Abstract
The Epilepsy Bioinformatics Study for Antiepileptogenic Therapy (EpiBioS4Rx) is an international, multicenter, multidisciplinary study aimed at preventing epileptogenesis (https://epibios.loni.usc.edu/). One of the study’s major objectives is the discovery of diagnostic, prognostic, and predictive plasma protein and microRNA (miRNA) biomarkers that are sensitive, specific, and translatable to the human condition. Epilepsy due to structural brain abnormalities, secondary to neurological insults such as traumatic brain injury (TBI), currently represents ~50% of all epilepsy cases. In the preclinical EpiBioS4Rx study, TBI was induced in adult male Sprague Dawley rats using a standardized protocol for lateral fluid-percussion injury. Whole blood was collected from the tail vein at baseline and 2, 9 and 30 days post-injury and processed for plasma separation. Biomaterial properties, sample preparation and integrity, and choice of analysis platform can significantly impact measured marker levels and, in turn, interpretation with respect to injury and/or other variables. We present here the results of procedural harmonization for the first 320 rats included in the EpiBioS4Rx study, from three international research centers, and preliminary proteomic and miRNA analyses. We also discuss experimental considerations for establishing rigorous quality controls with the goal of harmonizing operating procedures across study sites, and delivering high-quality specimens for preclinical biomarker discovery in a rat model of post-traumatic epilepsy (PTE).
Keywords: Case report form, Common data elements, Post-traumatic epilepsy, Quality control, Standardization, Traumatic brain injury
1. Introduction
Blood-based protein and microRNA (miRNA) biomarkers are noninvasive, cost-effective, and high throughput. However, reproducibility issues in biochemical assays continue to plague preclinical and clinical research, thereby hindering translation and implementation. Harmonization of various pre-analytical procedures (e.g., anesthesia, sample processing, storage), particularly in a multicenter study, is of the utmost importance because these, at least in part, relate to the miRNA and plasma protein biology itself (Mitchell et al., 2008; Grasedieck et al., 2012; Ge et al., 2014; Fortunato et al., 2014; van Vliet et al., 2017). Furthermore, without rigorous protocol standardization, validation, and state-of-the-art data analysis, it will be problematic to compare samples originating from different sites, let alone facilitate translation into clinical practice.
The first step in establishing a plasma protein and miRNA biomarker discovery pipeline is to establish methodologies for blood sampling and processing, as previously described by van Vliet et al. (2017) and detailed in this report. Our main objective was to harmonize protocols and procedures across three international study sites: the University of Eastern Finland (UEF; Kuopio, Finland), the University of Melbourne/Monash University (Melbourne, Australia), and the University of California, Los Angeles (UCLA; USA), and to provide an objective framework for plasma quality control using a two-pronged approach (spectrophotometric and antibody-based). We also completed pilot proteomic and miRNA studies to assess whether the degree of sample hemolysis can affect protein biomarker levels measured by reverse phase protein microarray (RPPM), and the expression of red blood cell (RBC) and platelet miRNA markers in low hemolysis samples.
2. Materials and Methods
2.1. Blood Collection and Processing
Animal care and use programs for the three EpiBioS4Rx study sites were approved prior to study commencement. For studies conducted at UEF, animal procedures were approved by the Animal Ethics Committee of the Provincial Government of Southern Finland and carried out in accordance with the guidelines of the European Community Council Directives 2010/63/EU. In Melbourne, animal procedures were approved by the Florey Animal Ethics Committee (ethics number 17–013 UM). At UCLA, all study protocols were approved by the University of California, Los Angeles Chancellor’s Animal Research Committee, which adheres to the Public Health Service Policy on Humane Care and Use of Laboratory Animals.
2.1.1. Data collection and recordkeeping.
The standardization of data collection across EpiBioS4Rx study sites is a principal component of harmonization and a prerequisite for accurate data analysis. The first effort in this direction was produced by the EPITARGET Consortium (https://www.epitarget.eu/cdes/). Common data elements were generated for all procedures using universal definitions and standardized case report forms, as recommended by the International League Against Epilepsy (ILAE) and EPITARGET (Harte-Hargrove et al., 2018; Lapinlampi et al., 2017). A template for blood sampling, plasma protein and RNA analysis can be found in Supplement 1.
2.1.2. Plasma preparation.
As of May 2018, a total of 320 adult male Sprague Dawley rats (UEF: 143, Melbourne: 97, and UCLA: 80) were randomized into the TBI group or sham-operated experimental control group (see Ekolle Ndode-Ekane et al. in this issue). Rats in each treatment group (UEF: 121 TBI, 22 sham; Melbourne: 78 TBI, 19 sham; UCLA: 63 TBI, 17 sham) were processed in subcohorts of about 10–15 animals each, and blood was sampled at 4 time points (baseline, days 2, 9, and 30 post-injury). Prior to venipuncture, rats were anesthetized with 5% isoflurane in an induction chamber, moved to the table, and kept unrestrained. Anesthesia was maintained through a nose mask (2% isoflurane). Blood was collected from the lateral tail vein in the distal one-third of the tail (https://www.nc3rs.org.uk/rat-tail-vein-non-surgical). If needed, the tail was warmed to dilate the blood vessels. This was done by immersing the tail in warm water at UEF and Melbourne. At UCLA, anesthetized rats were kept on a heating pad and the tail was placed in warm water for a few seconds prior to the blood draw. While extending the tail, a 23G butterfly needle (#367284, BD Vacutainer, BD Biosciences, Franklin Lakes, NJ) was inserted into the vein and blood was collected into a 0.5 mL K2EDTA tube (#365975, BD Microtainer, BD Biosciences) at room temperature (RT). Tubes were filled until the 0.5 mL line mark on the tube. Blood and anticoagulant were immediately mixed by inverting the tube 10 times to 180o and back. Sample tubes were then promptly centrifuged at 1300g (Centrifuge 5417R for UEF and UCLA, 5415R for Melbourne, Eppendorf Biotools, CA) for 10 min at +4°C to separate plasma from blood. After centrifugation, samples were placed on ice for initial quality control (QC) and aliquoting (Fig. 1).
Fig. 1.
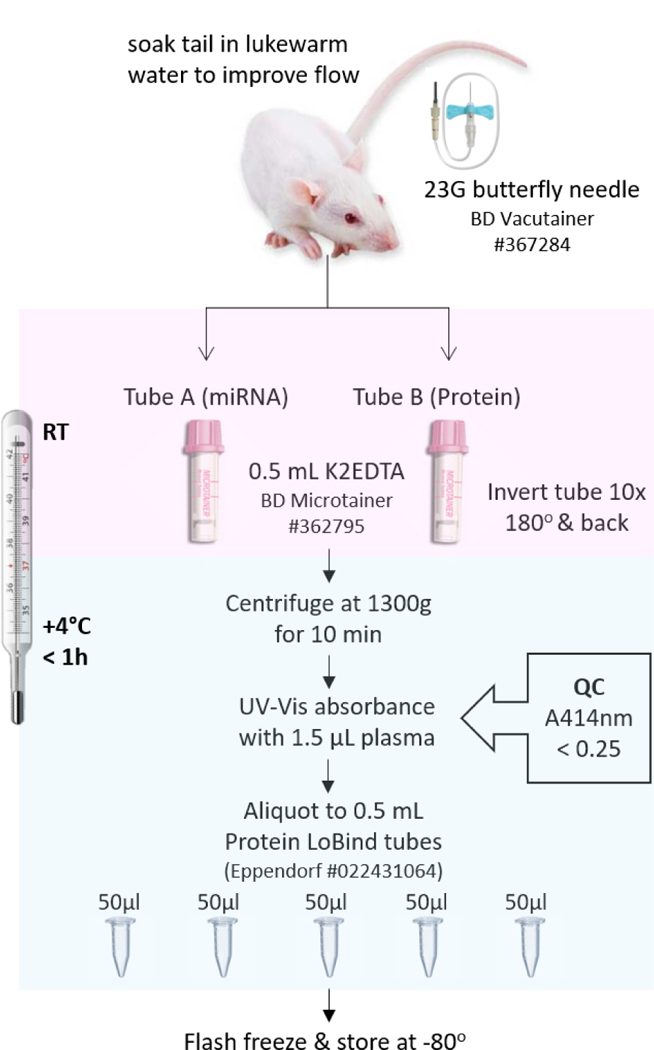
Overview of blood collection and plasma processing for proteomic and miRNA studies. Samples were kept at +4˚C for the duration of plasma processing including centrifuging, quality control, and aliquoting. Abbreviations: A414 nm, spectrophotometric absorbance at 414 nm; G, gauge; h, hour; K2EDTA, dipotassium ethylenediaminetetraacetic acid; min, minutes; QC, quality control; RT, room temperature.
