Abstract
Neuropeptide Y (NPY) is involved in multiple processes such as behavior, energy and bone metabolism. Previous studies have relied on global NPY depletion to examine its effects on bone. However, this approach is unable to distinguish the central or local source of NPY influencing bone. Our aim was to identify which cells within the skeleton express Npy and establish a model that will enable us to differentiate effects of NPY derived from different cell types. We have generated the NPY floxed (NPYflox) mice using CRISPR technology. By crossing the NPYflox mice with Hypoxanthine Phosphoribosyltransferase 1 (Hprt)-cre to generate a global knockout, we were able to validate and confirm loss of Npy transcript and protein in our global NPYKO.
Global deletion of NPY results in a smaller femoral cortical cross-sectional area (−12%) and reduced bone strength (−18%) in male mice. In vitro, NPY-deficient bone marrow stromal cells (BMSCs) showed increase in osteogenic differentiation detected by increases in alkaline phosphatase staining and bone sialoprotein and osteocalcin expression. Despite both sexes presenting with increased adiposity, female mice had no alterations in bone mass, suggesting that NPY may have sex-specific effects on bone. In this study we identified Npy expression in the skeleton and examined the effect of global NPY depletion to bone mass. The differential impact of NPY deletion in cortical and cancellous compartments along with differences in phenotypes between in vitro and in vivo, highlights the complex nature of NPY signaling, indicative of distinct sources that can be dissected in the future using this NPYflox model.
Keywords: neuropeptide Y (NPY), conditional mouse, bone, cortical bone, bone strength
Introduction
Over the past two decades, our knowledge of the interactions between bone and the nervous system has expanded. Following the discovery of hypothalamic regulation of bone mass, recent studies have focused on local regulation of the osteoblast and osteoclast lineage by neuropeptides [1–6]. Neuropeptide Y (NPY) is a 36 amino acid hydrophilic peptide that is highly abundant in the central and peripheral nervous systems [7, 8]. It is an orexigenic peptide that regulates metabolic processes through the hypothalamus, but also, has a role in peripheral tissues. NPY signaling is involved in a number of processes such as behavior, energy homeostasis, immune function and bone metabolism [8–11]. In addition to the nervous system, neuropeptides are expressed in peripheral tissues including bone and various cell types localized to the bone marrow environment [12–14].
NPY signaling is known to influence the skeleton [2, 3, 12, 15, 16]. NPY and its related peptides peptide YY (PYY) and pancreatic peptide (PP) signal through a family of 5 receptors known as Y receptors (Y1, Y2, Y4, Y5, y6). Hypothalamic regulation of bone is exerted by both Y1 and Y2 receptors and through the effects by peripheral sympathetic nervous system [2, 15, 17]. Deletion of Y1 and Y2 receptors in the brain revealed that NPY signaling inhibits osteogenesis [2, 15]. In addition, global deletion of NPY resulted in increased osteoblastic activity [16]. Interestingly, addition of NPY to the hypothalamic area in NPY global knockout mice resulted in only a partial rescue of the bone phenotype [16]. This indicated a potential role of peripheral sources of NPY as regulators of bone mass. As osteoblast lineage cells express NPY Y1 receptor, global deletion and bone directed deletion of Y1 targeting mature (Col2.3Cre) and early (Col3.6Cre) osteoblasts resulted in increased bone mass [4, 18].
To date, the source of NPY responsible for the activation of Y1 receptors on osteoblast lineage cells has not been defined. Besides Npy expression by sympathetic nerves in periosteum and bone marrow [19], our lab has previously demonstrated that Npy expression is highly enriched in osteocytes, in comparison to pre-osteoblasts, osteoprogenitors and osteoblasts [13]. Previous studies have relied on global deletion of NPY to examine its effects on the skeleton [16, 20–22], but there is a clear need to develop inducible cell type targeted conditional knockout mice for better definition of cell source and mechanisms of action of NPY. The aim of this study was to develop and characterize the NPY conditional knockout (NPYKO) mouse, which will facilitate cell-specific depletion of NPY.
Materials and Methods
Mouse strains and colony management
All procedures were approved by the UConn Health Institutional Animal Care and Use Committee and performed in an AAALAC accredited facility. Mice were group housed in ventilated cages with a 12hr light cycle (7am-7pm). The room temperature was maintained at 22°C. Water and irradiated rodent chow (Teklad 2918, Invigo, Indianapolis, IN) was provided ad libitum.
Generation of the NPY conditional knockout construct
Vectors pl452 and pl451 were designed with targeting arms to place a loxp site (pl452) upstream of the exon1 and frt-loxP site (pl451) downstream of exon 2. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) vectors using px330 were designed to cut in the locations to which the vectors would be targeted. A CRISPR plasmid was co-electroporated with the upstream targeting vector into mouse embryonic stem cells and were selected with G418. Colonies were picked and screened by polymerase chain reaction (PCR) for correct targeting. Positive clones were transfected with a CreIRESpuro for transient expression of Cre and cultured with puromycin for 2 days. Surviving clones were analyzed for complete recombination of the pl452 cassette leaving behind the upstream loxP. These clones were then co-electroporated with a CRISPR designed to cut downstream of the second exon along with a targeting vector to target a frt-neo-frt-loxP cassette. Clones were selected with G418, picked, and analyzed. The analysis was to both identify clones with targeting as well as clones with the targeting on the same allele as the upstream loxp site. Positive embryonic stem cell (ES) clones were aggregated with morula and transferred to recipient females. Pups were bred with a FLP mouse to recombine the frt-neo-frt-loxp leaving behind the downstream loxP site creating the floxed NPY mouse as represented in Figure 1A. These NPYflox mice were backcrossed on a C57BL/6J background for 8 generations.
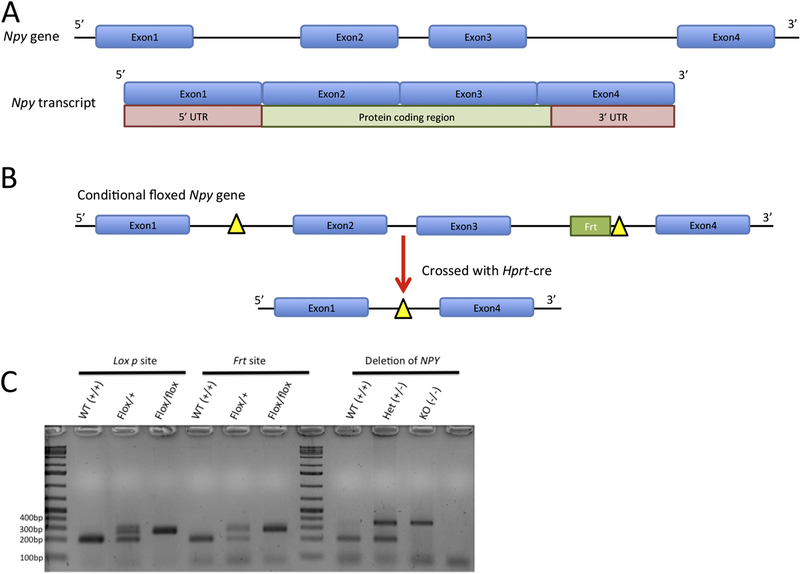
Figure 1: Generation of NPYflox and global NPYKO.
Structure and coding region of the NPY gene (A). Schematic diagram of the NPYflox allele and how the global NPYKO was generated (B). PCR demonstrating the incorporation of the loxP and Frt sites and validate NPY deletion in the global NPYKO (C). Expression was not detectable (ND) by qPCR.
