Abstract
Cyclotides are macrocyclic plant peptides characterized by a knotted arrangement of three disulfide bonds. They display a range of interesting bioactivities, including anti-HIV and insecticidal activities. More than 100 different cyclotides have been isolated from two phylogenetically distant plant families, the Rubiaceae and Violaceae. In this study we have characterized the cyclotides from Viola yedoensis, an important Chinese herb from the Violaceae family that has been reported to contain potential anti-HIV agents. From V. yedoensis five new and three known cyclotides were identified and shown to have anti-HIV activity. The most active of these is cycloviolacin Y5, which is one of the most potent of all cyclotides tested so far using in vitro XTT-based anti-HIV assays. Cycloviolacin Y5 is the most hydrophobic of the cyclotides from V. yedoensis. We show that there is a positive correlation between the hydrophobicity and the anti-HIV activity of the new cyclotides and that this trend tracks with their ability to disrupt membranes, as judged from hemolytic assays on human erythrocytes.
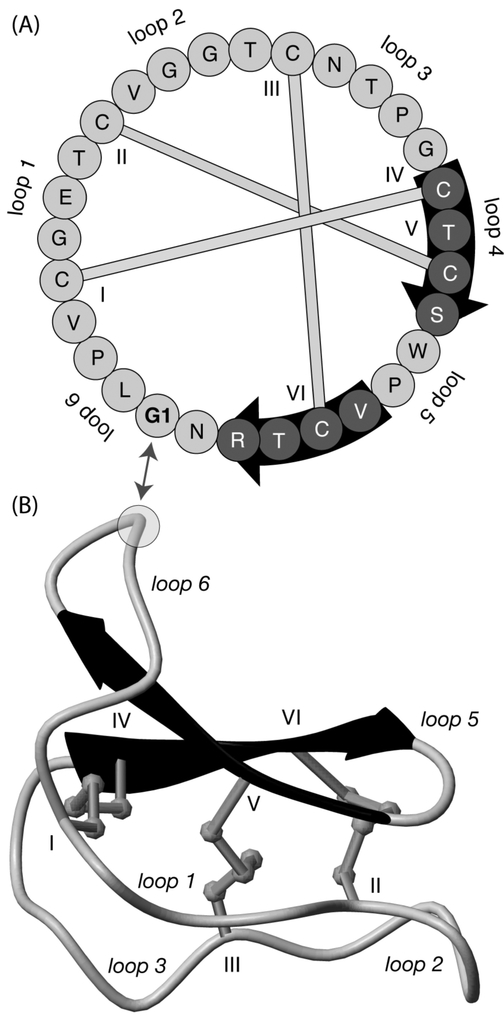
Cyclotides are a family of plant-derived peptides with unique structural properties.1,2 They are characterized by a topologically circular backbone, in which the ancestral N- and C-termini are joined seamlessly to form a continuous circle of peptide bonds. At their core is a cystine knot motif formed from six absolutely conserved Cys residues.1,3 The residues between the six Cys residues form six backbone “loops” that project from the conserved cystine knot core and are presumably responsible for their range of bioactivities. Figure 1 illustrates the structural features of kalata B1, the first characterized example of the cyclotide family, and highlights the cystine knot and backbone loops. The cyclotides have been divided into two subfamilies, Möbius and bracelet, depending on the presence or absence of a cis-Pro peptide bond in loop 5.1
Figure 1.
Sequence and structure (PDB ID: 1NB1) of the prototypic cyclotide kalata B1.47 Cyclotides have a well-defined three-dimensional structure, which is stabilized by a cystine knot and a cyclic peptide backbone. Cyclotides have six loops and six Cys residues, labeled I-VI. Panel A shows the sequence and disulfide connectivity of kalata B1. The numbering begins at the Gly residue in loop 6 and proceeds in a clockwise direction. The three-dimensional structure of kalata B1 is illustrated in panel B, with β-strands indicated with darkened arrows (also highlighted on the sequence in panel A).
