Abstract
The L proteins of rhabdoviruses, such as vesicular stomatitis virus (VSV) and rabies virus (RABV), possess an unconventional mRNA capping enzyme (GDP polyribonucleotidyltransferase, PRNTase) domain with a loop structure protruding into an active site cavity of the RNA-dependent RNA polymerase (RdRp) domain. Here, using complementary VSV and RABV systems, we show that the loop governs RNA synthesis and capping during the dynamic stop-start transcription cycle. A conserved tryptophan residue in the loop was identified as critical for terminal de novo initiation from the genomic promoter to synthesize the leader RNA and virus replication in host cells, but not for internal de novo initiation or elongation from the gene-start sequence for mRNA synthesis or pre-mRNA capping. The co-factor P protein was found to be essential for both terminal and internal initiation. A conserved TxΨ motif adjacent the tryptophan residue in the loop was required for pre-mRNA capping in the step of the covalent enzyme-pRNA intermediate formation, but not for either terminal or internal transcription initiation. These results provide insights into the regulation of stop-start transcription by the interplay between the RdRp active site and the dual-functional priming-capping loop of the PRNTase domain in non-segmented negative strand RNA viruses.
INTRODUCTION
GDP polyribonucleotidyltransferase (PRNTase, EC. 2.7.7.88) is an unconventional mRNA capping enzyme of rhabdoviruses, such as vesicular stomatitis virus (VSV), Chandipura virus and rabies virus (RABV), belonging to the Rhabdoviridae family in the Mononegavirales order (1–9). The enzyme carries out covalent catalysis in a unique 5′-phospho-RNA (pRNA) transfer reaction to generate a cap core structure (GpppA-) (1,4), which is strikingly different from the GMP transfer reaction catalyzed by eukaryotic and DNA viral mRNA capping enzymes (guanylyltransferases, EC. 2.7.7.50) (10,11). PRNTase is present as an enzymatic domain in rhabdoviral RNA-dependent RNA polymerase (RdRp) large (L) proteins, and shares five signature motifs A–E with PRNTase-like domains in L proteins of other non-segmented negative strand (NNS) RNA viruses belonging to the order Mononegavirales (e.g. Ebola, respiratory syncytial, measles) (5,7).
PRNTase Motif D consisting of histidine (VSV, H1227; RABV, H1241) and arginine (VSV, R1228; RABV, R1242) residues (also called HR motif) serves as a catalytic center (4). In the first step of the pRNA transfer reaction, a pair of electrons at the Nϵ2 position of the histidine residue in motif D nucleophilically attacks a 5′-α-phosphorus atom in 5′-triphosphorylated RNA (pppRNA) in an mRNA-start-sequence (pppAAC-)-dependent manner, resulting in the formation of a covalent enzyme-(histidyl-Nϵ2)–pRNA intermediate (called L-pRNA) (1,4). In addition to the histidine and arginine residues in motif D, a glycine (VSV, G1100; RABV, G1112) residue in motif A, threonine (VSV, T1157; RABV, T1170) residue in motif B, tryptophan (VSV, W1188; RABV, W1201) residue in motif C, and phenylalanine (VSV, F1269; RABV, F1285) and glutamine (VSV, Q1270; RABV, Q1286) residues in motif E were identified as essential for mRNA capping in the step of the L-pRNA intermediate formation (7,8). In the three-dimensional structure of the VSV L protein solved by cryo-electron microscopy (12), motifs A, B, C and E surround motif D to form the active site of the PRNTase domain (7). To produce the cap structure, the covalent L-pRNA intermediate transfers pRNA to GDP, the pRNA acceptor, in which the C-2-amino group of guanine and 2′- or 3′-hydroxyl group of ribose are critical for the transfer reaction (4,9). Currently, it remains unknown which amino acid residues specifically recognize GDP.
According to the single-entry stop-start transcription model (13,14), the VSV RdRp complex composed of the L and its co-factor P proteins initiates transcription at the 3′-end of the genomic RNA encapsidated with nucleo- (N) proteins (called N-RNA) to synthesize the leader RNA (LeRNA) of 47 nucleotides (nt). After LeRNA synthesis, the same RdRp complex sequentially transcribes internal genes (N, P, M, G and L) into five monocistronic mRNAs with a methylated 5′-cap structure (cap 1, m7GpppAm-) and 3′-poly(A) tail (15–17). A decrease in the efficiency of transcription reinitiation at each gene junction leads to the formation of a gradient in transcript abundance in the following order: Le > N > P > M > G > L. We have previously shown that cap-defective mutations in the PRNTase motifs, which abolish the covalent L-pRNA intermediate formation, induce aberrant termination and reinitiation of transcription within the first N gene using cryptic signals, releasing unusual 5′-triphosphorylated N mRNA fragments including a 5′-terminal fragment with residues 1–40 (N1–40), internal fragment with residues 41–68 (N41–68), and 3′-terminal fragment with residues 157–1326 and poly(A) tail (7,18). Frequent termination and reinitiation within the N gene by the cap-defective mutants cause a marked reduction in synthesis of downstream mRNAs as well as full-length N mRNA (7,18). Therefore, these observations suggest that the PRNTase domain serves as a key regulatory domain controlling stop-start transcription, and the successful production of full-length mRNAs requires the L-pRNA intermediate formation followed by pre-mRNA capping during mRNA chain elongation.
Primer-independent RdRps of double-strand RNA viruses [e.g. Φ6 phage (19), reovirus (20)], positive-strand RNA viruses [e.g., dengue virus (21), hepatitis C virus (22)], and segmented negative strand RNA viruses [e.g. influenza virus (23)] often possess a ‘priming loop’, which facilitates de novo initiation of transcription by priming an initiator nucleotide. Known priming loops are extended from different RdRp subdomains (thumb or palm) into their active sites and structurally diversified, but play similar roles in stabilizing the initiation complex formation. Interestingly, different RNA viral RdRps use distinct amino acid residues [e.g. tyrosine (19,22), serine (20), histidine (21), proline (23)] in their priming loops to interact with a purine base or phosphate group of the initiator nucleotide, suggesting that RNA viruses have evolved their own mechanisms of transcription initiation. Thus, understanding diversified mechanisms of de novo initiation by RdRps may aid developing specific antiviral drugs against them. In the unliganded (apo) state of VSV L (PDB id: 5A22) (12), a large loop structure, flanking PRNTase motif B, of the PRNTase domain is inserted into the active site cavity of the RdRp domain. By analogy to other viral primer-independent RdRps, the loop was suggested to be a counterpart of priming loops (12). However, it is not known whether the loop from the PRNTase domain plays any roles in rhabdoviral RNA biogenesis. In this study, we revealed that the loop serves dual functions in transcription initiation and pre-mRNA capping using VSV and RABV systems. Our experimental data combined with a structural model of a VSV transcription initiation complex provide insights into the intricate regulation of stop-start transcription by the flexible loop of the PRNTase domain in NNS RNA viruses.
