Short abstract
A total of 70 pregnant women were recruited for this study, including 35 early onset preeclampsia (PE) and 35 normal pregnant women. By RNA sequencing, the circular RNA (circRNAs) in placenta were identified. Differentially expressed circRNAs were bioinformatics analyzed with gene ontology, Kyoto Encyclopedia of Genes and Genomes, and circRNA–miRNA interaction prediction. Quantitative real time polymerase chain reaction(qRT-PCR) assay was used to verify the results. Compared with the normal pregnant women, there were 49 circRNAs differentially expressed in the placental tissue of PE women, including two circRNAs were up-regulated and 47 were down-regulated. Ten differentially expressed circRNAs were selected for validation by qRT-PCR, among which results of three circRNAs, circRNA_3286, circRNA_5593, and circRNA_3800, were consistent with the sequencing. According to the bioinformatic prediction, we speculate that these circRNAs may be involved in some cellular regulatory functions in PE through their targeted miRNAs. We also evaluated the expression of circRNA_3286 in plasma to be used as a potential biomarker for PE; in vitro Transwell assay shown transfected with si-circ-3286 significantly reduced invasion in HTR8/Svneo cells. In conclusion, we displayed a preliminary landscape of circRNA differential expression in PE and discussed their possible regulatory mechanisms. This study revealed a new insight into the molecular mechanism of PE.
Impact statement
The abnormal expression of many regulatory factors may be involved in the development of PE. circRNAs are proved to have a series of important biological functions; however, reports about circRNA and PE are rare. In this work, we evaluated the profile analysis of circRNAs in human placenta of PE by RNA-seq and found some newly differentially expressed circRNAs which might be involved in PE. Combined with bioinformatics analysis, their possible functions were preliminarily discussed.
Keywords: Preeclampsia, circRNA, miRNA, RNA sequencing, placenta, hsa_circ_0088227
Introduction
Preeclampsia (PE) is one of the main causes of maternal death. The global incidence rate of PE is 4.6%, while the incidence of eclampsia is 1.4%. PE can cause severe maternal and neonatal complications with multisystem disturbance syndrome.1,2 However, the exact pathogenesis remains unclear. With the deepening of the research on the pathogenesis of PE, more and more biological factors have aroused widespread concern, and their abnormal changes may be involved in the pathogenesis of PE. Especially in recent years, with the rapid development of molecular biology, the discovery of some small molecules may be able to show the features of disease more clearly.
As a kind of special non-coding RNA, circular RNAs (circRNAs) have the characteristics of evolutionary conservation, structural stability, tissue specificity, etc.3,4 Because of these biological features, circRNA has attracted wide attention in recent years. They play important roles in tumor, nervous system disease, diabetes, etc.5–7 CircRNAs serve its biological function as miRNA sponges and affect the expression of downstream genes.8,9 Some reports showed that there were also circRNAs in the human placenta and might be related to the occurrence of pregnancy complications.10 Qian’s11 group first reported that circRNAs might be involved in the invasion and apoptosis of trophoblast cells in the development of PE through acting on certain targeted miRNAs. However, their exact function about the development of PE remains largely unknown. It has attracted our interest in further research.
In the present study, we identified the different expression of circRNAs in placenta from early onset PE by RNA sequencing and investigated their biological function by bioinformatics analysis preliminarily. We hope to clarify the relationship between circRNAs and the occurrence of PE.
Materials and methods
Sample collection
During August 2016–June 2017, a total of 70 pregnant women undergone cesarean section from Changzhou Women and Children Health Hospital affiliated to Nanjing Medical University were recruited for this study, including 35 cases with early onset PE and 35 normal pregnant women. Table 1 shows their baseline characteristics. All pregnant women were excluded from primary hypertension, multiple pregnancy, chromosomal abnormalities, congenital malformations, and suspected perinatal infection. All the patients were consent informed.
Table 1.
The information of patients with PE and normal control groups.
| Parameter | PE (n = 35) | Control (n = 35) | P value |
|---|---|---|---|
| Age (years) | 30.41 ± 2.34 | 30.46 ± 2.01 | >0.05 |
| Systolic blood pressure (mmHg) | 160.05 ± 4.39 | 123.03 ± 4.46 | <0.01 |
| Diastolic blood pressure (mmHg) | 112.26 ± 4.88 | 81.91 ± 3.42 | <0.01 |
| Urine protein (g/24 h) | 3.82 ± 1.25 | 0.08 ± 0.07 | <0.01 |
| Gestational weeks | 35.07 ± 0.47 | 37.25 ± 1.19 | <0.01 |
| Neonatal weight (g) | 2442.62 ± 308.52 | 3233.71 ± 339.1 | <0.01 |
PE: preeclampsia.
Plasma samples were collected before cesarean section, and placental tissues were obtained within 10 min after cesarean section. We collected the central part of placenta and avoided infection, bleeding, and calcification. The tissues were cut into 1 g size, washed with normal saline, and dried with gauze to absorb the surface liquid. Then we cut them carefully into pieces and froze them in −80°C immediately after adding 1 mL Trizol. Three cases and three paired controls were chosen to use RNA sequencing analysis. The chosen circRNAs were further confirmed by qRT-PCR.