2.1.3. Primary plasma QC (hemoglobin absorbance).
Following centrifugation (Fig. 1), the supernatant (i.e., plasma) within the sampling tube was assessed for degree of hemolysis by measuring UV-Vis absorbance of hemoglobin at 414 nm (ND-1000, NanoDrop™, Thermo Fisher Scientific, Wilmington, DE). Plasma samples with absorbance values greater than 0.25 were considered unsuitable for further protein/miRNA evaluations (van Vliet et al., 2017). Samples were then aliquoted into Protein LoBind microcentrifuge tubes (#022431064, Eppendorf®), flash-frozen in dry ice, and stored at −80˚C until further use. All blood collection and plasma storage tubes were pre-labelled and QR-coded (Label Printer #BBP33, Brady Inc., Milwaukee, WI; Code Reader™ #CR1421, Code, Draper, UT) on the top and side for easier handling of large sample numbers.
2.1.4. Tube labelling and barcoding.
Labelling and barcoding procedures were harmonized across all sites using the following convention: Site ID+Rat ID+timepoint+TubeA/B+Aliquot number. In our case, site IDs were: 1 = University of Eastern Finland, 2 = Monash University, Melbourne, and 3 = UCLA. Rat IDs were comprised of unique three-digit numbers, and sampling time points covered blood collections before and after injury as follows: 00 = Baseline, 02 = 2 days post-injury, 09 = 9 days, 01 = 1 month. The A/B blood collection tube designation helped in purposing the samples to specific experiments (i.e., A = for miRNA analysis and B = for protein analysis) and in tracing back any downstream quality issues to a single tube. The final barcode/label included the aliquot number for respective A/B tubes.
2.1.5. Secondary plasma QC (hemoglobin concentration).
Pre-clinical plasma samples were quality controlled using the same hemoglobin variant, hemoglobin subunit alpha 1, used to QC clinical samples at the Biospecimen Exchange for Neurological Disorders (BioSEND; Indiana University, Bloomington, IN, USA) (Vespa et al., 2018). Secondary QC of rat plasma samples was conducted at the Uniformed Services University (USU; Bethesda, MD, USA) using RPPM as described in detail (section 2.2). Briefly, hemoglobin α levels in each sample were identified through specific antibody-antigen interactions. Binding of the primary antibody is detected using near-infrared fluorescent imaging and the signal is quantified and expressed as a measure of protein concentration (Fig. 4).
Fig. 4.
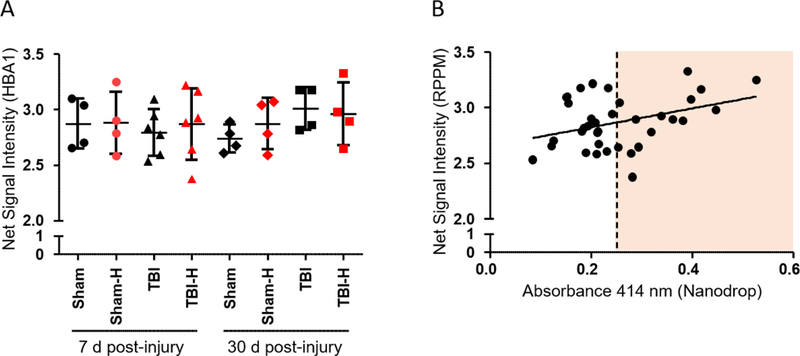
Hemoglobin α levels in duplicate low-hemolysis (black) vs. high-hemolysis (red) plasma samples. (A) Net signal intensity values represent hemoglobin α concentrations measured in sham and TBI rat samples using RPPM; data are presented as the mean ± SD. (B) Correlation between hemoglobin absorbance values measured at 414 nm and HBA1 antibody-based net signal intensity values in the same samples (Pearson Correlation: r = 0.328, p = 0.051). Abbreviations: d, days; H, high hemoglobin samples based on Nanodrop measurements; HBA1, hemoglobin subunit alpha 1; RPPM, reverse phase protein microarray; SD, standard deviation; TBI, traumatic brain injury.
2.1.6. Pilot plasma biomarker and miRNA studies (UEF rats only).
We completed preliminary experiments to investigate the effect of sample quality (i.e., the degree of hemolysis) on putative protein biomarkers measured by RPPM and miRNA markers measured by polymerase chain reaction (PCR). The plasma used for RPPM and PCR was obtained from a pilot cohort of UEF rats, both TBI and sham-operated controls. Details pertaining to injury and sham conditions can be found in Ekolle Ndode-Ekane et al. in this issue and as previously described (Kharatishvili et al., 2006; Huusko et al., 2013).
For proteomic analysis, we used duplicate (i.e., from the same rat) low-hemolysis and high-hemolysis plasma samples collected 7 days (TBI = 6, sham = 4) and 30 days (TBI = 4, sham = 4) after lateral fluid-percussion injury (FPI) or surgery only (sham-operated experimental controls). Rats were anesthetized with 5% isoflurane and decapitated. Trunk blood was collected into K2EDTA tubes (2 mL Vacutainer, BD Biosciences) and plasma was isolated as described in section 2.1.2.; samples were stored in 100 µL aliquots at −70°C until further processing. For miRNA analysis, we used serial tail vein samples (TBI = 4, sham = 3) collected at baseline, 2 days, 7 days, and 3 months post-injury or sham. Plasma was obtained as depicted in Fig. 1. However, we visually selected hemolysis-free plasma samples, as opposed to measuring hemoglobin absorbance at 414 nm, to further investigate trends in RBC contamination of plasma obtained using a standardized protocol.
2.2. Proteomics
2.2.1. Reverse Phase Protein Microarray (RPPM).
Sample preparation for RPPM analysis was performed as previously described in detail (Gyorgy et al., 2010; Kwon et al., 2011). Briefly, frozen plasma samples were thawed on ice and total protein concentrations were measured using a bicinchoninic acid assay (PI-23221, Thermo Scientific). Protein concentrations were standardized across all samples to a final concentration of 10 mg/mL and a total volume of 200 µL. Specifically, 50 µL of 4X SDS Sample Buffer (35% Glycerol, 8% SDS, 1X TBS, 1X Bond-Breaker TCEP Solution, 1X HALT, 0.0035% NaN3), 50 µL of 2X PBS, and T-per (typically 8–12 mg/mL for serum/plasma) were added to each sample to reach the specified concentration/volume. Samples were then denatured at 70°C for 10 min and transferred to a JANUS Varispan Integrator and Expanded Platform Workstation (PerkinElmer, Waltham, MA) to perform 8-step 1:2 serial dilutions using Dilution Buffer (3 parts Lysis Buffer and 1 part 4× SDS Sample Buffer) in 384-well microarray plates (#X7022, Molecular Devices, Sunnyvale, CA). The microarray plates were subsequently transferred into an Aushon 2470 Arrayer (Aushon Biosystems, Billerica, MA) where samples were printed (in duplicate) onto ONCYTE® AVID nitrocellulose film slides (#305177, Grace Bio-Labs, Bend, OR). The Aushon was set up with 16 pins and programmed for 1 deposition per spot. The spot diameter was set to 250 µm with spacing between dots at 500 µm on the x-axis and 560 µm on the y-axis. Wash time was set to 2 s without delays.
