Abstract
Background
Emerging evidence suggests a potential relationship between gut microbiota and the host response to chemotherapeutic drugs including 5-fluorouracil (5-Fu). Fusobacterium nucleatum (Fn) has been linked to the initiation and progression of colorectal cancer (CRC). Unfortunately, little was known about the relationship between Fn infection and chemotherapeutic efficacy. Here, we investigate the potential relationship between Fn infection and chemotherapeutic efficacy of 5-Fu in CRC.
Methods
Differentially expressed genes of CRC cell lines induced by Fn infection were analyzed based on a whole genome microarray analysis Then, we explored the relationship between upregulation of BIRC3 induced by Fn infection and chemoresistance to 5-Fu in vitro and in vivo. Furthermore, we dissected the mechanisms involved in Fn-induced BIRC3 expression. Finally, we investigated the clinical relevance of Fn infection, BIRC3 protein expression and chemoresistance to 5-Fu treatment in CRC patients.
Results
BIRC3 was the most upregulated gene induced by Fn infection via the TLR4/NF-κB pathway in CRC cells; Fn infection reduced the chemosensitivity of CRC cells to 5-Fu through upregulation of BIRC3 in vitro and in vivo. High Fn abundance correlated with chemoresistance in advanced CRC patients who received standard 5-Fu-based adjuvant chemotherapy after radical surgery.
Conclusions
Our evidence suggests that Fn and BIRC3 may serve as promising therapeutic targets for reducing chemoresistance to 5-Fu treatment in advanced CRC.
Electronic supplementary material
The online version of this article (10.1186/s13046-018-0985-y) contains supplementary material, which is available to authorized users.
Keywords: Fusobacterium nucleatum, BIRC3, 5-fluorouracil, Chemoresistance, Colorectal cancer
Background
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer-related death, accounting for 1.4 million new cases and 0.6 million deaths worldwide in 2012 [1]. In China, there were more than 0.37 million new cases of CRC that led to more than 0.19 million deaths in 2015 [2]. Recently, accumulating evidence has shown that intestinal bacteria dysbiosis is related to the initiation and development of CRC [3–5]. Moreover, the gut microbiota can also influence the efficacy of antitumor immunotherapy drugs [6–9] and have an impact on the treatment of CRC [10]. Interestingly, two studies have shown that bacterial metabolism can affect the host response to chemotherapeutic drugs including 5-fluorouracil (5-Fu) in C. elegans [11, 12]. Fusobacterium nucleatum (Fn), an anaerobic bacterium parasitic in the oral cavity, is increasingly linked to CRC [13–18]. However, few studies have focused on the impact of Fn on the treatment of CRC.
The inhibitor of apoptosis proteins (IAPs) are characterized by the presence of baculoviral IAP repeat (BIR) domains which are important for the binding and inhibition of caspases [19–21]. They can promote the survival of tumor cells and induce chemoresistance [22]. Therefore, IAPs have attracted wide attention as potential targets for cancer therapy [23]. BIRC3 is a member of the IAP family that can inhibit apoptosis by directly inhibiting the caspase cascade [24, 25]. BIRC3 can also contribute to chemoresistance in malignancies including CRC [26]. Our previous study using microarray analysis showed that Fn can significantly induce BIRC3 expression in CRC cell lines. Based on this finding, we hypothesize that the significant upregulation of BIRC3 expression induced by Fn might be responsible for chemoresistance in CRC.
In this study, we demonstrate that Fn infection reduced the chemosensitivity of CRC cells to 5-Fu through upregulation of BIRC3 in vitro and in vivo, and high Fn abundance correlates with chemoresistance in advanced CRC patients who received standard 5-Fu-based adjuvant chemotherapy after radical surgery. Our evidence suggests that Fn and BIRC3 may serve as promising therapeutic targets for reducing chemoresistance to 5-Fu treatment in advanced CRC.
Methods
Bacteria strains and cell lines
Fn strain ATCC 25586 was purchased from American Type Culture Collection (ATCC) and grown in Columbia blood agar (Sigma, USA) in an anaerobic bag (Merier, France) at 37 °C as previously described [15]. HCT116, HT29 and 293 T cells were obtained from GeneChem and cultured in DMEM-F12(Gibco, USA) supplemented with 10% FBS (Gibco) and 1% penicillin and streptomycin (Beyotime, China) at 37 °C in a humidified 5% CO2 atmosphere. For Fn infection assay, cells were cultured in medium without antibiotics and incubated with Fn at a multiplicity of infection (MOI) of 100:1 as previously described [27].
Patients and specimens
A total of 94 patients diagnosed with advanced CRC were included in this study. All the patients received standard 5-Fu-based adjuvant chemotherapy after radical surgery in Fudan University Cancer Center from 2007 to 2017. None of them received preoperative treatment. Ninety-four formalin-fixed paraffin-embedded (FFPE) CRC tissues were obtained from the pathological archives. Prognostic information was collected by the medical record system and telephone follow-up. The median follow-up time was 38.5 months, ranging from 7 to 132 months. During the follow-up period, 45 patients (47.8%) suffered from recurrence of the disease. Clinicopathological data of the patients are summarized in Table 1. Written informed consent was obtained from the patients, and the study was approved by the Ethics Committee of the hospital.
Table 1.
