Abstract
Cerebellar Purkinje cells (PCs) show conspicuous damages in many ataxic disorders. Targeted delivery of short nucleic acids, such as antisense oligonucleotides to PCs may be a potential treatment for ataxic disorders, especially spinocerebellar ataxias (SCAs), which are mostly caused by a gain of toxic function of the mutant RNA or protein. However, oligonucleotides do not cross the blood-brain barrier (BBB), necessitating direct delivery into the central nervous system (CNS) through intra-thecal, intra-cisternal, intra-cerebral ventricular, or stereotactic parenchymal administration. We have developed a novel liposome (100 to 200 nm in diameter) formulation, DCL64, composed of dipalmitoyl-phosphatidylcholine, cholesterol and poloxamer L64, which incorporates oligonucleotides efficiently (≥70%). Confocal microscopy showed that DCL64 was selectively taken up by brain microvascular endothelial cells by interacting with low density lipoprotein receptor (LDLr) family members on cell surface, but not with other types of lipid receptors such as caveolin or scavenger receptor class B type 1. LDLr family members are implicated in brain microvascular endothelial cell endocytosis/transcytosis, and are abundantly localized on cerebellar PCs. Intravenous administration of DCL64 in normal mice showed distribution of oligonucleotides to the brain, preferentially in PCs. Mice that received DCL64 showed no adverse effect on hematological, hepatic and renal functions in blood tests, and no histopathological abnormalities in major organs. These studies suggest that DCL64 delivers oligonucleotides to PCs across the BBB via intravenous injection with no detectable adverse effects. This property potentially makes DCL64 particularly attractive as a delivery vehicle in treatments of SCAs.
Keywords: blood brain barrier, liposomes, low density lipoprotein receptor, oligonucleotide, Purkinje cells, spinocerebellar ataxias
Introduction
Spinocerebellar ataxias (SCAs) are a group of autosomal dominant disorders manifested with ataxia. Most SCAs show neurodegeneration primarily involving cerebellar PCs [1–3]. Patients with SCAs typically have onset of ataxia in their adulthood followed by a relentless progression of the disease, often leading to devastating disability and death [4]. To date neither the U.S. Food and Drug Administration (FDA) nor the European Medicines Agency (EMA) has approved drugs to treat ataxia although clinical trials of a handful of drugs have shown symptomatic improvements. Disease-modifying drugs are being developed based on an increasing understanding of the pathogenic mechanisms. SCAs are typically caused by a gain of toxic function by mutant protein or RNA [3]. RNA interference (RNAi) is a particularly promising therapeutic strategy in SCAs because it intervenes in the pathway upstream of complex pathogenic cascades of the protein interactome [5]. There have been ongoing preclinical studies in SCA1, SCA2, SCA3, SCA6 and SCA7, showing encouraging results [6–11]. Therapeutic strategies using RNAi may also be applicable to non-polyQ SCAs caused by untranslated toxic RNA transcripts, such as SCAs 8, 10, 31 and 36 [12], and those caused by point mutations that lead to gain-of-function or dominant-negative effects of the mutant protein or even loss of function of the mutant protein [13].
Antisense oligonucleotides (ASOs), micro RNA (miRNA) and small interfering RNA (siRNA) are short nucleic acids which can be utilized to target specific mRNA with complementary nucleotide sequence. Depending on the design and the target, post-transcriptionally silencing the gene expression, modifying RNA splicing, or even upregulating gene expression can be achieved by degrading the target RNA, blocking RNA-protein interactions, or hindering translation [13–15]. Thus, ASOs, miRNA and siRNA are potential therapeutic agents for a wide variety of disorders. However, since ASOs, miRNAs and siRNA are large (6–12 kDa) negatively charged molecules, they cannot cross the blood brain barrier (BBB). The BBB is comprised of non-fenestrated brain capillaries which are formed by endothelial cells connected by tight junctions that effectively restrict transport of molecules from blood to brain [16]. Currently short nucleic acids are delivered into the central nervous system (CNS) through intra-thecal, intra-cisternal, intra-cerebral ventricular or stereotactic intra-tissue administration. Intravenous (iv) administration of short nucleic acids requires transporters or drug carriers that allow the nucleic acids to be delivered across the BBB. Conjugation of oligonucleotides to antibodies or peptides that bind to proteins expressed on brain microvascular endothelial cells (BMECs), such as transferrin receptors and insulin receptors [16–19] and coating of nanoparticles with polysorbate 80 [20–22] have shown their potential to carry oligonucleotides to the brain across the BBB by peripheral administration. However, potential toxicity, immunogenicity, alterations of specificity and functions of the oligonucleotide, manufacturing scalability and costs of oligonucleotides conjugation to peptides and antibodies remain concerns in clinical applications. While polysorbate 80 has been extensively used as safe food additives, no data is available for delivery of oligonucleotides [23].
BMECs express high levels of lipoprotein receptors [24–26]. The low-density lipoprotein receptor (LDLr) family, which includes the prototype family members, LDLr and LDLr-related protein 1 (LRP-1), has been used as brain delivery targets [27–30]. The Angiopep-2 peptide, derived from a family of Kunitz domain-derived peptides, was found to enhance transcytosis and brain parenchyma accumulation [31], and nanoparticles or polymers conjugated with the angiopep-2 peptide were able to deliver siRNA to U87 orthotopic gliomas [29,30]. The angiopep-2-based delivery is mediated by LRP-1 and, possibly, scavenger receptor class B type 1 (SR-B1) [29,31,32]. High expression of LDLr, LRP-1, and SR-B1 is found not only in BMECs but also in the cerebellum [33,34], and cerebellar PCs express high levels of LDLr and LRP-1 [33].
We developed a novel liposome formulation, DCL64, composed of dipalmitoyl-phosphatidylcholine (DPPC), cholesterol and poloxamer L64. DPPC was used to construct the liposomes because it is biocompatible and easy to manipulate. Cholesterol was added to increase the DPPC liposome affinity to lipoprotein receptors. L64 is a triblock polymer composed of polyethylene oxide-propylene oxide-polyethylene oxide. Göppert and Müller [35] demonstrated that apolipoproteins E, A-I and A-IV were adsorbed to poloxamer L64-containing nanoparticles upon plasma incubation. We hypothesize that liposomes, being lipid particles, will target lipoprotein receptors efficiently, and by binding to LDLr, LRP-1 and SR-B1, DCL64 will efficiently deliver nucleic acids to the cerebellum.
