Abstract
A core component in the cellular response to radiation occurs at the level of translational control of gene expression. Because a critical element in translation control is the availability of the initiation factor eIF4E, which selectively enhances the cap-dependent translation of mRNAs, we investigated a regulatory role for eIF4E in cellular radiosensitivity. eIF4E silencing enhanced the radiosensitivity of tumor cell lines but not normal cells. Similarly, pharmacological inhibition of eIF4E with ribavirin also enhanced tumor cell radiosensitivity. eIF4E attenuation did not affect cell cycle phase distribution or radiation-induced apoptosis, but it delayed the dispersion of radiation-induced γH2AX foci and increased the frequency of radiation-induced mitotic catastrophe. Radiation did not affect 4E-BP1 phosphorylation or cap-complex formation but it increased eIF4E binding to >1000 unique transcripts including many implicated in DNA replication, recombination and repair. Taken together, our findings suggest that eIF4E represents a logical therapeutic target to increase tumor cell radiosensitivity.
Keywords: Radiosensitization, eIF4E, mRNA translation, molecular target, post-transcriptional regulation
Introduction
Current approaches aimed at improving the efficacy of radiation as a cancer treatment modality involve the development and application of molecularly targeted radiosensitizers, a strategy that requires a thorough understanding of the fundamental processes comprising the cellular radioresponse. Along these lines, the post-translational modification of existing proteins, which is critical to cell survival after irradiation, has provided a rich source of potential targets for radiosensitization. More recently, radiation has also been shown to selectively regulate gene translation, a process that operates independently from transcription (1, 2). In contrast to the radiation-induced transcriptome, characteristics of the radiation-induced translational control of gene expression include a significant number of commonly affected genes among cell lines initiated from the same tissue and a correlation between the genes whose translational activity is affected by radiation and the expression of their corresponding protein. Whereas translational control appears to be a component of cellular radioresponse, whether molecules participating in this process serve as determinants of radiosensitivity has not been determined.
In eukaryotic cells the majority of translation occurs in a cap-dependent manner, which involves eIF4E binding to the 7-methyl guanosine (m7G) cap on the 5’ end of an mRNA resulting in the recruitment of eIF4G and eIF4A to form the eIF4F initiation complex and subsequently ribosome binding (3). This process is a final and rate-limiting step in translation initiation and is highly dependent on the availability of eIF4E. Elevated levels of eIF4E preferentially enhance the translation of mRNAs with long, highly structured 5’ untranslated regions (UTRs), which tend to encode proteins related to cell proliferation and survival such as c-myc, Bcl2, FGF-2, and survivin (4, 5). Moreover, eIF4E also promotes the nucelocytoplasmic shuttling of select mRNAs such as cyclin D and ornithine decarboxylase (ODC) with their increased cytoplasmic levels leading to increased translation (4, 5). Thus, via at least 2 mechanisms eIF4E plays a critical role in the regulating gene translation.
At the cellular level elevated eIF4E has been implicated in oncogenesis (6). Over-expression of eIF4E has been shown to drive the malignant transformation of primary human mammary epithelial cells (7) and immortalized rodent cells (8) with ectopic expression of eIF4E in animal models increasing the incidence of a variety of tumor types (9). Evaluation of biopsy and surgical specimens indicates that eIF4E expression is frequently elevated in a number of human cancers including breast, prostate, head and neck, and lung (4, 10). Increased eIF4E levels have also been associated with malignant progression (11) as well as poor therapeutic outcome (12, 13). Finally, in preclinical models inhibition of eIF4E activity results in cytotoxicity for tumor but not normal cells (12, 14). Given eIF4E’s function in the translational control of gene expression and data suggesting that it contributes to the neoplastic phenotype, we have defined the consequences of eIF4E knockdown on the radiosensitivity of tumor and normal cell lines. The data presented here indicate loss of eIF4E activity selectively enhances tumor cell radiosensitivity through an inhibition of DNA double strand break repair. In addition, radiation is shown to significantly increase the number of mRNAs bound to eIF4E.
Materials and Methods
Cell lines and treatments:
MDA-MB-231 (breast adenocarcinoma), A549 (lung adenocarcinoma) DU145 (prostate adenocarcinoma), and MRC9 (normal lung fibroblasts) were obtained from American Type Culture Collection (ATCC) and maintained in DMEM (MDA-MB-231), RPMI (A549), or MEM (DU145 and MRC9) media supplemented with 10% FBS (Invitrogen, Carlsbad CA). A549, DU145, and MRC9 were obtained from ATCC in 2010 and MDA-MB-231 cells in 2011. ATCC employs short tandem repeat DNA fingerprinting, karyotyping, and cytochrome C oxidase to authenticate cell lines. Primary human mammary epithelial cells (HMEC) were obtained from GIBCO in 2010 and maintained in complete Mammary Epithelial Growth Medium (Lonza). All cells were cultured less than 6 months after resuscitation. Cell cultures were maintained in an atmosphere of 5% CO2/95% air at 37°C. Ribavirin (Sigma-Aldrich) was dissolved in DMSO. Cell cultures were irradiated using 320 X-ray source (Precision XRay Inc.) at a dose rate of 2.3 Gy/min.
siRNA Transfection:
A pool of four siRNA duplexes (SMARTpool) targeted to eIF4E and a non-targeted siRNA pool (scramble) were purchased from Dharmacon Inc (Lafayette, CO). Transfection with the respective siRNA pool was carried out with cell cultures at 60–70% confluency using Dharmafect 1 transfection reagent (Dharmacon) per manufacturer’s protocol. All experiments were carried out 72 h post transfection.
Clonogenic Survival Assay:
To evaluate radiosensitivity cells were plated at clonal density in six well plates and irradiated 6h later. 10 to 14d after seeding, plates were stained with 0.5% crystal violet, the number of colonies determined, and the surviving fractions were calculated. Radiation survival curves were generated after normalizing for the cytotoxicity induced by eIF4E knock down or ribavirin treatment only. Data presented are the mean ± SE from at least 3 independent experiments.
