Summary
Bone-resorbing osteoclasts (OCs) are derived from myeloid precursors (MPs). Several transcription factors are implicated in OC differentiation and function; however, their hierarchical architecture and interplay are not well known. Analysis for enriched motifs in PU.1 and MITF chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) data from differentiating OCs identified eomesodermin (EOMES) as a potential novel binding partner of PU.1 and MITF at genes critical for OC differentiation and function. We were able to demonstrate using co-immunoprecipitation and sequential ChIP analysis that PU.1, MITF, and EOMES are in the same complex and present as a complex at OC genomic loci. Furthermore, EOMES knockdown in MPs led to osteopetrosis associated with decreased OC differentiation and function both in vitro and in vivo. Although EOMES is associated with embryonic development and other hematopoietic lineages, this is the first study demonstrating the requirement of EOMES in the myeloid compartment.
Subject Areas: Biological Sciences, Cell Biology, Developmental Biology, Omics
Graphical Abstract
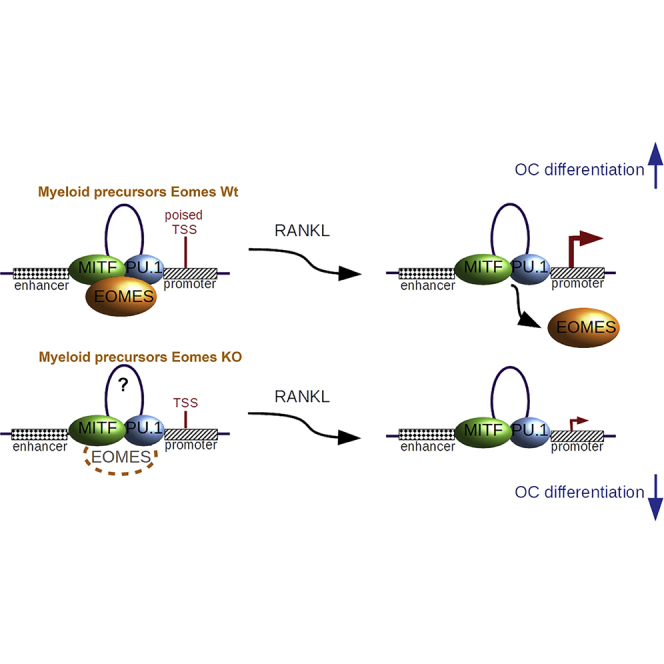
Highlights
-
•
Genomics and systems biology approaches can decipher transcription factor interactions
-
•
Motif analysis of MITF and PU.1 ChIP-seq peaks identified EOMES as a cofactor
-
•
This is the first report of EOMES' function in the myeloid compartment
-
•
EOMES knockdown results in reduced osteoclast formation and function
Biological Sciences; Cell Biology; Developmental Biology; Omics
Introduction
Hematopoiesis gives rise to a spectrum of cell types that are essential for normal immune and homeostatic functions throughout the body. Within this differentiation cascade, cells of the myeloid lineage give rise to bone-resorbing osteoclasts (OCs). In bones, OC differentiation from myeloid precursors (MPs) and subsequent OC function are regulated by microenvironmental cues from the surrounding stromal cells, in particular bone-forming osteoblasts. Downstream signaling through the CSF1/CSF1R and RANKL/RANK axes in the MPs, and later in the OCs, results in the activation of multiple signaling pathways and transcription factors (TFs) essential for OC differentiation, including PU.1, MITF, and NFATc1 (Hodgkinson et al., 1993, Takayanagi et al., 2002, Tondravi et al., 1997).
The ETS family member PU.1 is essential for myeloid lineage commitment (Iwasaki et al., 2005). We have shown that PU.1 and MITF physically and genetically interact to regulate a complex TF network regulating the course of OC differentiation, and ultimately OC function (Carey et al., 2018, Luchin et al., 2001, Sharma et al., 2007). Several of these PU.1-bound TF gene loci are present at the distal enhancer and “superenhancer” elements known to contain other TF-binding loci (Carey et al., 2018).
Eomesodermin (EOMES) was first identified as an essential developmental molecule controlling extraembryonic trophoblast formation (Ciruna and Rossant, 1999, Russ et al., 2000). EOMES has since been shown to be essential for (1) regulating mesoderm cell function and digit formation and (2) cerebral cortex development, patterning, and neurogenesis (Russ et al., 2000, Sessa et al., 2008, Sheeba and Logan, 2017). However, in postnatal life, EOMES has been predominately described along with the closely related T-box TF, T-bet, as a key regulator of CD8+ T cell and natural killer (NK) cell maturation and function (Gordon et al., 2012, Intlekofer et al., 2005, Pearce et al., 2003).
