Abstract
Membrane fission manifests during cell division, synaptic transmission, vesicular transport, and organelle biogenesis, yet identifying proteins that catalyze fission remains a challenge. Using a facile and robust assay system of supported membrane tubes in a microscopic screen that directly monitors membrane tube scission, we detect robust GTP- and ATP-dependent as well as nucleotide-independent fission activity in the brain cytosol. Using previously established interacting partner proteins as bait for pulldowns, we attribute the GTP-dependent fission activity to dynamin. Biochemical fractionation followed by mass spectrometric analyses identifies the Eps15-homology domain-containing protein1 (EHD1) as a novel ATP-dependent membrane fission catalyst. Together, our approach establishes an experimental workflow for the discovery of novel membrane fission catalysts.
The lipid bilayer is highly resilient to rupture and explains why it was selected over the course of evolution to serve a barrier function, yet membrane fission or the splitting of a membrane compartment is a central theme in biology that manifests in vesicular transport and organelle biogenesis. Membrane fission has been developed within a theoretical framework wherein a local application of curvature stress leads to constriction of a tubelike membrane intermediate that progresses to scission.1−3 Because these topological transformations require the bilayer to deviate from its preferred planar configuration, fission is an energetically unfavorable process and has given rise to the notion that cells could have evolved membrane fission catalysts (MFCs) to manage this process.4
An elaborate vesicular transport pathway manages the steady-state distribution of soluble and membrane proteins across different organelles. Genetic screens carried out in the 1990s identified the sec genes that brought to light some of the molecular players in this process.5 However, despite the ubiquitous nature of membrane fission, identifying MFCs remains a challenge in contemporary cell biology. Mutagenic screens carried out in the early 1970s identified dynamin,6 which since has emerged as the paradigmatic membrane fission apparatus.7−9 Remarkably, however, mouse embryonic fibroblasts lacking dynamin survive in culture for weeks and show defects only in a small set of vesicular transport pathways.10 Indeed, the elaborate organization of the endomembrane system is largely retained in cells lacking dynamin, suggesting the presence of alternate MFCs. Recent genomewide RNAi screens based on monitoring the transport of proteins across organelles have not yielded much success in identifying alternate MFCs.11 This is because defects in any of the subprocesses of protein sorting, membrane budding, and membrane fission produce similar phenotypes of loss of transport in such screens.
Furthermore, detecting membrane fission in a reconstituted setup has been a challenge because of the lack of facile and quantitative assays. Fission, leading to the release of transport intermediates, has been studied with purified cell membranes and protein-free liposomes, but their biochemical complexity makes it difficult to define the minimum machinery or catalyst required for fission.12,13 On the other hand, partial reconstitution of transport processes with cytosol, semipurified or purified coat, and accessory proteins on protein-free liposomes has required tedious electron microscopic analysis.14,15 Reports on protein-induced vesiculation of liposomes detected using electron microscopic or light scattering-based approaches have precluded further corroborative analysis using dynamic microscopy-based techniques due to their small size.13,16,17 In this regard, the recently described supported lipid bilayers with an excess reservoir (SUPER) are of limited utility because of the planar configuration of the membrane reservoir and the bulk nature of readouts.18 Together, these results emphasize the need to devise alternate strategies for identifying MFCs.
Materials and Methods
Supported Membrane Tubes (SMrT)
SMrT templates were prepared on either PEG400- or PEG8000-functionalized glass coverslips according to the method described in refs (19) and (20). Briefly, lipid stocks (Avanti Polar Lipids) were aliquoted into glass vials in the required proportions, diluted to a final total lipid concentration of 1 mM in chloroform, and stored at −80 °C. The p-Texas Red DHPE isomer was separated from a mixed isomer stock of Texas Red DHPE (Invitrogen) using thin-layer chromatography (TLC) on silica gel plates (Sigma) as described previously,21 except that methanol was used as the solvent phase. The probe was then extracted from the TLC plate using methanol. Lipid stocks were brought to room temperature before use. A small aliquot (∼1 nmol of total lipid) was spread on a freshly cleaned PEG-functionalized coverslip and kept under high vacuum for 5 min to remove traces of chloroform. The PEG-functionalized coverslip was sandwiched between an ITO-coated glass slide using a 0.1 mm silicone spacer and then fitted in a flow cell (FCS2 system, Bioptechs). The flow cell was filled with filtered and degassed 20 mM HEPES (pH 7.4) buffer with 150 mM NaCl and left undisturbed for 10 min at room temperature. SMrT templates were prepared by extrusion of the large vesicles, formed during hydration, to narrow membrane tubes by flowing excess buffer at high (∼30 mm·s–1 particle velocity inside the chamber) flow rates. Templates were judged ready for experiments when the entire membrane reservoir was extruded into tubes that remained pinned to the surface.
Fluorescence Microscopy
Fluorescence imaging was carried out on an Olympus IX71 inverted microscope equipped with a 100×, 1.4 NA oil-immersion objective. Fluorescent probes were excited with a stable light-emitting diode light source (Thor Laboratories), and fluorescence emission was collected through filters (Semrock) with excitation and emission wavelength band passes of 562 ± 40 and 624 ± 40 nm, respectively, on an Evolve 512 EMCCD camera (Photometrics).