MATERIALS AND METHODS
In vitro RNA synthesis
Viral proteins used for in vitro transcription were prepared as described in Supplementary Materials and Methods. In vitro first phosphodiester bond formation was performed with VSV L [0.15 μg, wild-type (WT) or mutant], P (40 ng) and N-RNA template (0.4 μg protein) for 1 h at 30°C in a transcription buffer (25 μl) containing 50 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 50 mM NaCl, 2 mM DTT, 0.2 mg/ml bovine serum albumin, 2 mM ATP, and 20 μM [α-32P]CTP (∼1 × 104 cpm/pmol). When oligo-RNA templates (0.2 μM, HPLC-purified) were used instead of N-RNA, 10 ng of VSV P was included in reaction mixtures. VSV P purified from insect cells was used rather than P from Escherichia coli unless otherwise mentioned. For RABV, L (0.15 μg, WT or mutant), P (20 ng) and oligo-RNA template (0.2 μM) were incubated in the transcription buffer for 2 h at 30°C. The reactions were stopped by adding 175 μl of RNA extraction buffer (6). 32P-Labeled RNAs are extracted with an equal volume of phenol/chloroform/isoamyl alcohol (25:24:1, v/v) and precipitated in the presence of glycogen with 5 volumes of ethanol as described by Ogino (6). The resulting pellet was rinsed with cold 90% ethanol, dried in a SpeedVac concentrator (Thermo Scientific), and dissolved in 10 μl of dephosphorylation buffer (Roche Applied Science). The substrates and products were digested with 10 units of calf intestinal alkaline phosphatase (CIAP, Roche Applied Science) at 37°C for 90 min. The reaction mixture was mixed with an equal volume of 96% formamide loading solution, heated at 95°C for 3 min, and analyzed by 20% urea-PAGE followed by autoradiography (6). Radioactive RNA bands were excised from gels, and their radioactivities were measured by liquid scintillation counting. The amounts of [α-32P]CMP incorporated into RNAs were calculated based on specific radioactivities of [α-32P]CTP. RNA products were identified as described in Supplementary Materials and Methods. Other in vitro transcription and capping assays were performed as described in (6–9) (see Supplementary Materials and Methods).
Mini-genome assay
The VSV mini-genome assay was performed with the pVSV-CAT2 (24), pBS-N (25), pBS-P (25) and pBS-L-Flag (7) [WT or mutant, derived from pBS-L (25)] plasmids as described previously (7). The reporter gene product and L were detected by CAT ELISA assay (Roche Applied Science) and Western blotting with anti-FLAG monoclonal antibody (Sigma-Aldrich), respectively, as described in (7).
Generation of recombinant VSVs
Recombinant (r) VSVs with the WT or mutant L gene were generated with the pVSV-L (18) [WT or mutant, derived from pVSVFL-2 (26)], pBS-N, pBS-P and pBS-L plasmids using the reverse genetics system (25) as described previously (18). The X-tremeGENE HP DNA Transfection Reagent (Roche Applied Science) was used for transfection. rVSVs were plaque-purified, and their L gene was sequenced as described in (18). Single-step growth curve experiments were performed as described in Supplementary Materials and Methods.
Modeling of the VSV terminal initiation complex
The protein coordinates corresponding to the VSV (PDB id: 5A22) (12) and Φ6 bacteriophage (PDB id: 1HI0) (19) polymerases were acquired from the RCSB (27). The two polymerase structures were overlaid by aligning their core polymerase coordinates with the aid of the secondary structure matching (SSM) tool in COOT (28). Nucleotides were added to the VSV structure and mutated to VSV specific nucleotides (3′-UGCU-5′ for the template strand and ATP and CTP for the first two residues of the initiation complex). The complete complex [VSV L, template RNA, nucleotides and divalent cations (2 Mg2+ and 1 Mn2+)] was energy minimized with PHENIX (29). To realign the priming loop with the modeled RNA, the priming loop was moved along the electron density (EMDB id: EMD-6337) from the cryo-EM studies of Liang et al. (12). Threading along the density was done to temper the movement of the loop, as the density represents an average of multiple conformations within the empty cavity (12). The new complex was energy minimized and the loop adjusted to eliminate any poor backbone torsions. The structure was minimized again. Structural images were generated using the PyMOL software [DeLano, W.L. (2002) The PyMOL Molecular Graphics System. http://www.pymol.org/.].
RESULTS
The loop extended from the VSV L PRNTase domain is required for RNA synthesis and mRNA capping
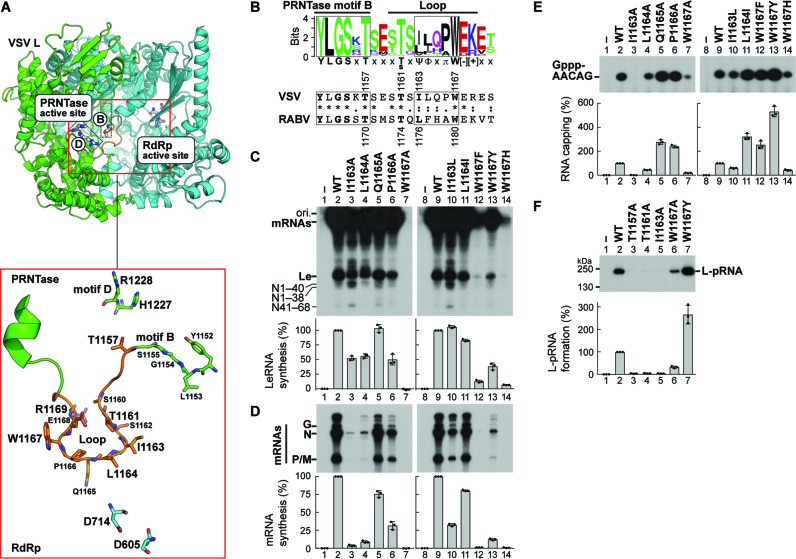
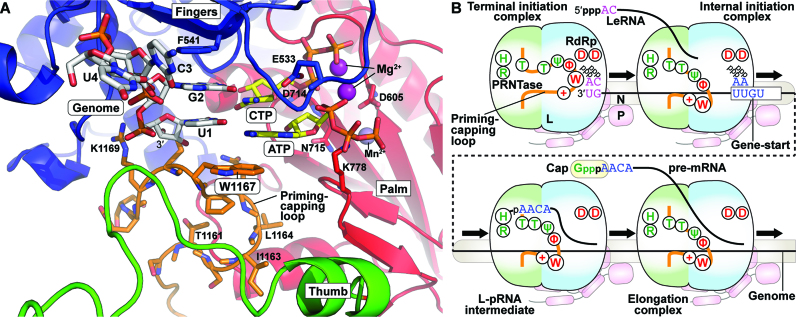
In the apo-VSV L (PDB id: 5A22) (12), the putative priming loop (residues 1160–1169), which flanks PRNTase motif B (residues 1152–1157), extends from the PRNTase domain into the RdRp active site cavity and completely blocks a putative RNA exit channel of the RdRp domain (Figure 1A). By comparing amino acid sequences of putative loops from 109 rhabdoviral L proteins (Supplementary Table S1), a consensus sequence, [T/S]xΨΦxπW[−][+] (Ψ, Φ, π, [−], [+] and x indicate aliphatic, hydrophobic, small, negatively and positively charged, and any amino acids, respectively), was found at the central region of the loop (Figure 1B).
Figure 1.
RNA synthesis and capping are controlled by distinct residues in a loop structure of the VSV L PRNTase domain. (A) A 3D structural model of the RdRp (cyan) and PRNTase (green) domains of VSV L is shown (top) in cartoon form with a close-up view of the large loop (orange) from the PRNTase domain is shown below. Top: Within the red box, each active site is labeled and active site residues are shown in stick model. B and D labels note PRNTase motifs B and D containing active site residues T1157 and H1227/R1228, respectively. Bottom: The loop with flanking residues is illustrated with surrounding PRNTase (T1157, H1227 and R1228) and RdRp (D605, D714) active site residues and residues on the loop shown in stick model. Models were generated from PDB id: 5A22. (B) A sequence logo [WebLogo (39)] for the loop flanking PRNTase motif B in rhabdoviral L proteins is shown with the corresponding sequences of VSV and RABV (numbers: amino acid positions). Ψ, Φ, π, [−], [+] and x indicate aliphatic, hydrophobic, small, negatively and positively charged, and any amino acids, respectively. (C, D) In vitro transcription was performed with VSV L [wild-type (WT) or mutant], P, and N-RNA. 32P-Labeled LeRNA and mRNAs (deadenylated) were analyzed by 20% (C) and 5% (D) urea-PAGE, respectively, followed by autoradiography. The positions of the origin (ori.) and previously identified transcripts are denoted. (E) In vitro capping was performed with pppAACAG, [α-32P]GDP and L (WT or mutant). Capped RNA products were analyzed by 20% urea-PAGE followed by autoradiography. (F) L (WT or mutant) was incubated with 32P-labeled pppAACAG. The resulting L-pRNA intermediate was analyzed by 7.5% SDS-PAGE. The graphs show relative activities for respective reactions, where radioactivities of respective products without (lanes 1 and 8) and with (lanes 2 and 9) of WT L were set to 0 and 100%, respectively. The dot-plots, columns, and error bars represent the individual normalized values, means, and standard deviations, respectively, from three independent experiments (n = 3).