In this study, we have demonstrated by motif-finding algorithms that the T-box TF EOMES is highly enriched in PU.1 and MITF co-bound peaks in MPs and OCs. Furthermore, EOMES physically interacts with PU.1 and MITF, co-binding these factors in a PU.1/MITF/EOMES complex at key OC-specific TF genomic loci, which we have previously shown to be bound by PU.1 and MITF (Carey et al., 2018). EOMES knockdown in murine MPs leads to decreased OC differentiation and function in vitro and in vivo. Although EOMES is associated with other hematopoietic lineages, this is the first study demonstrating the requirement of EOMES in OC differentiation, where it is a novel co-partner of PU.1 and MITF.
Results
EOMES Interacts with PU.1 and MITF to Regulate OC-Determining Gene Loci
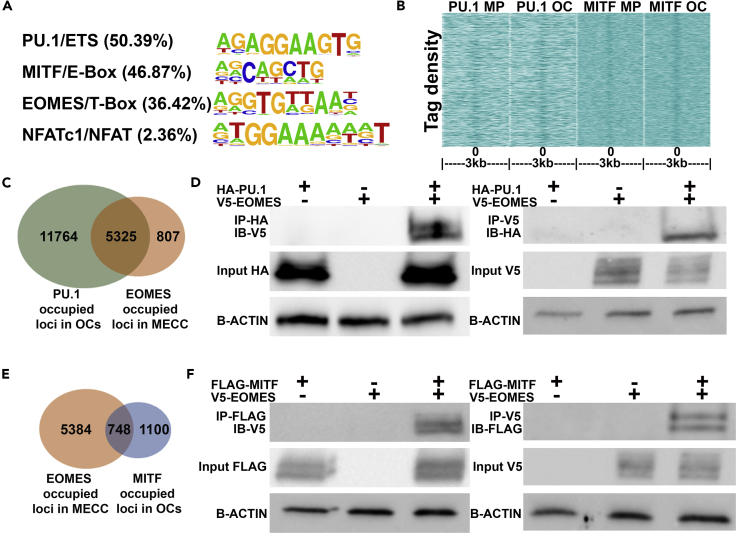
We previously performed PU.1 and MITF chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) in MPs and OCs. This revealed a conserved TF network regulating OC differentiation that is governed by PU.1 and MITF (Carey et al., 2018). To identify any novel binding partners that may also be important in establishing the OC TF network, we performed motif analysis of PU.1/MITF co-bound peaks in OCs. Apart from PU.1/ETS and MITF/E-Box motifs, a T-Box signature typical for EOMES/TBR2 was most enriched in PU.1/MITF co-bound peaks (Figure 1A). To evaluate the validity of these findings, we first analyzed a publicly available dataset of EOMES ChIP-seq from murine embryonic cerebral cortex (MECC) (Sessa et al., 2017). We evaluated the tag densities of both PU.1 and MITF in both MPs and OCs around EOMES-bound regions in MECC. These results indicated that the EOMES-occupied loci were associated with regions with higher PU.1 and MITF binding, as determined by K means clustering (Figure 1B). Furthermore, of the 17,089 genes associated with PU.1 binding in OCs, 31% of the genes were also associated with EOMES binding in MECC (Figure 1C). Tag densities of PU.1 occupancy from both MPs and OCs around MECC EOMES ChIP-seq peaks indicated that within 500 bp of EOMES peaks several PU.1 peaks may exist (Figure S1A). To examine whether PU.1 could physically interact with EOMES, we overexpressed V5-tagged EOMES with HA-tagged PU.1 in HEK293 cells followed by co-immunoprecipitation (coIP). Upon HA-PU.1 pull-down and probing for V5-EOMES by V5 antibody, an EOMES/PU.1 interaction was present (Figure 1D; left). In complementary experiments when EOMES complexes were pulled down with V5 antibody, a PU.1/EOMES interaction was also detected (Figure 1D; right). Next, we scrutinized whether MITF-occupied loci in OCs were also associated with EOMES-bound loci in MECC and found that 12% of the MITF-occupied sites overlapped with the EOMES-bound genes (Figure 1E), with the highest MITF tag density in MPs and OCs within 50 bases of EOMES peak centers in MECC (Figure S1B). FLAG-tagged MITF and V5-tagged EOMES overexpression in HEK293 cells followed by coIP also revealed an interaction between MITF and EOMES (Figure 1F).
