Abstract
Myosin regulatory light chain (RLC) phosphorylation is important for cardiac muscle mechanics/function as well as for the Ca2+-troponin/tropomyosin regulation of muscle contraction. This study focuses on the arginine to glutamine (R58Q) substitution in the human ventricular RLC (MYL2 gene), linked to malignant hypertrophic cardiomyopathy in humans and causing severe functional abnormalities in transgenic R58Q mice, including inhibition of cardiac RLC phosphorylation. Using a phosphomimic recombinant RLC variant where the phosphorylation site Ser-15 was substituted with an aspartic acid (S15D) and placed in the background of R58Q, we aimed to assess whether we could rescue/mitigate R58Q-induced structural/functional abnormalities in vitro. We show rescue of several R58Q-exerted adverse phenotypes in S15D-R58Q-reconstituted porcine cardiac muscle preparations. A low level of maximal isometric force observed for R58Q- versus WT-reconstituted fibers was restored by S15D-R58Q. Significant beneficial effects were also observed on the Vmax of actin-activated myosin ATPase activity in S15D-R58Q versus R58Q-reconstituted myosin, along with its binding to fluorescently-labeled actin. We also report that R58Q promotes the OFF state of myosin, both in reconstituted porcine fibers and in transgenic mouse papillary muscles, thereby stabilizing the super-relaxed state (SRX) of myosin, characterized by a very low ATP turnover rate. Experiments in S15D-R58Q-reconstituted porcine fibers showed a mild destabilization of the SRX state, suggesting an S15D-mediated shift in disordered-relaxed (DRX)↔SRX equilibrium towards the DRX state of myosin. Our study shows that S15D-phosphomimic can be used as a potential rescue strategy to abrogate/alleviate the RLC mutation-induced phenotypes and is a likely candidate for therapeutic intervention in HCM patients.
Keywords: Myosin Regulatory Light Chain (RLC), R58Q mutation, S15D-phosphorylation mimic, Super-relaxed state (SRX), Muscle contraction, Rescue of function
Graphical Abstract
Using a phosphomimic RLC variant where the phosphorylation site Ser-15 was substituted with an aspartic acid (S15D) and placed in the background of R58Q, we investigated whether we could rescue/mitigate R58Q-induced structural/functional abnormalities in vitro. A multitude of phosphomimic-induced functional rescue effects was observed, suggesting RLC phosphorylation as a major potential target for therapeutic intervention to alleviate genetic hypertrophic cardiomyopathies.
Introduction
Familial Hypertrophic Cardiomyopathy (HCM), a primary disorder of cardiac myocytes, is an autosomal dominant disease manifested by ventricular hypertrophy, myofibrillar disarray, myocardial fibrosis and sudden cardiac death (SCD), especially in young athletes [1, 2]. Genetic mutations that cause HCM primarily reside in β-myosin heavy chain (β-MHC) and myosin binding protein C (MyBP-C), but they are also present, with lower prevalence, in the myosin regulatory (RLC) and essential (ELC) light chains [3–5]. Both light chains stabilize the α-helical myosin neck region (lever arm), binding to their respective IQXXXRGXXXR motifs [6], but they also play important modulatory roles of tuning myosin cross-bridge kinetics and force-producing actin-myosin interactions. The N-terminal domain of the human ventricular RLC contains a divalent cation binding site and a myosin light chain kinase (MLCK)-specific phosphorylation site at Ser-15 [7]. A wealth of studies indicate that phosphorylation of myosin RLC is critical for the normal function of the heart, and that the myocardium containing dephosphorylated RLC has a reduced ability to produce force and maintain cardiac function at physiological levels (reviewed in [8]). Significantly depressed RLC phosphorylation was reported in heart failure (HF) patients [9] and was also observed in experimental animal models of cardiac disease [10–12]. Studies in mice showed that reduced RLC phosphorylation resulted in abnormal heart performance, presumably through morphological and/or myofibrillar functional alterations such as change in force, myofilament calcium sensitivity, ATPase activity and cross-bridge kinetics [13–16]. It has also been shown that overexpression of heart-specific small subunit of myosin light chain phosphatase (hHS-M21) in mice results in myocardial fibrosis, arrhythmias and HF, mainly due to the dephosphorylation of cardiac RLC and a contributing non-myosin RLC phosphorylation-dependent regulation [17]. The mechanistic basis for this phenomenon is likely the decreased ability of dephosphorylated myosin to efficiently execute the power stroke and generate enough force to maintain normal heart function compared with phosphorylated myosin containing physiological levels of RLC phosphorylation, i.e. ~0.4 mole Pi/mole RLC [13, 18].
Significant genetic heterogeneity of HCM as well as phenotypic variability with varying extent, distribution and severity of the disease have led to poor prediction of clinical outcomes [19]. It is therefore important to elucidate the etiology of HCM at the molecular level for better understanding of the disease mechanisms and for development of potential therapeutic targets. This study is focused on HCM-linked R58Q (Arg→Gln) mutation in the human cardiac RLC (MYL2 gene), identified by Flavigny and Hainque, in unrelated French families [20]. The patients presented with increased left ventricular (LV) wall thickness, abnormal ECG findings (with no mid LV obstruction) and occurrences of SCD [20]. R58Q was later found in German, Swedish and Italian populations and has been consistently associated with a phenotype of severe cardiomyopathy and cardiac death among family members [21–24].
In the three-dimensional structure of RLC, the R58Q mutation is located in the immediate extension of the exiting helix flanking the Ca2+ binding loop. Our previous studies showed an R58Q-induced decrease in RLC phosphorylation, as measured in rapidly frozen ventricular heart samples from transgenic R58Q mice [25]. In the present study, we aimed to determine whether the recombinant phosphomimic S15D-R58Q RLC variant containing aspartic acid substituted for serine-15 (S15D) [12, 26, 27] in the background of R58Q mutation, can rescue/alleviate the detrimental phenotypes observed for R58Q mutant alone. Our approach was based on our previous observation that the constitutive phosphorylation by S15D substitution of another HCM-linked D166V-RLC mutant was sufficient to prevent the development of hypertrophic cardiomyopathy in mice [12, 28]. Systolic and diastolic indices assessed via echocardiography and invasive hemodynamics demonstrated significant improvements of heart function in S15D-D166V mice compared with HCM-D166V mice [12]. Depressed maximal tension and abnormally high myofilament Ca2+ sensitivity observed in HCM-D166V hearts were abrogated in S15D-D166V mice. Further studies with low-angle X-ray diffraction suggested that altered myofilament structures in HCM-D166V mice were partially or fully reversed in S15D-D166V mice [12]. The central hypothesis underlying these observations and directing our current research is that RLC phosphomimic, when placed in the background of an HCM-causing mutation, may play important roles in rescuing/mitigating functional HCM-associated defects.
Here, we addressed this hypothesis and tested the beneficial effect of constitutive RLC phosphorylation using porcine cardiac muscle preparations depleted of endogenous RLC and reconstituted with phosphomimic S15D-R58Q variant. The results were compared to those obtained from R58Q mutant alone and WT-RLC. The study was performed at different levels of system organization (RLC protein, myosin, muscle fiber), allowing for a comprehensive characterization of the rescue effects via S15D-RLC pseudo-phosphorylation in vitro. We found a multitude of rescue of R58Q-induced adverse phenotypes, including rescue of inhibition of the actin-myosin interaction in rigor (no ATP) and in the actin-activated myosin ATPase activity by S15D-R58Q. Similar rescue effects were monitored in S15D-R58Q -reconstituted skinned porcine papillary muscles (PM) which demonstrated recovery of R58Q-diminished contractile force. To assess the effect of R58Q mutation on the stability of the sequestered, super-relaxed (SRX) state of myosin [29, 30], we performed mant-ATP chase experiments in both mouse fibers isolated from transgenic R58Q versus WT mice, as well as in RLC-depleted and mutant-reconstituted porcine PM. The results from both systems showed an R58Q-mediated stabilization of the SRX state of myosin. With a lack of available S15D-R58Q transgenic animal model and using mutant-reconstituted porcine PM, we demonstrated an S15D-R58Q –induced inhibition of the SRX state and a switch from the OFF to ON state of a small population of S15D-R58Q myosin heads.
Results
Effect of R58Q and S15D R58Q mutations on the secondary structure of human cardiac RLC
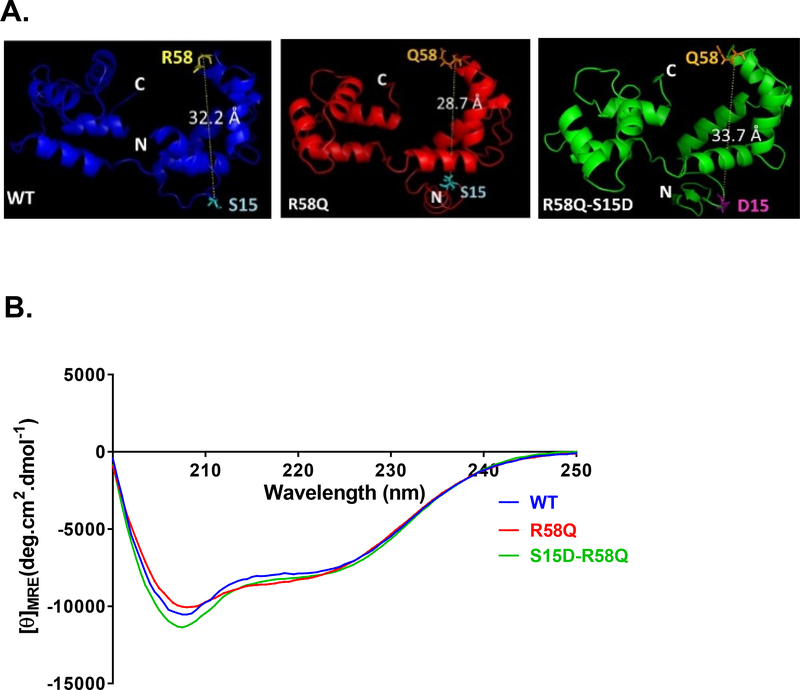
I-TASSER was used for the analysis of the five lowest-energy secondary structures of human ventricular RLC. The structures were computed using protein templates extracted from the Protein Data Bank (PDB). The following PDB structures with high similarity to RLC were used: PDB ID 3jvtB (chain B, calcium-bound scallop myosin regulatory domain (lever arm) with reconstituted complete light chains), PDB ID 2w4aA (chain B, isometrically contracting insect asynchronous flight muscle), PDB ID 1prwA (chain A, crystal structure of bovine brain Ca2+-calmodulin in a compact form), PDB ID 2mysB (chain B, myosin sub-fragment 1), PDB ID 4mvfA (chain A, crystal structure of plasmodium falciparum CDPK2 complexed with inhibitor staurosporine), PDB ID 4agrA (chain A, structure of a tetrameric galectin from Cinachyrella sp. -Ball sponge), PDB ID 4ik1A (chain A, high-resolution structure of Gcampj at pH 8.5) [31, 32]. Fig. 1A shows the predicted modeled structures of human ventricular RLCs (WT, R58Q and S15D-R58Q). Interestingly, the R58Q mutation results in a decrease in the apparent distance between the site of the mutation and Ser-15. In S15D-R58Q modeled structure, this distance is reversed to that observed for RLC-WT (UniProtKB P10916).
