Abstract
Recombinantly expressed proteins are susceptible to oxidation during expression, purification, storage and analysis; the residue most susceptible to oxidation is methionine. Methionine oxidation can be overestimated using current quantitative analysis methods because oxidation can occur during sample preparation, and researchers often do not use methods that account for this possibility. An experimental strategy had been developed previously to solve this problem through the use of an 18O labeled hydrogen peroxide reagent. However, the method did not address the analysis of peptides that contained multiple methionine residues. Herein, we develop and validate a new analysis method that uses theoretical isotope distributions and experimental spectra to quantify methionine oxidation that is present prior to sample preparation. The newly described approach is more rapid than the previously described method, and it needs only half the amount of protein for analysis. This method was validated using model proteins; then it was applied to the analysis of recombinant HIV-1 Env, the key protein in HIV vaccine candidates. While Met oxidation of this protein could not be analyzed using previous methods, the approach described herein was useful for determining the oxidation state of HIV-Env.
For TOC only:
INTRODUCTION
Methionine oxidation is an example of a post-translational modification (PTM) that requires careful monitoring when proteins are recombinantly expressed for biotherapeutic applications. This modification has been shown to be detrimental to protein folding and function, and it can cause immunogenic effects.1–4 Methionine oxidation has been linked to a number of diseases such as cataracts, rheumatoid arthritis, and Alzheimer’s.5,6 Met residues are susceptible to oxidation throughout the production of a recombinantly expressed protein. Different cell lines and expression conditions can affect the oxidation of a protein.7,8 Oxidation can be introduced during the steps required to purify the protein,9,10 and storage conditions have been shown to have an effect on methionine oxidation in proteins.11 Methionine oxidation can also be introduced during the steps required to analyze tryptic digests and during LC-MS analysis.12–14
Methionine oxidation has been quantified using liquid chromatography in conjunction with UV-Vis, mass spectrometry, or both detectors.15–19 However, the cited methods cannot be used to distinguish if oxidation occurs during the production of recombinant proteins (expression, purification, storage) or if it is an artifact introduced during protein digestion and analysis. Liu et al. have developed a method that accurately quantifies Met oxidation prior to sample preparation for LC-MS.14 Prior to tryptic digest the proteins are incubated with isotopically labeled H218O2. This procedure converts all unoxidized methionine residues into a labeled 18O form; all residues that were already oxidized remain present in their natural isotopic abundances. The relative quantities of the 16O peak and the 18O peak are then used to determine the amount of oxidation present prior to the labeling.14
In Liu’s work, the data analysis strategy requires researchers to accurately quantify the monoisotopic 16O peak and the 18O peak. The size of these peaks decreases with increasing peptide size, so the method is not well suited for measuring large analytes whose monoisotopic mass is not abundant. Furthermore, the data analysis approach is not directly applicable to peptides that contain multiple methionine residues due to the isotopic overlap of masses that are used to quantify the oxidation. The method also uses control samples that introduce additional degrees of experimental error, increase the required sample quantity, and decrease the throughput of the analyses.
Herein, we extend Liu’s original quantitative method by removing the need for a control sample and modifying the data analysis steps. Eliminating the control sample cuts the protein consumption and analysis time in half, and modifying the analysis steps allows researchers to analyze larger peptides, including those with multiple methionines. The oxidation of analytes with multiple methionines cannot be accurately quantified now by any existing method, but this kind of analysis is important because these species are present in many therapeutic proteins, including HIV-1 Env, an important recombinant protein that is a key component of many emerging HIV-1 vaccine strategies.20–22
The data analysis approach tested and validated herein compares theoretical and experimental spectra and minimizes the error between them to find the amount of native oxidation in peptides. A similar method was used to determine 15N enrichment in yeast samples.23 MacCoss et al. correlated experimentally generated spectra to a set of theoretically generated spectra using a Pearson correlation function. Results were comparable to what was measured using SIM-GC/MS, with the advantage that the isotopic enrichment was measured without standards,23 similarly our data analysis strategy eliminates the need for control samples, and because of this change, both the analysis time and protein consumption are cut in half.
The new data analysis technique was validated on an IgG monoclonal antibody and additionally applied to a protein that has multiple methionine residues on a single peptide, RNAse B. We then applied this method to HIV-1 Env, a recombinant form of the surface antigen on the HIV-1 virus.20–22 We analyzed the methionine residues and determined that the oxidation in the samples was low.