RNA extraction and sequencing
After sample collection, total RNA was extracted using RNA Extraction Kit (Qiagen) according to its protocol. Then 5 µg total RNA was treated by Ribo Zero Magnetic kit (Plant Leaf, MRZPL116, Epicentre) in order to remove ribosomal RNA. Three hundred nanogram ribosomal RNA-depleted RNA samples were interrupted by high temperature in a buffer containing two valence cations. The first strand of cDNA was reverse transcripted randomly by Superscript II (Invitrogen) using random primer (Takara). The second strand of cDNA chain was synthesized and dNTP was substituted by dUTP in the reaction system. The double-stranded cDNA was repaired and A was added to its 3 terminal end. Then Illumina Truseq adaptor was digested and purified by USER enzyme (NEB). Products were purified and digested, and amplified by Illumina P5 and P7 primers using PCR. The amplified products were separated by 2% agarose gel electrophoresis and the 350–500 bp bands were collected. This final product was the final library and was prepared for sequencing by Illumina HiSeq X Ten (Berry Genomics, Beijing, China).
Identification and quantification of human circRNAs
After sequencing, raw data were filtered, high quality clean data were obtained by removing the joint sequence and low quality reads. The sequence of clean data was compared with the reference genome, and mapped data were obtained. Based on mapped data, the quality of sequencing library was evaluated by inserting fragment length and randomness test. CircRNAs were predicted by CIRI12 and compared with the circRNAs in the circBase (http://www.circbase.org/). For the obtained clean reads, STAR was used to compare with the reference genome and the reference gene source ensemble GRCh37.fa (Hg 19.fa). Deseq13 software was used to standardize the number of junction reads of each sample circRNAs (basemean value was used to estimate the amount of expression). The different multiples were calculated and reads were tested, then the differentially expressed circRNAs were screened according to the test results. Fold changes (FCs)>2 and P < 0.05 were set as significant difference.
Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis
The differentially expressed circRNAs were subjected to gene ontology (GO) and KEGG pathway analyses based on their origin genes using Gene Ontology (http://www.geneontology.org/) and KOBAS software (KEGG Orthology-Based Annotation System).
CircRNA–miRNA interaction prediction
Since circRNA contains multiple miRNA binding sites, two prediction algorithms (Circinteractom and miRanda) were used to predict targeted miRNAs by circRNAs. The function of this part of circRNAs could be elucidated according to the functional annotation of the miRNA target genes.
qRT-PCR assay
We randomly selected 10 circRNAs from the results of RNA sequencing to qRT-PCR verification. After the total RNA was extracted as above, cDNA was obtained by reverse transcription Kit (Promega M-MLV), and qRT-PCR was performed according to the designed primers, part of which are shown in Table 2. The PCRs were run as follows: denaturation 95°C for 10 min, followed by amplification by 38 cycles of 95°C for 10 s and 60°C for 1 min.
Table 2.
Primer sequences of the circRNAs.
| 5′–3′ | 3′–5′ | |
|---|---|---|
| GAPDH | TGACTTCAACAGCGACACCCA | CACCCTGTTGCTGTAGCCAAA |
| circRNA_3800 | TCCAGTGTAAGCCCAGGGACA | CGCTGGCATTGAGGTTGTTGT |
| circRNA_5593 | GATGCCTGGGTGAAGAAGTA | CATGTGGACCTTGTTGTCTGT |
| circRNA_3286 | AATCTACACGCTGGATGAGC | AGGTGCGGACACTGGACTTA |
Cell culture and transfection
HTR-8/SVneo cells were obtained from the Chinese Academy of Sciences Committee (Shanghai, China) and cultured in 1640 with 10% FBS (Gibco, USA) at 37°C, 5% CO2. For the transfection of small interfering RNAs (siRNAs), cells (2 × 105) were seeded in six-well plates and transfected with 100 nM siRNA using Lipofectamine 3000 (Invitrogen, USA) after 24h. The sequences of siRNA for circRNA_3286 (si-circ-3286) was 5′-CACUCUGUCUACAGUGUAA-3′ (sense) and 5′-UUACACUGUAGACAGAGUG-3′ (antisense). The sequences of the negative control siRNAs (si-NC) were 5′-UUCUCCGAACGUGUCACGU-3′ (sense) and 5′-ACGUGACACGUUCGGAGAA-3′ (antisense). All these sequences were synthesized by Ribo Co., Ltd (Guangzhou, China).
Cell invasion assay
Transwell (Corning, USA) was assessed to evaluate the invasion capacity of the trophoblast cells. Upper chamber of Transwell was coated with Matrigel gel (BD, USA). HTR-8/SVneo cells were cultured in serum-free medium for 24 h, then the cell density was adjusted to 1 × 105 cells/mL, and 100 µL cell suspension was added to the top chamber, while 500 mL of culture medium containing 20% FBS was added to the lower chamber. After cultured in 37°C for 24 h, cells and the Matrigel on the top surface of the Transwell membrane were wiped off with cotton swab. The cells on the bottom surface of the membrane were fixed with cold paraformaldehyde and stained with 500 µL 0.1% crystal violet (Sigma, USA). Five fields on each membrane were randomly selected and the invaded cells were counted.
Statistical analysis
The data of qRT-PCR were analyzed by SPSS 19 software, they were expressed by mean and standard error, and the t test was used to compare the mean of the two sets of samples. The receiver operating characteristic (ROC) curve was established to evaluate the circRNAs’ diagnostic value by SPSS19. P < 0.05 was set as a statistical significance.