Figure 1.
A. Secondary structure predictions for HCRLC and mutants. Modeled structures of human ventricular RLC (MYL2) WT, R58Q and S15D-R58Q, using I-TASSER. Note that the distance between the C-α of R58 and S15 is changed in HCM-R58Q mutant and that the phosphomimic variant reverses this value to the level of WT. B. Effect of the R58Q mutation and S15D-R58Q phosphomimic on the CD spectra of human cardiac RLC. Far-UV CD was performed utilizing a 1-mm path quartz cell in a Jasco J-720 spectropolarimeter. Spectra were recorded at 190–250 nm with a bandwidth of 1 nm. [θ]MRE at 222 nm was used to calculate the α-helical content (fH) using the equation: [θ]222 = −30,300 fH − 2,340.
Circular Dichroism (CD) measurements
Far-UV Circular Dichroism spectroscopy was performed to detect any changes in the secondary structure of the human cardiac RLC-WT, R58Q and S15D-R58Q recombinant proteins. The effect of the R58Q mutation on the secondary structure of RLC-WT was quantified from changes in the mean residue ellipticity at 222 nm, which has been shown to be sensitive to changes in the α-helical content of the protein. As presented in Fig. 1B, R58Q mutation did not result in any significant changes in the CD spectrum, and the calculated α-helical content (in %±SD) was 18.74±0.87 compared to WT which was 18.01±0.43 and S15D-R58Q which was 18.69±0.12 (p>0.05, one-way ANOVA). The mean residue ellipticity, [θ]MRE at 222 nm was [θ]MRE = −8019.17±263.12 for the R58Q mutant versus [θ]MRE = −7797.14±131.02 for RLC-WT and [θ]MRE = −8001.70±37.09 for S15D-R58Q. Data are the average of 4 experiments (n=4), each consisting of 10 scans. The variation in the α-helical content of the RLC-WT and R58Q or S15D-R58Q was below 2% for the individual experiments.
Binding of porcine cardiac myosin reconstituted with RLCs to pyrene-labeled F-actin
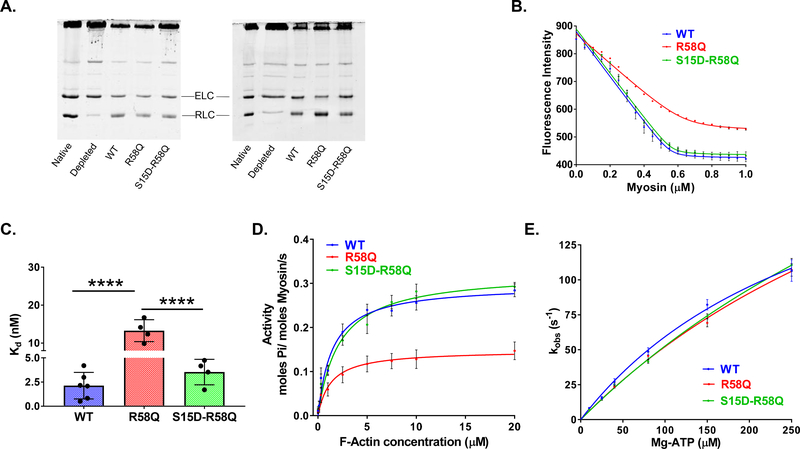
Porcine cardiac myosin was depleted of endogenous RLC and then reconstituted with recombinant RLC proteins. Treatment of the native porcine myosin resulted in depletion of approximately 80% RLC. Several batches of RLC-reconstituted myosin were used for this assay (Fig. 2A). Incubation of depleted myosin with recombinant RLC proteins resulted in up to ~87%, as judged by band intensities and the ratio of ELC to RLC in the reconstituted versus native myosin (Fig. 2A). The ability of RLC-depleted myosin, which underwent the extraction procedure, to bind recombinant RLC variants is quantified in Table 1.
Figure 2. Biochemical assays on RLC-depleted/reconstituted porcine cardiac myosin.
A. Representative SDS-PAGE of CDTA/Triton depleted and RLC reconstituted myosin. Two different batches of depleted and reconstituted porcine myosin are presented. B. Fluorescence-based binding of WT, R58Q and S15D-R58Q -reconstituted myosin to pyrene labeled F-actin. Actin at a concentration of 0.5 μM was titrated with increasing concentrations of reconstituted myosin (0.05–1.0 μM) under rigor (no ATP) conditions. Quenching of pyrene fluorescence on actin-myosin binding was recorded and the data fitted with a non-linear binding equation (see Methods). 4–6 independent titrations per group were performed. C. Plots of dissociation constants (Kd) for RLC-reconstituted myosins. Data from B. were plotted as bar graphs. Note the significantly higher Kd (lower affinity) for R58Q mutation, which is offset in the presence of S15D phosphomimic. ****p<0.0001 versus WT, S15D-R58Q for n = 4–6, by one-way ANOVA with Tukey’s multiple comparison test. D. Actin-activated myosin ATPase activity. Reconstituted myosins (at concentration of 1.1 μM) were incubated with increasing concentrations of actin (0.1–20 μM) and assayed for actin-activated myosin ATPase activity. The reactions were initiated with the addition of 2.5 mM ATP. Note that a significantly lower Vmax observed for R58Q mutation was rescued in the presence of the phosphomimic S15D-R58Q. Data are the average ± SEM of n = 12–17 independent experiments; ****p<0.0001 versus WT, S15D-R58Q; by one-way ANOVA with Tukey’s multiple comparison test. No significant differences in Km were observed among the groups. E. Stopped-flow measurements of actomyosin dissociation. Fast kinetic interactions between porcine myosin reconstituted with recombinant RLC proteins (WT, R58Q and S15D-R58Q) and pyrene-labeled F-actin. Dissociation rates (kobs) were plotted as a function of [Mg-ATP] = from 10–250 μM. The values of kobs ± SEM for each Mg-ATP concentration are presented in Table 4. No significant differences were found between the overall fits among the RLCs (Extra sum-of-squares F-test, p>0.05).
Table 1. Efficiency of RLC depletion and mutant RLC reconstitution in porcine cardiac myosin and porcine cardiac papillary muscles (PM).
Data are the average ± SEM.
| Depleted | WT | R58Q | S15D-R58Q | |
|---|---|---|---|---|
| Porcine Cardiac Myosin | ||||
| % RLC Present | 19.61 ± 1.78 | 87.20 ± 6.34 | 75.74 ± 9.00 | 86.88 ± 4.25 |
| n= # SDS-PAGE | 7 | 4 | 5 | 5 |
| Porcine Cardiac papillary muscles | ||||
| % RLC Present | 16.89 ± 1.17 | 61.31 ± 2.48 | 54.00 ± 4.02 | 64.50 ± 3.17 |
| n= # fibers | 9 | 13 | 13 | 12 |
Myosins reconstituted with R58Q, S15D-R58Q and WT RLCs were then tested for their interaction with actin. First, fluorescence-based binding assays of reconstituted myosins to pyrene-labeled F-actin were conducted under rigor (no ATP) conditions to assess changes induced by the R58Q mutation and its S15D-R58Q phosphomimic variant on the binding affinity. A decrease in the fluorescence intensity induced by the binding of myosin to F-actin was measured as a function of increasing myosin concentrations (Fig. 2B). Results showed a strong inhibition of the actin-myosin binding by R58Q mutant compared to WT reconstituted myosin and demonstrated by significantly higher apparent dissociation constant (Kd~13 nM) for R58Q versus control WT protein (Kd~2 nM) (Fig. 2C, Table 2). For the double S15D-R58Q mutant, the observed binding to F-actin with Kd~4 nM, was comparable to that obtained for WT control but was significantly different from R58Q mutant (p<0.0001, by one-way ANOVA with Tukey’s multiple comparison test) (Fig. 2C and Table 2). These results indicate the rescue in binding affinity by phosphomimic RLC in the background of R58Q mutation.
Table 2. Steady-state rigor (no ATP) binding of RLC depleted/reconstituted myosin to pyrene-labeled F-actin and actin-activated myosin ATPase activity.
Values are means ± SEM for n = # independent experiments.
| Parameter | WT | R58Q | S15D-R58Q |
|---|---|---|---|
| Steady-state rigor binding | |||
| Kd (nM) | 2.13 ± 0.56 | 13.26 ± 1.45**** | 3.53 ± 0.66 |
| n | 6 | 4 | 4 |
| ATPase activity | |||
| Vmax (s−1) | 0.32 ± 0.01 | 0.18 ± 0.02 **** | 0.36 ± 0.01 |
| Km (μM) | 2.56 ± 0.24 | 4.05 ± 0.65 | 3.72 ± 0.46 |
| n | 12 | 17 | 13 |
p<0.0001 versus WT, S15D-R58Q, one-way ANOVA with Tukey’s multiple comparison test.