Here we show that iv administration of DCL64 leads to oligonucleotide accumulation in cerebellar PCs. This study presents an important potential of DCL64 for clinical applications, especially in treatment of patients with SCAs by RNAi.
Materials and Methods
Animals
Two to three months old normal ICR mice (Envigo, Indianapolis, IN) were housed under SPF conditions at the University of Florida McKnight Brain Institute animal facility. All animals were euthanized by CO2 asphyxiation prior to tissue collection.
Cell culture
Human BMECs were obtained from Dr. Kwang Sik Kim (Johns Hopkins University) and cultured in Medium-199 (Lonza, Walkersville, MD) supplemented with 5% fetal bovine serum (FBS; Gibco, Waltham, MA), 5% NuSerum (Corning Life Sciences, Durham, NC), and 1% penicillin/streptomycin as described [36]. Murine embryonic NIH-3T3 fibroblasts (American Type Culture Collection, Manassas, VA) were grown in Dulbecco’s modified Eagles medium (Gibco) supplemented with 10% FBS and 1% penicillin/streptomycin.
Liposome preparation
DPPC and cholesterol were purchased from Avanti Polar Lipids (Alabaster, AL) while poloxamer L64 was purchased from Sigma-Aldrich Chemicals (St. Louis, MO). DPPC, cholesterol and L64 were mixed at 5:3:7 weight ratios in the presence of excess tertiary butanol (Thermo Fisher Scientific, Waltham, MA), frozen at −80°C overnight and lyophilized with a FreeZone 2.5 lyophilizer (Labconco, Kansas City, MO) and stored for up to 3 months at 20°C. Fluorescent DPPC or DCL64 liposomes were prepared by adding lissamine rhodamine B-labeled DPPC (Avanti Polar Lipids) to the lipid mixture at 5% weight ratio, frozen and lyophilized.
An 18-base nontoxic oligonucleotide, 5’-AGATGAACTTCAGGGTCA-3’, conjugated with Cyanine3 (Cy3) at the 5’ end was purchased from Trilink Biotechnologies (San Diego, CA). Bases 1, 2, 17, 18 were made of 2’-O-methyl modified phosphodiesters to enhance oligonucleotide stability while bases 3 to 16 were made of normal phosphodiesters. Oligonucleotides were mixed with DCL64 at a 1:10 weight ratio at room temperature. L64 and oligonucleotides were incubated for 30 min, followed by incubation with DPPC for 10 min, then with cholesterol for 10 min. The lipid oligonucleotide mixture was frozen overnight at −80°C and lyophilized.
Flow cytometric analysis
Fluorescent lipids were reconstituted with phosphate buffered saline (PBS) and incubated with BMECs at 200 µM for 24 h [36]. Cells were trypsinized and suspended in PBS containing 2% FBS. Cell suspensions were analyzed on an Accuri C6 flow cytometer (BD Biosciences, San Jose, CA) with a minimum cell count of 10,000 cells per sample. Experiments were repeated at least three independent times using triplicate samples.
Cryo electron microscopy
Liposomes incorporated with oligonucleotides were reconstituted with PBS at 1 mg/mL. Three-microliter aliquots were applied to C-flat holey carbon grids (Protochips, Inc. Morrisville, NC) and vitrified using a VitrobotTM Mark IV (FEI Co., Hillsboro, OR). The sample was examined using a 16-megapixel CCD camera (Gatan Inc., Pleasanton, CA) in a Tecnai (FEI Co.) G2 F20-TWIN Transmission Electron Microscope operated at a voltage of 200 kV using low dose conditions (~20 e/Å2). Images were recorded with a defocus of approximately 3 µm to improve contrast.
Co-localization of liposomes with lipid receptors
BMECs were plated in ibidi chambers (Martinsried, Germany). Two days later, BMECs were incubated with liposomal oligonucleotides (0.5 µg) at 37°C for 1 h. Cells were rinsed twice with PBS containing 2% bovine serum albumin and fixed with 4% paraformaldehyde for 10 min at room temperature. Slides were air dried and incubated at room temperature for 1 h with primary antibodies specific for LDLr, LRP-1, caveolin, or SR-B1 (Abcam, Cambridge, MA). Antibodies concentrations used for LDLr, LRP-1, caveolin, and SR-B1 were 24 µg/mL, 21 µg/mL, 40 µg/mL, and 28 µg/mL, respectively. Slides were then rinsed, incubated with Alexa 488-labeled secondary antibodies (Invitrogen, Carlsbad, CA) at room temperature for an additional hour before being counterstained with DAPI Fluoromount-G (Southern BioTech, Birmingham, AL). All co-localization experiments were repeated at least three independent times using duplicate samples.
To reduce the binding of liposomes to LDLr and LRP-1, BMECs were preincubated with peptides specific for LDLr (R&D Technologies, Minneapolis, MN) or LRP-1 (Abcam) at 37°C for 1 h before incubated with liposomal oligonucleotides for an additional hour. Cells were fixed and counterstained with DAPI. Quantification of red fluorescent oligonucleotide signals was done with Fiji ImageJ. Each image was processed using the “split channels” function, and the number of Cy3 signals in the red channel was then quantified using the “find maxima” function, set at a noise threshold of 80. Number of red fluorescent signals were normalized to cell number, and plotted. Two-tailed t-test showed that fluorescent signals were higher in the absence than in the presence of blocking peptides (p<0.005).
Intravenous administration of liposomes and tissue processing
Normal ICR mice were injected with 5 mg of DCL64 incorporated with 0.5 mg of Cy3-labeled oligonucleotides in 100 µL PBS (n=4). After 24 h, mice were euthanized by CO2 asphyxiation. Before their last breaths, mice were perfused with 4% paraformaldehyde in 10 mL PBS, followed with 10 mL PBS twice by intraaortic injections. Brains were dissected and fixed with 4% paraformaldehyde in PBS for 3–4 h at room temperature. They were then cryoprotected in 30% sucrose in PBS for 36–48 h at 4°C, embedded in optimal cutting temperature (OCT; Tissue-Tek, Sakura Finetek) and frozen in dry ice/isopentane mixture. Cryosections (20 µm; Leica Biosystems, Buffalo Grove, IL) were prepared and stored at 4°C.