Of the cyclotides that have been discovered so far, almost all have been isolated from plants of the Violaceae and Rubiaceae families.4–6 The discovery of the first cyclotide was prompted in part by reports of the uterotonic activity of an African medicinal tea made by boiling parts of the Rubiaceae plant Oldenlandia affinis. The active component of the tea was found to be a peptide, which was named kalata B1.7 The complete structural characterization of kalata B1 was not reported until some 20 years after the first reports of its bioactivity.8 Since then, a large number of cyclotides have been found in other members of the Rubiaceae family and more commonly in the Violaceae family, with cyclotides being found in every Violaceae plant that has so far been screened.4,9–11 A recent study on cyclotides from Viola hederacea proposed that there may be in excess of 9000 cyclotides in nature, suggesting that they are a large and potentially important family of plant peptides.9
Several members of the Violaceae plant family are key herbs used in traditional Chinese medicine. One such herb, Viola yedoensis (Makino),12 is a small perennial plant with violet flowers distributed in mainland China, Japan, and Korea.13 The dried whole plant (including the roots) is an important constituent of the traditional drug “Zi Hua Di Ding”, which is first boiled then ingested as a tea and used to treat toxic heat, swelling, carbuncles, sores, boils, snake bites, bronchitis, hepatitis, acute nephritis, appendicitis, and enteritis.14 Extracts from V. yedoensis have been shown to inhibit the growth of human immunodeficiency virus (HIV)12,15 and certain bacteria.16
Cyclotides have been reported to possess potent anti-HIV17,18 and weak antibacterial activity.19 Therefore, a study of cyclotides from V. yedoensis might provide insight into the active components of V. yedoensis extracts. Interestingly, cyclotides also have a range of other biological activities, including insecticidal,20,21 cytotoxic,22 neurotensin antagonistic,6 and hemolytic activities.19 The natural function of these peptides is probably defense-related, following findings that two cyclotides, kalata B1 and kalata B2, effectively inhibit the growth of larvae of the caterpillars Helicoverpa punctigera and Helicoverpa armigera, which are major cotton pests.20,21 In addition, cyclotides are remarkably stable, capable of tolerating harsh thermal, chemical, and enzymatic conditions.23 Their small size, range of biological activities, and stability make them ideal for potential agrochemical or pharmaceutical applications.24 In this paper, we characterize novel cyclotides from the Chinese violet, V. yedoensis, and assess their anti-HIV and hemolytic activity.
Results and Discussion
Traditional medicines have made a significant contribution to the western pharmacopeia, but the scientific basis for biological activity often lags behind anecdotal knowledge of their applications. To better understand the nature of the active components of the Chinese medicinal herb V. yedoensis, the suite of cyclotides produced by the plant was characterized. After the cyclotides were isolated using solvent extraction and purified using RP-HPLC, their sequences were obtained by tandem MS/MS and verified using quantitative amino acid analysis. As extracts from V. yedoensis have been previously reported to exhibit potent anti-HIV activity,12,15 anti-HIV assays were carried out on the most abundant cyclotides. The new cyclotides were also tested in hemolytic assays, which have been used in earlier studies as a marker for biological activity because cyclotides are believed to act through membrane interactions.25
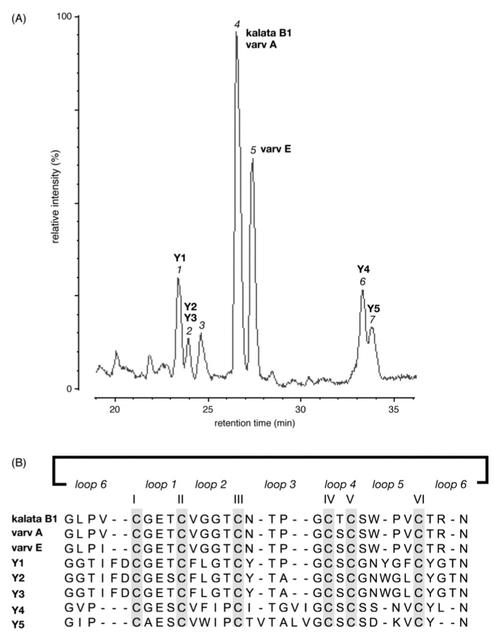
We have previously established a purification procedure for the isolation of kalata B1 from crude plant extracts, and this approach was used in the current study.20 Briefly, plant material was extracted into CH2Cl2/MeOH, purified with RP-HPLC, and characterized by MS.Similarapproacheshavebeenusedforarangeofcyclotides.6,18,26 Cyclotides are typically late-eluting on RP-HPLC and usually show a mass around 3 kDa. Masses that corresponded to potential cyclotides were isolated and sequenced using tandem MS, with this having been shown to be an effective method for characterizing cyclotide sequences.27 By way of example, cycloviolacin Y5 eluted with a retention time of 65 min. After reduction it was cleaved with trypsin and endoGluC and led to fragments with average masses of 1068 and 2072 Da. These fragments were sequenced with tandem MS and led to the sequence shown in Figure 2. The complete list of peptide fragments sequenced, including observed mass, enzymes used, and sequence coverage that allowed sequence elucidation, is supplied in the Supporting Information. From 1 kg of dried plant material, the approximate yield of kalata B1, varv A, varv E, and cycloviolacins Y1, Y2, Y3, Y4, and Y5 was 2.4 mg, 1.6 mg, 4.8 mg, 2 mg, 50 μg, 50 μg, 2.8 mg, and 1.4 mg, respectively.
Figure 2.