Clinicopathological characteristics of CRCs in according to Fn abundance or recurrence status
| Characteristics | All cases | Fn-low/negative | Fn-high | P value | Non-recurrence | Recurrence | P value |
|---|---|---|---|---|---|---|---|
| (n = 94) | (n = 73) | (n = 21) | (n = 49) | (n = 45) | |||
| Age | 0.849 | 0.646 | |||||
| ≥ 60 | 42(44.7%) | 33 (45.2%) | 9 (42.9%) | 23 (46.9%) | 19 (42.2%) | ||
| <60 | 52 (55.3%) | 40 (54.8%) | 12(57.1%) | 26 (53.1%) | 26(57.8%) | ||
| Sex | 0.936 | 0.323 | |||||
| Female | 41 (43.6%) | 32 (43.8%) | 9 (42.9%) | 19 (38.8%) | 22 (48.9%) | ||
| Male | 53 (56.4%) | 41 (56.2%) | 12 (57.1%) | 30 (61.2%) | 23 (51.1%) | ||
| Tumor location | 0.051 | 0.079 | |||||
| Rectum | 54 (57.4%) | 42 (57.5%) | 12 (57.1%) | 23 (46.9%) | 31 (68.9%) | ||
| Distal colon | 23 (24.5%) | 21 (28.8%) | 2 (9.5%) | 16 (32.7%) | 7 (15.6%) | ||
| Proximal colon | 17 (18.1%) | 10 (13.7%) | 7 (33.3%) | 10 (20.4%) | 7 (15.6%) | ||
| Tumor size | 0.156 | 0.131 | |||||
| >4 cm | 41 (43.6%) | 29 (39.7%) | 12(57.1%) | 25 (51.0%) | 16 (35.6%) | ||
| ≤ 4 cm | 53 (56.4%) | 44 (60.3%) | 9 (42.9%) | 24 (49.0%) | 29 (64.4%) | ||
| Differentiation | 0.840 | 0.573 | |||||
| Well/Moderate | 75 (79.8%) | 59 (80.8%) | 16 (76.2%) | 38 (77.6%) | 37 (82.2%) | ||
| Poor | 19 (20.2%) | 14 (19.2%) | 5 (23.8%) | 11 (22.4%) | 8 (17.8%) | ||
| Vascular invasion | 0.874 | 0.003 | |||||
| Positive | 30 (31.9%) | 23 (31.5%) | 7 (33.3%) | 9 (18.4%) | 21 (46.7%) | ||
| Negative | 64 (68.1%) | 50 (68.5%) | 14 (66.7%) | 40 (81.6%) | 24 (53.3%) | ||
| Neural invasion | 0.808 | 0.029 | |||||
| Positive | 22 (23.4%) | 18 (24.7%) | 4 (19.0%) | 7 (14.3%) | 15(33.3%) | ||
| Negative | 72 (76.6%) | 55 (75.3%) | 17 (81.0%) | 42 (85.7%) | 30 (6.7%) | ||
| TNM stage | 0.130 | 0.153 | |||||
| II | 45 (47.9%) | 38 (52.1%) | 7 (33.3%) | 20 (40.8%) | 25 (33.3%) | ||
| III | 49 (52.1%) | 35 (47.9%) | 14 (66.7%) | 29 (59.2%) | 20 (66.7%) | ||
| BIRC3 staining | 0.008 | ||||||
| Positive | 36 (38.3%) | 21 (28.8%) | 15 (71.4%) | 0.000 | 25 (51.0%) | 11 (71.4%) | |
| Negative | 58 (61.7%) | 52 (71.2%) | 6 (28.6%) | 24 (49.0%) | 34 (28.6%) | ||
| TLR4 staining | 0.036 | ||||||
| Positive | 50 (53.2%) | 35 (47.9%) | 15 (71.4%) | 0.057 | 21 (42.9%) | 29 (64.4%) | |
| Negative | 44 (46.8%) | 38 (52.1%) | 6 (28.6%) | 28 (57.1%) | 16 (35.6%) | ||
| Fn abundance | 0.014 | ||||||
| Fn-low/negative | 73 (77.7%) | 43(83.6%) | 30 (71.1%) | ||||
| Fn-high | 21 (22.2%) | 6 (16.4%) | 15 (28.9%) | ||||
Pearson Chi-square test
Quantitative real-time PCR
For detection of Fn abundance, genomic DNA (gDNA) was extracted from FFPE tissues with QIAamp DNA FFPE Tissue Kit (QIAGEN, Germany). The abundance of Fn was determined by detecting the 16S gene using qPCR. Each 10-μL reaction contained 80 ng of gDNA, 0.4 mM each primer and 1× final concentration of SYBR Green PCR Master Mix (Thermo Fisher Scientific, USA). Amplification was performed using the ABI Step One Plus Real-Time PCR System (Applied Biosystems, USA) under the following reaction conditions: 10 min at 95 °C, followed by 40 cycles of 95 °C for 15 s and at 60 °C for 1 min. The gene prostaglandin transporter (PGT) was used as the internal reference as previously described [13]. For detection of target gene expression, total RNA was isolated using TRIzol reagent (Invitrogen, USA), and 1 mg of total RNA was reverse transcribed using a reverse transcription kit (Promega, USA). Complementary DNA (cDNA) was amplified and quantified on the ABI Step One Plus Real-Time PCR System (Applied Biosystems) using SYBR Green (Takara, China) under the following reaction conditions: 30 s at 95 °C and 40 cycles of 5 s at 95 °C and 30 s at 60 °C. Q-PCR was performed in triplicate for each sample, and the 2-ΔΔCt method was used to assess the relative expression levels of target genes as previously described [28]. GAPDH served as an internal reference gene and all the primer sequences are given in Additional file 1: Table S1.
Microarray analysis
Total RNA was isolated from HCT116 cells infected with or without Fn for 24 h. Then, the probes were prepared by labeling the mRNA with Cy5-dUTP (for the control group) or with Cy3-dUTP (for the infected group) through reverse transcription. The labeled cDNA probes were hybridized to a Human Whole Genome Microarray (Zhuolibiotech, China) and scanned with an Axon 4000 scanner (Molecular Devices, Sunnyvale, CA, USA). Bioinformatics analysis was performed. The list of differentially expressed genes was filtered for fold changes > 2.0, and those genes were selected for further investigation.
Cell proliferation assay and drug cytotoxicity assay
The percentage of viable cells was determined by Cell Counting Kit-8 (CCK8; Yeasen, China) assay under different treatment conditions. Briefly, cells were seeded in 96-well plates with 100 μL of culture medium. Ten microliters of CCK8 solution was added to each well at the indicated time points and incubated at 37 °C for 2 h. The reaction product was assessed by measuring the optical absorbance at 450 nm using a spectrophotometer (Thermo Fisher Scientific, USA).
Western blot analysis
Total cellular protein was extracted and separated by 10% SDS-PAGE then transferred to polyvinylidene fluoride (PVDF) membranes. Membranes were blocked with 5% BSA in TBST for 1 h at room temperature and then incubated with the primary antibodies overnight at 4 °C, followed by incubation with secondary antibodies for 2 h at room temperature. The signals were detected using an enhanced chemiluminescence reagent (Millipore, USA). The information for all antibodies used is listed in Additional file 2: Table S2.
Immunohistochemical staining and immunofluorescence staining
For immunohistochemistry (IHC), an EnVision+ Dual Link Kit (Dako, USA) was used according to the manufacturer’s instructions. The staining process was performed as previously described [29]. All sample slides were scored separately by two pathologists according to the distribution, intensity and percentage of positive cells [30]. Briefly, the staining intensity was scored as 0 (negative), 1 (weak), 2 (medium) or 3 (strong). The extent of staining was scored as 0 (< 5%), 1 (5–25%), 2 (26–50%), 3 (51–75%) or 4 (> 75%). The scores of intensity and extent were multiplied to generate the immunoreactivity score. Final staining scores 0–4 and 5–12 were considered negative and positive, respectively.
Fn-induced NF-κB activation was detected using an NF-κB Activation Nuclear Translocation Assay Kit (Beyotime, China) according to the manufacturer’s instructions [31]. Briefly, cells were fixed and washed followed by incubation with blocking buffer for 1 h. Then, cells were incubated with the primary antibody against NF-κB P65 overnight in a humidified chamber at 4 °C before a Cy3-conjugated secondary antibody was applied for 1 h at room temperature. Finally, nuclei of cells were stained with DAPI for 5 min. Images were captured by laser confocal microscopy at an excitation wavelength of 350 nm for DAPI (blue) and 540 nm for Cy3 (red), and merged using software.
Apoptosis detection
The cell apoptosis ratio was determined by flow cytometric analysis with an Annexin V FITC/PI double stain assay (BD Biosciences, USA) according to the manufacturer’s instructions. Levels of apoptosis in the xenograft tumor tissues were determined by terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) technology using an Apoptosis Assay Kit (Beyotime, China).
Small interfering RNA (siRNA) silencing in vitro
SiRNAs were purchased from GenePharma (Shanghai, China). Transfection of siRNA was performed using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s protocol and nonspecific siRNAs were used as negative controls. The sequences of siRNA used in this study are shown in Additional file 1: Table S1.