Figure 1.
EOMES Interacts with PU.1 and MITF to Regulate OC-Determining Gene Loci
(A) Motifs enriched in MITF and PU.1 overlapped peaks in OCs with their percentage of occurrence.
(B) K means clustering diagram showing the density of PU.1 and MITF ChIP-seq fragments in MPs and OCs around EOMES-bound regions in murine embryonic cerebral cortex.
(C) Overlap of bound gene loci between EOMES ChIP-seq from MECC and PU.1 ChIP-seq in OCs.
(D) coIP experiments involving HA-PU.1 IP followed by V5-EOMES western blot in HEK293 cells overexpressing V5-tagged EOMES and HA-tagged PU.1 (left). Complementary experiments involving V5-EOMES immunoprecipitation (IP) followed by HA-PU.1 western blot in the same cells (right).
(E) Overlap of gene loci between EOMES ChIP-seq from MECC and MITF ChIP-seq in OCs.
(F) coIP experiments involving FLAG-MITF IP followed by V5-EOMES western blot in HEK293 cells overexpressing V5-tagged EOMES and FLAG-tagged MITF (left). Complementary experiments involving V5-EOMES IP followed by FLAG-MITF western blot in the same cells (right).
See also Figure S1.
Myeloid-Specific Knockdown of EOMES Disrupts the Expression of OC-Specific TFs
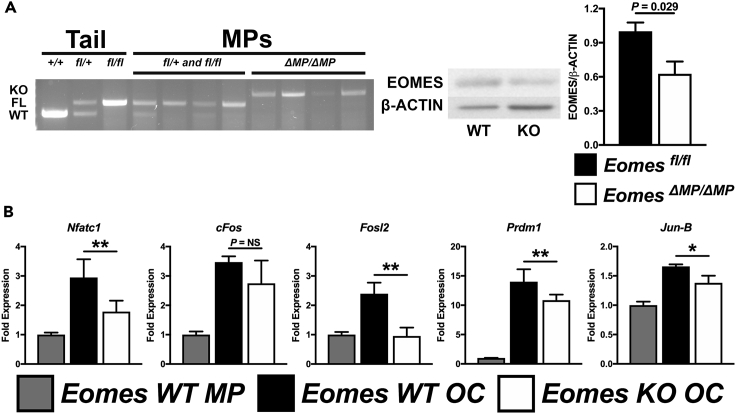
We have shown that the PU.1-MITF regulatory axis is a predominant regulator of the OC-specific TF network (Carey et al., 2018). To evaluate the molecular functions regulated by EOMES, we examined the molecular processes enriched in EOMES-associated genes from MECC using the ToppGene suite (Chen et al., 2009) and found that TF and promoter binding were highly enriched (Figure S2A). To verify if EOMES is essential for the expression of any of the TFs identified in the PU.1-MITF regulatory network, we used a tamoxifen-inducible, myeloid lineage-specific Csf1rTAMCre in combination with Eomesfl/fl mice to generate EomesΔMP/ΔMP mice with tamoxifen induction. EOMES knockdown in MPs was confirmed by both genotyping and western blot (Figure 2A). EomesΔMP/ΔMP mice showed a significant decrease in EOMES protein expression in MPs compared with controls (p = 0.029, Figure 2A). In vitro differentiation of MPs into OCs from EomesΔMP/ΔMP mice and control littermates revealed a significant reduction in the upregulation of pro-osteoclastogenic TFs, Nfatc1, Fosl2, Prdm1, and Jun-B when compared with controls (p < 0.05, Figure 2B). Although cFos upregulation in EomesΔMP/ΔMP OCs was reduced compared with controls, this did not reach statistical significance (p = 0.383, Figure 2B). In addition, TFs with known anti-osteoclastogenic effects, Irf8 and Bcl6, trended to be significantly increased in OCs following EOMES deletion (p = 0.074 and 0.060, respectively, Figure S2B).
Figure 2.