Generation of NPY global knockout mice
Male NPYflox mice were bred with female Hypoxanthine Phosphoribosyltransferase 1 (Hprt)-cre mice (129S1/Sv-Hprttm1(CAG-cre)Mnn/J that were backcrossed more than 10 generations on a C57/Bl6 background) to remove the floxed Npy gene in the embryo generating global knockout (Figure 1B). Upon confirmation of NPY deletion, this line was maintained by crossing heterozygous mice, to generate all three genotypes – wildtype (WT), heterozygous and knockout (NPYKO). Littermate WT and NPYKO mice were assessed for a phenotype.
RNA extraction and real time PCR
RNA was extracted from bone marrow stromal cell (BMSC) cultures using Trizol (Ambion, Foster City, CA) according to the manufacturer’s protocol and quantified using a Nanodrop One (Thermofisher, Waltham, MA). Depending on the RNA yield obtained, samples were screened for integrity either by loading 0.5ug into a 1.5% agarose gel and visualized using ethidium bromide or loaded onto an Experion Eukaryote Total RNA HighSensitivity chip (BioRad, Hercules, CA). RNA samples were DNAse treated (DNAse I, Cat. # 18068015, Invitrogen, Carlsbad, CA) and cDNA was synthesized using Improm-II Reverse Transcription kit (Cat. # A3800, Promega, Madison, WI). qPCR was run using the following Taqman (Applied Biosystems, Waltam, MA) probes: Npy (Mm03048253_m1), Y1 (Mm00650798_g1), Bsp (Mm00492555_m1) and Ocn (Mm03413826_mH). Genes were normalized to housekeeping gene Gapdh (Mm99999915_g1). Relative expression of target genes was calculated using the ∆∆Ct method.
Genotyping for the NPYfloxed allele and NPY deletion
DNA was extracted from tail tissue. Genotyping was performed using the specified primers as listed in Table 1 with the following run conditions: 95°C for 3 mins, 34 cycles (95°C for 30 sec, 58.7°C for 30 sec, 72°C for 30 sec), followed by 72°C for 3 mins. The products were subsequently loaded into a 2% agarose gel in Tris-acetate-EDTA (TAE) buffer and run for 40 minutes at 100V. The PCR product was stained with ethidium bromide and visualized using a ChemiDoc imaging system (BioRad) (Figure 1C).
Table 1.
PCR sequences for genotyping
| Primer pair | Sequence | Product size |
|---|---|---|
|
Lox p site NPYgtloxpF NPYgtloxpR |
GCCTCCGTGCCTCTTATCTT GATTTTGGAGGCAACAGAGC |
WT – 204 bp Mut – 270 bp |
|
Frt site NPYgtfrtF NPYgtfrtR |
CCAATCCTATCGCATCCCTA CTATCTGGAGGTGGCAGGTC |
WT – 198 bp Mut – 278 bp |
|
NPY deletion NPYgtloxpF NPYgtfrtF NPYgtfrtR |
GCCTCCGTGCCTCTTATCTT CCAATCCTATCGCATCCCTA CTATCTGGAGGTGGCAGGTC |
WT – 198 bp KO – 353 bp |
Immunohistochemistry
Brain samples were fixed in 4% paraformaldehyde (PFA) for 3 days, and then 30% sucrose at 4°C, prior to cryo-embedding. Tissues were cut at 7μm thickness on Cryotape (Cryofilm 2C, Section Lab, Japan) and cross-linked using Norland Optical Adhesive 61 (Norland Optical, Cranbury, NJ) onto glass slides. Sections were permeabilized with 0.3% Triton, peroxide treated (3% hydrogen peroxide), blocked with 5% rabbit serum, incubated with 1:5000 dilution of NPY antibody (Cat#:22940; Immunostar, Hudson, WI), 1:80 dilution of biotinylated secondary antibody and the rest of the staining was completed using the Vector peroxidase substrate kit DAB (Cat#: SK-4100; Vector Laboratories, Burlingame, CA). Sections were counter-stained with haematoxylin. Sections were mounted in 50% glycerol and imaged using a Zeiss Observer Z.1 microscope with an AxioCam MRc camera and Zen Pro 2012 software.
Tissue collection
To evaluate dynamic histomorphometry, mice were injected with calcein (10mg/kg Sigma-Aldrich, St. Louis, MO) nine days and alizarin complexone (30mg/kg Sigma-Aldrich, St. Louis, MO) two days before they were euthanized at 14 weeks of age. The right femur was fixed in PFA and stored in phosphate-buffered saline (PBS) for microCT, then transferred to 30% sucrose and frozen at −20°C prior to cryo-embedding. The left femur was wrapped in PBS-soaked gauze and was frozen at −20°C until torsion testing. The left gonadal fat pad was dissected-free, weighed and fixed in 4% PFA.
MicroCT
Femoral morphometry of 14 week-old mice was assessed using conebeam micro-focus X-ray computed tomography (μCT40, Scanco Medical AG, Switzerland). Serial tomographic images were acquired at 55 kV and 145 μA, collecting 1000 projections per rotation at 300 ms integration time. Three-dimensional 16-bit grayscale images were reconstructed using standard convolution back-projection algorithms with Shepp and Logan filtering, and rendered within a 16.4 mm field of view at a discrete density of 244,140 voxels/mm3 (isometric 16 μm voxels). Segmentation of bone was performed in conjunction with a constrained Gaussian filter to reduce noise, applying calibrated hydroxyapatite-equivalent density thresholds of 495 and 740 mg/cm3 for trabecular and cortical bone, respectively. Trabecular morphometry was measured in the distal femur metaphysis, defining a volumetric region of interest within the endocortical surface of a 1 mm span referenced 1 mm from the growth plate. Cortical morphometry was measured within a 600μm span, referenced 5.1 mm from the growth plate.
Mechanical testing (torsion)
For torsion testing, femurs were potted in methyl methacrylate (Orthodontic Resin, Dentisply Caulk Inc. Milford, DE) bone cement in aluminum jigs, allowed to rehydrate in PBS for one hour at room temperature and tested for biomechanical properties using outward rotation on a TestBench™ Torsion Testing system (Bose Corporation ElectroForce Systems Group, Eden Prairie, MN). Data aquisition rate was 10Hz, 1 degree/sec. Data were plotted as torque to rotation; bone strength (N.mm), stiffness (N.mm/deg) and twist to failure (deg) were determined.
Histomorphometry of bone
Cryosections (7μm) from undecalcified femurs were prepared as described above. The calcein/alizarin labels were evaluated under a dual filter using Osteomeasure (Osteometrics, Atlanta, GA). A second set were decalcified with 14% EDTA then stained with tartrate resistant acid phosphatase (TRAP) kit (Cat. #387A, Sigma-Aldrich, St. Louis, MO). TRAP sections were also evaluated using Osteomeasure. Histomorphometric measurements were obtained in a manner consistent with reported guidelines and nomenclature [23].
Whole body DXA
Mice were anaethetised with ketamine/xylaxine cocktail and whole body dual-energy X-ray absorptiometry (DXA) was performed on a Lunar Piximus II (GE, Fitchburg, WI). Each scan was analyzed so that the rectangular region of interest (ROI) began at the ankles of the forelimbs and ended at the ankles of the hind limbs. Both the head and tail were excluded from the ROI. The following measurements were derived from the whole body DXA scans: lean mass, fat mass, % fat mass, bone mineral density (BMD) and bone mineral content (BMC).