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were carried out using a ChIP Assay Kit (Beyotime, China) following by the manufacturer’s protocol. Briefly, cells were treated with 1% formaldehyde to cross-link DNA and proteins. Afterwards, the cell lysates were sonicated to generate chromatin fragments of 200–300 bp and immunoprecipitated with an anti-P65 antibody or IgG as a control. The region 5 kb upstream of BIRC3 gene was set as a negative control. The precipitated chromatin DNA was recovered and measured by qPCR. The percent input for each experiment was converted to fold change relative to an untreated control. Average fold changes from 3 independent experiments were then plotted. Primer sequences for the ChIP-qPCR assay are listed in Additional file 1: Table S1.
Dual luciferase reporter assay
The NF-κB P65 binding motif in the promoter region of BIRC3 was predicted by JASPAR (http://jaspar.genereg.net/). Two putative binding sites with the highest score were selected. Wild-type (WT) or mutated putative NF-κB P65 binding sites in the BIRC3 promoter region were amplified and cloned into the pGL3-Promoter vector (Additional file 3: Figure S1). 293 T cells were transfected with WT or mutated BIRC3 promoter construct and pcDNA-P65 or pcDNA-NC vector (GeneChem, China) using Lipofectamine 3000 (Invitrogen, USA). Luciferase activity was determined using a Dual Luciferase Reporter Assay Kit (Promega, USA) according to the manufacturer’s protocol.
Mouse model
For the xenograft experiments, 5-week-old male BALB/c nude mice (Chinese Academy of Sciences, Shanghai, China) were housed in laminar flow cabinets under specific pathogen-free conditions. To establish the CRC xenograft model., HCT116 (5 × 106 cells/0.5 ml) cells were subcutaneously injected into the right flank of the mice. One week after subcutaneous inoculation, mice were randomly divided into 4 groups for different sets of experiments (n = 5). Fn and siRNA were administered by multipoint intratumor injection every 3 days for a total of five injections using an in vivo transfection reagent (Entranster™-in vivo, Engreen, China) according to the instructions [32]. 5-Fu (Absin, China) was administered by an intraperitoneal injection of 5 mg/kg per mouse, twice per week for 2 weeks. Tumor volume (V) was calculated using the following formula: V = (a × b2)/2, where a and b represent the longer and shorter tumor diameters, respectively. After the 2-week treatment, all mice were sacrificed and the tumors were collected. The samples were fixed with formalin for IHC, TUNEL assay and hematoxylin and eosin (H&E) staining. The animal study was approved by the Committee on Animals Handling of Fudan University Shanghai Cancer Center.
Statistical analysis
Statistical analyses were performed using GraphPad Prism software 7.0 (GraphPad Software, La Jolla, CA). Data are presented as the means ± standard deviation. Unpaired Student t test or nonparametric Mann-Whitney test was applied to assess the statistical significance between groups with different treatment. Correlations were calculated according to Pearson Chi-square test All statistical tests were two-sided and p < 0.05 was considered statistically significant.
Results
Fn infection significantly induces expression of BIRC3 in CRC cell lines
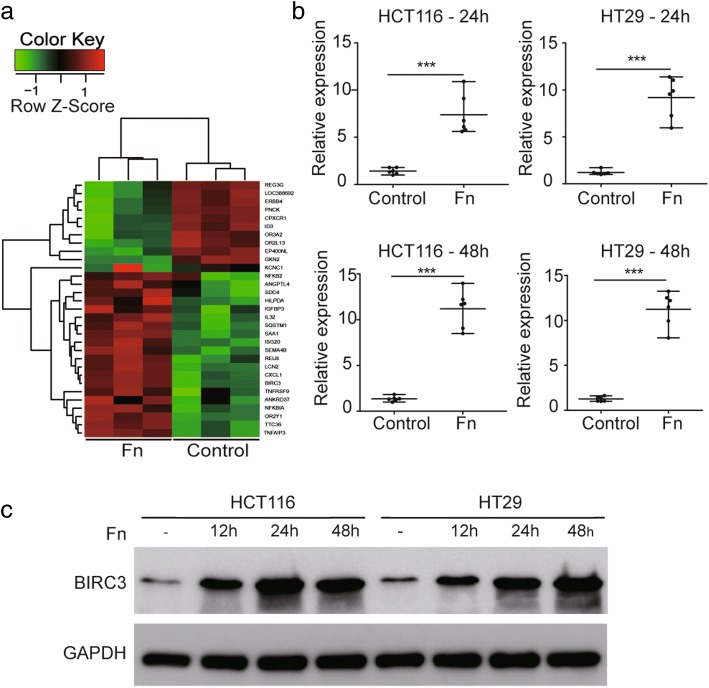
To assess the global effect of Fn infection in vitro, human CRC HCT116 cells were incubated with or without Fn for 24 h. Whole genome microarray analysis was performed and differentially expressed genes were analyzed. The result showed that BIRC3 was the gene with the highest expression (12-fold increase) induced by Fn infection compared with the control groups (Fig. 1a). To validate the upregulation of BIRC3 in CRC cell lines infected with Fn, qPCR and Western blot analysis were performed. The results showed that the mRNA levels of BIRC3 in HCT116 and HT29 cells were significantly upregulated by Fn infection for 24 h and 48 h respectively (Fig. 1b). Western blot analysis also showed that Fn infection increased the expression of BIRC3protein at different time intervals (Fig. 1c). As BIRC3 is a member of the IAP family [33], we also examined the protein levels of the other members of this family including BIRC2 (cIAP1), BIRC4 (XIAP) and BIRC5 (Survivin). The results showed that the protein levels of these genes was not affected by Fn infection (data not shown).
Fig. 1.
