Short abstract
An engineering perspective views cells as complex circuits that process inputs – drugs, environmental cues – to create complex outcomes – disease, growth, death – and this perspective has immense potential for drug development. Logical rules can describe the features of cells and reductionist approaches have exploited these rules for drug development. In contrast, the reductionist approach serially characterizes cellular components and develops a deep understanding of each component’s specific role. This approach underutilizes the full system of biomolecules relevant to disease pathology and drug effects. An engineering perspective provides the tools to understand and leverage the full extent of biological systems; applying both reverse and forward engineering, a strength of the engineering approach has demonstrated progress in advancing understanding of disease and drug mechanisms. Drug development lacks sufficient engineering specifications, or empirical models, of drug pharmacodynamic effects and future efforts to derive empirical models of drug effects will streamline this development. At this stage of progress, the scientist engineer is uniquely poised to solve problems in therapeutics related to modulating multiple diseases with a single or multiple therapeutic agents and identifying pharmacodynamics biomarkers with knowledge of drug pathways. This article underscores the value of these principles in an age where drug development costs are soaring and finding efficacious therapies is challenging.
Impact statement
Many untreated diseases are not monogenic and are instead caused by multiple genetic defects. Because of this complexity, computational, logical, and systems understanding will be essential to discovering novel therapies. The scientist engineer is uniquely disposed to use this type of understanding to advance therapeutic discovery. This work highlights benefits of the scientist engineer perspective and underscores the potential impact of these approaches for future therapeutic development. By framing the scientist engineer’s tool set and increasing awareness about this approach, this article stands to impact future therapeutic development efforts in an age of rising development costs and high drug attrition.
Keywords: Engineering, therapeutic discovery, systems biology, pharmacodynamics, molecular medicine, pathways, drug development
A landscape ripe for the scientist engineer
Drug development is costly and inefficient with less than 10% of drugs reaching the market.1 Limited efficacy or intolerable safety prevents these therapies from reaching patients. Therapeutic discovery has employed many paradigms for development. Some high-level themes include mechanism-agnostic drug selection by phenotypic outcome, optimized target binding with and without genetic evidence, and limited systems modeling for selecting nonobvious targets. Indeed, targets with genetic evidence are more successful overall,2,3 hinting at the importance of understanding at least some, biological mechanism. As the relatively ‘obvious’ or straightforward approaches fail to continually yield new therapies, the field will require alternative approaches. The emphasis of this article is the scientist engineer approach. The article begins with basic principles of this approach and highlights emerging impact of the scientist engineer approach in therapeutic development.
Logical rules can describe biology, but single rules are insufficient
At first, understanding that molecules give rise to phenotypes caused an increase in single-component associations, especially in Mendelian diseases; individual molecules were the main component of interest in a disease system. For instance, genetic changes to the hemoglobin protein caused sickle cell anemia,4,5 and BRCA1 mutational status was associated with poor prognosis.6,7 These discoveries advanced the field of molecular medicine and created many successful therapies. Yuri Lazebnik8 highlights the resulting relationship between biologists and the pharmaceutical industry in the quote ‘give me a target!.’ The quote reflects the role of biologists for studying, dissecting, and characterizing the disease process and then passing off the vulnerable molecular component to the pharmaceutical community to develop a therapeutic compound for this component. However, clean associations with single molecules and disease have not led to therapies for all diseases. For example, the classification, triple negative breast cancer, reflects that these cancers do not depend on the three, successfully drugged, components of other breast cancers.
Since that time, we have come to appreciate that many molecular components – genes, proteins, metabolites – contribute to complex biological phenotypes and that individual molecular medicine is insufficient for all diseases. This phenomenon has motivated a move to pathways-oriented discovery. In an engineering analogy, biologists have moved from single- to multi-component rules to explain disease. Sydney Brenner pursued the first wiring diagram of Caenorhabditis elegans, attempting to map out all relationships among the organism’s neurons. While an elegant idea, some have criticized the ‘connectome’ wiring diagram claiming that creation of the neuronal map has not improved understanding of the worm’s function and behavior. However, his analogy to electrical engineering wiring diagrams is a useful one and this analogy helps to organize the molecular components in biology.
The postgenomic era has shifted emphasis from discovering relevant cellular components to characterizing their emergent behaviors from their concerted interactions.9 The evolution in understanding and technology has influenced the fields of systems biology, systems pharmacology, systems bioengineering,10 and quantitative systems pharmacology (QSP).9 A central theme across these disciplines is that engineering can describe biology using logical abstractions, and that these abstractions can further understanding for therapeutic discovery.
