Abstract
Background
Long non‐coding RNAs play crucial roles in various biological activities and diseases. The role of long intergenic non‐coding RNA01638 (linc01638) in breast cancer, espe-cially in HER2-positive breast cancer, remains largely unknown.
Objective
To investigate the effect of linc01638 on tumorigenesis in HER2-positive breast cancer.
Methods
We first used qRT-PCR to detect linc01638 expression in HER2-positive breast cancer cells and tissues. Then we analyzed the effects of linc01638 expression in HER2-positive breast cancer cells through cell apoptosis assay, cell proliferation assay, colony formation assay, and cell invasion assay. We conducted mouse xenograft model to further confirm the role of linc01638 in HER2-positive breast cancer. Moreover, we used Western blot and IHC analysis to access the effect of linc01638 on DNMTs, BRCA1 and PTEN expressions in transplanted tumors.
Results
Linc01638 was found to be remarkably overexpressed in HER2-positive breast cancer cells and tissues. Suppression of linc01638 enhanced cell apoptosis, as well as inhibited the growth and in-vasiveness of HER2-positive breast cancer cells in vitro and tumor progression and metastasis in vivo. Furthermore, inhibition of linc01638 by shRNA attenuated expression of DNMT1, DNMT3a, and DNMT3b, and promoted expression of BRCA1 and PTEN in HER2-positive breast cancer cells and mouse xenograft models.
Conclusion
Linc01638 might be a promising biomarker and therapeutic target for treatment of HER2-positive breast cancer.
Keywords: Long non-coding RNA, Linc01638, DNMTs, HER2-positive breast cancer, tumorigenesis
1. Introduction
Breast cancer is the most common cancer and the leading cause of cancer-related death in females worldwide [1, 2]. Breast cancer is a heterogeneous disease and can be divided into five subtypes: luminal A, luminal B, HER2 (human epidermal growth factor receptor 2) over-expressing, basal-like, and unclassified [3-5]. The HER2-positive phenotype has been observed in about 15% of breast cancer patients [6]. It has been established that gene amplification and protein over expression of HER-2/neu on primary breast cancer indicate poor patient prognosis and decreased overall survival [7]. HER2-positive breast cancer is also related to unfavorable pathological parameters, such as poorly differentiation, high grade, high rates of cell proliferation and lymph-node involvement [8]. Over the past decade, thanks to the progress in the management of HER2-positive breast cancer, especially the application of HER2-targeted therapy [9], the overall survival (OS) of HER2-positive breast cancer has been prolonged [10]. Moreover, the mortality and recurrence of HER2-positive breast cancer have remarkably reduced [11, 12]. However, resistance to trastuzumab is a common and a major clinical dilemma in the treatment of HER2-positive breast cancer [13]. Relapse is noted among approximately 15% of patients after adjuvant chemotherapy combined with trastuzumab. Over 70% of patients with HER2-positive metastatic breast cancer acquire resistance to HER2-targeted therapy within 1 year [10, 14]. Also, over-expression of HER2 is associated with resistance to certain types of chemotherapy [8]. Such phenomena indicate that there might be other molecular targets that could be used as effective therapy for HER2-positive breast cancer.
Long non-coding RNAs (lncRNAs) are defined as functional non-coding RNA molecules larger than 200 bp in length [15, 16]. Numerous studies have revealed functions of lncRNAs in transcriptional, posttranscriptional and epigenetic gene regulation [17]. In addition, lncRNAs are involved in multiple human diseases, such as Alzheimer's disease [18], coronary artery disease [19] and autoimmune diseases [20]. In recent years, increasing studies focus on the link between dysregulations of lncRNAs and tumor. Aberrant expression of lncRNAs was reported to be attributed to the tumorigenesis of several types of human cancers, such as hepatocellular carcinoma [21], gastric cancer [22] and human urologic cancers [23-25]. Our previous study also indicated that lncRNAs act as a tumor activator in triple-negative breast cancer [26]. Furthermore, other studies have provided evidence that lncRNAs modulate breast cancer cell growth and apoptosis [27, 28]. However, the role of long intergenic non‐coding RNA01638 (linc01638) in breast cancer, especially in HER2-positive breast cancer, remains largely unclear.