Brain and Bacterial Cytosol Preparation
Brain cytosol was prepared from adult goat brains according to the method described in ref (22), with some modifications. Briefly, brains were cleaned of meninges and blood vessels and homogenized in a Waring blender in breaking buffer [25 mM Tris (pH 8.0), 500 mM KCl, 250 mM sucrose, 2 mM EGTA, and 1 mM DTT] with a protease inhibitor cocktail (Roche). The lysate was spun at 160000g for 2 h, and the supernatant was passed through a G-50 desalting column in 20 mM HEPES (pH 7.4) buffer with 150 mM KCl. BL21(DE3) cells grown to stationary phase in LB were pelleted and lysed by sonication in breaking buffer. The lysate was spun at 20000g for 1 h, and the supernatant was passed through a G-50 desalting column in 20 mM HEPES (pH 7.4) buffer with 150 mM KCl. Cytosol preparations were flash-frozen with 10% glycerol in liquid N2 and stored at −80 °C. The total protein content was estimated using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific).
Biochemical Fractionation
Brain cytosol was fractionated on a Q-sepharose column (GE Lifesciences) against a 150 to 1000 mM linear KCl gradient in 20 mM HEPES (pH 7.4) buffer. Fractions were dialyzed overnight against 20 mM HEPES (pH 7.4) buffer with 150 mM KCl. Fractions precipitated in 40% ammonium sulfate were resuspended in 20 mM HEPES (pH 7.4) buffer with 150 mM KCl and resolved on a Sephacryl S-300 size exclusion column (GE Lifesciences) in the same buffer. Western blots were performed using an EHD1-specific antibody (Abcam, catalog no. ab109311). Densitometric analysis of a Western blot of increasing amounts of purified recombinant EHD1 run on the same gel as the brain cytosol served as a standard for estimating the concentration of EHD1 in brain cytosol.
Mass Spectrometry
Mass spectrometric analysis of samples was carried out at the Taplin Biological Mass Spectrometry Facility at Harvard University (Cambridge, MA). Protein bands on a Coomassie-stained sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel were excised and sent to the facility in water. Samples were in-gel digested and analyzed according to procedures listed here (https://taplin.med.harvard.edu/home).
Protein Expression and Purification
cDNAs for human dynamin1 (Q05193) and human EHD1 (Q9H4M9) were cloned with N-terminal six-His and C-terminal StrepII tags in pET15B using polymerase chain reaction. mEGFP-LactC2 was a gift from S. Grinstein (Addgene plasmid 22852) and was cloned in the same vector. All proteins were expressed in BL21(DE3) grown in autoinduction medium (Formedium, Norfolk, U.K.) for 36–48 h at 18 °C. Cell pellets were stored frozen at −40 °C. For purification, frozen cell pellets were thawed in 20 mM HEPES (pH 7.4) buffer with 300 mM KCl and sonicated in an ice–water bath. Lysates were spun at 20000g, and the supernatant was incubated with HisPur Cobalt resin (Thermo Scientific) for 1 h at 4 °C. The resin was then poured into a PD-10 column, washed with 100 mL of the same buffer, and eluted with 20 mM HEPES (pH 7.4) buffer with 300 mM KCl and 150 mM imidazole. The elution was then applied to a 5 mL Streptactin column (GE Lifesciences) and washed with 20 mM HEPES (pH 7.4) buffer with 150 mM KCl. Bound protein was eluted in 20 mM HEPES (pH 7.4) buffer with 150 mM KCl and 2.5 mM desthiobiotin (Sigma). Purified proteins were spun at 100000g to remove aggregates before use in microscopic assays. All nucleotides were obtained from Jena Bioscience.
Data Analysis
SMrT templates represent an array of membrane tubes, and a 512 × 512 pixel microscope field typically showed 30–35 tubes. Templates formed on a PEG400-functionalized glass coverslip display a homogeneous size distribution, and each tube can be considered an isolated substrate for a membrane fission catalyst. On the basis of these considerations, we quantified membrane fission as tube scission probabilities by calculating the fraction of tubes (analyzed across multiple fields) showing cut ends, which was possible because the adherent PEG400-functionalized glass coverslip ensured that remnants of severed tubes were retained on the coverslip. The reported tube scission probabilities represent pooled analyses from one or two sets of experiments.
Results and Discussion
Re-Engineered SMrT Templates for a Fission Screen
On the basis of the rationale that narrow tubelike intermediates generated at late stages of typical vesicle budding reactions would best mimic substrates for potential MFCs, we employed our recently described SMrT templates19,20 (see Figure 1A) to screen for membrane fission activity in brain cytosol. The screen is based on analysis of cytosol fractions for their ability to sever membrane tubes. The arrayed design of these templates ensures a large sampling of fission events, and fission efficiency is quantified by scoring the number of tubes that show at least one cut after incubation with cytosol or its fractions. For the purpose of visualization, templates contained trace amounts (1 mol %) of a membrane charge- and curvature-insensitive fluorescent lipid probe, p-Texas Red DHPE.21,23 Because of these properties, the use of this probe in the membrane allows calculation of tube radii by using the planar supported lipid bilayer, formed at the site where the lipid is spotted (Figure 1A), as an in situ calibration standard.20
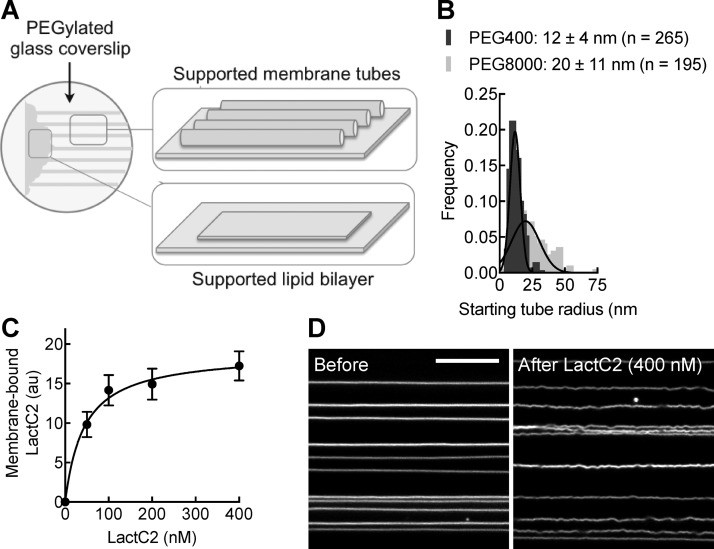
Figure 1.
