb and 156 kb in human and mouse cells, respectively). The first evidence of the correlation between oxidation and transcription comes from chromatin fractionation experiments showing that the levels of 8-oxodG in transcriptionally active euchromatin were approximately five-fold higher than in heterochromatin (15). Strikingly, however, long genes with high levels of 8-oxodG and γH2AX were among the least transcribed in MCF10A cells and MEFs, thus suggesting that transcription per se is not sufficient to explain the oxidatively-generated damage observed at those genes.
A growing list of evidence uncovered a new and unexpected tight connection between transcription and DNA damage to facilitate transcription and/or minimize the negative impact on genome stability deriving from transcription-replication clashes (24,63). Interestingly, we found that almost all the genes contained within the CFSs previously mapped at the molecular level in the human genome are among the long genes showing high levels of 8-oxodG and γH2AX within their gene body. CFSs were previously shown to contain very long, late-replicating, active genes (>300 kb), and to be poor in DNA replication origins (31,32,50,64), thus leading to hypothesize that CFSs are frequently under-replicated in S-phase and prone to DSB formation in the following mitosis (65–67), consistent with the observation that they accumulate DNA repair proteins (68). Alternative mechanisms of CFS instability have been suggested by recent reports showing that persistent R-loops impede replication at CFSs (69). High-throughput approaches to identify genomic regions harbouring recurrent DSBs in primary neural stem/progenitor cells allowed the identification of 27 recurrent DSB clusters, all occurring within the body of long, transcribed, and late-replicating genes, mostly only upon mild aphidicolin-induced replication stress (70). However, the role played by transcription stress in the generation of at least a subset of CFSs, cannot be excluded, as suggested by: i) the overlap of RECQL5 (the RNA Polymerase II-associated helicase)-dependent genomic rearrangements with both CFSs and the transcribed regions of long genes (71), ii) accumulation of Topoisomerase IIβ and DSBs markers at transcribed large genes to relieve torsional stress during transcription elongation (24), and iii) the correlation between gene length and γH2AX peak width (17).
We propose that accumulation of 8-oxodG at the DNA replication origins within the body of transcribed large genes is compatible with the increased sensitivity of persistent ssDNA, regardless of the mechanisms by which it is generated (e.g. collision between transcription and replication machineries, block of the leading-strand replication by a G4 structure (72)), to oxidation (29,30), or other kinds of lesion (73), as compared to dsDNA.
Genes characterized by co-occurrence of 8-oxodG and γH2AX enrichments showed great overlap when comparing human and mouse cells. Their transcription levels and gene length were also very similar in both species. Indeed, analysis of the few MEF-specific genes showed that they were significantly longer than their human orthologous genes and, the same was true for the MCF10A-specific genes, when compared to their mouse counterparts.
Phosphorylation of H2AX is known to play a key role in DDR and is required for the assembly of DNA repair proteins at sites containing DSBs (74). Co-enrichment of 8-oxodG and γH2AX within the body of long poorly-transcribed genes is consistent with the hypothesis that high steady-state levels of DNA oxidation contribute to the formation of constitutive endogenous DSBs and the inherent instability of these loci, as confirmed by their frequent association with recurrent deletions in cancer. These DSBs might be formed as secondary products during the processing of ssDNA lesions, such as dG or dA oxidation, deamination, alkylation, etc. Indeed, slower repair of 8-oxodG sites, with formation of abasic sites and nicks (single strand breaks, or SSB), as a consequence of OGG1 and APE1 enzymatic activities, respectively, might lead to DSB formation within genomic regions containing either closely opposed 8-oxodG sites, or individual 8-oxodG sites occurring in proximity to other kinds of lesion. Alternatively, isolated SSBs formed during the processing of 8-oxodG can be converted in DSBs during S-phase.
DATA AVAILABILITY
All sequencing data related to this study have been deposited to the NCBI Gene Expression Omnibus (GEO) under accession no. GSE100234.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr EV Avvedimento for helpful discussions and critical review of the manuscript, and Dr Carolina Fontanarosa for the technical assistance in the LC–MS/MS analysis, and Rossana Piccioni for the technical assistance in the preparation of libraries for OxiDIP-Seq. We thank Dr Bruno Amati for MEFs (3T9-MycER cells). S.A. thanks NAnA ONLUS for support (Francesca Martini Prize).