Calbindin staining
Mouse brain tissue slides were hydrated with PBS for 10 min, heated at 95°C in sodium citrate buffer for 30 min, and cooled at room temperature for 20 min. Then slides were washed with PBS three times, 10 min each, and blocked with normal goat serum at room temperature for 2 h. After removal of blocking buffer, slides were incubated with anti-calbindin antibody (Sigma-Aldrich) at 4°C overnight, washed with PBS three times before incubated with Alexa 594-labeled secondary antibodies (Invitrogen) at room temperature for 2 h. After washing with PBS, slides were air dried overnight before counterstained with DAPI Fluoromount-G (Southern BioTech).
Optical imaging
Confocal images were acquired using an Olympus IX2-DSU spinning disk confocal microscope (Tokyo, Japan) equipped with a Hamamatsu ORCA-AG camera (Hamamatsu City, Japan), or a Leica SP5 confocal microscope (Heidelberg, Germany). Fluorescent or H&E images were acquired using an Zeiss Axioplan 2 Upright Widefield Fluorescent Microscope (Carl Zeiss Jena GmbH, Germany) equipped with a QImaging Retiga 4000R Monochrome Camera (Surrey, BC, Canada). Images were resized for figures using Adobe Photoshop CS6.
The quantification of Cy3 fluorescent signals in cerebellum, forebrain and midbrain was done with Fiji ImageJ. Eight images each of cerebellum, forebrain and midbrain were quantified. Each image was first processed using the “split channels” function, and the number of Cy3 signals in the red channel was then quantified using the “find maxima” function, set at a noise threshold of 80.
Safety studies
In one safety experiment, 2 groups of 5 ICR mice were used. One group received DPPC liposomes while the other group received DCL64. Liposome doses were given at 5 mg lipids/mouse, which was approximately160 mg lipids/kg of mouse body weight. Blood samples, by performing facial vein bleeds 2 and 6 weeks post-injection, were analyzed for hematologic parameters, which include red blood cell (RBC) counts, white blood cell (WBC) counts, platelet counts, hemoglobin and hematocrit. Blood samples, obtained by performing facial vein bleeds 6 weeks post-injection, were analyzed for renal (blood urea nitrogen and serum creatinine levels) and hepatic biochemical functions (serum alanine aminotransferase and alkaline phosphatase levels). After the blood samples were collected at 6 weeks postinjection, mice were sacrificed by CO2 inhalation and necropsies were performed. Heart, lungs, kidney, spleen, liver and brain were removed from each mouse and fixed by immersion in 10% neutral buffered formalin (Fisher Scientific, Houston, TX, USA) immediately after autopsy. Specimens were processed for embedding in paraffin blocks, and sections (4 μm) were cut and stained with hematoxylin and eosin (H&E).
In another safety experiment, mice (5/group) were injected with DCL64 incorporated with oligonucleotides, once daily for 5 days. Mice were injected with a total of 5 mg liposome doses containing 0.5 mg oligonucleotides, which was approximately 160 mg lipids or 16.0 mg oligonucleotides per kg of mouse body weight. Additional mice were injected with a total of 10 mg liposome doses containing 1.0 mg oligonucleotides, which was approximately 320 mg liposomes or 32.0 mg oligonucleotides per kg of mouse body weight. Non-injected mice were used as controls. Hematological parameters, renal and hepatic biochemical functions were collected and analyzed. Necropsies were performed.
Results
Uptake of DCL64 by brain microvascular endothelial cells
We used liposomes to target lipoprotein receptors. We predicted that inclusion of cholesterol and poloxamer L64 in the liposome formulation would enhance the affinity of DPPC liposomes to lipoprotein receptors. Flow cytometric analysis indicated that DCL64 was efficiently taken up by BMECs compared to DPPC alone, suggesting that inclusion of cholesterol and L64 significantly increased liposome uptake by BMECs, approximately 25-fold higher (Fig 1). We also determined the uptake of DCL64 by NIH3T3 fibroblasts. The uptake of DCL64 by NIH3T3 fibroblasts was about 2-fold higher than that of DPPC (Fig 1). However, the fibroblasts uptake of DCL64 was ~16-fold lower than that of the BMECs (Fig 1).
Fig 1. Selective DCL64 liposomes uptake by brain microvascular endothelial cells (BMECs).
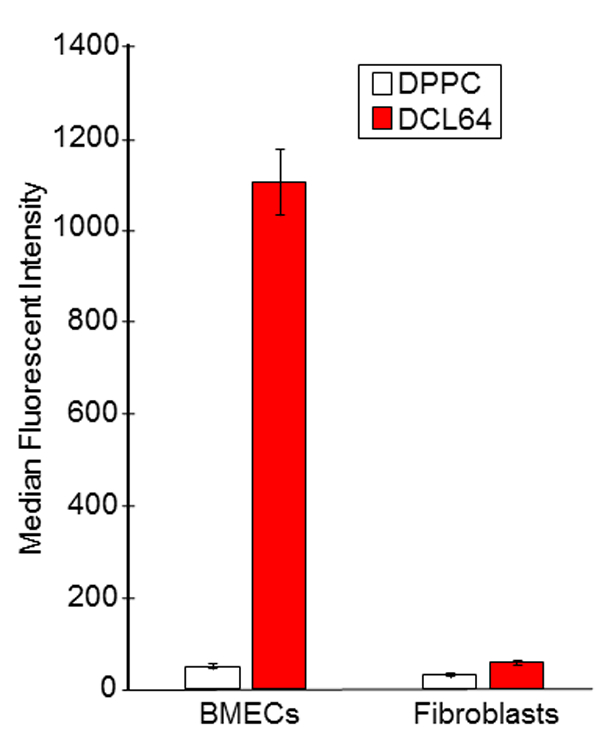
BMECs and fibroblasts were incubated with fluorescent DPPC liposomes (clear bars) or DCL64 liposomes (dark bars). Flow cytometric analysis indicated that incorporating cholesterol and poloxamer L64 into DPPC liposomes increased DCL64 uptake by BMECs but not by fibroblasts.
Cryo electron microscopy of oligonucleotide-incorporated DCL64
Next, we incorporated oligonucleotides into DCL64, and examined their ultrastructural features by cryo electron microscopy (Fig 2). The liposomes were individual spheroid structures and heterogeneous in size. Cryo electron microscopy showed that the size of DCL64-incorporated with oligonucleotides ranged between 100 to 200 nm in diameter (Fig 2).