SMrT screen to identify membrane fission catalysts. (A) Schematic of supported membrane tubes (SMrT) and a supported lipid bilayer (SLB). (B) Tube radius distribution on PEG400- and PEG8000-functionalized glass coverslips. Data represent means ± the standard deviation (SD) for the indicated numbers of tubes. (C) Plot showing membrane-bound mEGFP-LactC2 with increasing bulk protein concentrations. Data represent the mean ± SD for n ≥ 10 tubes analyzed for each protein concentration. (D) Micrographs of DOPC/DOPS/p-Texas Red DHPE SMrT templates (49:50:1 molar ratio) acquired in the Texas Red channel in the absence and presence of excess LactC2 domain. The scale bar is 10 μm.
Our previously described SMrT templates on PEG8000-functionalized glass coverslips formed with a lipid mix containing 15 mol % dioleoylphosphatidylserine (DOPS) in the background of dioleoylphosphatidylcholine (DOPC) produced tubes that showed a broad range of sizes (Figure 1B). Also, tubes that underwent scission were lost from the field of view because of the extremely passive surface. Together, these attributes complicate analysis of tube scission probabilities. To circumvent these problems, we tested a more adherent PEG400 surface, which required higher flow rates of buffer to extrude the membrane reservoir into tubes. The result of these modifications was a template that displayed thinner tubes, possibly in a state of higher membrane tension, and a more homogeneous size range (Figure 1B). Despite these characteristics, templates were remarkably stable to protein binding, even under conditions of extreme crowding on the membrane. This is evident from experiments carried out with the mEGFP-tagged LactC2 domain, which binds PS with high affinity,24,25 on templates formed of high (50 mol %) DOPS content. Estimating the mEGFP to p-Texas Red DHPE fluorescence ratio on tubes with increasing protein concentrations showed that binding reached saturation with an apparent Kd of 40 nM (Figure 1C), consistent with earlier estimates.25 At saturating concentrations of 400 nM, which on membranes containing 50 mol % PS should cause a high degree of protein crowding, tubes became less tense and flaccid but showed no signs of breakage or scission.
Membrane Fission Activity in Brain Cytosol
For the purpose of an initial screen, we incorporated 5 mol % phosphatidylinositol-4-phosphate (PI-4-P), a lipid enriched in intracellular membranes such as on the endocytic recycling compartment (ERC) and the Golgi apparatus,26 both of which represent organelles that release transport vesicles by as yet unidentified mechanisms, into the templates described above. We prepared a cytosol fraction from brain tissue and desalted it to remove small molecules such as nucleotides and other metabolites. Incubation of SMrT templates with 1 mg/mL desalted brain cytosol with a mixture of ATP and GTP (both at 1 mM) for 30 min at 37 °C revealed a large number of cuts (Figure 2A, white arrowheads). Templates incubated with brain cytosol alone or incubated with bacterial cytosol at similar protein concentrations with ATP and GTP showed no such effects (Figure 2A), indicating that cuts manifest from a regulated fission activity and not from mechanical instability or shearing of the tubes upon addition of a complex mix of proteins. Together, these results validate the use of SMrT templates in a screen for MFCs.
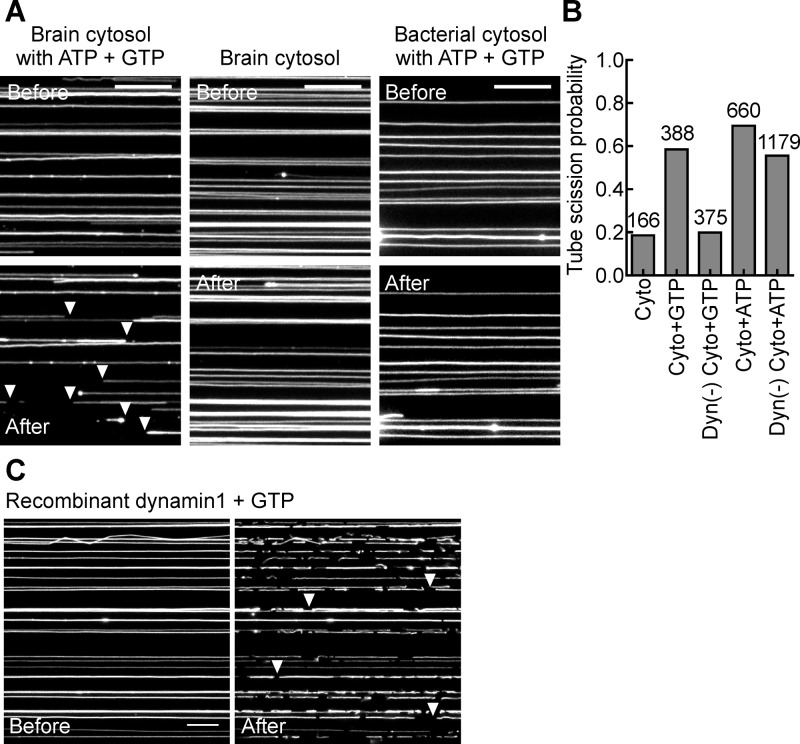
Figure 2.