BIRC3 expression is upregulated by Fn infection in CRC cell lines. a Hierarchical clustering of the differentially expressed gene heatmap between HCT116 cells and Fn-infected HCT116 cells using Microarray analysis. These genes were affected by changes > 2-fold with p values below 0.05. BIRC3 was the gene with the highest expression (12-fold increase) induced by Fn infection (n = 3). b The relative mRNA expression of the BIRC3 gene determined by qPCR in CRC cell lines infected with or without Fn for 24 h and 48 h respectively. Data are shown as individual values with the median and range from three independent experiments performed in triplicate (n = 6) (***p < 0.001 by unpaired Student t test). c Representative Western blot for BIRC3 protein and GAPDH protein extracted from CRC cells infected with Fn for 0 h, 12 h, 24 h and 48 h
Fn induces chemoresistance of CRC cells to 5-Fu via upregulation of BIRC3 in vitro
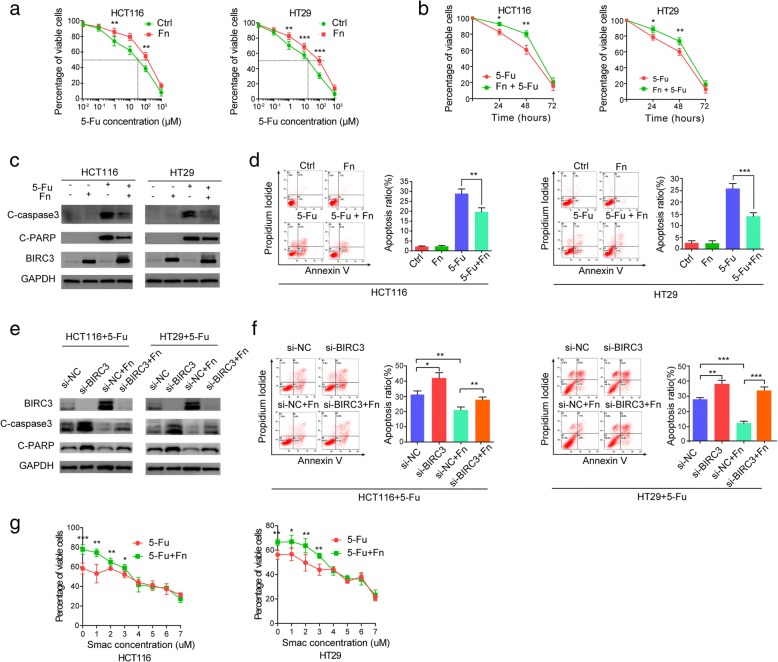
Since BIRC3 is a member of the IAP family that inhibits apoptosis by directly inhibiting caspase cascade [25], and can contribute to chemoresistance in malignancies including CRC [26, 34], we speculated that the upregulation of BIRC3 expression induced by Fn infection may lead to chemoresistance in CRC. To test this hypothesis, we initially determined whether Fn infection decreased cytotoxicity induced by 5-Fu in CRC cells. The results showed that Fn decreased cytotoxicity induced by 5-Fu in HCT116 and HT29 cells (Fig. 2a). We also found that the 50% inhibitory concentration (IC50) of 5-Fu in HCT116 and HT29 cells was approximately 30 μM and 20 μM, respectively (Fig. 2a). Based on this result, cells were incubated with or without Fn and then exposed to IC50 5-Fu and cell viability was assessed at the indicated time. Consistent with previous results, Fn infection significantly reduced cytotoxicity induced by 5-Fu treatment for 24 h or 48 h(Fig. 2b). To address whether Fn infection protects CRC cells from 5-Fu-mediated apoptosis, we cocultured HCT116 and HT29 cells with or without Fn in the presence or absence of IC50 5-Fu for 48 h. Western blotting showed that the cleavage of caspase3 (C-caspase3) and cleavage of PARP (C-PARP) was induced by 5-Fu, while this effect was attenuated by Fn infection (Fig. 2c). Flow cytometry analysis with Annexin V FITC/PI double stain assay was performed to assess the apoptosis ratio of each group. The results showed that 5-Fu-induced apoptosis was crippled by Fn infection (Fig. 2d). Taken together, these results suggest that Fn infection can reduce the chemosensitivity of CRC cells to 5-Fu in vitro.
Fig. 2.
Fn induces chemoresistance of CRC cells to 5-Fu via upregulation of BIRC3 in vitro. a Cells were incubated with or without Fn and then exposed to serial dilutions of 5-Fu for 48 h. Cell viability was determined by CCK8 assay. Data are presented as the percentage of viable cells (mean ± s.d.). The IC50 of 5-Fu in HCT116 and HT29 cells was approximately 30 μM and 20 μM respectively, as demonstrated by the dotted line. b Cells were incubated with or without Fn and then exposed to IC50 5-Fu. Cell viability was assessed by CCK8 assay at 0 h, 24 h, 48 h, and 72 h. Data are presented as the percentage of viable cells (mean ± s.d.). c-d Cells were incubated with or without Fn in the presence or absence of IC50 5-Fu for 48 h. Cleaved of caspase-3 (C-caspase-3), cleaved of PARP (C-PARP) and BIRC3 were detected by Western blot (c). Apoptosis was detected by flow cytometry. Data are presented as the ratio of apoptotic cells (mean ± s.d.). Representative quadrantal diagrams (left) and the corresponding histogram (right) from three independent experiments are shown (d). e-f Cells were transfected with specific siRNA for 24 h and then incubated with or without Fn in culture medium with IC50 5-Fu for an additional 48 h. Protein levels of BIRC3, C-caspase-3, and C-PARP were measured by Western blot (e). Apoptosis ration was detected with flow cytometry. Data are presented as the ratio of apoptotic cells (mean ± s.d.). Representative quadrantal diagrams (left) and the corresponding histogram (right) from three independent experiments are shown (f). g Cells were incubated with or without Fn and then exposed to gradient concentrations of a SMAC mimetic in the culture medium with IC50 5-Fu for 48 h. Cell viability was determined by CCK8 assay and the data are plotted. Data are presented as the percentage of viable cells (means±s.d.) from three independent experiments (*p < 0.05; **p < 0.01; ***p < 0.001 by unpaired Student t test)
To confirm the role of BIRC3 in the Fn-induced chemoresistance to 5-Fu, BIRC3 gene was silenced in CRC cells using specific siRNA. Cells were incubated with or without Fn and then exposed to IC50 5-Fu for 48 h. Western blotting showed that Fn infection crippled the level of C-caspase3 and C-PARP induced by 5-Fu, while this effect was abolished by silencing the BIRC3 gene in the cells (Fig. 2e). Under the same conditions, the apoptosis ratio of each group was detected with flow cytometry. We also observed that silencing of BIRC3 abrogated Fn-induced chemoresistance of the cells in response to 5-Fu (Fig. 2f). Furthermore, a small molecule antagonist of BIRC3 named SMAC mimetic (SM-406, Selleck, China) was used to antagonize the effect of BIRC3 [35, 36]. The results showed that the chemoresistance effect mediated by Fn gradually weakened with increasing of SMAC mimetic concentration in the cells (Fig. 2g). Altogether, these data strongly suggest that BIRC3 plays an essential role in the Fn-induced chemoresistance of CRC cells in response to 5-Fu in vitro.