Author contributions: S.A., B.M. and G.I.D. designed and conceived the study. S.A., G.D.P. and F.G. performed experiments. S.A. and G.S. and S.C. carried out bioinformatics and statistical analyses. T.C. supported bioinformatics analyses on Super Computing CINECA platforms and performed G4 analysis. P.P., A.A., B.M. and I.S. performed the LC–MS/MS experiments. L.L., P.G.P. and B.M. supervised as senior the study. G.I.D., S.A., L.L. and B.M. wrote the paper. All authors read and commented on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
AIRC IG13173 to B.M.; EC-FP7-ERC-InMec: 341131 and AIRC IG 17: IG-2017-20162 to P.G.P.; Epigenomics Flagship Project—EPIGEN, C.N.R. and from Grant MOVIE of the Rete delle Biotecnologie, Campania. Computer resource for this study was supported to SA from both CINECA ISCRA [IsC39_MSDDiM and IsC48_TraSMODa]; ELIXIR_ITA [ELIX_prj10]. Funding for open access charge: AIRC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Cooke M.S., Evans M.D., Dizdaroglu M., Lunec J.. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003; 17:1195–1214. [DOI] [PubMed] [Google Scholar]
- 2. Agnez-Lima L.F., Melo J.T.A., Silva A.E., Oliveira A.H.S., Timoteo A.R.S., Lima-Bessa K.M., Martinez G.R., Medeiros M.H.G., Di Mascio P., Galhardo R.S. et al. . DNA damage by singlet oxygen and cellular protective mechanisms. Mutat. Res. - Rev. Mutat. Res. 2012; 751:15–28. [DOI] [PubMed] [Google Scholar]
- 3. Shibutani S., Takeshita M., Grollman A.P.. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991; 349:431–434. [DOI] [PubMed] [Google Scholar]
- 4. Oka S., Nakabeppu Y.. DNA glycosylase encoded by MUTYH functions as a molecular switch for programmed cell death under oxidative stress to suppress tumorigenesis. Cancer Sci. 2011; 102:677–682. [DOI] [PubMed] [Google Scholar]
- 5. Cooke M.S., Evans M.D.. 8-Oxo-deoxyguanosine: reduce, reuse, recycle. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:13535–13536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Valavandis A., Vlachogianni T., Fiotakis C.. 8-hydroxy-2′ -deoxyguanosine (8-OHdG): A critical biomarker of oxidative stress and carcinogenesis. J. Environ. Sci. Heal. Part C. 2009; 27:120–139. [DOI] [PubMed] [Google Scholar]
- 7. Sova H., Jukkola-Vuorinen A., Puistola U., Kauppila S., Karihtala P.. 8-Hydroxydeoxyguanosine: a new potential independent prognostic factor in breast cancer. Br. J. Cancer. 2010; 102:1018–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bruner S.D., Norman D.P., Verdine G.L.. Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature. 2000; 403:859–866. [DOI] [PubMed] [Google Scholar]
- 9. Sidorenko V.S., Nevinsky G.A., Zharkov D.O.. Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair (Amst). 2007; 6:317–328. [DOI] [PubMed] [Google Scholar]
- 10. Robertson A.B., Klungland A., Rognes T., Leiros I.. Base excision repair: The long and short of it. Cell. Mol. Life Sci. 2009; 66:981–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perillo B., Ombra M.N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiariotti L., Malorni A., Abbondanza C., Avvedimento E. V. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008; 319:202–206. [DOI] [PubMed] [Google Scholar]
- 12. Amente S., Bertoni A., Morano A., Lania L., Avvedimento E.V., Majello B.. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010; 29:3691–3702. [DOI] [PubMed] [Google Scholar]
- 13. Amente S., Lania L., Avvedimento E.V., Majello B.. DNA oxidation drives Myc mediated transcription. Cell Cycle. 2010; 9:3002–3004. [DOI] [PubMed] [Google Scholar]
- 14. Li J., Braganza A., Sobol R.W.. Base excision repair facilitates a functional relationship between Guanine oxidation and histone demethylation. Antioxid. Redox Signal. 2013; 18:2429–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ba X., Bacsi A., Luo J., Aguilera-Aguirre L., Zeng X., Radak Z., Brasier A.R., Boldogh I.. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J. Immunol. 2014; 192:2384–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zarakowska E., Gackowski D., Foksinski M., Olinski R.. Are 8-oxoguanine (8-oxoGua) and 5-hydroxymethyluracil (5-hmUra) oxidatively damaged DNA bases or transcription (epigenetic) marks. Mutat. Res. - Genet. Toxicol. Environ. Mutagen. 2014; 764–765:58–63. [DOI] [PubMed] [Google Scholar]