Membrane fission activity in brain cytosol. (A) Effect of brain and bacterial cytosol with and without ATP and GTP on DOPC/DOPS/PI-4-P/p-Texas Red DHPE SMrT templates (79:15:5:1 molar ratio). Shown are images acquired in the Texas Red channel of the same microscope field before and after addition of cytosol. White arrowheads mark cut tubes. The scale bar is 10 μm. (B) Plots showing tube scission probability under the indicated conditions. The number of tubes sampled for each condition is indicated in the figure. (C) Micrographs of SMrT templates acquired in the Texas Red channel before and after incubation with recombinant dynamin1 (1 μM) and GTP (1 mM). White arrowheads mark cut tubes. The scale bar is 10 μm.
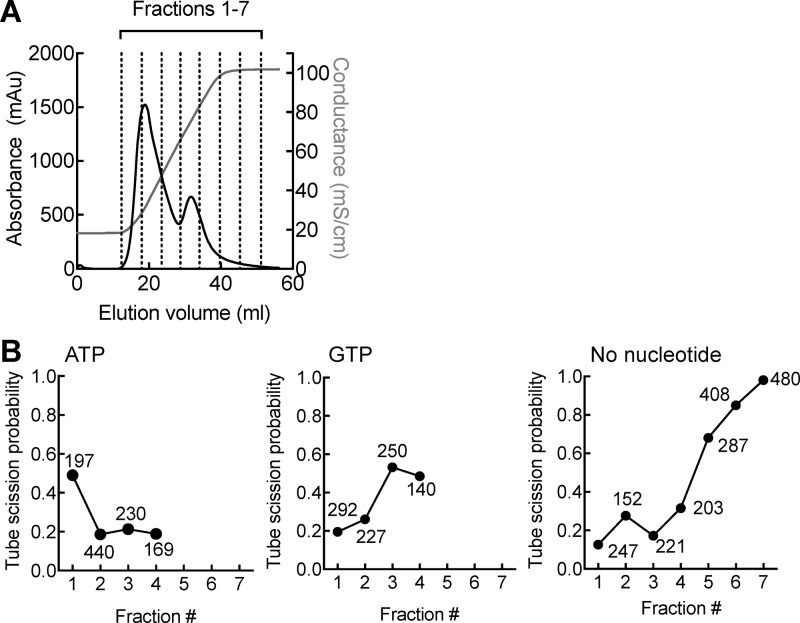
We then went on to systematically resolve the source of the fission activity in brain cytosol. Templates incubated with brain cytosol in the presence of 1 mM GTP showed a high tube fission probability (Figure 2B). We attribute this activity to dynamin, the canonical fission catalyst,7,8,27 because its depletion from cytosol using a GST-AmpIISH3 sepharose matrix as bait,28 significantly reduced fission activity to background levels (Figure 2B). Indeed, addition of recombinantly expressed and purified dynamin1 to the templates with GTP caused all of the tubes in these templates to undergo fission (Figure 2C). A previous study reported liposome vesiculation with brain cytosol in the presence of both GTP and ATP. While the GTP dependence was attributed to dynamin, the ATP dependence has remained unresolved.13 Remarkably, incubation of templates with cytosol and 1 mM ATP for 30 min at 37 °C revealed robust fission activity (Figure 2B), which remained so even with dynamin-depleted cytosol (Figure 2B). To further resolve the source of this activity, we separated proteins in brain cytosol by anion exchange chromatography into seven fractions (Figure 3A), which were then dialyzed against 20 mM HEPES (pH 7.4) buffer with 150 mM KCl and added to the templates at a total protein concentration of 0.1 mg/mL with or without nucleotides. The ATP-dependent fission activity was found enriched in fraction 1, while the GTP-dependent fission activity peaked at fraction 4 (Figure 3B). Chromatographic separation remarkably unraveled highly robust nucleotide-independent activity in fractions 5–7 (Figure 3B). Recent work using liposomes has suggested that a number of proteins can indeed display membrane destabilization or fission-like activity.17,29−33 It is possible that the nucleotide-independent fission activity could reflect functions of any one of these proteins and is currently being pursued in the lab. Importantly, because such nucleotide-independent activity is only weakly apparent in native unfractionated cytosol (Figure 2B), our results point to the possibility that such activity is kept under stringent control, perhaps by binding partners in cytosol.
Figure 3.
Resolution of fission activity in brain cytosol. (A) Plot showing results of anion exchange chromatography of brain cytosol and the fractions that were analyzed further. (B) Plots showing tube scission probability with anion exchange fractions under the indicated conditions. The number of tubes sampled for each condition is indicated in the figure.