The TLR4/NF-κB pathway regulates BIRC3 expression in CRC cells cocultured with Fn
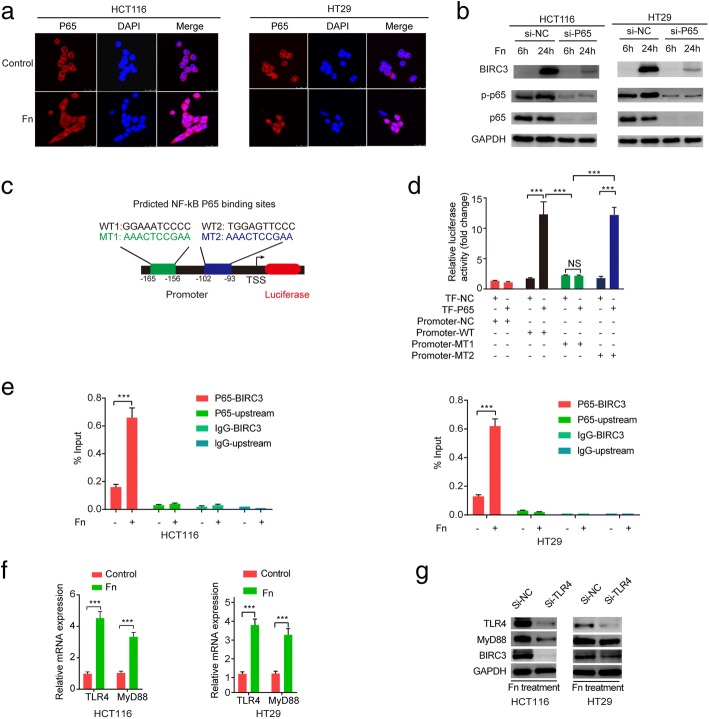
It was previously reported that Fn infection can activate the TLR4/NF-κB pathway in CRC cells [17, 37] and bioinformatics analysis of the microarray data also supported this conclusion. In addition, NF-κB activation induces the expression of a variety of target genes including BIRC3 [38, 39]. Based on this evidence, we speculated that the TLR4/NF-κB pathway may be involved in the Fn-induced upregulation of BIRC3. First, we observed NF-κB P65 nuclear translocation when the cells were incubated with Fn for 2 h. (Fig. 3a). Western blot analysis showed that Fn infection contributed to NF-κB P65 activation and upregulation of BIRC3, while silencing of NF-κB P65 abrogated the Fn-mediated upregulation of BIRC3 in the cells. (Fig. 3b). To examine the direct interaction between NF-κB P65 and the BIRC3 promoter region, a dual luciferase reporter assay was performed. The result showed that cotransfection of plasmids containing transcription factor P65 (TF-P65) and the wild-type (WT) pGL3-Promoter vector resulted in a significant increase in promoter activity compared to the transcription factor-negative (TF-NC) group (Fig. 3d), suggesting that NF-κB P65 positively regulates the activity of the BIRC3 promoter. To further explore the potential NF-κB P65 binding sites on the BIRC3 promoter region, two mutant type (MT) pGL3-Promoter vectors were generated (Fig. 3c and Additional file 3: Figure S1) and independently cotransfected with TF-P65 or TF-NC. We observed that cotransfection of the pGL3-MT2-Promoter plasmid resulted in the same effect on promoter activity as that with the pGL3-WT-Promoter plasmid, while this effect was not observed with the cotransfection of the pGL3-MT1-Promoter plasmid (Fig. 3d). Therefore, we may infer that the binding site is located 156 to 165 bp upstream of the transcription start site (TSS) (Fig. 3c). Furthermore, ChIP assays were performed for P65 in CRC cells. We found that recruitment of P65 to the BIRC3 promoter region was enhanced by Fn infection in the cells (Fig. 3e). To investigate whether TLR4 and MyD88 are involved in the Fn-mediated upregulation of BIRC3, we first confirmed that Fn infection can upregulate the levels of TLR4 and MyD88 transcripts in the cells. (Fig. 3f). Next, cells were transfected with specific siRNA (si-TLR4 or si-NC) for 24 h followed by coculture with Fn for additional 48 h. Western blotting analysis showed that silencing of TLR4 crippled Fn-mediated upregulation of BIRC3 in the cells.(Fig. 3g). Collectively, these results support the hypothesis that the Fn induces BIRC3 expression through TLR4/NF-κB pathway in CRC cells.
Fig. 3.
Fn induces BIRC3 expression through TLR4/NF-kb pathway. a Cells were cocultured with or without Fn for 2 h followed by incubation with an antibody against NF-κB P65 and a Cy3 fluorescein-conjugated secondary antibody, and nuclei were stained with DAPI. b Cells were transfected with specific siRNA (si-P65 or si-NC) for 24 h followed by cocultured with Fn for an additional 6 h or 24 h. Protein levels of BIRC3, p-P65, and P65 were measured by Western blot. c Schematic representation of the location of two predicted NF-κB P65 binding motifs in the BIRC3 promoter region. WT: wild-type; MT1: mutant type 1; MT2: mutant type 2. d pGL3-Promoter reporter plasmids (NC, WT, MT1, MT2) and pcDNA3.1-TF-cDNA vectors (TF-NC, TF-P65) were generated. Dual luciferase reporter assay were carried out according to different combinations. TF: transcription factor; NS: nonsignificant. e Cells were cocultured with or without Fn for 2 h. Then, ChIP assays were carried out with an anti-P65 antibody or rabbit IgG. qPCR was then carried out for the BIRC3 promoter region or the region 5 kb upstream of BIRC3 gene. f Cells were co-cultured with or without Fn for 24 h. The relative mRNA level of TLR4 and MyD88 was detected by qPCR. g Cells were transfected with specific siRNA (si-TLR4 or si-NC) for 24 h followed by co-cultured with Fn for an additional 48 h. Protein levels of TLR4, MyD88, and BIRC3 were measured by Western blot. (***p < 0.001 by unpaired Student’s t-test)
Fn induces chemoresistance of CRC cells in response to 5-Fu in vivo
A subcutaneous xenograft model was used to further explore whether Fn induces chemoresistance in CRC cells and to determine the role of BIRC3 in this effect in vivo. Four groups were designed according to the purpose of this experiment. The results showed that tumor growth was significantly decreased by 5-Fu treatment while this effect was crippled by Fn infection in vivo (Fig. 4a-b). Interestingly, silencing of BIRC3 partially attenuated the Fn-mediated chemoresistance of tumors in response to 5-Fu in vivo (Fig. 4a-b). These findings were further confirmed by TUNEL assay (Fig. 4c-d). To explore the relationship between Fn infection and the expression level of TLR4 and BIRC3 in vivo, IHC was carried out in tumor sections. We observed that Fn infection significantly upregulated the expression levels of TLR4 and BIRC3 (Fig. 4c-d). In conclusion, these data indicate that BIRC3 plays an essential role in the Fn-mediated chemoresistance of CRC cells in response to 5-Fu in vivo.
Fig. 4.