Results
Identification and quantification of human circRNAs
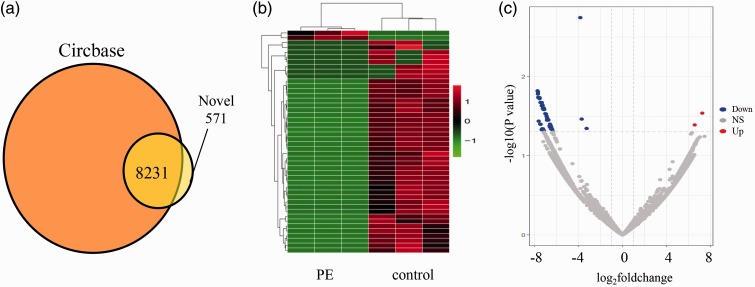
Compared with the information of human circRNAs in CircBase, we had identified 8802 circRNAs in the placenta. According to the CircBase, 8231 circRNAs had been reported, while 571 circRNAs were newly discovered (Figure 1(a)). Heat map (Figure 1(b)) and Volcano plots (Figure 1(c)) revealed the different expression profiles of circRNAs between PE women and normal control group. Compared with the normal pregnant women, there were 49 circRNAs (FC > 2 and P < 0.05) differentially expressed in the placental tissue of PE women, including two circRNAs were up-regulated and 47were down-regulated. Their information are shown in Table 3.
Figure 1.
Differentially expressed circRNA in PE and control group. (a) The obtained circRNAs were compared with the circRNAs in the known database. (b) Heat map presentation of the expression profiles of the circRNAs. Each row represents one circRNA, and each column represents one sample. Red color indicates up-regulation, green indicates downregulation. (c) Differentially expressed circRNAs were displayed by Volcano plots. Gray dots indicated circRNAs with no significant difference. Red dots indicated significant up-regulated circRNAs, blue dots indicated significant down-regulated. NS: Not significant. (A color version of this figure is available in the online journal.)
Table 3.
Statistics of differentially expressed transcripts.
| ID name | circBase name | Gene symbol | chrom | log2FoldChange | Regulation | p value |
|---|---|---|---|---|---|---|
| circ_4437 | hsa_circ_0000044 | PUM1 | 1 | −7.278 | Down | 0.0456 |
| circ_8695 | hsa_circ_0002669 | DOCK1 | 10 | −7.095 | Down | 0.0260 |
| circ_8310 | hsa_circ_0002968 | MAPK8 | 10 | −7.319 | Down | 0.0233 |
| circ_6991 | hsa_circ_0005379 | GDI2 | 10 | −7.645 | Down | 0.0183 |
| circ_8432 | hsa_circ_0096364 | SHANK2 | 11 | −7.660 | Down | 0.0157 |
| circ_5124 | hsa_circ_0028319 | TMEM116 | 12 | −6.411 | Down | 0.0461 |
| circ_8124 | hsa_circ_0007723 | CCDC91 | 12 | −7.440 | Down | 0.0212 |
| circ_1473 | hsa_circ_0004985 | ARID2 | 12 | −6.734 | Down | 0.0352 |
| circ_4792 | hsa_circ_0000417 | CPSF6 | 12 | −6.827 | Down | 0.0335 |
| circ_5240 | hsa_circ_0002886 | TMTC2 | 12 | −7.598 | Down | 0.0365 |
| circ_780 | hsa_circ_0002814 | HERC2 | 15 | −6.491 | Down | 0.0428 |
| circ_3223 | hsa_circ_0002466 | TTBK2 | 15 | −7.601 | Down | 0.0184 |
| circ_967 | hsa_circ_0035472 | RNF111 | 15 | −7.210 | Down | 0.0456 |
| circ_7167 | hsa_circ_0002226 | ETFA | 15 | −6.716 | Down | 0.0354 |
| circ_7423 | hsa_circ_0000642 | ZFAND6 | 15 | −6.844 | Down | 0.0355 |
| circ_4958 | hsa_circ_0003559 | XYLT1 | 16 | −6.519 | Down | 0.0416 |
| circ_730 | hsa_circ_0042170 | NCOR1 | 17 | −6.457 | Down | 0.0437 |
| circ_5391 | hsa_circ_0007990 | PGAP3 | 17 | −6.996 | Down | 0.0294 |
| circ_8090 | hsa_circ_0000854 | ZCCHC2 | 18 | −7.665 | Down | 0.0166 |
| circ_2523 | hsa_circ_0000857 | ZNF236 | 18 | −7.130 | Down | 0.0250 |
| circ_8563 | hsa_circ_0006209 | CTDP1 | 18 | −7.266 | Down | 0.0457 |
| circ_4390 | hsa_circ_0006670 | SIPA1L3 | 19 | −6.799 | Down | 0.0344 |
| circ_8122 | – | LIMS2 | 2 | −6.605 | Down | 0.0418 |
| circ_1698 | hsa_circ_0058092 | FN1 | 2 | −6.413 | Down | 0.0460 |
| circ_3042 | hsa_circ_0001103 | SERPINE2 | 2 | −7.326 | Down | 0.0468 |