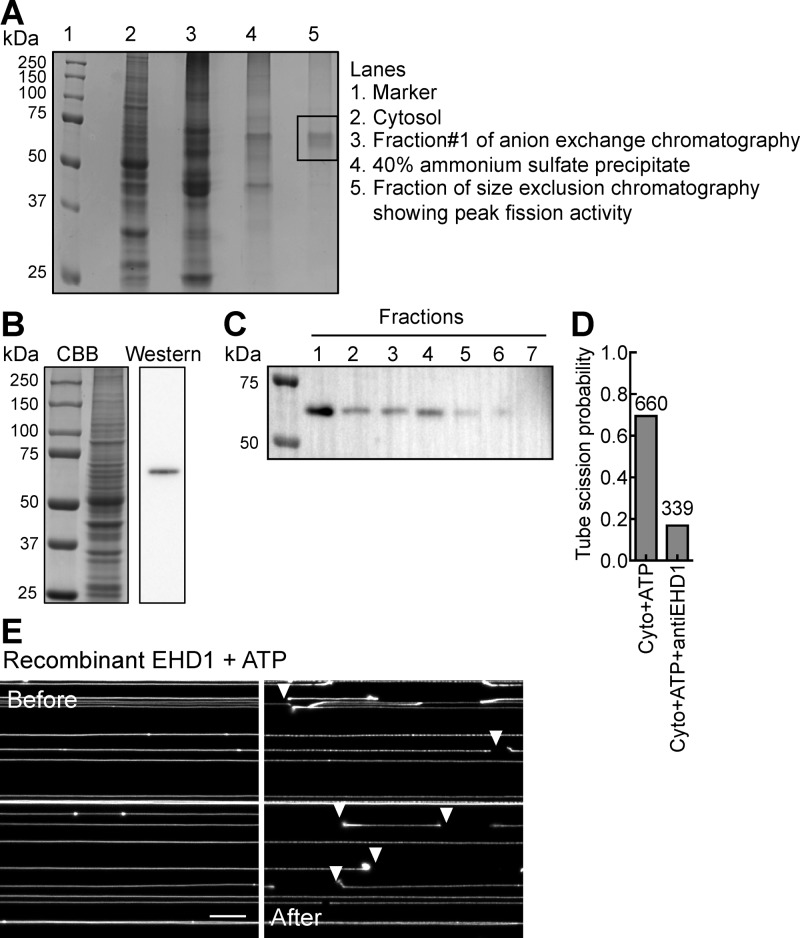
To identify the source of the ATP-dependent membrane fission activity, fraction 1 was further concentrated by a 40% ammonium sulfate precipitation and resolved using size exclusion chromatography. Further analysis showed that this activity was enriched in a fraction that yields bands at ∼60 kDa on a 10% SDS–PAGE gel (Figure 4A). Mass spectrometric analysis of this band (marked by the box) identified a total of 208 proteins (Table S1); 183 of these displayed unique peptide sequences (Table S2). Gene ontology annotation of the 183 proteins using DAVID34 revealed 25 putative ATP-binding proteins (Table S3), among which were few peripherally associated membrane proteins such at the ATPase that contains a C-terminal EH domain (EHD1). EHD proteins belong to the dynamin superfamily.35−38 Mammals have four paralogs (EHD1–4), which display ∼70% amino acid identity. Despite such a high degree of sequence similarity, EHD proteins are distributed to diverse compartments in mammalian cells. EHD1 and EHD3 are localized predominantly to the ERC; EHD2 is present at the plasma membrane, and EHD4 is localized to a Rab5-positive early endocytic compartment.35,39,40 Studies carried out in model organisms that have a single ortholog of EHD, most similar to EHD1 in mammals, have revealed functions associated with recycling of receptors.41,42 Furthermore, mice lacking a single paralog EHD1 display embryonic lethality due to defects in neural tube development arising from aberrant sonic hedgehog signaling and formation of primary cilia.43,44 Importantly, recent reports indicate a capacity for EHD1 to sever ERC tubules in an ATP-dependent manner in semipermeabilized cells45,46 and that its knockdown leads to an expansion of the previously vesicular ERC in long membrane tubules.47 Together, these observations prompted us to further investigate this class of proteins, despite the small number of peptides found in the mass spectrometric analysis (see Table S1).
Figure 4.
Identification of the ATP-dependent fission catalyst. (A) CBB-stained 10% SDS–PAGE gel of various fractions showing the band analyzed using mass spectrometry. Lane 5 in the gel shows the fraction from size exclusion chromatography showing peak fission activity. The boxed region was cut out and sent for mass spectrometric analysis. (B) CBB-stained SDS–PAGE gel (left) of brain cytosol and the corresponding Western blot (right) using an EHD1-specific antibody. (C) Western blot of anion exchange fractions using the same antibody. (D) Plot showing tube scission probability with brain cytosol with ATP in the absence and presence of the EHD-specific antibody. The number of tubes sampled for each condition is indicated in the figure. (E) Micrographs of SMrT templates acquired in the Texas Red channel before and after incubation with recombinant EHD1 (1 μM) and ATP (1 mM). White arrowheads mark cuts on the tubes. The scale bar is 10 μm.
A Western blot analysis of brain cytosol using an EHD1-specific antibody revealed a single band at ∼60 kDa (Figure 4B) that appeared enriched in fraction 1 of the anion exchange chromatography gel (Figure 4C). Our efforts at depleting EHD1 using a GST-NPF-bound sepharose matrix as bait were unsuccessful, possibly because of the low binding affinity reported for this interaction.48 Quantitative Western blot analysis using purified recombinant EHD1 as a standard revealed brain cytosol to contain 0.4 ng of EHD1/μg of total protein. Using this estimate, we added a 3-fold molar excess of the EHD1-specific antibody (assuming molecular masses of 60 kDa for EHD1 and 150 kDa for IgG) directly to brain cytosol and passed it onto templates with ATP. Remarkably, the presence of the antibody reduced the ATP-dependent fission activity to background levels (Figure 4D), similar to that seen in the absence of ATP (Figure 2B). To validate these results, we purified recombinantly expressed human EHD1 and added it to templates with ATP. These experiments revealed that EHD1 alone is sufficient to cause fission (Figure 4E). These results indicate that the ATP-dependent membrane fission activity in brain cytosol originates primarily from EHD1. The mechanism by which EHD1 manages membrane remodeling and fission will be the subject of another report.