Actin activated myosin ATPase assays
To measure the effect of mutation and pseudo-phosphorylation of RLC in the background of mutation on ATPase enzymatic activity, F-actin activated myosin ATPase activity assays were performed on porcine cardiac myosin preparations reconstituted with human recombinant RLC proteins: WT, R58Q and S15D-R58Q (Fig. 2D, Table 2). Steady-state actin activated myosin ATPase activity was determined as a function of increasing F-actin concentrations. ATPase isotherms were obtained by plotting the ATPase activity versus [F-actin], with data points expressed as averages ± SEM of n=12–17 experiments. The data were fitted to the Michaelis–Menten equation yielding the Vmax (maximal ATPase activity) and Km (Michaelis-Menten constant) parameters. Our results revealed a significantly lower Vmax (in s−1) of R58Q-reconstituted myosin compared to WT RLC myosin (Table 2, p<0.0001, by one-way ANOVA with Tukey’s multiple comparison test). Interestingly, phosphomimetic S15D-R58Q double mutant was able to restore maximal ATPase activity to a level similar to that obtained for WT-reconstituted myosin (Table 2, no statistical difference between S15D-R58Q and WT myosin). The Km values did not significantly vary among all tested proteins (Table 2).
Stopped-flow measurements
To further characterize interaction of reconstituted porcine myosins with actin, we measured the dissociation rates (kobs) of myosin (at concentration 0.25 μM) bound to pyrene-labeled F-actin (0.25 μM) stabilized by phalloidin (0.25 μM) (Fig. 2E). The time course of the change in pyrene fluorescence was monitored as a function of ATP concentrations. Myosins were stoichiometrically mixed with pyrene-F-actin (2 heads of myosin per actin monomer), in a 1:1 volume ratio with increasing concentrations of [Mg-ATP] = from 10–250 μM in a stopped-flow apparatus. An increase in the fluorescence intensity of pyrene was monitored as a function of time as the myosin heads dissociated from actin on the addition of Mg-ATP. Fig. 2E presents dissociation rates (kobs)-[Mg-ATP] dependence for WT, R58Q and S15D-R58Q RLCs. The observed dissociation rates (kobs in s−1) derived from the averaged fluorescence traces and fitted with a single exponential are presented in Table 3. Compared to WT, R58Q mutant-reconstituted myosin showed significantly slower transition rate at 150 μM MgATP while S15D-R58Q-reconstituted myosin demonstrated rates greater than that for R58Q (Table 3). No other notable differences were noted for any other MgATP concentrations and we believe that despite a change at 150 μM, R58Q does not alter dissociation rates compared to WT. Comparison of independent fits for different RLCs-reconstituted myosin with a global fit (sharing K1 and Kmax) did not reveal any significant differences between the overall fits among all tested RLCs (Fig. 2E, Extra sum-of-squares F-test, p>0.05).
Table 3. Actin-myosin dissociation rates (kobs in s−1).
Values are means ± SEM for n = # independent titrations of actin-myosin complex with [Mg-ATP]; 7–13 transients were averaged for each run.
| System | [MgATP] | |||||
|---|---|---|---|---|---|---|
| 10 μM | 25 μM | 40 μM | 80 μM | 150 μM | 250 μM | |
| WT | 8.25±0.44 n=19 | 15.65±0.96 n=11 | 27.27±2.03 n=18 | 48.89±2.57 n=21 | 82.18±3.78 n=18 | 106.63±7.76 n=13 |
| S15D-R58Q | 7.29±0.43 n=19 | 14.69±0.89 n=16 | 25.30±1.31 n=23 | 41.52±1.24 n=22 | 73.99±1.76 n=35 | 110.65±4.67 n=33 |
| R58Q | 7.26±0.31 n=12 | 15.13±0.67 n=10 | 23.61±1.05 n=20 | 45.64±2.58 n=28 | 69.19±2.85** n=27 | 107.11±4.50 n=31 |
p<0.01 versus WT, one-way ANOVA with Tukey’s multiple comparison test.
Steady-state force measurements
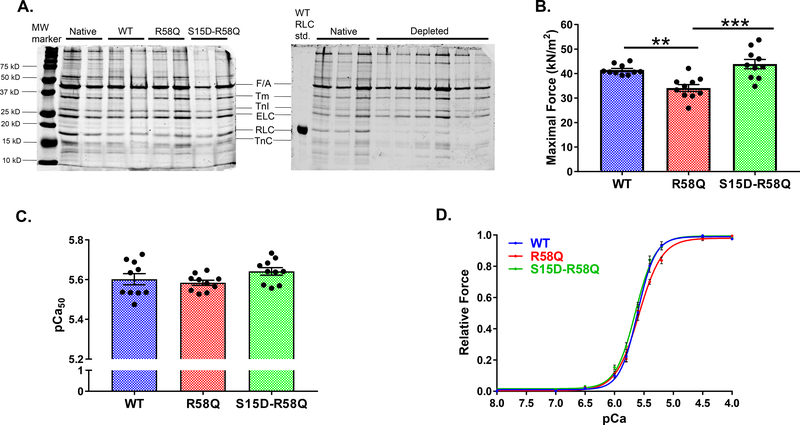
Measurements of steady-state force generation and calcium sensitivity of force were performed on skinned PM strips depleted of RLC and reconstituted with RLC mutant proteins (Fig. 3, Table 4). Fig. 3A shows representative SDS-PAGE images of CDTA-depleted and RLC/TnC (Troponin C) -reconstituted porcine PM. Reconstitution with TnC was necessary to make sure that the changes in force were not due to TnC deficiency [33]. On average, ~17% of porcine RLC remained in the fibers, which were subsequently incubated with recombinant RLC proteins. The reconstitution levels achieved in RLC-depleted and mutant-reconstituted PM for WT, R58Q and S15D-R58Q are shown in Table 1. Percent reconstitution was calculated according to SDS-PAGE band densities of the RLC and ELC proteins (Fig. 3A). The ELC/RLC ratios were compared with those of control untreated muscle strips. ELC protein was used as a loading control as it was not affected by the RLC extraction/reconstitution procedure.
Figure 3. Measurements of steady-state force in skinned porcine PM strips.
A. Representative SDS–PAGE of WT, R58Q and S15D-R58Q reconstituted fibers (left), native and CDTA/Triton RLC-depleted porcine papillary muscle strips (right). B. Maximal tension at pCa4. C. Ca2+-sensitivity of force development. D. Effect of pseudo-phosphorylation on force-pCa. Porcine PM strips were reconstituted with the recombinant RLC constructs: WT, R58Q and S15D-R58Q. Significantly depressed maximal force with no changes in Ca2+-sensitivity of force was observed for R58Q-reconstituted fibers compared with WT. Reconstitution with S15D-R58Q increased maximal tension to the level observed for WT fibers. Data are the average ± SEM of n = 10 fibers per group. **p<0.01 (R58Q versus WT), †††p<0.001 (S15D-R58Q versus R58Q) by one-way ANOVA with Tukey’s multiple comparison test.
Table 4. Maximal tension (Fmax) and Ca2+ sensitivity of force development and the super-relaxed state (SRX) measurements in RLC-depleted/reconstituted skinned porcine cardiac papillary muscle (PM) strips as well as in skinned cardiac PM fibers from transgenic R58Q versus WT mice.
Values are means ± SEM for n = # fibers.
| Parameter | WT | R58Q | S15D-R58Q | |
|---|---|---|---|---|
| Measurements of steady-state force in porcine PM | ||||
| Fmax (in kN/m2) | 41.47 ± 0.65 | 34.08 ± 1.42 ** | 43.88 ± 1.90 ††† | |
| Ca2+-sensitivity, pCa50 | 5.60 ± 0.03 | 5.58 ± 0.01 | 5.64 ± 0.02 | |
| Hill coefficient, nH | 3.35 ± 0.46 | 2.32 ± 0.22 | 2.66 ± 0.15 | |
| n = # fibers | 10 | 10 | 10 | |
| SRX in porcine reconstituted PM | ||||
| P1 | 74.18 ± 2.57 | 60.13 ± 2.65 ** | 70.50 ± 2.90† | |
| T1 (s) | 5.95 ± 0.69 | 6.60 ± 0.75 | 5.51 ± 0.81 | |
| P2 | 25.82 ± 2.57 | 39.87 ± 2.65 ** | 29.51 ± 2.90† | |
| T2 (s) | 56.43 ± 11.88 | 77.69 ± 19.87 | 69.73 ± 13.04 | |
| n = # fibers | 17 | 18 | 16 | |
| SRX in R58Q transgenic mouse PM | ||||
| WT | R58Q | |||
| P1 | 61.89 ± 3.09 | 44.66 ± 4.44 * | ||
| T1 (s) | 5.71 ± 1.35 | 3.89 ± 0.64 | ||
| P2 | 37.81 ± 3.00 | 53.01 ± 4.42 * | ||
| T2 (s) | 269.90 ± 43.96 | 170.80 ± 29.90 | ||
| n= # fibers | 7 | 11 | ||
p<0.05,
p<0.01 versus WT;
p<0.05,
p<0.001 versus R58Q, one-way ANOVA with Tukey’s multiple comparison test (porcine system) and t-test (transgenic mice). P1 and P2 - populations of myosin heads in the fast and slow (respectively) phase of ATP turnover; T1 and T2 - nucleotide turnover times (T, lifetime) in the fast and slow (respectively) phase of ATP turnover.
A significantly lower maximal isometric force per cross-section of muscle (in kN/m2) was observed for R58Q-reconstituted porcine muscle preparations (~34) compared to WT (~41) with p<0.01 by one-way ANOVA (Table 4). Upon reconstitution with the phosphomimetic S15D-R58Q, porcine muscle strips demonstrated rescue mechanism, manifested by a significantly higher and similar to WT level of maximal tension (~44) (Fig. 3B). The pCa50 values, presented in Fig. 3C and Table 4, were not significantly different among the three groups (Fig. 3D).
Measurements of nucleotide turnover rate and the super-relaxed (SRX) state using fluorescent mantATP chase assay
We also assessed the effect of R58Q mutation on the SRX state of myosin in porcine PM fibers (Fig. 4) and in transgenic mouse papillary muscles (Fig. 5). With a lack of available S15D-R58Q transgenic animal model, the effect of S15D-R58Q double mutant was investigated in RLC-depleted and S15D-R58Q-reconstituted porcine PM fibers.