EXPERIMENTAL
Materials and Reagents
Trizma hydrochloride, trizma base, acetic acid, HPLC grade acetonitrile, methanol, ethanol, 4-vinyl pyridine, isotopically labeled hydrogen peroxide (H218O2), formic acid, Ribonuclease B (RNAse B), Immunoglobulin G (IgG), and bovine serum albumin (BSA) were purchased from Sigma (St. Louis, MO). Molecular weight cutoff filters (MWCO) were purchased form EMD Millipore (Billerica, MA). Water was purified using a Millipore Direct-Q3 Water Purification System (Billerica, MA). Sequencing grade trypsin and Peptide:N-Glycosidase F (PNGase F) from Elizabethkingia meningosepticum were obtained from Promega (Madison, WI) and Calbiochem (San Diego, CA), respectively. The HIV-1 Env protein, CH505 gp150, was expressed and purified as described previously.24
Sample Preparation
Proteins were prepared in 50 mmol tris buffer at a concentration of 1 mg/mL and incubated with an equal volume of 1% H218O2 for 2 hours at room temperature in the dark; control samples were oxidized under the same conditions with H2O2. Samples were buffer exchanged with 50 mmol tris buffer using 50 kDa (IgG and gp150) or 3 kDa (RNAse B) molecular weight cutoff filters (MWCO), centrifuging for 20 min. at 14,000 × g. Following buffer exchange, samples were alkylated for one hour at room temperature in the dark using vinylpyridine at a concentration ten times the molar excess of the protein, then deglycosylated for one week using 1 μL of PNGase F at a concentration of 1500 units/μL at 37 °C. Finally, proteins were digested for 18 hours at 37 °C using trypsin at a trypsin to protein ratio of 1:25 (w/w).
LC-MS
The tryptic digests were separated on an Acquity HPLC system (Waters, Milford, MA) coupled to an LTQ Velos Pro Mass Spectrometer (ThermoScientific, San Jose, CA). Mobile Phase A was 99.9% water and 0.1 % formic acid. Mobile Phase B was 99.9% acetonitrile and 0.1% formic acid. A 5 μL sample was injected at a concentration of 1 pmol/μL and separated on a 15 × 0.32 mm Aquasil C18 column, at a flow rate of 7 μL/min. Peptides were eluted using the following gradient: the column was equilibrated to 2% B prior to injection; the gradient was ramped to 40% B over 30 min, then ramped to 60% B for 15 min; the gradient was finally increased to 90% B and held for 10 min to wash the column; it was re-equilibrated at 2% B for 20 min. Samples were infused into ESI ion source, maintained at a potential of 3kV and a capillary temperature of 250 °C. The mass spectrometer was operated in the positive ion mode from m/z 400 to 2000. The resolution was 30,000 at m/z 400.
Data analysis
For all analyses: MS ion counts were quantified using a mass width of 10 ppm and the retention time corresponding to the full width at half maximum (FWHM) of the most abundant isotopic peak; however, for broad peaks, the maximum retention time was 90 seconds. Different peak areas were quantified, depending on the analysis approach implemented, since the existing method (used for validation) and the new method (described herein) make use of different peaks. In validation studies, the 16O and 18O peak areas were measured, and then adjusted to compensate for contributions from reagent impurity and isotopic overlap, as described before.14 Oxidation was calculated by dividing the adjusted 18O area by the sum of the adjusted 18O and 16O areas.