Our study therefore has uncovered a novel fission catalyst, and we believe this is the first reported to utilize ATP for fission. The identification of a novel MFC EHD1, second only to dynamin that has a rich 30-year history, validates our approach of using SMrT templates to expand the repertoire of MFCs. As stated earlier, detecting membrane fission in vitro has been a challenge because of the lack of quantitative assays, which we overcome in this report by the use of SMrT templates. Considering that the assay is carried out in a flow cell and that the readout of the screen is binary, the combined use of microfluidics and automated image analysis routines should dramatically improve the throughput of such a screen. Importantly, the screen is easily amenable to further tuning with a change in the type of lipids displayed on the tubes as well as by the artificial recruitment of any desired adaptor protein on the membrane as described previously,49 which should facilitate discovery of membrane remodeling functions by proteins that otherwise engage the membrane with low affinity. Together, we foresee these developments to expand and systematically catalog proteins that display an intrinsic tendency to remodel and sever membranes.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.8b00925.
Descriptions of Tables S1–S3 (PDF)
Table listing proteins identified and annotated from the mass spectrometric analysis (Table S1) (XLSX)
Table listing proteins identified and annotated from the mass spectrometric analysis (Table S2) (XLSX)
Table listing proteins identified and annotated from the mass spectrometric analysis (Table S3) (XLSX)
Author Contributions
S.C.K. and T.J.P. designed the microscopic screen. S.C.K., K.R., and S.B. performed all experiments and along with T.J.P. analyzed data. T.J.P. wrote the manuscript.
S.C.K. thanks the Council of Scientific and Industrial Research (CSIR) for a graduate fellowship. K.R. and S.B. thank IISER Pune for student fellowships. T.J.P. acknowledges funding support from the Department of Biotechnology-Wellcome Trust India Alliance (Grant IA/S/16/2/502708) and the Howard Hughes Medical Institute (HHMI). T.J.P. is an International Research Scholar of the HHMI.
The authors declare no competing financial interest.
Supplementary Material
References
- Frolov V. A.; Escalada A.; Akimov S. A.; Shnyrova A. V. (2015) Geometry of membrane fission. Chem. Phys. Lipids 185, 129–140. 10.1016/j.chemphyslip.2014.07.006. [DOI] [PubMed] [Google Scholar]
- Kozlovsky Y.; Kozlov M. M. (2003) Membrane fission: model for intermediate structures. Biophys. J. 85, 85–96. 10.1016/S0006-3495(03)74457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassereau P.; Jin R.; Baumgart T.; Deserno M.; Dimova R.; Frolov V. A.; Bashkirov P. V.; Grubmüller H.; Jahn R.; Risselada H. J.; Johannes L.; Kozlov M. M.; Lipowsky R.; Pucadyil T. J.; Zeno W. F.; Stachowiak J. C.; Stamou D.; Breuer A.; Lauritsen L.; Simon C.; Sykes C.; Voth G. A.; Weikl T. R. (2018) The 2018 biomembrane curvature and remodeling roadmap. J. Phys. D: Appl. Phys. 51, 343001. 10.1088/1361-6463/aacb98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J.; Kozlov M. M. (2006) How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 7, 9–19. 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- Schekman R. (2002) SEC mutants and the secretory apparatus. Nat. Med. 8, 1055–1058. 10.1038/nm769. [DOI] [PubMed] [Google Scholar]
- Suzuki D. T.; Grigliatti T.; Williamson R. (1971) Temperature-sensitive mutations in Drosophila melanogaster. VII. A mutation (para-ts) causing reversible adult paralysis. Proc. Natl. Acad. Sci. U. S. A. 68, 890–893. 10.1073/pnas.68.5.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid S. L.; Frolov V. A. (2011) Dynamin: Functional Design of a Membrane Fission Catalyst. Annu. Rev. Cell Dev. Biol. 27, 79–105. 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- Praefcke G. J. K.; McMahon H. T. (2004) The dynamin superfamily: universal membrane tubulation and fission molecules?. Nat. Rev. Mol. Cell Biol. 5, 133–147. 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Ferguson S. M.; De Camilli P. (2012) Dynamin, a membrane-remodelling GTPase. Nat. Rev. Mol. Cell Biol. 13, 75–88. 10.1038/nrm3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson S.; Raimondi A.; Paradise S.; Shen H.; Mesaki K.; Ferguson A.; Destaing O.; Ko G.; Takasaki J.; Cremona O.; O'Toole E.; De Camilli P. (2009) Coordinated Actions of Actin and BAR Proteins Upstream of Dynamin at Endocytic Clathrin-Coated Pits. Dev. Cell 17, 811–822. 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G. D.; Swetha M. G.; Kumari S.; Lakshminarayan R.; Dey G.; Mayor S. (2009) Analysis of Endocytic Pathways in Drosophila Cells Reveals a Conserved Role for GBF1 in Internalization via GEECs. PLoS One 4, e6768. 10.1371/journal.pone.0006768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwako I.; Schmid S. L. (2005) Clathrin-Coated Vesicle Formation from Isolated Plasma Membranes. Methods Enzymol. 404, 503–511. 10.1016/S0076-6879(05)04044-9. [DOI] [PubMed] [Google Scholar]