Immunoblotting and antibodies:
Cells were lysed in 50mM Tris-HCl (pH 7.5), 150mM NaCl, 2mM EDTA, 2mM EGTA, 25mM NaF, 25mM β-glycerophosphate, 0.2% Triton X-100, 0.3% NP-40, and 0.1mM sodium orthovanadate (for cytoplasmic proteins), or 50mM Tris-HCL (ph 8.0), 1% SDS, and 10mM EDTA (for nuclear proteins); supplemented with 1x phosphatase inhibitor cocktails II and III (Sigma-Aldrich), and 1x HALT protease inhibitor cocktail (Thermo Scientific) for 15 minutes on ice. Total protein was quantified using BCA protein assay (Thermo Scientific); separated by SDS-PAGE; transferred to PVDF (Millipore) and probed with the indicated antibodies. Bands were visualized using Pierce ECL Western Blotting Substrate (Thermo Scientific). Anti-eIF4E, anti-CHK1, anti-4E-BP-1, anti-phospho-eIF4E S209, anti-phospho-4E-BP-1 T37/46, and anti-phospho-4E-BP-1 S65 antibodies were purchased from Cell Signaling Technology. Anti-β-actin and anti-eIF4G antibodies were obtained from Sigma-Aldrich and BD Biosciences, respectively. Anti-Rad51 and anti-Rad17 antibodies were purchased from Santa Cruz Biotechnologies. Donkey-anti-rabbit and sheep-anti-mouse Horseradish Peroxidase conjugated secondary antibodies were purchased from GE Healthcare.
Cell Cycle Analysis:
Cell cycle phase distribution was determined by flow cytometric analysis. Cells were trypsinized, fixed with 70% ethanol, stained with Guava Cell Cycle Reagent (Millipore), and analyzed with the Guava EasyCyte flow cytometer (Millipore).
Apoptotic Cell Death:
Cells undergoing apoptosis were quantified according to annexin V staining (Annexin V Apoptosis Detection Kit, BD Biosciences). Briefly, for each treatment condition cells were resuspended in 1x Annexin V Binding Buffer and incubated with Annexin V-Cy5 antibody in the dark at room temperature. Hoechst 33258 was added for live/dead discrimination and samples analyzed by flow cytometry (BD Biosciences LSRII flow cytometer).
Immunofluorescent analysis of γH2AX foci:
To visualize foci, cells, grown in chamber slides, were fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked with 1% bovine serum albumin (BSA) in PBS containing 5% goat serum. The slides were incubated with antibody to phospho-H2AX (Millipore) followed by incubation with goat-anti-mouse-Alexa488 (Invitrogen) and mounted with Prolong gold anti-fade reagent containing 4’, 6-diamidino-2-phenylindole (Invitrogen) to visualize nuclei. Cells were analyzed on a Zeiss upright fluorescent microscope.
Mitotic Catastrophe:
Cells, grown in chamber slides, were fixed with a 10% neutral buffered formalin solution and incubated with antibody to α-tubulin (Sigma-Aldrich) followed by incubation with goat anti-mouse with Alexa-488 antibody and mounted with Prolong gold anti-fade reagent containing 4’, 6-diamidino-2-phenylindole. Cells with nuclear fragmentation, defined as the presence of two or more distinct nuclear lobes within a single cell were classified as being in mitotic catastrophe.
Cap-Binding Assay:
eIF4F cap complex formation was measured using m7-GTP batch chromatography (15). Briefly, cells were lysed in 20mM Tris-HCl (pH 7.4), 150mM NaCl, 1mM EDTA, 1mM EGTA, 1mM β-glycerophosphate, 1mM sodium orthovanadate, 1% Triton X-100, 0.2mM PMSF, 1x phosphatase inhibitor cocktails II and III (Sigma-Aldrich), and 1x HALT protease inhibitor cocktail (Thermo Scientific) for 15m on ice. 400μg of lysate were pre-cleared for 1h at 4°C then incubated with m7-GTP Sepharose 4B beads (GE Healthcare) overnight at 4°C. Beads were washed three times with lysis buffer; bound protein was eluted, denatured, and then separated using SDS-PAGE followed by immunoblotting for eIF4G and eIF4E.
RIP-Chip and Microarray Analysis:
The RIP-Chip kit and anti-eIF4E antibody were obtained from MBL International (Woburn, Ma); the procedure was performed in biological triplicate according to manufacturer’s protocol. Briefly, 107 cells were washed followed by lysis and isolation of the cytoplasmic fraction, which was then pre-cleared with Protein-A sepharose beads at 4°C for 1h. The lysates were then split into equal parts; half was incubated with eIF4E conjugated Protein-A sepharose beads and half was incubated with IgG conjugated Protein-A sepharose beads (negative control). The RNA associated with each type of bead was then eluted and isolated.
The isolated RNA was amplified using GeneChip 3’ IVT Express Kit (Affymetrix) and hybridized to GeneChip Human genome U133A 2.0 array chips (Affymetrix) per manufacturer’s protocol. Using Affymetrix Expression Console, Mas5 normalization was performed on all data sets. An expression cutoff of p ≤ 0.05 was implemented to filter all data. The negative control expression values (IgG) were subtracted from their respective sample counterparts on a probeset basis; the three replicates were then averaged. Probesets that had fold increase ≥ 1.5 (radiation to control) or went from an expression value less than or equal to 0 before radiation to positive expression value after radiation (not bound to bound) were then further analyzed by Ingenuity Pathway Analysis. The data have been deposited in NCBI’s Gene Expression Omnibus (16) and are accessible through GEO Series accession number GSEXXXX.