To define the function(s) of the loop of VSV L, we examined the effects of single alanine-substitutions for residues 1159–1171 on in vitro RNA synthesis and capping (Figure 1C–E and Supplementary Figures S1 and S2). VSV RdRp reconstituted with wild-type (WT) L and P transcribed the N-RNA template into LeRNA and mRNAs (Figure 1C and D, lanes 2 and 9). However, as reported for cap-defective mutations (e.g. T1157A in motif B) (7), the I1163A (Figure 1C and D, lane 3) and T1161A (Supplementary Figure S2A and B) mutations more significantly diminished mRNA synthesis than LeRNA synthesis and triggered production of the aberrant N mRNA fragments, such as N1–40 and N41–68 (7,18). The L1164A mutation also reduced mRNA synthesis activity to a greater extent than LeRNA synthesis activity, but did not induce production of the N mRNA fragments (Figure 1C and D, lane 4). The I1163, L1164, and T1161 residues could be functionally replaced, in part, with closely similar amino acids in mRNA synthesis (Figure 1D, lanes 10 and 11; Supplementary Figure S2B). In addition, a basic amino acid at position 1169 was necessary for efficient RNA synthesis (Supplementary Figure S2A and B). Furthermore, the W1167A mutant was found to be completely inert in RNA synthesis (Figure 1C and D, lane 7), whereas W1167 mutants with other aromatic amino acid substitutions showed transcription activities (Y >> F > H), although to significantly lesser extents than WT (lanes 12–14). The I1163A (Figure 1E, lane 3; Figure 1F, lane 5) and T1161A mutants (see Supplementary Figure S2C; Figure 1F, lane 4) were defective in capping in the step of the L-pRNA intermediate formation. In contrast, the W1167 residue was not essential for RNA capping, although the W1167A and W1167Y mutations reduced and enhanced capping activity, respectively (Figure 1E, lanes 7 and 13; Figure 1F, lanes 6 and 7). These results suggest that different residues in the loop regulate RNA synthesis and/or capping in distinct steps.
The W1167 residue in the loop of the VSV L PRNTase domain is required for VSV gene expression and propagation in host cells
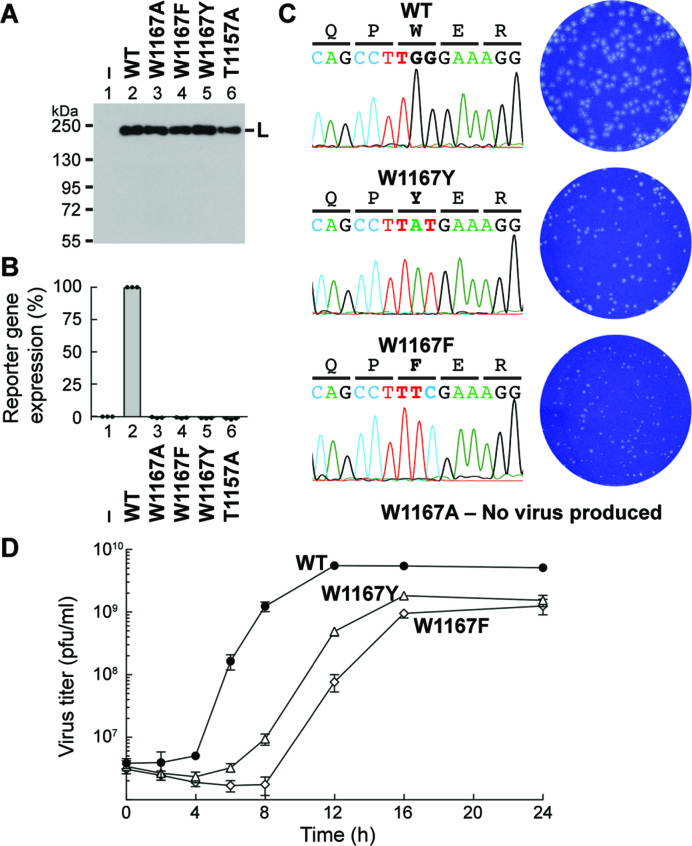
To investigate the role of the W1167 residue of VSV L in viral replication in host cells, we performed the VSV mini-genome assay using the plasmids expressing the negative-sense mini-genome with a reporter gene (CAT) (24) and three replication proteins (N, P and L) (Figure 2A and B). Plasmids expressing Flag-tagged WT and cap-defective T1157A mutant (7) L proteins were used as positive and negative controls, respectively. Although the W1167A, W1167F, and W1167Y mutants (Figure 2A, lanes 3–5) were expressed in transfected cells to similar levels as WT L (lane 2), none of them supported reporter gene expression from the mini-genome at detectable levels (Figure 2B, columns 3–5).
Figure 2.
The W1167 residue in VSV L is essential for VSV gene expression and propagation in host cells. (A, B) The VSV mini-genome assay was performed with plasmids expressing N, P, and C-terminal FLAG-tagged L (WT or mutant). L proteins expressed in the transfected cells were detected by Western blotting with anti-FLAG antibody (A). The blot is a representative of three independent experiments. Lane 1 indicates no L plasmid. The graph shows relative expression levels of a reporter gene product (CAT), where expression levels in cells transfected without (column 1) and with (column 2) the WT L plasmid were set to 0 and 100%, respectively (B). The reporter gene product was not detected in lysates of cells expressing the indicated W1167 mutants as well as T1157A (negative control) (7) in the three independent experiments. (C) Effects of the indicated mutations on rVSV generation were examined. When rVSVs were viable, the mutation sites in rVSV genomes were sequenced (left). Plaque phenotypes of WT and mutant rVSVs were compared (right). (D) BHK-21 cells were infected with rVSV harboring the WT (closed circles), W1167Y (open triangles) or W1167F (open diamonds) L gene at a multiplicity of infection of five plaque-forming unit (pfu) per cell and cultured for 24 h. Single-step growth curves were obtained by titration of the rVSVs in the culture supernatants harvested at the indicated time points by a plaque assay. Symbols and error bars represent the means of titers and standard deviations, respectively (n = 3).
To further investigate the role of W1167 in VSV replication, we attempted to generate recombinant (r) VSVs with the W1167 mutations in the L gene using the reverse genetics system (25). Multiple attempts to generate rVSVs with the W1167A mutation were unsuccessful, indicating that the alanine mutation is lethal to VSV. In contrast, rVSVs possessing the W1167F or W1167Y mutation in the L gene could be generated, but displayed small plaque phonotypes when compared to the WT virus (Figure 2C), suggesting that the substitutions of these aromatic amino acids for W1167 render VSV capable of growing, although very slowly, in host cells. Single-step growth curve experiments (Figure 2D) revealed that the duration of eclipse phases for the W1167F (∼8 h) and W1167Y (∼6 h) viruses was longer than that for the WT virus (∼4 h). The W1167Y and W1167F viruses reached their maximum titers at 16 and 24 h post-infection, respectively, which were 3–4-fold lower than that of the WT virus at 12 h post-infection. These results indicate that the W1167F virus is more severely attenuated than the W1167Y virus, consistent with the RNA synthesis activities of the W1167F and W1167Y L mutants in vitro (see Figure 1C and D).