- 17. Lin C., Yang L., Tanasa B., Hutt K., Ju B. gun, Ohgi K., Zhang J., Rose D.W., Fu X.D., Glass C.K. et al. . Nuclear Receptor-Induced chromosomal proximity and DNA Breaks underlie specific translocations in cancer. Cell. 2009; 139:1069–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bunch H., Lawney B.P., Lin Y.-F., Asaithamby A., Murshid A., Wang Y.E., Chen B.P.C., Calderwood S.K.. Transcriptional elongation requires DNA break-induced signalling. Nat. Commun. 2015; 6:10191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Madabhushi R., Gao F., Pfenning A.R., Pan L., Yamakawa S., Seo J., Rueda R., Phan T.X., Yamakawa H., Pao P.C. et al. . Activity-Induced DNA breaks govern the expression of neuronal Early-Response genes. Cell. 2015; 161:1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Puc J., Kozbial P., Li W., Tan Y., Liu Z., Suter T., Ohgi K.A., Zhang J., Aggarwal A.K., Rosenfeld M.G.. Ligand-dependent enhancer activation regulated by topoisomerase-I activity. Cell. 2015; 160:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Singh I., Ozturk N., Cordero J., Mehta A., Hasan D., Cosentino C., Sebastian C., Krüger M., Looso M., Carraro G. et al. . High mobility group protein-mediated transcription requires DNA damage marker γ-H2AX. Cell Res. 2015; 25:837–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schwer B., Wei P.-C., Chang A.N., Kao J., Du Z., Meyers R.M., Alt F.W.. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:2258–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beato M., Wright R.H., Vicent G.P.. DNA damage and gene transcription: accident or necessity. Cell Res. 2015; 25:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fong Y.W., Cattoglio C., Tjian R.. The intertwined roles of transcription and repair proteins. Mol. Cell. 2013; 52:291–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ohno M., Miura T., Furuichi M., Tominaga Y., Tsuchimoto D., Sakumi K., Nakabeppu Y.. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006; 16:567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yoshihara M., Jiang L., Akatsuka S., Suyama M., Toyokuni S.. Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 2014; 21:603–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clark D.W., Phang T., Edwards M.G., Geraci M.W., Gillespie M.N.. Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radic. Biol. Med. 2012; 53:51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ding Y., Fleming A.M., Burrows C.J.. Sequencing the Mouse Genome for the Oxidatively Modified Base 8-Oxo-7,8-dihydroguanine by OG-Seq. J. Am. Chem. Soc. 2017; 139:2569–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sagelsdorff P., Lutz W.K.. Sensitivity of DNA and Nucleotides to Oxidation by Permanganate and Hydrogen Peroxide. Mech. Models Toxicol. Arch. Toxicol. Suppl. 1987; 11:84–88. [DOI] [PubMed] [Google Scholar]
- 30. Cerutti P.A. Friedberg EC, Hanawalt PC. Measurement of thymidine damage induced by oxygen radical species. DNA Repair. 1981; Decker, NY: 57–68. [Google Scholar]
- 31. Helmrich A., Ballarino M., Tora L.. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011; 44:966–977. [DOI] [PubMed] [Google Scholar]
- 32. Helmrich A., Ballarino M., Nudler E., Tora L.. Transcription-replication encounters, consequences and genomic instability. Nat. Struct. Mol. Biol. 2013; 20:412–418. [DOI] [PubMed] [Google Scholar]
- 33. Ambrosio S., Di Palo G., Napolitano G., Amente S., Dellino G.I., Faretta M., Pelicci P.G., Lania L., Majello B.. Cell cycle-dependent resolution of DNA double-strand breaks. Oncotarget. 2016; 7:4949–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raiber E.A., Beraldi D., Ficz G., Burgess H.E., Branco M.R., Murat P., Oxley D., Booth M.J., Reik W., Balasubramanian S.. Genome-wide distribution of 5-formylcytosine in embryonic stem cells is associated with transcription and depends on thymine DNA glycosylase. Genome Biol. 2012; 13:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M. et al. . DNA methylation on N6-adenine in mammalian embryonic stem cells. Nature. 2016; 532:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu T., Pan Y., Kao S.Y., Li C., Kohane I., Chan J., Yankner B.A.. Gene regulation and DNA damage in the ageing human brain. Nature. 2004; 429:883–891. [DOI] [PubMed] [Google Scholar]