The mammalian DNA methyltransferase (DNMTs) family consists of two main groups: maintenance methyltransferases (DNMT1) and de novo methyltransferases (DNMT3a and DNMT3b) [26, 29]. They are essential for mammalian development and play important roles in various cellular processes, including chromatin structure, X chromosome inactivation and genome stability [30]. Disruption of DNMTs may lead to chromosome instability and cancer progression [31]. For example, DNMT3a and DNMT3b may be relevant to cervical cancer progression [32]. DNMT1 was found to be involved in acquired resistance to chemotherapy in lung cancer [33]. In breast cancer, elevated expression of DNMTs might be an adverse factor, contributing to lymph node metastasis, advanced stage, ER negativity, PR negativity, and HER-2 over-expression. However, the mechanisms and pathways of DNMTs in HER2-positive breast cancer are not yet completely clear and should be further explored.
In this study, we determined the expression of linc01638 in HER2-positive breast cancer cells and tissues. Subsequently, we examined the effects of linc01638 on HER2-positive breast cancer both in vitro and in vivo, and investigated the regulation of DNMTs by linc01638. The results of this study suggest that linc01638 might be a potential biomarker and therapeutic target for HER2-positive breast cancer treatment.
2. Materials and Methods
2.1. Cell Lines and Cell Culture
Human breast cancer cell lines (MDA-MB-231, MCF-7, T47D, BT549, BT483, BT20, BT474 and SKBR3) and non-tumor mammary epithelial cell lines (184A and MCF-10A) were obtained from the American Type Culture Collection (Manassas, VA, USA) and passaged in our laboratory for less than six months after thawing frozen aliquots. All cells were maintained according to the supplier’s instructions. Before use, all cell lines were authenticated by short-tandem repeat DNA profiling and were found to be free of mycoplasma infection.
2.2. Clinical Samples
A total of 50 pairs of human HER2-positive breast cancer tissues and their adjacent normal mammary tissues were subjected to quantitative real-time PCR (qRT-PCR) analysis. Resected cancerous tissues and paired matched normal mammary tissues were immediately stored in RNAlater (Ambion).
This study was approved by the Ethics Committee of Sun Yat-Sen University Cancer Center Health Authority. The collection and use of tissues followed the procedures that were in accordance with the ethical standards formulated in the Helsinki Declaration.
2.3. RNA Isolation and qRT-PCR
Total RNA from the cells was extracted with the TRIzol reagent(Invitrogen, Carlsbad, CA, USA). Real-time PCR was performed with SYBR Green (Biorad) in the CFX96 Touch Real-Time PCR Detection System (Bio-Rad) to determine the relative expression levels of the target genes.
2.4. Flow Cytometric Analysis of Apoptosis
Annexin V/propidium iodide (PI) staining and flow cytometry were performed according to the manufacturer’s instructions using an Annexin V-fluorescein Isothiocyanate Apoptosis Detection Kit (KeyGen Biotech, Nanjing, China).
2.5. Cell Proliferation Assay
Cells were plated on 6-well plates at the desired concentrations. Cell numbers were counted after 1, 2, 3, and 4 days of incubation using a Coulter Counter (Beckman Coulter, Fullerton, USA) in triplicate.
2.6. Colony Formation Assay
6-well plates were covered with a layer of 0.6% agar in medium supplemented with 20% fetal bovine serum (FBS). The cells were prepared in 0.3% agar and seeded in triplicate. When the plates were incubated at 37 °C for 2 weeks, the colonies were counted.