Myeloid-Specific Knockdown of Eomes Disrupts the Expression of OC-Specific TFs
(A) Knockdown of EOMES specifically in MPs (EomesΔMP/ΔMP) was confirmed by (1) DNA genotyping PCR (with historical genomic tail-tip extracted DNA as reference) and (2) western blot analysis (with quantification, n = 2) following 3 days treatment with tamoxifen using myeloid-lineage-specific Csf1rTAMCre in combination with Eomesfl/fl mice when compared with Eomesfl/fl, Eomesfl/+, or Eomes+/+ controls.
(B) Evaluation of key pro-osteoclastogenic TFs by quantitative real-time RT-PCR (n = 2–6). For all figures and subfigures, data are represented as mean ± SD for all bar graphs and all images are representative. *p < 0.05, **p < 0.01. KO, knockout; WT, wild-type.
See also Figure S2.
The PU.1-MITF-EOMES Complex Drives Nfatc1 Expression by Association with the Looping Peak within the Nfatc1 Superenhancer
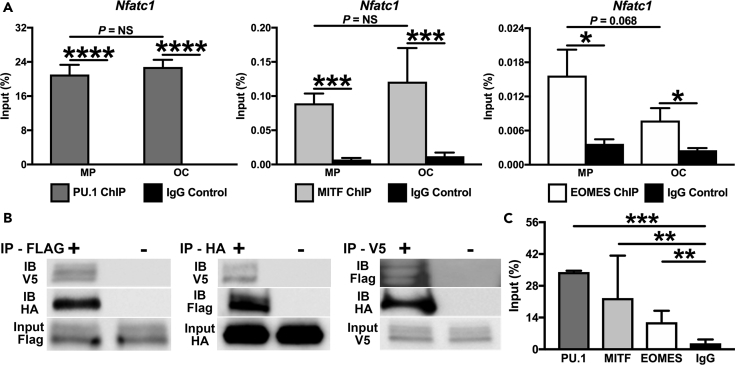
We have recently identified a conserved intronic enhancer in the OC master regulator Nfatc1 that requires PU.1 to loop back to the proximal promoter by means of changes in chromatin conformation (Carey et al., 2018). Thus we evaluated whether EOMES directly binds to this Nfatc1 enhancer co-bound by PU.1 and MITF (Figure 3A). There was significant enrichment of PU.1, MITF, and EOMES at the Nfatc1 enhancer in both MPs and OCs when compared with IgG controls (p < 0.05). Interestingly, whereas PU.1 and MITF occupancy is sustained at Nfatc1 as MPs differentiated into OCs (Figure 3A), EOMES is present at a lower abundance in OCs when compared with MPs (p = 0.068, Figure 3A).
Figure 3.
The PU.1-MITF-EOMES Complex Drives Nfatc1 Expression by Association with the Looping Peak within the Nfatc1 Superenhancer
(A) Conventional ChIP of PU.1, MITF, and EOMES binding at the conserved intronic enhancer of Nfatc1 in MPs and OCs (n = 2–3).
(B) HA-PU.1, FLAG-MITF, or V5-EOMES immunoprecipitation (IP) followed by HA-PU.1, FLAG-MITF, or V5-EOMES western blot in HEK293 cells co-transfected with FLAG-MITF, HA-PU.1, and V5-EOMES.
(C) Sequential ChIP using PU.1, MITF, and EOMES antibodies following initial PU.1 ChIP in MPs (ReChIP) at the looping peak of Nfatc1. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Next, we verified whether EOMES could represent a subunit of the PU.1-MITF complex by co-expression of FLAG-MITF, HA-PU.1, and V5-EOMES in HEK293 cells. In a series of complimentary experiments, when the total cell lysate was pulled down with (1) FLAG-MITF and probed for V5-EOMES and HA-PU.1 (left), (2) HA-PU.1 and probed for V5-EOMES and FLAG-MITF (middle), or (3) V5-EOMES and probed for FLAG-MITF and HA-PU.1 (right), all signals were detected in the immunoprecipitations (Figure 3B). These results indicated that PU.1, MITF, and EOMES all interact and can remain in a complex together. Previously, we have shown by ReChIP analysis that PU.1 and MITF can simultaneously occupy specific regions of chromatin (Hu et al., 2007). We again employed ReChIP to determine if EOMES can also interact with PU.1 and MITF while all occupy a regulatory site near the Nfatc1 gene. For ReChIP, soluble chromatin from MPs was pulled down using the PU.1 antibody. After dissociation of the chromatin complex from the antibody, sequential ChIP was performed using PU.1, MITF, and EOMES antibodies. Using qRT-PCR with primers designed at the looping peak of Nfatc1, we observed that ∼34% of the first immunoprecipitate (input for the second ChIP) was pulled down with the PU.1 antibody (Figure 3C). MITF was able to pull down ∼23% of the first immunoprecipitate, and EOMES was able to pull down ∼12%, all significantly more than the IgG control (∼2.7%) (p < 0.01, Figure 3C). This result further indicates that PU.1, MITF, and EOMES can all interact in MPs and do so to regulate the expression of OC TF network genes like Nfatc1.