| circ_6515 | hsa_circ_0004164 | THAP4 | 2 | −7.421 | Down | 0.0401 |
| circ_7224 | hsa_circ_0002795 | SPAST | 2 | −6.934 | Down | 0.0297 |
| circ_4399 | hsa_circ_0008440 | EVA1A | 2 | 7.233 | Up | 0.0289 |
| circ_2262 | hsa_circ_0001148 | RBM39 | 20 | −7.325 | Down | 0.0216 |
| circ_4899 | hsa_circ_0001222 | FBXO7 | 22 | −7.464 | Down | 0.0398 |
| circ_1687 | hsa_circ_0121643 | HSPBAP1 | 3 | 6.546 | Up | 0.0408 |
| circ_3811 | hsa_circ_0001369 | KLHL24 | 3 | −6.880 | Down | 0.0317 |
| circ_5754 | – | LOC100507487 | 4 | −6.384 | Down | 0.0464 |
| circ_5428 | hsa_circ_0075161 | NSD1 | 5 | −7.698 | Down | 0.0151 |
| circ_5050 | hsa_circ_0079492 | BZW2 | 7 | −7.519 | Down | 0.0190 |
| circ_6525 | hsa_circ_0134318 | GLI3 | 7 | −6.570 | Down | 0.0398 |
| circ_8679 | hsa_circ_0079363 | DAGLB | 7 | −7.133 | Down | 0.0480 |
| circ_8641 | hsa_circ_0136108 | CSGALNACT1 | 8 | −6.398 | Down | 0.0462 |
| circ_980 | hsa_circ_0001798 | SPIDR | 8 | −7.237 | Down | 0.0231 |
| circ_3058 | hsa_circ_0002798 | STK3 | 8 | −6.584 | Down | 0.0434 |
| circ_3421 | – | PAPPA | 9 | −7.171 | Down | 0.0260 |
| circ_7147 | hsa_circ_0088214 | PAPPA | 9 | −7.318 | Down | 0.0213 |
| circ_4046 | hsa_circ_0088249 | PAPPA | 9 | −6.467 | Down | 0.0460 |
| circ_3286 | hsa_circ_0088227 | PAPPA | 9 | −3.823 | Down | 0.0018 |
| circ_6886 | hsa_circ_0008732 | BNC2 | 9 | −7.226 | Down | 0.0251 |
| circ_8540 | hsa_circ_0091581 | GPC3 | X | −6.303 | Down | 0.0495 |
| circ_2396 | hsa_circ_0007108 | ZFX | X | −7.512 | Down | 0.0185 |
| circ_5593 | – | ACSS1 | 11 | −3.253 | Down | 0.0452 |
| circ_3800 | – | SHANK2 | 10 | −3.698 | Down | 0.0344 |
Functional analysis of differentially expressed circRNAs
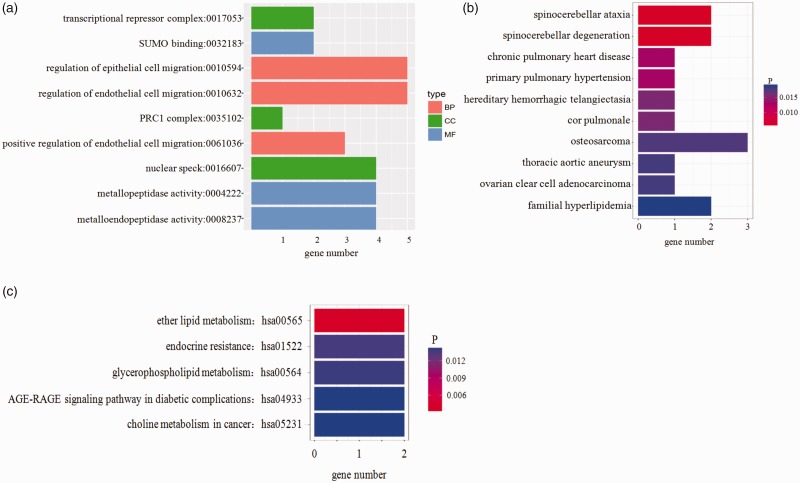
GO is an international standard classification system for gene function. The differentially expressed circRNAs’ origin genes were analyzed by GO and KEGG enrichment (Figure 2(a) and (c)). We found these differentially expressed circRNAs might be associated with GO functional annotation of transcriptional repressor complex, SUMO binding, regulation of epithelial/endothelial cell migration, etc. Meanwhile, the most enriched KEGG pathways included ether lipid metabolism, endocrine resistance, glycerophospholipid metabolism. Disease enrichment result (Figure 2(b)) shows that they might involve in spinocerebellar ataxia, spinocerebellar degeneration, chronic pulmonary heart disease, etc.
Figure 2.
GO and KEGG enrichment terms of differentially expressed circRNA source genes. (a) GO enrichment terms, (b) disease enrichment result, and (c) KEGG enrichment terms. The x axis indicates the numbers of circRNAs annotated. PRC1, Polycomb repressive complex 1; SUMO, Small ubiquitin-like modifiers. (A color version of this figure is available in the online journal.)
qRT-PCR assay
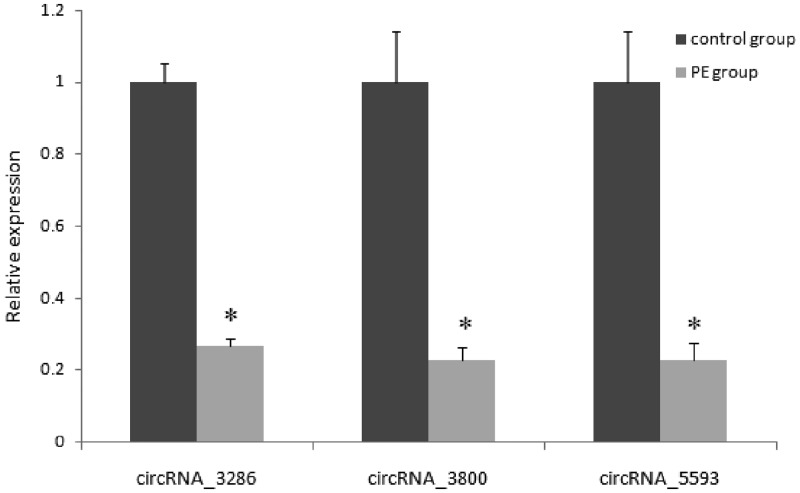
In order to validate the RNA sequencing results and study the possible mechanism of circRNAs in the development of PE, 10 circRNAs were randomly selected to detect by qRT-PCR. Compared with the control group, the expression of three circRNAs (circRNA_3286, circRNA_5593, and circRNA_3800) in placenta (35 cases of PE and 35 cases of normal) was decreased significantly (Figure 3), which is consistent with RNA sequencing data, their primer sequences are shown in Table 2.