LC-MS profile and sequence of cyclotides characterized from V. yedoensis. Panel A shows the LC-MS profile of the crude extract run at a 1%/min gradient of solvent 2 (90% CH3CN, 0.1% HCO2H): solvent 1 (0.1% HCO2H) starting at 15% solvent 2. Each major peak is labeled sequentially according to elution time, and the characterized cyclotides are labeled in bold. The sequences of the identified cyclotides are shown in panel B. Peak 3 was difficult to purify and gave multicomponent MS/MS spectra, which were difficult to interpret, and it was not examined further. Cyclotides have three disulfide bonds, and the disulfide connectivity is likely to be conserved across all cyclotides. The disulfide connectivity of the prototypic cyclotide, kalata B1, is shown in Figure 1.
Several of the isolated cyclotides proved to be known compounds, which were established by comparison of the experimentally derived sequences with a database of cyclic proteins, CyBase, which currently contains 95 naturally occurring cyclotides.28 Using the search facilities of CyBase three of the eight sequences were identified as kalata B1, varv A, and varv E. Overall the study reinforces the fact that cyclotides seem to be ubiquitous in plants from the Violaceae family. Kalata B1 appears to be a particularly prevalent cyclotide and is present in several species from the Violaceae and Rubiaceae families.
Analysis of the new cyclotide sequences from V. yedoensis shows that there are localized regions of conservation. The most conserved residues include the six Cys residues, the Glu in loop 1, a Thr in loop 3, and an Asn in loop 6. The six conserved Cys residues are required to form the characteristic cystine knot of the cyclotides, which in combination with the cyclic backbone largely accounts for their exceptional stability.23 The Glu in loop 1 is also highly conserved across members of the cyclotide family, with only one known instance of its substitution (to an Asp) reported recently.29 This Glu residue and the Thr in loop 3 are believed to participate in a hydrogen bond network that stabilizes the cyclotide fold.30 The Asn in loop 6 has been implicated in the biosynthetic pathway of the cyclotides and is situated at the C-terminal cyclization point of cyclotide precursor proteins.11,20,31–33 Apart from these conserved residues, other regions show considerable variation, which supports the view that the cyclotide scaffold is a natural combinatorial template and well suited for the design of peptide drugs with novel bioactivies.30
Previous studies have reported that the medicinal herb V. yedoensis has anti-HIV activity,12,15 but these studies did not identify the active compound. Assays on a range of cyclotides, including circulins A-F18 and palicourein,34 have shown that cyclotides demonstrate anti-HIV activity, and so we tested the new cyclotides for this activity. Table 1 shows that all of the assayed cyclotides from V. yedoensis were active against HIV. This activity, coupled with the ability of cyclotides to resist degradation following boiling and ingestion, suggests that cyclotides are likely to be important constituents of the anti-HIV activity of V. yedoensis extracts. With the exception of cycloviolacin Y1, all of the tested cyclotides were more active than the prototypic cyclotide kalata B1.
Table 1.
Anti-HIV Activity of Cyclotides from V. yedoensis
| peptide | EC50a (μM) | IC50b (μM) |
|---|---|---|
| kalata Bl | 0.66 | 5.7 |
| varv E | 0.35 | 4.0 |
| cycloviolacin Yl | 1.2 | >4.5 |
| cycloviolacin Y4 | 0.12 | 1.7 |
| cycloviolacin Y5 | 0.04 | 1.8 |
Values for HIV-infected cell cultures.
Values for uninfected cell cultures.
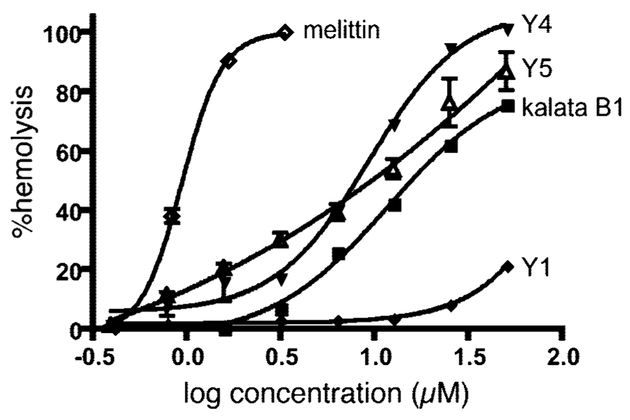
A number of cyclotides have been reported to have hemolytic activity, and they are believed to interact with membranes in vivo.20 To compare the membrane-disrupting activity of the cyclotides from V. yedoensis with previously reported cyclotides, hemolytic assays were performed, and the results are shown in Figure 3. The well-known hemolytic peptide melittin was used as a positive control and for comparison of the absolute levels of hemolysis under the conditions studied.35 Cycloviolacins Y4 and Y5 are the most hemolytic of the new cyclotides, both being more potent than the prototypic cyclotide kalata B1. By contrast, cycloviolacin Y1 is substantially less active than all of the other cyclotides tested.
Figure 3.
