Abstract
Glaucoma is a common optic neuropathy that is characterized by the progressive degeneration of axons and the loss of retinal ganglion cells (RGCs). Glaucoma is one of the leading causes of irreversible blindness worldwide. Current glaucoma treatments only slow the progression of RGCs loss. Induced pluripotent stem cells (iPSCs) are capable of differentiating into all three germ layer cell lineages. iPSCs can be patient-specific, making iPSC-derived RGCs a promising candidate for cell replacement. In this review, we focus on discussing the detailed approaches used to differentiate iPSCs into RGCs.
Keywords: glaucoma, retinal ganglion cells, induced pluripotent stem cells, differentiation
INTRODUCTION
Glaucoma is a common optic neuropathy and one of the leading causes of irreversible blindness in the world. It is characterized by the progressive degeneration of axons and the loss of retinal ganglion cells (RGCs). Approximately 111.8 million people will be affected by glaucoma by 2040[1]. The main risk factor for glaucomatous optic neuropathy is high intraocular pressure (IOP). Current glaucoma treatments that primarily target increased IOP, including topical eyedrops, laser treatment, and surgeries, but only slow the progression of RGC loss[2]. There are an increasing number of recent reports on the development of neuroprotective therapies for glaucoma, that could be used as adjunctive treatments to lower IOP[3]. These studies investigated neuroprotective strategies, including the delivery of a neurotrophic factor, the molecular application of anti-apoptosis and anti-inflammation treatments, and the reduction of oxidative stress[4]. However, the clinical application is partially limited, because long-term maintenance of the supplementation is difficult and there is a possibility of compulsory repeated interventions. Recently, cell-based therapy has been demonstrated to be effective for the treatment of several diseases[5]–[8]. The cells used for these therapies include mesenchymal stem cells (MSCs), dental pulp stem cells, embryonic stem cells (ESCs), and induced pluripotent stem cells (iPSCs) and so on. In 2006, Takahashi and Yamanaka[9] successfully reprogrammed mouse and adult fibroblasts to a pluripotent state by using four transcription factors (Oct3/4, Sox2, Klf4, and c-Myc) that are involved in the pluripotency maintenance in ESCs. The resulting cells, iPSCs, could form colonies that are morphologically similar to ESCs and are capable of differentiating into all three germ layer cell lineages. Since iPSCs could be reprogrammed by the somatic cells of a patient, they could maintain the total unique genomic information of each individual. These patient-derived iPSCs could serve as a perfect in vitro model for genetic disease studies and thus have a promising role in the development of personalized treatment. These factors have resulted in an explosion of studies that attempt to exploit the reprogramming of somatic cells into iPSCs, reporting various modified protocols designed to improve the reprogramming efficiency and facilitate clinical application. For example, instead of retroviruses, multiple studies have used plasmid[10]–[11], miRNA[12], and protein[13] as transcript factors delivery vectors to prevent the risk of insertional mutagenesis of the host cells. Other reports indicate that the addition of small molecules, such as valproic acid (VPA)[14], AZA5-aza-cytidine (AZA)[15], butyrate[16], vitamin C[17], transforming growth factor-β (TGF-β) receptor inhibitor (A-83-01)[18]–[19], MEK inhibitor (PD325901)[18]–[19], GSK3β inhibitor (CHIR99021)[18]–[19], and ROCK inhibitor (HA-100)[18]–[19] could enhance reprogramming efficiency and even replace the use of certain transcription factors in iPSCs generation protocols. Table 1 shows several examples of the experimental features of protocols to transform somatic cells into iPSCs. Insight is required regarding how to induce iPSCs to differentiate into the specialized cell fate of interest. An increasing number of reports have indicated that iPSCs could be differentiated into RGCs, photoreceptors, and retinal pigment epithelium (RPE) under appropriate conditions[20]–[21]. The current review provides a perspective on the key methods that led to the differentiation of RGCs, and divulged the problems that must be solved before the iPSCs-derived RGCs could fulfill its potential in medical applications, such as the mechanisms of pathology, screening treatment drugs, and development of cell-based and patient-specific therapies targeting glaucoma and other optic neuropathies.
Table 1. Examples of experimental features from somatic cell to iPSCs.
| Cell source | Culture condition | Reprogramming factors | Delivery method | Time length (d) | Efficiency (%) | Reference |
| Mouse fibroblasts | Cell feeder | Oct3/4, Sox2, C-myc, Klf4 | Retrovirus | N/A | 0.02 | [9] |
| Human fibroblasts | Cell feeder | Oct4, Sox2, Nanog, Lin28 | Lentivirus | 15 | 0.022 | [22] |
| Human fibroblasts | Cell feeder | Oct3/4, Sox2, Klf4, Sall4 | Retrovirus | 25 | 0.014 | [23] |
| Human blood mononuclear cells | Cell feeder | Oct4, Sox2, Klf4, C-myc, Lin28 | Plasmid | 14 | 0.05 | [11] |
| Human fibroblasts | Cell feeder | Oct3/4, Sox2, Klf4, Glis1 | Retrovirus | 22 | 0.17 | [24] |
| Human fibroblasts | Cell feeder | Oct3/4, Sox2, Klf4, L-myc, Lin28, P53 | Plasmid, shRNA | 14 | 0.03 | [25] |
| Human keratinocytes | Cell feeder | Oct4, Sox2, Klf4, C-myc, Rem2 | Retrovirus | 12 | 0.2 | [26] |
| Human fibroblasts | Cell feeder | Oct4, Sox2, Myc, Klf4, Sv40 large T antigen | Lentivirus | 14 | 0.46 | [27] |
| Human fibroblasts | Cell feeder | Oct3/4, Sox2, C-myc, Klf4 | Adenovirus | 25 | 0.0002 | [28] |
| Mouse and human fibroblasts | Cell feeder | Oct3/4, Sox2, C-myc, Klf4 | 2A peptide | 9 | 2.5 | [13] |
| Human fibroblasts | Cell feeder | Oct3/4, Sox2, C-myc, Klf4 | PiggyBac transposon | 14 | 0.01 | [13] |
| Human fibroblasts | Cell feeder | Oct3/4, Sox2, C-myc, Klf4 | PiggyBac transposon | 10-14 | 0.03 | [29] |
| Human adipose stromal cells | Cell feeder | Oct4, Sox2, Nanog, Lin28 | Minicircle DNA | 18 | 0.0075 | [30] |
| Human fibroblasts | Xeno-free, feeder-free | Oct3/4, Sox2, Klf4, L-myc, Lin28, TP53, A-83-01, sodium butyrate, CHIR99021 | Plasmids, shRNA, small molecules | 16 | 0.1 | [10] |
| Mouse fibroblasts | Cell feeder | Mir-200c, Mir-302s, Mir-369s | MiRNA | 15 | 0.1 | [12] |
| Human fibroblasts | Cell feeder | Mir-200c, Mir-302s, Mir-369s | MiRNA | 20 | 0.002 | [12] |
| Human fibroblasts | Cell feeder | Oct4, Sox2, VPA | Retrovirus, small molecules | N/A | 0.01 | [31] |
| Human fibroblasts | Cell feeder | Oct4, Sox2, Klf4, VPA | Retrovirus, small molecules | N/A | 1 | [31] |
| Human bone mesenchymal stromal cell | Feeder-free | Oct4, Sox2, Nanog, Lin28, Sv40lt, Klf4, C-myc, A-83-01, PD325901, CHIR99021, HA-100 | Plasmids, small molecules | N/A | N/A | [18] |
| Mouse fibroblasts | Cell feeder | Oct4, tranylcypromine, VPA, CHIR99021, 616452 | Lentivirus, small molecules | 18d | 0.03 | [32] |
| Human fibroblasts | Xeno-free, feeder-free | Oct3/4, Sox2, C-myc, Klf4 | Lentivirus | 7-14d | 0.3 | [33] |
iPSCs: Induced pluripotent stem cells; VPA: Valproic acid.