2.2.2. Immunochemical detection.
After overnight desiccation at 4°C, the printed slides were rehydrated with three 1X TBS washes, air dried, and then blocked with a solution of 1X Azure Protein-Free Blocking Buffer (1-part 5X Azure Protein-Free Blocking Buffer (#AC2112, Azure Biosystems, Dublin, CA) and 4 parts deionized water). After blocking, slides were washed three times with 1X TBST and once with 1X TBS and then incubated overnight at 4°C with the primary antibody solutions and a cover slip (#25×60I-M-5439–001-LS, mSeries LifterSlip; Thermo Fisher Scientific, Waltham, MA). The primary antibodies were diluted in 1X Azure Protein-Free Blocking Buffer as follows: hemoglobin subunit alpha 1:150 (HBA1; Abcam, ab92492), glial fibrillary acidic protein 1:1000 (GFAP; Abcam, ab7260), microtubule-associated protein tau 1:100 (Tau; Cell Signaling, 4019), phosphorylated tau (Ser202) 1:20 (p-Tau; Cell Signaling, 11834), aquaporin-4 1:20 (AQP4; Abcam, ab46182), 4-hydroxynonenal Michael Adducts 1:100 (HNE; EMD Millipore, 393207), cytosolic phospholipase-A2 1:50 (cPLA2; R&D Systems, AF6659), matrix metalloproteinase-9 1:500 (MMP9; Abcam, ab38898), and fibrinogen 1:250 (FGB; Santa Cruz, sc69775). All primary antibodies were pretested for specificity by western blot.
The following day, slides were washed three times with 1X TBST and once with 1X TBS and then incubated with the appropriate secondary antibody solution for 1 h at RT. Secondary antibodies, Goat anti-Rabbit IgG (H&L) Superclonal™ Alexa Fluor® 790 (#A27041) and Goat anti-Mouse IgG (H&L) Superclonal™ Alexa Fluor® 790 (#A28182) from Thermo Fisher, were diluted 1:10,000 with 1X Azure Protein-Free Blocking Buffer. After three washes each with 1X TBST and 1X TBS, the slides were air dried and subsequently scanned in an InnoScan 710-IR microarray scanner (Innopsys, Carbonne, France).
2.2.3. Data acquisition and analysis.
Scanner fluorescence data were imported into a Microsoft Excel-based bioinformatics program. After normalizing net signal intensities to respective secondary antibodies to correct for local background noise, points indiscernible from background were excluded (SNR < 2, Net Fluorescence < 5). Net fluorescence values for each sample (8 dilutions printed in triplicate per block) were used to calculate the first and third quartiles and interquartile range; outliers above and below 1.5 of the interquartile range were excluded. Triplicate net fluorescence values for each individual block were then averaged and categorized according to experimental ID and group.
2.3. miRNA
Total RNA extraction from plasma samples was performed using the Exiqon miRCURY™ RNA Isolation Kit for biofluids (#300112, Exiqon, Vedbæk, Denmark). We used 50 µL of plasma as the starting sample and added nuclease-free water (#AM9937, Ambion, Life Technologies, Carlsbad, CA) for a final volume of 200 µL. At the end of the protocol, RNA samples were eluted in 25 µL of nuclease-free water. Next, the TaqMan® small RNA assay (TaqMan® MicroRNA Reverse Transcription Kit, #4366596, Applied Biosystems, Foster City, CA) was used to perform miRNA-specific reverse transcription (RT). The cDNA synthesis reaction master mix was designed according to protocol [https://tools.thermofisher.com/content/sfs/manuals/cms_042167.pdf]. The 15 µL RT reaction was then prepared by combining 7 µL of the master mix with 3 µL of 5X miRNA-specific RT primer (miR-23a, miR-451, miR-425; Applied Biosystems) and 5 µL of the undiluted template RNA. The reaction components were thoroughly mixed and loaded into the thermal cycler (Bio-Rad T100™ thermal cycler, CA). Components of the following qPCR setup included: miRNA-specific 20X PCR primers (Applied Biosystems), undiluted cDNA template obtained from the TaqMan RT reactions, and TaqMan Universal PCR Master Mix (2X) with UNG (#4440038, Applied Biosystems). In qPCR, each sample was run in duplicate with suitable no-template controls (NTCs). Thermal cycling conditions were performed according to the StepOnePlusTM Real-Time PCR System guidelines, and the results were analyzed with StepOne Software v2.1 (Applied Biosystems).
2.4. Statistical analysis
All data were analyzed using IBM SPSS Statistics software 23. Tests were two-tailed with α = 0.05.
Site comparisons.
Study site differences in harmonization parameters were assessed using the Kruskal-Wallis test, followed by the Mann-Whitney U test for all pairwise comparisons.
Protein biomarker analysis.
Correlation between hemoglobin absorbance values and hemoglobin concentration (RPPM) was assessed using Pearson Correlation. To meet the assumption of normality, data was natural log-transformed and visually inspected using frequency histograms in relation to the normal curve prior to analysis. Biomarker concentrations in duplicate low- and high-hemolysis plasma samples (i.e., from the same rat) were compared using a Wilcoxon test for paired samples.
MicroRNA analysis.
Differences in hemolysis between groups and time points were evaluated using the Kruskal-Wallis test. Correlation between Δ (miR-23a - miR-451) and miR-425 was assessed using Pearson correlation.
3. Results
3.1. Harmonization of blood collection and processing procedures
We selected several quantitative parameters to compare the degree of harmonization across study sites. These include consistency in the timing of blood collections and administration of anesthesia, plasma quality as determined by spectrophotometric absorbance, and the impact of personnel training (i.e., a learning effect) on the yield and quality of collected plasma.
Sampling time.
We found some variation in the timing of blood collections across sites (Fig. 2A). At UEF, blood collections were about equally divided between 2 intervals, 6:00–9:59 and 10:00–13:59, while all blood collections at UCLA were performed between 10:00 and 13:59. Most blood collections at Melbourne took place between 6:00 and 9:59 with some at 2 additional time points. Despite greater variation in scheduling at UEF and Melbourne, all blood samples were collected during animals’ active phase. Rats are housed on a 12-h light/12-h dark cycle at the three study sites; lights on from 07:00 to 19:00 at UEF and Melbourne, and from 06:00 to 18:00 at UCLA.
Fig. 2.
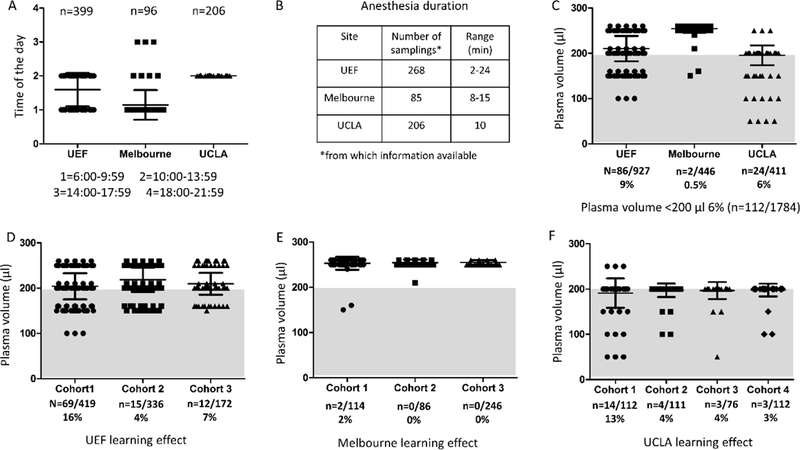
Parameters used to assess harmonization between study sites. (A) Time of day when blood samples were drawn. UCLA uses a standard time (i.e., midday) to collect all blood samples. More variation was observed at UEF and Melbourne. (B) Anesthesia duration during blood sampling. (C) Plasma volume obtained per draw at each study sites. Overall, 6% of samples (n = 112/1784) collected so far have less than 200 µL of stored plasma. The number of samples falling in that category varied between sites as it was 9% of all samples in UEF, 0.5% in Melbourne, and 6% in UCLA. (D-F) Learning effect in the plasma volumes collected at each site (D, UEF; E, Melbourne; F, UCLA). All sites showed improvement in obtaining > 200 µL of plasma in later cohorts. Abbreviations: SD, standard deviation; UEF, University of Eastern Finland; UCLA, University of California, Los Angeles.