Hemolytic activity of cyclotides from V. yedoensis. Hemolytic activities of kalata B1, cycloviolacin Y1, cycloviolacin Y4, cycloviolacin Y5, and melittin were measured for human erythrocytes. The HD50 values (and 95% confidence intervals) were calculated using Prism software to be 11.7 μM (10.2–13.4 μM) for kalata B1, 9.3 μM (7.7–11.1 μM) for cycloviolacin Y4, 8.7 μM (7.4–10.2 μM) for cycloviolacin Y5, and 0.94 μM (0.90–0.97 μM) for melittin. Melittin is a well-known hemolytic agent that was used as a positive control and gave an activity consistent with literature values.
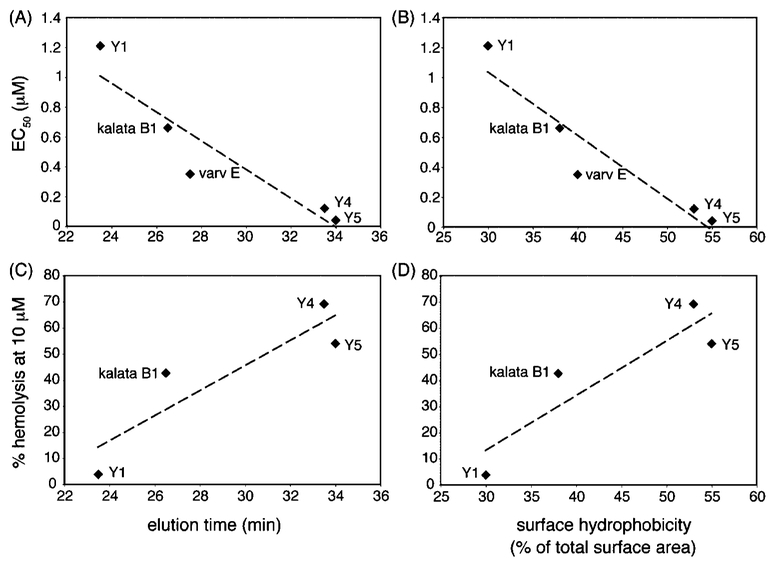
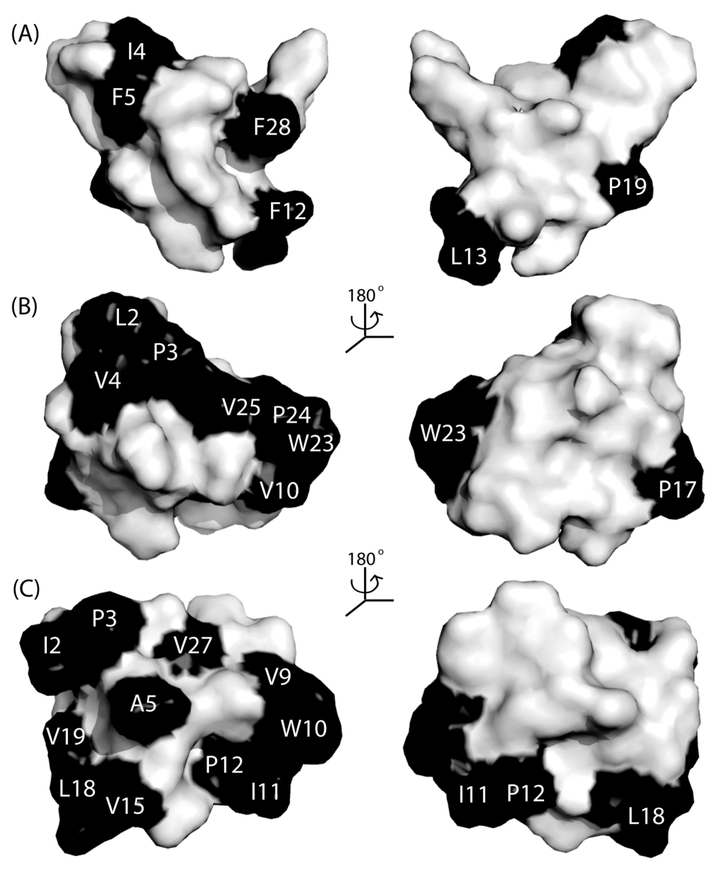
As shown in Figure 4, there appears to be a correlation between cyclotide bioactivity and hydrophobicity. In the anti-HIV assays, cycloviolacin Y1, with the earliest elution time, showed the weakest activity, and cycloviolacin Y5, which eluted last by RP-HPLC, exhibited the most potent anti-HIV activity. In fact, cycloviolacin Y5 is one of the most potent of the reported anti-HIV-active cyclotides, with an EC50 (40 nM) value similar to that of the cyclotides circulin A and B.36 The anti-HIV activity also correlates well with the predicted surface hydrophobicity of the cyclotides. The structures of cycloviolacins Y1 and Y5 were modeled against the solution structures of tricyclon A and cycloviolacin O1, respectively. The similarity between the predicted structures and their templates was verified using HR NMR chemical shift comparisons, which is provided in the Supporting Information. Figure 5 shows a comparison of surface representations of cycloviolacin Y1, kalata B1, and cycloviolacin Y5. Cycloviolacin Y5, the most anti-HIV-active cyclotide, has the largest proportion of hydrophobic residues on its surface.