- 37. Ostuni R., Piccolo V., Barozzi I., Polletti S., Termanini A., Bonifacio S., Curina A., Prosperini E., Ghisletti S., Natoli G.. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013; 152:157–171. [DOI] [PubMed] [Google Scholar]
- 38. Patel R.K., Jain M.. NGS QC toolkit: A toolkit for quality control of next generation sequencing data. PLoS One. 2012; 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Langmead B., Trapnell C., Pop M., Salzberg S.. 2C- Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quinlan A.R., Hall I.M.. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu S., Grullon S., Ge K., Peng W.. Spatial clustering for identification of chip-enriched regions (SICER) to map regions of histone methylation patterns in embryonic stem cells. Methods Mol. Biol. 2014; 1150:97–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of Lineage-Determining transcription factors prime cis-Regulatory elements required for macrophage and B Cell Identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ye T., Krebs A.R., Choukrallah M.A., Keime C., Plewniak F., Davidson I., Tora L.. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic. Acids. Res. 2011; 39:e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang W., Loganantharaj R., Schroeder B., Fargo D., Li L.. PAVIS: a tool for peak annotation and visualization. Bioinformatics. 2013; 29:3097–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. D’Antonio M., D’Onorio De Meo P., Pallocca M., Picardi E., D’Erchia A.M., Calogero R.A., Castrignanò T., Pesole G.. RAP: RNA-Seq analysis pipeline, a new cloud-based NGS web application. BMC Genomics. 2015; 16:S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Anders S., Pyl P.T., Huber W.. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015; 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Le Tallec B., Koundrioukoff S., Wilhelm T., Letessier A., Brison O., Debatisse M.. Updating the mechanisms of common fragile site instability: How to reconcile the different views. Cell. Mol. Life Sci. 2014; 71:4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bignell G.R., Greenman C.D., Davies H., Butler A.P., Edkins S., Andrews J.M., Buck G., Chen L., Beare D., Latimer C. et al. . Signatures of mutation and selection in the cancer genome. Nature. 2010; 463:893–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sahakyan A.B., Chambers V.S., Marsico G., Santner T., Di Antonio M., Balasubramanian S.. Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 2017; 7:14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wamer W.G., Wei R.R.. In vitro photooxidation of nucleic acids by ultraviolet A radiation. Photochem. Photobiol. 1997; 65:560–563. [DOI] [PubMed] [Google Scholar]
- 54. Pelle E., Huang X., Mammone T., Marenus K., Maes D., Frenkel K.. Ultraviolet-B-induced oxidative DNA base damage in primary normal human epidermal keratinocytes and inhibition by a hydroxyl radical scavenger. J. Invest. Dermatol. 2003; 121:177–183. [DOI] [PubMed] [Google Scholar]
- 55. Cadet J., Douki T.. Oxidatively generated damage to DNA by UVA radiation in cells and human skin. J. Invest. Dermatol. 2011; 131:1005–1007. [DOI] [PubMed] [Google Scholar]
- 56. Greinert R., Volkmer B., Henning S., Breitbart E.W., Greulich K.O., Cardoso M.C., Rapp A.. UVA-induced DNA double-strand breaks result from the repair of clustered oxidative DNA damages. Nucleic Acids Res. 2012; 40:10263–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wäster P.K., Ollinger K.M.. Redox-dependent translocation of p53 to mitochondria or nucleus in human melanocytes after UVA- and UVB-induced apoptosis. J. Invest. Dermatol. 2009; 129:1769–1781. [DOI] [PubMed] [Google Scholar]