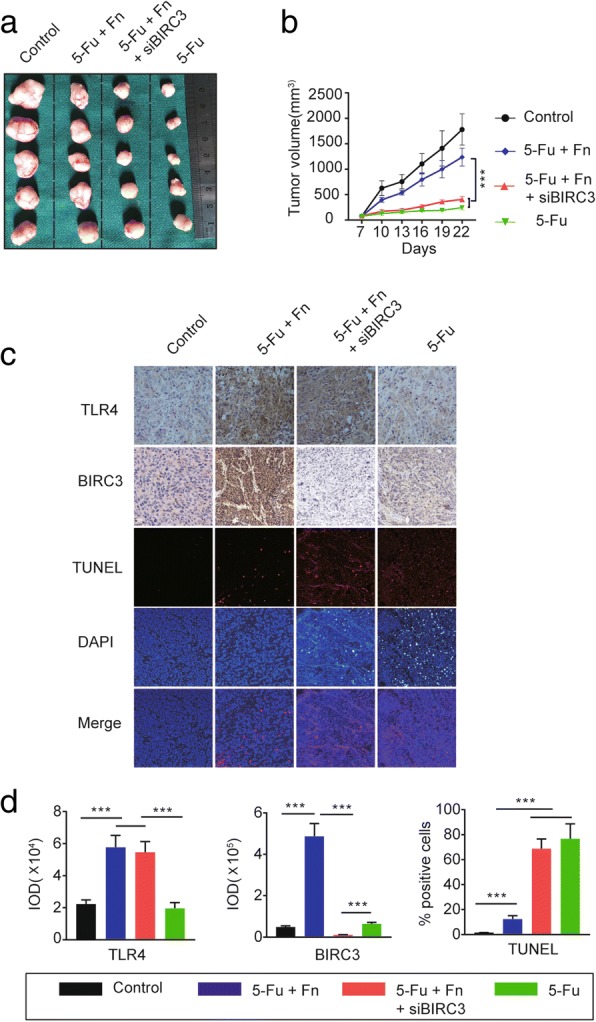
Fn induces chemoresistance of CRC cells in response to 5-Fu in vivo. a After the 2-week treatment, mice under different conditions were sacrificed and the tumors were collected and photographed. b Tumor sizes were measured every 3 days and tumor growth curves were established. Data are presented as tumor volume (mean ± s.d.; ***p < 0.001 by nonparametric Mann-Whitney test). c Immunohistochemical staining was carried out to detect the proteins expression levels of TLR4 and BIRC3, and a TUNEL assay was performed to detect tumor cell apoptosis in xenograft tumor tissues. d Integrated optical density (IOD) values were obtained using Image-Pro Plus software (Media Cybernetics, Rockville, MD, USA) and used to quantify the protein expression levels of TLR4 and BIRC3. Data are presented as IOD values. (mean ± s.d.; ***p < 0.001 by nonparametric Mann-Whitney test). As for TUNEL assay, data are presented as the percentage of TUNEL- positive cells per high-power field of the various tumor sections. (mean ± s.d.; ***p < 0.001 by nonparametric Mann-Whitney test)
High Fn abundance correlates with poor recurrence-free survival (RFS) in advanced CRC
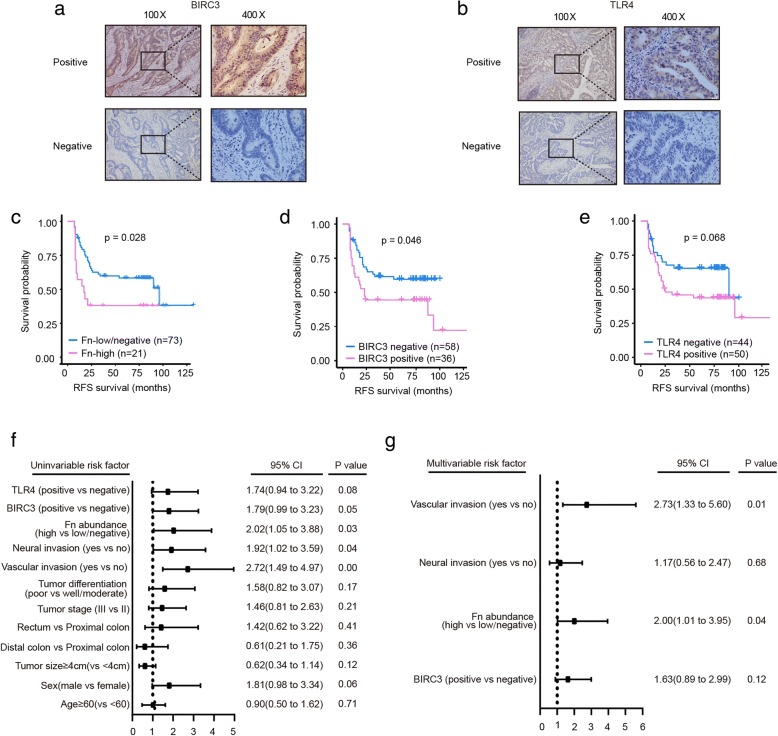
To investigate whether Fn infection and its downstream target genes can affect the efficacy of 5-Fu-based chemotherapy in CRC patients, we initially detected the Fn abundance and BIRC3 and TLR4 expression levels in FFPE CRC tissues from patients who were pathologically diagnosed with advanced CRC and received standard 5-Fu-based adjuvant chemotherapy after radical surgery. The distribution of Fn abundance is shown in Additional file 4: Figure S2. We set a cut-off value of 0.1 (2-ΔCt) for Fn abundance and divided the patients into the Fn-low/negative group and the Fn-high group as previously described [40]. Based on this division, we identified 22.3% of patients as having a high amount of Fn. The expression level of TLR4 and BIRC3 in FFEP tissues were determined by IHC assay. and divided into positive or negative status. Representative IHC images of BIRC3 and TLR4 proteins in FFEP CRC tissues were shown in Fig. 5a (BIRC3) and Fig. 5b (TLR4). As shown in Table 1, BIRC3 expression level was positively correlated with Fn abundance (71.4% in the Fn-high group vs. 28.8% in the Fn-low/negative group; p < 0.001). We also observed a trend of higher TLR4 expression in the Fn-high group (71.4%) than in the Fn-low/negative group (47.9%), although there was no statistical significance (p = 0.057). In line with our previous in vitro findings, these results suggested that Fn infection and its downstream target genes TLR4 and BIRC3 are clinically relevant in CRC patients.
Fig. 5.
High Fn abundance correlates with poor RFS in advanced CRC. a-b Representative IHC images of BIRC3 (a) and TLR4 (b) proteins in FFEP CRC tissues. c-e RFS was compared between patients with high and low/negative abundances of Fn (c), between the BIRC3 -positive group and BIRC3-negative group (d), and between the TLR4-positive group and TLR4-negative group (e) using Kaplan-Meier analysis and the log-rank test. f-g Univariate (f) and multivariable (g) analyses were performed in the 94 patients included. CI: confidence interval
Moreover, high Fn abundance (p = 0.014), high levels of TLR4 (p = 0.036) and BIRC3 (p = 0.008) proteins were more likely detected in patients with recurrence, compared with patients without recurrence.(Table 1). In addition, vascular invasion (p = 0.003) and neural invasion (p = 0.029) positively correlated with recurrence status (Table 1). Kaplan-Meier analysis showed that high amount of Fn (Fig. 5c, p = 0.028) and high level of BIRC3 expression (Fig. 5d, p = 0.046) correlated with poor RFS. However, the level of TLR4 expression was independent of RFS (Fig. 5e, p = 0.068). Univariate Cox regression analyses for RFS were performed and the results showed that the risk of recurrence was associated with high Fn abundance (HR = 2.02; 95% CI = 1.05 to 3.88; p = 0.03), positive vascular invasion (HR = 2.72; 95% CI = 1.49 to 4.97; p = 0.00) and positive neural invasion (HR = 1.92; 95% CI = 1.02 to 3.59; p = 0.04) (Fig. 5f). In the multivariate Cox regression analysis for RFS, the risk of recurrence was correlated with high amount of Fn (HR = 2.00; 95% CI = 1.01 to 3.95; p = 0.04) and positive vascular invasion (HR = 2.73; 95% CI = 1.33 to 5.60; p = 0.01) (Fig. 5g). Taken together, these results demonstrate that high Fn abundance is an independent risk factor for recurrence in advanced CRC patients who received standard 5-Fu-based adjuvant chemotherapy after radical surgery, and further support that upregulation of BIRC3 expression induced by Fn might be responsible for chemoresistance in CRC.