Anesthesia duration.
The length of anesthesia during blood sampling was approximately 10 minutes, on average, at each study site (Fig. 2B). However, these values ranged from 2–24 min (median: 5 min) at UEF and 8–15 min at Melbourne. At UCLA, the induction of anesthesia was approximately 5 min after which blood was drawn (an additional 5 min). Anesthesia was terminated at the 10-min mark in all blood collections at UCLA, complete or otherwise.
Absorbance spectrophotometry.
A comparison of hemoglobin absorbance values at 414 nm was also conducted across the three sites (Fig. 3). The average absorbance value for collected plasma was 0.14 ± 0.06 in UEF, 0.13 ± 0.06 in Melbourne, and 0.20 ± 0.12 in UCLA. Degree of hemolysis, as indicated by Nanodrop absorbance values and visual inspection, was set as follows: A414 nm < 0.25 for low hemolysis samples and A414 nm ≥ 0.25 for high hemolysis samples. Based on these criteria, 4% (38/916), 3% (13/447), and 16% (66/409) of samples were above the set threshold in UEF, Melbourne, and UCLA, respectively. Overall, the plasma samples from UCLA had higher absorbance values as compared to UEF or Melbourne (p < 0.001, Mann-Whitney U). Since blood samples were obtained at 4 distinct time points (baseline, 2, 9, and 30 days), we compared absorbance values at the different time points. We found a small, albeit significant increase in A414 nm values at the 30-day time point compared to all other time points (p < 0.05). Accordingly, 9% of samples were disqualified at this time point compared to 6–7% at all other time points.
Fig. 3.
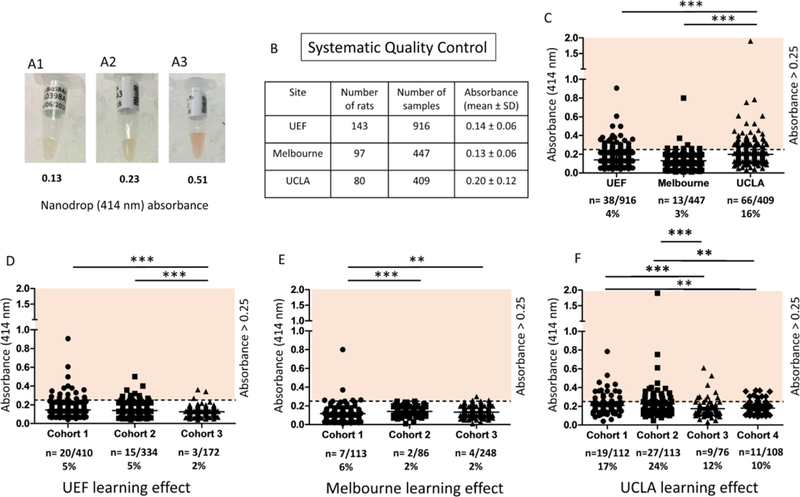
Systematic quality control of rat plasma using Nanodrop spectrophotometer measured hemoglobin absorbance. (A1–3) Representative examples of rat plasma. For each sample, absorbance was measured by UV-Vis module at 414 nm wavelength. (A1) Nanodrop measured absorbance was 0.13 representing good plasma quality. (A2) Nanodrop measured absorbance was 0.23 bordering on upper limit of inclusion criteria (< 0.25). (A3) Nanodrop measured absorbance was 0.51 representing hemolysis in the sample, also visible by reddish color. (B) A table showing the number of rats (UEF n = 143, Melbourne n = 97, and UCLA n = 80) and samples (UEF n = 916, Melbourne n = 447, and UCLA n = 409) included in the EpiBioS4Rx Multicenter Study as of May 2018. (C) A comparison of the three study sites where hemoglobin absorbance was assessed. The percentage of high hemolysis samples (absorbance ≥ 0.25) per site is indicated below the name. (D) The learning effect in UEF cohorts 1–3. (E) The learning effect in Melbourne cohorts 1–3. (F) The learning effect in UCLA cohorts 1–4. Statistically significant differences (Mann-Whitney U) are indicated as follows: **p < 0.01 and ***p < 0.001. Abbreviations: SD, standard deviation; UEF, University of Eastern Finland; UCLA, University of California, Los Angeles.
Learning effect.
Blood sampling and plasma aliquoting at each center were performed as listed in Table 1. While inter-person variation would have been interesting to explore, most samples at UEF were processed by the same individual. As more data is collected, the learning effect between individuals at each study site will also be compared. Nonetheless, we assessed the learning effect in plasma collection and processing between different animal cohorts at each site and found that the volume of plasma collected improved over time (Fig. 2D–F), such that all sites showed improvement in obtaining > 200 µL of plasma in later cohorts. Interestingly, when comparing plasma volumes obtained at the different time points, the 30-day time point had, on average, 9 µL less of plasma than the 2-day time point (p < 0.01).
Table 1.
Venipuncture and blood processing at each study site.
| UEF | Melbourne | UCLA | |
|---|---|---|---|
| Blood draw | Person 1: 90% Person 2: 10% Person 3: once |
Person 1: 50% Person 2: 30% Person 3: 20% |
Person 1: 40% Person 2: 40% Person 3: 20% |
| Aliquoting of plasma | Person 1: 95% Person 2: 2.5% Person 3: 2.5% |
Person 1: 25% Person 2: 25% Person 3: 25% Person 4: 25% |
Person 1: 60% Person 1: 60% Person 3: 20% |
Consistent with the learning effect in plasma volume, we also found significant improvement in plasma quality at the three study sites (Fig. 3D–E). At UEF, cohort 3 had statistically lower absorbance values as compared to cohorts 1 and 2 (p < 0.001, Mann-Whitney U). Melbourne cohorts 2 and 3 had statistically lower absorbance values compared to cohort 1 (p < 0.001 and p < 0.01, respectively; Mann-Whitney U). Similarly, UCLA cohorts 3 and 4 had statistically lower absorbance values compared to cohort 1 (p < 0.001 and p < 0.01, respectively; Mann-Whitney U) and to cohort 2 (p < 0.001 and p < 0.01, respectively; Mann-Whitney U).
3.2. Proteomic analysis
Increased hemolysis of RBCs may interfere with antibody-based analysis and affect plasma protein concentrations. Using duplicate low- and high-hemolysis plasma samples from 18 UEF rats, we measured hemoglobin α concentrations using RPPM (Fig. 4A). We found that high-hemolysis samples generally showed greater variance in hemoglobin concentrations compared to low-hemolysis samples, but there was no clear correlation between hemoglobin absorbance values obtained at UEF and hemoglobin α levels measured by RPPM at USU (Fig. 4B). Using previously tested protein markers, we found that hemolysis did not increase the plasma levels of any proteins included in the analysis (Fig. 5). However, higher hemoglobin α levels resulted in significantly lower p-Tau values in one high hemolysis sample (p < 0.05, paired sample comparison). Importantly, in the absence of baseline or naïve plasma samples (and a higher n per group) in our pilot study, we are unable to rule out the effect of TBI and/or sham surgery on protein marker levels relative to hemoglobin concentrations.
Fig. 5.