FACTORS INVOLVED IN THE DIFFERENTIATION OF RGCS FROM IPSCS
iPSCs provide a widely available, non-ethically disputed, and almost infinite source of pluripotent cells, providing a new paradigm in regenerative medicine stem cell maintenance and differentiation. Recently, several studies have demonstrated differentiation of reprogrammed iPSCs into fully functional RGCs. Table 2 shows a summary of experimental features of the transformation of iPSCs into RGCs. During of the RGCs differentiation process, intrinsic and extrinsic factors play key roles. Here, we reviewed factors that contribute in the differentiation of RGCs.
Table 2. Summary of experimental features from iPSCs to RGCs.
| Cell type | Differentiation procedure | Differentiation culture medium | Genetic modification | Culture condition | RGCs markers | Time length (d) | Efficiency | Electrophysiological analysis | Transplantation | Reference |
| hiPSC | EBs, neurosphere, RGCs | ES medium, DAPT | Gelatin pretreated, laminin coated | Brn3a, Islet-1 Atoh7, γ-syn, Thy-1 | 21 | 27.836%±2.56% | + | N/A | [34] | |
| hiPSC | Neural, retina-like structures, RGCs | Proneural medium, FGF2, DAPT | Xeno-free, coating-free | Brn3a, Pax6, CALRETININ | 21 | N/A | N/A | N/A | [20] | |
| miPSC | EBs, RGCs | Neuronal medium, Dkk1, Noggin, DAPT | Math5 overexpression | Coating-free | Brn3b, Islet-1, Thy1, γ-syn | 8 | 10% | N/A | N/A | [35] |
| miPSC | EB, neural progenitors, RGCs | NIM, NEM, RDM | PDL-coated, laminin coated | Brn3b, Math5 | 50 | 14.3%±3.439% | + | N/A | [21] | |
| hiPSC | EBs, OV-like structures, RGCs | RDM, RMM, matrigel | Three-dimensional culture, cell-feeder | Brn3b, Atoh7, Islet1, γ-syn, Thy1 | 26-29 | N/A | + | N/A | [36] | |
| hiPSC | EBs, OV-like structures, RGCs | NIM, RDM | NA | Brn3, Map2 | 70 | 49.13%±2.95% | N/A | N/A | [37] | |
| hiPSC | EBs, neurospheres, RGCs | NIM, FBS, RDM, BDNF | Coating-free | Brn3, Islet1, Map2 | 40 | 36.1%±1.7% | + | N/A | [38] | |
| hiPSC | EBs, neurospheres, RGCs | NIM, RDM | Synthemax plates, feeder-free, xeno-free | Brn3 | 60 | 15% | N/A | N/A | [39] | |
| hiPSC | EBs, neural aggregates, RGCs | EB medium, Dkk1, Lefty A, Noggin, RDM, DAPT | Atoh7 overexpression | Matrigel coated | Brn3b, Islet1, Atoh7 | >16 | 23% | N/A | N/A | [40] |
| hiPSC | EBs, RPCs, RGCs | EB medium, differentiation medium, DAPT | Synthemax plates | Brn3b, Map2, Thy1, Atoh7 | 60 | N/A | N/A | N/A | [41] | |
| miPSC | EBs, OV like structures, RGCs | RDM, RMM, matrigel | Cell-feeder, PDL/laminin coated | Brn3b, Math5, Islet1, γ-syn, Thy1 | 21 | 30% | + | N/A | [42] | |
| hiPSC | Neural aggregates, RGCs | mTeSR1 medium, NIM, RDM | Matrigel coated | Brn3b, Tuj1, Thy1 | 14 | 10% | + | Rabbit, Rhesus monkey safe | [43] | |
| hiPSC | Neural rosettes, RGCs | Basal medium, Shh, FGF, DAPT, CNTF | PDL/Laminin matrigel coated | Brn3b, Atoh7, Isl1 | 15 | 27.92%±13.38% | + | N/A | [44] | |
| miPSC | EBs, RPCs, RGCs | NIM, NEM, RDM, Noggin, DKK1, Shh, FGF, DAPT | PDL/Laminin matrigel coated | Brn3b, Atoh7, Islet1, Thy1.2 | 15 | 13.3%±1.5% | + | Rat safe | [45] | |
| miPSC | EBs, RGCs | EB medium, NIM, RDM, DAPT | Matrigel coated, PLGA scaffold | Brn3b, islet1, NF145 | 12 | 37% | N/A | N/A | [46] |
hiPSC: Human iPSC; miPSC: Mouse iPSC; EB: Embryoid bodies; RGCs: Retinal ganglion cell; OV-like structures: Optic vesicle-like structures; RPCs: Retinal progenitor cells; ES: Embryonic stem; FBS: Fetal bovine serum; DAPT: N-[N-(3, 5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butylester; FGF: Fibroblast growth factor; NIM: Neural induction medium; RDM: Retinal differentiation medium; CNTF: Cilliary neurotrophic factor; RMM: Retinal maturation medium; BDNF: Brain-derived neurotrophic factor; NEM: Neural expansion medium; PDL: Poly-D-lysine; N/A: Not applicable.