- Kinuta M.; Yamada H.; Abe T.; Watanabe M.; Li S.-A.; Kamitani A.; Yasuda T.; Matsukawa T.; Kumon H.; Takei K. (2002) Phosphatidylinositol 4,5-bisphosphate stimulates vesicle formation from liposomes by brain cytosol. Proc. Natl. Acad. Sci. U. S. A. 99, 2842–2847. 10.1073/pnas.261715599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K.; Haucke V.; Slepnev V.; Farsad K.; Salazar M.; Chen H.; De Camilli P. (1998) Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell 94, 131–141. 10.1016/S0092-8674(00)81228-3. [DOI] [PubMed] [Google Scholar]
- Matsuoka K.; Orci L.; Amherdt M.; Bednarek S. Y.; Hamamoto S.; Schekman R.; Yeung T. (1998) COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell 93, 263–275. 10.1016/S0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Lee M. C. S.; Orci L.; Hamamoto S.; Futai E.; Ravazzola M.; Schekman R. (2005) Sar1p N-Terminal Helix Initiates Membrane Curvature and Completes the Fission of a COPII Vesicle. Cell 122, 605–617. 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Boucrot E.; Pick A.; Çamdere G.; Liska N.; Evergren E.; McMahon H. T.; Kozlov M. M. (2012) Membrane Fission Is Promoted by Insertion of Amphipathic Helices and Is Restricted by Crescent BAR Domains. Cell 149, 124–136. 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil T. J.; Schmid S. L. (2010) Supported Bilayers with Excess Membrane Reservoir: A Template for Reconstituting Membrane Budding and Fission. Biophys. J. 99, 517–525. 10.1016/j.bpj.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar S.; Kamerkar S. C.; Pucadyil T. J. (2015) A high-throughput platform for real-time analysis of membrane fission reactions reveals dynamin function. Nat. Cell Biol. 17, 1588–1596. 10.1038/ncb3254. [DOI] [PubMed] [Google Scholar]
- Dar S.; Kamerkar S. C.; Pucadyil T. J. (2017) Use of the supported membrane tube assay system for real-time analysis of membrane fission reactions. Nat. Protoc. 12, 390–400. 10.1038/nprot.2016.173. [DOI] [PubMed] [Google Scholar]
- Jung H.; Robison A. D.; Cremer P. S. (2009) Detecting Protein–Ligand Binding on Supported Bilayers by Local pH Modulation. J. Am. Chem. Soc. 131, 1006–1014. 10.1021/ja804542p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.; Huang B.; Graham M.; Raimondi A.; Heuser J. E.; Zhuang X.; De Camilli P. (2010) Coupling between clathrin-dependent endocytic budding and F-BAR-dependent tubulation in a cell-free system. Nat. Cell Biol. 12, 902–908. 10.1038/ncb2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh W.-T.; Hsu C.-J.; Capraro B. R.; Wu T.; Chen C.-M.; Yang S.; Baumgart T. (2012) Langmuir 28, 12838–12843. 10.1021/la302205b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen M. H.; Graversen H.; Fedosov S. N.; Petersen T. E.; Rasmussen J. T. (2000) Functional Analyses of Two Cellular Binding Domains of Bovine Lactadherin †. Biochemistry 39, 6200–6206. 10.1021/bi992221r. [DOI] [PubMed] [Google Scholar]
- Shao C.; Novakovic V. A.; Head J. F.; Seaton B. A.; Gilbert G. E. (2008) Crystal structure of lactadherin C2 domain at 1.7A resolution with mutational and computational analyses of its membrane-binding motif. J. Biol. Chem. 283, 7230–7241. 10.1074/jbc.M705195200. [DOI] [PubMed] [Google Scholar]
- Roth M. G. (2004) Phosphoinositides in Constitutive Membrane Traffic. Physiol. Rev. 84, 699–730. 10.1152/physrev.00033.2003. [DOI] [PubMed] [Google Scholar]
- Antonny B.; Burd C.; De Camilli P.; Chen E.; Daumke O.; Faelber K.; Ford M.; Frolov V. A.; Frost A.; Hinshaw J. E.; Kirchhausen T.; Kozlov M. M.; Lenz M.; Low H. H.; McMahon H.; Merrifield C.; Pollard T. D.; Robinson P. J.; Roux A.; Schmid S. (2016) Membrane fission by dynamin: what we know and what we need to know. EMBO J. 35, 2270–2284. 10.15252/embj.201694613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge P.; Vallis Y.; McMahon H. T. (1997) Inhibition of receptor-mediated endocytosis by the amphiphysin SH3 domain. Curr. Biol. 7, 554–560. 10.1016/S0960-9822(06)00254-5. [DOI] [PubMed] [Google Scholar]