2.7. Cell Invasion Assay
Cell invasion was evaluated using transwell. Cells in serum-free medium were added to the upper chamber of the transwell (8 μm pore size, BD Biosciences, New Jersey, USA), and 10% fetal bovine serum was added to the lower chamber. After 48 hours of incubation, invasive cells adhering to the lower membrane of the inserts were stained with crystal violet, counted, and imaged.
2.8. Western Blot
Protein was extracted from SK-BR3 cell using RIPA lysis buffer with a proteinase inhibitor. Proteins in the lysates were separated by 12% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinyli- denedifluoride (PVDF) membrane (Millipore, USA). To block the nonspecific binding, membranes were incubated at room temperature for 1 h with 5% skim milk powder. Subsequently, the membranes were incubated for 12 h at 4 °C with antibodies against DNMT1, DNMT3a, DNMT3b, BRCA1, and PTEN purchased from Santa Cruz Biotechnology, (Santa Cruz, CA). The target proteins were detected by chemiluminescence. β-actin was used as a protein loading control.
2.9. Mouse Xenograft Model
All the animal procedures were performed in accordance with institutional guidelines. Ethical approval was obtained
from the Institute Research Ethics Committee of Sun Yat-Sen University Cancer Center.
SK-BR3 cells that stably transfected with sh-linc01638 or sh-control were collected and suspended in PBS at a concentration of 1 × 106 cells/ml. Afterward, the cells were inoculated subcutaneously into the dorsal flanks of each nude mouse with 100 µl of cancer cell suspension (four in each group). Tumor size was measured every four days. After 28 days, the mice were sacrificed, necropsies were performed, and the tumors were weighed.
To assay the effect of linc01638 on experimental metastasis, 1 × 105of SK-BR3 cells infected with sh-linc01638 or sh-control were injected into the tail vein of the nude mouse (four in each group). Necropsies were performed after 28 days. The number of microscopic metastases in the lung of each HE-stained section from individual mice were analyzed by morphological observation.
2.10. Statistical Analysis
Comparisons between groups were analyzed with Student’s t-test, one-way ANOVA, and x2 tests. All differences were statistically significant at the P< 0.05 level. Statistical analyses were performed using SPSS16.0 software.
3. Results
3.1. Increased Expression of Linc01638 in HER2-positive Breast Cancer Cells and Tissues
We analyzed the expression of linc01638 in various mammary cell lines by qRT-PCR. Linc01638 expression was found to be increased in the HER2-positive breast cancer cells compared to non-tumor mammary epithelial cells (Fig. 1A). Furthermore, linc01638 expression was assessed in 50 pairs of HER2-positive breast cancer tissues and their matched normal tissues using qRT-PCR. Among 50 pairs samples, 40 (80%) showed overexpression of linc01638 (Fig. 1B). These results indicated thatlinc01638 might be a potential biomarker for HER2-positive breast cancer.
Fig. (1).
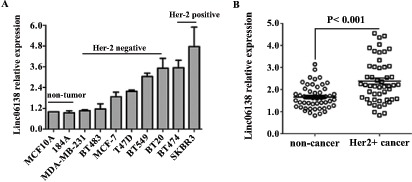
Increased expression of linc01638 in HER2-positive breast cancer cells and tissues. (A) Linc01638 mRNA expression was analyzed by qRT-PCR in two non-tumor cell lines, six HER2-negative tumor cell lines, and two HER2-positive tumor cell lines. Linc01638 expression was normalized on the expression of β-actin. Error bars represent standard deviations (SD) from three replicates in one experiment. (B) Linc01638 mRNA levels was determined by qRT-PCR in 50 pairs of HER2-positive tissues and adjacent normal tissues.
3.2. Linc01638 Knockdown Enhances Cell Apoptosis
We investigated the role of linc01638 on cell apoptosis in HER2-positive breast cancer cells using Annexin V-FITC/PI staining-based FACS analysis. SK-BR3 cells with a relatively high expression of linc01638 (Fig. 1A) were transfected with either linc01638 shRNA or control shRNA. As shown in Fig. (2A), linc01638 knockdown dramatically increased cell apoptosis in SK-BR3 cells. The apoptosis of cells in sh-linc01638-treated group considerably increased than that of the shRNA control group.