Figure 3.
qRT-PCR verification of differentially expressed circRNAs in PE and control placenta. *P < 0.05. PE: preeclampsia.
CircRNA–miRNA interaction analysis
CircRNAs act as target molecules of miRNAs; the interaction analysis of circRNA–miRNA can help to resolve the function and mechanism of circRNA as miRNA sponges, so we elucidated the function of this part of circRNAs according to the miRNA target genes. According to the prediction results, the miRNAs interacted with circRNA_3286, circRNA_5593, and circRNA_3800 are shown in Table 4.
Table 4.
Differentially expressed circRNAs and their targeted miRNAs.
| circRNA | miRNA | circRNA | miRNA | circRNA | miRNA |
|---|---|---|---|---|---|
| circRNA_3800 | hsa-miR-1208 | circRNA_3286 | hsa-miR-1183 | circRNA_5593 | hsa-miR-2278 |
| hsa-miR-1256 | hsa-miR-1224-5p | hsa-miR-3141 | |||
| hsa-miR-1538 | hsa-miR-1306-5p | hsa-miR-326 | |||
| hsa-miR-296-5p | hsa-miR-337-3p | hsa-miR-330-5p | |||
| hsa-miR-335 | hsa-miR-384 | hsa-miR-3687 | |||
| hsa-miR-4488 | hsa-miR-421 | hsa-miR-634 | |||
| hsa-miR-4767 | hsa-miR-516b | hsa-miR-661 | |||
| hsa-miR-634 | hsa-miR-5193 | hsa-miR-937-5p | |||
| hsa-miR-661 | hsa-miR-616 | ||||
| hsa-miR-6784-5p | hsa-miR-711 | ||||
| hsa-miR-744-5p | hsa-miR-766 | ||||
| hsa-miR-934 |
Evaluation of circRNA _3286 in plasma of PE patients
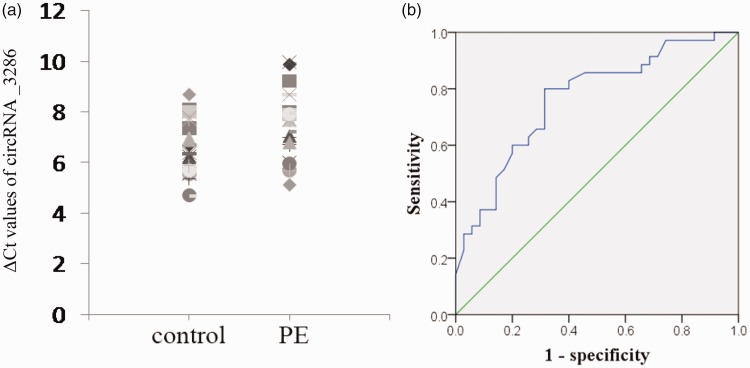
We evaluated the expression of these three circRNAs validated by qRT-PCR in plasma to be used as candidate biomarkers for PE. In all plasma samples, circRNA_3286 had a detection rate of 100% (Figure 4(a)), while the other two circRNAs (circRNA_5593 and circRNA_3800) had detection rates below 20% in both PE and control plasma samples, therefore, they were excluded from our subsequent analyses. ROC curves were obtained from the data of all 35 plasma of pregnant women with PE and 35 normal controls; circRNA_3286 showed an area under ROC curve of 0.764 (95% CI: 0.652–0.875, P < 0.01), sensitivity of 80%, and specificity of 68.6% (Figure 4(b)).
Figure 4.
Evaluation of circRNA _3286 expression in plasma of PE patients. (a) circRNA _3286 expression in plasma was presented as ΔCt values normalized to GAPDH; larger ΔCt indicates lower expression. (b) ROC curves were obtained from the data of all 35 plasma of pregnant women with PE and 35 normal controls. PE: preeclampsia. (A color version of this figure is available in the online journal.)
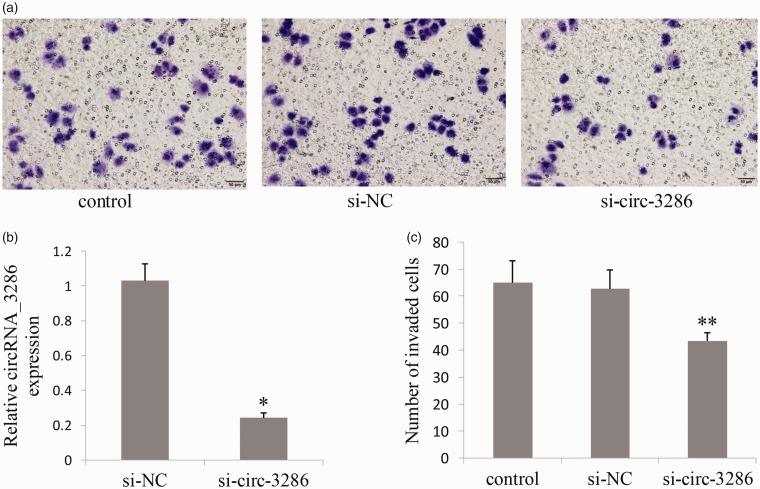
si- circ_3286 inhibited cell invasion in HTR8/Svneo cells
Transwell assay was conducted to further demonstrate the role of circRNA_3286 in HTR8/Svneo cell invasion. Transfected with si-circ-3286 significantly reduced circRNA _3286 expression (Figure 5(b)) and cell invasion (Figure 5(a) and (c)). The number of cells migrating through the Transwell membrane in si-circ-3286 group was less than that in control and si-NC group (p<0.01).