Results
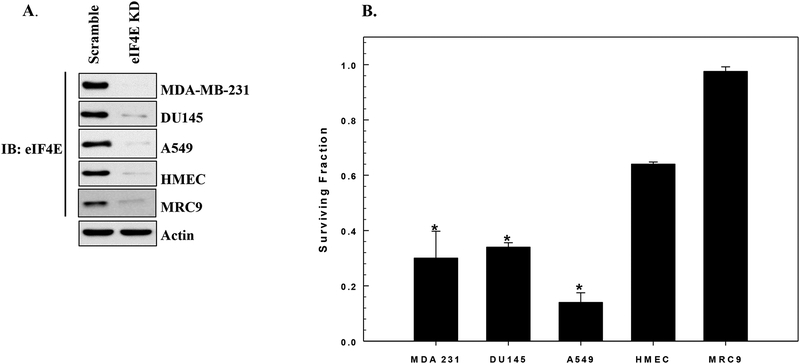
To determine whether eIF4E plays a role in determining radiosensitivity 3 tumor lines (MDA-MB-231, breast carcinoma; DU145, prostate carcinoma; A549, lung carcinoma) and 2 normal cell lines (HMEC mammary epithelial and MRC9 lung fibroblasts) were evaluated using the clonogenic survival assay. Each cell line was treated with siRNA specific to eIF4E or non-targeted siRNA; 72h after transfection cultures seeded at clonal density for survival analysis. As shown in Figure 1A, siRNA to eIF4E reduced eIF4E protein levels significantly when compared to non-targeted siRNA. The effects of eIF4E knockdown alone on the survival of each cell line are shown in Figure 1B. eIF4E knockdown significantly reduced clonogenic survival of all three tumor lines. As compared to the tumor cells, eIF4E knockdown induced significantly less cytotoxicity in the normal cell lines. These results are consistent with previous reports showing that tumor cells are more dependent on eIF4E for survival than normal cells (14, 17).
Figure 1:
Effect of eIF4E knockdown on clonogenic cell survival. Cultures were transfected with siRNA specific to eIF4E (eIF4E KD) or non-targeted siRNA (Scramble). A) Representative immunoblots from each cell line showing extent of eIF4E protein reduction 72h after transfection. B) 72h post-transfection cells were plated at specified densities and colony-forming efficiency was determined 10–14 days later. Surviving fractions for eIF4E KD cells were calculated after normalizing to the surviving fraction obtained for cells receiving the scrambled siRNA. Values shown represent the means ± SE for 3–4 independent experiments. *p < 0.04 according to Student’s t test (all tumor cell lines compared to HMEC).
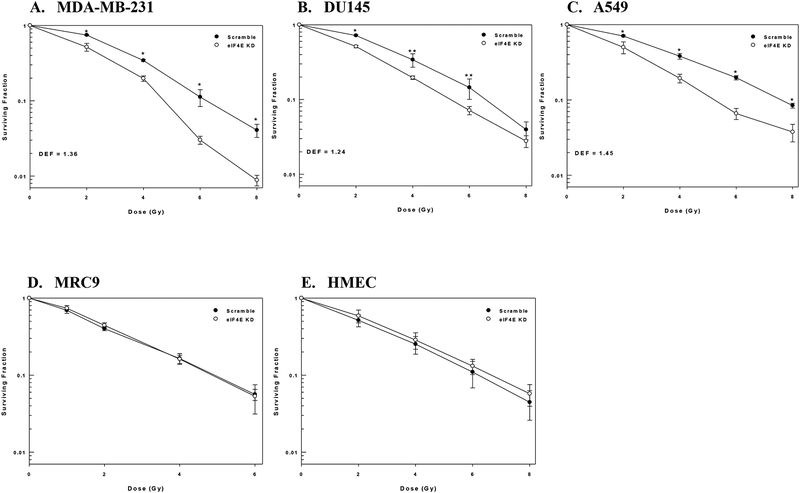
The effects of eIF4E knockdown on cellular radiosensitivity are shown in Figure 2. For this study cells were treated as described above, trypsinized and irradiated 6h after seeding. Treatment with siRNA to eIF4E resulted in an increase in the radiosensitivity of each of the 3 tumor cell lines as compared to non-targeted siRNA (Figure 2A–C). The dose enhancement factors at a surviving fraction of 0.1 (DEFs) for MDA-MB-231, DU145, and A549 were 1.34, 1.24, and 1.44, respectively. The same experiment was performed on the two normal cell lines (Figure 2D–E). In contrast to the tumor cell lines, eIF4E knockdown had no effect on the radiosensitivity of the two normal cell lines. These results suggest that eIF4E contributes to survival after irradiation of tumor but not normal cells.
Figure 2:
The effects of eIF4E knockdown on cellular radiosensitivity. A) MDA-MB-231, B) A549, C) DU145, D) MRC9, and E) HMEC cells were transfected with non-targeted siRNA (Scramble) or siRNA specific for eIF4E (eIF4E KD). 72h post-transfection cells were plated, allowed to attach for 6h, and irradiated. Colony-forming efficiency was determined 10–14 days later and survival curves were generated after normalizing for cell killing from siRNA alone. DEFs were calculated at a surviving fraction of 0.1. Values shown represent the mean ± SE for 3–4 independent experiments. * p < 0.05; ** p < 0.1 according to Student’s t test.