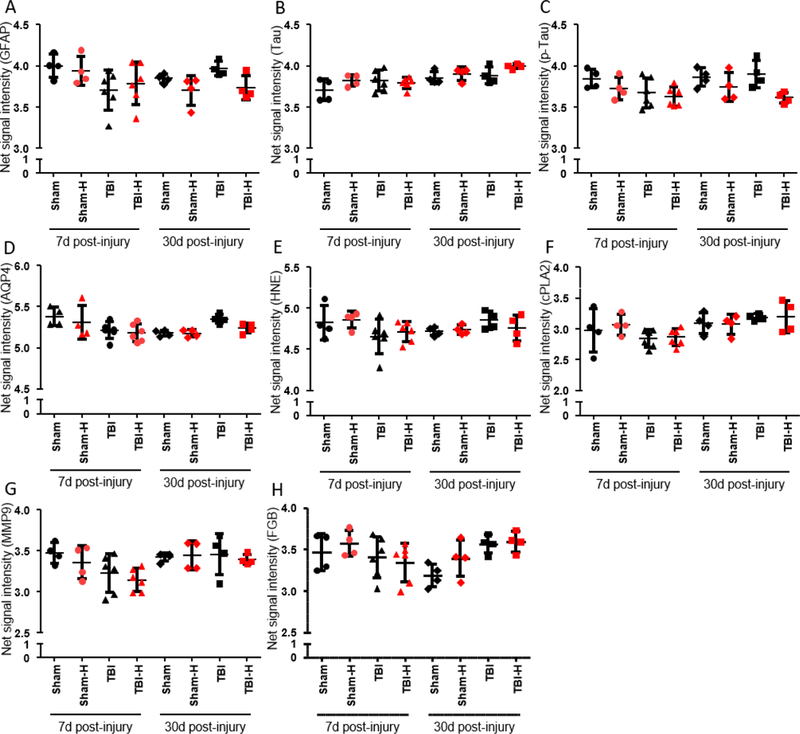
Plasma levels of select proteins measured in duplicate low-hemolysis (black) vs. high-hemolysis (red) samples collected at 7 and 30 days post-TBI or sham. Hemolysis did not appear to increase the levels of any tested marker. However, the hemoglobin content of high-hemolysis plasma samples may have interfered with protein measurements, resulting in lower values in high-hemolysis samples relative to their low-hemolysis counterparts. (A) Glial fibrillary acidic protein (GFAP) is an intermediate filament with diverse functions in the CNS, such as cytostructural integrity. (B) Tau is associated with microtubules in the nervous system. (C) Hyperphosphorylation of Tau (pTau) results in the protein’s dissociation from microtubules, compromising structure and, in turn, function. P-Tau values were significantly lower in one high-hemolysis sample (p < 0.05, Wilcoxon test). (D) Aquaporin-4 (AQP4) is a water-selective channel protein predominantly found in the brain. Under physiological and pathological conditions, AQP4 plays an important role in water homeostasis. (E) 4-Hydroxynonenal (HNE) is a byproduct of lipid peroxidation, indicative of oxidative stress. (F) Cytosolic phospholipase-A2 (cPLA2) becomes catalytically active in the presence of free Ca2+ as seen in stimulated cells; cPLA2 hydrolyzes cellular phospholipids whose breakdown products contribute to inflammation and degeneration. (G) Matrix metalloproteinase-9 (MMP9) plays an essential role in local proteolysis of the extracellular matrix. (H) Fibrinogen (FGB) was the first hemostatic factor to be described as a target for miRNAs (Fort et al., 2010). FGB levels increase in the acute phase of the injury and in inflammatory states. Data are presented as the mean ± SD. Abbreviations: d, days; H, high hemoglobin samples based on Nanodrop measurements; SD, standard deviation; TBI, traumatic brain injury.
3.3. miRNA Analysis
We performed a qPCR experiment using visually-inspected, hemolysis-free plasma samples to investigate the effect of RBC and platelet contamination on miRNA markers. We found that all plasma samples from both groups had RBC-enriched miR-23a and miR-451 levels below the recommended cut-off limit of 5 (Fig. 6A); there were no statistically significant differences in plasma hemolysis levels between sham and TBI rats. However, we found a correlation between Δ (miR-23a - miR-451), a hemolysis marker, and miR-425, a platelet marker, indicating that even in visibly good quality samples RBC and platelet contamination go hand in hand (Fig. 6B; r = −0.510, p < 0.01).
Fig. 6.
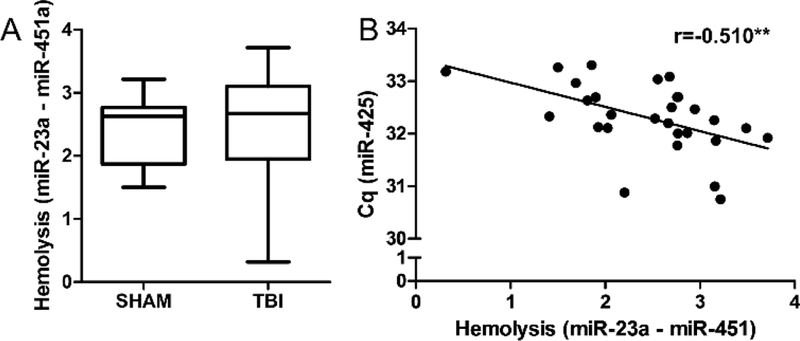
MicroRNA hemolysis marker correlates with platelet contamination in low-hemolysis plasma samples. (A) Differences in plasma miRNAs miR-23a and miR-451 indicate low hemolysis (Δ < 5). There were no statistically significant differences in the level of hemolysis between sham (n = 3) and TBI rats (n = 4) at any of the measured time points (baseline, 2 d, 7 d, and 3 months post-TBI); data from the different time points were pooled after analysis as shown in panel A. (B) Pearson’s correlation indicated that the higher the ratio of miR-451a to miR-23a, the higher the miR-425 content in tested samples (r = - 0.541, **p < 0.01). A lower cycle value (Cq, y-axis) in the PCR analysis represents a higher expression level. Abbreviations: Cq, quantification cycle, r = Pearson correlation coefficient; TBI, traumatic brain injury. Whiskers in panel A represent min and max values.
4. Discussion
4.1. Harmonization of blood collection and processing procedures
Blood collection and processing procedures described here were aimed at minimizing preanalytical variation to ensure the highest quality of biomaterial for preclinical biomarker analysis. In preparing our SOPs and this interim analysis, we considered all experimental details: from choice of anticoagulant to wet lab techniques. While EDTA and citrate are both acceptable anticoagulants for downstream molecular techniques, such as qRT-PCR, heparin significantly inhibits subsequent PCR (Kroh et al., 2010). A standard needle gauge was selected for venipuncture to minimize hemolysis; smaller bore needles (i.e., higher gauge number) may impact the turbulence (stress) for RBCs as they are collected. In contrast, using too large a needle may damage the vein and increase the flow rate of blood, creating turbulence within the needle and the collection tube. Proper pipetting is also critical for preventing contamination of the supernatant (plasma) by the cellular pellet (Kroh et al., 2010).
To that effect, we found that practice significantly improved technique, resulting in a higher volume of plasma following centrifugation and improved sample quality (lower A414 nm values). Training led to more precise venipuncture and faster blood draws, which resulted in less coagulation in the catheter line and tube (i.e., blood is mixed with EDTA sooner), greater plasma yield, and easier pipetting. Finally, consistency in the scheduling of blood collections, to target biomarker discovery from the point of impact to the evolution of the secondary injury process, is particularly important for the acute, 2-day post-injury time point. Furthermore, by standardizing the time of day blood is sampled at each site, we aimed to reduce diurnal variation in tested biomarker levels.
Blood plasma was selected for protein and miRNA biomarker discovery for several reasons. Unlike serum, plasma contains all components of the blood soluble phase (excluding cells and platelets) as well as proteins representing various physiological and pathological processes, which can be of predominantly peripheral rather than CNS origin (Anderson and Anderson, 2002). In the context of biomarker discovery, the complexity of the plasma proteome has its advantages and disadvantages. Advantages include the diversity of proteins, reflecting both health and disease conditions, ease of collection, and increased sample volume at each timepoint. Disadvantages include the very low abundance of diagnostically important proteins relative to albumin, which constitutes approximately 55% of total protein content (Anderson and Anderson, 2002; Zaias et al., 2009). While serum is ideal for both protein and miRNA analysis, the coagulation cascade (in serum preparation) has been found to affect the spectrum of extracellular miRNAs measured in blood (Teruel-Montoya et al., 2015; Wang et al., 2012). Nonetheless, hemolysis (i.e., the breakdown of erythrocytes and release of hemoglobin and other intracellular components) can also impact the quality of plasma and interfere with miRNA analysis and antibody-based proteomics (Sowemimo-Coker, 2002).