DIFFERENTIATION OF RETINA PROGENITOR CELLS
The vertebrate eye is formed via coordinated interactions between the neuroepithelium, the surface ectoderm, and the extraocular mesenchyme, which originate from the neural crest and the mesoderm[47]. Following the eye field formation, the neuroepithelium of the ventral forebrain evaginates, thus forming bilateral optic vesicles (OVs). After undergoing invagination, OVs compose distinct ocular tissues of the neural retina, the RPE, and the optic stalk[47]. During these processes, the differentiation and the fate determination of retinal cells are strictly controlled at the molecular level by cell-intrinsic transcription factors and are also influenced by cell-extrinsic signals. Previous studies show that a group of eye field transcription factors (EFTFs) are expressed in a specific region, the anterior neural plate. The EFTFs include ET, Chx10, Rax (also known as Rx), Pax6, Six3, Six6 (also known as Optx2), Lhx2, and Otx2[48]. Otx2 is required for RPE specification during eye development[49] and a group of genes encoding homeobox-containing transcription factors are thought to be at the top of the gene regulatory network during neural retina formation, such as Chx10, Pax6, Six3, Six6, and Rx[50]–[51]. These homeobox genes are expressed in all neuroblasts at the beginning of retinogenesis and are required for the specification of RGCs, as well as other retinal cell types.
The paired-like homeobox gene Chx10 is the earliest specific marker of neural retinal progenitor cells that is expressed in the presumptive neural retina and functions to repress the expression of the microphthalmia-associated transcription factor (Mitf) in the distal optic vesicle[52]. Mitf is a basic helix-loop-helix (HLH) transcription factor that acts as a master regulator of RPE development and is essential for the acquisition and the maintenance of RPE cells[52]. Mutations in Chx10 cause the ocular retardation phenotype in mice[53], suggesting that Chx10 plays critical roles in neural retinal development.
Pax6 is a paired-like homeobox gene that has maintained a high level of conservation throughout the evolution of the eye[51]. Studies have demonstrated that Pax6 is critical during the early stages of eye development[54]. Additionally, Pax6 can has the ability to directly activate the basic HLH transcription factor Ath5, the most important transcription factor in RGCs specification[55]. Loss-of-function mutations in human Pax6 could result in could ocular syndrome aniridia[56], suggesting that Pax6 plays a key role in eye formation.
Six3 and Six6 are closely related members of the Six-homeodomain family. Human Six3 mutation could result in microphthalmia and severe malformation of the brain. Mutation in Six6 is also associated with bilateral anophthalmia[57]. These effects suggest that both Six3 and Six6 play important roles during retinal determination.
Rx is initially expressed throughout the anterior neural plate and later throughout the neural retina[51]. Mutations in both alleles of the mice Rx gene result in an inability to develop OVs, and mutation of the human gene is associated with anophthalmia and sclerocornia[58]. The Rx function is important during neural retina development.
Overall, these EFTFs perform roles during retina development and could be used as markers for the retina progenitor cells (RPCs) to monitor the iPSC differentiation process. In addition to these intrinsic factors, various neurotropic factors and pathways have been implicated in retina cell specification and differentiation. Elucidation of these extrinsic signaling pathways could allow researchers to more efficiently differentiate iPSCs into RGCs. These pathways include fibroblast growth factor (FGF), insulin-like growth factor (IGF), bone morphogenetic protein (BMP), nodal, and Wnt signaling pathways. These pathways all regulate the development of the neural retina, where the FGF and the IGF provide positive regulation, and the BMP, the nodal, and the Wnt signaling pathways serve as negative regulatory factors[49],[52],[59]–[60].
RETINAL GANGLION CELL DIFFERENTIATION
RGCs are the first neuronal cell type to emerge in the developing retina of vertebrates. The specification and the differentiation procedures are regulated by a group of transcription factors, including the Ath5, the Notch, and the Brn3 factors.
Ath5 (Atoh7 in humans, Math5 in mice, lakritz in zebrafish, Cath5 in chicks, and Xath5 in Xenopus) is a basic HLH transcription factor. Its expression coincides with the emerging of RGCs, where it plays a key role in the genetic regulation of RGC fate determination. Ath5 regulates the expression of the POU-domain transcription factor Brn3b and the LIM-homeodomain factor Isl1, RGC-specific differentiating transcription factors[61]. Math5 loss-of-function in mice and the lakritz mutation in zebrafish result in the nearly complete absence of RGCs, but overexpression of Cath5 in chicks and Xath5 in Xenopus promoted RGCs production[62]–[63], suggesting the important and irreplaceable role that Ath5 plays in RGC differentiation.
The Notch pathway is a negative regulator of RGC production that is activated by the binding of a cell-surface Notch to ligands. Notch activity in mice is down regulated just prior to the differentiation of the RGCs during normal eye development[63]. Pax6 directly activates Ath5, and Notch signaling inhibits Ath5 expression[62]. Altering Notch expression with antisense oligonucleotides in the chick retina results in an increased RGC number, where the expression of the constitutively active Notch decreased ganglion cell numbers[64]. Together, the opposing activities of Pax6 and Notch determine the accurate expression of Ath5 in a subset of cells that are proficient for RGC specification.
The class IV POU domain transcription factor Brn3b (also called Pou4f2) is downstream of Ath5 and one of the earliest markers for RGC differentiation[65]. Although remains unclear at present if Ath5 directly regulates Brn3b, Brn3b expression is significantly reduced in the Math5-null retina[66]. During mouse retinal development, the Brn3b gene is expressed in a large set of post-mitotic ganglion cell precursors and is required for both their early and terminal differentiation[67]. Brn3b-null mice exhibit programmed cell death of approximately 70% of newly formed RGCs and axon growth defects[68]. Brn3a and Brn3c (also known as Pou4f1 and Pou4f3), are two members of the class IV POU domain transcription factors. Similar to Brn3b, both Brn3a and Brn3c are expressed in differentiated RGCs during mouse retinogenesis, but two days after the onset of Brn3b expression[69]. Studies have shown that the loss function of Brn3a and Brn3c in mice does not cause retinal defects but observed some promotion of RGC axon development in the absence of Brn3b in mice and chicks. The relationship between these factors remains unclear[70]. The Brn3 factors play important roles in RGCs differentiation, although their roles remain only partially understood.