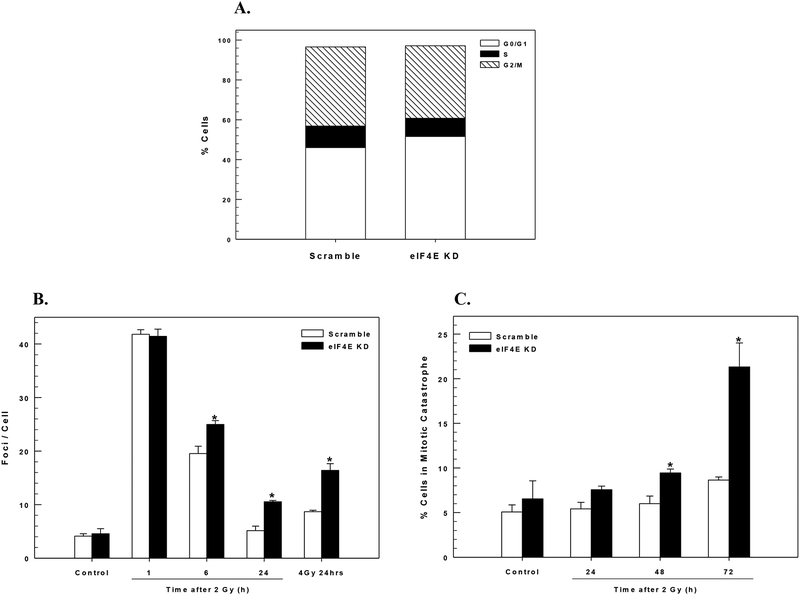
To investigate the mechanism responsible for the tumor cell radiosensitization induced by eIF4E knockdown we focused on MDA-MB-231 cells. Given that eIF4E has been reported to influence translation of a number of proteins involved in cell cycle regulation (18), a reduction in eIF4E levels could result in cell cycle phase redistribution. Because such an effect can be a critical factor in determining radiosensitivity, flow cytometry was used to determine the cell cycle distribution in MDA-MB-231 cells after eIF4E knockdown. As shown in Figure 3A the cell cycle phase distribution pattern was not significantly altered at 72h after exposure to eIF4E siRNA as compared to non-targeted siRNA. These results indicated that redistribution of cells into a radiosensitive phase of the cell cycle does not account for eIF4E knockdown-mediated enhancement in radiation-induced cell killing. eIF4E knockdown has been shown to induce apoptosis in breast cancer cell lines (19). To determine whether the increase in radiosensitivity resulting from eIF4E knockdown was due to an enhancement of radiation-induced apoptosis, we determined Annexin V staining at 24 and 48h after exposure to 6 Gy for cells exposed to siRNA to eIF4E and non-target siRNA. As expected for a solid tumor cell line, radiation alone did not induce a significant apoptotic response, and this response was not significantly enhanced with eIF4E knockdown (data not shown). These results indicate that apoptosis is not the mechanism of cell death following radiation in eIF4E deficient cells.
Figure 3:
Mechanism of radiosensitization by eIF4E knockdown. In the following experiments MDA-MB-231 cells were transfected with siRNA specific to eIF4E (eIF4E KD) or non-targeted siRNA (Scramble). All experiments were carried out 72 hours post-transfection. A) Cell cycle phase distribution was determined. Values represent the mean of three independent experiments. B) Cells were irradiated with 2 or 4Gy and collected at the specified time; γH2AX foci were counted in at least 50 cells per condition. Values shown represent the means ± SE for 3 independent experiments, *p < 0.04 according to Student’s t test (eIF4E KD compared to scramble). C) Cells were irradiated (2 Gy) and collected at the specified time points. Cells were classified as being in mitotic catastrophe by the presence of nuclear fragmentation, which was defined as a single cell containing two or more distinct nuclear lobes. At least 50 cells per condition were scored. Values represent the mean ± SE for 3 independent experiments. *p< 0.04
The critical lesion responsible for radiation-induced cell death is the DNA double strand break (DSB). Because γH2AX foci correspond to radiation-induced DSBs and their dispersal correlates with DSB repair (20, 21), the effects of eIF4E knockdown on radiation-induced γH2AX were evaluated in MDA-MB-231 cells (Figure 3B). At 1h after exposure to 2 Gy no difference in foci levels was detected between control cells (non-targeted siRNA) and cells in which eIF4E was knocked down, suggesting that eIF4E levels have no effect on the initial level of radiation-induced DSBs. However, at 6 and 24h after irradiation (2 Gy) the number of γH2AX foci remaining in the eIF4E knockdown cells was significantly greater than in control cells. Additionally, a significant level of γH2AX foci retention was observed in eIF4E deficient cells 24 h after 4 Gy when compared to non-targeted siRNA treated cells. These data suggest that eIF4E knockdown results in an inhibition of radiation-induced DNA DSB repair.
Given the apparent inhibition of DSB repair and no increase in radiation-induced apoptosis after eIF4E knockdown, we hypothesized that the mechanism of cell death involved an increase in radiation-induced mitotic catastrophe. Cells with nuclear fragmentation, defined as the presence of two or more distinct nuclear lobes within a single cell, were classified as being in mitotic catastrophe. As shown in Figure 3C, eIF4E knockdown resulted in a significant increase in the percentage of cells undergoing mitotic catastrophe at 48 and 72h after exposure to 2 Gy. These results suggest the increase in radiosensitivity following eIF4E knockdown involves the inhibition of DSB repair after radiation, which then contributes to an increase in the number of cells undergoing mitotic catastrophe.
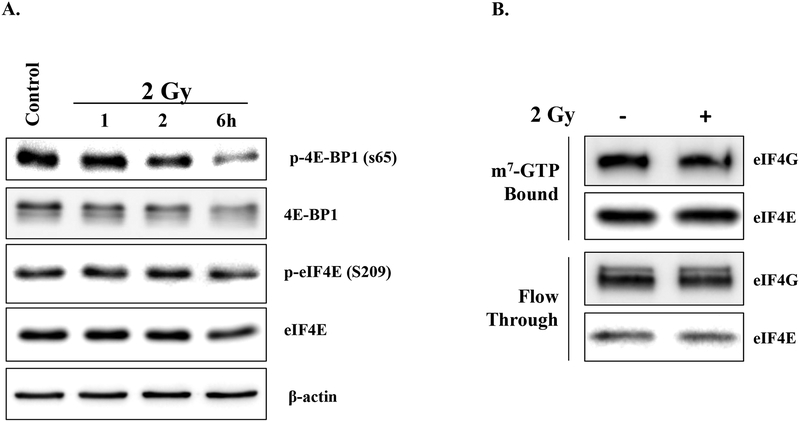
A critical regulator of eIF4E is 4E-BP1, which binds to eIF4E preventing its interaction with eIF4G and subsequently eIF4F complex formation (22). Phosphorylation of 4E-BP1 releases eIF4E resulting in eIF4F formation and cap-dependent translation (23); it has been reported that exposure of normal human cell lines to 8 Gy induces 4E-BP1 phosphorylation (24). However, exposure of MDA-MB-231 cells to 2 Gy under conditions used for clonogenic survival analysis (Figures 1–2) did not increase 4E-BP phosphorylation (Figure 4A). Post-translational activation of eIF4E via phosphorylation at S209 has also been shown to influence eIF4E activity (25); radiation had no effect on eIF4E phosphorylation (Figure 4A). m7-GTP batch chromatography is a standard approach for assessing eIF4F cap-complex formation (15, 24). Consistent with the lack of effect on 4E-BP1 and eIF4E phosphorylation, radiation had no effect on cap-complex formation, as evidenced by the lack of a change in bound eIF4G levels (Figure 4B). These results suggest that radiation does not increase the overall activity of eIF4E or cap-dependent translation initiation in general.