Yuri Lazebnik described the ebb and flow of biological research through the analogy of fixing a radio: hype generates understanding of some new biological process, leading to the creation of new wiring diagrams, the pursuit for a magical cure, and eventual frustration at discovered contradictions in the diagrams and the failure of the potential miracle cure. An interpretation is that biological phenotypes are complicated, and it is unclear if biologists have the tools to decipher complex disease. Molecular biomedical research has amassed a tremendous amount of knowledge of the individual radio components, and many fields are now positioned to exploit these siloed discoveries at a systems level to understand their contribution to the full, disease, radio system.
Tuning biology is difficult when the wiring diagrams are wrong
George E.P. Box said, ‘all models are wrong, some are useful,’ and in the age of big data, the possible parts list for building biological models is increasing. This challenges biologists to broadly consider their disease system of interest when assembling parts of the model, but practicality still constrains the number of components that can be carefully tuned and manipulated. In a systems-wide study of resistance to tyrosine kinase inhibitors, Wilson et al.11 demonstrated that exogenous growth factors could confer drug resistance to cancers treated with targeted antigrowth therapies. For instance, addition of neuregulin (NRG1) to human epidermal growth factor receptor 2 (HER2)-amplified cancers caused resistance to the anti-HER2 therapy, lapatinib, or addition of hepatocyte growth factor (HGF) to epidermal growth factor receptor (EGFR)-mutated cancers caused resistance to the anti-EGFR therapy, erlotinib. In these contexts, discovering driver components was sufficient for an initial therapeutic discovery, but incomplete for a complete remission of disease. To a biologist, this finding suggests that growth factors are redundant in function; to an engineer, this suggests that the wiring diagrams of growth factor pathways are incomplete.
In other engineering contexts, engineers pick parts to match a specification. If you are building a smartphone, you need a material that can withstand heat, protect the hardware, and ideally survive an occasional bath in sunscreen or pool water. When selecting a therapeutic compound, scientists and engineers can specify the requirements, but it is difficult to select the compound to do the job. Janes et al.10 captured this idea when explaining the systems biologists approach to biology. Compared to creating a nose cone for an airplane, an aerospace engineer uses Fourier’s laws to describe how a material will behave in a certain environment. A biological engineer does not always have the same rules to describe how a drug will affect a patient. Rather, the biological engineer often tweaks, adapts, and applies existing paradigms in a mutual learning and application process – somewhere between forward and reverse engineering.10 Achieving useful biological specifications will require iterative improvements to current wiring diagrams.
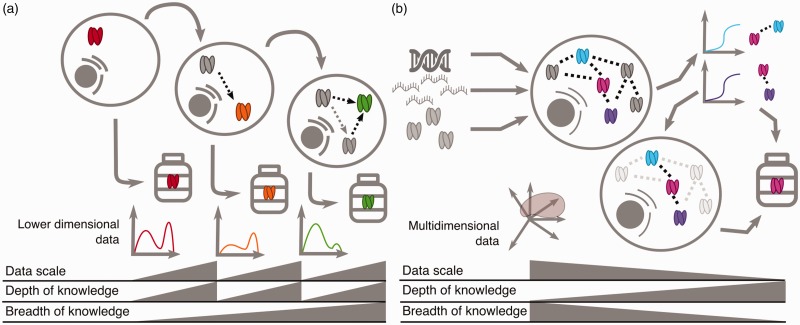
Reductionist scientists and scientist engineers approach creation of biological wiring diagrams with different approaches. Reductionist scientists serially drill into a disease. They characterize single components using low-dimensional data and develop a deep sense of the component’s role in disease. Complexity is added to the system as each component is rigorously investigated (Figure 1(a)). The scientist engineer begins with larger cohorts of data, often at multiple time points, doses, and treatment scales, and constructs an initial abstraction of their model system. Later, dedicated experiments refine the model and inform therapeutic development (Figure 1(b)). The scientist engineer applies current knowledge while maintaining a healthy skepticism of its completeness. It is not sufficient to build upon the existing components; one must also explore and expand that component list in the process of building representative systems. Both perspectives will continue to inform therapeutic discovery as we update our understanding of the disease radio.
Figure 1.
A scientist engineer and reductionist biologist make different contributions to biological understanding. (a) The reductionist approach: a reductionist scientist serially perturbs a system and evolves a system schematic by characterizing individual component. At each phase, the reductionist scientist studies each component in greater depth. (b) The scientist engineer approach: a scientist engineer embraces the complexity of the system and starts with ‘omics’ level measurements to first derive a low-resolution map. At this early stage, the scientist engineer has many modeling techniques at their disposal including machine learning, statistical, and differential equations modeling. Later modeling tests the map and dedicated experiments refine relationships in the map. The scientist engineer first embraces complexity and then investigates single components in depth as needed. Iterative cycles of dedicated experiments update model assumptions and lead to new discoveries; only one cycle is depicted. In both examples, gray triangles indicate the relative magnitude of data scale, depth, and breadth of knowledge.