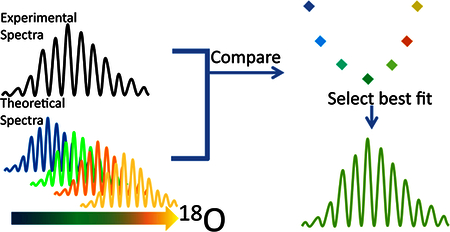
For the data analysis strategy developed herein, henceforth referred to as MOLIEE (Methionine Oxidation Least Isotopic Error Equivalency), all m/z’s across an isotopic envelope were used for the analysis. Experimental fractional isotopic abundances were found using peak areas calculated in Xcalibur, and these were compared to theoretical values. Theoretical fractional isotopic abundances for spectra with varying amounts of 18O incorporation were generated using IDCalc, which is available at https://proteome.gs.washington.edu/software/IDCalc/. The theoretical and experimental fractional abundances of a given m/z were compared by taking the absolute value of the difference between the theoretical and experimental fractional abundances at each m/z and dividing this value by the theoretical fractional abundance. This procedure was repeated for all m/z’s across an isotopic envelope, and the results for all the m/z’s were averaged, giving the average relative error (ARE) between the theoretical and experimental spectra for a given theoretical 18O content. The goal of this comparison - between the theoretical and the actual spectra - was to identify the theoretical content of 18O that minimized the average relative error between the two spectra, so that the percent of 18O in the experimental spectrum could be determined. Therefore, the abundance of 18O was adjusted in the theoretical spectra, and the ARE was plotted against the 18O content of the theoretical spectra, so that a minimum ARE could be determined. In order to eliminate potential errors caused by the limited dynamic range of the detector, only peaks that had an experimental fractional isotopic abundance greater than 1% were considered in the calculations.
Results and Discussion
The goal of this study is to develop a method to measure Met oxidation for large peptides and those with multiple Mets. The overall approach is to improve upon an existing published method, from Liu et al.,14 by using their sample preparation method and adding a novel data analysis strategy that extends the utility of their method. The previously described sample preparation strategy is optimal already because it eliminates the risk of over-estimating oxidation present in samples by labeling all unoxidized Met residues with 18O using H218O2. Figure 1 shows the sample preparation method. After LC-MS analysis of the oxidized, digested peptides, the amount of oxidation present prior to sample preparation can be quantified by comparing Met’s that are 18O labeled to those that are oxidized in natural isotopic abundances. Liu et al’s methods provide a new means to accurately quantify the oxidation present in recombinantly expressed proteins, but the data analysis strategy is limited to simple analytes because the methods they use to account for isotopic overlap using control standards become unfeasible as the number of methionine residues on a peptide increases.
Figure 1:
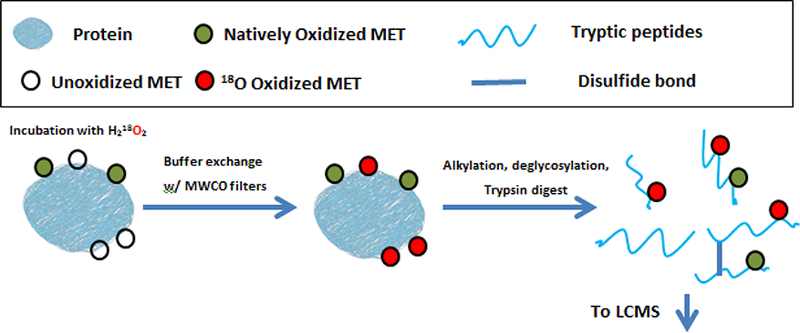
Sample preparation strategy for analysis of oxidized methionine residues.