- 58. Pastukh V.M., Roberts J., Clark D.W., Bardwell G.C., Patel M., Al-Mehdi A.-B., Borchert G., Gillespie M.N.. An oxidative DNA ‘Damage’ and repair mechanism localized in the VEGF promoter is important for Hypoxia-induced VEGF mRNA expression. Am. J. Physiol. - Lung Cell. Mol. Physiol. 2015; 309:L1367–L1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. LeTallec B., Millot G., Blin M., Brison O., Dutrillaux B., Debatisse M.. Common fragile site profiling in epithelial and erythroid cells reveals that most recurrent cancer deletions lie in fragile sites hosting large genes. Cell Rep. 2013; 4:420–428. [DOI] [PubMed] [Google Scholar]
- 60. Debatisse M., Le Tallec B., Letessier A., Dutrillaux B., Brison O.. Common fragile sites: Mechanisms of instability revisited. Trends Genet. 2012; 28:22–32. [DOI] [PubMed] [Google Scholar]
- 61. Dellino G.I., Cittaro D., Piccioni R., Luzi L., Banfi S., Segalla S., Cesaroni M., Mendoza-Maldonado R., Giacca M., Pelicci P.G.. Genome-wide mapping of human DNA-replication origins: Levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res. 2013; 23:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cayrou C., Ballester B., Peiffer I., Fenouil R., Coulombe P., Andrau J.C., Van Helden J., Méchali M.. The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Res. 2015; 25:1873–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gaillard H., Aguilera A.. Transcription as a threat to genome integrity. Annu. Rev. Biochem. 2016; 85:291–317. [DOI] [PubMed] [Google Scholar]
- 64. Helmrich A., Stout-Weider K., Hermann K., Schrock E., Heiden T.. Common fragile sites are conserved features of human and mouse chromosomes and relate to large active genes. Genome Res. 2006; 16:1222–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Duda H., Arter M., Gloggnitzer J., Teloni F., Wild P., Blanco M.G., Altmeyer M., Matos J.. A mechanism for controlled breakage of Under-replicated chromosomes during mitosis. Dev. Cell. 2016; 39:740–755. [DOI] [PubMed] [Google Scholar]
- 66. Naim V., Wilhelm T., Debatisse M., Rosselli F.. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 2013; 15:1008–1015. [DOI] [PubMed] [Google Scholar]
- 67. Ying S., Minocherhomji S., Chan K.L., Palmai-Pallag T., Chu W.K., Wass T., Mankouri H.W., Liu Y., Hickson I.D.. MUS81 promotes common fragile site expression. Nat. Cell Biol. 2013; 15:1001–1007. [DOI] [PubMed] [Google Scholar]
- 68. Harrigan J.A., Belotserkovskaya R., Coates J., Dimitrova D.S., Polo S.E., Bradshaw C.R., Fraser P., Jackson S.P.. Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 2011; 193:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Madireddy A., Kosiyatrakul S.T., Boisvert R.A., Herrera-Moyano E., García-Rubio M.L., Gerhardt J., Vuono E.A., Owen N., Yan Z., Olson S. et al. . FANCD2 facilitates replication through common fragile sites. Mol. Cell. 2016; 64:388–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wei P.C., Chang A.N., Kao J., Du Z., Meyers R.M., Alt F.W., Schwer B.. Long neural genes harbor recurrent DNA break clusters in neural Stem/Progenitor cells. Cell. 2016; 164:644–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Saponaro M., Kantidakis T., Mitter R., Kelly G.P., Heron M., Williams H., Söding J., Stewart A., Svejstrup J.Q.. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014; 157:1037–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schiavone D., Jozwiakowski S.K., Romanello M., Guilbaud G., Guilliam T.A., Bailey L.J., Sale J.E., Doherty A.J.. PrimPol is required for replicative tolerance of g quadruplexes in vertebrate cells. Mol. Cell. 2016; 61:161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chan K., Sterling J.F., Roberts S.A., Bhagwat A.S., Resnick M.A., Gordenin D.A.. Base damage within Single-Strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genet. 2012; 8:e1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bassing C.H., Alt F.W.. H2AX may function as an anchor to hold broken chromosomal DNA ends in close proximity. Cell Cycle. 2004; 3:149–153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data related to this study have been deposited to the NCBI Gene Expression Omnibus (GEO) under accession no. GSE100234.