Combined forward and reverse engineering advances understanding of disease and therapeutic mechanisms
Using an engineering approach, Fey et al.12 discovered that c-Jun N-Terminal kinase (JNK) activation, or response to stress, could distinguish between neuroblastoma patients with poor or good prognosis. To discover the relationship between prognosis and cellular ability to activate JNK response, they first assembled the cellular signaling components of this pathway, incorporated patient expression data, and then investigated weaknesses in this pathway associated with prognosis. The approach highlighted a harmony between forward and reverse engineering approaches: by initially reverse engineering the neuroblastoma signaling network, they were later able to forward engineer the model to discover that cancers prevent the stress response.
Hass et al.13 also used an engineering approach to attribute receptor dimerization patterns to explain drug resistance and sensitivity. The group first experimentally measured cellular proliferation in response to various ligands and then employed a signaling model combined with bagged decision tree analysis to discern which signaling features distinguished proliferation between cell lines. In this reverse engineering phase, the group learned that receptor hetero- and homo- dimerization patterns were the most responsible for differences in proliferation between cancer cell lines. They further applied their model in a forward engineering context and incorporated cancer patient receptor expression data to confirm that these signaling features are correlated with antigrowth factor receptor therapies. For instance, their model confirmed that non-small cell lung carcinoma (NSCLC) is dependent on EGFR signaling and that these patients are responsive to anti-EGFR inhibitors. The model predicted that NSCLC cells are only weakly dependent on HGF signaling; the group hypothesized that NSCLC patients were showing weak response to therapies that block the HGF receptor, Met, because of the weak dependency of NSCLC on HGF signaling.13
Where serial, reductionist approaches are insufficient, an engineer’s perspective has advanced cancer biology using cell signaling. Traditional biomarker and over-expression analyses have not found molecular liabilities for malignant melanoma and triple-negative breast cancer. Instead, Miller et al. investigated the signaling dynamics of a cancer cell in the presence of mitogen-activated protein kinase kinase 1 (MEK) inhibitors. The group discovered that MEK inhibition decreased shedding of the cell surface proteins MET proto-oncogene, receptor tyrosine kinase, Erb-B2 Receptor Tyrosine Kinase 2 (HER2), and AXL receptor tyrosine kinase. Increased surface levels of these proteins enabled bypass signaling and continued growth in the presence of targeted therapy.14 Updating the ‘wiring diagrams’ for cancer signaling gave the authors a novel perspective on this system.
Gianchandani et al.15 applied the rules of linear programming for understanding the regulatory network in Escherichia coli. They extended previous work in regulatory matrices that used matrix representations to describe cellular responses to environmental stimuli. In this extended work, the authors demonstrated that these regulatory matrices could accommodate genome-scale models of E. coli and subsequently describe the complex, gene regulatory behaviors of E. coli. These results emphasized that a broad range of engineering tools have utility for advancing biological understanding.
This idea was more thoroughly detailed in a white paper, where Sorger et al.9 discussed the importance of QSP as an academic discipline that better informs future translational decisions. The authors explained the potential for quantitative and systems pharmacology to identify ‘on-target’ toxicities and validate the idea that drug perturbations can influence many disease and physiological phenotypes. Thus far, many of the examples provided embody the themes of QSP and validate the utility of this approach. These approaches have uncovered a set of best practices – or engineering specifications – for therapeutic development, opened avenues for new perspectives on development, and identified possibilities for further interdisciplinary work.