We extend the methods herein by using the same H218O2 labeling strategy but a unique data analysis approach, which is better suited for more complex analytes, MOLIEE (methionine oxidation least isotopic error equivalency). MOLIEE calculates Met oxidation using the entire isotopic envelope. By contrast, Liu et als14 methods calculate the oxidation using only the first and third peaks in the isotopic cluster of the experimental spectra, and these peaks decrease in size as the size of the peptide increases. Furthermore, as the number of methionine residues on a peptide increase, the complexity of isotopic overlap in a peptide increases and the previously published method becomes unfeasible. MOLIEE is also uniquely suited for samples with multiple methionine residues. Data collected herein are analyzed using both data analysis strategies shown in Figure 2; the oxidation quantified in both strategies is compared to validate MOLIEE using the previously published data analysis strategy.14
Figure 2:
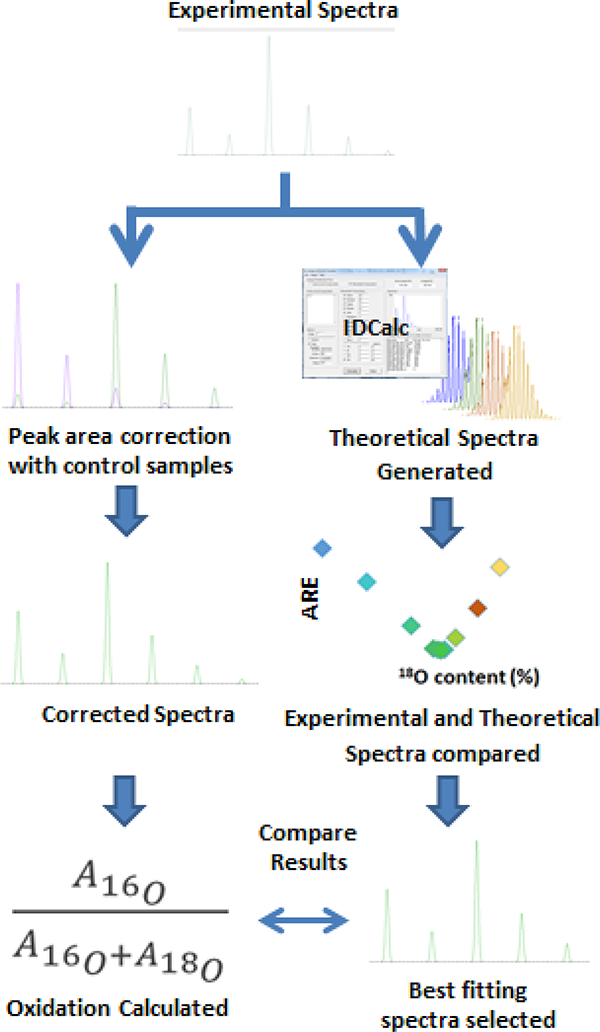
Method validation for single methionine containing peptides. Mass spectra were generated and data were analyzed using a previously published data analysis method17 (left) and MOLIEE (right). Results were compared to validate MOLIEE.
The first model protein used for method validation was IgG, a monoclonal antibody with two conserved methionine residues in the Fc region. IgG was chosen as a model because monoclonal antibodies are a very important class of biotherapeutics, and the oxidation of their methionines can lead to protein degredation, which erodes product quality.25 Upon tryptic digest of IgG, one of the Met’s of interest resides on a 7 residue peptide, and the other resides on a more complex peptide that includes a disulfide bond. These residues were chosen for analysis because they are known to be susceptible to oxidation, and they have been characterized in a number of studies.4,11,14 The results comparing Met oxidation for the two peptides in IgG are shown in Table 1. The first peptide, DTLMISR, was oxidized to a minor extent with both analysis methods measuring less than 10% oxidation. The second peptide was oxidized to a slightly higher degree, with both methods reporting about 16% oxidation. The results from both peptides, summarized in Table 1, suggest that the two data analysis strategies agree with each other within a reasonable margin of error.
Table 1:
Validation studies in model proteins comparing data analysis methods
| Protein | Peptide or disulfide | Peak area quantitation | MOLIEE |
|---|---|---|---|
| IgG | DTLMISR | 7.5 ± 0.3 | 9.7± 0.2 |
| IgG | NQVSLTCLVK WQQGNVFSCSVMHEALHNHYTQK |
16.0 ± 4.2 | 15.8 ± 1.4 |
| RNAse B | QHMDSSTSAASSSNYCNQMMK NGQTNCYQSYSTMSITDCR NVACK |
N/A | 2.6 ± 0.1 |
Next, MOLIEE was used to analyze Met oxidation in RNAse B to demonstrate its applicability to peptides with multiple methionine residues. This protein contains four Met residues; when the protein is proteolytically cleaved with trypsin, all four Mets reside within two peptides that are connected by a disulfide bond. The oxidation in RNAse B, using the data analysis strategy described herein, was found to be 2.6 +/−0.1%. (See Table 1.) The previously described data analysis strategy could not be used to quantify Met oxidation in RNAse B because the isotopic overlap resulting from the multiple oxidized residues cannot feasibly be accounted for in that strategy.