Figure 4:
The effect of radiation on eIF4E activation. A) MDA-MB-231 cells were irradiated (2 Gy) and collected at the specified times and subjected to immunoblot analysis. Actin was used as a loading control. B) m7-GTP affinity chromatography was performed on MDA-MB-231 cells that were irradiated and collected 1h after 2 Gy, and compared to unirradiated counterparts. m7-GTP bound and unbound proteins (flow through) were resolved via SDS-PAGE followed by immunoblot analysis. eIF4E was used as a loading control. Blots are representative of two independent experiments.
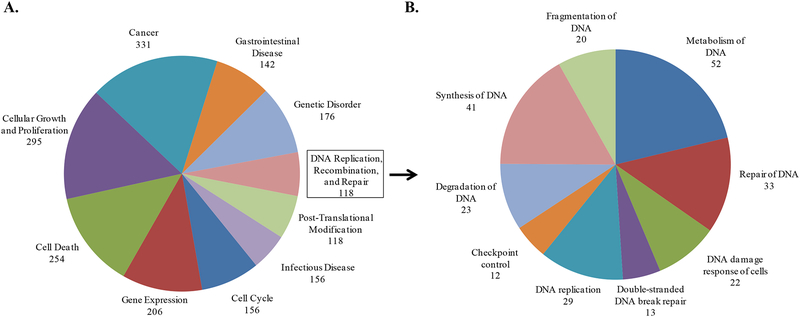
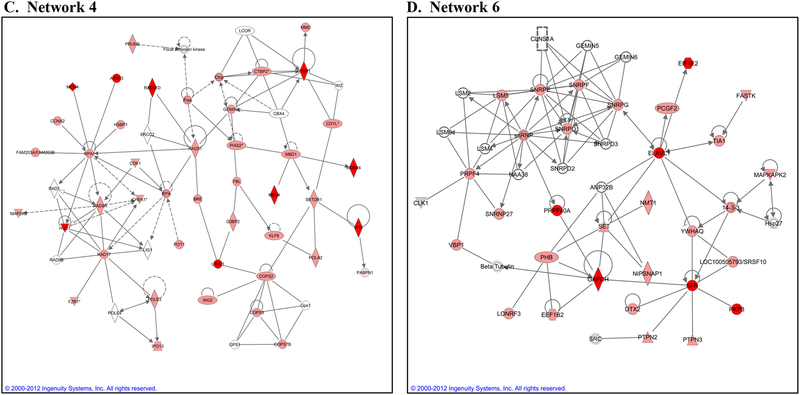
As noted in the Introduction, eIF4E selectively stimulates the translation initiation of certain mRNA subpopulations. To further investigate the role of eIF4E in cellular radioresponse, we determined whether radiation influences the mRNAs bound to eIF4E using RIP-Chip analysis (RNA-Binding protein immunoprecipitation followed by microarray analysis of the bound mRNAs). In this experiment, MDA-MB-231 cells were irradiated (2 Gy), 6h later cytoplasmic lysates were collected and eIF4E was immunoprecipitated. RNA was then eluted from the immunoprecipitated eIF4E and subjected to microarray analysis, which was compared to the same process performed on unirradiated cells. In this analysis irradiation was found to increase the eIF4E binding of 1124 unique transcripts (either fold increase ≥ 1.5 or not bound to bound as described in Materials in Methods). The full list of genes is shown in Supplemental Table S1. These transcripts were then subjected to Ingenuity Pathway Analysis (IPA), which distributes genes into networks defined by known interactions and then matches these networks with specific biologically significant pathways. The top ten biological functions associated with the eIF4E bound mRNAs are shown in Figure 5A. The specific functions of the genes contained within the DNA Replication, Recombination, and Repair category are further delineated (Figure 5B) and shown to encompass many aspects of the DNA damage response, including DSB repair and checkpoint control. To illustrate the interactions between the mRNA whose binding to eIF4E was affected by radiation, the top ten networks and their associated functions are shown in Table 1. Whereas there are numerous functions associated with these networks, of particular interest with respect to radiosensitivity is Network 4, which includes genes associated with DNA Replication, Recombination and Repair. Notably, this network contains several hub proteins: Rad17, Rad51, and CHEK1 each of which influences several other proteins. Network 6, which involves genes participating in RNA post-transcriptional processing is shown in Figure 5D; it also includes several hub proteins (e.g., ELAVL1, snRNP, and PRPF4). This network illustrates eIF4E’s capacity to modulate the post-transcriptional regulation of gene expression both directly, as an RNA Binding Protein (RBP), and indirectly through its influence on other proteins involved in post-transcriptional mRNA processing. The data presented in Figure 5 indicate that genes targeted by eIF4E after irradiation are not a random collection, but instead are functionally related mRNA subsets.