Engineering specifications for therapeutic development
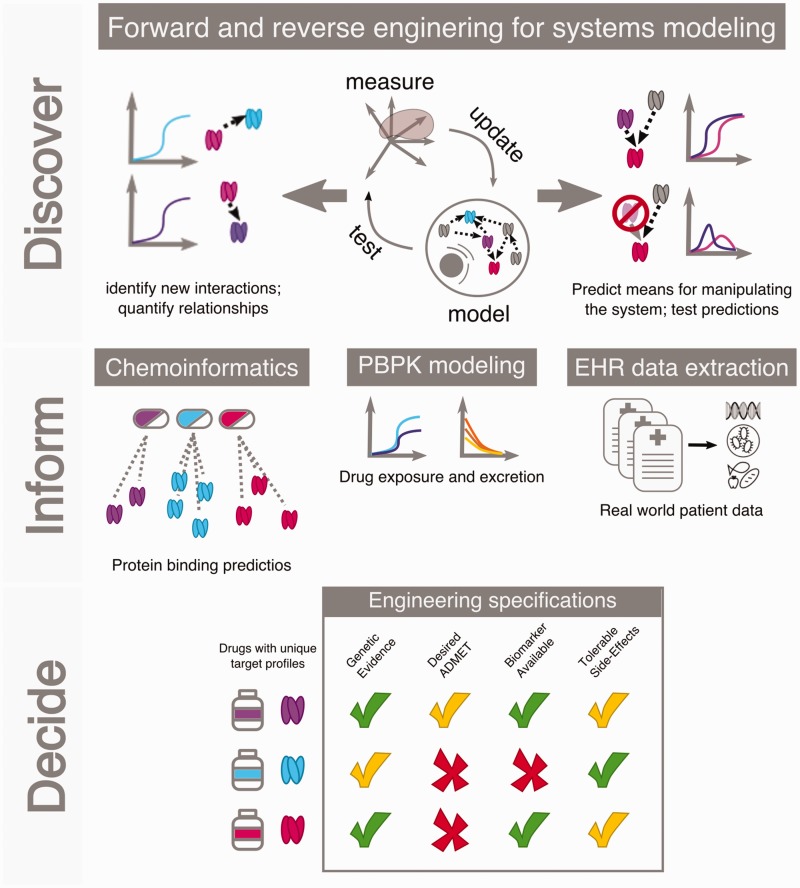
Empirical models and product specifications are key tools for making design choices in other engineering disciplines. Stress–strain curves describe material elasticity under loads and when designing a product, material scientists can consult these empirical models to select a material with desired performance. These principles are applicable to drug discovery; disposition science has characterized how the body modifies a drug and these specifications have influenced therapeutic design parameters such as the type of drug, dosing, and route of administration. However, we lack sufficient empirical models for drug pharmacodynamics or how the drug modifies the body. Industry has relied on ‘indirect’ response models for many years, such as observing a desired phenotypic endpoint (e.g. lower blood pressure, reduced blood glucose) to develop effective, if not optimal, therapies. Mitigating disease phenotypes is necessary but often these effects come at the cost of undesirable side effects. This target-agnostic approach does not allow for consideration or prioritization of target molecules. QSP models can integrate all available information and bring mechanistic insights into target selection and characterization. For instance, adverse event (AE) characterization is one opportunity where QSP models could improve target selection: learning empirical models of adverse outcomes would enable future development efforts to prioritize drug targets with roles in disease pathways and deprioritize targets involved in AE pathways. Further development of these empirical models could enable patient-forward design where engineers consider the specific necessities of a particular patient – both single and multiple diseases – and design compounds to these patient needs. These models will be continuously evolving, but without a scientist engineer perspective, it will be intractable to decipher the interplay of these systems for optimal therapeutic design. Here we reinforce the importance of absorption, distribution, metabolism, excretion, and toxicity (ADMET) principles and highlight four trends that will influence future decisions about therapeutic compounds. Figure 2 summarizes this approach.
Figure 2.
The scientist engineer approach to therapeutic discovery. The approach leverages quantitative modeling and employs forward and reverse engineering principles to derive understanding of the disease system (top). Informatics approaches, such as chemoinformatics or EHR studies, and PBPK models inform the druggability, tunability (the extent to which a target can be modulated), or relevance of a drug target (middle). Final decisions aggregate these data and assess desired specifications when selecting a lead compound (bottom). EHR: electronic health record; PBPK: physiologically based pharmacokinetic.
ADMET science characterizes drug pharmacokinetics
ADMET science has established routine tests and knowledge about how the body processes and affects drugs.16 These fundamentals are essential to any new development pipeline and still constrain the selection of lead compounds. However, the scientist engineer approach could augment how these models are applied. For instance, pharmacokinetic models currently aid in projection of the anticipated human dose and researchers use these models before transitioning to clinical trials. Though, in many biological settings, partial or incomplete target engagement can be sufficient for altering a disease phenotype. For instance, therapeutically targeting the serotonin system is promising for mitigating movement deficit in Parkinson’s patients.17 The therapy, buspirone, is only a partial agonist of the serotonin system and demonstrates efficacy in mitigating movement deficit.17 In these contexts, as we reverse engineer the model and parameters, we can expand ADMET science to better represent the pharmacodynamics of altering disease biology.