The data in Table 1 demonstrate that MOLIEE is capable of analyzing large peptides; however, the accuracy of MOLIEE in analyzing peptides containing multiple methionine residues could not be assessed through comparison to the previous data analysis strategy. Therefore, further experiments were conducted with RNAse B in order to validate the new method’s accuracy. Normal hydrogen peroxide and 18O labeled hydrogen peroxide were mixed in different ratios and used to oxidize RNAse B to simulate different amounts of oxidation that could be present prior to sample preparation; a diagram of the experiment is shown in Figure 3. Three samples were prepared with different ratios of labeled and unlabeled hydrogen peroxide. The expected values of oxidation in the samples were 77%, 54%, and 31%, and the values calculated using MOLIEE were 75%, 49%, and 33%, respectively. In this case, the three samples showed low deviations from the true values, with two of the three samples showing a 2% error, and the highest error being a 5% deviation for the sample containing 54% oxidation. It is likely that the greatest source of error in these experiments came from preparing the mixtures of H216O2 and H218O2 and not from instrumental variations that would lead to an incorrect assessment of the isotopic distribution of the sample. Technical replicates had very tight precision, with deviations never exceeding 2% error, and this high degree of precision was also observed in the earlier validation experiment described in Table 1. Furthermore, the experiment in Figure 3 demonstrates that the method does not appear to introduce experimental bias. These validation data would suggest that a reliable estimate of the uncertainty of the measurement would be +/− 2% oxidation in most cases. In sum, these results indicate that the newly developed method is uniquely suited for the accurate quantitation of oxidation in analytes that contain multiple methionine residues.
Figure 3:
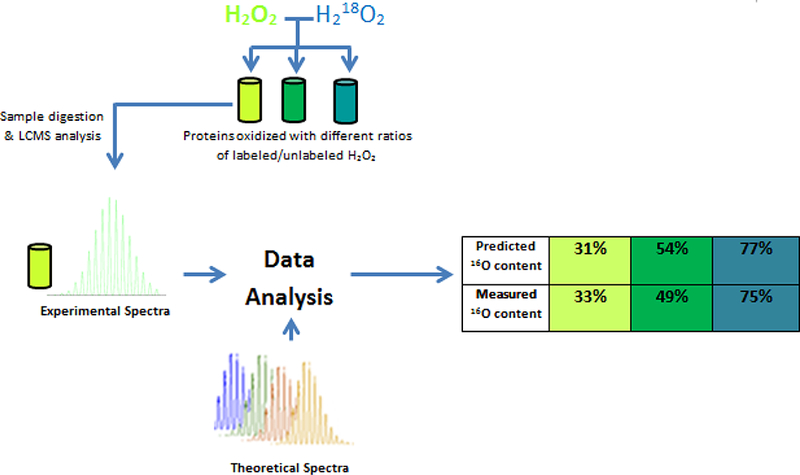
Different ratios of labeled and unlabeled hydrogen peroxide were used to oxidize RNAse B, and peptides were analyzed using MOLIEE to verify that the new data analysis method could accurately measure oxidation in peptides with multiple methionine residues.
Application: Met Oxidation in HIV-1 Env.
After method validation was completed, the approach described herein was applied to a pharmaceutically valuable protein, HIV-1 Env, which is the surface antigen on the HIV-1 virus. The oxidation in ten tryptic peptides containing 14 Met residues was quantified using MOLIEE, including four peptides with multiple Mets. Nine of the Met residues showed negligible oxidation, less than or equal to one percent. Minor oxidation between four and six percent was quantified in three tryptic peptides containing five Met residues. The error in the measurement is estimated to be +/− 2% oxidation, based on all the validation experiments carried out prior to the Env samples. A summary of the peptides, their oxidation, and the associated error can be found in Table 2. This is the first published assessment of site-by-site Met oxidation on any HIV-Env protein, and this work demonstrates that the native form of this protein is not oxidized substantially. Met oxidation analysis can therefore be used as one method to profile an Env protein’s quality during protein production for immunogenicity studies and vaccine trials. MOLIEE was capable of quantifying oxidation in multiple-methionine containing peptides residues present in Env, while previously established methods cannot be used for this analysis. MOLIEE also eliminates the need for control samples, minimizing the consumption of precious protein samples.