Islet-class factor Islet-1 (also known as Isl1) is a LIM-homeodomain factor that is co-expressed with Brn3b in post-mitotic and differentiating RGCs and is also downstream of Ath5 in the gene regulatory network of RGC development[61]. Islet-1 and Brn3b exhibit essentially identical retinal expression patterns during the early stages of development and are the earliest known transcription factors that are specifically expressed in developing RGCs[71]. Retina-specific depletion of Islet-1 in mice results in the apoptosis of a majority of RGCs, as well as and in RGC axon guidance defects[61]. The specification and differentiation of RGCs occur via a stepwise process that involves a hierarchical gene regulatory network. These factors could be used as reliable markers for RGCs and as targets to differentiate RGCs.
γ-synuclein is highly expressed in the axons and the cytoplasm of RGCs, at the same time as Brn3a in the human retina[72]. The co-expression of Brn3a and γ-synuclein could be used as a marker of RGCs. Barnstable and Drager[73] found that Thy-1 antigen could be a ganglion cell-specific marker in rodent retina. Therefore, all of these factors could be used as RGCs markers.
CULTURE PROCEDURES AND CONDITIONS INVOLVED IN RGC DIFFERENTIATION FROM IPSCS
With detailed understanding of the molecular steps involved in RGC differentiation, investigators successfully differentiated iPSCs into RGC-like cells. Although at first the levels of expressed RGC-specific markers such as Ath5, Brn3b, Thy1, and Islet1 were not promising, significant progress has been made in generating RGC-like cells later.
CULTURE PROCEDURES
The culture and the differentiation of iPSCs nearly mimic normal eye development. The embryoid bodies (EBs) form first, followed by formation of OV-like structures or neurospheres. EBs is cellular aggregates that consist of a mixture of endodermal, mesodermal, and ectodermal cells, which are representative of the three primary germ layers in development. OV-like structures or neurospheres are proliferative cellular aggregates of neural progenitor cells that have the ability to differentiate into neurons or glia. Initially, studies demonstrated that dynamic behavior and critical development of the checkpoints in RGCs induction is an essential mean to confirm the establishment of the RGCs lineage and mimic normal eye development. Several reports indicate that these protocols require multiple steps and trained handling, which could generate undetectable variations and would not be compatible with the manufacturing process required for a therapeutic approach that might require large-scale production of cells. Reichman et al[20] developed a method that bypassed the EBs formation and used exogenous molecules, coating, or matrigel to simplify and shorten the procedure for RGC differentiation from hiPSCs. In this process, the neural retina-like (NR-like) structures formed first, and then developed into RGCs. Other reports demonstrated that mouse iPSCs-derived EBs express RPCs markers hierarchically and differentiate into RGCs directly[35],[46]. Regardless of the step that was bypassed, these simple protocols allowed RGCs generation that were similar to native RGCs at the molecular, biochemical, and functional levels. The development of protocols that are easy-to-operate and less risky represents an important step towards effective clinical application. However, it should be noticed that those differentiation protocols that have been simplified and skip some steps do have drawbacks. First, these RGCs were largely examined the phenotype finally acquired but not the mechanisms by which they were acquired. Second, these RGCs didn't recapitulate the hierarchical regulatory mechanisms for RGC differentiation, thus predicting stability of the acquired phenotype might be affected[45].
CULTURE CONDITIONS
It is common to modify genes and/or use certain supplements and molecules to mimic in vivo signal pathways during RGC differentiation. N2, B27, and FGF2 are commonly added to the basal culture medium in various combinations to promote neural retina differentiation. FGF could play a role in patterning the domains of the optic vesicle and could be required for the Chx10 expression in the presumptive neural retina[74].
Nodal and BMPs are members of the TGF-β superfamily, where their pathways play key roles in antagonizing the neural anterior default differentiation program in hESCs. This demonstrates that the antagonists of the Nodal or the BMPs pathway, such as Lefty and Noggin, could be added to an RPC differentiation culture medium to ensure low levels of Nodal or BMPs and to enhance neural differentiation of iPSCs[52],[60]. The Wnt signaling pathway negatively regulates neurite extension in the mouse retina, so it is not surprising that the Wnt antagonist Dickkopf (Dkk) could be added to the RPC differentiation culture medium to promote the neural differentiation of iPSCs[35],[59]. The Gdf8 (a member of the TGF-β superfamily) and Hedgehog (Hh, shh in mouse) proteins are regulated by Brn3b and are extracellular signaling molecules involved in the cell non-autonomous function of RGCs, where the RGCs regulate their own production and RPC proliferation[68]. These factors help to promote neural retina differentiation from iPSCs when added to the RPC differentiation culture medium.
In addition to supplements in the culture medium, genetic modification is another strategy used to promote RGC differentiation. During RGC differentiation, Ath5 plays an irreplaceable role, so either direct or indirect genetic modification of Ath5 could be an effective way to increase the induction of RGCs. Chen et al[35] found that over expression of Math5 in mouse activated RGC-related genes in iPSCs, including Brn3b, Islet-1, and Thy1, indicating that Math5 overexpression stimulates the differentiation of iPSCs into RGCs. Deng et al[40] demonstrated that overexpression of Atoh7 promoted RGC specification in the hiPSC-derived RPCs. Alternatively, strategies may exploit the upstream factors. For example, the Notch pathway activity is downregulated before RGC differentiation during retinogenesis, where it has been proven to inhibit Ath5 expression. Studies have explored Notch pathway inhibitors as a method to increase the expression of Ath5. The most commonly used inhibitor is N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenyl-glycine-t-butylester (DAPT)[20],[35],[46]. Riazifar et al[34] even showed that a single chemical, DAPT, could induce iPSCs to differentiate into functional RGCs.
Similar to the iPSC reprogramming procedures, the xenogeneic products for RGC growth and differentiation are essential prerequisites that enable the clinical application of iPSCs-derived RGCs. Sridhar et al[39] first demonstrated a xeno-free approach to differentiate hiPSCs into RGCs. Synthemax plates and nonxenogeneic Nutristem medium were used to maintain iPSCs and used mTESR1/Nutristem, Synthemax plates, and retinal differentiation medium to induce neurospheres and RGCs. Possible graft rejection risks might be caused by the use of animal products or other undefined components during routine cell culturing, so the Xeno-free approach is an attractive promising alternative. Conventional two dimensional (2D) cultures have been engineered for attachment and proliferation of iPSCs. Alternatively, three-dimensional (3D) culture system is gaining popularity, as it has the ability of self-organization, requires fewer extrinsic growth factors, and the intrinsic pattern appears to be similar to the normal eye development[36]. Existing 3D culture techniques include suspension culture, cell encapsulation in gels and cell culture in scaffolds[75]–[76]. Hallam et al[77] showed that light responsive retinal organoids derived from iPSC lines can be generated at the scale needed for pharmacology and drug screening purposes by 3D culture techniques. As RGCs lie in the innermost layer of neural retina, the techniques may explore the functional axons[43].