Figure 5:
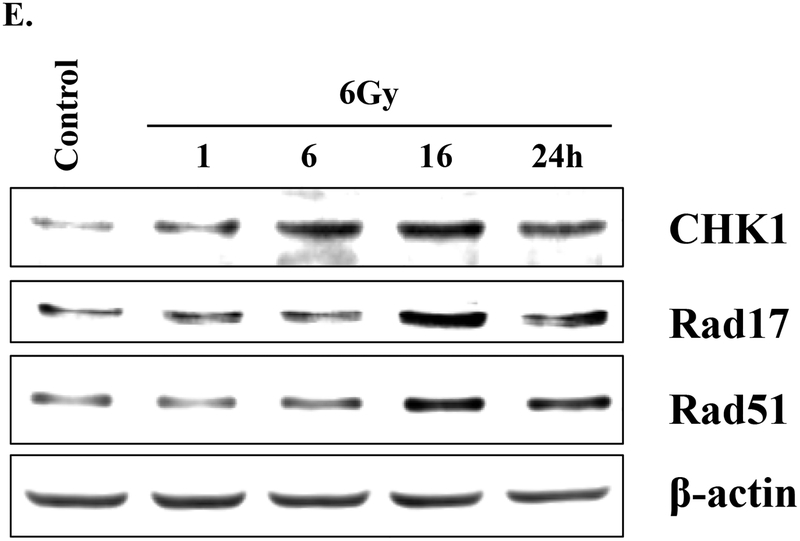
Rip Chip analysis of the effects of radiation on eIF4E mRNA clients. MDA-MB-231 cells were irradiated (2 Gy) and collected 6 hours later. eIF4E was immunoprecipitated, RNA bound to eIF4E was isolated and subjected to microarray analysis and mRNAs whose binding to eIF4E after irradiation were classified using IPA. A) The top ten biological functions (containing 100 or more genes) of the mRNAs whose binding to eIF4E was increased by radiation. B) The biological functions of the mRNAs (with greater than 10 genes) within the DNA Replication, Recombination, and Repair category are further delineated. C) Network 4 and D) Network 6 are shown with dark red indicating not bound to bound and lighter red indicating fold increase ≥ 1.5. E) Immunoblot analysis of DNA Damage response related proteins predicted by RIP-Chip analysis to be induced by radiation. MDA-MB-231 cells were radiated (6 Gy) and collected at the specified times. Actin was used as a loading control. Blots are representative of two independent experiments.
Table 1:
Functions associated with the top ten networks for genes who were increasingly bound to eIF4E after radiation (2Gy 6h) in MDA-MB-231 cells.
| ID | Score | Focus Molecules | Top Functions |
|---|---|---|---|
| 1 | 46 | 33 | Genetic Disorder, Skeletal and Muscular Disorders, Neurological Disease |
| 2 | 44 | 33 | Cancer, Cellular Movement, Connective Tissue Development and Function |
| 3 | 44 | 32 | Cancer, Infectious Disease, Respiratory Disease |
| 4 | 42 | 31 | DNA Replication, Recombination, and Repair, Cell Cycle, Gene Expression |
| 5 | 39 | 30 | Cellular Function and Maintenance, Cellular Compromise, Tissue Development |
| 6 | 39 | 30 | RNA Post-Transcriptional Modification, Dermatological Diseases and Conditions, Genetic Disorder |
| 7 | 37 | 29 | Post-Translational Modification, Cellular Movement, Cell Cycle |
| 8 | 35 | 28 | Amino Acid Metabolism, Small Molecule Biochemistry, Cellular Assembly and Organization |
| 9 | 33 | 27 | Post-Translational Modification, Protein Degradation, Protein Synthesis |
| 10 | 32 | 27 | Endocrine System Development and Function, Lipid Metabolism, Molecular Transport |
Given eIF4E’s role in cap-dependent translation, an increase in the binding of a given mRNA to eIF4E would be expected to result in an increase in its corresponding protein product. Thus, to investigate the functional significance of the RIP-Chip analysis, we determined the effects of radiation on the levels of three of the hub proteins from Network 4 (CHK1, Rad17, and Rad51), proteins with established roles in the DNA damage response (26–28). MDA-MB-231 cells were irradiated (6 Gy) and collected for protein analysis at times out to 24h. As shown in Figure 6E, the levels of CHK1, Rad17, and Rad51 were increased after irradiation, consistent with a correlation between the mRNAs whose binding to eIF4E was increased after irradiation and the increase in their corresponding protein.
Figure 6:
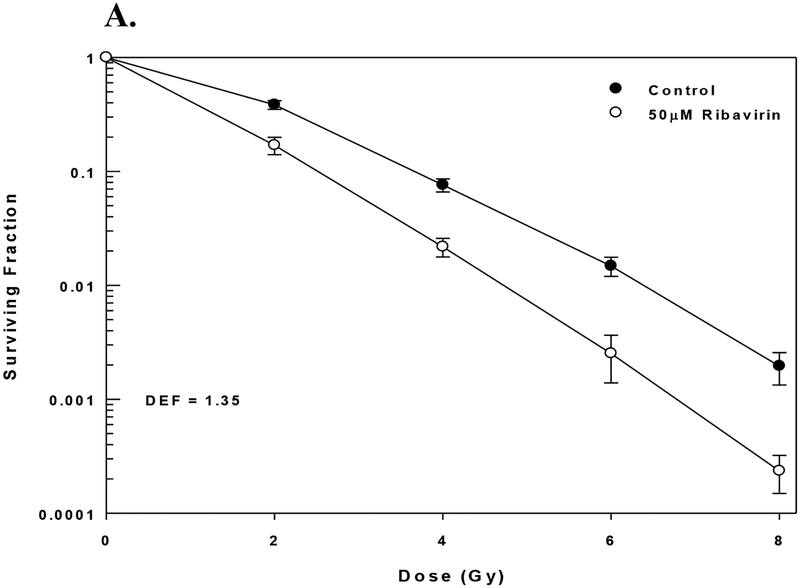
Effects of ribavirin on radiosensitivity. A) MDA-MB-231 cells were plated for clonogenic survival analysis and treated with 50 μM ribavirin for 1h, followed by radiation. Ribavirin was left on for the duration of the clonogenic assay. Values represent the mean ± SE for 3 independent experiments.
Because the data presented above suggest that eIF4E may serve as a target for radiosensitization, we determined the effects of ribavirin on the radiosensitivity of MDA-MD-231 cells. Whereas initially described as an anti-viral therapy, recent laboratory studies have shown that ribavirin inhibits eIF4E activity (17, 29) providing a basis for clinical trials as an anti-neoplastic treatment. To test whether pharmacological inhibition of eIF4E results in similar radiosensitization to eIF4E knockdown, MDA-MB-231 cells were plated for clonogenic survival analysis, treated with 50 μM ribavirin, a concentration that inhibits eIF4E activity in breast cancer cells (12), for 1h and irradiated. Ribavirin treatment alone reduced the surviving fraction to 0.30 ± 0.07, similar to that induced by eIF4E knockdown. As shown in Figure 6 this ribavirin treatment protocol enhanced the radiosensitivity of MDA-MB-231 cells with a DEF of 1.35. These results suggest that targeting eIF4E may be a valid strategy for radiosensitization.