Fig. (2).
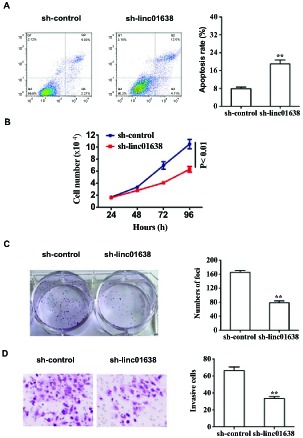
Linc01638 knockdown enhances cell apoptosis and attenuates cell growth and invasion abilities in HER2-positive breast cancer cells. SK-BR3 cells were transfected with linc01638 shRNA or nonspecific control shRNA. (A) Cell apoptosis assay. Apoptosis in SK-BR3 was evaluated by flow cytometry 48 h after shRNA transfection using an Annexin V-FITC/PI-staining (left). The apoptosis rate was shown (right). (B) Cell proliferation assay. SK-BR3 cells were seeded in 6-well plates after transfection, and cell numbers was counted daily for 4 days. (C) Colony formation assay. Representative images are presented (left). The quantification of colonies stained with crystal violet was shown (right). (D) Cell invasion assay. The invasion activity was determined by a transwell assay at 48 h post-transfection. All data represent the means ± standard deviations (SD) of three separate independent experiments (**, P<0.01).
3.3. Linc01638 Knockdown Attenuates Cell Growth and Invasion Abilities in HER2-positive Breast Cancer Cells
To investigate the function of linc01638 in HER2-positive breast cancer cell growth, we employed cell proliferation and colony formation assays. We found that compared with the control cells, the knockdown of linc01638 significantly attenuated the proliferation of SK-BR3 cells (Fig. 2B), as well as inhibited cell clonogenicity, resulting in a marked reduction in colony size (Fig. 2C). We also conducted a transwell assay to identify the effect of linc01638 on cell invasion. Our results showed that linc01638 knockdown significantly suppressed the invasiveness of SK-BR3 cells (Fig. 2D).
3.4. Linc01638 Knockdown Inhibits Tumor Progression and Metastasis in HER2-positive Breast Cancer in vivo
We studied the effect of linc01638 on HER2-positive breast cancer growth in vivo through a xenograft model in nude mice. SK-BR3 cells transfected with sh-linc01638 or nonspecific shRNA control were subcutaneously injected into the flanks of nude mice, and the xenografts were harvested 28 days later. We observed that the tumor size of mice transfected with linc01638 shRNA was smaller, and the tumor weight was significantly lighter than that of the control group (Fig. 3A). We also assessed the effect of linc01638 on tumor metastasis in vivo. To do that, we injected SK-BR3 cells transfected with sh-linc01638 or
Fig. (3).
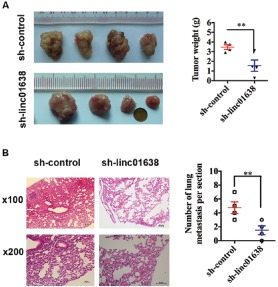
Linc01638 knockdown inhibits tumor progression and metastasis in HER2-positive breast cancer. (A) Xenografts were harvested 28 days after tumor cell injection. Pictures of the tumor xenografts were photographed (left), and the weights of the tumors were analyzed (right). (B) Experimental metastasis in mouse xenograft models. SK-BR3 cells treated with linc01638 shRNA or the control injected into the tail vein of nude mice. After 4 weeks, the mice were sacrificed, and their lung tissues were dissected and stained by hematoxylin and eosin (HE). The micrometastases in the HE-stained sections of individual mice were calculated. Each group had 4 mice. Bar=50μm. All data are shown as means ± SD. **, P<0.01.
nonspecific shRNA control into the nude mice via tail vein. After 28 days, the mice were sacrificed, and their lung tissues were dissected. As shown in (Fig. 3B), HE staining showed that a significantly lower number of macroscopic lung metastases was observed in cells infected with sh-linc01638 than those transfected with control shRNA. Taken together, these observations indicated that linc01638 could enhance tumorigenesis and metastasis in vivo.