Sectioning of adipose tissue
Following fixation, the gonadal fat pad was dehydrated in an ethanol gradient and then placed into xylene prior to embedding in paraffin. 5µm sections were cut on a microtome and stained with Mayers haematoxylin (Polyscientific R&D; Bay Shore, NY) and eosin (Shandon Eosin Y alcoholic #6766007; Thermofisher).
Bone marrow stromal cell (BMSC) cultures
Bone marrow cells were flushed from femora and tibiae from female mice. The cells were plated at a seeding density of 1 × 107 cells per well in a 6 well plate in alpha minimal essential media (α-MEM) media (Gibco, Billings, MT) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 5U Penicillin/Streptomycin (P/S). Osteogenic media consisted of α-MEM, 10% FBS, P/S, 50μg/mL ascorbic acid and 8mM β-glycerophosphate. Alkaline leukocyte phosphatase (ALP) staining was performed using ALP staining kit (Cat. #86R, Sigma-Aldrich, St. Louis, MO). The results of the staining were imaged using a scanner, Epson Perfection 1240 (Seiko Epson Corporation, Nagano Japan).
Calvarial osteoblast cultures
Calvariae were isolated from P5 pups. Parietal bones were digested at 37°C in 0.05% trypsin and collagenase P (0.5mg/mL) in PBS for 30 mins at a time; discarding the first fraction and keeping the subsequent 3 fractions. Cells were plated at a density of 2 × 105 cells per well in a 6 well plate and were changed to osteogenic media on Day 7. RNA was extracted on Day 4, 7, 11 and 14 of culture.
Bone marrow mononuclear cell cultures (BMM)
BMMs were prepared as previously described [24]. Bone marrow cells were prepared and cultured in α-MEM media with 10% FBS overnight on tissue culture. Non-adherent cells were then layered over the Ficoll Hypaque (GE Healthcare, Piscataway, NJ, USA) and centrifuged to remove red blood cells and stromal cells. Cells were cultured in macrophage-colony stimulating factor (M-CSF) alone or with stimulation by Receptor activator of nuclear factor kappa-Β ligand (RANKL) (both at 30 ng/ml; R&D Systems, Minneapolis, MN) for the days indicated with medium change on day 3.
Statistical analyses
All data were expressed as means ± standard error of the mean (SEM). Differences between groups were assessed by t-tests or one-way ANOVAs and followed by post hoc Tukey tests, if applicable. The statistical test used for each graph is stated in the figure legend. Statistical analyses were performed using Prism 6 (GraphPad software, CA, USA).
Results
NPY and Y1 expression in bone tissue
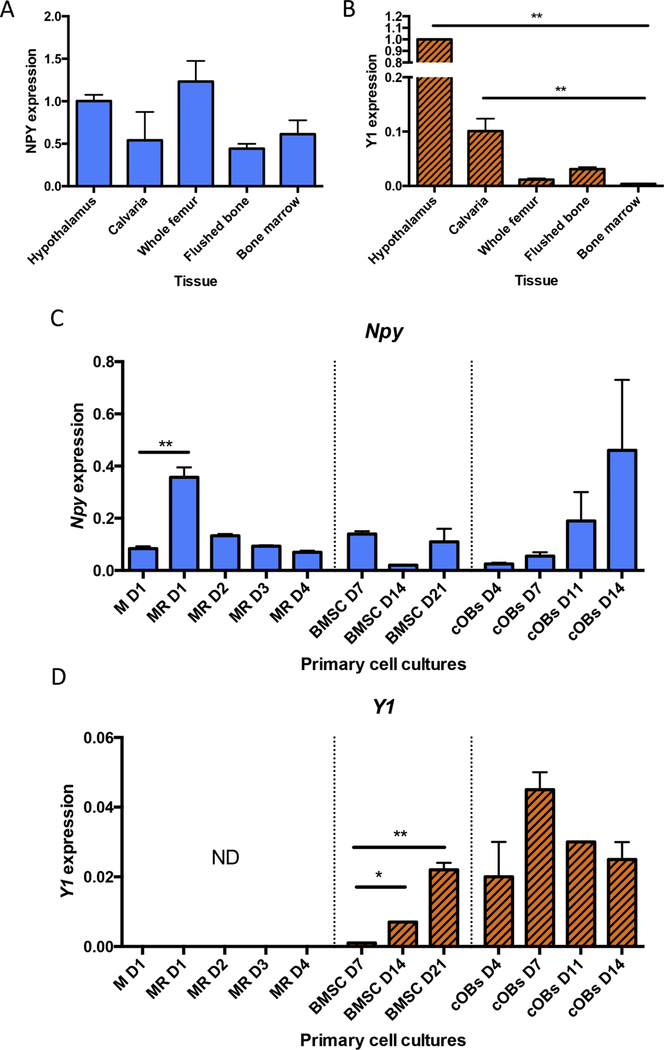
We evaluated the presence of NPY and Y1 expression in murine skeletal tissue. Hypothalamic RNA from WT mice was included as a positive control for both probes (Figure 2A,B). Npy expression was detected in calvaria, whole femur, flushed bone (bone with periosteum removed and bone marrow flushed out), and bone marrow at levels similar to what was detected in the hypothalamus (Figure 2A). Y1 expression was detected in calvaria, whole femur and flushed bone; we were unable to detect Y1 expression in bone marrow (Figure 2B). Interestingly, flushed bone showed an enrichment of Y1 expression compared to whole femur and bone marrow.
Figure 2: NPY and Y1 are locally expressed in bone tissue.
NPY (A) and Y1 expression (B) in hypothalamus and bone tissue. NPY (C) and Y1 (D) expression in primary cell cultures of bone marrow monocytes/macrophages (BMMs) stimulated with mCSF only (day 1) and in response to mCSF and RANKL stimulation (M+R; days 1–4), bone marrow stromal cells (BMSCs) undergoing differentiation (days 7, 14, 21) and calvarial osteoblasts (cOB) (day 4, 7, 11, 14). Mean±SEM. All samples were normalized to hypothalamic levels. Expression was not detectable (ND) by qPCR. Statistical analyses: one-way ANOVA, where *p<0.05 and **p<0.01. BMM cultures – n = 3; BMSC culture – n = 2; calvarial osteoblasts – n =2.
To further investigate Npy and Y1 expression within specific cell types present in bone, we isolated several primary skeletal cell types from WT mice. Npy expression was detected in bone marrow monocytes and osteoclast cultures across all times points (Figure 2C); most notably, initial stimulation with RANKL increased Npy expression by 4 fold. Expression of Npy was identified in bone marrow stromal cell (BMSC) cultures that were undergoing differentiation with osteogenic media (ascorbic acid and β-glycerophosphate) (Figure 2C). However, these cultures are heterogeneous in their preparation as they are prepared by culturing flushed bone marrow cells [25]. Thus, NPY in BMSC cultures is not necessarily arising from BMSCs or osteoblasts per se. Increasing levels of Npy expression were detected in calvarial osteoblast cultures between day 4 and Day 14 (Figure 2C). Y1 receptor expression was not detected in bone marrow mononuclear cells (BMMs) (Figure 2D). Thus, BMMs may contribute as a source of NPY, but are unable to respond to NPY stimulation. Y1 expression was found as BMSCs differentiated into osteoblasts and higher Y1 expression was observed in calvarial osteoblast cultures (Figure 2D). Thus, we were able to demonstrate that NPY is produced in skeletal tissues and cells of the osteoblast lineage express the Y1 receptor reinforcing the notion that they respond to changes in NPY levels.