The W1167 residue in the loop of the VSV L PRNTase domain is essential for terminal de novo initiation
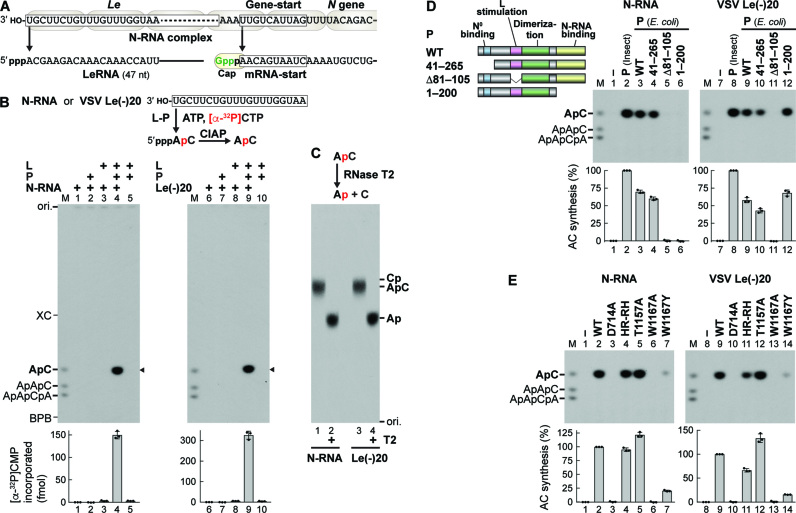
VSV RdRp initiates transcription de novo at the 3′-terminal Le promoter to synthesize LeRNA (14,30,31) and sequentially at internal gene-start sequences to generate 5′-capped mRNAs (15–17,32) (Figure 3A). To analyze the mechanism of terminal de novo initiation, we established a first phosphodiester bond formation (AC synthesis) assay with ATP, [α-32P]CTP, L, P and N-RNA or a 20-nt oligo-RNA template with the Le promoter sequence [VSV Le(–)20] (Figure 3B). After transcription, the resulting products and remaining substrates were digested with calf intestinal alkaline phosphatase (CIAP), allowing us to separate 32P-labeled ApC from Pi by 20% urea–PAGE as described by Emerson (14). A mixture of 5′-hydroxyl-ApC, ApApC and ApApCpA was run in M lanes. It should be noted that shorter 5′-hydroxyl-oligonucleotides migrate slower than longer 5′-hydroxyl-oligonucleotides within the range of 2–5 nt on denaturing polyacrylamide gels (14) (see Supplementary Figure S3). Using this assay, we demonstrated that both L and P are necessary for de novo initiation from either N-RNA or VSV Le(–)20 (Figure 3B, lanes 4 and 9; Supplementary Figure S4). The 32P-labeled products, both of which co-migrate with ApC, could be digested with RNase T2 into mononucleotides that co-migrate with adenosine 3′-phosphate, but not cytidine 3′-phosphate, on the TLC plate (Figure 3C, lanes 2 and 4). Thus, the products were unambiguously identified as ApC.
Figure 3.
The tryptophan residue in the loop of VSV L is required for terminal de novo initiation of transcription. (A) VSV LeRNA and capped N mRNA are synthesized from the 3′-terminal Le promoter and internal gene-start sequence, respectively. (B) In vitro transcription was performed with ATP, [α-32P]CTP, L, P, and/or template [N-RNA or an oligo-RNA with the VSV Le promoter sequence, VSV Le(–)20]. Calf intestine alkaline phosphatase (CIAP)-resistant RNA products were analyzed by 20% urea-PAGE followed by autoradiography. The positions of 5′-hydroxyl oligo-RNA markers (M lane), xylene cyanol FF (XC), and bromophenol blue (BPB) are shown. The graphs indicate amounts of [α-32P]CMP incorporated into RNAs (marked by arrowheads). (C) CIAP-resistant RNA products (from N-RNA or Le(–)20) were incubated with or without RNase T2, and then analyzed along with internal markers (ApC, Cp, or Ap) by PEI-cellulose TLC. (D) Schematic structures of VSV P and its deletion mutants are shown with the indicated functional domains. In vitro AC synthesis was performed with L, P (from insect cells or E. coli, WT or mutant), and template [N-RNA or VSV Le(–)20]. The graphs show relative activities of P proteins, where radioactivities of AC synthesized without (lanes 1 and 7) and with (lanes 2 and 8) WT P (insect) were set to 0 and 100%, respectively. (E) In vitro AC synthesis was performed with L (WT or mutant), P (insect), and template [N-RNA or VSV Le(–)20]. Relative AC synthesis activities of L mutants were expressed as percentages of the WT activities. The dot-plots, columns, and error bars represent the individual values, means, and standard deviations, respectively (n = 3).
As shown in Figure 3D, VSV P possesses a series of functional domains. Using bacterially-expressed P (WT) and its deletion mutants, we showed that both the L-stimulatory (33) (Figure 3D, lane 5) and N-RNA-binding (34) (lane 6) domains are essential for AC synthesis from N-RNA, whereas the latter is dispensable for AC synthesis from VSV Le(–)20 (lane 12). As shown in Figure 3E, in either template, AC synthesis activity was abrogated by a mutation(s) of the RdRp active site [D714A (4,18), lanes 3 and 10], but not the PRNTase active site [HR-RH (H1227R plus R1228H) (4,18), lanes 4 and 11; T1157A (7), lanes 5 and 12], in VSV L. The W1167A mutant was totally inactive in AC synthesis (lanes 6 and 13), whereas the W1167Y mutant retained 16–20% of the WT activities (lanes 7 and 14), indicating that an aromatic residue at position 1167 is critical for terminal de novo initiation. However, our inability to develop an RNA chain elongation assay or internal de novo initiation assay for the VSV L protein hampered further studies on a role(s) of W1167 in other steps of transcription (see Discussion).