Figure 4.
Effect of hydrophobicity on cyclotide bioactivity. The anti-HIV activities (panels A and B) and hemolytic activities (panels B and C) of kalata B1, varv E, cycloviolacin Y1, cycloviolacin Y4, and cycloviolacin Y5 were correlated with hydrophobicity, which was measured experimentally using RP-HPLC (retention times from Figure 2) and theoretically using surface area calculations. The surface hydrophobicity was calculated by modeling the cyclotides using MODELLER.40
Figure 5.
Surface representations of selected cyclotides from V. yedoensis. Surface representations are shown for cycloviolacin Y1 (A), kalata B1 (B), and cycloviolacin Y5 (C). Hydrophobic residues (Ala, Leu, Ile, Val, Trp, Phe, Pro) are shaded black.
Interestingly, the correlation between anti-HIV activity and simple content of hydrophobic amino acids for the new cyclotides was poor. Specifically, the hydrophobic content for a particular cyclotide was calculated using eq 1 (Experimental Section), which is based on the number of each amino acid type in the sequence and the hydrophobic index of the amino acid as measured by a hydrophobicity scale, such as the Kyte-Doolittle scale.37 The poor correlation with simple hydrophobic content, but excellent correlation with hydrophobic surface area, suggests that the presentation of the amino acids on the cyclotide framework is more important than the simple amino acid composition.
A similar correlation between hydrophobicity and bioactivity can be seen from the hemolytic assays. Cycloviolacin Y1, which is the least hydrophobic cyclotide assayed, gave the lowest hemolytic activity, and cycloviolacin Y5, which is the most hydrophobic, showed the highest hemolytic activity. This relationship between structure and hemolytic activity was reported in a previous study38 that examined the surface properties of several cyclotides, including cycloviolacin H4, which possesses the most potent hemolytic activity of the cyclotides published to date, and tricyclon A,39 with virtually no hemolytic activity.
In summary, we have shown that the Chinese medicinal herb V. yedoensis contains a suite of new and known cyclotides. The purified cyclotides exhibit anti-HIV activity and are likely to be responsible for the reported anti-HIV activity of V. yedoensis. Furthermore, the correlation between hemolytic/anti-HIV activity and hydrophobic surface area seen in this study reinforces the view that the mode of action of cyclotides involves interactions with membranes.
Experimental Section
General Experimental Procedures.
HPLC was carried out on either a Waters 600 controller system equipped with a Waters 484 tunable absorbance detector or an Agilent 1100 series system with a UV detector at variable wavelengths of 215, 254, and 280 nm. Masses were analyzed on a Micromass LCT mass spectrometer equipped with an electrospray ionization source. For MALDI-TOF MS analysis, the instrument used was a Voyager DE-STR mass spectrometer (Applied Biosystems); 200 shots per spectra were acquired in positive ion reflector mode. The laser intensity was set to 2300, the accelerating voltage to 20 000 V, and the grid voltage to 64% of the accelerating voltage; the delay time was 165 ns. The low mass gate was set to 500 Da. Data were collected between 500 and 5000 Da. Calibration was undertaken using a peptide mixture obtained from Sigma Aldrich (MSCal1). Nanospray MS/MS analysis was conducted on a QStar mass spectrometer, a capillary voltage of 900 V was applied, and the spectra were acquired at m/z 400–2000 for TOF spectra and m/z 60–2000 for product ion spectra. The collision energy for peptide fragmentation was varied between 10 and 50 V, depending on the size and charge of the ion. The Analyst software program was used for data acquisition and processing. NMR sample analysis was done on a Bruker ARX 600 spectrometer, and data were processed using TOPSPIN (Bruker).
Plant Material.
Dried plant material of V. yedoensis was purchased from a local Chinese herbalist in Brisbane, Australia, in May 2005. A voucher sample is held at the Institute for Molecular Bioscience, University of Queensland (room 2.025, curated by D. Craik).
Isolation and Purification of Cyclotides from V. yedoensis.
Following a previously established procedure,20 dried plant material was ground in a kitchen blender and left overnight in 1:1 CH2Cl2/MeOH. After plant debris was removed using a cotton plug filter, and distilled water was added to promote the separation of the aqueous partition that was collected with the residual methanol evaporated in vacuo.