Fig 2. Cryo electron micrographs of DCL64 liposomes.
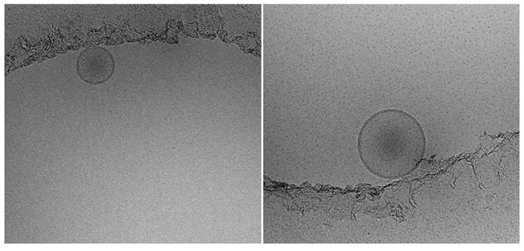
DCL64 liposomes incorporated with oligonucleotides were prepared and cryo electron micrographs were taken. Scale bar: ―――― = 100 microns.
Liposome binding to LDLr family
BMECs are known to express LDLr, LRP-1, caveolin, and SR-B1 [24–26]. We propose that the avid uptake of DCL64 by BMECs is mediated by these lipoprotein/lipid receptors. This is because apolipoproteins E, A-I, and A-IV, which are ligands of LDLr, LRP-1, and SR-B1, are adsorbed to L64. Cholesterol binds to caveolin. Confocal microscopy was utilized to track oligonucleotide and lipoprotein/lipid receptor signals in BMECs. BMECs, in the absence of DCL64, did not show fluorescence (Fig 3A, top panel, left). BMECs incubated with DCL64-incorporated oligonucleotides showed red fluorescence from the Cy3-oligonucleotide cargo (Fig 3A, top panel, right). To determine the uptake mechanism of DCL64 liposomes, antibodies targeted to LDLr, LRP-1, caveolin, or SR-B1 were added to BMECs after incubation with DCL64-incorporated oligonucleotides. Red oligonucleotide signals were found to co-localize with the green LDLr and LRP-1 signals (Fig 3A, middle panels), indicating that DCL64 was bound to LDLr and LRP-1. However, there was very little overlap between the red oligonucleotide signals and the green caveolin or SR-B1 signals (Fig 3A, bottom panels), indicating that DCL64 did not co-localize with caveolin or SR-B1.
Fig 3. Co-localization of DCL64 liposomes with LDLr family.
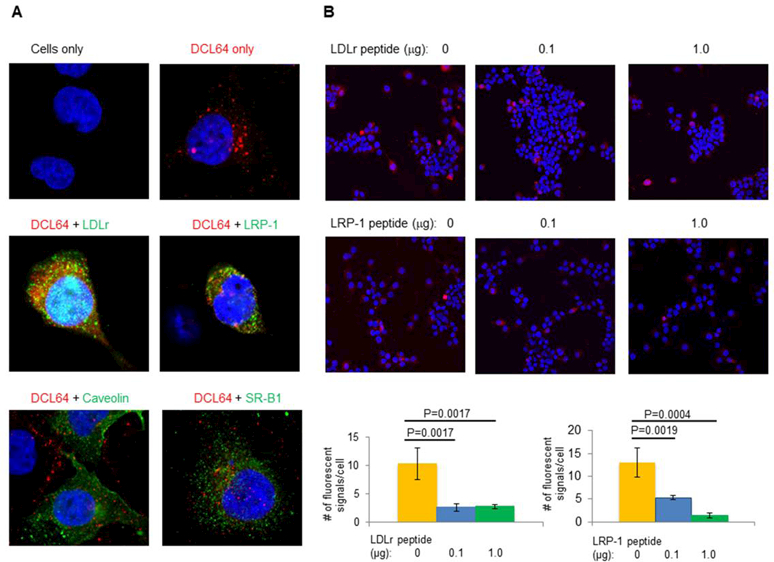
(A) DCL64 liposomes incorporated with oligonucleotides (Cy3) co-localized with LDLr and LRP-1 (Alexa 488 green), but not with caveolin or SR-B1 (Alexa 488 green). (Top left) BMECs only; (top right) BMECs incubated with DCL64; (middle) BMECs incubated with DCL64 followed by antibodies specific for LDLr (left) or LRP-1 (right); (bottom) BMECs incubated with DCL64 followed by antibodies specific for caveolin (left) or SR-B1 (right). (B) LDLr and LRP-1 blocking peptides decreased the binding of fluorescent DCL64 liposomes to BMECs. BMECs were preincubated with LDLr or LRP-1 blocking peptides before incubated with DCL64; (left) 0 µg blocking peptides; (center) 0.1 µg blocking peptides; (right) 1.0 µg blocking peptides. Scale bars: (Panel A) — = 10 microns; (Panel B) ――― = 20 microns.
We then determined whether BMEC uptake of DCL64 could be reduced by LDLr or LRP-1 blocking peptides. Compared to untreated cells, binding of DCL64 to BMECs was reduced when BMECs were pre-incubated with increasing amounts of LDLr-blocking peptides (Fig 3B, top row) or LRP-1-blocking peptides (Fig 3B, middle row). But, caveolin-blocking peptides did not affect DCL64 binding to BMECs (Supplementary Fig 1). These studies indicate that LDLr and LRP-1, but not caveolin or SR-B1, are essential for the BMEC uptake of DCL64.
Intravenous DCL64 injection led to oligonucleotides accumulation in brain and did not have adverse effects in mice
We determined whether iv administration of DCL64 could deliver an oligonucleotide cargo to the cerebellum. We focused on cerebellum as this is the part of the brain where we plan for future targeted therapeutic intervention. PCs in cerebellum were evident by H&E staining (Fig 4A, left). Fluorescent signal was not observed in noninjected mice cerebella (Fig 4A, right), but fluorescent oligonucleotides were found within the cerebella 4 h (Fig 4B, right) and 24 h (Fig 4C, right) post iv injection. DCL64 was localized to PCs which were identified by calbindin staining on the cryosections adjacent to the DCL64 cryosection slides (Fig 4B-C, left), and in dendrites in the molecular layer (Supplementary Fig 2). Fluorescent oligonucleotides were also found elsewhere in the brain including the forebrain (Fig 4D, left) and the midbrain (Fig 4D, right) 24 h post injection. But oligonucleotide signal was higher in the cerebellum than in the forebrain or midbrain (Fig 4E).
Fig 4. Intravenous injection of DCL64 liposomes led to oligonucleotide accumulation in brain, including PCs.