Spectrophotometry provides a quick and easily reproducible means to QC samples prior to freezing. However, it is important to note that the optical absorption of hemoglobin differs if the molecule is bound to oxygen (oxyhemoglobin) or is unbound (deoxyhemoglobin)(Prahl, 1999). Similarly, absorption varies if hemoglobin is in the methemoglobin state (i.e., when Fe2+ is oxidized to Fe3+) and is unable to bind oxygen (Prahl, 1999). Oxyhemoglobin exhibits absorbance peaks at 414 nm, 541 nm, and 576 nm, whereas deoxyhemoglobin absorbance peaks at 431 nm (Kirschner et al., 2011). Therefore, absorbance at the wavelength used in our study (414 nm) is associated with free oxyhemoglobin (Kirschner et al., 2011; Shah et al., 2016), with the caveat that hemoglobin absorbance levels in plasma may be affected by albumin, bilirubin, and other plasma proteins (Sowemimo-Coker, 2002).
The lack of correlation between hemoglobin absorbance values and hemoglobin α levels measured by RPPM are likely related to assay differences and what is being measured (optical absorbance vs. antigen-antibody interaction), the volume of plasma in which the hemoglobin was suspended (1.5 µL for Nanodrop vs. picoliters for RPPM), and assay-related differences in plasma processing. Nanodrop measurements were obtained using never-frozen, unprocessed plasma samples while RPPM requires the denaturation of samples for subsequent immobilization/printing. Therefore, the former likely represents oxyhemoglobin levels (structure affects spectral absorbance), while the latter represents all hemoglobin α levels (assuming all exposed epitopes were Ab-bound). Despite the lack of correlation between the two methods, using more than one objective measure for evaluating biosample quality has its merits. Spectrophotometry as a primary QC measure can ensure that severely contaminated samples are not included in downstream studies. RPPM as a secondary QC measure can be used to guide biomarker data analysis (e.g., hemoglobin concentration as a covariate). Importantly, even in the apparent absence of significant RBC hemolysis a sample can still have platelet contamination.
The difference between miRNA markers miR-23a and miR-451 has been found to be a sensitive and reliable indicator of sample hemolysis (Blondal et al., 2013; Shah et al., 2016). While these two studies utilized human blood samples, this marker has also been shown to correlate well with spectrophotometric absorbance values in rat plasma samples (van Vliet et al., 2017). Our pilot miRNA analysis further emphasizes the need for rigorous quality assessment methods and proper exclusion criteria for preclinical samples (reviewed by van Vliet et al., 2017), as plasma samples below the current suggested limits for hemolysis can still have concurrent contamination by RBC- and platelet-specific miRNAs. Furthermore, diurnal oscillations have been found in miRNAs (Kinoshita et al., 2014; Heegaard et al., 2016) and in proteins, including those involved in hemostasis (Cohen et al., 1978; Soulban and Labrecque, 1989). Therefore, the standardization of blood collections is not only important for avoiding contamination from blood cells, but also for eliminating variation in biomarker levels due to diurnal patterns and anesthesia duration.
Anesthesia is another major confounding factor in preclinical studies because most volatile inhalation anesthetics (i.e., isoflurane, halothane, desflurane), like other intravenous anesthetic drugs (e.g., propofol), have been shown to exert neuroprotective effects following various CNS traumas (Schifilliti et al., 2010). Anesthesia-related neuroprotection can modify the extent and/or onset of injury-induced pathologies, such as inflammation, degeneration, and apoptosis, by potentially changing the secondary injury process after TBI. A recent study has shown evidence for isoflurane preventing acquired epilepsy in a rat model of temporal lobe epilepsy (Bar-Klein et al., 2016), suggesting that the levels of blood- and CSF-based biochemical markers may also be affected. Anesthesia may impact the glymphatic system and waste clearance in the brain. Recent evidence shows enhanced clearance that is dose- and compound-specific (Benveniste et al., 2017; Gakuba et al., 2018). Therefore, the objective is to standardize the length of anesthesia (as closely as possible) across sites during surgery and FPI, as well as blood collections as to not confound future comparisons of the different biochemical outcome measures.
4.2. Biochemical assay considerations
Antibody-based analytical platforms have been invaluable for the advancement of biomedical research and clinical diagnostics. One such platform, reverse phase protein microarray (RPPM) or array (RPPA), has made its mark in the past decade as the most cost-effective immunoassay for measuring proteins in various biopsy specimens, tissues and biofluids, including blood (serum and plasma), urine, and cerebrospinal fluid (Grote et al., 2008; Lu et al., 2016). RPPM is a sensitive, quantitative, and high-throughput screening method that is well-suited for protein biomarker discovery (Nishizuka et al., 2016). Consistent with other antibody-based assays, RPPM is dependent on the availability of high-quality antibodies. Unlike therapeutic and diagnostic antibodies, there are currently no regulations or best practices for research antibody validation. Therefore, commercially available antibodies significantly vary in specificity and sensitivity. Different antigens, isotypes, and purification methods (among other variables) can produce disparate results under identical experimental conditions. Therefore, in the absence of standardized application-specific antibody testing, careful selection and validation of antibodies is critical for proteomic studies.
Primary antibodies must be evaluated based on multiple criteria prior to purchasing (Weller, 2018). These include species reactivity, clonality, identity and relative size of immunogen (i.e., the target), binding selectivity or crossreactivity, tested applications (e.g., immunohistochemistry, western blot), the availability of blocking peptide, and customer reviews (if available). Selected antibodies are then pre-tested using western blot prior to use in RPPM (Gyorgy et al., 2010). Lot-to-lot variations in antibody specificity and efficacy are common, therefore repurchases of the same antibody are also tested regardless of clonality. Ideally, the same biomaterial (e.g., plasma, serum, or tissue homogenate), antibodies (primary and secondary), and blocking buffer are used for the western blot as well as RPPM. Background information on the protein of interest (i.e., tissue-specific expression) is helpful for determining the proper positive and negative controls to be included in the analysis. The western blot should show a single band at the expected molecular weight. However, it is not unusual for the molecular weight to vary due to post-translational modifications or for additional bands to appear due to the presence of different isoforms. Therefore, close attention should be paid to binding selectivity (as indicated in vendor’s documentation), and if the blocking peptide is available for purchase, then primary antibodies should be tested with and without the blocking peptide (Agoston et al., 2009; Gyorgy et al., 2010; Signore et al., 2017).
For miRNA biomarker discovery, small RNA sequencing from animals after epilepsy phenotyping expectedly provides tens of regulated miRNAs when epilepsy, no-epilepsy, and control groups are compared. Bioinformatics analysis can reveal miRNA target gene networks and molecular functions. Validation of candidate miRNAs in rat plasma can be done using real-time PCR (RT-PCR) or droplet digital PCR (ddPCR). Baseline and serial sampling of plasma provides opportunity to see expression patterns of miRNAs over time during epileptogenesis and to identify optimal post-TBI timepoints for assessment. Based on these analyses, the top candidates (5–10) can be selected for further analysis in a separate well-powered animal cohort and later in clinical adoption.
To date, previous studies have presented a high frequency of contradictory data regarding plasma and serum miRNA profiles and their usability as disease biomarkers for epilepsy (reviewed in Pitkänen et al., 2018). Interestingly, our previous bioinformatics analysis (Pitkänen et al., 2018) highlighted “miRNA clusters” in which each separate miRNA may signal the same pathologic condition (e.g., inflammation, dysregulated transcription or protein synthesis, or neurodegeneration). This suggests a novel approach to concentrate not only to one or several miRNAs, but also to other miRNAs on differential expression gene lists based on their predicted or validated targets. Through the literature, expression fold-changes reported in preclinical miRNA biomarker studies are relatively low (Gorter et al., 2014; Roncon et al., 2015) creating problems for power calculations and statistical analysis.