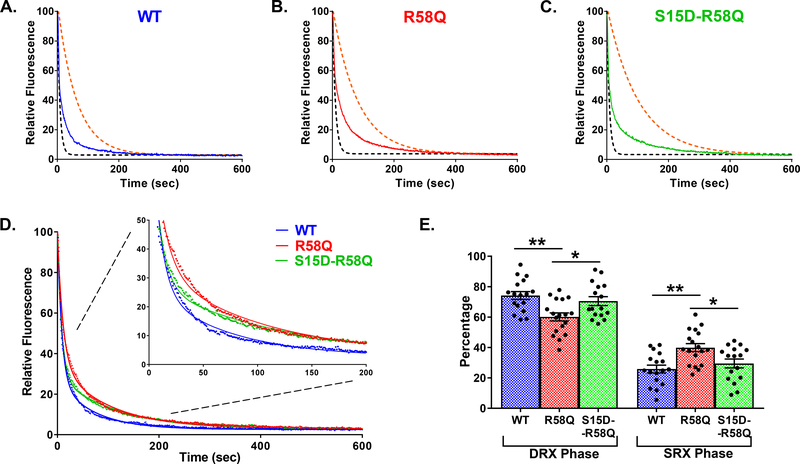
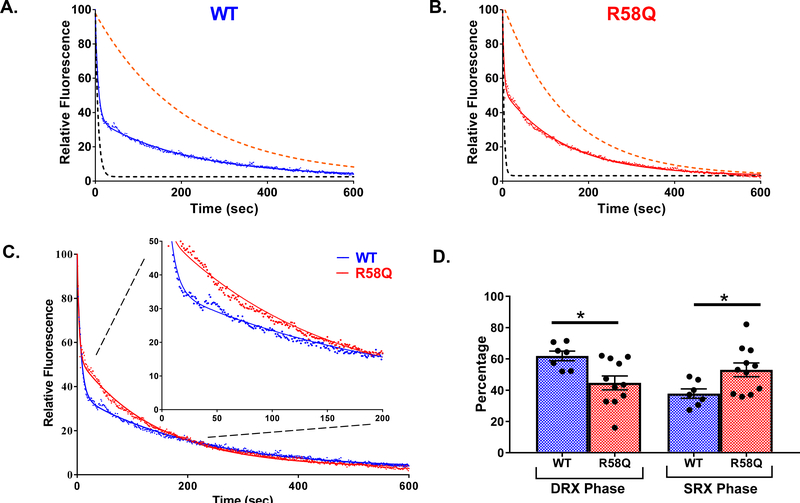
Figure 4. Single nucleotide turnover studies in reconstituted porcine PM fibers.
Fluorescence decays of mant-ATP release in skinned porcine papillary muscle fibers reconstituted with WT (blue trace, A), R58Q (red trace, B), and S15D-R58Q (green trace, C) recombinant proteins. The fibers were first incubated in 250 μM mant-ATP and subsequently chased with a solution containing 4 mM ATP. Decay traces were fitted to a double-exponential equation, yielding DRX and SRX lifetimes (T) and population proportion (P). Simulated single-exponential dashed curves represent DRX (black) and SRX (orange) phases for each recombinant protein. D. Summary of experimental ATP-chase curves for R58Q, S15D-R58Q and WT reconstituted fibers. Decay traces were fitted to double-exponential equation. The inset plots present the SRX phase from 0–200 s. E. Percentages of myosin heads in the DRX (P1) versus SRX (P2) states. R58Q fibers showed a significantly increased P2 population compared to WT and S15D-R58Q significantly decreased P2 compared to R58Q mutant. Data are the average ± SEM of n = 16–18 fibers. **p<0.01 vs. WT and *p<0.05 vs. R58Q; by one-way ANOVA with Tukey’s multiple comparison test.
Figure 5. Single nucleotide turnover studies in PM fibers from transgenic WT versus R58Q mice.
Fluorescence decays of mant-ATP release in transgenic WT (blue trace, A) and R58Q (red trace, B). The fiber was first incubated in 250 μM mant-ATP and subsequently chased with a solution containing 4 mM ATP. Decay traces were fitted to a double-exponential equation, yielding DRX and SRX rates. Simulated single-exponential dashed curves represent DRX (black) and SRX (orange) phases for WT and R58Q fiber. C. Overlay of WT and R58Q decay traces fitted to double-exponential equation with inset plots. D. Percentages of myosin heads in the DRX (P1) versus SRX (P2) states. R58Q fibers showed a significantly increased P2 population compared to WT. Data are the average ± SEM of n = 7–11 fibers. *p<0.05 versus WT; by student’s t-test.
Skinned porcine cardiac PM fibers, RLC-depleted and reconstituted with recombinant WT, R58Q, S15D-R58Q proteins, were incubated in a relaxing solution containing 250 μM mant-ATP. Then the fibers were mounted in a flow cell of the IonOptix apparatus and subjected to nucleotide exchange assay using 4 mM dark ATP -relaxing solution (Fig. 4). Decay in fiber fluorescence was recorded as the mant-nucleotide was released from myosin and replaced by dark ATP [34]. Figs. 4A-D show the time-dependent average fluorescence decay curves for WT (blue trace), R58Q (red trace) and S15D-R58Q (green trace) during the chase phase with non-fluorescent ATP. The decay of fluorescence intensity was fitted to the following double-exponential function:
where P1 and P2 are the relative fiber fluorescence fractions (amplitudes) of the two states: DRX (disordered-relaxed) and SRX (super-relaxed) and T1 and T2 depict their respective lifetimes (in seconds) (Fig. 4 and Table 4). The P1 and P2 values (in %) illustrate the population of myosin heads in DRX and SRX states, respectively.
Simulated single-exponential dashed curves for the fast phase (black) and slow phase (orange) for each protein were derived from fluorescence decay rates (1/T) obtained from fitting the experimental double-exponential curves to single-exponential fits (Fig. 4). As shown in Table 4, R58Q-reconstituted porcine fibers significantly increased the proportion of myosin heads in the SRX state with P2=40% for R58Q compared to 26% for WT fibers. The presence of S15D in the background of the mutation (S15D-R58Q) led to destabilization of SRX and the shift of myosin heads toward the DRX state with P2=30% (Fig. 4E, Table 4). No significant differences in lifetimes for both phases among WT, R58Q and S15D R58Q were observed.
Studies in transgenic R58Q versus WT PM mouse fibers (Fig. 5A-D) confirmed the R58Q-induced stabilization of the SRX state of myosin with P2= 53% in R58Q compared with P2=38% in WT mice (Table 4). The differences in P1 and P2 for R58Q versus WT animals were statistically significant (Figs. 5D, Table 4), providing evidence for the mutation-induced promotion of the sequestered, SRX state and a decrease in the population of myosin heads existing in the DRX state.
Discussion
In order to investigate the potential role of RLC pseudo-phosphorylation in rescuing and/or mitigating an HCM mutation-induced abnormalities in vitro, we used recombinant phosphomimetic S15D-R58Q double mutant that was reconstituted in RLC-depleted porcine cardiac muscle preparations and tested for its functional implications compared to R58Q mutant alone. Our results support an S15D-dependent novel rescue strategy to alleviate a detrimental cardiac phenotype associated with the R58Q mutation in MYL2, found in HCM patients of different ethnic descent to cause a phenotype of severe cardiomyopathy and SCD [20–24].
In the RLC molecule, the R58Q mutation replaces the bulky and positively charged arginine with an uncharged but polar glutamine. The mutation lies in the N-terminal domain of the RLC, in close proximity to the cation binding site and to Ser-15 phosphorylation site. Based on I-TASSER modeling, the R58Q mutation results in a decrease in the apparent distance between the mutation and Ser-15 sites, an effect that is normalized with the introduction of S15D in the background of R58Q mutation. Far UV CD spectra did not reveal any significant changes in the α-helical content as a result of R58Q mutation or its phosphomimic mutation compared to WT RLC. In the absence of loss/gain of α-helical content, it is possible that the extra negative charge from the aspartic acid residue of S15D is able to change the conformation of the Ca2+ binding loop or the region flanking it, suggesting an allosteric communication between the phosphorylation and Ca2+ binding sites [7]. As we previously showed, R58Q mutant completely abolished Ca2+ binding to non-phosphorylated form of RLC and MLCK-induced phosphorylation of R58Q resulted in restoration of Ca2+ binding with a simultaneous increase in α-helical content [7].
Porcine cardiac muscle preparations containing physiologically relevant β-MHC isoform were reconstituted with human cardiac RLC mutant proteins to further assess biochemical implications of the HCM-causing mutation and the relevance of the S15D-mediated rescue. Assessing the ability of myosin to strongly bind to actin (under rigor conditions), we show a decrease in the binding affinity because of R58Q mutation compared with WT, and rescue of binding to actin by S15D-R58Q -reconstituted myosin. This clearly suggests that pseudo-phosphorylation of RLC can play a role in restoration of actin-myosin rigor-state binding. We also evaluated the consequence of the mutation(s) on myosin’s enzymatic properties and its interaction with actin. Actin-activated myosin ATPase activity assays showed a significant decrease in maximal ATPase activity (Vmax) as a result of R58Q mutation compared with WT myosin. This represents a reduced rate constant of the transition from the weakly-bound to strongly-bound myosin cross-bridges, most likely conferring a lower myosin duty ratio compared with either WT or S15D-R58Q mutant. Decreased Vmax may also suggest an R58Q-mediated reduction in the number of functionally accessible myosin heads that are capable of interacting with actin. Consistent with this notion, assessment of myosin’s ability to produce contractile force in reconstituted porcine papillary muscle strips revealed an R58Q-dependent decrease in maximal isometric tension without any significant change in myofilament Ca2+ sensitivity. Pseudo-phosphorylation of R58Q could rescue both the ATPase activity and isometric force to the level of WT.
A small but not significant S15D-mediated increase in the Ca2+ sensitivity of force was observed between R58Q and S15D-R58Q reconstituted preparations. Similar results were obtained in our earlier study between WT and S15D reconstituted porcine muscle preparations [27]. However, much bigger increase in the Ca2+ sensitivity of force in these studies was produced by HCM-linked D166V mutation [26, 27], most likely due to the hypercontractile behavior of D166V mutant. The presence of phosphomimetic mutation in the background of D166V (S15D-D166V) was able to bring the Ca2+ sensitivity back to that observed for WT reconstituted strips. Hence, we believe that pseudophosphorylation (in the background of a disease-causing mutation) will not always tend to increase but rather normalize calcium sensitivity for optimal cardiac performance. Therefore, S15D substitution, by virtue of introducing a negative charge at the phosphorylation site, may be able to either abrogate possible steric constraints introduced by 58Q residue in the conformation-sensitive region of the myosin-lever arm [35], or introduce structural alterations that may be purely coincidental with the phosphomimetic nature of the mutation, and this largely remains hypothetical and subject of further research.