TRANSPLANTATION AND CONNECTION
In vivo experiment, Chen et al[35] successfully transplanted iPSC-derived RGCs into the mice vitreous chamber, but found almost no cells migrated into the retina. It was thought that this could be due to the normal retinal environment that acts as a barrier for graft integration and the formation of functional synapses. However, Parameswaran et al[45] indicated that following transplantation of the iPSC-derived RGCs in the rat model of ocular hypertension, cells were incorporated into the host RGC layer and expressed RGC-specific markers without producing tumors. Hertz et al[78] demonstrated that RPC-derived RGCs exhibit a similar capacity for integration as developing primary RGCs, where they appear to form a lower number of presynaptic punctae in both normal and optic nerve axotomy rats models. Although the effects of cell transplantation therapy are not clearly understood, cell transplantation could offer a feasible strategy for neuroprotective and cell-replacement therapy.
The function of the RGCs as projection neurons of the retina is dependent on their ability to form contacts with central targets. An important challenge for iPSC-derived RGCs transplantation is for these cells to integrate in the ganglion cell layer (GCL) and to also develop functional axons that connect to the optic nerve and possibly form connections with the brain. iPSC-derived RGCs with functional axons have been successfully established[34],[36],[42]. Teotia et al[44] showed that iPSC-derived RGCs in vitro expressed a series of guidance molecules that could guide the axons in the retina, at the optic chiasm, and even in the central targets. Tissue engineering is now used for treatment of glaucoma and other retinal diseases by using a medical polymer biomaterial as a substrate for cell seeding and delivery. For example, the use of electrospun[79], Netrin-1[80], or 3D-printed scaffolds[81] have been used to control RGCs neurite growth. Li et al[43] transplanted of an engineered iPSC-derived RGCs scaffold biomaterial of biodegradable poly (lactic-co-glycolic acid) (PLGA) into the retinal surfaces of rabbits and rhesus monkeys, which resulted in dendritic arbors, prominent axons, neurite networks, and altered electrophysiological properties. The cell-scaffold could be relevant for potential clinical applications within tissue engineering therapy and retinal surface delivery, however, the vitrectomy needed to guarantee effective adhesive remains a challenge.
PERSPECTIVES AND CONCLUSIONS
Additional optimization of the process to generate iPSC-derived RGCs is required for clinical applications. First, the primitive hiPSCs used to differentiate into RGCs should be patient-specific and should be reprogrammed under feeder-free, xeno-free, and integration-free conditions to avoid the risk of graft rejection and genotoxicity. Second, the medium devoid of xenogeneic components should be used for RGCs differentiation when specifying RPCs and RGCs. Third, high-efficiency, simple and reproducible protocols should be established, where predictable variations should be minimized during each step. Fourth, the hiPSC-derived RGCs should be easily translatable to an in vivo environment, and the brain connection should be established.
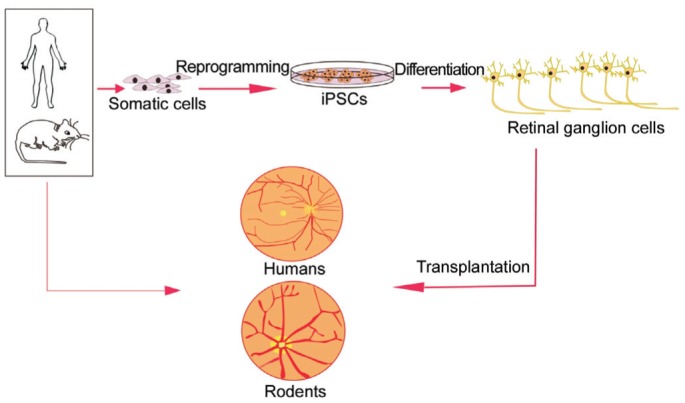
In conclusion, some significant challenges still exist, such as the risk of teratocarcinoma formation. A lack of robust and highly reproducible differentiation protocols and accurate axon guidance still exists. However, iPSC-derived RGCs remain a potentially life-changing tool for the study of RGCs biology and the eventual personalized treatment for glaucoma and other optic nerve diseases (Figure 1).
Figure 1. Flow diagram of personalized treatment for glaucoma and other optic nerve diseases.
Acknowledgments
The authors thank Dr. Yong Zeng for helping to draw the Figure 1.
Foundation: Supported by “555 Talent Plan” Grant from the Changsha National High-Tech Industrial Development Zoom.
Conflicts of Interest: Ji SL, None; Tang SB, None.