Figure 5.
si- circ_3286 inhibited cell invasion in HTR8/Svneo cells. (a) Transfected with si-circ-3286 significantly reduced HTR8/Svneo cell invasion. (b) Transfected with si-circ-3286 reduced circRNA _3286 expression. (c) The number of cells migrating through the Transwell membrane in si-circ-3286 group was less than that in control and si-NC group. *P<0.05, **P<0.01. si-NC: negative control siRNA. (A color version of this figure is available in the online journal.)
Discussion
In the past, circRNAs were thought to be a class of low abundant RNA molecules formed by the splicing of exons.14 Recently, with the rapid development of bioinformatics and the constant innovation of high-throughput sequencing technology, more and more functional circRNAs have been identified.3,15 Accumulating evidence indicated that circRNAs had a series of important biological functions, such as the miRNA sponges.6 They acted as competitive endogenous RNAs through effectively adsorbing miRNAs and regulating their target genes. Research about circRNAs is mainly focused on tumor, and they might become a kind of new biomarkers.16–18 However, there were few reports about the relationship between circRNAs and pregnancy. Maass’s group detected 63 circRNAs in human placenta. By function prediction, they thought some circRNAs might be related to pregnancy complications, such as early onset PE, fetal growth restriction, and infection during pregnancy.10
It is well known that PE is a serious and common pregnancy complication, but its pathogens remain to be elucidated. It is believed that it may involve immune maladaptation, inadequate placental development and trophoblast invasion, placental ischemia, oxidative stress and thrombosis, and so on. It is generally recognized that all the symptoms of PE are related to the placenta, while the fetus is not a necessary condition. Because in some cases of ectopic pregnancy, the symptoms of PE would continue to exist except for the placenta being removed, even if the fetus was dead.2 Recently, the discovery of small molecules in the placenta may be able to reveal the essence of PE more accurately. For example, He’s group reported that clusters of lncRNAs were aberrantly expressed in PE placenta, which might play a partial or key role in PE development. Especially they put forward that LOC391533, LOC284100, and CEACAMP8 might contribute to the mechanism underlying PE.19 However, the research focus on circRNAs and PE is extremely rare.
In our study, we successfully discovered 49 differentially expressed circRNAs in the PE placenta by RNA sequencing. Their biological functions were predicted by bioinformatics analysis. Meanwhile, three circRNAs (circRNA_3286, circRNA_5593, and circRNA_3800) were selected to confirm the results of the RNA-seq and explore the possible mechanism of PE. Analogously, Qian et al.11 also found the differentially expressed circRNAs in the placenta of PE patients by microarrays. Compared with their chip technology, RNA sequencing technology has many advantages. For example, the gene chip reflects the transcriptional expression level by judging the strength of the hybridization signal indirectly, so it is easy to be interfered by background signal and interleaving hybridization. At the same time, microarray cannot be used for detecting of low-abundance transcription. Without special probes, RNA sequencing can detect all transcripts in cells with higher resolution and coverage, in this way new transcripts can be discovered.
As we know, circRNAs have a series of important biological functions, such as the miRNA sponges.6 They act as competitive endogenous RNAs through effectively adsorbing miRNAs and regulating their target genes. CircRNAs can interact with RNA-binding proteins to affect the expression of their parent gene mRNAs.20,21 On the other hand, the complementary pairing between introns can form a balance between linear RNA and circRNA, which would influence the expression of mRNA and even protein translation.20,22
Among these three circRNAs verified by qRT-PCR, we preliminarily discussed the possible mechanisms for their participation in PE. We found circRNA_3286 was overlapped in the PAPPA gene which encodes a pregnancy-related protein. It is well known that the circulating concentrations of PAPPA were proved to be associated with the development of PE.23,24 This is a reminder that circRNA_3286 might be involved in the pathogenesis of PE. Through bioinformatics analysis, we found circRNA_3286 was targeted by miR-1224-5p, miR-1306-5p, miR-711, and miR-5193 etc. Qian et al.25 reported exogenous expression of miR-1224-5p in glioma cells suppressed proliferation and invasion and promoted apoptosis, and it was decreased in high-grade gliomas. Studies suggested that the function of the placental trophoblast cells has an important role in the occurrence of PE, such as differentiation, proliferation, migration, and invasion.26–28 Moreover, TGFβ1 signaling pathway was thought to be involved in the occurrence and development of PE,29 while miR-1224-5p was reported to act as tumor-suppressive role via the TGFβ1/Smad3 signaling pathway.30 So we speculated that miR-1224-5p might also play its role in inhibiting trophoblast cell proliferation, invasion, and induce apoptosis via TGFβ1/Smad3 signaling pathway in PE. Based on the above theory, we think that circRNA_3286 may be involved in the differentiation, proliferation, migration, and invasion of PE placenta through miR-1224-5p. TGFβ1 signaling pathway may be its important mode of action. Meanwhile, it is believed that cardiovascular disease was closely related to PE,31,32 mainly in the vascular endothelial injury. MiR-1306-5p was recently reported positively associated with adverse clinical outcome in heart failure,33,34 which had the binding sites of circRNA_3286. MiR-711 was another target of circRNA_3286 and it was reported to be associated with proliferation and apoptosis in tumor.35,36 In addition, circRNA_5593 and circRNA_3800 were two unreported circRNAs. They also had target miRNAs, such as miR-744-5p, miR-1538, miR-2278, miR-3687 and they were gradually found to play an important role in the life process.37–40 Then we evaluated the expression of these three circRNAs in plasma to be regarded as potential biomarkers for prediction of PE, among which the expression data of circRNA_3286 might serve as a candidate potential biomarker for diagnosis of PE. In recent years, more and more studies indicated that the abnormal recasting of spiral arteries and the defect of trophoblast cell invasion play an important role in the pathogenesis of PE.27,41 In order to explore whether circRNA_3286 affects the invasion of trophoblast cells, Transwell assay was conducted in HTR8/SVneo cells. We found that transfected with si-circ-3286 significantly inhibited the cell invasion capability. This result suggested that abnormal expression of circRNA_3286 may be involved in the pathogenesis of PE.