Myocardium can rapidly downregulate energy output to very low levels during times of stress (e.g. cardiomyopathy mutation), ischemia, or hypoxia [36]. Quantitative epi-fluorescence studies, measuring single nucleotide turnovers, have revealed a new form of highly inhibited state of myosin termed as the super-relaxed state (SRX), with extremely low ATP turnover rates (> 100 s) [29]. In SRX, the myosin heads are highly ordered and interact with one another along the axis of the thick filament forming structures known as the interacting heads motifs (IHMs) [37]. This recently discovered SRX state of myosin motor was shown to be disrupted by myosin RLC phosphorylation [29, 38]. Study from M. Irving laboratory using polarized fluorescence experiments in rat ventricular trabeculae has shown that RLC phosphorylation can shift the equilibrium between perpendicular ON and parallel OFF states toward more ON state of myosin [39]. Using bifunctional sulforhodamine labelled RLCs exchanged into demembranated rat right ventricular trabeculae, the authors further showed that order fluorescence polarization parameter <P2> was significantly higher for R58Q under both relaxing (pCa 9) and full calcium activation (pCa 4.3) conditions, suggesting that R58Q mutation induces a more parallel OFF orientation of the myosin heads [40]. They also showed that MLCK-dependent phosphorylation of R58Q RLC-exchanged trabeculae led to an increase by about ~30% of maximum active isometric force. While this supports the notion that R58Q promotes an OFF state of the thick filaments, it does not provide information about the effect of R58Q mutation on the number of functional myosin heads that are available for actin interaction and ATP utilization and on the population of myosin head existing in SRX versus DRX state [40]. Our mant-ATP chase experiments in both reconstituted porcine and transgenic fibers indicate that R58Q mutation stabilizes the slow SRX phase of myosin compared to WT, and thereby decreases the number of “working” myosin heads that are able to participate in the force-generating state, without significantly affecting the turnover rate of slow/fast proportion compared to WT. Owing to an unavailability of transgenic S15D-R58Q mice, phosphomimetic experiments were limited to reconstituted porcine fibers where we showed an S15D-mediated shift in equilibrium from SRX to DRX compared to R58Q mutant, thereby increasing the proportion of functionally active myosin heads available for the interaction with actin. This effect was in accord with the reduced ATPase activity and diminished isometric force observed for R58Q and their recovery in the presence of the phosphomimetic substitution in the background of mutation. Karabina et al. showed that in the presence of load, porcine myosin bearing the R58Q mutation had largely reduced actin sliding velocity and reduced force production ability compared to WT and that these defects were restored upon mutant phosphorylation [41].
Unlike many HCM mutations, R58Q does not fall into a typical “hypercontractile” category, as it does not weaken the SRX-IHM state to produce more functionally accessible heads [37]. A common mechanism via which different sarcomeric proteins may induce the development of HCM has yet to be identified. For example, functional effects of different HCM-causing mutations in the MYH7 gene on force generation, ATPase and myofilament Ca2+-sensitivity were shown to be quite different, with the force-pCa relationship shifted to the left (hypercontractile) or right (hypocontractile) compared with WT [42]. Similarly, studies with HCM-causing myosin head converter domain mutations suggested a decreased intrinsic force for some of the MYH7 HCM mutations suggesting their hypocontractile behavior [43]. Our recent work on myosin ELC mouse models of HCM demonstrated that two different HCM mutations exerted opposite effects on myosin power output compared to WT ELC and yet led to the same HCM disease in vivo in mice [44]. Along the same lines, recent work from the Houdusse group has shown that various HCM mutations can have disparate effects on the myosin motor function and yet result in similar HCM phenotypes [45]. The question of how different myosin mutations impair the function of myosin leading to the HCM disease will have to be addressed in future high resolution structural studies.
Concluding remarks
We demonstrate here the feasibility of a robust in vitro molecular rescue strategy to mitigate/prevent functional abnormalities associated with hypertrophic cardiomyopathy-causing R58Q mutation. We propose that R58Q mutation, by removing the positive charge, may significantly alter the electrostatic interactions in the RLC-MHC hinge interface and thus impair the myosin cross-bridges in their interaction with actin to produce force and muscle contraction. We hypothesize that S15D-R58Q phosphomimic works by virtue of producing a unique structural balance enabling the pseudo-phosphorylated R58Q-myosin to abrogate the majority of detrimental biomechanical phenotypes in vitro. We also report on R58Q-mediated stabilization of the SRX population of myosin heads and on S15D-R58Q-induced shift in the SRX↔DRX equilibrium toward the DRX state in which myosin heads can readily interact with thin filaments and produce force.
Overall, our findings strongly support and validate our previous in vitro reports showing phosphomimic rescue in HCM-causing D166V mutation where majority of functional and structural abnormalities were alleviated by S15D in the background of D166V [26, 27]. These in vitro results gave basis to testing the S15D phosphimimic-mediated rescue of HCM phenotype in vivo [12]. We showed that S15D pseudo-phosphorylation of D166V-RLC mutant was sufficient to prevent the development of hypertrophic cardiomyopathy in mice [12, 28]. Future studies will explore and evaluate the therapeutic potential of pseudo-phosphorylation of R58Q-RLC in vivo to rescue its HCM phenotype. The work described in this report is a necessary first step toward applying the S15D strategy in more clinical settings for the management of cardiomyopathies that, among other symptoms, are manifested by depressed levels of RLC phosphorylation.
Materials and Methods
All animal procedures and experiments were performed in accordance with the “Guide for the Care and Use of Laboratory Animals” (NIH Publication 85–23, revised 2011). All the protocols were approved by the Institutional Animal Care and Use Committee at the University of Miami Miller School of Medicine. UM has an Animal Welfare Assurance on file with the Office of Laboratory Animal Welfare (OLAW), NIH. The assurance number is #A-3224–01, effective November 24, 2015. IACUC protocol # is 18–110-LF. Mice were euthanized through CO2 inhalation followed by cervical dislocation. Transgenic (Tg) mouse models with the human ventricular RLC WT L2 (expressing 100% of human cardiac WT-RLC) and R58Q line-8 (L8, expressing 100% of human cardiac R58Q-RLC) mutant were generated at the University of Miami as previously described [46].
Mutation, expression and purification of wild-type (WT) human cardiac RLC and the HCM mutants
WT human cardiac RLC cDNA was cloned via reverse transcription-polymerase chain reaction using primers based on the published cDNA RLC sequence (GenBank™ Accession No. AF020768) using standard methods as described previously [46]. RLC mutants (R58Q and S15D-R58Q) were generated using overlapping sequential polymerase chain reaction as described earlier [27]. Briefly, WT and mutant cDNAs were constructed with an NcoI site upstream of the N-terminal ATG and a BamHI site following the stop codon to facilitate ligation into the NcoI–BamHI cloning site of the pET-3d (Novagen) plasmid vector and transformation into DH5α cloning host bacteria for expression. The cDNAs were transformed into BL21 expression host cells and all proteins were expressed in large (16 liters) cultures. Purification was achieved using an S-Sepharose column (equilibrated with 2 M urea, 20 mM sodium citrate, 0.1mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), 0.02% NaN3, pH 6.0), followed by a Q-Sepharose column (equilibrated with 2 M urea, 25 mM Tris–HCl, 0.1 mM PMSF, 1mM DTT, 0.02% NaN3, pH 7.5). The proteins were eluted with a salt gradient of 0–450 mM NaCl and the final purity was evaluated using 15% SDS–PAGE.
Preparation of porcine cardiac myosin
Porcine cardiac myosin was purified as described in Pant et al. [47] Briefly, ventricular muscle was chilled on ice, washed with ice-cold distilled H2O and minced, followed by extraction on ice with stirring for 1.5 h in Edsall–Weber solution (0.012 M Na2CO3, 0.04 M NaHCO3, and 0.6 M KCl, pH 9.0) (300 ml/100 g of muscle). Then, the homogenate was centrifuged at 13,000g for 20 min, and the supernatant was precipitated with 13 volumes of water containing 1mM EDTA (ethylenediaminetetraacetic acid) and 1mM DTT. After centrifugation at 13,000g for 10 minutes, the pellet was resuspended in buffer containing 0.5M KCl, 20mM MOPS (3-(Nmorpholino) propanesulfonic acid) (pH 7.0), 1 mM DTT, and 10 mM MgATP and centrifuged at 186,000g for 1.5 h. Supernatant containing native porcine cardiac myosin was precipitated with 13 volumes of ice cold water and 1 mM DTT and centrifuged at 8,000g for 10 minutes. The pellets were left on ice overnight. Then, the pellet containing porcine cardiac myosin was dissolved in the above buffer and clarified by centrifugation at 186,000g for 1.5 h, mixed with glycerol (1:1), and stored at −20°C until used.
Depletion of endogenous RLC from porcine cardiac myosin and reconstitution with human cardiac WT or RLC mutants
Endogenous RLC was depleted from porcine cardiac myosin by treatment with 1% Triton X-100 and 5 mM CDTA (1, 2-cyclohexylenedinitrilotetraacetic acid), pH 8.5, as described previously [47]. Depleted myosins were then resuspended in a buffer composed of 0.4 M KCl, 50 mM MOPS, 2 mM MgCl2, pH 7.0, and 1 mM DTT, mixed in a 1:3 molar ratio with recombinant human cardiac RLCs (WT, R58Q or S15D-R58Q), and dialyzed against the same buffer at 4°C for 2 h. RLC-reconstituted myosins were then dialyzed against 5 mM DTT H2O overnight. Precipitated myosin–RLC complexes were then collected by centrifugation (2×8,000g for 10 minutes) and resuspended in myosin buffer composed of 0.4 M KCl, 10 mM MOPS, pH 7.0, and 1 mM DTT. Samples were taken for SDS–PAGE analysis before they were mixed 1:1 with glycerol and stored at −20°C until needed for experiments. At least three-four isolated preparations of recombinant RLC-reconstituted myosins were used for all the assays involving reconstituted porcine myosin.