- Snead W. T.; Hayden C. C.; Gadok A. K.; Zhao C.; Lafer E. M.; Rangamani P.; Stachowiak J. C. (2017) Membrane fission by protein crowding. Proc. Natl. Acad. Sci. U. S. A. 114, E3258–E3267. 10.1073/pnas.1616199114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczkowski J. E.; Fromme J. C. (2014) Structural Basis for Membrane Binding and Remodeling by the Exomer Secretory Vesicle Cargo Adaptor. Dev. Cell 30, 610–624. 10.1016/j.devcel.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaruttini N.; Redondo-Morata L.; Colom A.; Humbert F.; Lenz M.; Scheuring S.; Roux A. (2015) Relaxation of Loaded ESCRT-III Spiral Springs Drives Membrane Deformation. Cell 163, 866–879. 10.1016/j.cell.2015.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.; Navarro M. V.; Peng G.; Molinelli E.; Lin Goh S.; Judson B. L.; Rajashankar K. R.; Sondermann H. (2009) Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc. Natl. Acad. Sci. U. S. A. 106, 12700–12705. 10.1073/pnas.0902974106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renard H.-F.; Simunovic M.; Lemière J.; Boucrot E.; Garcia-Castillo M. D.; Arumugam S.; Chambon V.; Lamaze C.; Wunder C.; Kenworthy A. K.; Schmidt A. A.; McMahon H. T.; Sykes C.; Bassereau P.; Johannes L. (2015) Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature 517, 493–496. 10.1038/nature14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W.; Sherman B. T.; Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Daumke O.; Lundmark R.; Vallis Y.; Martens S.; Butler P. J. G.; McMahon H. T. (2007) Architectural and mechanistic insights into an EHD ATPase involved in membrane remodelling. Nature 449, 923–927. 10.1038/nature06173. [DOI] [PubMed] [Google Scholar]
- Caplan S.; Naslavsky N.; Hartnell L. M.; Lodge R.; Polishchuk R. S.; Donaldson J. G.; Bonifacino J. S. (2002) A tubular EHD1-containing compartment involved in the recycling of major histocompatibility complex class I molecules to the plasma membrane. EMBO J. 21, 2557–2567. 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B. D.; Caplan S. (2008) Mechanisms of EHD/RME-1 Protein Function in Endocytic Transport. Traffic 9, 2043–2052. 10.1111/j.1600-0854.2008.00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naslavsky N.; Caplan S. (2011) EHD proteins: key conductors of endocytic transport. Trends Cell Biol. 21, 122–131. 10.1016/j.tcb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blume J. J.; Halbach A.; Behrendt D.; Paulsson M.; Plomann M. (2007) EHD proteins are associated with tubular and vesicular compartments and interact with specific phospholipids. Exp. Cell Res. 313, 219–231. 10.1016/j.yexcr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Sharma M.; Naslavsky N.; Caplan S. (2008) A Role for EHD4 in the Regulation of Early Endosomal Transport. Traffic 9, 995–1018. 10.1111/j.1600-0854.2008.00732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.; ZHANG Y.; Paupard M. C.; Lin S. X.; Hall D. H.; Hirsh D. (2001) Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 3, 573–579. 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Olswang-Kutz Y.; Gertel Y.; Benjamin S.; Sela O.; Pekar O.; Arama E.; Steller H.; Horowitz M.; Segal D. (2009) Drosophila Past1 is involved in endocytosis and is required for germline development and survival of the adult fly. J. Cell Sci. 122, 471–480. 10.1242/jcs.038521. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S.; Rainey M. A.; Arya P.; Dutta S.; George M.; Storck M. D.; McComb R. D.; Muirhead D.; Todd G. L.; Gould K.; Datta K.; Waes J. G.-V.; Band V.; Band H. (2016) Endocytic recycling protein EHD1 regulates primary cilia morphogenesis and SHH signaling during neural tube development. Sci. Rep. 6, 20727. 10.1038/srep20727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q.; Insinna C.; Ott C.; Stauffer J.; Pintado P. A.; Rahajeng J.; Baxa U.; Walia V.; Cuenca A.; Hwang Y.-S.; Daar I. O.; Lopes S.; Lippincott-Schwartz J.; Jackson P. K.; Caplan S.; Westlake C. J. (2015) Early steps in primary cilium assembly require EHD1/EHD3-dependent ciliary vesicle formation. Nat. Cell Biol. 17, 531–531. 10.1038/ncb3155. [DOI] [PubMed] [Google Scholar]
- Cai B.; Giridharan S. S. P.; Zhang J.; Saxena S.; Bahl K.; Schmidt J. A.; Sorgen P. L.; Guo W.; Naslavsky N.; Caplan S. (2013) Differential roles of C-terminal Eps15 homology domain proteins as vesiculators and tubulators of recycling endosomes. J. Biol. Chem. 288, 30172–30180. 10.1074/jbc.M113.488627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B.; Xie S.; Caplan S.; Naslavsky N. (2014) GRAF1 forms a complex with MICAL-L1 and EHD1 to cooperate in tubular recycling endosome vesiculation. Front. Cell Dev. Biol. 2, 22. 10.3389/fcell.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.; Uchida Y.; Wang J.; Matsudaira T.; Nakagawa T.; Kishimoto T.; Mukai K.; Inaba T.; Kobayashi T.; Molday R. S.; Taguchi T.; Arai H. (2015) Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 34, 669–688. 10.15252/embj.201489703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun A.; Pinyol R.; Dahlhaus R.; Koch D.; Fonarev P.; Grant B. D.; Kessels M. M.; Qualmann B. (2005) EHD proteins associate with syndapin I and II and such interactions play a crucial role in endosomal recycling. Mol. Biol. Cell 16, 3642–3658. 10.1091/mbc.e05-01-0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil T. J.; Holkar S. S. (2016) Comparative analysis of adaptor-mediated clathrin assembly reveals general principles for adaptor clustering. Mol. Biol. Cell 27, 3156–3163. 10.1091/mbc.e16-06-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.