Generation and validation of NPYflox and global NPYKO models
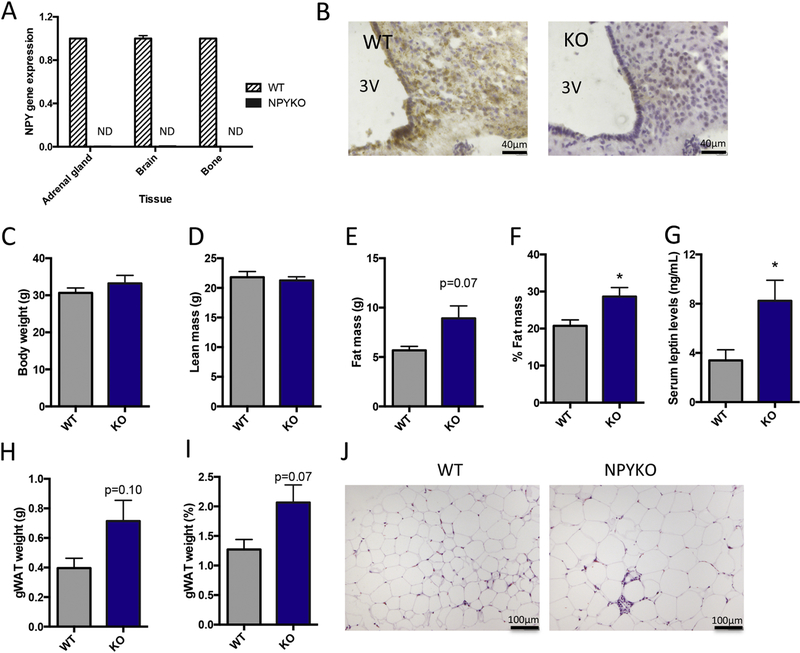
The Npy gene in the mouse is located on chromosome 6 and consists of 4 exons. The protein-coding region of the Npy transcript is made up of exon 2, exon 3 and the first part of exon 4 (Figure 1A). The NPY floxed model was generated using CRISPR technology by inserting a loxP site between Exon 1 and Exon 2 and an Frt-loxP site between Exon 3 and Exon 4 to flank all but two protein-coding amino acids (Figure 1B; for full details, see Materials and Methods). Global NPY deficient mice (NPYKO) were generated by crossing Hprt-cre mice with the NPYflox mice (Figure 1B). The insertion of the loxP site (≈70bp) and frt site (≈278bp) for the NPYflox and recombination of the NPY locus for the global NPYKO (≈353bp) were verified by PCR (Figure 1C). Deletion of exons 2 and 3 in the NPYKO resulted in a loss of Npy gene expression in the adrenal gland, brain and bone tissues (Figure 3A). The highest expression of Npy occurs in the arcuate nucleus [7, 8]. Therefore, we confirmed absence of the protein in the arcuate nucleus of NPYKO mice by immunohistochemistry (Figure 3B). Thus, we were able to establish the first NPY conditional knockout mouse and generate a global NPY-deficient mouse using our conditional knockout.
Figure 3: Validation of global NPYKO and evaluation of body composition.
Gene expression of NPY in adrenal gland, brain and bone (A), n = 3. Immunohistochemistry for NPY in arcuate nucleus (B). The third ventricle is marked (3V). Body weight of male mice at 14 weeks of age (C). Whole body DXA data: Lean mass (D), fat mass (E) and % fat mass (F). Serum leptin levels as determined by ELISA (G). Dissected gonadal white adipose tissue weight (H) and expressed as % of body weight (I). Representative images of haematoxylin and eosin staining of gonadal white adipose tissue (J). Mean±SEM. Statistical analysis: Students t-test (C-I), where *p<0.05. Body weight and DXA analyses: Male WT (n=5); Male NPYKO (n=7).
Increased adiposity in male NPY-deficient mice
NPY is implicated in the regulation of whole body energy homeostasis; Y1 deficient mice develop adult-onset obesity at around 12–15 weeks of age [15, 26]. In our global NPY-deficient model, at 14 weeks of age, no difference in body weight (Figure 3C) or lean mass (Figure 3D) was present in male WT and NPYKO mice. An increase in fat mass (Figure 3E, p=0.07) and %fat mass (Figure 3F) was observed in male NPYKO mice. Serum leptin levels were correlated with fat mass (Males R2=0.64, p=0.06; Females: R2=0.76, p<0.01). We identified that our NPY-deficient mice had increased leptin levels compared to WT mice in both genders (Figure 3G), consistent with the observed increase in fat mass (Figure 3E,F). The relationship between fat mass and leptin levels was not altered in NPYKO mice, suggesting that in our model NPY deficiency does not directly affect circulating leptin levels. There was also a trend towards increased gonadal fat pad weight in male mice (Figure 3H, p=0.10; Figure 3I, p=0.07). Increased adipocyte size was observed in the gonadal fat pads of NPYKO mice (Figure 3J). Altogether, we identified an increase in adiposity in NPYKO mice, suggesting that whole body energy metabolism is altered in this global NPY-deficient model.
Male NPY-deficient mice have a smaller cross-sectional area and impaired bone strength
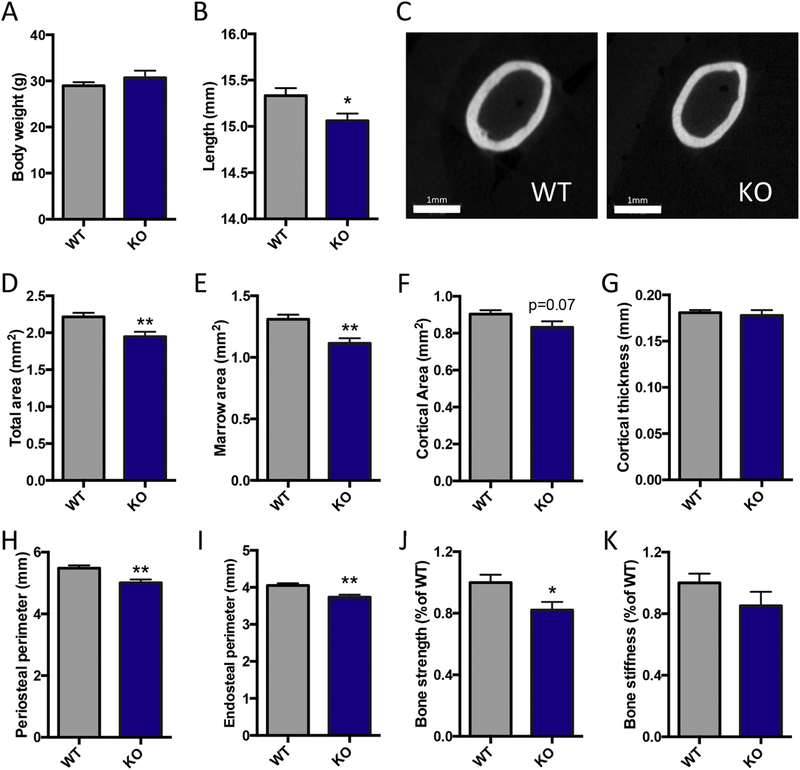
Male NPYKO mice had no difference in body weight at 14 weeks of age (Figure 4A). Femoral length was shorter by 2% in NPYKO mice in comparison to WT (Figure 4B). We assessed for changes in cortical bone mass using microCT and bone strength using torsion testing. In males, NPYKO mice had significant reductions in total and marrow area by 12% and 15% respectively (Figure 4C-E). There was a trend towards reduced cortical area in male mice (Figure 4F, p=0.07), but no difference in cortical thickness (Figure 4G). This corresponded with reductions in periosteal and endosteal perimeters in male mice (Figure 4H,I). Changes in cortical cross-sectional area can impact bone strength [27]; the computed indices of strength generated from the microCT data implied that male NPYKO bones would be weaker in terms of bending properties (Suppl. table 1). Our torsion testing of WT and NPYKO femurs identified that NPYKO mice had weaker bones as they had an 18% reduction in bone strength in comparison to WT mice (Figure 4J). No significant difference in bone stiffness was found between WT and NPYKO (Figure 4K), suggesting that the reduction observed reduction in strength may be associated with the change in cross-sectional area. Altogether, this data shows that male NPYKO mice have a smaller cross-sectional area in comparison to WT mice and impaired bone strength.