Preparation of skeletal muscle F-actin
Rabbit skeletal muscle F-actin was prepared as described previously [27]. Briefly, extraction of rabbit skeletal acetone powder was achieved with G-actin buffer consisting of 2 mM Tris-HCl (pH 8.0), 0.2 mM Na2ATP, 0.5 mM β-mercaptoethanol, 0.2 mM CaCl2, and 0.0005% NaN3 at a ratio of 20 ml/g for 30 minutes with stirring on ice. The extract was then clarified by centrifugation at 7,000g at 4°C for 1 h. The pellet was discarded, and the supernatant was adjusted to a final concentration of 40 mM KCl, 2 mM MgCl2 and 1 mM Na2ATP (pH 8.0). F-actin was allowed to polymerize for 2 h at 4°C. The KCl concentration was slowly increased to a final concentration of 0.6 M, and the solution was slowly stirred on ice for 30 minutes to remove any possible traces of tropomyosin-troponin. F-actin was collected by ultracentrifugation at 160,000g at 4°C for 1.5 h. The supernatant was discarded, and the F-actin pellets were re-dissolved in a buffer consisting of 10 mM MOPS (pH 7.0) and 40 mM KCl and stored in liquid nitrogen until further use.
Labeling of F-actin with pyrene
Rabbit skeletal actin was fluorescently labeled with Pyrene lodoacetamide (PIA) as described in Muthu et al. [27]. Briefly, 20–40 μM F-actin was incubated at room temperature, in the dark, for 16 hours with a 1.5 molar excess of N-(1-pyrene) iodoacetamide (Invitrogen, Molecular Probes) in a buffer containing 10 mM MOPS, 1 mM MgCl2 and 40 mM KCl at pH 7.0. Then, the reaction was quenched with 1 mM DTT and the preparation was centrifuged at 1000g for 1 h. F-actin was dialyzed against G-actin buffer (2 mM Tris-HCl pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 1mM DTT) to remove excess of pyrene and was allowed to polymerize overnight to form F-actin. The resulting molar ratio of pyrene/F-actin was 0.87 determined using the molar extinction coefficient, ε344(pyrene)=22,000 M−1cm−1.
Binding of porcine myosin reconstituted with RLC to pyrene labeled F-actin
Pyrene-labeled F-actin at a concentration of 0.5 μM (stabilized by 0.5 μM phalloidin) was titrated with porcine myosin reconstituted with WT, R58Q and S15D R58Q RLCs, from 0.05–1 μM, in 0.05 μM increments until it reached stoichiometrical saturation. Fluorescence measurements were done using a JASCO 6500 Spectrofluorometer. PIA was excited at 340 nm and fluorescence was collected at 407 nm. The titration data were fitted to the following quadratic equation to obtain the binding constant (Kd) and stoichiometry (n):
where m1 = initial signal (fluorescence intensity at 0 μM myosin), m2 = maximal amplitude (fluorescence intensity at saturating myosin concentrations), n = stoichiometry of binding (mole of myosin per mole of actin), and x = total concentration of added myosin.
Stopped flow measurements
Porcine cardiac myosins reconstituted with WT, R58Q and S15D R58Q recombinant RLC proteins at concentrations of 0.25 μM were mixed with 0.25 μM pyrene-labeled F-actin (stabilized by 0.25 μM phalloidin) in rigor buffer containing 0.4 M KCl, 1 mM DTT, and 10 mM MOPS at pH 7.0. The complexes were mixed in a 1:1 ratio (vol/vol) with increasing concentrations of Mg-ATP (10–250 μM) dissolved in the same buffer. MgATP-dependent dissociation of RLC-reconstituted myosins from labeled F-actin was observed by monitoring the time course of the change in pyrene fluorescence. Measurements were performed at 21°C using a BioLogic (Claix, France) model SFM-20 stopped-flow instrument outfitted with a Berger ball mixer and an FC-8 observation cuvette. Data were collected and digitized using a JASCO 6500 Fluorometer with an estimated dead time of 3.5 ms. Pyrene-F-actin was excited at 347 nm, and emission was monitored at 404 nm using monochromators set at 20-nm bandwidths. Typically, 7–13 stopped-flow records were averaged and fit to a single exponential equation to obtain the apparent rate constant, kobs (s−1), at a given Mg-ATP concentration. Observed myosin dissociation rates were plotted as a function of [Mg-ATP] and fitted to the following hyperbolic equation:
where K1 is the association constant for Mg-ATP to XBs (in M−1), which is the reciprocal of the dissociation constant (1/K1); S is the concentration of Mg-ATP (expressed in M); and k+2 is the maximal observed rate constant at saturating Mg-ATP (expressed in s−1), which reflect the cross-bridge detachment [48].
Actin activated myosin ATPase assays
Actomyosin interaction kinetics was assessed using the F-actin-activated myosin ATPase activity assays. Reconstituted porcine myosin preparations, previously stored 1:1 in glycerol, were precipitated with 13 volumes of ice-cold 2 mM DTT H2O and collected by centrifugation at 2×8,000g for 10 minutes. Myosin pellets were resuspended in 0.4 M KCl, 10 mM MOPS and 1 mM DTT at pH 7.0, and then dialyzed overnight at 4°C against the same buffer. Myosin concentrations were determined using Coomassie Plus protein assay (Pierce, Rockford, IL, USA). Myosin at a final concentration of 1.1 μM was titrated with increasing amounts of rabbit skeletal F-actin (in μM): 0.1, 0.3, 1, 2.5, 5, 7.5, 10 and 20 [27]. The assays were performed in triplicate on 96-well microplates in a 120 μl reaction volume containing 25 mM imidazole, 4 mM MgCl2, 1 mM EGTA and 1 mM DTT at pH 7.0. The final KCl salt concentration was 76.7 mM. The reactions were initiated with the addition of 2.5 mM ATP with mixing in a Jitterbug incubator shaker and allowed to proceed for 20 minutes at 30°C. Termination of the reaction was achieved by addition of ice-cold trichloroacetic acid (TCA) at a final concentration of 4%. Precipitated proteins were cleared by centrifugation, and the inorganic phosphate was determined according to the Fiske and Subbarrow method [49]. Data were analyzed using the Michaelis–Menten equation, yielding Vmax and Km [50].
Skinned porcine papillary muscle fibers
Freshly isolated porcine hearts were placed in oxygenated physiological salt solution of 140 mM NaCl, 4 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 1.8 mM NaH2PO4, 5.5 mM glucose, and 50 mM HEPES buffer at pH 7.4. Papillary muscles were isolated from the left ventricles, dissected into muscle bundles of about 20 × 3 mm, and were chemically skinned in a 50% glycerol, 50% pCa 8 solution (10−8 M [Ca2+], 1 mM [Mg2+], 7 mM EGTA, 5 mM [Mg–ATP2+], 20 mM imidazole, pH 7.0, 15 mM creatine phosphate; ionic strength = 150 mM adjusted with potassium propionate) containing 1% Triton X-100 for 24 h at 4°C. Then, the bundles were transferred to the same solution without Triton X-100 and stored at −20 °C until further use.
CDTA-extraction of endogenous RLC from skinned cardiac muscle strips and reconstitution with human cardiac WT or RLC mutants
Endogenous RLC depletion was achieved in small muscle strips (~100 μm wide and ~1.4 mm long), isolated from glycerinated papillary muscle bundles, in a buffer containing: 5 mM CDTA, 1% Triton X-100, 50 mM KCl, 40 mM Tris, 0.6 mM NaN3, 0.2 mM PMSF at pH 8.4, supplemented with protease inhibitor cocktail. The muscle strips were attached to the arms of a force transducer and incubated in this solution for 5 minutes at room temperature followed by incubation in the fresh solution of the same composition for another 30 minutes. The extent of RLC extraction was determined by analysis of the samples runs on SDS–PAGE. Since depletion of the endogenous RLC may result in partial extraction of the endogenous Troponin C (TnC), cardiac TnC along with the RLC WT or mutants were added back into the CDTA-treated strips. Reconstitution of the RLC-depleted strips with porcine cardiac TnC and RLC-WT or R58Q or S15D-R58Q mutants was performed in muscle strips mounted to the force transducer in pCa 8 solution containing 40 μM RLC of interest and 15 μM TnC at room temperature. The solution of TnC was included in the reconstitution protein mixture during the first 30 minutes of incubation. It was followed by a 30 minutes incubation of muscle strips with fresh 40 μM RLC proteins only. Reconstituted muscle strips were then washed in pCa 8 buffer and subjected to force/pCa and SRX measurements. The extent of RLC extraction and then reconstitution was quantified by densitometry analysis of SDS–PAGE gels.
ATP chase experiments
Nucleotide exchange experiments were performed in: (1) Porcine papillary muscle fibers, depleted and reconstituted with recombinant WT, R58Q, S15D-R58Q RLC proteins and (2) Transgenic R58Q, WT mouse cardiac papillary muscles (S15D-R58Q mice model was not available). Transgenic RLC-WT and RLC-R58Q mice were euthanized using CO2 inhalation, and the hearts were rapidly excised and rinsed in pCa8 solution supplemented with 15% glycerol and 30 mM 2,3-butanedione monoxime (BDM). Intact papillary muscles were dissected from the left ventricle and the fibers were chemically skinned with 1% Triton X-100 and 50/50 (%) pCa 8 and glycerol overnight at −4°C. This removal of membranes allows modulation of the myofilament chemical environment.
The skinned transgenic mouse PM muscle strips were manually dissected into small fiber bundles having a diameter of 80–100 μm. The same diameter was also observed for RLC-depleted and mutant-reconstituted porcine PM used in the mant-ATP experiments. The fibers were washed with pCa 8 solution followed with three washes with basic rigor buffer containing 120 mM KAcetate, 5 mM MgAcetate, 2.5 mM K2HPO4, 2.5 mM KH2PO4, and 50 mM 3-(n-morpholino) propanesulfonic acid, pH 6.8, with fresh 2 mM DTT. Then, the fibers were taped firmly on a glass slide and covered with a cover slip forming a simple flow chamber and were incubated for several minutes with 250 μM mant-ATP (in rigor buffer) until maximum fluorescence was achieved. The chase experiments were performed using Ionoptix Calcium and Contractility Recording System equipped with hyperSwitch dual excitation light source (with sub-millisecond switching times) and inverted fluorescence microscope with MyoCam-S™ digital variable field rate CCD video system. Excitation was achieved via the 340 nm filter with a 12 nm bandpass and emission was recorded by 510 nm filter with a 40 nm bandpass. Subsequently, mant-ATP was chased with 4 mM unlabeled ATP (in rigor buffer) that resulted in a decrease in fluorescence intensity as the hydrolyzed mant-ATP was exchanged for ATP. The sarcomere length of fibers varied from 1.9–2.1 μm and was not found to influence the fluorescence decays. Fluorescence intensity, I, as a function of time, t, was fit using a two-exponential decay function (see Results) with P1 and P2 depicting the amplitudes of fiber fluorescence fractions characterized by the fast T1 and slow T2 fluorescence lifetimes. Fitting was done using a nonlinear least-squares algorithm in Graphpad Prism version 7.