Genetic evidence improves therapeutic efficacy
Rational therapeutic design, grounded in understanding of disease etiology has markedly improved drug discovery and will continue to influence successful therapeutics.2 Drugs with genetic evidence supporting the intended target represent ∼2% of all drugs in early discovery and represent ∼8% in drugs that are eventually approved.2 PCSK9 inhibitors are one such success story where detailed understanding of cholesterol metabolism and identification of functional variants that conferred bonus health to certain patients yielded a breakthrough therapy with improved efficacy over existing treatments. Routine testing of genetic evidence supporting disease target selection will improve drug efficacy.3
Biomarkers are associated with improved overall efficacy
The term ‘biomarkers’ is an umbrella term covering a range of biomolecules used to influence clinical decision making.18 These biomarkers can influence therapeutic selection and dosing, among other clinical decisions. As of 2016, a meta-analysis of clinical development success rates for 2005–2015 found that drugs with biomarkers had a 25.9% likelihood of reaching the market whereas drugs without biomarkers had an 8.4% likelihood.1 An earlier investigation found that the proportion of early stage (i.e. phase I or phase II) clinical trials with biomarkers was less than 15% and the proportion of late stage (i.e. phase III and phase IV) clinical studies with biomarkers was less than 10%.19 Drug biomarkers are associated with overall success but are not yet mainstream.
Side effect profiles will preclude targets from development
Pathways analysis has greatly improved identification and characterization of disease targets. Cancer therapies are a salient example where knowledge of dysregulated growth pathways led to therapeutic innovations to alter these growth outcomes. Similarly, pathways-level investigations of AE pathways will characterize drug targets for their role in unintended side effects. Already, multiple approaches studying drug-induced peripheral neuropathy,20 rhabdomyolysis,21 and Stevens–Johnson syndrome22 demonstrated that certain protein targets are more liable to induce undesirable effects.
The culmination of these engineering specifications informs the scientist engineer approach (Figure 2). In this approach, the scientist engineer uses a blend of dedicated modeling and experimental techniques to gain systems understanding (Discover). The scientist engineer uses data-driven fields, such as chemoinformatics, physiologically based pharmacokinetic (PBPK), and electronic health record (EHR) data to inform and often, revise, their systems models (Inform). The culmination of this approach is developing therapeutic decisions using knowledge of the relevant engineering specifications (Decide). In practice, this process is less definitive and the Discover and Inform phases can be iterated or blended depending on the biological problem. The scientist engineer approach is motivated by the pursuit of mechanistic understanding of the fundamental biological principles governing a system. Depending on the biological system and the data available, the tools of the scientist engineer overlap with data scientists, statisticians, and computer scientists. The scientist engineer can be conversant in machine learning, big data, and simulations as needed to understand their system.
Multicomponent and multidisease manipulation will improve future therapeutics
Small molecules throw another wrench into the therapeutic development process. It turns out that biology contains a fair bit of redundancy and even if properly optimized, a small molecule will bind many proteins beyond the original intended target. Recent estimates predict that drugs bind an average of 329 proteins (or a median of 38),23 making the idea of optimizing a small molecule for a central disease component unachievable; the drug will bind the disease protein and many other proteins in the cell. As example of the consequences of this binding promiscuity, Tatonetti et al.24,25 show that a combination of paroxetine and pravastatin interacts to increase blood glucose. Paroxetine is an antidepressant and pravastatin is a therapy for reducing blood pressure; neither of these compounds is marketed for metabolic disease but alters metabolic processes when used together. If we view the drug combination simply as an example of engaging more protein binding partners, we learn that multiprotein engagement can have profound outcomes on biology.26 In this example, patients experienced effects on depression, blood pressure, and blood glucose. This is further supported by work from Molecular Health and from the FDA Adverse Event Reporting System. FDA scientists used Molecular Health’s software to extract and identify drug targets associated with AEs such as Stevens–Johnson Syndrome.27 The AEs associated with molecular targets further emphasize the multifaceted outcomes of perturbing these biological components. Another interpretation of high binding promiscuity is the possibility that small molecules can be leveraged for multiple disease indications simultaneously.
Chemoinformatics approaches and high-throughput experimentation will improve our understanding of small molecules’ multicomponent effects, and systems biology will interpret the effects of multiprotein binding. One such chemoinformatic approach, the pocketFEATURE algorithm identified similar binding pockets to predict ligand off-target binding.28 Application of the pocketFEATURE algorithm predicted novel protein binding partners responsible for drug side effects. Using a ligand-based approach, Keiser et al.29 developed an approach to functionally link proteins based on the similarity of their ligands. They later applied this method to predicting off-target protein binding of FDA-approved drugs using ligand similarity profiles to identify the new targets.30 High-throughput screens remain a workhorse for early discovery16 of protein to drug binding. These screens are complemented with quantitative analyses of relationships between chemical structure and activity for making decisions about the disposition of a drug within the body.16 These techniques lay the foundation for understanding the full phenome affected by small molecule interventions.