Figure 4: Male NPYKO femurs have a smaller cortical cross-sectional area and reduced bone strength.
Body weight (A) and femoral length (B) of mice at 14 weeks of age. Representative images of WT and NPYKO in mid-diaphyseal region of the femur (C). MicroCT analysis: total area (D), marrow area (E), cortical area (F), cortical thickness (G) periosteal perimeter (H) and endosteal perimeter (I). Torsion testing: bone strength (J) and bone stiffness (K). Mean±SEM. Statistical analysis: Students t-test (A, B, D-J), where *p<0.05 and **p<0.01. Body weight, femoral length and microCT analyses: male WT (n=14); male NPYKO (n=13). Torsion testing: male WT (n=8); male NPYKO (n=8).
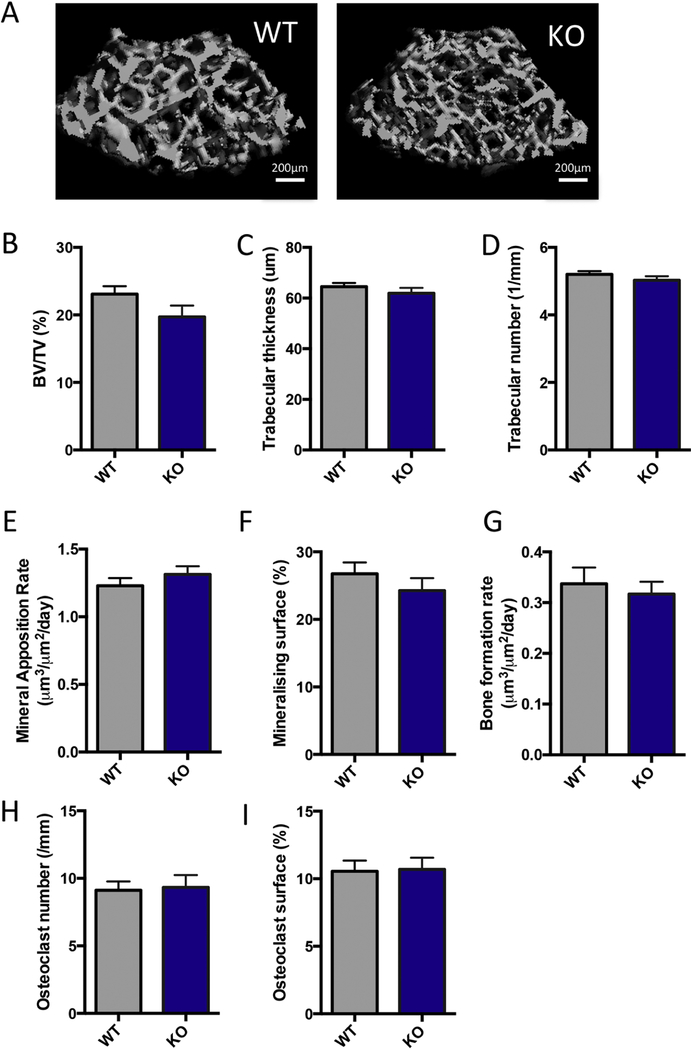
Consistent with the reduction in cross-sectional bone area observed in the cortical region of interest (ROI), a reduction in total bone volume and tissue volume were observed in the cancellous ROI (Figure 5A). However, the bone volume to tissue volume ratio (BV/TV) between WT and NPYKO mice was not significantly different (Figure 5B). No significant differences in trabecular thickness and trabecular number were identified (Figure 5C,D). Histomorphometric analysis of dynamic labeling in the cancellous region indicated that there was no change in mineral apposition rate, mineralizing surface or bone formation rate (Figure 5E,F,G). NPY-deficiency did not affect osteoclast number or surface in NPYKO mice (Figure 5H,I); consistent with our data showing that osteoclasts do not express the Y1 receptor (Figure 2D). The effects of NPY deficiency primarily reduce bone length, cortical cross-sectional area and bone strength.
Figure 5: NPY-deficiency does not affect cancellous bone.
Representative images from male WT and NPYKO mice (A). MicroCT analysis: BV/TV (B), trabecular thickness (C) and trabecular number (D). Dynamic histomorphometry: mineral apposition rate (E), mineralizing surface (F), and bone formation rate (G). TRAP staining: osteoclast number (H) and osteoclast surface (I). Mean±SEM. Statistical analysis: Students t-test (B-I). Male WT (n=14); Male NPYKO (n=13).
Sex-specific effects of NPY on the skeleton
Sexual dimorphism has been observed in metabolic studies with other NPY models [11, 28–30]. Therefore, we examined female mice of the same age to determine whether NPY-deficiency had the same effect in male and female mice. At 14 weeks of age, a significant increase in body weight was observed in female NPYKO mice in comparison to WT mice (Table 2). This increase in body weight was attributed to both an increase in lean mass and fat mass (Table 2). Thus, increased adiposity was observed in both sexes, however the metabolic effects of NPY-deficiency in female mice were greater in magnitude in comparison to male. Leptin levels were elevated in female NPYKO mice corresponding with increased fat mass (Table 2). Femoral length had a trend towards a reduction in female NPYKO mice (Table 2, p=0.14), similar to the observation in male mice. However, no changes in bone cross-sectional area, cortical or cancellous bone mass were identified in female NPYKO mice (Table 2). The presence of changes in body composition similar to male NPYKO mice and the absence of a bone phenotype in female mice suggest that there may be sex-specific effects of NPY on the skeleton.
Table 2.
Characteristics of female NPYKO mice (Mean ± SEM)
| Measurement | WT n=7 |
NPYKO n=6 |
|---|---|---|
| Metabolic | ||
| Body weight (g) | 20.87±0.18 | 26.22±1.78** |
| Lean mass (g) | 15.23±0.11 | 16.78±0.61* |
| Fat mass (g) | 3.30±0.18 | 7.18±1.26** |
| % Fat mass | 17.7±0.9 | 29.2±3.0* |
| Leptin levels (ng/mL) | 2.26±0.4 | 6.63±1.7* |
| Bone | ||
| Femur length | 14.97±0.08 | 14.75±0.12 |
| Total area (mm2) | 1.67±0.02 | 1.66±0.05 |
| Marrow area (mm2) | 0.98±0.01 | 0.97±0.04 |
| Cortical area (mm2) | 0.69±0.01 | 0.69±0.02 |
| Cortical thickness (mm) | 0.158±0.003 | 0.161±0.002 |
| Periosteal perimeter (mm) | 4.67±0.04 | 4.65±0.07 |
| Endosteal perimeter (mm) | 3.50±0.03 | 3.48±0.06 |
| BV/TV (%) | 4.8±0.8 | 5.0±0.8 |
| Trabecular thickness (μm) | 49.0±1.1 | 48.3±1.6 |
| Trabecular number (1/mm) | 3.3±0.1 | 3.4±0.1 |
Student’s t-test
p<0.05
p<0.01 when compared to WT.