Steady-state force measurements
Porcine papillary muscle strips (~1.4 mm in length and 100 μm in diameter), reconstituted with recombinant RLC proteins, were attached to a force transducer of the Guth muscle research system, placed in a 1 ml cuvette and freshly skinned in 1% Triton X-100 dissolved in pCa 8 buffer for 30 minutes to remove the remaining membrane and ECM (extracellular matrix) proteins. Next, the fibers were washed 3 times for 5 minutes in pCa 8 buffer and their lengths were adjusted to remove the slack. This procedure resulted in sarcomere length of 2.1–2.2 μm as judged by the first order optical diffraction pattern as described in Yuan et al.[12]. Then the maximal steady-state force was measured in pCa 4 solution ([Ca2+] =10−4 M). Maximal tension readings (in pCa 4) were taken before and after the force-pCa curve, averaged and expressed in kN/m2 with the cross-sectional area assumed to be circular. The measurement of fiber diameter was taken at ∼3 points along the fiber length with an SZ6045 Olympus microscope (zoom ratio of 6.3:1, up to ×189 maximum magnification) and averaged. Maximal steady-state force (pCa 4) was determined for all RLC recombinant proteins: WT, R58Q and S15D-R58Q reconstituted in RLC-depleted skinned porcine cardiac muscle strips. All mechanical experiments on glycerinated skinned papillary muscle fibers were carried out at room temperature (22°C).
Ca2+ -dependence of force development
After the initial steady-state force was determined, the muscle strips were relaxed in pCa 8 buffer and were then exposed to solutions of increasing Ca2+ concentration from pCa 8 to pCa 4. The level of force was measured in each “pCa” solution. Data were analyzed using the Hill equation [51], yielding the pCa50 (free Ca2+ concentration which produces 50% of the maximal force) and nH (Hill coefficient). The pCa50 represents the measure of Ca2+ sensitivity of force and the nH is the measure of myofilament cooperativity.
Secondary structure prediction of RLCs
The secondary structure predictions were conducted with the I-TASSER online server from Zhang lab at the University of Michigan (http://zhanglab.ccmb.med.umich.edu/I-TASSER/). In brief, the amino acid sequences of WT, R58Q and S15D R58Q RLCs were compared against template proteins of similar structure chosen from the PDB library. The full-length protein was assembled from excised fragments and simulated into the lowest energy model using specific algorithms. The confidence of each predicted model structure was presented as C-score, ranging from −5 to 2, which was proportional to the quality of prediction [31, 32]. The predicted structures were then modeled using PyMOL molecular visualization system allowing determination of the distances, in Angstroms (Å), between C-α of the neighboring amino acids.
CD Measurements
Circular dichroism (CD) spectroscopy, a form of light absorption spectroscopy that measures the difference in absorbance of right and left circularly polarized light, is very sensitive to the secondary structure of polypeptides and proteins. Far-UV circular dichroism spectra were acquired using a 1-mm path quartz cell in a Jasco J-720 spectropolarimeter. Spectra were recorded at 195–250 nm with a bandwidth of 1 nm at a speed of 50 nm/minutes and a resolution of 0.2 nm. Analysis and processing of data were done using the Jasco system software (Windows Standard Analysis, version 1.20). Ten scans were averaged, and base line buffer optical activities were subtracted with no numerical smoothing. Mean residue ellipticity ([θ]MRE, in degrees·cm2/dmol) for the spectra were calculated (utilizing the same Jasco system software) using the following equation:
[θ] is the measured ellipticity in millidegrees, Cr is the mean residue molar concentration, and l is the path length in cm. The α-helical content for each protein was calculated using the following standard equation for [θ] at 222 nm:
fHis the fraction of α-helical content (fH× 100, expressed in %). The measurements were performed at 22°C in 30 mM NaCl, and 0.3 mM EGTA, 0.7 mM MgCl2, 3 mM Tris-HCl at pH 7.4. Spectra are presented as the mean residue ellipticity.
SDS–PAGE
Preparations of control, CDTA-depleted and WT, R58Q and S15D-R58Q reconstituted porcine cardiac myosins and skinned muscle strips were run on 15% SDS–PAGE. Bands were visualized by Coomassie blue G-250. Respective RLC protein bands were quantified using ImageJ (imagej.nih.gov/ij). RLC depletion and/or reconstitution were quantified from the band intensity of the RLC and ELC proteins according to the equation:
where RLCnat and ELCnat depict the level of RLC and ELC in control untreated muscle strips. ELC protein was used as a loading control.
Statistical analyses
All values are shown as means ± SD (standard deviation) or ± SEM (standard error of the mean). Statistically significant differences between two groups were determined using an unpaired Student’s t-test and comparisons between multiple groups were performed using one-way ANOVA and the Tukey’s multiple comparison test. Extra-sum-of-squares F test was used to compare the goodness-of-fit of nested models for stopped-flow data curves (GraphPad Prism 7). Significance was defined as *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
Acknowledgements
This work was supported by the National Institutes of Health [Grant number HL123255 (DSC)]; and the American Heart Association [Grant number 17PRE33650085 (SY)].
Nonstandard Abbreviations:
- BDM
2,3-butanedione monoxime
- CDTA
(1, 2-cyclohexylenedinitrilotetraacetic acid)
- DRX
disordered relaxed state
- DTT
dithiothreitol
- E2
estradiol
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol tetraacetic acid
- ELC
essential light chain
- HCM
hypertrophic cardiomyopathy
- HF
heart failure
- K1
association constant for MgATP to XBs
- k+2
maximal observed rate constant at saturating MgATP
- kobs
observed rate constant of fluorescence increase in stopped flow experiments
- m1
initial signal (fluorescence intensity at 0 μM myosin)
- m2
maximal amplitude (fluorescence intensity at saturating myosin concentrations)
- β-MHC
β-myosin heavy chain
- MLCK
myosin light chain kinase
- MOPS
(3-(N-morpholino) propanesulfonic acid)
- nH
Hill coefficient
- P1
relative fiber fluorescence fraction in disordered relaxed state
- P2
relative fiber fluorescence fraction in super relaxed state
- Pi
inorganic phosphate
- PIA
pyrene iodoacetamide
- PMSF
phenylmethylsulfonyl fluoride
- RLC
regulatory light chain
- S
Mg-ATP concentration
- SCD
sudden cardiac death
- SRX
super relaxed state
- T1
nucleotide turnover lifetime in disordered relaxed state
- T2
nucleotide turnover lifetime in super relaxed state
- TCA
trichloroacetic acid
- Tg
transgenic
- Tm
tropomyosin
- TnC
troponin C
- TnI
troponin I
- WT
wild-type
- XB
cross-bridge
Footnotes
Conflicts of interest: None
References
- 1.Alcalai R, Seidman JG & Seidman CE (2008) Genetic Basis of Hypertrophic Cardiomyopathy: From Bench to the Clinics, J Cardiovasc Electrophysiol. 19, 104–110. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ (2002) Hypertrophic cardiomyopathy: a systematic review, JAMA. 287, 1308–20. [DOI] [PubMed] [Google Scholar]
- 3.Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, Rayment I, Sellers JR, Fananapazir L & Epstein ND (1996) Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle, Nat Genet. 13, 63–9. [DOI] [PubMed] [Google Scholar]
- 4.Muthu P, Huang W, Kazmierczak K & Szczesna-Cordary D (2012) Functional Consequences of Mutations in the Myosin Regulatory Light Chain Associated with Hypertrophic Cardiomyopathy, In: Veselka J (Ed). Cardiomyopathies – From Basic Research to Clinical Management. Ch. 17. InTech, Croatia, pp 383–408. [Google Scholar]
- 5.Szczesna-Cordary D (2003) Regulatory light chains of striated muscle myosin. Structure, function and malfunction, Curr Drug Targets Cardiovasc Haematol Disord. 3, 187–97. [DOI] [PubMed] [Google Scholar]
- 6.Rayment I, Rypniewski WR, Schmidt-Base K, Smith R, Tomchick DR, Benning MM, Winkelmann DA, Wesenberg G & Holden HM (1993) Three-dimensional structure of myosin subfragment-1: a molecular motor, Science. 261, 50–8. [DOI] [PubMed] [Google Scholar]
- 7.Szczesna D, Ghosh D, Li Q, Gomes AV, Guzman G, Arana C, Zhi G, Stull JT & Potter JD (2001) Familial hypertrophic cardiomyopathy mutations in the regulatory light chains of myosin affect their structure, Ca2+ binding, and phosphorylation, J Biol Chem. 276, 7086–92. [DOI] [PubMed] [Google Scholar]
- 8.Yadav S & Szczesna-Cordary D (2017) Pseudophosphorylation of cardiac myosin regulatory light chain: a promising new tool for treatment of cardiomyopathy, Biophys Rev. 9, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Velden J, Papp Z, Zaremba R, Boontje NM, de Jong JW, Owen VJ, Burton PBJ, Goldmann P, Jaquet K & Stienen GJM (2003) Increased Ca2+-sensitivity of the contractile apparatus in end-stage human heart failure results from altered phosphorylation of contractile proteins, Cardiovasc Res. 57, 37–47. [DOI] [PubMed] [Google Scholar]