Discussion
Based on γH2AX data, the mechanism through which eIF4E influences tumor cell radiosensitivity appears to involve DNA DSB repair. It is unlikely that this translation initiation factor directly participates in the DSB repair process suggesting that the mechanism involves an aspect of the post-transcriptional regulation of gene expression. We have previously shown that radiation affects the translation of certain subsets of mRNAs through recruitment of existing mRNAs to and away from polysomes (1, 2). The RIP-Chip results presented here showing that radiation enhances the binding of eIF4E to specific mRNA subpopulations is consistent with the radiation-induced translational control of gene expression. Moreover, a major subset of the mRNAs whose eIF4E binding was increased by radiation corresponded to those coding for proteins involved in DNA Replication, Recombination and Repair and Cell Cycle, which could then play a role in determining radiosensitivity. A role for radiation-induced gene translation in the cell survival response is suggested by the recent work by Singh et.al. showing that DNA DSBs are generated not only from the initial radiation deposition, but also from chemical processing occurring for hours after exposure to radiation (30). In this situation a requirement for the rapid increase in DNA repair proteins may contribute to the recovery process. However, based on the experiments using siRNA to knockdown eIF4E (Figure 2), it is not possible to determine whether the tumor cell radiosensitization was the result of eliminating the radiation-induced enhancement in gene translation and/or changes in mRNA translation that are induced before irradiation. Along these lines, the eIF4E inhibitor ribavirin enhanced MDA-MD-231 cells radiosensitivity when given 1h before irradiation (Figure 6). Clearly, the mechanisms through which the reduction of eIF4E levels affect radiation-induced tumor cell killing require additional investigation.
Whereas knockdown of eIF4E levels induced radiosensitization of tumor cells, the same procedure had no effect on the radiosensitivity of normal cell lines. This tumor selectivity may involve the increased dependence of tumor cells on eIF4E activity. For both ribavirin and an antisense oligonucleotide (ASO) to eIF4E, tumor cells are more sensitive in terms of cytoxicity than normal cells (12, 14). Consistent with these previous findings knockdown of eIF4E in the current study reduced survival of the tumor cell lines to a greater degree than on the normal cells. eIF4E serves as a funnel point (31) for a number of oncogenic pathways reflecting the consequences of activation of RTKs along with Ras and PI3K pathways (5, 32, 33). The elevated eIF4E availability under these circumstances then putatively enhances the translation selectively and disproportionally of genes mediating cell proliferation and survival and other processes contributing to the neoplastic phenotype (34). It would seem that many of the eIF4E dependent genes whose translation is increased in tumor cells may also contribute to the ability of the cell to survive after a variety of insults including radiation.
Whereas the mechanisms remain to be completely defined, in the study described here knockdown of eIF4E was shown to enhance the radiosensitivty of 3 human tumor lines while having no effect on the radiosensitivity of 2 normal cell lines. These data suggest that eIF4E provides a tumor selective target for radiosensitization. Because laboratory data has already indicated that eIF4E contributes to the neoplastic phenotype, strategies for targeting eIF4E are being investigated at the preclinical and clinical setting. One approach is the use of an ATP-active site inhibitor of mTOR. In contrast to allosteric mTOR inhibitors, i.e. rapalogs, the active site inhibitors completely inhibit mTORC1 function, preventing the phosphorylation of the mTOR substrate 4E-BP1, which prevents release of eIF4E and limits its availability for eIF4F formation (35). An additional approach has been the development of small molecule inhibitors of the eIF4E-eIF4G interaction, which prevent complete formation of the eIF4F cap-complex (36). Inhibiting eIF4E expression with an eIF4E ASO has been shown to reduce eIF4E levels and to inhibit tumor cell growth in preclinical models (14). Finally, there has been considerable pre-clinical data evaluating ribavirin as an eIF4E activity inhibitor (17, 29). The mTOR active site inhibitors, ribavirin, and the eIF4E ASO are currently in clinical trials both as single agents (34, 37, 38), as well as in combination with chemotherapy (39). The data presented in the current study showing that reduced eIF4E expression selectively enhances tumor radiosensitivity supports the clinical evaluation of these eIF4E-targeting strategies in combination with radiotherapy.