Myeloid-Specific EOMES Deletion Decreased OC Differentiation and Function In Vivo and In Vitro
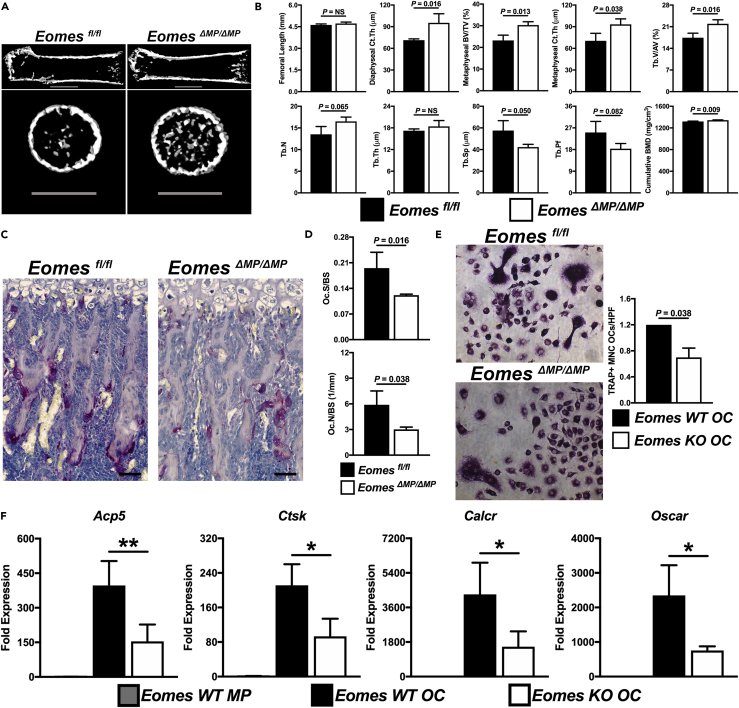
EomesΔMP/ΔMP mice possessed increased bone mass when compared with control Eomesfl/fl mice upon micro-computed tomographic analysis (Figure 4A). Most notably, there was a significant increase in bone volume/total volume in the distal femoral metaphysis (p = 0.013) and cortical thickness in both the distal femoral metaphysis (p = 0.038) and diaphysis (p = 0.016) (Figure 4B). In addition, EomesΔMP/ΔMP mice displayed a significant increase in cumulative bone mineral density (BMD) (p = 0.009) and trabecular volume/all bone volume (p = 0.016), a trend toward a significant increase in trabecular number (p = 0.065), and a decrease in trabecular spacing (p = 0.050, Figure 4B). The statistical trend and decrease in trabecular pattern factor (p = 0.082) in EomesΔMP/ΔMP mice indicates greater trabecular connectivity (Figure 4B). Based on our MP-specific deletion of EOMES, these results suggest that decreased osteoclastic bone resorption is responsible for the bone phenotype observed in EomesΔMP/ΔMP mice. In further support of this hypothesis, histomorphometric analysis revealed a significant reduction in both OC number (p = 0.038) and OC surface (p = 0.016) per bone surface in EomesΔMP/ΔMP mice compared with controls (Figures 4C and 4D). In vitro differentiation of OCs derived from EomesΔMP/ΔMP mice showed a significant reduction in the formation of TRAP-positive multinucleated OCs (p = 0.038, Figures 4E and 4A) and significant decrease in the expression of the OC marker genes Acp5, Ctsk, Calcr, and Oscar (p < 0.05, Figure 4F).
Figure 4.
Myeloid-Specific Eomes Deletion Decreased OC Differentiation and Function In Vivo and In Vitro
(A) Femoral micro-computed tomographic (μCT) cross-sectional images in the craniocaudal view and the distal femoral metaphysis in the proximodistal view from 8-day-old EomesΔMP/ΔMP mice and Eomesfl/fl controls. Scale bars, 1 mm.