Off-label drug use supports the idea that drugs influence multiple diseases. Drugs tested and approved for one disease indication often end up used for another disease.31 Bioinformatics methods can extract these off-label uses from EHRs32 and in silico methods can predict where marketed drugs may affect additional diseases.33 These data sources improve our understanding of drug action and provide an opportunity to learn how therapeutics affect multiple diseases and how to apply these principles to drug repurposing efforts. Repeated evidence has shown that patients and diseases are not one-dimensional and that drugs are not one-dimensional; this presents an opportunity to develop drugs aware of their multidimensional effects instead of for single-disease effects. In this multidisease paradigm of drug action, one can imagine specifying a compound that mitigates diabetes, does not affect cholesterol, and improves depression.
Novel experimental systems will be essential to test multicomponent and multidisease therapeutic strategies predicted by engineering approaches. Cokol et al.34 designed and validated the diagonal measurement of n-way drug combinations (DiaMOND) method to look for tuberculosis combination treatments. The group realized that testing n-way combinations was expensive and inefficient. They designed a testing methodology using a limited set of doses and combinations to determine synergy, additivity, or antagonism between n-way drug combinations and discovered novel two-, three-, and four-way treatments for tuberculosis.34 Interestingly, they also discovered that pairwise interactions are only partially predictive of n-way combinations. The fact that higher order combinations are not entirely predicted by their subcomponents further necessitates a scientist engineer approach to manage and interpret these complex drug interactions.
Pathways analysis can inform drug pharmacodynamics and biomarker selection
Drug effects extend beyond the target of interest and thus, discovering pathways models is an important feature of understanding drug effects. Guney et al. performed an in silico screening approach to identify drug repurposing opportunities. In developing this method, they related the drug-binding proteins of approved drugs to genes associated with the disease the drug intended to treat.33 In this process, they discovered that drug targets are not always directly or closely associated with the genes of the intended-to-treat disease. This suggests one or both of two outcomes – the drug binds alternative, unknown proteins or the effects of drug binding are long range and are propagated by signaling pathways. The scientist engineer approach is particularly well suited for discovering long-range interactions and will complement work to identify additional drug-binding proteins.
A pathways perspective could identify gene variants that predispose a patient to a poor or enhanced drug outcome for pharmacogenomics research. Genotyping for warfarin dosing is an iconic example of pharmacogenomics where CYP2C9, VKORC1, CYP4F2, and rs12777823 status affect the prescribed warfarin dose.35 To date, these associations are largely discovered through large GWAS studies followed by dedicated experimental validation, and indeed there is a great need for more studies investigating the genetic dependencies of therapeutic compounds.36 However, because pathways contain the molecular components of drug and disease pathways, these pathway components may also become a source of pharmacogenomic genes.
Conclusions
Approaching biological problems from a scientist engineer perspective will improve future drug development endeavors. The QSP field embraces the contributions of the scientist engineer and has already demonstrated impact on understanding of disease systems and mechanisms of drug action. The contribution and skills of the scientist engineer are poorly defined and evolving. Addressing the evolving definition of systems biology, Janes et al. define systems bioengineering as a discipline using engineering design in combination with dedicated experiments for understanding biological systems. They eloquently highlight the forward and reverse engineering theme related to the scientist engineer approach: ‘those driven by technology […] favor induction to generate hypotheses after collecting and analyzing large sets of observations.’10 In the context of this review, a systems biologist is just one of many specific subdisciplines that embody the principles of the scientist engineer. We have highlighted the importance of the combined forward and reverse perspectives as fundamental to the scientist engineer and posited two areas in which this approach will be uniquely successful: considering therapeutics as tools for managing multiple conditions and identifying drug and patient biomarkers.
To successfully consider new therapeutics, the scientist engineer will need to be conversant in many of the computational and biological subdisciplines. Innovation will require tools from systems biology, computational biology, bioinformatics, statistics, and engineering if we are to change the nature of therapeutic development.
ACKNOWLEDGMENTS
The author would like to thank Russ Altman and Douglas Lauffenburger for helpful discussions which shaped this perspective and Emily Flynn for reading this manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING
JLW was supported by NIH LM007033 and a grant from Pfizer Inc. This publication was made possible by Grant Number U01FD004979 from the FDA, which supports the UCSF-Stanford Center of Excellence in Regulatory Sciences and Innovation. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the HHS or FDA.