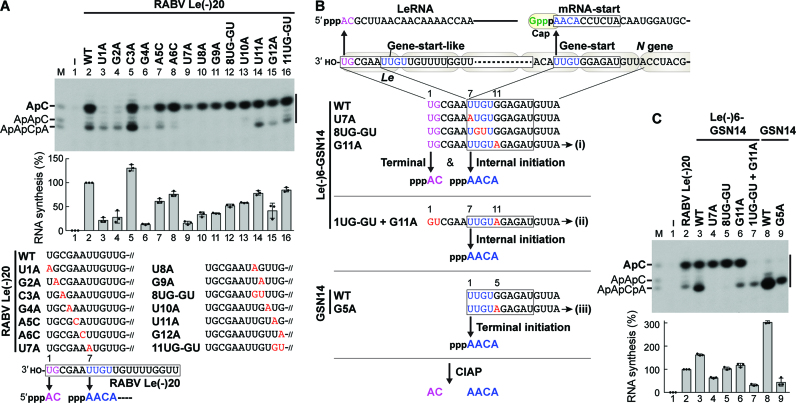
A putative loop in the RABV L PRNTase domain plays dual roles in transcription and capping
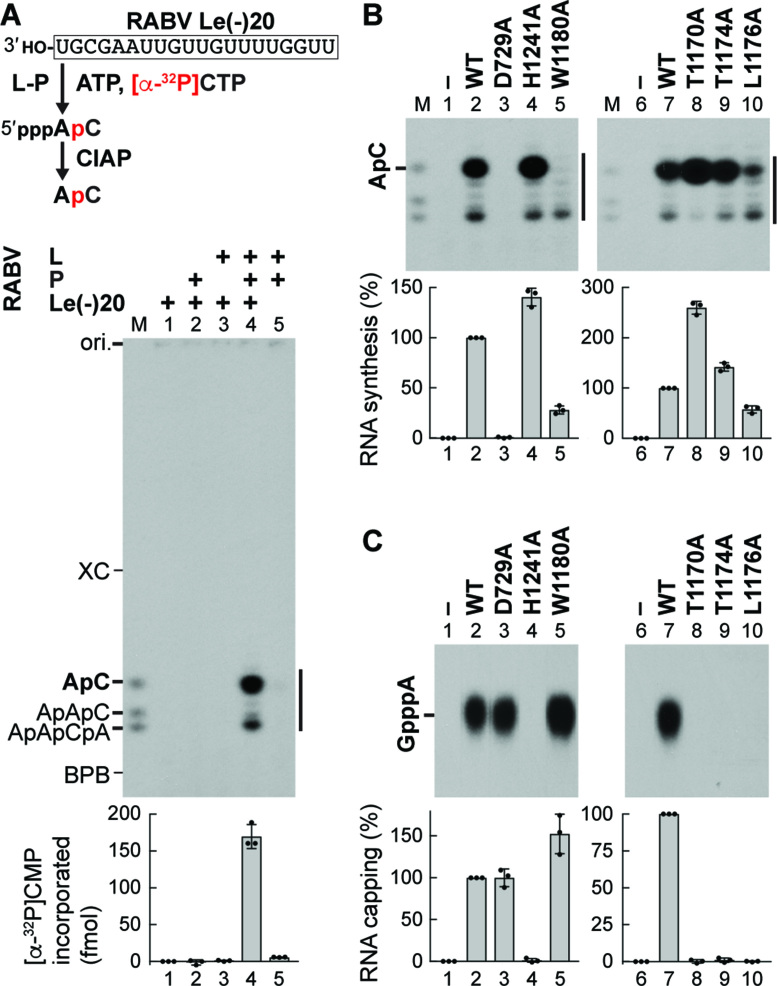
To further explore potential roles of putative priming loops of rhabdoviral L proteins in each step of RNA biogenesis, we used the L protein of RABV, an important human pathogen, as another model system. Since RABV and VSV belong to taxonomically distinct groups (the Lyssavirus and Vesiculovirus genera, respectively), we were particularly interested in whether the RABV counterpart of the putative VSV priming loop (see Figure 1B) has evolutionarily conserved functions in transcription and capping. To investigate whether the putative priming loop in the RABV L PRNTase domain is involved in de novo initiation, we developed an AC synthesis system with RABV L and P using a 20-nt oligo-RNA template with the RABV Le promoter sequence [RABV Le(–)20] (Figure 4 and Supplementary Figures S5 and S6). Similar to VSV RdRp, both RABV L and P were required for AC synthesis (Figure 4A, lane 4). Unexpectedly, RABV RdRp synthesized heterogeneous longer transcripts in addition to AC. The RdRp (D729A) and PRNTase (H1241A) active site mutants (8) were defective in RNA synthesis (Figure 4B, lane 3) and capping (Figure 4C lane 4), respectively. Interestingly, an alanine mutation of W1180 (RABV counterpart of VSV W1167) appeared to diminish AC synthesis, but not synthesis of the heterogeneous transcripts (Figure 4B, lane 5) or RNA capping (Figure 4C, lane 5). In contrast, alanine mutations of T1170 (8), T1174, and L1176 (RABV counterparts of VSV T1157, T1161, and I1163, respectively) abolished RNA capping (Figure 4C, lanes 8–10), but not RNA synthesis (Figure 4B, lanes 8–10). These results indicate that the putative priming loop in the RABV PRNTase domain also participates in RNA synthesis and capping.
Figure 4.
A putative loop structure in the PRNTase domain of RABV L plays dual roles in RNA synthesis and capping. (A) In vitro transcription was performed with ATP, [α-32P]CTP, RABV L, P and/or an oligo-RNA with the RABV Le promoter sequence [RABV Le(–)20]. Transcripts were analyzed as in Figure 3B. The graph indicates amounts of [α-32P]CMP incorporated into RNAs (marked by the vertical line). (B, C) WT and mutant RABV L proteins were subjected to in vitro AC synthesis with P and RABV Le(–)20 (B) or capping with pppAACAC and [α-32P]GDP (C). In panel C, nuclease P1 and CIAP-resistant products were analyzed along with the standard GpppA cap by PEI-cellulose TLC. Relative RNA synthesis and capping activities of RABV L mutants were expressed as percentages of the WT activities. The dot-plots, columns, and error bars represent the individual normalized values, means, and standard deviations, respectively (n = 3).
The RABV N gene-start sequence and gene-start-like sequence in the RABV Le promoter serve as internal transcription initiation signals
Since RABV RdRp appeared to synthesize AC and the heterogeneous transcripts from terminal and internal initiation sites in RABV Le(–)20, these initiation sites were mapped (Figure 5A and Supplementary Figure S6D). Mutations of U1 or G2 (Figure 5A, lanes 3 and 4) abolished synthesis of AC, but not the heterogeneous transcripts, whereas mutations of U7, U8, and/or G9 completely abrogated synthesis of the heterogeneous transcripts, but not AC (lanes 9–12). The A6C and U10A mutations decreased levels of heterogeneous transcripts, which co-migrate with AACA and AAC, respectively, without affecting AC synthesis (lanes 8 and 13), whereas the G4A mutation negatively affected synthesis of all the transcripts (lane 6). Furthermore, RABV RdRp was found to produce a transcript(s), which co-migrates with ApApCpA as a major product, besides AC from RABV Le(–)20 with the U11A mutation (lane 14), suggesting that RdRp transcribed the internal region at positions from 7 to 10.
Figure 5.
The RABV N gene-start sequence serves as internal and terminal initiation signals for RABV RdRp. (A) In vitro oligo-RNA synthesis was performed with RABV L, P, and the indicated oligo-RNA templates as in Figure 4A. Relative RNA synthesis activities of the templates were expressed as percentages of the activity of RABV Le(–)20 (WT) (lane 2). Partial oligo-RNA sequences (mutated bases shown in red) are written in the 3′ to 5′ direction. Two independent transcription initiation sites were identified at positions 1 and 7 in RABV Le(–)20 (bottom). (B) RABV LeRNA and capped N mRNA are synthesized from the 3′-terminal Le promoter and internal gene-start sequence, respectively. The gene-start-like sequence in the RABV Le(–)20 template functions as an internal initiation site (see panel A). To examine whether the N gene-start sequence serves as internal and/or terminal initiation sites, the indicated oligo-RNAs with the N gene-start sequence were designed. The Le(–)6-GSN14 G11A, Le(–)6-GSN14 1UG-GU + G11A, and GSN14 G5A templates are called templates (i), (ii), and (iii), respectively, in Figure 6. (C) The indicated templates were subjected to transcription with ATP and [α-32P]CTP, and RABV RdRp (L and P) as in Figure 4A. Relative RNA synthesis activities of the templates were expressed as percentages of the activity of RABV Le(–)20 (lane 2).