3.5. Linc01638 Affects the Expression of DNMTs, BRCA1 and PTEN in HER2-positive Breast Cancer
To assess the effect of linc01638 on DNMTs signaling pathway and BRCA1/PTEN expression, we transfected sh-linc01638 or nonspecific shRNA control into SK-BR3 cells and conducted Western blot analysis. We observed that inhibition of linc01638 attenuated the expression of DNMT1, DNMT3a, and DNMT3b and promoted expression of BRCA1 and PTEN (Fig. 4A). We also used the mouse xenograft model and IHC to further confirm the expression of DNMT1, DNMT3a, DNMT3b, BRCA1 and PTEN expression in tumor xenografts. As shown in Fig. (4B), the expression of DNMT1, DNMT3a, and DNMT3b was down-regulated and the expression of BRCA1 and PTEN was up-regulated in SK-BR3 cells infected with sh-linc01638. These findings indicated that linc01638 can stimulated DNMTs signaling pathway and inhibited BRCA1/PTEN expression in HER2-positive breast cancer, therefore promoted tumorigenesis.
Fig. (4).
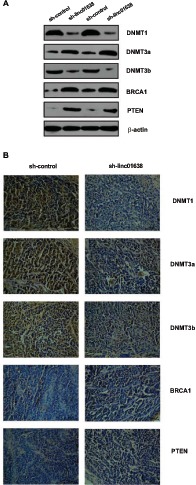
Linc01638 affects expression of DNMTs, BRCA1 and PTEN in HER2-positive breast cancer. (A) The protein expression of DNMT1, DNMT3a, DNMT3b, BRCA1, and PTEN in SK-BR3 detected by Western blot. β-actin was used as the loading control. (B) Immunohistochemistry assay of DNMT1, DNMT3a, DNMT3b, BRCA1, and PTEN expressions in tumor xenografts.
4. Discussion
It has been well established that lncRNAs participated in the pathogenesis of breast cancer, such as estrogen-induced cell proliferation [34], endocrine resistance [35] and cell apoptosis [36]. Several lncRNAs are identified as tumor-driving oncogenic lncRNAs, while some are regarded as tumor suppressive lncRNAs in breast cancer [37]. However, few studies have focused on the dysregulation of lncRNAs in the HER2-positive subtype breast cancer [38] and the role of linc01638 on such subtype of breast cancer has not yet been studied. It is critical to understand the link between linc01638 and HER2-positive breast cancer. Our study showed that linc01638 was overexpressed in HER2-positive breast cancer cells and tissues. Based on the results of our study, linc01638 may be a new molecular biomarker and might have potential therapeutic value for HER2-positive breast cancer treatment.
To better understand the function of linc01638 in HER2-positive breast cancer, we examined the effects of linc01638 on HER2-positive breast cancer growth and metastasis both in vitro and in vivo. We found that the inhibition of linc01638 expression induced cell apoptosis and attenuated the proliferation and invasiveness of the HER2-positive breast cancer cells. In our in vivo study, we found that downregulation of linc01638 inhibited tumor progression and metastasis in HER2-positive breast cancer. Consistent with a previous study [38], our results demonstrated that linc01638 is a tumor activator in HER2-positive breast cancer. It is worth being further explored to determine the mechanisms of linc01638 in breast cancer development and progression.