Table 2:
Quantified oxidation using MOLIEE in methionine containing peptides from HIV-1 Env, the uncertainty of the analysis is estimated to be ±2%
| Peptide | Measured oxidation |
|---|---|
| SAVGMGAVFLGFLGAAGSTMGAASITLTVQAR | <1% |
| NDMVDQMHEDVISLWDQSLKPCVK | 5% |
| EVHNVWATHACVPTDPNPQEMVLK | 1% |
| AMYAPPIAGDITCISDITGLLLTR | <1% |
| TYGDIWDDMTW MQWER | 4% |
| TYMADSTDMADSTETDSTR | <1% |
| DNTETFRPGGGNMK | <1% |
| QIINMWQEVGR | <1% |
| DVTENFNMWK | <1% |
| AIEAQQHMLK | 6% |
Conclusion
We have developed and validated a new data analysis method, MOLIEE, to quantify methionine oxidation in proteins. This method provides improved reproducibility when measuring large peptides and is uniquely suitable for quantifying analytes with multiple methionine residues. The method was applied to HIV-1 Env, and the quantified Met oxidation was low.
Acknowledgements:
We would like to thank Drs. Barton F. Haynes and S. Munir Alam at the Duke Human Vaccine Institute for kindly providing the Env protein for these studies. We would also like to thank our funders, NIH grant R01AI125093, R01AI094797 and T32-GM008359.
References
- 1.Lu HS, Fausset PR, Narhi LO, Horan T, Shinagawa K, Shimamoto G, Boone TC. “Chemical Modification and Site-Directed Mutagenesis of Methionine Residues in Recombinant Human Granulocyte Colony-Stimulating Factor: Effect on Stability and Biological Activity,” Arch. Biochem. Biophys 1999, 362, 1–11. [DOI] [PubMed] [Google Scholar]
- 2.Luo X, Uehara H, Shacter E. “Taurine Chloramine-Induced Inactivation of Cofilin Protein Through Methionine Oxidation,” Free Radic. Biol. Med 2014, 75, 84–94. [DOI] [PubMed] [Google Scholar]
- 3.Barb AW, Prestegard JH. “NMR Analysis Demonstrates Immunoglobulin G N-glycans are Accessible and Dynamic,” Nat. Chem. Biol 2011, 7(3), 147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaza-Bulseco G, Faldu S, Hurkmans K, Chumsae C, Liu H. “Effect of Methionine Oxidation of a Recombinant Monoclonal Antibody on the Binding Affinity to Protein A and Protein G,” J. Chromatogr. B 2008, 870, 55–62. [DOI] [PubMed] [Google Scholar]
- 5.Castegna A, Aksenova Mi, Aksenova Ma, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesvery WR, Butterfield DA. “Proteomic Identification of Oxidatively Modified Proteins in Alzheimer’s Disease Brain. Part I: Creatine Kinase BB, Glutamine Synthase, and Ubiquitin Carboxy-Terminal Hydrolase L-1,” Free Radic. Biol. Med 2002, 33(4), 562–571. [DOI] [PubMed] [Google Scholar]
- 6.Garner MH, Spector A. “Selective Oxidation of Cysteine and Methionine in Normal and Senile Cataractous Lenses,” Proc. Natl. Acad. Sci. U.S.A 1980, 77(3), 1274–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berti PJ, Ekiel I, Lindahl P, Abrahamson M, Storer AC. “Affinity Purification and Elimination of Methionine Oxidation in Recombinant Human Cystatin C,” Protein Expr. Purif. 1997, 11, 111–118. [DOI] [PubMed] [Google Scholar]
- 8.Lam XM, Yang JY, Cleland JL, “Antioxidants for Prevention of Methionine Oxidation in Recombinant Monoclonal Antibody HER2,” J. Pharm. Sci 1997, 86(11), 1250–1255. [DOI] [PubMed] [Google Scholar]
- 9.Krishnamurthy R, Madurawe RD, Bush KD, Lumpkin JA. “Conditions Promoting Metal-Catalyzed Oxidations During Immobilized Cu-lminodiacetic Acid Metal Affinity-Chromatography,” Biotechnol. Prog 1995, 11, 643–650. [Google Scholar]
- 10.Sun G, Anderson VE. “Prevention of Artifactual Protein Oxidation Generated During Sodium Dodecyl Sulfate-gel Electrophoresis,” Electrophoresis. 2004, 25, 959–965. [DOI] [PubMed] [Google Scholar]