The sequence of the suggested internal initiation signal (UUGU, residues 7–10) in the RABV Le promoter was found to be identical to the gene-start sequences, which act as internal initiation signals for mRNA synthesis in the RABV genome (Figure 5B). Thus, we examined whether the N gene-start sequence serves as an initiation signal for RABV RdRp when placed internally or terminally in oligo-RNAs. First, the 6-nt Le sequence [Le(–)6] was fused to the 14-nt N gene-start sequence (GSN14) to create a hybrid Le(–)6-GSN14 template. As a result, transcripts, which co-migrate with AC and AACA, were synthesized from Le(–)6-GSN14 (Figure 5C, lane 3). Although an expected transcript synthesized from the N gene-start sequence in the presence of ATP and CTP was AACACC, 5′-hydroxyl-oligonucleotides with 4–6 nt (e.g. AACA, AACAC, AACACC) were not separable in the gel. The U7A (lane 4) or 8UG-GU (lane 5) mutation in Le(–)6-GSN14 abolished synthesis of the putative internal transcripts, while the G11A mutation appeared to terminate internal transcription at position 10 (lane 6). Using a combination of the 1UG-GU and G11A mutations, we demonstrated that synthesis of putative AACA is independent of AC synthesis from the 3′-end (lane 7). Furthermore, GSN14 WT (lane 8) and G5A (lane 9) appeared to act as templates for terminal initiation. By identifying RNA products synthesized from selected templates, RABV Le(–)20, Le(–)20 U11A, Le(–)6-GSN14 G11A, and GSN14 G5A (see Supplementary Figures S7-S9), we concluded that RABV RdRp produces AC from the terminal RABV Le promoter and AACA-started RNAs from the internal or terminal N gene-start sequence as well as from the internal gene-start-like sequence in the RABV Le promoter.
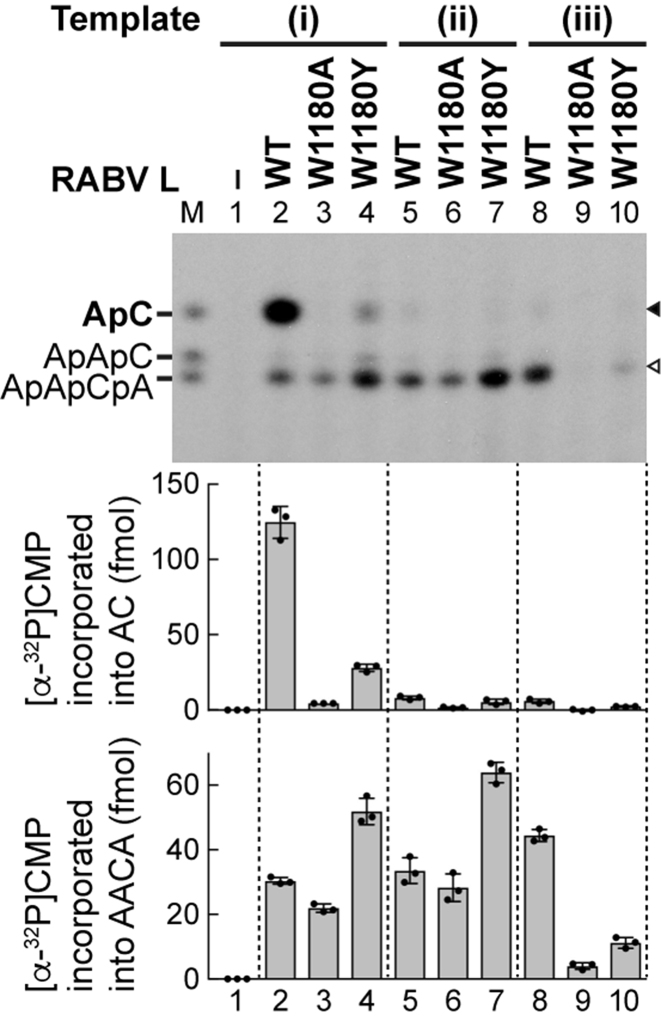
The tryptophan residue conserved in rhabdoviral PRNTase domains is essential for terminal de novo initiation, but not for internal de novo initiation or elongation
Based on the findings shown in Figure 5, we selected unique RABV templates: (i) Le(–)6-GSN14 G11A with the terminal and internal initiation signals for synthesis of AC and AACA, respectively, (ii) Le(–)6-GSN14 1UG-GU + G11A with the internal initiation signal for synthesis of AACA, and (iii) GSN14 G5A with the terminal initiation signal for synthesis of AACA, to investigate the roles of W1180 of RABV L in terminal and internal initiation (Figure 6). Consequently, we discovered that the W1180A mutation abolishes terminal de novo initiation to synthesize either AC from template (i) (lane 3, upper band) or AACA from template (iii) (lane 9, lower band), whereas it only modestly affects internal initiation to synthesize AACA from template (i) or (ii) (lanes 3 and 6, lower band). Similar to the VSV system (see Figure 3E), the W1180Y mutant exhibited weak, but significant, terminal de novo initiation activity to synthesize AC from template (i) (Figure 6, lane 4, upper band) or AACA from template (iii) (lane 10, lower band), and showed internal de novo initiation activity to synthesize AACA from template (i) or (ii) (lanes 4 and 7, lower band) higher than that of WT L (lanes 2 and 5, lower band). These results indicate that W1180 is obligately required for terminal de novo initiation even if the gene-start sequence is placed at the 3′-end of template, but is not necessary for internal initiation or elongation.
Figure 6.
The tryptophan residue in the loop of the RABV PRNTase domain is essential for terminal de novo initiation, but not for internal initiation. WT and mutant RABV L proteins were subjected to transcription with ATP, [α-32P]CTP, P and the indicated templates (see Figure 5B). Amounts of [α-32P]CMP incorporated into AC and AACA (marked by closed and open arrowheads, respectively) are shown in the graphs. The dot-plots, columns, and error bars represent the individual values, means, and standard deviations, respectively (n = 3).
DISCUSSION
In this study, we uncovered that the flexible loop structure extended from the PRNTase domain of the L proteins of rhabdoviruses, VSV and RABV, plays dual roles in transcription and pre-mRNA capping. The conserved tryptophan residue in the loop is required for terminal de novo transcription initiation, but not for internal de novo initiation, elongation, or pre-mRNA capping, whereas the TxΨ motif in the loop is essential for pre-mRNA capping in the step of the L-pRNA intermediate formation, but not for de novo transcription initiation. Furthermore, we directly demonstrated, for the first time, that the co-factor P proteins of VSV and RABV are required for de novo transcription initiation with respective L proteins (see Supplementary Discussion). Thus, this study provides new insights into how rhabdoviruses comprising a large group of NNS RNA viruses have evolved their unique RNA biosynthesis machineries to carry out each step of RNA synthesis and capping.
It has been reported that an aromatic amino acid residue (e.g. Y, H) in a priming loop of several primer-independent RdRps plays a critical role in stabilizing their terminal de novo initiation complexes (19,21,22). In the crystal structure of the dsRNA phage Φ6 RdRp in complex with a 3′-CC-ended template and initiator and incoming GTP molecules (PDB id: 1HI0) (19), a tyrosine residue in a loop structure of its C-terminal domain stacks the guanine ring of the initiator GTP (see Supplementary Figure S10). Similarly, we speculate that the indole side chain of the tryptophan residue of VSV L binds the adenine ring of the initiator ATP via a π- π stacking interaction in a terminal initiation complex (see structural models in Figure 7A and Supplementary Figures S10A and S11). The VSV L cryo-EM studies of Liang et al. (12) were performed in absence of RNA. So, to evaluate this hypothesis, the VSV terminal initiation complex was modeled based on alignment of the VSV L and the Φ6 RdRp initiation complex. RNA from the aligned Φ6 RdRp initiation complex was placed in the VSV L structure and mutated. The priming loop was, then, rearranged to align W1167 with the initiator ATP and minimized. This could be achieved with a minimal movement of the residues in the loop. The total distance shift of W1167 was 6.6 angstroms (CA–CA, see Supplementary Figure S11). In the cryo-EM studies (12), the priming loop is noted to be flexible, potentially due to absence of the template RNA and nucleotides. Given this, the reasonable movement of the priming loop in our model is consistent with those structural studies (12). As an added point, the newly threaded priming loop still lies along the averaged density from the original EM studies (12).