In vitro cultures of NPYKO BMSCs show increased osteoblastic differentiation
BMSCs were isolated from female WT and NPYKO mice (Figure 6A). Increased alkaline phosphatase staining was observed in NPYKO cultures (Figure 6B). In WT cultures, Npy expression decreased with differentiation at Day 14 and Day 21; and as expected, was absent in the NPYKO BMSCs (Figure 6C). Y1 receptor expression in WT BMSCs increased on days 14 and 21 following differentiation (Figure 6D). Y1 expression was unaltered during the differentiation of NPYKO BMSCs (Figure 6D). As expected, bone sialoprotein (Bsp) and osteocalcin (Ocn) levels increased in WT mice with differentiation (Figure 6E,F). In comparison, BMSCs from NPYKO mice had greater expression of Bsp and Ocn on day 14 and day 21 of culture, indicating that NPY-deficient BMSCs have enhanced differentiation (Figure 6E,F). Cultures derived from male mice showed the same increase in ALP staining and increased mineralization (data not shown). These in vitro experiments are consistent with reports in other NPY models [3, 12, 15] and suggest that the skeletal phenotype observed in global NPY deficiency may be complex due to the role of NPY in multiple systems (i.e. central and peripheral nervous systems) and also its local production in bone.
Figure 6: BMSC cultures from NPYKO mice in osteogenic media have increased differentiation.
Schematic diagram of BMSC culture and time points (A). Alkaline phosphatase staining of Day 7 BMSCs (B); one representative experiment is shown, from n=4 independent biological replicates. Time course of gene expression of neuropeptide Y (C), Y1 receptor (D), bone sialoprotein (E) and osteocalcin (F). Expression was not detectable (ND) by qPCR. One representative experiment is shown, n = 2 independent biological experiments.
Discussion
To better understand the sources of NPY and its receptors in bone tissues and bone derived cell cultures; we completed a comprehensive analysis of their expression. While we confirmed Npy expression in more mature osteoblast lineage cells, a high level of expression was detected in BMMs induced by RANKL. Y1 receptor expression was detected in mature osteoblasts but not in BMMs, thus suggesting that BMMs may be a source of NPY but not respond to NPY stimulation. We have generated a novel murine model allowing for conditional deletion of NPY. By crossing our NPY floxed mice with Hprt-cre to generate a global knockout, we validated and confirmed loss of Npy transcript and protein in our global NPYKO. Male NPYKO mice had a smaller cross-section in the femur and reduced bone strength. In vitro NPY-deficient BMSCs showed enhanced differentiation. Altogether, this work confirms Npy expression in the skeleton and by generating a conditional knockout we have been able to establish a model to dissect the contribution of NPY from specific cell types.
The skeletal phenotype of reduced cortical cross section and impaired bone strength determined using our global NPYKO differs from other NPY-deficient models previously published. Elefteriou et al (2003) examined the NPY deficient model (NPYtm1RPA) on a 129SV/C57BL6 background whereby exon 2 of the Npy gene was replaced by a LacZ-neo fusion cassette. These mice had no differences in cancellous bone mass, but only vertebral bone mass was examined [31]. Wang et al (2016) also utilized the NPYtm1RPA mouse model on a 129SV background and reported significant increases in both femoral cortical and cancellous bone mass [21]. The global NPYKO model (Npytm1(cre)Tkar) on a mixed 129SV/C57BL6 background has been frequently used in studies by Baldock et al. [16, 17, 22, 32]; this model was generated by knocking-in a cre construct into exon 2 of the Npy locus, which resulted in a null allele [11]. These studies demonstrated that NPY deficiency results in increased cortical bone mass. Effects on cancellous bone in this model are context-dependent. The construction of each of these global NPYKO mice has been different, and each involved insertion of genes into the locus (lacZ-neo cassette or cre-recombinase) which may affect the phenotype. Differing genetic backgrounds of the mice may also account for the differences in the effects on bone mass [33–35].
In addition, the complexity of the NPYKO model may be attributed to a combination of Y receptor deficiency phenotypes since we have removed the ligand from all tissues. The Y1 receptor knockout mouse displays a phenotype of adult-onset obesity and increases in cortical bone mass [15]. In addition to the significant increases in bone formation rate, global Y1 receptor knockout mice have increased osteoclast surface [15]. This has been recapitulated in peripheral Y1 antagonist BIBO3304-treated mice [36]. Recently, Sousa et al. (2016) suggested that osteoclasts from Y1 deficient mice have reduced ability to resorb bone, and thus, this coupled to increased bone formation, led to the observed phenotype of increased bone mass in Y1 deficient mice [37]. Mice with a specific deletion of the Y1 receptor from osteoblasts and osteocytes have no changes in adiposity and an increase in cortical bone mass [4]. These mice have no changes in osteoclastic parameters suggesting that NPY signaling through the osteoblast is not influencing osteoclasts. Baldock et al. (2002) reported increases in cortical and cancellous bone mass in their Y2KO maintained on a mixed C57/Bl6–129Sv background [2]. However, Mikic et al. (2008) observed in their Y2KO on a mixed 129Sv-Balb/C background that these mice had smaller cortical cross-sections, less cortical bone area and lower structural strength [38]. Y6-deficient mice have reduced cortical and cancellous bone mass, which was associated with an increase in osteoclast progenitors leading to increased resorption and a decrease in osteoblast progenitors leading to reduced osteoblast activity [39]. Altogether, the differences observed with each of these models demonstrate the complexity involved between NPY and its receptors and the impact that central signaling external to bone can have on the skeleton.
Both sexes had significant increases in adiposity; however, we only observed a bone phenotype in male NPYKO mice. Sexual dimorphism has been observed with other NPY models [11, 28–30]. While the majority of studies focusing on bone have reported the same phenotype between both sexes [15, 16, 39], there has been evidence that NPY-Y1 signaling increases bone mass in the presence of androgens as no differences in bone mass were found between WT and Y1KO mice that had undergone gonadectomy [29]. Estrogen is known to affect bone remodeling [40] and estrogen deficiency increases sympathetic activity [41]. Since NPY can inhibit sympathetic activity, this may have subsequent effects on bone. A metabolic study has shown that NPY ablated the reduction in energy expenditure and increase in fat mass induced by short-term estrogen deficiency [42]. These studies suggest that androgens and estrogen may be factors involved in understanding the sex-specific skeletal differences we observed.
Npy is expressed in the nervous system and also in the periphery. Here, we have observed Npy expression in bone and primary cells (BMMs, BMSCs and calvarial osteoblasts). While local overexpression of NPY in osteoblasts using the Col1a1 2.3kb reporter reduces cortical bone mass [3], there has not been any assessment of local endogenous NPY production. By generating the conditional NPY mouse, we will be able to explore the effect of local NPY deficiency by targeting different cell types using specific cre models. The targeted nature of these future models will assist in separating the multitude of effects observed in our global NPYKO. It is likely that some of the phenotype (i.e. adiposity) observed in the NPYKO is attributed to changes in central NPY signaling. The observed effect on bone mass in our NPYKO model may be arising from direct loss of NPY signaling, or through indirect effects such as the influence of adipose tissue on bone. Increased adiposity can lead to increases in cortical bone mass due to the positive effects of load bearing, but at the same time increases in sympathetic tone, inflammatory cytokines and oxidative stress which can negatively impact bone mass [22, 43, 44]. The effects of other tissues on bone mass may explain the difference we observed between our in vivo and in vitro studies. While bone mass was impacted by global NPY deficiency, further work is required to dissect and determine the contribution of individual NPY sources.