- 11.Chumsae C, Gaza-Bulseco G, Sun J, Liu H. “Comparison of Methionine Oxidation in Thermal Stability and Chemically Stressed Samples of a Fully Human Monoclonal Antibody,” J. Chromatogr. B 2007, 850, 285–294. [DOI] [PubMed] [Google Scholar]
- 12.Zang L, Carlage T, Murphy D, Frenkel R, Bryngelson P, Madsen M, Lyubarskaya Y. “Residual Metals Cause Variability in Methionine Oxidation Measurements in Protein Pharmaceuticals Using LC-UV/MS Peptide Mapping,” J. Chromatogr. B 2012, 895–896, 71–76. [DOI] [PubMed] [Google Scholar]
- 13.Cook KD, Chen M. “Oxidation Artifacts in the Electrospray Mass Spectrometry of Aβ Peptide,” Anal. Chem 2007, 79, 2031–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Ponniah G, Neill A, Patel R, Andrien B, “Accurate Determination of Protein Methionine Oxidation by Stable Isotope Labeling and LC-MS Analysis,” Anal. Chem 2013, 85, 11705–11709. [DOI] [PubMed] [Google Scholar]
- 15.Sharp JS, Becker JM, Hettich RL. “Protein Surface Mapping by Chemical Oxidation: Structural Analysis by Mass Spectrometry,” Anal. Biochem 2003, 313, 216–225. [DOI] [PubMed] [Google Scholar]
- 16.Duenas ET, Keck R, Devos A, Jones AJS, Cleland JL, “Comparison between Light Induced and Chemically Induced Oxidation of rhVEGF,” Pharm. Res 2001, 18(10), 1455–1460. [DOI] [PubMed] [Google Scholar]
- 17.Bjellaas T, Holm A, Molander P, T∅rnes JA, Greibrokk T, Lundanes E. “Trace Determination of Peptides in Water Samples Using Packed Capillary Liquid Chromatography with UV and MS Detection and Characterization of Peptide Oxidation Products by MS,” Anal. Bioanal.Chem 2004, 378, 1021–1030. [DOI] [PubMed] [Google Scholar]
- 18.Brock JWC, Ames JM, Thorpe SR, Baynes JW. “Formation of Methionine Sulfoxide during Glycoxidation and Lipoxidation of Ribonuclease A,” Arch. Biochem. Biophys 2007, 457(2), 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houde D, Kauppinen P, Mhatre R, Lyubarskaya Y. “Determination of Protein Oxidation by Mass Spectrometry and Method Transfer to Quality Control,” J. Chromatogr. A 2006, 1123, 189–198. [DOI] [PubMed] [Google Scholar]
- 20.Haynes BF. “New Approaches to HIV Vaccine Development,” Curr. Opin. Immunol 2015, 35, 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Gils MJ, Sanders RW. “Broadly Neutralizing Antibodies against HIV-1: Templates for a Vaccine,” Virology. 2013, 435, 46–56. [DOI] [PubMed] [Google Scholar]
- 22.Sanders RW, Moore JP. “Native-like Env Trimers as a Platform for HIV-1 Vaccine Design,” Immunol. Rev 2017, 275, 161–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacCoss MJ, Wu CC, Matthews DE, Yates III JR. “Measurement of the Isotope Enrichment of Stable Isotope-Labeled Proteins Using High-Resolution Mass Spectra of Peptides,” Anal. Chem 2005, 77, 7646–7653. [DOI] [PubMed] [Google Scholar]
- 24.Go EP, Ding H, Zhang S, Ringe RP, Nicely N, Hua D, Steinbock RT, Golabek M, Alin J, Alam SM, Cupo A, Haynes BF, Kappes JC, Moore JP, Sodroski JG, Desaire H. “A Glycosylation Benchmark Profile for HIV-1 Envelope Glycoprotein Production Based on Eleven Env Trimers,” J. Virol 2017, 91(9), e02428–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang R, Jain T, Lynaugh H, Nobrega RP, Lu X, Boland T, Burnina I, Sun T, Caffry I, Brown W, Zhi X, Lilov A, Xu Y. “Rapid Assessment of Oxidation via Middle-Down LCMS Correlates with Methionine Side-Chain Solvent-Accessible Surface Area for 121 Clinical Stage Monoclonal Antibodies.” mAbs. 2017, 9(4), 646–653. [DOI] [PMC free article] [PubMed] [Google Scholar]