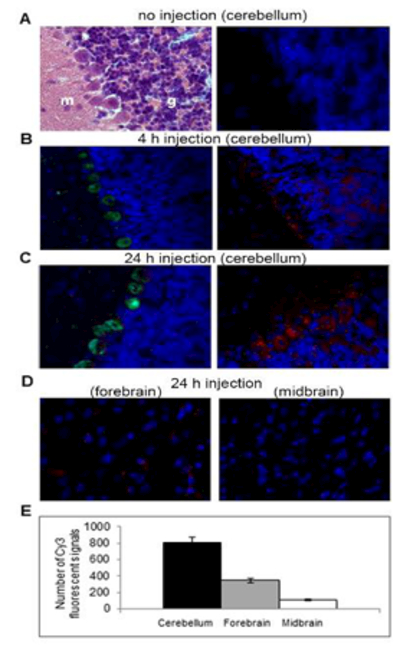
Mice were injected with fluorescent DCL64 and euthanized 4 or 24 h postinjection. (A, left) H&E stain showing the molecular layer (m), granule cell layer (g) and PC layer in between; (A, right) fluorescent micrograph of the same region as the H&E stain in noninjected mouse. (B) Detection of fluorescent oligonucleotide 4 h post DCL64 injection in cerebellum; (left) calbindin staining [pseudo-colored in green] and (right) fluorescent oligonucleotide [red color]. (C) Detection of fluorescent oligonucleotide [in red] 24 h post DCL64 injection in cerebellum; (left) calbindin staining [pseudo-colored in green] and (right) red fluorescent oligonucleotide. Red fluorescence is consistently found in the cytoplasm of PCs. (D) Detection of fluorescent oligonucleotide [in red] 24 h post DCL64 injection in forebrain and midbrain; (left) forebrain and (right) midbrain. Note that red fluorescence signals in these cells are less diffuse and often punctate (D) compared with PCs shown in (C). The results at 4 h post DCL64 injection was similar (data not shown). Scale bars: (Panels A, C, D) — = 10 microns; (Panel B) – = 10 microns. (E) Quantification of red fluorescent oligonucleotide signals in cerebellum, forebrain and midbrain was done with Fiji ImageJ. Eight images each of cerebellum, forebrain and midbrain were processed and quantified. Two-tailed ttest showed that fluorescent signals were higher in cerebellum than forebrain (p<0.05), and midbrain (p<0.005).
We also determined if the DCL64 could have an adverse effect on mice. Intravenous injections of control DPPC or DCL64 liposomes did not affect mouse hematological parameters, hepatic biochemical functions or renal biochemical functions (Tables 1 and 2). The hematological parameters, renal biochemical functions and hepatic biochemical functions were very similar between noninjected mice and mice injected with up to 10 mg of DCL64-incorporated oligonucleotides (Tables 3 and 4). Histopathology of major organs, such as liver, kidneys, spleen, lung, heart and brain, was normal in all animals. These mice did not display any behavioral abnormalities suggestive of brain dysfunctions such as abnormal gait, seizures or seizure-like activities, paresis, spasticity or hind limb clasping [37,38].
Table 1. DCL64 liposomes did not affect mice hematological parameters.
| Weeks postinjection |
Liposomes | Dose (mg) | RBC (106/µL) |
Hemoglobin (g/dL) | Hematocrit (%) | Platelets (103/µL) | WBC (103/µL) |
|---|---|---|---|---|---|---|---|
| 2 | DPPCa | 5 | 11.4±0.6 | 14.9±0.8 | 58.6±2.7 | 1247.4±204.4 | 6.3±0.9 |
| 2 | DCL64a | 5 | 11.5±0.8 | 15.7±0.6 | 62.5±4.2 | 1326.4±150.9 | 7.3±1.4 |
| 6 | DPPCa | 5 | 10.8±0.7 | 14.3±0.7 | 57.4±4.8 | 1321.6±233.2 | 6.0±1.6 |
| 6 | DCL64a | 5 | 10.8±0.6 | 14.5±0.2 | 58.2±1.5 | 1264.4±147.7 | 5.8±1.5 |
Five mice were used.
Table 2. DCL64 liposomes did not affect mice renal or hepatic biochemical functions.
| Weeks
postinjection |
Liposomes | Dose (mg) | BUN (mg/dL) |
Creatinine (mg/dL) | Alanine transferase (%) | Alkaline phosphatase (IU/L) |
|---|---|---|---|---|---|---|
| 6 | DPPCa | 5 | 26.6±2.3 | 0.3±0.1 | 106.0±39.1 | 20.0±20.1 |
| 6 | DCL64a | 5 | 29.2±8.7 | 0.2±0.1 | 112.2±60.0 | 30.8±24.0 |
Five mice were used.
Table 3. DCL64 liposomes incorporated with oligonucleotides did not affect mice hematological parameters.
| Weeks
postinjection |
Treatment | Dose (mg) | RBC
(106/µL) |
Hemoglobin (g/dL) | Hematocrit (%) | Platelets (103/µL) | WBC
(103/µL) |
|---|---|---|---|---|---|---|---|
| 2 | Noninjecteda | 0 | 11.1±0.8 | 14.4±1.3 | 57.3±3.5 | 1039.4±228.6 | 6.7±1.8 |
| 2 | DCL64a | 5 | 10.6±0.3 | 14.1±0.6 | 55.8±2.0 | 1443.0±157.4 | 7.3±1.1 |
| 2 | Noninjectedb | 0 | 10.9±1.3 | 15.5±1.0 | 56.5±5.7 | 1127.5±359.6 | 6.3±0.4 |
| 2 | DCL64b | 10 | 10.8±0.7 | 15.5±1.0 | 54.2±3.6 | 1112.0±205.7 | 5.0±1.5 |
| 6 | Noninjecteda | 0 | 10.4±0.7 | 12.4±1.0 | 51.3±3.9 | 844.0±412.6 | 6.5±1.6 |
| 6 | DCL64a | 5 | 11.2±0.3 | 13.1±0.2 | 60.0±1.4 | 1254.6±142.2 | 6.0±0.5 |
| 6 | Noninjectedb | 0 | 10.9±0.8 | 14.3±0.7 | 56.9±2.5 | 1103.6±219.1 | 5.9±0.7 |
| 6 | DCL64b | 10 | 10.9±0.7 | 13.9±0.7 | 56.6±2.1 | 1084.0±171.3 | 6.7±0.4 |
Five mice were used.
Five mice were used.