(B) μCT quantification of bone morphometric variables (n = 3). BMD, bone mineral density; BV/TV, bone volume/total volume; Ct.Th, cortical thickness; Tb.V/AV, trabecular volume/all bone volume; Tb.N, trabecular number; Tb.Sp, trabecular spacing; Tb.Pf, trabecular pattern factor; Tb.Th, trabecular thickness.
(C) Tartrate-resistant acid phosphatase (TRAP) staining (in violet) with hematoxylin counterstain to identify OCs in the distal femoral metaphysis of mice in (A). Scale bars, 50 μm.
(D) Histomorphometric quantification of Oc.S/BS and Oc.N/BS from (C) (n = 3). Oc.N/BS, OC number per bone surface; Oc.S/BS, OC surface per bone surface.
(E and F) (E) Representative images of in vitro TRAP-positive multinucleated OC formation from Csf1rTAMCre;Eomesfl/fl and Eomesfl/fl mice and quantification per high-powered field (HPF) (n = 2). (F) RT-PCR analysis of genes regulating OC differentiation and function in Eomesfl/fl (WT) MPs and OCs and EomesΔMP/ΔMP (Eomes KO) OCs after differentiation (n = 2–6). *p < 0.05, **p < 0.01. KO, knockout; WT, wild-type.
Discussion
EOMES is the earliest expressed T-box gene during development (Ciruna and Rossant, 1999). Constitutive or conditional EOMES deletion has demonstrated its crucial role in lineage commitment, differentiation, and maintenance of multiple stem and progenitor cell populations (Pimeisl et al., 2013). The functionality of EOMES in post-natal life is best described in the context of the immune system, where it is a key regulator of cell-mediated immunity and T cell function (Intlekofer et al., 2005, Pearce et al., 2003). Here, the essential role of EOMES is evident in that it is a disease susceptibility loci for multiple diseases linked with T cell dysfunction (Berndt et al., 2016, Kim et al., 2015).
One of the key insights obtained in our study is the discovery that EOMES is a functional partner of PU.1 and MITF in regulating the TF network governing OC differentiation. Although EOMES is capable of inducing the full array of mesodermal cell types and genes (Sheeba and Logan, 2017), expression and functionality of EOMES in bone-resorbing OCs, or any cell of the myeloid lineage for that matter, has not been reported. Our results confirm that like the master myeloid regulator PU.1 (Carey et al., 2018), EOMES modulates OC differentiation by promoting the expression of pro-osteoclastogenic TFs and inhibiting the expression of TFs that negatively regulate OC differentiation, albeit to a lesser magnitude than PU.1. Our in silico analysis indicated high enrichment of the EOMES-binding motif in PU.1-MITF co-bound loci. Furthermore, when PU.1 and MITF tag densities were mapped against EOMES ChIP-seq peaks from MECC (Sessa et al., 2017), both PU.1 and MITF tags were enriched within 50 bp of the EOMES peak centers.
Our in silico results led us to query whether EOMES could interact with PU.1, MITF, or both. No physical interaction between EOMES and either MITF or ETS family TFs, like PU.1, has been previously reported. However, previous work has shown that EOMES regulates the expression of genes regulated by (1) MITF, such as granzyme B and interferon-γ, and (2) ETS1 and MEF (ETS family members), including perforin in certain lymphoid cell populations (Glimcher et al., 2004). Interestingly, in CD8+ T cells, different chromatin conformations are associated with ETS family member and EOMES binding (Moskowitz et al., 2017). In the naive cell state, there is increased accessibility by ETS members, and in the memory cell state, an open conformation allows more ready access by EOMES (Moskowitz et al., 2017). In our study, we were able to demonstrate in the myeloid compartment that the ETS family TF, PU.1, and MITF, are in a complex with EOMES, which is found at the looping peak of the Nfatc1 enhancer. These results indicate that in the looping peak of Nfatc1, EOMES associates with the PU.1-MITF complex in at least a subset of MPs. Considering that only a subset of MPs differentiate to form OCs in vivo, EOMES might be more specifically present only in those MPs committing to the OC precursor population. Previous work has shown that EOMES changes its binding profile during transition from one cell type to another, suggesting that its function may change at different points of differentiation (Tsankov et al., 2015). Conventional ChIP revealed that like PU.1 and MITF, EOMES is recruited and bound to a key TF locus essential for OC differentiation. Interestingly, we detected a statistical trend toward a lower abundance of EOMES binding on Nfatc1 in OCs when compared with MPs. This suggests that EOMES is more essential in establishing chromatin dynamics in MPs than in differentiating OCs.