Non-coding RNA has been shown to be important in targeting mammalian DNMTs [39]. Moreover, abnormal DNMTs activities are relevant to strong Her-2 expression and anti-Her-2 therapy [40, 41]. Thus, we investigated the effect of linc01638 on DNMTs signaling pathway by Western blot. We found that linc01638 knockdown inhibited expression of DNMT1, DNMT3a, and DNMT3b and promoted expression of BRCA1 and PTEN. In addition, IHC confirmed the same effect of linc01638 on DNMTs signaling pathway in tumor xenografts. Collectively, these findings provided some new insights into the role of linc01638 in breast cancer, especially in association with the tumorigenesis and cancer progression in HER2-positive breast cancer. We showed that linc01638 is involved in HER2-positive breast cancer, partly related to DNMTs dsyregulation. However, further studies are warranted to elucidate the mechanisms of linc01638 on DNMTs in HER2-positive breast cancer.
Conclusion
In summary, the present study showed that linc01638 was over-expressed in HER2-positive breast cancer cells and tumor tissues. The inhibition of linc01638 by shRNA impaired HER2-positive breast cancer cell growth and invasiveness. It also triggered cell apoptosis in vitro and inhibited tumor growth in vivo by targeting DNMTs. Our data revealed that linc01638 plays a crucial role in HER2-positive breast cancer, especially in tumor growth and metastasis. Taken together, linc01638 might be a promising biomarker and therapeutic target for treatment of HER2-positive breast cancer.
Acknowledgements
This work was supported by funds from the National Natural Science Foundation of China (81372133, Xiangsheng Xiao; 81302318, Xinhua Xie).
Ethics Approval and Consent to Participate
This study was approved by the Ethics Committee of Sun Yat-Sen University Cancer Center Health Authority.
Human and Animal Rights
All humans procedures were in accordance with the standards set forth in the Declaration of Helsinki principles of 1975, as revised in 2008 (http://www.wma.net/en/20activities/10ethics/10helsinki/).
The reported experiments for animals were in accordance with the standards set forth in the 8th Edition of Guide for the Care and Use of Laboratory Animals (http://grants.nih.gov/grants/olaw/Guide-for-the-care-and-use-of-laboratory-animals.pdf) published by the National Academy of Sciences, The National Academies Press, Washington DC, United States of America?.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Figueroa-Magalhaes M.C., Jelovac D., Connolly R., Wolff A.C. Treatment of HER2-positive breast cancer. Breast. 2014;23(2):128–136. doi: 10.1016/j.breast.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prat A., Pineda E., Adamo B., Galván P., Fernández A., Gaba L., Díez M., Viladot M., Arance A., Muñoz M. Clinical implications of the intrinsic molecular subtypes of breast cancer. Breast. 2015;24(Suppl. 2):S26–S35. doi: 10.1016/j.breast.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 4.Abramson V.G., Lehmann B.D., Ballinger T.J., Pietenpol J.A. Subtyping of triple-negative breast cancer: implications for therapy. Cancer. 2015;121(1):8–16. doi: 10.1002/cncr.28914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey L.A., Perou C.M., Livasy C.A., Dressler L.G., Cowan D., Conway K., Karaca G., Troester M.A., Tse C.K., Edmiston S., Deming S.L., Geradts J., Cheang M.C., Nielsen T.O., Moorman P.G., Earp H.S., Millikan R.C. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 6.Slamon D.J., Clark G.M., Wong S.G., Levin W.J., Ullrich A., McGuire W.L. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 7.Slamon D.J., Godolphin W., Jones L.A., Holt J.A., Wong S.G., Keith D.E., Levin W.J., Stuart S.G., Udove J., Ullrich A. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 8.Burstein H.J. The distinctive nature of HER2-positive breast cancers. N. Engl. J. Med. 2005;353(16):1652–1654. doi: 10.1056/NEJMp058197. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed S., Sami A., Xiang J. HER2-directed therapy: current treatment options for HER2-positive breast cancer. Breast Cancer. 2015;22(2):101–116. doi: 10.1007/s12282-015-0587-x. [DOI] [PubMed] [Google Scholar]