Discussion
Fluorouracil-based adjuvant chemotherapy is a standard treatment for advanced CRC patients [41]. 5-Fu chemoresistance is a major challenge and the prognosis for CRC patients can be very poor due to recurrence of disease [42]. However, the potential mechanisms involved in the resistance to chemotherapeutic drugs including 5-Fu are not fully understood. It has been reported that drug absorption disorder, changes in drug targets, activation of DNA repair pathways, inhibition of apoptosis, and the tumor microenvironment are involved in drug resistance [43, 44]. The mechanism of 5-Fu resistance is associated with the activity of enzymes involved in the modulation of 5-Fu metabolism, such as thymidylate synthase, thymidine phosphorylase and dihydropyrimidine dehydrogenase [41]. Microsatellite instability status also correlates with chemosensitivity to 5-Fu [45]. Interestingly, increasing evidence suggests that gut microbiota may affect the efficacy of antitumor drugs [10, 46]. In this study, we initially found that BIRC3 was the most upregulated gene induced by Fn infection in CRC cell lines. Moreover, BIRC3 is a member of the IAP family that inhibits apoptosis by directly inhibiting the caspase cascade [24, 25], which has potential to aid in the treatment of CRC [19]. As inhibition of apoptosis is an important mechanism of drug resistance [47], we speculated that the upregulation of BIRC3 induced by Fn may be associated with chemoresistance. It is well acknowledged that 5-Fu-based chemotherapy is the principal postoperative treatment for advanced CRC [48]. Therefore, we hypothesized that Fn infection and upregulation of BIRC3 may be involved in resistance to 5-Fu. To test this hypothesis, we first demonstrated that Fn infection reduced the chemosensitivity of CRC cells to 5-Fu in vitro and in vivo. Next, we confirmed that this biological effect depends on the upregulation of BIRC3. Furthermore, we dissected the mechanisms involved in Fn-induced BIRC3 expression, which showed that the TLR4/NF-κB pathway regulates BIRC3 expression in CRC cells that are cocultured with Fn. Finally, to correspond to the aforementioned findings, we investigated the clinical significance of Fn infection and its downstream target genes. The results showed that a high amount of Fn was an independent risk factor for recurrence in advanced CRC patients.
Many studies have reported that overexpression of BIRC3 is associated with chemoresistance in the treatment of malignancies. Karasawa et al. found that BIRC3-positive patients had a shorter disease-free survival after 5-Fu-based chemotherapy [26] and suggested that BIRC3 can serve as a therapeutic target in CRC and other malignancies [49]. Krajewska et al. found that elevated expression of BIRC3 significantly correlated with shorter overall survival in early-stage CRC [50]. In addition, overexpression of BIRC3 significantly correlated with resistance to several chemotherapeutic drugs, including 5-Fu, in pancreatic cancer cells [34]. Upregulation of BIRC3 induced by IL-1β results in chemoresistance to doxorubicin in breast cancer cells [51]. Consistent with these findings, our results showed that a high level of BIRC3 induced by Fn infection contributed to chemoresistance to 5-Fu in vitro, and correlated with poor RFS in CRC patients. A recent study reported that Fn can mediate chemoresistance by autophagy pathway activation in CRC [52]. In line with this, our study observed similar phenomena elucidated with different mechanism. Another study found that Fn can migrate with CRC cells to metastatic sites and that CRC patients may benefit from eradication of Fn with antibiotic treatment [53]. Therefore, we may infer that Fn as a part of metastatic tumor plays a role in the chemoresistance to 5-Fu treatment and contributes to disease recurrence. However, there are some limitations in this research. First, this study just focuses on Fn enriched in tumor tissues and the indirect effect of Fn on 5-Fu. It is still unknown that whether Fn inhabiting in intestinal lumen of CRC patients can directly affect the efficacy of 5-Fu. Second, subcutaneous xenograft cannot simulate intestinal microenvironment very well. So, further study is needed to investigate the direct interactions between Fn and chemotherapeutic drugs.
Conclusions
In conclusion, our study initially found that Fn infection reduces the chemosensitivity of CRC cells to 5-Fu through the upregulation of BIRC3, and that the TLR4/NF-κB pathway regulates BIRC3 expression in CRC cells cocultured with Fn (Fig. 6). Targeting Fn infection and BIRC3 may be promising in the treatment of advanced CRC patients who received 5-Fu-based chemotherapy after radical surgery, and may optimize the current treatment strategy of CRC.
Fig. 6.
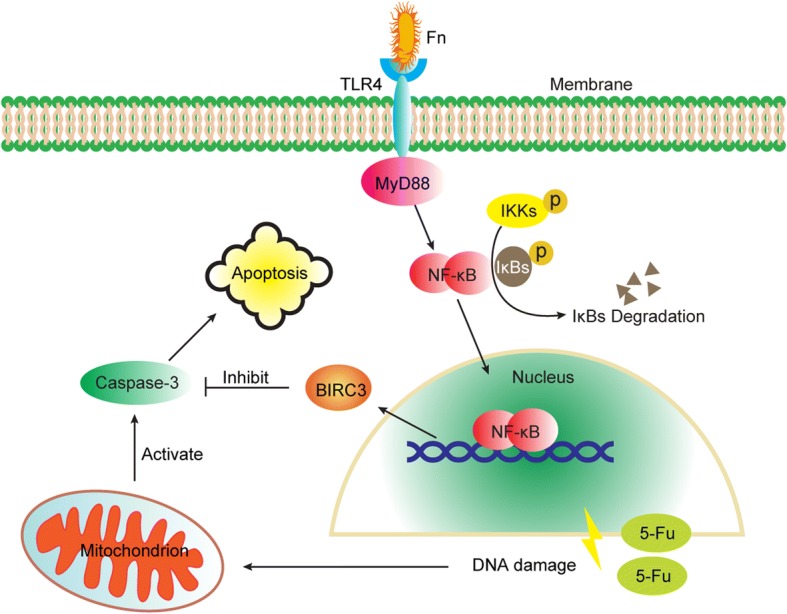
Schematic diagram of the mechanism by which Fn protects CRC cells from 5-Fu-mediated apoptosis via upregulation of BIRC3 expression
Additional files
Table S1. Primer sequences of qPCR and ChIP-qPCR and siRNA sequences used in the study. (XLSX 10 kb)
Table S2. Information of antibody used in the study. (XLSX 9 kb)
Figure S1. The peak map of DNA sequencing about the constructions of wild-type and mutant BIRC3 promoters. (PDF 712 kb)
Figure S2. Distribution of Fn abundance in CRC patients (n = 94). The relative abundance of Fn in FFPE CRC tissues was determined by 2-ΔCt. The patients were ranked according to abundance of Fn. A cut-off value of 0.1 was set to distinguish the high Fn abundance group (n = 21) from the low/negative Fn abundance group(n = 73). (TIF 508 kb)
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No.81001069). Fudan Outstanding Young Talent Training Plan (No.YJYQ201601), and Shanghai Pujiang Program (No.17PJD007).
Availability of data and materials
All data are included in the manuscript and the additional files.