References
- 1.Biotechnology Innovation Organization. Clinical development success rates 2006–2015 Biotechnology Innovation Organization 2016, pp.1–28
- 2.Nelson MR, Tipney H, Painter JL, Shen J, Nicoletti P, Shen Y.Floratos A, Sham PC, Li MJ, Wang J, Cardon LR, Wittaker J, Sanseau P. The support of human genetic evidence for approved drug indications. Nat Genet 2015; 47:1–7 [DOI] [PubMed] [Google Scholar]
- 3.Nelson MR, Johnson T, Warren L, Hughes AR, Chissoe SL, Xu C-F. Waterworth DM. The genetics of drug efficacy: opportunities and challenges. Nat Rev Genet 2016; 17:197–206 [DOI] [PubMed] [Google Scholar]
- 4.Pauling L, Itano HA, Singer SJ, Wells IC. Sickle cell anemia, a molecular disease. Science 1949; 110:543–8 [DOI] [PubMed] [Google Scholar]
- 5.Swensen JJ, Agarwal AM, Esquilin JM, Swierczek S, Perumbeti A, Hussey D.Lee M, Joiner CH, Pont-Kingdon G, Lyon E, Prchal JT. Sickle cell disease resulting from uniparental disomy in a child who inherited sickle cell trait. Blood 2010; 116:2822–5 [DOI] [PubMed] [Google Scholar]
- 6.Zhu Y, Wu J, Zhang C, Sun S, Zhang J, Liu W, Huang J, Zhang Z. BRCA mutations and survival in breast cancer: an updated systematic review and meta-analysis. Oncotarget 2016; 7:70113–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keeney MG, Couch FJ, Visscher DW, Lindor NM. Non-BRCA familial breast cancer: review of reported pathology and molecular findings. Pathology 2017; 49:363–70 [DOI] [PubMed] [Google Scholar]
- 8.Lazebnik Y. Can a biologist fix a radio? – or, what I learned while studying apoptosis. Cancer Cell 2002; 2:179–82 [DOI] [PubMed] [Google Scholar]
- 9.National Institutes of Health. Quantitative and systems pharmacology in the post-genomic era: new approaches to discovering drugs and understanding therapeutic mechanisms, https://www.nigms.nih.gov/Training/Documents/SystemsPharmaWPSorger2011.pdf (2011, accessed 6 June 2018)
- 10.Janes KA, Chandran PL, Ford RM, Lazzara MJ, Papin JA, Peirce SM, Saucerman JJ, Lauffenburger DA. An engineering design approach to systems biology. Integr Biol 2017; 9:574–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, Ribas A, Li J, Moffat J, Sutherlin DP, Koeppen H, Merchant M, Neve R, Settleman J. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature 2012; 487:505–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fey D, Halasz M, Dreidax D, Kennedy SP, Hastings JF, Rauch N, Munoz AG, Pilkington R, Fischer M, Wastermann F, Kolch W, Kholodenko BN, Croucher DR. Signaling pathway models as biomarkers: patient-specific simulations of JNK activity predict the survival of neuroblastoma patients. Sci Signal 2015; 8:ra130. [DOI] [PubMed] [Google Scholar]
- 13.Hass H, Masson K, Wohlgemuth S, Paragas V, Allen JE, Sevecka M, Pace E, Timmer J, Stelling J, MacBeath G, Schoeberl B, Raue A. Predicting ligand-dependent tumors from multi-dimensional signaling features. NPJ Syst Biol Appl 2017; 3:783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller MA, Oudin MJ, Sullivan RJ, Wang SJ, Meyer AS, Im H, Frederick DT, Tadros J, Griffith LG, Lee H, Weissleder R, Flaherty KT, Gertler FB, Lauffenburger DA. Reduced proteolytic shedding of receptor tyrosine kinases is a post-translational mechanism of kinase inhibitor resistance. Cancer Discov 2016; 6:382–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gianchandani EP, Joyce AR, Palsson BØ, Papin JA. Functional states of the genome-scale Escherichia coli transcriptional regulatory system. PLoS Comput Biol 2009; 5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanders JM, Beshore DC, Culberson JC, Fells JI, Imbriglio JE, Gunaydin H.Haidle AM, Labroli M, Mattioni BE, Sciammetta N, Shipe WD, Sheridan RP, Suen LM, Verras A, Walji A, Joshi EM, Bueters T. Informing the selection of screening hit series with in silico absorption, distribution, metabolism, excretion, and toxicity profiles. J Med Chem 2017; 60:6771–80 [DOI] [PubMed] [Google Scholar]
- 17.Eskow KL, Gupta V, Alam S, Park JY, Bishop C. The partial 5-HT1A agonist buspirone reduces the expression and development of l-DOPA-induced dyskinesia in rats and improves l-DOPA efficacy. Pharmacol Biochem Behav 2007; 87:306–14 [DOI] [PubMed] [Google Scholar]