- 10.Slamon D.J., Leyland-Jones B., Shak S., Fuchs H., Paton V., Bajamonde A., Fleming T., Eiermann W., Wolter J., Pegram M., Baselga J. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 11.Berry D.A., Cronin K.A., Plevritis S.K., Fryback D.G., Clarke L., Zelen M., Mandelblatt J.S., Yakovlev A.Y., Habbema J.D.F., Feuer E.J. Effect of screening and adjuvant therapy on mortality from breast cancer. N. Engl. J. Med. 2005;353(17):1784–1792. doi: 10.1056/NEJMoa050518. [DOI] [PubMed] [Google Scholar]
- 12.Perez E.A., Romond E.H., Suman V.J., Jeong J.H., Sledge G., Geyer C.E., Jr, Martino S., Rastogi P., Gralow J., Swain S.M., Winer E.P. Trastuzumab plus adjuvant chemotherapy for human epidermal growth factor receptor 2-positive breast cancer: planned joint analysis of overall survival from NSABP B-31 and NCCTG N9831. J. Clin. Oncol. 2014;32(33):3744–3752. doi: 10.1200/JCO.2014.55.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumler I., Tuxen M.K., Nielsen D.L. A systematic review of dual targeting in HER2-positive breast cancer. Cancer Treat. Rev. 2014;40(2):259–270. doi: 10.1016/j.ctrv.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Nahta R., Esteva F.J. Herceptin: mechanisms of action and resistance. Cancer Lett. 2006;232(2):123–138. doi: 10.1016/j.canlet.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 15.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Feng Y., Hu X., Zhang Y., Zhang D., Li C., Zhang L. Methods for the study of long noncoding RNA in cancer cell signaling. Methods Mol. Biol. 2014;1165:115–143. doi: 10.1007/978-1-4939-0856-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hauptman N., Glavac D. Long non-coding RNA in cancer. Int. J. Mol. Sci. 2013;14(3):4655–4669. doi: 10.3390/ijms14034655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., Laurent G.S., III, Kenny P.J., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Boerwinkle E. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sigdel K.R., Cheng A., Wang Y., Duan L., Zhang Y. The emerging functions of long noncoding RNA in immune cells: autoimmune diseases. J. Immunol. Res. 2015;2015:848790. doi: 10.1155/2015/848790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X., Xie X., Xiao Y.F., Xie R., Hu C.J., Tang B., Li B.S., Yang S.M. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360(2):119–124. doi: 10.1016/j.canlet.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 22.Shao Y., Ye M., Jiang X., Sun W., Ding X., Liu Z., Ye G., Zhang X., Xiao B., Guo J. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120(21):3320–3328. doi: 10.1002/cncr.28882. [DOI] [PubMed] [Google Scholar]
- 23.Wang L., Fu D., Qiu Y., Xing X., Xu F., Han C., Xu X., Wei Z., Zhang Z., Ge J., Cheng W. Genome-wide screening and identification of long noncoding RNAs and their interaction with protein coding RNAs in bladder urothelial cell carcinoma. Cancer Lett. 2014;349(1):77–86. doi: 10.1016/j.canlet.2014.03.033. [DOI] [PubMed] [Google Scholar]
- 24.Peter S., Borkowska E., Drayton R.M., Rakhit C.P., Noon A.P., Chen W., Catto J.W. Identification of differentially expressed long noncoding RNAs in bladder cancer. Clin. Cancer Res. 2014;20(20):5311–5321. doi: 10.1158/1078-0432.CCR-14-0706. [DOI] [PubMed] [Google Scholar]