We demonstrated that MP-specific EOMES deletion disrupted the PU.1-MITF TF network and resulted in osteopetrosis in vivo, a finding consistent with decreased OC differentiation and function. Historically, EOMES knockout mice arrest at the blastocyst stage, resulting in early embryonic lethality (Russ et al., 2000). Lineage-specific EOMES deletion within the hematopoietic hierarchy using Vav-Cre has been reported (Gordon et al., 2012). However, whereas there was a global reduction in NK cells, which are of lymphoid origin, no investigation on changes within the myeloid lineage were reported (Gordon et al., 2012).
In summary, the work presented here is the first evidence of EOMES having a functional role in any cell of the myeloid lineage, where EOMES is a novel binding partner of PU.1 and MITF and works in conjunction with these factors to regulate the OC TF network.
Limitations of the Study
Although the systems biology approaches we used to identify EOMES as a partner of PU.1 and MITF are robust, they also give rise to several other “hits” due to the vast number of ETS DNA-binding domains. We filtered most of the ETS motifs as a subset of binding regions associated with PU.1, which is an ETS factor. An additional limitation is that due to the lack of reagents, the temporal kinetics of PU.1, MITF, and EOMES recruitment to target loci was not established in the current study.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
The authors are grateful to the core facilities involved in this work, OSU ULAR and MUSC DLAR, for animal care and Jason Bice of the OSU-CCC Solid Tumor Biology Histology Core for tissue preparation. Authors are grateful to Dr. Lyudmyla I. Sharma for the conceptualization and creation of graphical abstract. This work was supported by NIH-NIAMS Grant 2R01AR044719-15A (M.C.O. and S.S.M.).
Author Contributions
Conceptualization, M.C.O., and S.M.S.; Methodology, H.A.C., J.F.C., M.C.O., and S.M.S; Investigation, H.A.C., B.E.H., D.J.S., K.A.T., and S.M.S; Formal Analysis, B.E.H., T.J.R., R.E.T., J.F.C., and S.M.S; Visualization, B.E.H., D.J.S., and S.M.S; Writing – Original Draft, H.A.C., B.E.H., and S.M.S; Writing – Review & Editing, B.E.H., M.C.O., J.F.C., and S.M.S.; Funding Acquisition, M.C.O. and S.M.S.; Resources, M.C.O., J.F.C., R.E.T., and S.M.S.; Supervision, M.C.O. and S.M.S.
Declaration of Interests
The authors declare no competing interests.
Published: January 25, 2019
Footnotes
Supplemental Information includes Transparent Methods and two figures and can be found with this article online at https://doi.org/10.1016/j.isci.2018.12.018.
Contributor Information
Michael C. Ostrowski, Email: ostrowsk@musc.edu.
Sudarshana M. Sharma, Email: sharmas@musc.edu.
Data and Software Availability
Raw images of immunoprecipitation and western blot data used in this has been deposited to Mendeley depository and can be accessed at URL:http://dx.doi.org/10.17632/wdv72kxgjg.1.