However, there were limitations in this study. First, the samples number was small, especially for RNA sequencing. Second, the bioinformatics analysis was superficial, and it was not enough to suggest the role of circRNAs in the development of PE. In addition, the number of differentially expressed circRNAs which we discovered were not very many. The obtained circRNAs were defined mainly by bioinformatic prediction, which has its own limitation on accuracy and sensitivity. Our study on the possible mechanism of related circRNAs is only at the initial stage; additional in-depth function of these placenta-related circRNAs should be investigated in vivo/in vitro for a more comprehensive understanding in future study.
In conclusion, our study displayed a preliminary landscape of circRNA differential expression in PE. These circRNAs may be involved in the changes of trophoblast function of PE placenta, especially circRNA_3286. This study revealed a new insight into the molecular mechanism of PE.
ACKNOWLEDGMENTS
We thank all of the project participants for their contributions.
Authors’ contributions
WZ, HW, and BY carried out the assays and participated in designing the study. BY and HW carried out clinical consultation. XW, WL, JK, and FZ carried out laboratory tests and performed the statistical analysis. WZ and BY conceived the study, participated in its design and coordination, and drafted the manuscript.
DECLARATION OF CONFLICTING INTERESTS
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
This work was supported by the Project funding for the training of high level health professionals in Changzhou (2016CZLJ013) and Changzhou science and technology support project (Social Development CE20175021).
Ethics approval
The study design and protocol were reviewed and approved by the ethics committee of Changzhou Maternity and Child Health Hospital affiliated to Nanjing Medical University.
References
- 1.Abalos E, Cuesta C, Grosso AL, Chou D, Say L. Global and regional estimates of preeclampsia and eclampsia: a systematic review. Eur J Obstet Gynecol Reprod Biol 2013; 170:1–7 [DOI] [PubMed] [Google Scholar]
- 2.Maynard SE, Venkatesha S, Thadhani R, Karumanchi SA. Soluble Fms-like tyrosine kinase 1 and endothelial dysfunction in the pathogenesis of preeclampsia. Pediatr Res 2005; 57:1R. [DOI] [PubMed] [Google Scholar]
- 3.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-338 [DOI] [PubMed] [Google Scholar]
- 4.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, William F, Marzlu FF, Norman ES. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lukiw WJ. Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 2013; 4:307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Christian K, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–8 [DOI] [PubMed] [Google Scholar]
- 7.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015; 5:12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One 2016; 11:e0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao YF. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 ‘sponge’ in human cartilage degradation. Sci Rep 2016; 6:22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC, Rajewsky N. A map of human circular RNAs in clinically relevant tissues. J Mol Med 2017; 95:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qian Y, Lu Y, Rui C, Qian Y, Cai M, Jia R. Potential significance of circular RNA in human placental tissue for patients with preeclampsia. Cell Physiol Biochem 2016; 39:1380. [DOI] [PubMed] [Google Scholar]
- 12.Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015; 16:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anders S, Huber W. Differential expression of RNA-Seq data at the gene level – the DESeq package. 2013. http://www.bioconductor.org/packages/devel/bioc/vignettes/DESeq/inst/doc/DESeq.pdf
- 14.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 1976; 73:3852–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoffelen R, Jimenez MI, Dierckxsens C. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015; 444:132–6 [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma. J Cancer Res Clin Oncol 2017; 143:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S. Circular RNA expression in basal cell carcinoma. Epigenomics 2016; 8:619–32 [DOI] [PubMed] [Google Scholar]
- 19.He X, He Y, Xi B, Zheng J, Zeng X, Cai Q, Ouyang Y, Wang C, Zhou X, Huang H, Deng W, Xin S, Huang Q, Liu H. LncRNAs expression in preeclampsia placenta reveals the potential role of LncRNAs contributing to preeclampsia pathogenesis. PLoS One 2013; 8:e81437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55–66 [DOI] [PubMed] [Google Scholar]
- 21.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015; 10:170–7 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792–806 [DOI] [PubMed] [Google Scholar]
- 23.Poon LC, Maiz N, Valencia C, Plasencia W, Nicolaides KH. First-trimester maternal serum pregnancy-associated plasma protein-A and pre-eclampsia. Ultrasound Obstet Gynecol 2010; 33:23–33 [DOI] [PubMed] [Google Scholar]