Table 4. DCL64 liposomes incorporated with oligonucleotides did not affect mice renal or hepatic biochemical functions.
| Weeks postinjection |
Treatment | Dose (mg) | BUN (mg/dL) |
Creatinine (mg/dL) | Alanine transferase (%) | Alkaline phosphatase (IU/L) |
|---|---|---|---|---|---|---|
| 6 | Noninjecteda | 0 | 28.2±3.0 | 0.3±0.0 | 129.8±60.7 | 36.2±24.9 |
| 6 | DCL64a | 5 | 26.8±3.4 | 0.3±0.0 | 126.8±73.0 | 37.6±18.0 |
| 6 | Noninjectedb | 0 | 30.8±2.5 | 0.3±0.1 | 68.8±14.1 | 58.8±22.5 |
| 6 | DCL64b | 10 | 29.5±3.5 | 0.3±0.1 | 55.5±12.3 | 88.3±18.5 |
Five mice were used.
Five mice were used.
Discussion
Here we show that the novel DCL64 formulation allows oligonucleotide delivery to cerebellar PCs upon iv administration. The data suggest that this formulation has potential to improve the clinical applicability of RNAi technologies in treatments of SCAs. The delivery of ASO, siRNA and miRNA is currently dependent on intrathecal, intracerebral ventricular, intracisternal, or stereotactic brain parenchymal injection. Even with these direct delivery methods to the CNS it is essential that sufficient amounts of RNAi reagents are taken up specifically by cells affected by the disease, which is often not the case. Activity of ASO and siRNA has been shown in cell culture by transfection, using cationic polyplexes. However, cationic DNA polyplexes do not cross the BBB and do not allow gene delivery to the brain [39]. Also, cationic polyplexes could induce erythrocyte aggregation and be trapped in the capillaries of lung tissues, causing pulmonary embolism [40,41]. Thus, the capability of DCL64 to deliver ASO to brain cells, preferentially to cerebellar PCs, provides an important edge to advance RNAi therapeutic development for patients suffering from SCAs.
DPPC, cholesterol, and poloxamer L64 of DCL64 are lipid components that potentially serve as targeting ligands to lipoprotein receptors. The exact delivery mechanism of DCL64 is unclear but its avid uptake by BMECs (Fig 1), blocking of the uptake by LDLr- and LRP-1-specific peptides (Fig 3B0, and its co-localization with LDLr and LRP-1 on BMECs (Fig 3A) suggest that the uptake mechanism involves LDLr family members. The mechanism is likely to be similar to that of nanoparticles coated with surfactant polysorbate 80 (Tween 80), which targets LDLr and SR-B1 and has been used to deliver chemotherapeutics to brain tumors as well as brain-derived neurotropic factor to the brain [20–22,42]. Through apolipoproteins E and A-I, which are ligands of LDLr and SRB-1, the polysorbate 80-coated nanoparticles mimic lipoprotein particles and are taken up by brain endothelial cells via receptor-mediated endocytosis and cross the BBB. Similarly, HDL particles carrying siRNA deposited the cargo in BMECs upon iv administration [27], and LDLr and apolipoprotein E were essential for this as siRNA delivery was significantly reduced in mice deficient in LDLr or apolipoprotein E [27].
Furthermore, poloxamers P85 and P188 have been used to enhance drug delivery to the brain. Poloxamer P85 was conjugated to leptin [43] while P188 was used to coat nanoparticles [44]. The amount of apolipoprotein E adsorbed to the poloxamer-stabilized nanoparticles appeared to be inversely correlated with the number of polyethylene oxide units, and L64 showed the highest adsorption of apolipoprotein E [35]. Apolipoproteins A-I and A-IV were also adsorbed to L64 poloxamer-stabilized nanoparticles [35], suggesting that they could bind to SR-B1. Nonetheless, there was very little co-localization of DCL64 with SR-B1 (Fig 3A). This is likely because the formulation and the preparation of the DCL64 liposome and the poloxamer-stabilized nanoparticles are very different in coating surfactants, core lipids, and entrapped drug [36]. Earlier, we have demonstrated that iv injection deposited liposomes composed of dioleoylphosphatidylcholine, poloxamer P188, and cholesterol (DOPC/P188/Cholesterol) into myelinated peripheral nerves as well as choroid plexus epithelia of brain lateral ventricles [36]. The components of the DOPC/P188/Cholesterol and DCL64 are similar but they co-localize with different receptors. The DOPC/P188/Cholesterol liposomes are associated with lipid rafts which are enriched with caveolins [36], but DCL64 is not co-localized with caveolin (Fig 3A). The different lipid components likely affect the protein adsorption patterns to the liposomes, which likely contribute to different receptors co-localization and brain distribution patterns.
We did not expect DCL64 to have adverse effects in vivo. This is because DPPC and cholesterol are widely abundant, naturally occurring, and non-immunogenic lipids. The safety of various poloxamer polymers has been extensively studied. Poloxamers are relatively non-toxic to animals, with LD50 values reported from 5 to 35 g/kg [45]. Nonetheless, since the LDLr family is expressed ubiquitously, it is necessary to determine if DCL64 could produce adverse effects in vivo in further preclinical studies. The lipid and oligonucleotide doses studied here are similar to those used in our previous reports [46,47], which are the bases of our ongoing liposomal ASOs clinical trials. Complete blood count including platelet count, serum liver transaminase levels, blood urea nitrogen and creatinine levels in mice that received injections of empty DCL64 liposomes and DCL64 liposomal oligonucleotides showed no evidence of hematological, hepatic or renal adverse effects.
In summary, the DCL64 liposome described here provide a promising delivery vehicle of short nuclei acids to ataxia, dystonia, neurobehavioral disorders and other neurological disorders caused by cerebellar pathology. The cost and manufacturing of DCL64 is feasible and easily scalable. The ability of DCL64 to enter cerebellar PCs through systemic administration offers a potential advantage over existing formulations.
Conclusions
We developed a novel DCL64 liposome formulation that is avidly taken up by BMECs, primarily via LDLr and LRP-1. Intravenous administration of DCL64 led to oligonucleotide accumulation in cerebellar PCs. This study presents an important potential of DCL64 for clinical applications, especially in treatment of patients with SCAs.
Supplementary Material
Acknowledgments
We are grateful to the technical support provided by University of Florida (UF) ICBR Cytometry Core, ICBR Electron Microscopy Core, MBI-UF Cell & Tissue Analysis Core, UF Molecular Pathology Core, and UF Animal Care Services. This study was supported by UF Opportunity Grant to ATA and TA.