Supplementary Material
Footnotes
Conflict of Interest: none
References
- 1.Lu X, de la Pena L, Barker C, Camphausen K, Tofilon PJ. Radiation-induced changes in gene expression involve recruitment of existing messenger RNAs to and away from polysomes. Cancer Res. 2006;66:1052–61. [DOI] [PubMed] [Google Scholar]
- 2.Kumaraswamy S, Chinnaiyan P, Shankavaram UT, Lu X, Camphausen K, Tofilon PJ. Radiation-induced gene translation profiles reveal tumor type and cancer-specific components. Cancer Res. 2008;68:3819–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh AC, Truitt ML, Ruggero D. Oncogenic AKTivation of translation as a therapeutic target. Br J Cancer. 2011;105:329–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Culjkovic B, Borden KL. Understanding and Targeting the Eukaryotic Translation Initiation Factor eIF4E in Head and Neck Cancer. J Oncol. 2009;2009:981679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–4. [DOI] [PubMed] [Google Scholar]
- 6.Mamane Y, Petroulakis E, Rong L, Yoshida K, Ler LW, Sonenberg N. eIF4E--from translation to transformation. Oncogene. 2004;23:3172–9. [DOI] [PubMed] [Google Scholar]
- 7.Larsson O, Li S, Issaenko OA, Avdulov S, Peterson M, Smith K, et al. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res. 2007;67:6814–24. [DOI] [PubMed] [Google Scholar]
- 8.Lazaris-Karatzas A, Montine KS, Sonenberg N. Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5’ cap. Nature. 1990;345:544–7. [DOI] [PubMed] [Google Scholar]
- 9.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, et al. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–6. [DOI] [PubMed] [Google Scholar]
- 10.Graff JR, Konicek BW, Lynch RL, Dumstorf CA, Dowless MS, McNulty AM, et al. eIF4E activation is commonly elevated in advanced human prostate cancers and significantly related to reduced patient survival. Cancer Res. 2009;69:3866–73. [DOI] [PubMed] [Google Scholar]
- 11.De Benedetti A, Harris AL. eIF4E expression in tumors: its possible role in progression of malignancies. Int J Biochem Cell Biol. 1999;31:59–72. [DOI] [PubMed] [Google Scholar]
- 12.Pettersson F, Yau C, Dobocan MC, Culjkovic-Kraljacic B, Retrouvey H, Puckett R, et al. Ribavirin treatment effects on breast cancers overexpressing eIF4E, a biomarker with prognostic specificity for luminal B-type breast cancer. Clin Cancer Res. 2011;17:2874–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen CN, Hsieh FJ, Cheng YM, Lee PH, Chang KJ. Expression of eukaryotic initiation factor 4E in gastric adenocarcinoma and its association with clinical outcome. J Surg Oncol. 2004;86:22–7. [DOI] [PubMed] [Google Scholar]
- 14.Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Yue P, Deng X, Ueda T, Fukunaga R, Khuri FR, et al. Protein phosphatase 2A negatively regulates eukaryotic initiation factor 4E phosphorylation and eIF4F assembly through direct dephosphorylation of Mnk and eIF4E. Neoplasia. 2010;12:848–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kentsis A, Topisirovic I, Culjkovic B, Shao L, Borden KL. Ribavirin suppresses eIF4E-mediated oncogenic transformation by physical mimicry of the 7-methyl guanosine mRNA cap. Proc Natl Acad Sci U S A. 2004;101:18105–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J Cell Biol. 2006;175:415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong K, Wang R, Wang X, Lin F, Shen JJ, Gao P, et al. Tumor-specific RNAi targeting eIF4E suppresses tumor growth, induces apoptosis and enhances cisplatin cytotoxicity in human breast carcinoma cells. Breast Cancer Res Treat. 2009;113:443–56. [DOI] [PubMed] [Google Scholar]
- 20.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, et al. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, et al. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 2010;9:662–9. [DOI] [PubMed] [Google Scholar]
- 22.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–45. [DOI] [PubMed] [Google Scholar]
- 23.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Braunstein S, Badura ML, Xi Q, Formenti SC, Schneider RJ. Regulation of protein synthesis by ionizing radiation. Mol Cell Biol. 2009;29:5645–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010;107:14134–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan MA, Parsels LA, Zhao L, Parsels JD, Davis MA, Hassan MC, et al. Mechanism of radiosensitization by the Chk1/2 inhibitor AZD7762 involves abrogation of the G2 checkpoint and inhibition of homologous recombinational DNA repair. Cancer Res. 2010;70:4972–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Post S, Weng YC, Cimprich K, Chen LB, Xu Y, Lee EY. Phosphorylation of serines 635 and 645 of human Rad17 is cell cycle regulated and is required for G(1)/S checkpoint activation in response to DNA damage. Proc Natl Acad Sci U S A. 2001;98:13102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao H, Luoto KR, Meng AX, Bristow RG. The receptor tyrosine kinase inhibitor amuvatinib (MP470) sensitizes tumor cells to radio- and chemo-therapies in part by inhibiting homologous recombination. Radiother Oncol. 2011;101:59–65. [DOI] [PubMed] [Google Scholar]
- 29.Tan K, Culjkovic B, Amri A, Borden KL. Ribavirin targets eIF4E dependent Akt survival signaling. Biochem Biophys Res Commun. 2008;375:341–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh SK, Wang M, Staudt C, Iliakis G. Post-irradiation chemical processing of DNA damage generates double-strand breaks in cells already engaged in repair. Nucleic Acids Res. 2011;39:8416–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armengol G, Rojo F, Castellvi J, Iglesias C, Cuatrecasas M, Pons B, et al. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67:7551–5. [DOI] [PubMed] [Google Scholar]
- 32.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18:39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zindy P, Berge Y, Allal B, Filleron T, Pierredon S, Cammas A, et al. Formation of the eIF4F translation-initiation complex determines sensitivity to anticancer drugs targeting the EGFR and HER2 receptors. Cancer Res. 2011;71:4068–73. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh AC, Ruggero D. Targeting eukaryotic translation initiation factor 4E (eIF4E) in cancer. Clin Cancer Res. 2010;16:4914–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dowling RJ, Topisirovic I, Fonseca BD, Sonenberg N. Dissecting the role of mTOR: lessons from mTOR inhibitors. Biochim Biophys Acta. 2010;1804:433–9. [DOI] [PubMed] [Google Scholar]
- 36.Moerke NJ, Aktas H, Chen H, Cantel S, Reibarkh MY, Fahmy A, et al. Small-molecule inhibition of the interaction between the translation initiation factors eIF4E and eIF4G. Cell. 2007;128:257–67. [DOI] [PubMed] [Google Scholar]
- 37.Assouline S, Culjkovic B, Cocolakis E, Rousseau C, Beslu N, Amri A, et al. Molecular targeting of the oncogene eIF4E in acute myeloid leukemia (AML): a proof-of-principle clinical trial with ribavirin. Blood. 2009;114:257–60. [DOI] [PubMed] [Google Scholar]
- 38.Hong DS, Kurzrock R, Oh Y, Wheler J, Naing A, Brail L, et al. A phase 1 dose escalation, pharmacokinetic, and pharmacodynamic evaluation of eIF-4E antisense oligonucleotide LY2275796 in patients with advanced cancer. Clin Cancer Res. 2011;17:6582–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ClinicalTrials.gov [database on the Internet]. Bethesda (MD): National Library of Medicine (US); 2000. - [cited 2012 January 1]; Available from: http://clinicaltrials.gov/ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.