Supplemental Information
References
- Berndt S.I., Camp N.J., Skibola C.F., Vijai J., Wang Z., Gu J., Nieters A., Kelly R.S., Smedby K.E., Monnereau A. Meta-analysis of genome-wide association studies discovers multiple loci for chronic lymphocytic leukemia. Nat. Commun. 2016;7:10933. doi: 10.1038/ncomms10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey H.A., Hildreth B.E., 3rd, Geisler J.A., Nickel M.C., Cabrera J., Ghosh S., Jiang Y., Yan J., Lee J., Makam S. Enhancer variants reveal a conserved transcription factor network governed by PU.1 during osteoclast differentiation. Bone Res. 2018;6:8. doi: 10.1038/s41413-018-0011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B.G., Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mech. Dev. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- Glimcher L.H., Townsend M.J., Sullivan B.M., Lord G.M. Recent developments in the transcriptional regulation of cytolytic effector cells. Nat. Rev. Immunol. 2004;4:900–911. doi: 10.1038/nri1490. [DOI] [PubMed] [Google Scholar]
- Gordon S.M., Chaix J., Rupp L.J., Wu J., Madera S., Sun J.C., Lindsten T., Reiner S.L. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson C.A., Moore K.J., Nakayama A., Steingrimsson E., Copeland N.G., Jenkins N.A., Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hu R., Sharma S.M., Bronisz A., Srinivasan R., Sankar U., Ostrowski M.C. Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors. Mol. Cell. Biol. 2007;27:4018–4027. doi: 10.1128/MCB.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Iwasaki H., Somoza C., Shigematsu H., Duprez E.A., Iwasaki-Arai J., Mizuno S., Arinobu Y., Geary K., Zhang P., Dayaram T. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Bang S.Y., Lee H.S., Cho S.K., Choi C.B., Sung Y.K., Kim T.H., Jun J.B., Yoo D.H., Kang Y.M. High-density genotyping of immune loci in Koreans and Europeans identifies eight new rheumatoid arthritis risk loci. Ann. Rheum. Dis. 2015;74:e13. doi: 10.1136/annrheumdis-2013-204749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchin A., Suchting S., Merson T., Rosol T.J., Hume D.A., Cassady A.I., Ostrowski M.C. Genetic and physical interactions between Microphthalmia transcription factor and PU.1 are necessary for osteoclast gene expression and differentiation. J. Biol. Chem. 2001;276:36703–36710. doi: 10.1074/jbc.M106418200. [DOI] [PubMed] [Google Scholar]
- Moskowitz D.M., Zhang D.W., Hu B., Le Saux S., Yanes R.E., Ye Z., Buenrostro J.D., Weyand C.M., Greenleaf W.J., Goronzy J.J. Epigenomics of human CD8 T cell differentiation and aging. Sci. Immunol. 2017;2:1–13. doi: 10.1126/sciimmunol.aag0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce E.L., Mullen A.C., Martins G.A., Krawczyk C.M., Hutchins A.S., Zediak V.P., Banica M., DiCioccio C.B., Gross D.A., Mao C.A. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Pimeisl I.M., Tanriver Y., Daza R.A., Vauti F., Hevner R.F., Arnold H.H., Arnold S.J. Generation and characterization of a tamoxifen-inducible Eomes(CreER) mouse line. Genesis. 2013;51:725–733. doi: 10.1002/dvg.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ A.P., Wattler S., Colledge W.H., Aparicio S.A., Carlton M.B., Pearce J.J., Barton S.C., Surani M.A., Ryan K., Nehls M.C. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Sessa A., Ciabatti E., Drechsel D., Massimino L., Colasante G., Giannelli S., Satoh T., Akira S., Guillemot F., Broccoli V. The Tbr2 molecular network controls cortical neuronal differentiation through complementary genetic and epigenetic pathways. Cereb. Cortex. 2017;27:3378–3396. doi: 10.1093/cercor/bhw270. [DOI] [PubMed] [Google Scholar]
- Sessa A., Mao C.A., Hadjantonakis A.K., Klein W.H., Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S.M., Bronisz A., Hu R., Patel K., Mansky K.C., Sif S., Ostrowski M.C. MITF and PU.1 recruit p38 MAPK and NFATc1 to target genes during osteoclast differentiation. J. Biol. Chem. 2007;282:15921–15929. doi: 10.1074/jbc.M609723200. [DOI] [PubMed] [Google Scholar]
- Sheeba C.J., Logan M.P. The roles of T-box genes in vertebrate limb development. Curr. Top. Dev. Biol. 2017;122:355–381. doi: 10.1016/bs.ctdb.2016.08.009. [DOI] [PubMed] [Google Scholar]
- Takayanagi H., Kim S., Koga T., Nishina H., Isshiki M., Yoshida H., Saiura A., Isobe M., Yokochi T., Inoue J. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev. Cell. 2002;3:889–901. doi: 10.1016/s1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- Tondravi M.M., McKercher S.R., Anderson K., Erdmann J.M., Quiroz M., Maki R., Teitelbaum S.L. Osteopetrosis in mice lacking haematopoietic transcription factor PU.1. Nature. 1997;386:81–84. doi: 10.1038/386081a0. [DOI] [PubMed] [Google Scholar]
- Tsankov A.M., Gu H., Akopian V., Ziller M.J., Donaghey J., Amit I., Gnirke A., Meissner A. Transcription factor binding dynamics during human ES cell differentiation. Nature. 2015;518:344–349. doi: 10.1038/nature14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.