REFERENCES
- 1.Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014;121(11):2081–2090. doi: 10.1016/j.ophtha.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Fan BJ, Wiggs JL. Glaucoma: genes, phenotypes, and new directions for therapy. J Clin Invest. 2010;120(9):3064–3072. doi: 10.1172/JCI43085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang EE, Goldberg JL. Glaucoma 2.0: Neuroprotection, neuroregeneration, neuroenhancement. Ophthalmology. 2012;119(5):979–986. doi: 10.1016/j.ophtha.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danesh-Meyer HV. Neuroprotection in glaucoma: recent and future directions. Curr Opin Ophthalmol. 2011;22(2):78–86. doi: 10.1097/ICU.0b013e32834372ec. [DOI] [PubMed] [Google Scholar]
- 5.Thomas KE, Moon LD. Will stem cell therapies be safe and effective for treating spinal cord injuries? Br Med Bull. 2011;98(1):127–142. doi: 10.1093/bmb/ldr013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abematsu M, Tsujimura K, Yamano M, Saito M, Kohno K, Kohyama J, Namihira M, Komiya S, Nakashima K. Neurons derived from transplanted neural stem cells restore disrupted neuronal circuitry in a mouse model of spinal cord injury. J Clin Invest. 2010;120(9):3255–3266. doi: 10.1172/JCI42957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia JM, Mendonca L, Brant R, Abud M, Regatieri C, Diniz B. Stem cell therapy for retinal diseases. World J Stem Cells. 2015;7(1):160–164. doi: 10.4252/wjsc.v7.i1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mead B, Berry M, Logan A, Scott RA, Leadbeater W, Scheven BA. Stem cell treatment of degenerative eye disease. Stem Cell Res. 2015;14(3):243–257. doi: 10.1016/j.scr.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Hu W, He Y, Xiong Y, Lu H, Chen H, Hou L, Qiu Z, Fang Y, Zhang S. Derivation, expansion, and motor neuron differentiation of Human-Induced pluripotent stem cells with non-integrating episomal vectors and a defined xenogeneic-free culture system. Mol Neurobiol. 2016;53(3):1589–1600. doi: 10.1007/s12035-014-9084-z. [DOI] [PubMed] [Google Scholar]
- 11.Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J, Cheng L. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21(3):518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8(6):633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458(7239):771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26(7):795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454(7200):49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, Yusa K, Bradley A, Meyers DJ, Mukherjee C, Cole PA, Cheng L. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28(4):713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, Chen K, Li Y, Liu X, Xu J, Zhang S, Li F, He W, Labuda K, Song Y, Peterbauer A, Wolbank S, Redl H, Zhong M, Cai D, Zeng L, Pei D. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6(1):71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Megges M, Oreffo RO, Adjaye J. Episomal plasmid-based generation of induced pluripotent stem cells from fetal femur-derived human mesenchymal stromal cells. Stem Cell Res. 2016;16(1):128–132. doi: 10.1016/j.scr.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, Chau KF, Vodyanik MA, Jiang J, Jiang Y. Efficient feeder-free episomal reprogramming with small molecules. PLoS One. 2011;6(3):e17557. doi: 10.1371/journal.pone.0017557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reichman S, Terray A, Slembrouck A, Nanteau C, Orieux G, Habeler W, Nandrot EF, Sahel JA, Monville C, Goureau O. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc Natl Acad Sci U S A. 2014;111(23):8518–8523. doi: 10.1073/pnas.1324212111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parameswaran S, Balasubramanian S, Babai N, Qiu F, Eudy JD, Thoreson WB, Ahmad I. Induced pluripotent stem cells generate both retinal ganglion cells and photoreceptors: therapeutic implications in degenerative changes in glaucoma and age-related macular degeneration. Stem Cells. 2010;28(4):695–703. doi: 10.1002/stem.320. [DOI] [PubMed] [Google Scholar]
- 22.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 23.Tsubooka N, Ichisaka T, Okita K, Takahashi K, Nakagawa M, Yamanaka S. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells. 2009;14(6):683–694. doi: 10.1111/j.1365-2443.2009.01301.x. [DOI] [PubMed] [Google Scholar]
- 24.Maekawa M, Yamaguchi K, Nakamura T, Shibukawa R, Kodanaka I, Ichisaka T, Kawamura Y, Mochizuki H, Goshima N, Yamanaka S. Direct reprogramming of somatic cells is promoted by maternal transcription factor Glis1. Nature. 2011;474(7350):225–229. doi: 10.1038/nature10106. [DOI] [PubMed] [Google Scholar]
- 25.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka K, Shibata T, Kunisada T, Takahashi M, Takahashi J, Saji H, Yamanaka S. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8(5):409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 26.Edel MJ, Menchon C, Menendez S, Consiglio A, Raya A, Izpisua Belmonte JC. Rem2 GTPase maintains survival of human embryonic stem cells as well as enhancing reprogramming by regulating p53 and cyclin D1. Genes Dev. 2010;24(6):561–573. doi: 10.1101/gad.1876710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mali P, Ye Z, Hommond HH, Yu X, Lin J, Chen G, Zou J, Cheng L. Improved efficiency and pace of generating induced pluripotent stem cells from human adult and fetal fibroblasts. Stem Cells. 2008;26(8):1998–2005. doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- 28.Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27(11):2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- 29.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. PiggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458(7239):766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narsinh KH, Jia F, Robbins RC, Kay MA, Longaker MT, Wu JC. Generation of adult human induced pluripotent stem cells using nonviral minicircle DNA vectors. Nat Protoc. 2011;6(1):78–88. doi: 10.1038/nprot.2010.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26(11):1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Zhang Q, Yin X, Yang W, Du Y, Hou P, Ge J, Liu C, Zhang W, Zhang X, Wu Y, Li H, Liu K, Wu C, Song Z, Zhao Y, Shi Y, Deng H. Generation of iPSCs from mouse fibroblasts with a single gene, Oct4, and small molecules. Cell Res. 2011;21(1):196–204. doi: 10.1038/cr.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu HF, Chai C, Lim TC, Leong MF, Lim JK, Gao S, Lim KL, Wan AC. A defined xeno-free and feeder-free culture system for the derivation, expansion and direct differentiation of transgene-free patient-specific induced pluripotent stem cells. Biomaterials. 2014;35(9):2816–2826. doi: 10.1016/j.biomaterials.2013.12.050. [DOI] [PubMed] [Google Scholar]