Compliance with Ethical Standards
Disclosure of potential conflicts of interest:
ATA is an employee and stock holder of Bio-Path Holdings, Inc. TA is Adjunct Professor of Baylor College of Medicine, and receives grants from the NIH (R01 NS083564), the National Ataxia Foundation (NAF), the Myotonic Dystrophy Foundation (MDF), and the Marigold Foundation, and has been supported by Biohaven Pharmaceuticals and Ionis Pharmaceuticals for clinical trials of their drugs, and by Pacific Biosciences for symposium honoraria. TA also serves on the Medical and Research Advisory Board of the NAF, and the Scientific Advisory Board of the MDF. ATA and TA filed a US patent application (Serial # 14/390,584).
Research involving animals:
The animal research in this work was approved by the UF Institutional Animal Care and Use Committee (IACUC #201307878).
Research involving human participants:
This work does not involve human subject research.
References
- 1.Bird TD. Hereditary Ataxia Overview. In: Pagon RA, Adam MP, Ardinger HH, et al. , editors. GeneReviews® [Internet]. Seattle: University of Washington, Seattle; 1998. [updated 2016]. [PubMed] [Google Scholar]
- 2.Shakkottai VG, Fogel BL. Clinical neurogenetics: autosomal dominant spinocerebellar ataxia. Neurol Clin 2013;31:987–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matilla-Dueñas A, Ashizawa T, Brice A, Magri S, McFarland KN, Pandolfo M, et al. Consensus paper: pathological mechanisms underlying neurodegeneration in spinocerebellar ataxias. Cerebellum 2014;13:269–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobi H, du Montcel ST, Bauer P, Giunti P, Cook A, Labrum R, et al. Long-term disease progression in spinocerebellar ataxia types 1, 2, 3, and 6: a longitudinal cohort study. Lancet Neurol 2015;14:1101–8. [DOI] [PubMed] [Google Scholar]
- 5.Bushart DD, Murphy GG, Shakkottai VG. Precision medicine in spinocerebellar ataxias: treatment based on common mechanisms of disease. Ann Transl Med 2016;4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keiser MS, Monteys AM, Corbau R, Gonzalez-Alegre P, Davidson BL. RNAi prevents and reverses phenotypes induced by mutant human ataxin-1. Ann Neurol 2016;80:754–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scoles DR, Meera P, Schneider MD, Paul S, Dansithong W, Figueroa KP, et al. Antisense oligonucleotide therapy for spinocerebellar ataxia type 2. Nature 2017;544:362–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toonen LJ, Schmidt I, Luijsterburg MS, van Attikum H, van Roon-Mom WM. Antisense oligonucleotide-mediated exon skipping as a strategy to reduce proteolytic cleavage of ataxin-3. Sci Rep 2016;6:35200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodríguez-Lebrón E, Costa MD, Luna-Cancalon K, Peron TM, Fischer S, Boudreau RL, et al. Silencing Mutant ATXN3 Expression Resolves Molecular Phenotypes in SCA3 Transgenic Mice. Mol Ther 2013;21:1909–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyazaki Y, Du X, Muramatsu S, Gomez CM. An miRNA-mediated therapy for SCA6 blocks IRES-driven translation of the CACNA1A second cistron. Sci Transl Med 2016;8:347ra94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramachandran PS, Boudreau RL, Schaefer KA, La Spada AR, Davidson BL. Nonallele specific silencing of ataxin-7 improves disease phenotypes in a mouse model of SCA7. Mol Ther 2014;22:1635–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang N, Ashizawa T. RNA toxicity and foci formation in microsatellite expansion diseases. Curr Opin Genet Dev 2017;44:17–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khorkova O, Hsiao J, Wahlestedt C. Oligonucleotides for upregulating gene expression. Pharm Pat Anal 2013;2:215–29. [DOI] [PubMed] [Google Scholar]
- 14.Fiszer A, Olejniczak M, Switonski PM, Wroblewska JP, Wisniewska-Kruk J, Mykowska A, et al. An evaluation of oligonucleotide-based therapeutic strategies for polyQ diseases. BMC Mol Biol 2012;13:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corey DR. Synthetic Nucleic Acids and Treatment of Neurological Diseases. JAMA Neurol 2016;73:1238–1242. [DOI] [PubMed] [Google Scholar]
- 16.Lichota J, Skjørringe T, Thomsen LB, Moos T. Macromolecular drug transport into the brain using targeted therapy. J Neurochem 2010;113:1–13. [DOI] [PubMed] [Google Scholar]
- 17.Mathupala SP. Delivery of small-interfering RNA (siRNA) to the brain. Expert Opin Ther Pat 2009;19:137–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Souza EB, Cload ST, Pendergrast PS, Wah DWY. Novel therapeutic modalities to address nondrugable protein interaction targets. Neuropsychopharmacol 2009;34:142–58. [DOI] [PubMed] [Google Scholar]
- 19.Pardridge WM. shRNA and siRNA delivery to the brain. Adv Drug Deliv Rev 2007;59:141–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreuter J, Gelperina S. Use of nanoparticles for cerebral cancer. Tumori 2008;94:271–77. [DOI] [PubMed] [Google Scholar]
- 21.Petri B, Bootz A, Khalansky A, Hekmatara T, Müller R, Uhl R, et al. Chemotherapy of brain tumour using doxorubicin bound to surfactant-coated poly(butyl cyanoacrylate) nanoparticles: revising the role of surfactants. J Control Release 2007;117:51–8. [DOI] [PubMed] [Google Scholar]
- 22.Khalin I, Alyautdin R, Wong TW, Gnanou J, Kocherga G, Kreuter J. Brain-derived neurotrophic factor delivered to the briain using poly (lactide-co-glycolide) nanoparticles improves neurological and cognitive outcome in mice with traumatic brain injury. Drug Deliv 2016;23:3520–28. [DOI] [PubMed] [Google Scholar]
- 23.Pardridge WM. The blood-brain barrier: bottleneck in brain drug development. Neurotherapeutics 2005;2:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Méresse S, Delbart C, Fruchart JC, Cecchelli R. Low-density lipoprotein receptor on endothelium of brain capillaries. J Neurochem 1989;53:340–5. [DOI] [PubMed] [Google Scholar]
