Abstract
This paper presents optical coherence tomography (OCT) images of the human retina and manual delineations of eight retinal layers. The data includes 35 human retina scans acquired on a Spectralis OCT system (Heidelberg Engineering, Heidelberg, Germany), 14 of which are healthy controls (HC) and 21 have a diagnosis of multiple sclerosis (MS). The provided data includes manually delineation of eight retina layers, which were independently reviewed and edited. The data presented in this article was used to validate automatic segmentation algorithms (Lang et al., 2013).
Specifications table
| Subject area | Ophthalmology |
| More specific subject area | Human retina, Multiple sclerosis |
| Type of data | Optical coherence tomography |
| How data was acquired | Spectral Domain OCT using Spectralis OCT system |
| Data format | Raw and Processed |
| Experimental factors | Human retina without pretreatment |
| Experimental features | The structure of the human retina was examined with SD-OCT |
| Data source location | The Johns Hopkins Hospital, Baltimore, MD 21287 USA |
| Data accessibility | Public download |
| Related research article | A. Lang, A. Carass, M. Hauser, E.S. Sotirchos, P.A. Calabresi, H.S. Ying, and J.L. Prince, “Retinal layer segmentation of macular OCT images using boundary classification”, Biomedical Optics Express, 4(7):1133–1152. |
Value of the data
-
•
This is currently the largest public data set of manually delineated layers of the human retina from OCT scans.
-
•
The data are fully delineated so thickness analysis and algorithm comparison can be performed.
-
•
The data can also be used for training and validation of segmentation algorithms.
1. Data
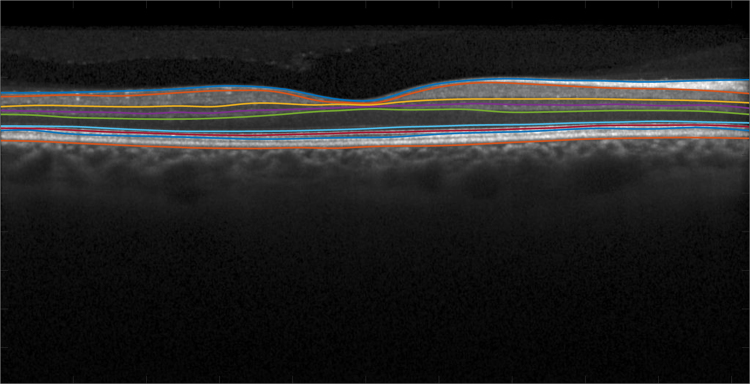
The data presented in this article was used to validate automatic segmentation algorithms [1], [2], [3]. The data comprise scans of the right eye of 35 subjects scanned on a Spectralis OCT system (Heidelberg Engineering, Heidelberg, Germany). The cohort comprises 14 healthy controls (HC) and 21 patients with multiple sclerosis (MS); complete demographic information is included in Table 1. All the scans were manually delineated once using internally developed software. For each subject, we provide a retinal OCT image consisting of 49 B-scans and 9 layer boundaries delineated in every B-scan. An example B-scan and the manual delineation is shown in Fig. 1. The provided layers are listed in Table 2. The data is available for download from: http://iacl.jhu.edu/Resources.
Table 1.
Demographic details for the data. The top line is the information of the entire data set, subsequent lines are specific to that category. N (M/F) denotes the number of patients and the male/female ratio, respectively. Age is the mean age (and standard deviation) in years at scan time. The key for the table is: HC – Healthy Controls; MS – Multiple Sclerosis.
| Dataset | N (M/F) | Age Mean (SD) |
|---|---|---|
| ALL | 35 (6/29) | 39.49 (10.94) |
| HC | 14 (2/12) | 35.77 (13.03) |
| MS | 21 (4/17) | 41.97 (8.77) |
Fig. 1.
An example B-scan images, showing the included manual delineations.
Table 2.
Manually delineated retina layers.
| Layer name | Abbreviation |
|---|---|
| Retina nerve fiber layer | RNFL |
| Ganglion cell layer and inner plexiform layer | GCL+IPL |
| Inner nuclear layer | INL |
| Outer plexiform layer | OPL |
| Outer nuclear layer | ONL |
| Inner photoreceptor segments | IS |
| Outer photoreceptor segments | OS |
| Retinal pigment epithelium | RPE |
2. Experimental design, materials, and methods
The Spectralis scanner׳s automatic real-time function is used to acquire the scans. Each B-scan was averaged at least 12 images at the same location and the signal-to-noise ratio of the final averaged scans was at least 20 dB. A macular cube scan (20° × 20°) was acquired with 49 B-scans, each B-scan consists of 1024 A-scans, and each A-scan has 496 pixels. The B-scan resolution varied slightly between subjects, the lateral resolution (between A-scans) has a mean over all the subjects of 5.8 µm (±0.2) and the axial resolution (between two pixels in an A-scan) is 3.9 µm (±0.0). The through-plane distance (slice separation) has a mean of 123.6 µm (±3.6) between images, resulting in an imaging area of approximately 6 × 6 mm2. The volume data was exported from the scanner using the.vol file format. To facilitate data use, we also provide scripts for reading the.vol files into Matlab. The manual delineations are saved in the.mat file which can be directly imported into Matlab.