- 34.Riazifar H, Jia Y, Chen J, Lynch G, Huang T. Chemically induced specification of retinal ganglion cells from human embryonic and induced pluripotent stem cells. Stem Cells Transl Med. 2014;3(4):424–432. doi: 10.5966/sctm.2013-0147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen M, Chen Q, Sun X, Shen W, Liu B, Zhong X, Leng Y, Li C, Zhang W, Chai F, Huang B, Gao Q, Xiang AP, Zhuo Y, Ge J. Generation of retinal ganglion-like cells from reprogrammed mouse fibroblasts. Invest Ophthalmol Vis Sci. 2010;51(11):5970–5978. doi: 10.1167/iovs.09-4504. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka T, Yokoi T, Tamalu F, Watanabe S, Nishina S, Azuma N. Generation of retinal ganglion cells with functional axons from human induced pluripotent stem cells. Sci Rep. 2015;5:8344. doi: 10.1038/srep08344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sridhar A, Ohlemacher SK, Langer KB, Meyer JS. Robust differentiation of mRNA-Reprogrammed human induced pluripotent stem cells toward a retinal lineage. Stem Cells Transl Med. 2016;5(4):417–426. doi: 10.5966/sctm.2015-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohlemacher SK, Sridhar A, Xiao Y, Hochstetler AE, Sarfarazi M, Cummins TR, Meyer JS. Stepwise differentiation of retinal ganglion cells from human pluripotent stem cells enables analysis of glaucomatous neurodegeneration. Stem Cells. 2016;34(6):1553–1562. doi: 10.1002/stem.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sridhar A, Steward MM, Meyer JS. Nonxenogeneic growth and retinal differentiation of human induced pluripotent stem cells. Stem Cells Transl Med. 2013;2(4):255–264. doi: 10.5966/sctm.2012-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng F, Chen M, Liu Y, Hu H, Xiong Y, Xu C, Liu Y, Li K, Zhuang J, Ge J. Stage-specific differentiation of iPSCs toward retinal ganglion cell lineage. Mol Vis. 2016;22:536–547. [PMC free article] [PubMed] [Google Scholar]
- 41.Tucker BA, Solivan-Timpe F, Roos BR, Anfinson KR, Robin AL, Wiley LA, Mullins RF, Fingert JH. Duplication of TBK1 stimulates autophagy in iPSC-derived retinal cells from a patient with normal tension glaucoma. J Stem Cell Res Ther. 2014;3(5):161. doi: 10.4172/2157-7633.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka T, Yokoi T, Tamalu F, Watanabe S, Nishina S, Azuma N. Generation of retinal ganglion cells with functional axons from mouse embryonic stem cells and induced pluripotent stem cells. Invest Ophthalmol Vis Sci. 2016;57(7):3348–3359. doi: 10.1167/iovs.16-19166. [DOI] [PubMed] [Google Scholar]
- 43.Li K, Zhong X, Yang S, Luo Z, Li K, Liu Y, Cai S, Gu H, Lu S, Zhang H, Wei Y, Zhuang J, Zhuo Y, Fan Z, Ge J. HiPSC-derived retinal ganglion cells grow dendritic arbors and functional axons on a tissue-engineered scaffold. Acta Biomater. 2017;54:117–127. doi: 10.1016/j.actbio.2017.02.032. [DOI] [PubMed] [Google Scholar]
- 44.Teotia P, Chopra DA, Dravid SM, Van Hook MJ, Qiu F, Morrison J, Rizzino A, Ahmad I. Generation of functional human retinal ganglion cells with target specificity from pluripotent stem cells by chemically defined recapitulation of developmental mechanism. Stem Cells. 2017;35(3):572–585. doi: 10.1002/stem.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parameswaran S, Dravid SM, Teotia P, Krishnamoorthy RR, Qiu F, Toris C, Morrison J, Ahmad I. Continuous non-cell autonomous reprogramming to generate retinal ganglion cells for glaucomatous neuropathy. Stem Cells. 2015;33(6):1743–1758. doi: 10.1002/stem.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xie BB, Zhang XM, Hashimoto T, Tien AH, Chen A, Ge J, Yang XJ. Differentiation of retinal ganglion cells and photoreceptor precursors from mouse induced pluripotent stem cells carrying an Atoh7/Math5 lineage reporter. PLoS One. 2014;9(11):e112175. doi: 10.1371/journal.pone.0112175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuhrmann S. Eye morphogenesis and patterning of the optic vesicle. Curr Top Dev Biol. 2010;93:61–84. doi: 10.1016/B978-0-12-385044-7.00003-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuber ME, Gestri G, Viczian AS, Barsacchi G, Harris WA. Specification of the vertebrate eye by a network of eye field transcription factors. Development. 2003;130(21):5155–5167. doi: 10.1242/dev.00723. [DOI] [PubMed] [Google Scholar]
- 49.Nishihara D, Yajima I, Tabata H, Nakai M, Tsukiji N, Katahira T, Takeda K, Shibahara S, Nakamura H, Yamamoto H. Otx2 is involved in the regional specification of the developing retinal pigment epithelium by preventing the expression of sox2 and fgf8, factors that induce neural retina differentiation. PLoS One. 2012;7(11):e48879. doi: 10.1371/journal.pone.0048879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marquardt T, Gruss P. Generating neuronal diversity in the retina: one for nearly all. Trends Neurosci. 2002;25(1):32–38. doi: 10.1016/s0166-2236(00)02028-2. [DOI] [PubMed] [Google Scholar]
- 51.Chow RL, Lang RA. Early eye development in vertebrates. Annu Rev Cell Dev Biol. 2001;17:255–296. doi: 10.1146/annurev.cellbio.17.1.255. [DOI] [PubMed] [Google Scholar]
- 52.Muller F, Rohrer H, Vogel-Hopker A. Bone morphogenetic proteins specify the retinal pigment epithelium in the chick embryo. Development. 2007;134(19):3483–3493. doi: 10.1242/dev.02884. [DOI] [PubMed] [Google Scholar]
- 53.Bone-Larson C, Basu S, Radel JD, Liang M, Perozek T, Kapousta-Bruneau N, Green DG, Burmeister M, Hankin MH. Partial rescue of the ocular retardation phenotype by genetic modifiers. J Neurobiol. 2000;42(2):232–247. doi: 10.1002/(sici)1097-4695(20000205)42:2<232::aid-neu7>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 54.Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105(1):43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 55.Riesenberg AN, Le TT, Willardsen MI, Blackburn DC, Vetter ML, Brown NL. Pax6 regulation of Math5 during mouse retinal neurogenesis. Genesis. 2009;47(3):175–187. doi: 10.1002/dvg.20479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dansault A, David G, Schwartz C, Jaliffa C, Vieira V, de la Houssaye G, Bigot K, Catin F, Tattu L, Chopin C, Halimi P, Roche O, Van Regemorter N, Munier F, Schorderet D, Dufier JL, Marsac C, Ricquier D, Menasche M, Penfornis A, Abitbol M. Three new PAX6 mutations including one causing an unusual ophthalmic phenotype associated with neurodevelopmental abnormalities. Mol Vis. 2007;13:511–523. [PMC free article] [PubMed] [Google Scholar]