- 25.Dehouck B, Dehouck MP, Fruchart JC, Cecchelli R. Upregulation of the low density lipoprotein receptor at the bood-brain barrier: intercommunications between brain capillary endothelial cells and astrocytes. J Cell Biol 1994;126:465–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dehouck B, Fenart L, Dehouck MP, Pierce A, Torpier G, Cecchelli R. A new function for the LDL receptor: transcytosis of LDL across the blood-brain barrier. J Cell Biol 1997;138:877–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuwahara H, Nishina K, Yoshida K, Nishina T, Yamamota M, Saito Y, et al. Efficient in vivo delivery of siRNA into brain capillary endothelial cells along with endogenous lipoprotein. Mol Ther 2011;19:2213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang ZZ, Li JQ, Wang ZZ, Dong DW, Qi XR. Tumor-targeting dual peptides-modified cationic liposomes for delivery of siRNA and docetaxel to gliomas. Biomaterials 2014;35:5226–39. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Hao Y, Li H, Zhao Y, Meng D, Li D, et al. Co-delivery of doxorubicin and siRNA for glioma therapy by a brain targeting system: angiopep-2-modified poly (lactic-co-glycolic acid) nanoparticles. J Drug Target 2015;23:832–46. [DOI] [PubMed] [Google Scholar]
- 30.An S, He D, Wagner E, Jiang C. Peptide-like polymers exerting effective glioma-targeted siRNA delivery and release for therapeutic application. Small 2015;11:5142–50. [DOI] [PubMed] [Google Scholar]
- 31.Demeule M, Régina A, Ché C, Poirier J, Hguyen T, Gabathuler R, et al. Identification and design of peptides as a new drug delivery system for the brain. J Pharmacol Exp Ther 2008;324:1064–72. [DOI] [PubMed] [Google Scholar]
- 32.Srimanee A, Regberg J, Hallbrink M, Vajragupta O, Langel U. Role of scavenger receptors in peptide-based delivery of plasmid DNA across a blood-brain barrier model. Int J Pharm 2016;500:128–35. [DOI] [PubMed] [Google Scholar]
- 33.Kim DH, Iijima H, Goto K, Sakai J, Ishii H, Kim HJ, et al. Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J Biol Chem 1996;271:8373–80. [DOI] [PubMed] [Google Scholar]
- 34.Srivastava RA, Jain JC. Scavenger receptor class B type I expression and elemental analysis in cerebellum and parietal cortex regions of the Alzheimer’s disease brain. J Neurol Sci 2002;196:45–52. [DOI] [PubMed] [Google Scholar]
- 35.Göppert TM, Müller RH. Protein adsorption patterns on poloxamer- and poloxamine-stabilized solid lipid nanoparticles (SLN). Eur J Pharm Biopharm 2005;60:361–72. [DOI] [PubMed] [Google Scholar]
- 36.Lee S, Ashizawa AT, Kim KS, Falk DJ, Notterpek L. Liposomes to target peripheral neurons and Schwann cells. PLoS One 2013;8:e78724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.White M, Xia G, Gao R, Wakamiya M, Sarkar PS, McFarland K, et al. Transgenic mice with SCA10 pentanucleotide repeats show motor phenotype and susceptibility to seizure: a toxic RNA gain-of-function model. J Neurosci Res 2012;90:706–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kn McFarland, Ashizawa T Transgenic models of spincerebellar ataxia type 10: modeling a repeat expansion disorder. Genes (Basel) 2012;3:481–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boado RJ. Blood-brain barrier transport of non-viral gene and RNAi therapeutics. Pharm Res 2007;24:1772–87. [DOI] [PubMed] [Google Scholar]
- 40.Sakurai F, Nishioka T, Yamashita F, Takakura Y, Hashida M. Effects of erythrocytes and serum proteins on lung accumulation of lipoplexes containing cholesterol or DOPE as a helper lipid in the single-pass rat lung perfusion system. Eur J Pharm Biopharm 2001;52:165–72. [DOI] [PubMed] [Google Scholar]
- 41.Ogris M, Brunner S, Schuller S, Kircheis R, Wagner E. PEGylated DNA/transferrin_PEI complexes: reduced interaction with blood components, extended circulation in blood and potential for systemic gene delivery. Gene Ther 1999;6:595–605. [DOI] [PubMed] [Google Scholar]
- 42.Sempf K, Arrey T, Gelperina S, Schorge T, Meyer B, Karas M, et al. Adsorption of plasma proteins on uncoated PLGS nanoparticles. Eur J Pharm Biopharm 2013;85:53–60. [DOI] [PubMed] [Google Scholar]
- 43.Price TO, Farr SA, Yi X, Vinogradov S, Batrakova E, Banks WA, et al. Transport across the blood-brain barrier of pluronic leptin. J Pharmacol Exp Ther 2010;333:253–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gelperina S, Maksimenko O, Khalansky A, Vanchugova L, Shipulo E, Abbasova K, et al. Drug delivery to the brain using surfactant-coated poly (lactide-co-glycolide) nanoparticles: influence of the formulation parameters. Eur J Pharm Biopharm 2010;74:157–63. [DOI] [PubMed] [Google Scholar]
- 45.Singh-Joy SD, McLain VC. Safety assessment of poloxamers 101, 105, 108, 122, 123, 124, 181, 182, 183, 184, 185, 188, 212, 215, 217, 231, 234, 235, 237, 238, 282, 284, 288, 331, 333, 334, 335, 338, 401, 402, 403, and 407, poloxamer 105 benzoate, and poloxamer 182 dibenzoate as used in cosmetics. Int J Toxicol 2008;27:93–128. [DOI] [PubMed] [Google Scholar]
- 46.Gutiérrez-Puente Y, Tari AM, Stephens C, Rosenblum M, Guerra RT, Lopez-Berestein G. Safety, pharmacokinetics, and tissue distribution of liposomal P-ethoxy antisense oligonucleotides targeted to Bcl-2. J Pharmacol Exp Ther 1999;291:865–69. [PubMed] [Google Scholar]
- 47.Tari AM, Gutiérrez-Puente Y, Stephens C, Sun T, Rosenblum M, Belmont J, et al. Liposome-incorporated Grb2 antisense oligodeoxynucleotide increases the survival of mice bearing bcr-abl-positive leukemia xenografts. Int J Oncol 2007;31:1243–50. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