Figure 7.
The priming-capping loop of the rhabdoviral PRNTase domain governs terminal de novo initiation and pre-mRNA capping during stop-start transcription. (A) The VSV L terminal initiation complex was modeled based on the bacteriophage Φ6 initiation complex. RNA was modeled based on alignment of the core polymerases of VSV (PDB id: 5A22) and bacteriophage Φ6 (PDB id: 1HI0). The priming-capping loop (orange carbon backbone) was adjusted minimally within the cavity and the VSV L protein was energy minimized with the VSV specific RNA [genomic RNA - (3′-UGCU-5′), white carbon backbone], initial (ATP) and incoming (CTP) nucleotides (yellow carbon backbone), the two catalytic Mg2+ ions (purple) and Mn2+ (obscured) to yield the model. RdRp subdomains are individually colored: palm (red), fingers (blue) and thumb (green). Catalytic aspartates, D605 and D714, are shown on the palm. W1167 π-stacks with the initiator ATP. F541 sits stacked inline with 3′-template nucleotides, U1 and G2. (B) A model is presented for the dual roles of the priming-capping loop of the rhabdoviral L protein in terminal de novo initiation and mRNA capping. Amino acid residues in the loop required for transcription and mRNA capping are shown in red and green, respectively (see Figure 1B). The P protein is associated with the L protein and N-RNA, and essential for de novo initiation.
Since VSV RdRp did not show any template-dependent primer extension activity when oligo-RNAs of 2–5 nt were used as primers (data not shown), we were not able to examine whether the tryptophan mutations affect RNA chain elongation by using a primer extension assay. On the other hand, RdRps of other NNS RNA viruses, such as human respiratory syncytial virus (pneumovirus) (35), Nipah virus (paramyxovirus) (36), and Ebola virus (filovirus) (37), have been reported to extend short primers in template-dependent manners. One possibility is that the apo state of VSV L cannot accept external primers to form elongation complexes due to steric hindrance with the large priming loop extended into the RdRp active site cavity (12). Unexpectedly, we found that RABV RdRp initiates transcription from the gene-start-like sequence in the RABV Le(–)20 template to generate transcripts with 5–13 nt (Figure 4 and Supplementary Figure S9), although its biological role is currently unknown. Importantly, this finding led us to establish the assay for RABV RdRp using the templates containing the internal RABV N gene-start sequence to monitor internal de novo initiation and subsequent RNA chain elongation (Figure 5), and eventually to demonstrate that the W1180 residue in RABV L is not essential for internal initiation or elongation (Figure 6). However, our attempts to develop an internal de novo initiation assay using oligo-RNA templates for VSV RdRp have so far been unsuccessful. Thus, we were not able to analyze whether the W1167 residue in VSV L is required for internal initiation from the VSV gene-start sequence. On the other hand, we could not investigate the effects of the T1174A and L1176A mutations in RABV L on its intermediate formation activity due to the unavailability of an in vitro assay. Nevertheless, combining the findings obtained using the complementary assay systems for VSV and RABV allowed us to understand the precise roles of the loop in transcription and capping by rhabdoviral L proteins.
RABV and VSV RdRps showed AC synthesis activities without externally added templates, although to much lesser degrees than template-dependent AC synthesis activities (see Supplementary Figure S12). In either RABV or VSV, alanine substitutions for the tryptophan residues in the loop abolished their AC synthesis activities without external templates, whereas tyrosine substitutions rendered them partially active. However, this activity by VSV RdRp was significantly decreased in the presence of RNases T1 and A, suggesting that 3′-UG-ended cellular RNAs co-purified with VSV L from insect cells might serve as templates (Supplementary Figure S13). In contrast, RABV RdRp synthesized AC even in the presence of RNases T1 and A, suggesting that RABV RdRp may have an intrinsic AC synthesis activity, similar to the AG synthesis activity of dengue virus RdRp (21). These observations suggest that the tryptophan residue of RABV L is required for template-independent AC formation, but further validation studies are necessary. On the other hand, the mutations of the tryptophan residues in VSV and RABV L proteins did not affect their binding to respective P proteins (Supplementary Figures S14 and S15). Based on the biochemical data combined with the structural model of the VSV terminal initiation complex (Figure 7A and Supplementary Figures S10A and S11), we suggest that the tryptophan residues are essential for binding of the initiator ATP to the RdRp active sites to form the terminal initiation complexes, although we cannot rule out the possibility that they interact with the 3′-terminal U residues of their genomes. High-resolution structural analyses of their terminal de novo initiation complexes are necessary to provide the structural basis for further understanding the role of the tryptophan residues.
Taken together, our findings suggest a new model in which temporally and mechanistically distinct events during the stop-start transcription cycle are controlled by the loop structure (hereafter, priming-capping loop) of the rhabdoviral PRNTase domain (Figure 7B). In the terminal initiation complex, the tryptophan residue in the priming-capping loop governs terminal de novo initiation for synthesis of LeRNA and, by extension, the genome and anti-genome. Since the RNA exit channel of the RdRp domain is obstructed with the priming-capping loop in the apo form of VSV L (12), the priming-capping loop must be retracted for RNA chain elongation. After LeRNA synthesis, RdRp reinitiates transcription at the gene-start sequence, but no longer requires the tryptophan residue for internal de novo initiation. It is interesting to note that a proline residue in a priming loop of the influenza virus RdRp is essential for de novo initiation from the 3′-end of the genomic promoter, but not from the internal initiation site in the anti-genomic promoter (23). Therefore, rhabdoviruses and influenza virus use similar strategies to carry out terminal and internal de novo initiation, although products of the latter reactions (mRNAs and pppApG primer, respectively) were totally different. The priming-capping loop may undergo a further structural rearrangement to form the L-pRNA intermediate. In this step, the TxΨ motif in the loop together with the threonine residue in PRNTase motif B may play a critical role in recognizing the 5′-end of pre-mRNA. The pRNA moiety is subsequently transferred from the intermediate to GDP to form capped pre-mRNA during mRNA chain elongation. Thus, this study offers insights into the control of RNA biosynthesis by PRNTase domains that have specifically evolved in NNS RNA viruses.
Since amino acid sequences between PRNTase motifs B and C are highly diversified among NNS RNA viruses belonging to different families in the order Mononegavirales (Supplementary Figure S16), it is currently not possible to predict whether other viruses use these regions as priming-capping loops. Plant rhabdoviruses and pneumoviruses possess conserved aromatic residues (Y or F) in their putative loops, suggesting that these residues serve as priming amino acid residues for terminal de novo initiation. However, aromatic amino acid residues (e.g. W, F, Y, H) are not conserved in putative loops of paramyxoviral and filoviral PRNTase domains. Therefore, it would be necessary to elucidate the molecular mechanisms of transcription initiation by respective L proteins and to identify key elements in their PRNTase domains required for transcription initiation, if any. Cressey et al. (38) have recently developed an in vitro transcription assay to monitor terminal and internal de novo initiation with respiratory syncytial viral RdRp. Thus, it is feasible to analyze whether a tyrosine residue in the putative priming-capping loop of respiratory syncytial viral RdRp is involved in de novo initiation. Stop-start transcription is one of unique gene expression strategies established by NNS RNA viruses, and appears to be regulated in various steps with the PRNTase domains. Further biochemical and structural studies will reveal common and distinct mechanisms of transcriptional controls and pre-mRNA capping by NNS RNA viral PRNTase domains, and provide the basis for designing anti-viral agents targeting these unique domains.