One approach to tackle the problem is to perform a multicenter study, making it possible to increase animal numbers. Knowledge of the correct phenotype of the animal is of the utmost importance to decrease variance within one group and wrong negative or positive cases. Bioinformatics pipeline and choice for cut-off limit in differential gene expression analysis can create very different result with possible problems in validation phases. Our experience has shown that determining suitable endogenous controls for quantifying circulating miRNAs in a rat is very challenging especially as plasma easily contains miRNA-residuals (amount varies sample to sample) from the red blood cells and platelets damaged during sample processing and storage. It is important to consider that data normalization can lead to a highly variable outcome, and it can even change upregulation to downregulation of the miRNA of interest.
4.3. Preliminary proteomic analysis
The effect of hemolysis on plasma protein concentrations poses important considerations for sample preparation, quality control, analysis, and biomarker selection. Using previously tested proteins of interest, we found that higher hemolysis did not increase the plasma levels of any proteins included in the analysis, likely due to the normalization of total protein concentrations across all samples as part of our RPPM protocol. Conversely, higher hemoglobin levels appeared to interfere with measured protein levels. Specifically, we found a significant decrease in p-Tau levels in one high hemolysis sample (p < 0.05, paired sample comparison).
Tau is an intrinsically disordered protein, and the lack of a well-defined structure makes it highly susceptible to intra- and intermolecular interactions (Avila et al., 2016). Phosphorylation at specific epitopes has been suggested to induce the opening of the transiently folded tau structure (Bibow et al., 2011), thereby increasing the likelihood of dimerization/aggregation and interference by other molecules (e.g., hemoglobin). Another possibility is that hemolysis is accompanied / associated with increased proteolytic activity in the affected plasma samples. If the specific epitopes of the protein of interest are damaged, or partly damaged due to increased proteolytic activity, protein biomarker values can be affected. Furthermore, in the absence of baseline or naïve plasma samples (and a higher n per group), we are unable to rule out the effect of TBI and/or sham surgery on plasma protein levels relative to hemoglobin concentrations.
While injury and time-related effects on biomarker levels are beyond the scope of this discussion, it is important to note that the “immediacy” of a protein marker measured in plasma can significantly vary depending on the molecule’s site of origin (e.g., cranial vs. extracranial) and collection site (e.g., retro-orbital vs. tail vein sample) (Anderson and Anderson, 2002). Furthermore, the time of day when blood is collected can also impact protein levels. The circadian clock proteins BMAL1, CLOCK, and REV-ERBα have been found to regulate critical aspects of immunity (Curtis et al., 2014), including the repression of key inflammatory molecules such as monocyte chemokine ligand-2, interleukin-1β (IL-1β), interleukin-6, tumor necrosis factor-α, and interferon-γ (Nguyen et al., 2013). Increased IL-1β levels were previously found to be associated with epileptogenesis (Diamond et al., 2014). Therefore, diurnal variations (if any) in marker levels should be accounted for in biomarker discovery.
Conclusion
Stringent harmonization of protocols and procedures is essential, particularly for complex multicenter studies such as EpiBioS4Rx. Our analysis of select harmonization parameters and plasma quality shows that protocol standardization and personnel training can significantly improve the quantity and quality of samples for biomarker discovery.
Supplementary Material
Highlights.
Our analysis of harmonization parameters and plasma sample quality indicates:
Personnel training can significantly improve plasma quality for biomarker discovery
High-hemolysis samples show more variable hemoglobin protein concentrations
Hemoglobin content may interfere with protein biomarkers measured in plasma
RBC miRNAs correlate with platelet contamination even in low-hemolysis samples
Acknowledgements
This work was supported by the National Institute of Neurological Disorders and Stroke (NINDS) Centers without Walls [grant number U54 NS100064].
Footnotes
This article is part of a special issue ‘Discovery of diagnostic biomarkers for post-traumatic epileptogenesis – an interim analysis of procedures in preclinical multicenter trial EpiBios4Rx’
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Agoston DV, Gyorgy A, Eidelman O, Pollard HB, 2009. Proteomic biomarkers for blast neurotrauma: targeting cerebral edema, inflammation, and neuronal death cascades. J Neurotrauma 26(6): p. 901–11. [DOI] [PubMed] [Google Scholar]
- 2.Anderson NL, Anderson NG, 2002. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867. [DOI] [PubMed] [Google Scholar]
- 3.Avila J, Jiménez JS, Sayas CL, Bolós M, Zabala JC, Rivas G, Hernández F, 2016. Tau Structures. Front Aging Neurosci 8:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-Klein G, Klee R, Brandt C, Bankstahl M, Bascuñana P, Töllner K, Dalipaj H, Bankstahl JP, Friedman A, Löscher W, 2016. Isoflurane prevents acquired epilepsy in rat models of temporal lobe epilepsy. Ann Neurol 80(6):896–908. [DOI] [PubMed] [Google Scholar]
- 5.Benveniste H, Lee H, Ding F, Sun Q, Al-Bizri E, Makaryus R, Probst S, Nedergaard M, Stein EA, Lu H, 2017. Anesthesia with Dexmedetomidine and Low-dose Isoflurane Increases Solute Transport via the Glymphatic Pathway in Rat Brain When Compared with High-dose Isoflurane. Anesthesiology Vol.127, 976–988. doi: 10.1097/ALN.0000000000001888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibow S, Ozenne V, Biernat J, Blackledge M, Mandelkow E, Zweckstetter M, 2011. Structural impact of proline-directed pseudophosphorylation at AT8, AT100, and PHF1 epitopes on 441-residue tau. J. Am. Chem. Soc 133(40):15842–5. doi: 10.1021/ja205836j. Epub 2011 September 15. [DOI] [PubMed] [Google Scholar]
- 7.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, Dahlsveen IK, 2013. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods 59:S1–S6. [DOI] [PubMed] [Google Scholar]
- 8.Cohen M, Simmons DJ, Joist HJ, 1978. Diurnal hemostatic changes in the rat. Thromb. Res 12(6):965–71. [DOI] [PubMed] [Google Scholar]
- 9.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA, 2014. Circadian clock proteins and immunity. Immunity 40(2):178–86. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Diamond ML, Ritter AC, Failla MD, Boles JA, Conley YP, Kochanek PM, Wagner AK, 2014. IL-1beta associations with posttraumatic epilepsy development: a genetics and biomarker cohort study. Epilepsia 55, 1109–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Epilepsy Bioinformatics Study for Antiepileptogenic Therapy (EpiBioS4Rx) https://epibios.loni.usc.edu/
- 12.Fort A, Borel C, Migliavacca E, Antonarakis SE, Fish RJ, Neerman-Arbez M, 2010. Regulation of fibrinogen production by microRNAs. Blood 116: 2608–15. [DOI] [PubMed] [Google Scholar]; Fortunato O, Boeri M, Verri C, Conte D, Mensah M, Suatoni P, Pastorino U, Sozzi G, 2014. Assessment of circulating microRNAs in plasma of lung cancer patients. Molecules 19, 3038–3054. 10.3390/molecules19033038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gakuba C, Gaberel T, Goursaud S, Bourges J, Di Palma C, Quenault A, Martinez de Lizarrondo S, Vivien D, Gauberti M, 2018. General Anesthesia Inhibits the Activity of the “Glymphatic System”. Theranostics 8(3): 710–722. doi: 10.7150/thno.19154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z, 2014. miRNA in plasma exosome is stable under different storage conditions. Molecules 19, 1568–1575. 10.3390/molecules19021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorter JA, Iyer A, White I, Colzi A, van Vliet EA, Sisodiya S, Aronica E, 2014. Hippocampal subregion-specific microRNA expression during epileptogenesis in experimental temporal lobe epilepsy. Neurobiol Dis 62:508–20. doi: 10.1016/j.nbd.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Grasedieck S, Schöler N, Bommer M, Niess JH, Tumani H, Rouhi A, Bloehdorn J, Liebisch P, Mertens D, Döhner H, Buske C, Langer C, Kuchenbauer F, 2012. Impact of serum storage conditions on microRNA stability. Leukemia 26, 2414–2416. 10.1038/leu.2012.106. [DOI] [PubMed] [Google Scholar]