Automatic segmentation methods evaluated using this dataset can be found in Refs. [1], [2], [3], [4], [5], [6] and other works using this data, in whole or part, include [7], [8], [9], [10], [11], [12].
Acknowledgments
This work was supported by the NIH under NEI grant R01-EY024655 (PI: J.L. Prince) and NINDS grant R01-NS082347 (PI: P.A. Calabresi).
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.12.073.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Lang A., Carass A., Hauser M., Sotirchos E.S., Calabresi P.A., Ying H.S., Prince J.L. Retinal layer segmentation of macular OCT images using boundary classification. Biomed. Opt. Express. 2013;4(7):1133–1152. doi: 10.1364/BOE.4.001133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carass A., Lang A., Hauser M., Calabresi P.A., Ying H.S., Prince J.L. Multiple-object geometric deformable model for segmentation of Macular. OCT. Biomed. Opt. Express. 2014;5(4):1062–1074. doi: 10.1364/BOE.5.001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.He Y., Carass A., Yun Y., Zhao C., Jedynak B.M., Solomon S.D., Saidha S., Calabresi P.A., Prince J.L. Fetal, Infant and Ophthalmic Medical Image Analysis. Springer; 2017. Towards topological correct segmentation of macular OCT from cascaded FCNs; pp. 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Y. He, A. Carass, Y. Yun, C. Zhao, B.M. Jedynak, S.D. Solomon, P.A. Calabresi, J.L. Prince, Topology Guranteed Segmentation of the Human Retina from OCT Using Convolutional Neural Networks. arXiv preprint 〈arXiv:1803.05120〉 (2018).
- 5.Y. Liu, A. Carass, S.D. Soloman, S. Saidha, P.A. Calabresi, J.L. Prince, Multi-layer fast level set segmentation for macular OCT, in: Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), 2018, pp. 1445–1448. [DOI] [PMC free article] [PubMed]
- 6.Lang A., Carass A., Bittner A.K., Ying H.S., Prince J.L. Improving graph based segmentation for severe pathology in Retinitis Pigmentosa patients. Proc. SPIE Med. Imaging (SPIE-MI 2017) 2017 doi: 10.1117/12.2254849. (pp. 10137-10137-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.A. Lang, A. Carass, A.K. Bittner, H.S. Ying, J.L. Prince, Intensity inhomogeneity correction of macular OCT using N3 and retinal flatspace, in: Proceedings of the IEEE 13th International Symposium on Biomedical Imaging (ISBI 2016), 2016, pp. 197–200. [DOI] [PMC free article] [PubMed]
- 8.Lang A., Carass A., Jedynak B.M., Solomon S.D., Saidha S., Calabresi P.A., Prince J.L. Intensity inhomogeneity correction of SD-OCT data using macular flatspace. Med. Image Anal. 2018;43:85–97. doi: 10.1016/j.media.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhargava P., Lang A., Al-Louzi O., Carass A., Prince J.L., Calabresi P.A., Saidha S. Applying an open-source segmentation algorithm to different OCT devices in multiple sclerosis patients and healthy controls. Mult. Scler. Int. 2015 doi: 10.1155/2015/136295. (Article ID 136295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen M., Lang A., Ying H.S., Calabresi P.A., Prince J.L., Carass A. Analysis of Macular OCT images using deformable registration. OCT. Biomed. Opt. Express. 2014;5(7):2184–2195. doi: 10.1364/BOE.5.002196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.M. Chen, A. Lang, E. Sotirchos, H.S. Ying, P.A. Calabresi, J.L. Prince, A. Carass, Deformable registration of Macular OCT using A-mode scan similarity, in: Proceedings of the IEEE 10th International Symposium on Biomedical Imaging (ISBI 2013), 2013, pp. 476–479. [DOI] [PMC free article] [PubMed]
- 12.Reaungamornrat S., Carass A., He Y., Saidha S., Calabresi P.A., Prince J.L. Inter-scanner variation independent descriptors for constrained diffeomorphic registration of retinal OCT. Proc. SPIE Med. Imaging (SPIE-MI 2018) 2018 doi: 10.1117/12.2293790. (pp. 10574-10574-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material