After an initial purification using a C18 flash column, several steps of reversed-phase high-performance liquid chromatography (RP-HPLC) were employed to purify the cyclotides present. Preparative RP-HPLC was performed on a Waters 600 controller system equipped with a Waters 484 tunable absorbance detector. The crude sample was loaded onto a Phenomenex Jupiter C18 column (250 × 22 mm, 5 μm, 300 Å) and eluted at a flow rate of 8 mL/min with 1% buffer B (90% HPLC grade CH3CN in H2O/0.09% CF3COOH) per min gradient. Semi-preparative RP-HPLC was performed on an Agilent 1100 series system with variable-wavelength detector and a Phenomenex Jupiter C18 column (250 × 10 mm, 5 μm, 300 Å). Analytical RP-HPLC was performed using a Phenomenex Jupiter C18 column (250 × 4.6 mm, 5 μm, 300 Å). Masses were analyzed on a Micromass LCT mass spectrometer equipped with an electrospray ionization source.
LC-MS Analysis.
A sample (100 μL) of the product from the C18 flash column purification was used in the LC-MS, which was performed on an Agilent 1100 Series HPLC system with Phenomenex Jupiter column (C18, 150 × 2 mm, 5 μm) run at 200 μL/min. Peptides were eluted with a 1%/min gradient (of 90% CH3CN/0.1% HCO2H) from 15% to 75%, which was coupled directly to an Applied Biosystems QStar Pulsar mass spectrometer.
Reduction of Peptides and MALDI-MS Analysis.
To ca. 6 nmol of peptide in 20 μL of 0.1 M NH4HCO3 (pH 8.0) was added 1 μL of 0.1 M tris(2-carboxyethyl)phosphine, and the solution was incubated at 65 °C for 10 min. The reduction was confirmed by MALDI-TOF MS after desalting using Ziptips (Millipore), which involved several washing steps followed by elution in 10 μL of 80% CH3CN (0.5% HCO2H). The desalted samples were mixed in a 1:1 ratio with matrix consisting of a saturated solution of α-cyano-4-hydroxycinnamic acid (CHCA) in 50% CH3CN (0.5% HCO2H).
Enzymatic Digestion and Nanospray MS/MS Sequencing.
To the reduced peptide, trypsin, chymotrypsin, or a combination of endoGluC and trypsin was added, to give a final peptide-to-enzyme ratio of 50:1. The trypsin incubation was allowed to proceed for 1 h, the chymotrypsin digestion was allowed to proceed for 3 h, while for the combined digestion trypsin was added initially for 1 h followed by the addition of endoGluC and a further 3 h incubation. The digestions were quenched by the addition of an equal volume of 0.5% HCO2H and desalted using Ziptips (Millipore). Samples were stored at 4 °C prior to analysis. The fragments resulting from the digestion were examined first by MALDI-TOF MS followed by nanospray MS/MS analysis on a QStar mass spectrometer. The MS/MS were examined and sequenced on the basis of the presence of both b- and y-series of ions present (N- and C-terminal fragments).
Anti-HIV Assays.
Using a previously described in vitro XTT-based anti-HIV assay,34 the most abundant cyclotides purified from V. yedoensis were tested to examine their effect on virus-induced killing in HIV-infected cultures.
Homology Modeling.
Structures of cycloviolacin Y1, varv E, cycloviolacin Y4, and cycloviolacin Y5 were predicted using Modeler 8v1,40 a restraint-based structure generation program. Altogether 100 models of each cyclotide were generated, and the lowest energy structure, according to Modeler 8v1, was selected and evaluated. Cycloviolacin Y1 was modeled on tricyclon A (PDB ID: 1YP8), varv E was modeled on kalata B1 (PDB ID: 1NB1), and cycloviolacin Y4 and Y5 were modeled on cycloviolacin O1 (PDB ID: 1NBJ).
NMR Sample Analysis.
Samples of cycloviolacin Y1 and Y5 were dissolved in 90% H2O/10% D2O to a concentration of 1 mM at pH 3. An additional 100 μL of CD3CN was added to the sample of cycloviolacin Y1 to increase solubility. Spectra were recorded on a Bruker ARX 600 spectrometer at a sample temperature of 298 K. For resonance assignment a set of two-dimensional TOCSY, with a mixing period of 80 ms, and NOESY, with mixing times of 200 ms, were recorded. All two-dimensional spectra were collected over 4096 data points in the f2 dimension and 512 increments in the f1 dimension and processed using TOPSPIN (Bruker). Chemical shifts were referenced to water at4.75 ppm.
Hydrophobic Content Estimation.
Several hydrophobicity scales were used to predict the hydrophobic content of a given cyclotide. These scales were the Kyte-Doolittle scale,37 the Engelman scale,41 the Eisenberg scale,42 the Hopp-Woods scale,43 the Cornette scale,44 the Rose scale,45 and the Janin scale.46 For each scale, a value v(x) is given for each amino acid x. Therefore, for a protein sequence, where n(x) gives the number of amino acid x in the sequence, the hydrophobic content, H, is given by
| (1) |
Hemolytic Assays.