- 10.Scruggs SB, Hinken AC, Thawornkaiwong A, Robbins J, Walker LA, de Tombe PP, Geenen DL, Buttrick PM & Solaro RJ (2009) Ablation of ventricular myosin regulatory light chain phosphorylation in mice causes cardiac dysfunction in situ and affects neighboring myofilament protein phosphorylation, J Biol Chem. 284, 5097–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheikh F, Ouyang K, Campbell SG, Lyon RC, Chuang J, Fitzsimons D, Tangney J, Hidalgo CG, Chung CS, Cheng H, Dalton ND, Gu Y, Kasahara H, Ghassemian M, Omens JH, Peterson KL, Granzier HL, Moss RL, McCulloch AD & Chen J (2012) Mouse and computational models link Mlc2v dephosphorylation to altered myosin kinetics in early cardiac disease, J Clin Invest. 122, 1209–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan CC, Muthu P, Kazmierczak K, Liang J, Huang W, Irving TC, Kanashiro-Takeuchi RM, Hare JM & Szczesna-Cordary D (2015) Constitutive phosphorylation of cardiac myosin regulatory light chain prevents development of hypertrophic cardiomyopathy in mice, Proc Natl Acad Sci U S A. 112, E4138–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Shelton JM, Richardson JA, Kamm KE & Stull JT (2008) Myosin regulatory light chain phosphorylation attenuates cardiac hypertrophy, J Biol Chem. 283, 19748–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerrick WGL, Kazmierczak K, Xu Y, Wang Y & Szczesna-Cordary D (2009) Malignant familial hypertrophic cardiomyopathy D166V mutation in the ventricular myosin regulatory light chain causes profound effects in skinned and intact papillary muscle fibers from transgenic mice, FASEB J. 23, 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren SA, Briggs LE, Zeng H, Chuang J, Chang EI, Terada R, Li M, Swanson MS, Lecker SH, Willis MS, Spinale FG, Maupin-Furlowe J, McMullen JR, Moss RL & Kasahara H (2012) Myosin light chain phosphorylation is critical for adaptation to cardiac stress, Circulation. 126, 2575–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toepfer CN, West TG & Ferenczi MA (2016) Revisiting Frank-Starling: regulatory light chain phosphorylation alters the rate of force redevelopment (ktr ) in a length-dependent fashion, J Physiol. 594, 5237–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arimura T, Muchir A, Kuwahara M, Morimoto S, Ishikawa T, Du CK, Zhan DY, Nakao S, Machida N, Tanaka R, Yamane Y, Hayashi T & Kimura A (2018) Overexpression of heart-specific small subunit of myosin light chain phosphatase results in heart failure and conduction disturbance, Am J Physiol Heart Circ Physiol. 314, H1192–H1202. [DOI] [PubMed] [Google Scholar]
- 18.Chang AN, Mahajan P, Knapp S, Barton H, Sweeney HL, Kamm KE & Stull JT (2016) Cardiac myosin light chain is phosphorylated by Ca2+/calmodulin-dependent and -independent kinase activities, Proc Natl Acad Sci U S A. 113, E3824–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marian AJ & Braunwald E (2017) Hypertrophic Cardiomyopathy: Genetics, Pathogenesis, Clinical Manifestations, Diagnosis, and Therapy, Circ Res. 121, 749–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flavigny J, Richard P, Isnard R, Carrier L, Charron P, Bonne G, Forissier JF, Desnos M, Dubourg O, Komajda M, Schwartz K & Hainque B (1998) Identification of two novel mutations in the ventricular regulatory myosin light chain gene (MYL2) associated with familial and classical forms of hypertrophic cardiomyopathy, J Mol Med (Berl). 76, 208–14. [DOI] [PubMed] [Google Scholar]
- 21.Kabaeva ZT, Perrot A, Wolter B, Dietz R, Cardim N, Correia JM, Schulte HD, Aldashev AA, Mirrakhimov MM & Osterziel KJ (2002) Systematic analysis of the regulatory and essential myosin light chain genes: genetic variants and mutations in hypertrophic cardiomyopathy, Eur J Hum Genet. 10, 741–8. [DOI] [PubMed] [Google Scholar]
- 22.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet J-P, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M & EUROGENE Heart Failure Project (2003) Hypertrophic cardiomyopathy: Distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy, Circulation. 107, 2227–2232, and erratum (2004), 109(25), p. 3258 [DOI] [PubMed] [Google Scholar]
- 23.Morner S, Richard P, Kazzam E, Hellman U, Hainque B, Schwartz K & Waldenstrom A (2003) Identification of the genotypes causing hypertrophic cardiomyopathy in northern Sweden, J Mol Cell Cardiol. 35, 841–9. [DOI] [PubMed] [Google Scholar]
- 24.Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E, Ommen SR, Theis JL, Vaubel RA, Re F, Armentano C, Poggesi C, Torricelli F & Cecchi F (2008) Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy, Mayo Clin Proc. 83, 630–8. [DOI] [PubMed] [Google Scholar]
- 25.Abraham TP, Jones M, Kazmierczak K, Liang H-Y, Pinheiro AC, Wagg CS, Lopaschuk GD & Szczesna-Cordary D (2009) Diastolic dysfunction in familial hypertrophic cardiomyopathy transgenic model mice, Cardiovasc Res. 82, 84–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muthu P, Kazmierczak K, Jones M & Szczesna-Cordary D (2012) The effect of myosin RLC phosphorylation in normal and cardiomyopathic mouse hearts, J Cell Mol Med. 16, 911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muthu P, Liang J, Schmidt W, Moore JR & Szczesna-Cordary D (2014) In Vitro Rescue Study of a Malignant Familial Hypertrophic Cardiomyopathy Phenotype by Pseudo-Phosphorylation of Myosin Regulatory Light Chain, Arch Biochem Biophys. 552–553, 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granzier HL & de Tombe PP (2015) Myosin light chain phosphorylation to the rescue, Proc Natl Acad Sci U S A. 112, 9148–9149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hooijman P, Stewart MA & Cooke R (2011) A new state of cardiac Myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart, Biophys J. 100, 1969–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart MA, Franks-Skiba K, Chen S & Cooke R (2010) Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers, Proc Natl Acad Sci U S A. 107, 430–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roy A, Kucukural A & Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction, Nat Protoc. 5, 725–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Yan R, Roy A, Xu D, Poisson J & Zhang Y (2015) The I-TASSER Suite: protein structure and function prediction, Nat Methods. 12, 7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szczesna-Cordary D, Guzman G, Ng SS & Zhao J (2004) Familial hypertrophic cardiomyopathy-linked alterations in Ca2+ binding of human cardiac myosin regulatory light chain affect cardiac muscle contraction, J Biol Chem. 279, 3535–42. [DOI] [PubMed] [Google Scholar]
- 34.McNamara JW, Li A, Smith NJ, Lal S, Graham RM, Kooiker KB, van Dijk SJ, Remedios CGD, Harris SP & Cooke R (2016) Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes, J Mol Cell Cardiol. 94, 65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nag S, Sommese RF, Ujfalusi Z, Combs A, Langer S, Sutton S, Leinwand LA, Geeves MA, Ruppel KM & Spudich JA (2015) Contractility parameters of human beta-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function, Sci Adv. 1, e1500511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Depre C & Vatner SF (2007) Cardioprotection in stunned and hibernating myocardium, Heart Fail Rev. 12, 307–17. [DOI] [PubMed] [Google Scholar]
- 37.Anderson RL, Trivedi DV, Sarkar SS, Henze M, Ma W, Gong H, Rogers CS, Gorham JM, Wong FL, Morck MM, Seidman JG, Ruppel KM, Irving TC, Cooke R, Green EM & Spudich JA (2018) Deciphering the super relaxed state of human beta-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers, Proc Natl Acad Sci U S A. 115, E8143–E8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naber N, Cooke R & Pate E (2011) Slow myosin ATP turnover in the super-relaxed state in tarantula muscle, J Mol Biol. 411, 943–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kampourakis T & Irving M (2015) Phosphorylation of myosin regulatory light chain controls myosin head conformation in cardiac muscle, J Mol Cell Cardiol. 85, 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kampourakis T, Ponnam S & Irving M (2018) Hypertrophic cardiomyopathy mutation R58Q in the myosin regulatory light chain perturbs thick filament-based regulation in cardiac muscle, J Mol Cell Cardiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karabina A, Kazmierczak K, Szczesna-Cordary D & Moore JR (2015) Myosin regulatory light chain phosphorylation enhances cardiac beta-myosin in vitro motility under load, Arch Biochem Biophys. 580, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brenner B, Seebohm B, Tripathi S, Montag J & Kraft T (2014) Familial hypertrophic cardiomyopathy: functional variance among individual cardiomyocytes as a trigger of FHC-phenotype development, Front Physiol. 5, 392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawana M, Sarkar SS, Sutton S, Ruppel KM & Spudich JA (2017) Biophysical properties of human beta-cardiac myosin with converter mutations that cause hypertrophic cardiomyopathy, Sci Adv. 3, e1601959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Yuan CC, Kazmierczak K, Szczesna-Cordary D & Burghardt TP (2018) Single cardiac ventricular myosins are autonomous motors, Open Biol. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robert-Paganin J, Auguin D & Houdusse A (2018) Hypertrophic cardiomyopathy disease results from disparate impairments of cardiac myosin function and auto-inhibition, bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Xu Y, Kerrick WGL, Wang Y, Guzman G, Diaz-Perez Z & Szczesna-Cordary D (2006) Prolonged Ca2+ and force transients in myosin RLC transgenic mouse fibers expressing malignant and benign FHC mutations, J Mol Biol. 361, 286–299. [DOI] [PubMed] [Google Scholar]
- 47.Pant K, Watt J, Greenberg M, Jones M, Szczesna-Cordary D & Moore JR (2009) Removal of the cardiac myosin regulatory light chain increases isometric force production, FASEB J. 23, 3571–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloemink M, Deacon J, Langer S, Vera C, Combs A, Leinwand L & Geeves MA (2014) The hypertrophic cardiomyopathy myosin mutation R453C alters ATP binding and hydrolysis of human cardiac beta-myosin, J Biol Chem. 289, 5158–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fiske CH & Subbarow Y (1925) The colorimetric determination of phosphorus, J Biol Chem. 66, 375–400. [Google Scholar]
- 50.Hanson KR, Ling R & Havir E (1967) A computer program for fitting data to the Michaelis-Menten equation, Biochem Biophys Res Commun. 29, 194–197. [DOI] [PubMed] [Google Scholar]
- 51.Hill TL, Einsenberg E & Greene LE (1980) Theoretical model for the cooperative equilibrium binding of myosin subfragment-1 to the actin-troponin-tropomyosin complex., Proc Natl Acad Sci. 77 3186–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]