There are a few limitations of this study. The skeleton is constantly being remodeled. During development there is rapid growth and modeling occurring, followed by the attainment of peak bone mass and further remodeling. We examined the effect of NPY deficiency at a single time point that represents adult animals nearing peak bone mass. It is possible that examining mice at different ages may help to elucidate the role of NPY during different stages of bone modeling and remodeling. While our focus was on the skeletal phenotype of these mice, our analysis of the changes in fat mass were limited and had a small n. It is difficult to gauge why these NPYKO mice have increased adiposity from these observations, thus further examination of the metabolic phenotype (such as food intake, energy expenditure, etc.) is needed to elucidate the mechanisms involved.
We have developed the NPY conditional mouse and generated a global NPY deficient model to examine the impact of NPY signaling on bone. Our study has identified that global NPY deficiency regulates bone mass and in this case, leads to smaller bones with impaired strength. Notably, the findings of this study clearly demonstrate that global deficiency contributes to the overall phenotype through various mechanisms and this may explain the discrepancy we observed between our in vivo and ex vivo results. Since Npy is expressed in both the nervous system and skeletal tissues and influences energy metabolism, our work in this study generating and characterizing the NPY floxed mouse will facilitate the selective depletion of NPY from different cell populations in vivo to dissect the contribution of central and local NPY signaling in the future.
Supplementary Material
Highlights:
Identification of Npy and Y1 expression in bone and primary bone cultures.
Generation of novel NPYflox mouse model.
Male global NPY deficient mice (NPYKO) have reduced femoral cortical cross-section and impaired mechanical strength.
Sex-specific effects of NPY on bone; female mice have no skeletal phenotype.
In vitro BMSCs show increased osteogenesis.
Acknowledgements
We would like to thank Dr. Sun-Kyeong Lee (UConn Health) for provision of cDNA to assess NPY and Y1 levels in BMMs in response to RANKL stimulation. We would also like to thank Dr. Mark Kronenberg (UConn Health) for provision of RNA from calvarial osteoblast culture time courses.
Supported by: This work has been supported by NIH/NIAMS grants AR055607, AR055607-07S1 and REP funds by UConn to I.K.
Footnotes
Disclosures
All authors have no conflicts of interest regarding this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ducy P, Amling M, Takeda S, Priemel M, Schilling AF, Beil FT, Shen J, Vinson C, Rueger JM, Karsenty G. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell 2000;100: 197–207. [DOI] [PubMed] [Google Scholar]
- [2].Baldock PA, Sainsbury A, Couzens M, Enriquez RF, Thomas GP, Gardiner EM, Herzog H. Hypothalamic Y2 receptors regulate bone formation. J Clin Invest 2002;109: 915–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Matic I, Matthews BG, Kizivat T, Igwe JC, Marijanovic I, Ruohonen ST, Savontaus E, Adams DJ, Kalajzic I. Bone-specific overexpression of NPY modulates osteogenesis. J Musculoskelet Neuronal Interact 2012;12: 209–18. [PMC free article] [PubMed] [Google Scholar]
- [4].Lee NJ, Nguyen AD, Enriquez RF, Doyle KL, Sainsbury A, Baldock PA, Herzog H. Osteoblast specific Y1 receptor deletion enhances bone mass. Bone 2011;48: 461–7. [DOI] [PubMed] [Google Scholar]
- [5].Lee NJ, Nguyen AD, Enriquez RF, Luzuriaga J, Bensellam M, Laybutt R, Baldock PA, Herzog H. NPY signalling in early osteoblasts controls glucose homeostasis. Mol Metab 2015;4: 164–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Horsnell H, Baldock PA. Osteoblastic Actions of the Neuropeptide Y System to Regulate Bone and Energy Homeostasis. Curr Osteoporos Rep 2016;14: 26–31. [DOI] [PubMed] [Google Scholar]
- [7].Schwartz TW, Fuhlendorff J, Kjems LL, Kristensen MS, Vervelde M, O’Hare M, Krstenansky JL, Bjornholm B. Signal epitopes in the three-dimensional structure of neuropeptide Y. Interaction with Y1, Y2, and pancreatic polypeptide receptors. Ann N Y Acad Sci 1990;611: 35–47. [DOI] [PubMed] [Google Scholar]
- [8].Blomqvist AG, Herzog H. Y-receptor subtypes--how many more? Trends Neurosci 1997;20: 294–8. [DOI] [PubMed] [Google Scholar]
- [9].Wee NK, Kulkarni RN, Horsnell H, Baldock PA. The brain in bone and fuel metabolism. Bone 2016;82: 56–63. [DOI] [PubMed] [Google Scholar]
- [10].Wheway J, Herzog H, Mackay F. The Y1 receptor for NPY: a key modulator of the adaptive immune system. Peptides 2007;28: 453–8. [DOI] [PubMed] [Google Scholar]
- [11].Karl T, Duffy L, Herzog H. Behavioural profile of a new mouse model for NPY deficiency. Eur J Neurosci 2008;28: 173–80. [DOI] [PubMed] [Google Scholar]
- [12].Igwe JC, Jiang X, Paic F, Ma L, Adams DJ, Baldock PA, Pilbeam CC, Kalajzic I. Neuropeptide Y is expressed by osteocytes and can inhibit osteoblastic activity. J Cell Biochem 2009;108: 621–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Paic F, Igwe JC, Nori R, Kronenberg MS, Franceschetti T, Harrington P, Kuo L, Shin DG, Rowe DW, Harris SE, Kalajzic I. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone 2009;45: 682–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alves CJ, Alencastre IS, Neto E, Ribas J, Ferreira S, Vasconcelos DM, Sousa DM, Summavielle T, Lamghari M. Bone Injury and Repair Trigger Central and Peripheral NPY Neuronal Pathways. PLoS One 2016;11: e0165465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Baldock PA, Allison SJ, Lundberg P, Lee NJ, Slack K, Lin EJ, Enriquez RF, McDonald MM, Zhang L, During MJ, Little DG, Eisman JA, Gardiner EM, Yulyaningsih E, Lin S, Sainsbury A, Herzog H. Novel role of Y1 receptors in the coordinated regulation of bone and energy homeostasis. J Biol Chem 2007;282: 19092–102. [DOI] [PubMed] [Google Scholar]
- [16].Baldock PA, Lee NJ, Driessler F, Lin S, Allison S, Stehrer B, Lin EJ, Zhang L, Enriquez RF, Wong IP, McDonald MM, During M, Pierroz DD, Slack K, Shi YC, Yulyaningsih E, Aljanova A, Little DG, Ferrari SL, Sainsbury A, Eisman JA, Herzog H. Neuropeptide Y knockout mice reveal a central role of NPY in the coordination of bone mass to body weight. PLoS One 2009;4: e8415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Baldock PA, Lin S, Zhang L, Karl T, Shi Y, Driessler F, Zengin A, Hormer B, Lee NJ, Wong IP, Lin EJ, Enriquez RF, Stehrer B, During MJ, Yulyaningsih E, Zolotukhin S, Ruohonen ST, Savontaus E, Sainsbury A, Herzog H. Neuropeptide y attenuates stress-induced bone loss through suppression of noradrenaline circuits. J Bone Miner Res 2014;29: 2238–49. [DOI] [PubMed] [Google Scholar]