- 57.Gallardo ME, Lopez-Rios J, Fernaud-Espinosa I, Granadino B, Sanz R, Ramos C, Ayuso C, Seller MJ, Brunner HG, Bovolenta P, Rodríguez de Córdoba S. Genomic cloning and characterization of the human homeobox gene SIX6 reveals a cluster of SIX genes in chromosome 14 and associates SIX6 hemizygosity with bilateral anophthalmia and pituitary anomalies. Genomics. 1999;61(1):82–91. doi: 10.1006/geno.1999.5916. [DOI] [PubMed] [Google Scholar]
- 58.Voronina VA, Kozhemyakina EA, O'Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS, Mathers PH. Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet. 2004;13(3):315–322. doi: 10.1093/hmg/ddh025. [DOI] [PubMed] [Google Scholar]
- 59.Ouchi Y, Tabata Y, Arai K, Watanabe S. Negative regulation of retinal-neurite extension by beta-catenin signaling pathway. J Cell Sci. 2005;118(19):4473–4483. doi: 10.1242/jcs.02575. [DOI] [PubMed] [Google Scholar]
- 60.Sakuma R, Ohnishi Yi Y, Meno C, Fujii H, Juan H, Takeuchi J, Ogura T, Li E, Miyazono K, Hamada H. Inhibition of Nodal signalling by Lefty mediated through interaction with common receptors and efficient diffusion. Genes Cells. 2002;7(4):401–412. doi: 10.1046/j.1365-2443.2002.00528.x. [DOI] [PubMed] [Google Scholar]
- 61.Pan L, Deng M, Xie X, Gan L. ISL1 and BRN3B co-regulate the differentiation of murine retinal ganglion cells. Development. 2008;135(11):1981–1990. doi: 10.1242/dev.010751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schneider ML, Turner DL, Vetter ML. Notch signaling can inhibit Xath5 function in the neural plate and developing retina. Mol Cell Neurosci. 2001;18(5):458–472. doi: 10.1006/mcne.2001.1040. [DOI] [PubMed] [Google Scholar]
- 63.Nelson BR, Gumuscu B, Hartman BH, Reh TA. Notch activity is downregulated just prior to retinal ganglion cell differentiation. Dev Neurosci. 2006;28(1-2):128–141. doi: 10.1159/000090759. [DOI] [PubMed] [Google Scholar]
- 64.Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121(11):3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- 65.Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93(9):3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128(13):2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prasov L, Glaser T. Dynamic expression of ganglion cell markers in retinal progenitors during the terminal cell cycle. Mol Cell Neurosci. 2012;50(2):160–168. doi: 10.1016/j.mcn.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mu X, Beremand PD, Zhao S, Pershad R, Sun H, Scarpa A, Liang S, Thomas TL, Klein WH. Discrete gene sets depend on POU domain transcription factor Brn3b/Brn-3.2/POU4f2 for their expression in the mouse embryonic retina. Development. 2004;131(6):1197–1210. doi: 10.1242/dev.01010. [DOI] [PubMed] [Google Scholar]
- 69.Xiang M. Requirement for Brn-3b in early differentiation of postmitotic retinal ganglion cell precursors. Dev Biol. 1998;197(2):155–169. doi: 10.1006/dbio.1998.8868. [DOI] [PubMed] [Google Scholar]
- 70.Liu W, Khare SL, Liang X, Peters MA, Liu X, Cepko CL, Xiang M. All Brn3 genes can promote retinal ganglion cell differentiation in the chick. Development. 2000;127(15):3237–3247. doi: 10.1242/dev.127.15.3237. [DOI] [PubMed] [Google Scholar]
- 71.Wu F, Kaczynski TJ, Sethuramanujam S, Li R, Jain V, Slaughter M, Mu X. Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc Natl Acad Sci U S A. 2015;112(13):E1559–1568. doi: 10.1073/pnas.1421535112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Surgucheva I, Weisman AD, Goldberg JL, Shnyra A, Surguchov A. Gamma-synuclein as a marker of retinal ganglion cells. Mol Vis. 2008;14:1540–1548. [PMC free article] [PubMed] [Google Scholar]
- 73.Barnstable CJ, Dräger UC. Thy-1 antigen: a ganglion cell specific marker in rodent retina. Neuroscience. 1984;11(4):847–855. doi: 10.1016/0306-4522(84)90195-7. [DOI] [PubMed] [Google Scholar]
- 74.Rowan S, Chen CM, Young TL, Fisher DE, Cepko CL. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development. 2004;131(20):5139–5152. doi: 10.1242/dev.01300. [DOI] [PubMed] [Google Scholar]
- 75.Zweigerdt R, Olmer R, Singh H, Haverich A, Martin U. Scalable expansion of human pluripotent stem cells in suspension culture. Nature Protocols. 2011;6(5):689–700. doi: 10.1038/nprot.2011.318. [DOI] [PubMed] [Google Scholar]
- 76.Leong MF, Lu HF, Lim TC, Du C, Ma NKL, Wan ACA. Electrospun polystyrene scaffolds as a synthetic substrate for xeno-free expansion and differentiation of human induced pluripotent stem cells. Acta Biomaterialia. 2016;46:266–277. doi: 10.1016/j.actbio.2016.09.032. [DOI] [PubMed] [Google Scholar]
- 77.Hallam D, Hilgen G, Dorgau B, Zhu L, Yu M, Bojic S, Hewitt P, Schmitt M, Uteng M, Kustermann S, Steel D, Nicholds M, Thomas R, Treumann A, Porter A, Sernagor E, Armstrong L, Lako M. Human-induced pluripotent stem cells generate light responsive retinal organoids with variable and nutrient-dependent efficiency. Stem Cells. 2018;36(10):1535–1551. doi: 10.1002/stem.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hertz J, Qu B, Hu Y, Patel RD, Valenzuela DA, Goldberg JL. Survival and integration of developing and progenitor-derived retinal ganglion cells following transplantation. Cell Transplant. 2014;23(7):855–872. doi: 10.3727/096368913X667024. [DOI] [PubMed] [Google Scholar]
- 79.Kador KE, Montero RB, Venugopalan P, Hertz J, Zindell AN, Valenzuela DA, Uddin MS, Lavik EB, Muller KJ, Andreopoulos FM, Goldberg JL. Tissue engineering the retinal ganglion cell nerve fiber layer. Biomaterials. 2013;34(17):4242–4250. doi: 10.1016/j.biomaterials.2013.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kador KE, Alsehli HS, Zindell AN, Lau LW, Andreopoulos FM, Watson BD, Goldberg JL. Retinal ganglion cell polarization using immobilized guidance cues on a tissue-engineered scaffold. Acta Biomater. 2014;10(12):4939–4946. doi: 10.1016/j.actbio.2014.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kador KE, Grogan SP, Dorthé EW, Venugopalan P, Malek MF, Goldberg JL, D'Lima DD. Control of retinal ganglion cell positioning and neurite growth: combining 3D printing with radial electrospun scaffolds. Tissue Eng Part A. 2016;22(3-4):286–294. doi: 10.1089/ten.tea.2015.0373. [DOI] [PMC free article] [PubMed] [Google Scholar]