DATA AVAILABILITY
The atomic coordinates of the model of the VSV L terminal initiation complex (id: ma-5k432) have been deposited on Model Archive (www.modelarchive.org).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Amiya K. Banerjee (Cleveland Clinic), Makoto Sugiyama (Gifu University), Naoto Ito (Gifu University), Sue A. Moyer (University of Florida) and John K. Rose (Yale University) for generous gifts of the materials.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Case Western Reserve University; National Institutes of Health [AI093569 to T.O., AI116738 to T.J.G.]. Funding for open access charge: Case Western Reserve University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ogino T., Banerjee A.K.. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell. 2007; 25:85–97. [DOI] [PubMed] [Google Scholar]
- 2. Ogino T., Banerjee A.K.. Formation of guanosine(5′)tetraphospho(5′)adenosine cap structure by an unconventional mRNA capping enzyme of vesicular stomatitis virus. J. Virol. 2008; 82:7729–7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ogino T., Banerjee A.K.. The HR motif in the RNA-dependent RNA polymerase L protein of Chandipura virus is required for unconventional mRNA-capping activity. J. Gen. Virol. 2010; 91:1311–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ogino T., Yadav S.P., Banerjee A.K.. Histidine-mediated RNA transfer to GDP for unique mRNA capping by vesicular stomatitis virus RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:3463–3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ogino T., Banerjee A.K.. An unconventional pathway of mRNA cap formation by vesiculoviruses. Virus Res. 2011; 162:100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ogino T. In vitro capping and transcription of rhabdoviruses. Methods. 2013; 59:188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neubauer J., Ogino M., Green T.J., Ogino T.. Signature motifs of GDP polyribonucleotidyltransferase, a non-segmented negative strand RNA viral mRNA capping enzyme, domain in the L protein are required for covalent enzyme-pRNA intermediate formation. Nucleic Acids Res. 2016; 44:330–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ogino M., Ito N., Sugiyama M., Ogino T.. The rabies virus L protein catalyzes mRNA capping with GDP polyribonucleotidyltransferase activity. Viruses. 2016; 8:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ogino M., Ogino T.. 5′-Phospho-RNA acceptor specificity of GDP polyribonucleotidyltransferase of vesicular stomatitis virus in mRNA capping. J. Virol. 2017; 91:e02322-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Furuichi Y., Shatkin A.J.. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000; 55:135–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 2001; 66:1–40. [DOI] [PubMed] [Google Scholar]
- 12. Liang B., Li Z., Jenni S., Rahmeh A.A., Morin B.M., Grant T., Grigorieff N., Harrison S.C., Whelan S.P.. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015; 162:314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Testa D., Chanda P.K., Banerjee A.K.. Unique mode of transcription in vitro by vesicular stomatitis virus. Cell. 1980; 21:267–275. [DOI] [PubMed] [Google Scholar]
- 14. Emerson S.U. Reconstitution studies detect a single polymerase entry site on the vesicular stomatitis virus genome. Cell. 1982; 31:635–642. [DOI] [PubMed] [Google Scholar]
- 15. Abraham G., Rhodes D.P., Banerjee A.K.. The 5′ terminal structure of the methylated mRNA synthesized in vitro by vesicular stomatitis virus. Cell. 1975; 5:51–58. [DOI] [PubMed] [Google Scholar]
- 16. Abraham G., Banerjee A.K.. Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:1504–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ball L.A., White C.N.. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 1976; 73:442–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ogino T. Capping of vesicular stomatitis virus pre-mRNA is required for accurate selection of transcription stop-start sites and virus propagation. Nucleic Acids Res. 2014; 42:12112–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Butcher S.J., Grimes J.M., Makeyev E.V., Bamford D.H., Stuart D.I.. A mechanism for initiating RNA-dependent RNA polymerization. Nature. 2001; 410:235–240. [DOI] [PubMed] [Google Scholar]
- 20. Tao Y., Farsetta D.L., Nibert M.L., Harrison S.C.. RNA synthesis in a cage–structural studies of reovirus polymerase lambda3. Cell. 2002; 111:733–745. [DOI] [PubMed] [Google Scholar]
- 21. Selisko B., Potisopon S., Agred R., Priet S., Varlet I., Thillier Y., Sallamand C., Debart F., Vasseur J.J., Canard B.. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 2012; 8:e1002912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Appleby T.C., Perry J.K., Murakami E., Barauskas O., Feng J., Cho A., Fox D. 3rd, Wetmore D.R., McGrath M.E., Ray A.S. et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science. 2015; 347:771–775. [DOI] [PubMed] [Google Scholar]
- 23. Te Velthuis A.J., Robb N.C., Kapanidis A.N., Fodor E.. The role of the priming loop in influenza A virus RNA synthesis. Nat. Microbiol. 2016; 1:16029. [DOI] [PubMed] [Google Scholar]
- 24. Grdzelishvili V.Z., Smallwood S., Tower D., Hall R.L., Hunt D.M., Moyer S.A.. A single amino acid change in the L-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 2005; 79:7327–7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawson N.D., Stillman E.A., Whitt M.A., Rose J.K.. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:4477–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schnell M.J., Buonocore L., Whitt M.A., Rose J.K.. The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J. Virol. 1996; 70:2318–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E.. The Protein Data Bank. Nucleic Acids Res. 2000; 28:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Lohkamp B., Scott W.G., Cowtan K.. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Colonno R.J., Banerjee A.K.. A unique RNA species involved in initiation of vesicular stomatitis virus RNA transcription in vitro. Cell. 1976; 8:197–204. [DOI] [PubMed] [Google Scholar]
- 31. Morin B., Rahmeh A.A., Whelan S.P.. Mechanism of RNA synthesis initiation by the vesicular stomatitis virus polymerase. EMBO J. 2012; 31:1320–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stillman E.A., Whitt M.A.. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J. Virol. 1999; 73:7199–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rahmeh A.A., Morin B., Schenk A.D., Liang B., Heinrich B.S., Brusic V., Walz T., Whelan S.P.. Critical phosphoprotein elements that regulate polymerase architecture and function in vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14628–14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Green T.J., Luo M.. Structure of the vesicular stomatitis virus nucleocapsid in complex with the nucleocapsid-binding domain of the small polymerase cofactor, P. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:11713–11718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Deval J., Fung A., Stevens S.K., Jordan P.C., Gromova T., Taylor J.S., Hong J., Meng J., Wang G., Dyatkina N. et al. Biochemical effect of resistance mutations against synergistic inhibitors of RSV RNA polymerase. PLoS One. 2016; 11:e0154097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jordan P.C., Liu C., Raynaud P., Lo M.K., Spiropoulou C.F., Symons J.A., Beigelman L., Deval J.. Initiation, extension, and termination of RNA synthesis by a paramyxovirus polymerase. PLoS Pathog. 2018; 14:e1006889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tchesnokov E.P., Raeisimakiani P., Ngure M., Marchant D., Gotte M.. Recombinant RNA-dependent RNA polymerase complex of Ebola virus. Sci. Rep. 2018; 8:3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cressey T.N., Noton S.L., Nagendra K., Braun M.R., Fearns R.. Mechanism for de novo initiation at two sites in the respiratory syncytial virus promoter. Nucleic Acids Res. 2018; 46:6785–6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Crooks G.E., Hon G., Chandonia J.M., Brenner S.E.. WebLogo: a sequence logo generator. Genome Res. 2004; 14:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates of the model of the VSV L terminal initiation complex (id: ma-5k432) have been deposited on Model Archive (www.modelarchive.org).