- [18].Lee NJ, Doyle KL, Sainsbury A, Enriquez RF, Hort YJ, Riepler SJ, Baldock PA, Herzog H. Critical role for Y1 receptors in mesenchymal progenitor cell differentiation and osteoblast activity. J Bone Miner Res 2010;25: 1736–47. [DOI] [PubMed] [Google Scholar]
- [19].Tabarowski Z, Gibson-Berry K, Felten SY. Noradrenergic and peptidergic innervation of the mouse femur bone marrow. Acta Histochem 1996;98: 453–7. [DOI] [PubMed] [Google Scholar]
- [20].Park MH, Lee JK, Kim N, Min WK, Lee JE, Kim KT, Akiyama H, Herzog H, Schuchman EH, Jin HK, Bae JS. Neuropeptide Y Induces Hematopoietic Stem/Progenitor Cell Mobilization by Regulating Matrix Metalloproteinase-9 Activity Through Y1 Receptor in Osteoblasts. Stem Cells 2016;34: 2145–56. [DOI] [PubMed] [Google Scholar]
- [21].Wang FS, Lian WS, Weng WT, Sun YC, Ke HJ, Chen YS, Ko JY. Neuropeptide Y mediates glucocorticoid-induced osteoporosis and marrow adiposity in mice. Osteoporos Int 2016;27: 2777–2789. [DOI] [PubMed] [Google Scholar]
- [22].Wee NKY, Enriquez RF, Nguyen AD, Horsnell H, Kulkarni R, Khor EC, Herzog H, Baldock PA. Diet-induced obesity suppresses cortical bone accrual by a neuropeptide Y-dependent mechanism. International Journal of Obesity 2018. [DOI] [PubMed]
- [23].Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR, Parfitt AM. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 2013;28: 2–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yeon Won H, Hwan Mun S, Shin B, Lee SK. Contradictory Role of CD97 in Basal and Tumor Necrosis Factor-Induced Osteoclastogenesis In Vivo. Arthritis Rheumatol 2016;68: 1301–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chang MK, Raggatt LJ, Alexander KA, Kuliwaba JS, Fazzalari NL, Schroder K, Maylin ER, Ripoll VM, Hume DA, Pettit AR. Osteal tissue macrophages are intercalated throughout human and mouse bone lining tissues and regulate osteoblast function in vitro and in vivo. J Immunol 2008;181: 1232–44. [DOI] [PubMed] [Google Scholar]
- [26].Zhang L, Macia L, Turner N, Enriquez RF, Riepler SJ, Nguyen AD, Lin S, Lee NJ, Shi YC, Yulyaningsih E, Slack K, Baldock PA, Herzog H, Sainsbury A. Peripheral neuropeptide Y Y1 receptors regulate lipid oxidation and fat accretion. Int J Obes (Lond) 2010;34: 357–73. [DOI] [PubMed] [Google Scholar]
- [27].van der Meulen MC, Jepsen KJ, Mikic B. Understanding bone strength: size isn’t everything. Bone 2001;29: 101–4. [DOI] [PubMed] [Google Scholar]
- [28].Zammaretti F, Panzica G, Eva C. Sex-dependent regulation of hypothalamic neuropeptide Y-Y1 receptor gene expression in moderate/high fat, high-energy diet-fed mice. J Physiol 2007;583: 445–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Allison SJ, Baldock PA, Enriquez RF, Lin E, During M, Gardiner EM, Eisman JA, Sainsbury A, Herzog H. Critical interplay between neuropeptide Y and sex steroid pathways in bone and adipose tissue homeostasis. J Bone Miner Res 2009;24: 294–304. [DOI] [PubMed] [Google Scholar]
- [30].Han R, Li A, Li L, Kitlinska JB, Zukowska Z. Maternal low-protein diet up-regulates the neuropeptide Y system in visceral fat and leads to abdominal obesity and glucose intolerance in a sex- and time-specific manner. FASEB J 2012;26: 3528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Elefteriou F, Takeda S, Liu X, Armstrong D, Karsenty G. Monosodium glutamate-sensitive hypothalamic neurons contribute to the control of bone mass. Endocrinology 2003;144: 3842–7. [DOI] [PubMed] [Google Scholar]
- [32].Wong IP, Nguyen AD, Khor EC, Enriquez RF, Eisman JA, Sainsbury A, Herzog H, Baldock PA. Neuropeptide Y is a critical modulator of leptin’s regulation of cortical bone. J Bone Miner Res 2013;28: 886–98. [DOI] [PubMed] [Google Scholar]
- [33].Bouxsein ML, Myers KS, Shultz KL, Donahue LR, Rosen CJ, Beamer WG. Ovariectomy-induced bone loss varies among inbred strains of mice. J Bone Miner Res 2005;20: 1085–92. [DOI] [PubMed] [Google Scholar]
- [34].Turner CH, Hsieh YF, Muller R, Bouxsein ML, Baylink DJ, Rosen CJ, Grynpas MD, Donahue LR, Beamer WG. Genetic regulation of cortical and trabecular bone strength and microstructure in inbred strains of mice. J Bone Miner Res 2000;15: 1126–31. [DOI] [PubMed] [Google Scholar]
- [35].Li CY, Schaffler MB, Wolde-Semait HT, Hernandez CJ, Jepsen KJ. Genetic background influences cortical bone response to ovariectomy. J Bone Miner Res 2005;20: 2150–8. [DOI] [PubMed] [Google Scholar]
- [36].Sousa DM, Baldock PA, Enriquez RF, Zhang L, Sainsbury A, Lamghari M, Herzog H. Neuropeptide Y Y1 receptor antagonism increases bone mass in mice. Bone 2012;51: 8–16. [DOI] [PubMed] [Google Scholar]
- [37].Sousa DM, Conceicao F, Silva DI, Leitao L, Neto E, Alves CJ, Alencastre IS, Herzog H, Aguiar P, Lamghari M. Ablation of Y1 receptor impairs osteoclast bone-resorbing activity. Sci Rep 2016;6: 33470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mikic B, Zhang M, Webster E, Rossmeier K. Effect of y2 receptor deletion on whole bone structural behavior in mice. Anat Rec (Hoboken) 2008;291: 14–8. [DOI] [PubMed] [Google Scholar]
- [39].Khor EC, Yulyaningsih E, Driessler F, Kovacic N, Wee NKY, Kulkarni RN, Lee NJ, Enriquez RF, Xu J, Zhang L, Herzog H, Baldock PA. The y6 receptor suppresses bone resorption and stimulates bone formation in mice via a suprachiasmatic nucleus relay. Bone 2016;84: 139–147. [DOI] [PubMed] [Google Scholar]
- [40].Khosla S, Oursler MJ, Monroe DG. Estrogen and the skeleton. Trends Endocrinol Metab 2012;23: 576–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gomes HL, Graceli JB, Goncalves WL, dos Santos RL, Abreu GR, Bissoli NS, Pires JG, Cicilini MA, Moyses MR. Influence of gender and estrous cycle on plasma and renal catecholamine levels in rats. Can J Physiol Pharmacol 2012;90: 75–82. [DOI] [PubMed] [Google Scholar]
- [42].Zengin A, Nguyen AD, Wong IP, Zhang L, Enriquez RF, Eisman JA, Herzog H, Baldock PA, Sainsbury A. Neuropeptide Y mediates the short-term hypometabolic effect of estrogen deficiency in mice. Int J Obes (Lond) 2013;37: 390–8. [DOI] [PubMed] [Google Scholar]
- [43].Cao JJ. Effects of obesity on bone metabolism. J Orthop Surg Res 2011;6: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cao JJ, Picklo MJ. N-acetylcysteine supplementation decreases osteoclast differentiation and increases bone mass in mice fed a high-fat diet. J Nutr 2014;144: 289–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.