Peptides were dissolved in water and serially diluted in phosphate-buffered saline (PBS) to give 20 mL test solutions in a 96-well U-bottomed microtiter plate (Nunc). Human type A red blood cells (RBCs) were washed with PBS and centrifuged at 4000 rpm for 60 s in a microcentrifuge several times until a clear supernatant was obtained. A 0.25% suspension of washed RBCs in PBS was added (100 μL) to the peptide solutions. The plate was incubated at room temperature for 1 h and centrifuged at 150 g for 5 min. Aliquots of 100 μL were transferred to a 96-well flat-bottomed microtiter plate (Falcon), and the absorbance was measured at 405 nm with an automatic Multiskan Ascent plate reader (Labsystems). The level of hemolysis was calculated as the percentage of maximum lysis (1% Triton X-100 control) after adjusting for minimum lysis (PBS control). Synthetic melittin (Sigma) was used for comparison.
Supplementary Material
Footnotes
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- (1).Craik DJ; Daly NL; Bond T; Waine CJ Mol. Biol 1999, 294, 1327–1336. [DOI] [PubMed] [Google Scholar]
- (2).Craik DJ; Daly NL; Mulvenna J; Plan MR; Trabi M Curr. Protein Pept. Sci 2004, 5, 297–315. [DOI] [PubMed] [Google Scholar]
- (3).Pallaghy PK; Nielsen KJ; Craik DJ; Norton RS Protein Sci. 1994, 3, 1833–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Goransson U; Luijendijk T; Johansson S; Bohlin L; Claeson PJ Nat. Prod 1999, 62, 283–286. [DOI] [PubMed] [Google Scholar]
- (5).Gustafson KR; Walton LK; Sowder RCJ; Johnson DG; Pannell LK; Cardellina JH II; Boyd MR J. Nat. Prod 2000, 63, 176–178. [DOI] [PubMed] [Google Scholar]
- (6).Witherup K; Bogusky M; Anderson P; Ramjit H; Ransom R; Wood T; Sardana MJ Nat. Prod 1994, 57, 1619–1625. [DOI] [PubMed] [Google Scholar]
- (7).Gran L Lloydia. 1973, 36, 174–178. [PubMed] [Google Scholar]
- (8).Saether O; Craik DJ; Campbell ID; Sletten K; Juul J; Norman DG Biochemistry 1995, 34, 4147–4158. [DOI] [PubMed] [Google Scholar]
- (9).Trabi M; Svangard E; Herrmann A; Goransson U; Claeson P; Craik DJ; Bohlin LJ Nat. Prod 2004, 67, 806–810. [DOI] [PubMed] [Google Scholar]
- (10).Simonsen SM; Sando L; Ireland DC; Colgrave ML; Bharathi R; Goransson U; Craik DJ Plant Cell 2005, 17, 3176–3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ireland DC; Colgrave ML; Nguyencong P; Daly NL; Craik DJ J. Mol. Biol 2006, 357, 1522–1535. [DOI] [PubMed] [Google Scholar]
- (12).Chang RS; Yeung HW AntiViral Res. 1988, 9, 163–175. [DOI] [PubMed] [Google Scholar]
- (13).Makino T Bot. Mag. Tokyo 1912, 26, 148–151. [Google Scholar]
- (14).Qin P; Chen QP; Wang BR; Shi LW In Species Systematization and Quality Evaluation of Commonly Used Chinese Tradtitional Drugs; Lou ZQ, Qin P, Eds.; Beijing Medical University and Beijing Union Medical University: Beijing, 1995; pp 565–670. [Google Scholar]
- (15).Ngan F; Chang RS; Tabba HD; Smith KM AntiViral Res. 1988, 10, 107–16. [DOI] [PubMed] [Google Scholar]
- (16).Xie C; Kokubun T; Houghton PJ; Simmonds MS J. Phytother. Res 2004, 18, 497–500. [DOI] [PubMed] [Google Scholar]
- (17).Daly NL; Gustafson KR; Craik DJ FEBS Lett. 2004, 574, 69–72. [DOI] [PubMed] [Google Scholar]
- (18).Gustafson KR; McKee TC; Bokesch HR Curr. Protein Pept. Sci 2004, 5, 331–340. [DOI] [PubMed] [Google Scholar]