- 18.Wilson JL, Altman RB. Biomarkers: delivering on the expectation of molecularly driven, quantitative health. Exp Biol Med 2018; 243:313–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi K, Masuda S, Kimura H. Analyzing global trends of biomarker use in drug interventional clinical studies. Drug Discov Ther 2012; 6:1–6 [PubMed] [Google Scholar]
- 20.Hur J, Guo AY, Loh WY, Feldman EL, Bai JPF. Integrated systems pharmacology analysis of clinical drug-induced peripheral neuropathy. CPT: Pharmacomet Syst Pharmacol 2014; 3:e114–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hur J, Liu Z, Tong W, Laaksonen R, Bai JPF. Drug-induced rhabdomyolysis: from systems pharmacology analysis to biochemical flux. Chem Res Toxicol 2014; 27:421–32 [DOI] [PubMed] [Google Scholar]
- 22.Hur J, Zhao C, Bai JPF. Systems pharmacological analysis of drugs inducing Stevens–Johnson syndrome and toxic epidermal necrolysis. Chem Res Toxicol 2015; 28:927–34 [DOI] [PubMed] [Google Scholar]
- 23.Zhou H, Gao M, Skolnick J. Comprehensive prediction of drug-protein interactions and side effects for the human proteome. Sci Rep 2015; 5:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tatonetti NP, Fernald GH, Altman RB. A novel signal detection algorithm for identifying hidden drug-drug interactions in adverse event reports. J Am Med Inform Assoc 2012; 19:79–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatonetti NP, Denny JC, Murphy SN, Fernald GH, Krishnan G, Castro V, Yue P, Tsau PS, Kohane I, Roden DM, Altman RB. Detecting drug interactions from adverse-event reports: interaction between paroxetine and pravastatin increases blood glucose levels. Clin Pharmacol Ther 2011; 90:133–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keiser MJ, Irwin JJ, Shoichet BK. The chemical basis of pharmacology. Biochem Am Chem Soc 2010; 49:10267–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burkhart KK, Abernethy D, Jackson D. Data mining FAERS to analyze molecular targets of drugs highly associated with Stevens-Johnson syndrome. J Med Toxicol 2015; 11:265–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T, Altman RB. Using multiple microenvironments to find similar ligand-binding sites: application to kinase inhibitor binding. PLoS Comput Biol 2011; 7:e1002326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol 2007; 25:1197–206 [DOI] [PubMed] [Google Scholar]
- 30.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, Whaley R, Glennon RA, Hert J, Thomas KLH, Edwards DD, Shoichet BK, Roth BL. Predicting new molecular targets for known drugs. Nature 2009; 462:175–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hussain SA, Abbas AN, Alhadad HA, Al-Jumaili AA, Abdulrahman ZS. Physician-pharmacist agreement about off-label use of medications in private clinical settings in Baghdad, Iraq. Pharm Pract 2017; 15:979–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jung K, LePendu P, Chen WS, Iyer SV, Readhead B, Dudley JT, Shah NH. Automated detection of off-label drug use. PLoS One 2014; 9:e89324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guney E, Menche J, Vidal M, Barabási A-L. Network-based in silico drug efficacy screening. Nat Commun 2016; 7:10331 [Mismatch] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cokol M, Kuru N, Bicak E, Larkins-Ford J, Aldridge BB. Efficient measurement and factorization of high-order drug interactions in Mycobacterium tuberculosis. Sci Adv 2017; 3:e1701881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson JA, Caudle KE, Gong L, Whirl-Carrillo M, Stein CM, Scott SA, Lee MT, Gage BF, Kimmel SE, Perera MA, Anderson JL, Pirmohamed M, Klein TE, Limdi NA, Cavallari LH, Wadelius M. Clinical pharmacogenetics implementation consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing. Clin Pharmacol Ther 2017; 89:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giacomini KM, Yee SW, Mushiroda T, Weinshilboum RM, Ratain MJ, Kubo M. Genome-wide association studies of drug response and toxicity: an opportunity for genome medicine. Nat Rev Drug Discov 2017; 16(1):1 [DOI] [PMC free article] [PubMed] [Google Scholar]