- 25.Hirata H., Hinoda Y., Shahryari V., Deng G., Nakajima K., Tabatabai Z.L., Ishii N., Dahiya R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75(7):1322–1331. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J., Shuang Z., Zhao J., Tang H., Liu P., Zhang L., Xie X., Xiao X. Linc00152 promotes tumorigenesis by regulating DNMTs in triple-negative breast cancer. Biomed. Pharmacother. 2018;97:1275–1281. doi: 10.1016/j.biopha.2017.11.055. [DOI] [PubMed] [Google Scholar]
- 27.Tuo Y.L., Li X.M., Luo J. Long noncoding RNA UCA1 modulates breast cancer cell growth and apoptosis through decreasing tumor suppressive miR-143. Eur. Rev. Med. Pharmacol. Sci. 2015;19(18):3403–3411. [PubMed] [Google Scholar]
- 28.Sun M., Gadad S.S., Kim D.S., Kraus W.L. Discovery, annotation, and functional analysis of long noncoding RNAs controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol. Cell. 2015;59(4):698–711. doi: 10.1016/j.molcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luczak M.W., Jagodzinski P.P. The role of DNA methylation in cancer development. Folia Histochem. Cytobiol. 2006;44(3):143–154. [PubMed] [Google Scholar]
- 30.Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005;6(8):597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 31.Jin B., Robertson K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013;754:3–29. doi: 10.1007/978-1-4419-9967-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poomipark N., Flatley J.E., Hill M.H., Mangnall B., Azar E., Grabowski P., et al. Methyl Donor Status Influences DNMT Expression and Global DNA Methylation in Cervical Cancer Cells. Asian Pac. J. Cancer Prev. 2016;17(7):3213–3222. [PubMed] [Google Scholar]
- 33.Poomipark N., Flatley J.E., Hill M.H., Mangnall B., Azar E., Grabowski P., Powers H.J. Epigenetics and miRNA as predictive markers and targets for lung cancer chemotherapy. Cancer Biol. Ther. 2015;16(7):1056–1070. doi: 10.1080/15384047.2015.1046023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun H., Wang G., Peng Y., Zeng Y., Zhu Q.N., Li T.L., Cai J.Q., Zhou H.H., Zhu Y.S. H19 lncRNA mediates 17beta-estradiol-induced cell proliferation in MCF-7 breast cancer cells. Oncol. Rep. 2015;33(6):3045–3052. doi: 10.3892/or.2015.3899. [DOI] [PubMed] [Google Scholar]
- 35.Hayes E.L., Lewis-Wambi J.S. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015;17:40. doi: 10.1186/s13058-015-0542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickard M.R., Williams G.T. Regulation of apoptosis by long non-coding RNA GAS5 in breast cancer cells: implications for chemotherapy. Breast Cancer Res. Treat. 2014;145(2):359–370. doi: 10.1007/s10549-014-2974-y. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y., Sharma S., Watabe K. Roles of lncRNA in breast cancer. Front. Biosci. 2015;7:94–108. doi: 10.2741/s427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F., Lyu S., Dong S., Liu Y., Zhang X., Wang O. Expression profile analysis of long noncoding RNA in HER-2-enriched subtype breast cancer by next-generation sequencing and bioinformatics. OncoTargets Ther. 2016;9:761–772. doi: 10.2147/OTT.S97664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Denis H., Ndlovu M.N., Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Rep. 2011;12(7):647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou W., Jiang Z., Liu N., Xu F., Wen P., Liu Y., Zhong W., Song X., Chang X., Zhang X., Wei G. Down-regulation of CXCL12 mRNA expression by promoter hypermethylation and its association with metastatic progression in human breast carcinomas. J. Cancer Res. Clin. Oncol. 2009;135(1):91–102. doi: 10.1007/s00432-008-0435-x. [DOI] [PubMed] [Google Scholar]
- 41.Dou S., Yao Y.D., Yang X.Z., Sun T.M., Mao C.Q., Song E.W., Wang J. Anti-Her2 single-chain antibody mediated DNMTs-siRNA delivery for targeted breast cancer therapy. J. Control. Release. 2012;161(3):875–883. doi: 10.1016/j.jconrel.2012.05.015. [DOI] [PubMed] [Google Scholar]