- (19).Tam J; Lu Y; Yang J; Chiu K Proc. Natl. Acad. Sci. U.S.A 1999, 96, 8913–8918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Jennings C; West J; Waine C; Craik D; Anderson M Proc. Natl. Acad. Sci. U.S.A 2001, 98, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jennings CV; Rosengren KJ; Daly NL; Plan M; Stevens J; Scanlon MJ; Waine C; Norman DG; Anderson MA; Craik DJ Biochemistry 2005, 44, 851–860. [DOI] [PubMed] [Google Scholar]
- (22).Svangard E; Goransson U; Hocaoglu Z; Gullbo J; Larsson R; Claeson P; Bohlin LJ Nat. Prod 2004, 67, 144–147. [DOI] [PubMed] [Google Scholar]
- (23).Colgrave ML; Craik DJ Biochemistry 2004, 43, 5965–5975. [DOI] [PubMed] [Google Scholar]
- (24).Craik DJ; Simonsen S; Daly NL Curr. Opin. Drug Discovery Dev 2002, 5, 251–260. [PubMed] [Google Scholar]
- (25).Kamimori H; Hall K; Craik DJ; Aguilar MI Anal. Biochem 2005, 337, 149–153. [DOI] [PubMed] [Google Scholar]
- (26).Goransson U; Svangard E; Claeson P; Bohlin L Curr. Protein Pept. Sci 2004, 5, 317–329. [DOI] [PubMed] [Google Scholar]
- (27).Goransson U; Broussalis AM; Claeson P Anal. Biochem 2003, 318, 107–117. [DOI] [PubMed] [Google Scholar]
- (28).Mulvenna JP; Wang C; Craik DJ Nucleic Acids Res. 2006, 34, D192–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Plan MR; Goransson U; Clark RJ; Daly NL; Colgrave ML; Craik DJ ChemBioChem 2007, 8, 1001–1011. [DOI] [PubMed] [Google Scholar]
- (30).Craik DJ; Cemezar M; Wang CK; Daly NL Biopolymers 2006, 84, 250–266. [DOI] [PubMed] [Google Scholar]
- (31).Dutton JL; Renda RF; Waine C; Clark RJ; Daly NL; Jennings CV; Anderson MA; Craik DJ J. Biol. Chem 2004, 279, 46858–46867. [DOI] [PubMed] [Google Scholar]
- (32).Gunasekera S; Daly NL; Anderson MA; Craik DJ IUBMB Life 2006, 58, 515–524. [DOI] [PubMed] [Google Scholar]
- (33).Saska I; Gillon AD; Hatsugai N; Dietzgen RG; Hara-Nishimura I; Anderson MA; Craik DJ J. Biol. Chem 2007, 282, 29721–29728. [DOI] [PubMed] [Google Scholar]
- (34).Bokesch HR; Pannell LK; Cochran PK; Sowder II RC; McKee TC; Boyd MR J. Nat. Prod 2001, 64, 249–250. [DOI] [PubMed] [Google Scholar]
- (35).Barry DG; Daly NL; Clark RJ; Sando L; Craik DJ Biochemistry 2003, 42, 6688–6695. [DOI] [PubMed] [Google Scholar]
- (36).Gustafson KR; Sowder RC; Henderson LE; Parsons IC; Kashman Y; Cardellina JH; McMahon JB; Buckheit RW; Pannell LK; Boyd MR J. Am. Chem. Soc 1994, 116, 9337–9338. [Google Scholar]
- (37).Kyte J; Doolittle RF J. Mol. Biol 1982, 157, 105–132. [DOI] [PubMed] [Google Scholar]
- (38).Chen B; Colgrave ML; Wang C; Craik DJ J. Nat. Prod 2006, 69, 23–28. [DOI] [PubMed] [Google Scholar]
- (39).Mulvenna JR; Sando L; Craik DJ Structure 2005, 13, 691–701. [DOI] [PubMed] [Google Scholar]
- (40).Fiser A; Do RK; Sali A Protein Sci. 2000, 9, 1753–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Engelman DM; Steitz TA; Goldman A Annu. ReV. Biophys. Biophys. Chem 1986, 15, 321–353. [DOI] [PubMed] [Google Scholar]
- (42).Eisenberg D; Schwarz E; Komaromy M; Wall RJ Mol. Biol 1984, 179, 125–142. [DOI] [PubMed] [Google Scholar]
- (43).Hopp TP; Woods KR Mol. Immunol 1983, 20, 483–489. [DOI] [PubMed] [Google Scholar]
- (44).Cornette JL; Cease KB; Margalit H; Spouge JL; Berzofsky JA; DeLisi CJ Mol. Biol 1987, 195, 659–685. [DOI] [PubMed] [Google Scholar]
- (45).Rose GD; Geselowitz AR; Lesser GJ; Lee RH; Zehfus MH Science 1985, 229, 834–838. [DOI] [PubMed] [Google Scholar]
- (46).Janin J Nature 1979, 277, 491–492. [DOI] [PubMed] [Google Scholar]
- (47).Rosengren KJ; Daly NL; Plan MR; Waine C; Craik DJ J. Biol. Chem 2003, 278, 8606–8616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.