- 24.Canini S, Prefumo F, Pastorino D, Crocetti L, Afflitto CG, Venturini PL, De Biasio P. Association between birth weight and first-trimester free beta-human chorionic gonadotropin and pregnancy-associated plasma protein A. Fertil Steril 2008; 89:174–8 [DOI] [PubMed] [Google Scholar]
- 25.Qian J, Li R, Wang YY, Shi Y, Luan WK, Tao T, Zhang JX, Xu YC, You YP. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem 2015; 403:33. [DOI] [PubMed] [Google Scholar]
- 26.Li X. Expressions of transcription factors Smad4 and NF-κB in preeclampsia placenta tissue and exploration of its relationship with expressions of apoptotic and invasive genes. J Hainan Med Univ 2015; 10:79–82 [Google Scholar]
- 27.Roland CS, Hu J, Ren CE, Chen H, Li J, Varvoutis MS, Leaphart LW, Byck DB, Zhu X, Jiang SW. Morphological changes of placental syncytium and their implications for the pathogenesis of preeclampsia. Cell Mol Life Sci 2016; 73:365–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia RZ, Rui C, Li JY, Cui XW, Wang X. CDX1 restricts the invasion of HTR-8/SVneo trophoblast cells by inhibiting MMP-9 expression. Placenta 2014; 35:450–4 [DOI] [PubMed] [Google Scholar]
- 29.Chen Q, Chen L, Liu B, Vialli C, Stone P, Ching LM, Chamley L. The role of autocrine TGFbeta1 in endothelial cell activation induced by phagocytosis of necrotic trophoblasts: a possible role in the pathogenesis of pre-eclampsia. J Pathol 2010; 221:87–95 [DOI] [PubMed] [Google Scholar]
- 30.Yao X, Cui X, Wu X, Xu P, Zhu W, Chen X, Zhao T. Tumor suppressive role of miR-1224-5p in keloid proliferation, apoptosis and invasion via the TGF-Î21/Smad3 signaling pathway. Biochem Biophys Res Commun 2017;495:713–720 [DOI] [PubMed]
- 31.Ramsay JE, Stewart F, Greer IA, Sattar N. Microvascular dysfunction: a link between pre-eclampsia and maternal coronary heart disease. BJOG 2003; 110:1029–31 [PubMed] [Google Scholar]
- 32.Ghosseindoha C, Neer JV, Wissink B, Breetveld NM, Windt LJD, Dijk APJV, van der Vlugt MJ, Janssen MC, Heidema WM, Scholten RR, Spaanderman ME. Pre-eclampsia: an important risk factor for asymptomatic heart failure. Ultrasound Obstet Gynecol 2017; 49:143. [DOI] [PubMed] [Google Scholar]
- 33.Boven NV, Kardys I, Vark LCV, Akkerhuis KM, Ronde MWJD, Khan MAF, Merkus D, Liu Z, Voors AA, Asselbergs FW, van den Bos EJ, Boersma E, Hillege H, Duncker DJ, Pinto YM, Postmus D. Serially measured circulating microRNAs and adverse clinical outcomes in patients with acute heart failure. Eur J Heart Failure 2017;20:89-96 [DOI] [PubMed]
- 34.Bayes-Genis A, Lanfear DE, De Ronde MWJ, Lupón J, Leenders JJ, Liu Z, Zuithoff NPA, Eijkemans MJC, Zamora E, De Antonio M, Zwinderman AH, Pinto-Sietsma SJ, Pinto YM. Prognostic value of circulating microRNAs on heart failure‐related morbidity and mortality in two large diverse cohorts of general heart failure patients. Eur J Heart Failure 2017;20:67–75 [DOI] [PubMed]
- 35.Zhu SX, Tong XZ, Zhang S. Expression of miR-711 and mechanism of proliferation and apoptosis in human gastric carcinoma. Oncol Lett 2017; 14:4505–10 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Liao A, Tan G, Chen L, Zhou W, Hu H. RASSF1A inhibits gastric cancer cell proliferation by miR-711- mediated downregulation of CDK4 expression. Oncotarget 2016; 7:5842–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ertekin S, Dinç E, Ayaz L, Tamer L. Evaluation of circulating miRNAs in wet age-related macular degeneration. Mol Vis 2014; 20:1057–66 [PMC free article] [PubMed] [Google Scholar]
- 38.Daniel R, Wu Q, Williams V, Clark G, Guruli G, Zehner Z. A panel of microRNAs as diagnostic biomarkers for the identification of prostate cancer. Int J Mol Sci 2017; 18:1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Todeschini P, Salviato E, Paracchini L, Ferracin M, Petrillo M, Zanotti L, Tognon G, Gambino A, Calura E, Caratti G, Martini P, Beltrame L, Maragoni L, Gallo D, Odicino FE, Sartori E, Scambia G, Negrini M, Ravaggi A, D’Incalci M, Marchini S, Bignotti E, Romualdi C. Circulating miRNA landscape identifies miR-1246 as promising diagnostic biomarker in high-grade serous ovarian carcinoma: a validation across two independent cohorts. Cancer Lett 2016; 388:320–7 [DOI] [PubMed] [Google Scholar]
- 40.Kaymaz BT Günel NS Ceyhan MÇetintaş VB, Özel B,Yandım MK Kıpçak S Aktan Ç Gökbulut AA Baran Y Can BK.. Revealing genome-wide mRNA and microRNA expression patterns in leukemic cells highlighted “hsa-miR-2278” as a tumor suppressor for regain of chemotherapeutic imatinib response due to targeting STAT5A. Tumor Biol 2015; 36:7915–7927 [DOI] [PubMed] [Google Scholar]
- 41.Crosley EJ, Elliot MG, Christians JK, Crespi BJ. Placental invasion, preeclampsia risk and adaptive molecular evolution at the origin of the great apes: evidence from genome-wide analyses. Placenta 2013; 34:127–32 [DOI] [PubMed] [Google Scholar]