- 17.Grote T, Siwak DR, Fritsche HA, Joy C, Mills GB, Simeone D, Whitcomb DC, Logsdon CD, 2008. Validation of reverse phase protein array for practical screening of potential biomarkers in serum and plasma: accurate detection of CA19–9 levels in pancreatic cancer. Proteomics 8(15): p. 3051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gyorgy AB, Walker J, Wingo D, Eidelman O, Pollard HB, Molnar A, Agoston DV, 2010. Reverse phase protein microarray technology in traumatic brain injury. J Neurosci Methods 192(1): p. 96–101. [DOI] [PubMed] [Google Scholar]
- 19.Harte-Hargrove LC, French JA, Pitkänen A, Galanopoulou AS, Whittemore V, Scharfman HE, 2017. Common data elements for preclinical epilepsy research: Standards for data collection and reporting. A TASK3 report of the AES/ILAE Translational Task Force of the ILAE. Epilepsia 58 Suppl 4:78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heegaard NH, Carlsen AL, Lilje B, Ng KL, Rønne ME, Jørgensen HL, Sennels H, Fahrenkrug J, 2016. Diurnal Variations of Human Circulating Cell-Free Micro-RNA. PLoS One 11(8):e0160577. doi: 10.1371/journal.pone.0160577. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huusko N, Römer C, Ndode-Ekane XE, Lukasiuk K, Pitkänen A, 2015. Loss of hippocampal interneurons and epileptogenesis: a comparison of two animal models of acquired epilepsy. Brain Struct Funct 220(1):153–91. doi: 10.1007/s00429-013-0644-1. [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita C, Aoyama K, Matsumura N, Kikuchi-Utsumi K, Watabe M, Nakaki T, 2014. Rhythmic oscillations of the microRNA miR-96–5p play a neuroprotective role by indirectly regulating glutathione levels. Nat Commun 5:3823. doi: 10.1038/ncomms4823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kharatishvili I, Nissinen JP, McIntosh TK, Pitkänen A, 2006. A model of posttraumatic epilepsy induced by lateral fluid-percussion brain injury in rats. Neuroscience 140(2):685–97. [DOI] [PubMed] [Google Scholar]
- 24.Kirschner MB, Kao SC, Edelman JJ, Armstrong NJ, Vallely MP, van Zandwijk N, Reid G, 2011. Haemolysis during Sample Preparation Alters microRNA Content of Plasma. PLOS ONE 6, e24145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroh EM, Parkin RK, Mitchell PS, Tewari M, 2010. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods 50(4): 298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon SK, Kovesdi E, Gyorgy AB, Wingo D, Kamnaksh A, Walker J, Long JB, Agoston DV, 2011. Stress and traumatic brain injury: a behavioral, proteomics, and histological study. Front Neurol 2:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lapinlampi N, Melin E, Aronica E, Bankstahl JP, Becker A, Bernard C, Gorter JA, Gröhn O, Lipsanen A, Lukasiuk K, Löscher W, Paananen J, Ravizza T, Roncon P, Simonato M, Vezzani A, Kokaia M, Pitkänen Asla. 2017. Common data elements and data management: Remedy to cure underpowered preclinical studies. Epilepsy Research, 129, 87–90. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N,Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M, 2008. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A 105, 10513–10518. 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishizuka SS, Mills GB, 2016. New era of integrated cancer biomarker discovery using reverse-phase protein arrays. Drug Metab Pharmacokinet 31(1): p. 35–45. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A, 2013. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science 341, 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitkänen A, Ndode-Ekane XE, Lapinlampi N, Puhakka N, 2018. Epilepsy biomarkers – Toward etiology and pathology specificity. Neurobiol Dis pii: S0969–9961(18)30145–1. doi: 10.1016/j.nbd.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prahl S, 1999. Optical Absorption of Hemoglobin http://omlc.org/spectra/hemoglobin/index.html
- 33.Roncon P, Soukupovà M, Binaschi A, Falcicchia C, Zucchini S, Ferracin M, Langley SR, Petretto E, Johnson MR, Marucci G, Michelucci R, Rubboli G, Simonato M, 2015. MicroRNA profiles in hippocampal granule cells and plasma of rats with pilocarpine-induced epilepsy - comparison with human epileptic samples. Sci. Rep 5, 14143. doi: 10.1038/srep14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schifilliti D, Grasso G, Conti A, Fodale V, 2010. Anaesthetic-Related Neuroprotection Intravenous or Inhalational Agents? CNS Drugs 24 (11): 893–907. doi: 1172-7047/10/0011-0893 [DOI] [PubMed] [Google Scholar]
- 35.Shah JS, Soon PS, Marsh DJ, 2016. Comparison of Methodologies to Detect Low Levels of Hemolysis in Serum for Accurate Assessment of Serum microRNAs. PLOS ONE 11, e0153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Signore M, Manganelli V, Hodge A, 2017. Antibody validation by western blotting. Methods Mol Biol 1606:51–70. [DOI] [PubMed] [Google Scholar]
- 37.Soulban G, Labrecque G, 1989. Circadian rhythms of blood clotting time and coagulation factors II, VII, IX and X in rats. Life Sci 45(25):2485–9. [DOI] [PubMed] [Google Scholar]
- 38.Sowemimo-Coker SO, 2002. Red blood cell hemolysis during processing. Transfus. Med. Rev 16, 46–60. [DOI] [PubMed] [Google Scholar]
- 39.Teruel-Montoya R, Rosendaal FR, Martınez C, 2015. MicroRNAs in hemostasis. J. Thromb. Haemost 13: 170–81. DOI: 10.1111/jth.12788 [DOI] [PubMed] [Google Scholar]
- 40.van Vliet EA, Puhakka N, Mills JD, Srivastava PK, Johnson MR, Roncon P, Das Gupta S, Karttunen J, Simonato M, Lukasiuk K, Gorter JA, Aronica E, Pitkänen A, 2017. Standardization procedure for plasma biomarker analysis in rat models of epileptogenesis: focus on circulating microRNAs. Epilepsia 58(12):2013–2024. doi: 10.1111/epi.13915. [DOI] [PubMed] [Google Scholar]
- 41.Vespa PM, Shrestha V, Abend N, Agoston D, Au A, Bell MJ, Bleck TP, Blanco MB, Claassen J, Diaz-Arrastia R, Duncan D, Ellingson B, Foreman B, Gilmore EJ, Hirsch L, Hunn M, Kamnaksh A, McArthur D, Morokoff A, O’Brien T, O’Phelan K, Robertson CL, Rosenthal E, Staba R, Toga A, Willyerd FA, Zimmermann L, Yam E, Martinez S, Real C, Engel J Jr.; EpiBioS4Rx Study Group, Agoston D, Au A, Bell MJ, Bleck TP, Branch C, Buitrago Blanco M, Bullock R, Burrows BT, Claassen J, Clarke R, Cloyd J, Coles L, Crawford K, Diaz-Arrastia R, Duncan D, Ellingson B, Engel J, Foreman B, Galanopoulou A, Gilmore E, Grohn O, Harris N, Hartings J, Lawrence H, Hunn M, Jette N, Johnston L, Jones N, Kanner A, McArthur D, Monti M, Morokoff A, Moshe S, Mowrey W, O’Brien T, O’Phelan K, Pitkanen A, Raman R, Robertson C, Rosenthal E, Shultz S, Snutch T, Staba R, Toga A, Van Horn J, Vespa P, Willyerd F, Zimmermann L, 2018. The epilepsy bioinformatics study for anti-epileptogenic therapy (EpiBioS4Rx) clinical biomarker: Study design and protocol. Neurobiol Dis pii: S0969–9961(18)30311–5. doi: 10.1016/j.nbd.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K, Yuan Y, Cho J-H, McClarty S, Baxter D, Galas DJ, 2012. Comparing the MicroRNA Spectrum between Serum and Plasma. PLoS ONE 7(7): e41561. doi: 10.1371/journal.pone.0041561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weller M, 2018. Ten Basic Rules of Antibody Validation. Anal Chem Insights 13:1177390118757462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Julia Z, Martha M, Carolyn C, David Y, Norman HA, 2009. Reference values for serum proteins of common laboratory rodent strains. J Am Assoc Lab Anim Sci 48(4): 387–390. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