Abbreviations
- 5-Fu
5-fluorouracil
- ATCC
American Type Culture Collection
- CCK8
Cell Counting Kit-8
- ChIP
Chromatin immunoprecipitation assay
- CRC
Colorectal cancer
- FFPE
Formalin-fixed paraffin-embedded
- Fn
Fusobacterium nucleatum
- gDNA
Genomic DNA
- IAP
Inhibitor of apoptosis protein
- IC50
50% inhibitory concentration
- IHC
Immunohistochemistry
- MOI
Multiplicity of infection
- MT
Mutant type
- PGT
Gene prostaglandin transporter
- qPCR
Quantitative real-time polymerase chain reaction
- RFS
Recurrence-free survival
- siRNA
Small interfering RNA
- TF-NC
Transcription factor-negative control
- TF-P65
Transcription factor-P65
- TSS
Transcription start site
- TUNEL
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling
- WT
Wild-type
Authors’ contributions
SC and YM conceived the experiments. SZ, YY, WW and BG conducted the experiments. SZ and GC analyzed and interpreted the data. SZ wrote the manuscript and prepared the figures. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Fudan Cancer Hospital and in accordance with the ethical standards formulated in the Declaration of Helsinki. Informed consent was obtained from all patients included in the study. All animal studies were approved by the Committee on Animals Handling of Fudan University Shanghai Cancer Center.
Consent for publication
All authors approved of the manuscript and consented to its publication.
Competing interests
The authors declare that have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sheng Zhang, Email: zhangs16@fudan.edu.cn.
Yongzhi Yang, Email: yzyangsjtu@msn.com.
Wenhao Weng, Email: wengwenhao2010@163.com.
Bomin Guo, Email: gbm1205@126.com.
Guoxiang Cai, Email: gxcai2015@163.com.
Yanlei Ma, Phone: +86 21 64175590, Email: yanleima@fudan.edu.cn.
Sanjun Cai, Phone: +86 21 64175590, Email: sanjuncai@hotmail.com.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Garcia-Castillo V, Sanhueza E, McNerney E, Onate SA, Garcia A. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J Med Microbiol. 2016;65(12):1347–1362. doi: 10.1099/jmm.0.000371. [DOI] [PubMed] [Google Scholar]
- 4.Wu N, Yang X, Zhang R, Li J, Xiao X, Hu Y, Chen Y, Yang F, Lu N, Wang Z, et al. Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb Ecol. 2013;66(2):462–470. doi: 10.1007/s00248-013-0245-9. [DOI] [PubMed] [Google Scholar]
- 5.Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol. 2014;12(10):661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 6.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zitvogel L, Ayyoub M, Routy B, Kroemer G. Microbiome and Anticancer Immunosurveillance. Cell. 2016;165(2):276–287. doi: 10.1016/j.cell.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Routy B, Gopalakrishnan V, Daillere R, Zitvogel L, Wargo JA, Kroemer G. The gut microbiota influences anticancer immunosurveillance and general health. Nat Rev Clin Oncol. 2018;15(6):382–396. doi: 10.1038/s41571-018-0006-2. [DOI] [PubMed] [Google Scholar]
- 10.Coleman OI, Nunes T. Role of the microbiota in colorectal Cancer: updates on microbial associations and therapeutic implications. Biores Open Access. 2016;5(1):279–288. doi: 10.1089/biores.2016.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott TA, Quintaneiro LM, Norvaisas P, Lui PP, Wilson MP, Leung KY, Herrera-Dominguez L, Sudiwala S, Pessia A, Clayton PT, et al. Host-microbe co-metabolism dictates Cancer drug efficacy in C. elegans. Cell. 2017;169(3):442–456. doi: 10.1016/j.cell.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Gonzalez AP, Ritter AD, Shrestha S, Andersen EC, Yilmaz LS, Walhout AJM. Bacterial metabolism affects the C. elegans response to Cancer chemotherapeutics. Cell. 2017;169(3):431–441. doi: 10.1016/j.cell.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mima K, Nishihara R, Qian ZR, Cao Y, Sukawa Y, Nowak JA, Yang J, Dou R, Masugi Y, Song M, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980. doi: 10.1136/gutjnl-2015-310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, et al. Binding of the Fap2 protein of fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum increases proliferation of colorectal Cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152(4):851–866. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Cai S, Ma Y. Association between fusobacterium nucleatum and colorectal cancer: Progress and future directions. J Cancer. 2018;9(9):1652–1659. doi: 10.7150/jca.24048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura K, Fujibuchi W, Ishida K, Naitoh T, Ogawa H, Ando T, Yazaki N, Watanabe K, Haneda S, Shibata C, et al. Inhibitor of apoptosis protein family as diagnostic markers and therapeutic targets of colorectal cancer. Surg Today. 2011;41(2):175–182. doi: 10.1007/s00595-010-4390-1. [DOI] [PubMed] [Google Scholar]
- 20.Deveraux QL, Reed JC. IAP family proteins--suppressors of apoptosis. Genes Dev. 1999;13(3):239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 21.Altieri DC. Survivin and IAP proteins in cell-death mechanisms. Biochem J. 2010;430(2):199–205. doi: 10.1042/BJ20100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang CY, Cusack JC, Jr, Liu R, Baldwin AS., Jr Control of inducible chemoresistance: enhanced anti-tumor therapy through increased apoptosis by inhibition of NF-kappaB. Nat Med. 1999;5(4):412–417. doi: 10.1038/7410. [DOI] [PubMed] [Google Scholar]
- 23.LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27(48):6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- 24.Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS., Jr NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 1998;281(5383):1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 25.Park SM, Yoon JB, Lee TH. Receptor interacting protein is ubiquitinated by cellular inhibitor of apoptosis proteins (c-IAP1 and c-IAP2) in vitro. FEBS Lett. 2004;566(1–3):151–156. doi: 10.1016/j.febslet.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 26.Karasawa H, Miura K, Fujibuchi W, Ishida K, Kaneko N, Kinouchi M, Okabe M, Ando T, Murata Y, Sasaki H, et al. Down-regulation of cIAP2 enhances 5-FU sensitivity through the apoptotic pathway in human colon cancer cells. Cancer Sci. 2009;100(5):903–913. doi: 10.1111/j.1349-7006.2009.01112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendes RT, Nguyen D, Stephens D, Pamuk F, Fernandes D, Van Dyke TE, Kantarci A. Endothelial cell response to fusobacterium nucleatum. Infect Immun. 2016;84(7):2141–2148. doi: 10.1128/IAI.01305-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Huang L, Zhai E, Cai S, Lin Y, Liao J, Jin H, Peng S, Xu L, Chen M, Zeng Z. Stress-inducible Protein-1 promotes metastasis of gastric cancer via Wnt/beta-catenin signaling pathway. J Exp Clin Cancer Res. 2018;37(1):6. doi: 10.1186/s13046-018-0676-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L, Shi S, Guo Z, Zhang X, Han S, Yang A, Wen W, Zhu Q. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS One. 2013;8(6):e65539. doi: 10.1371/journal.pone.0065539. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Xu Z, Lin S, Wu W, Tan H, Wang Z, Cheng C, Lu L, Zhang X. Ghrelin prevents doxorubicin-induced cardiotoxicity through TNF-alpha/NF-kappaB pathways and mitochondrial protective mechanisms. Toxicology. 2008;247(2–3):133–138. doi: 10.1016/j.tox.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Wang F, Zhang P, Ma Y, Yang J, Moyer MP, Shi C, Peng J, Qin H. NIRF is frequently upregulated in colorectal cancer and its oncogenicity can be suppressed by let-7a microRNA. Cancer Lett. 2012;314(2):223–231. doi: 10.1016/j.canlet.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 33.Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, Shiels H, Hardwick JM, Thompson CB. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. EMBO J. 1996;15(11):2685–2694. doi: 10.1002/j.1460-2075.1996.tb00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopes RB, Gangeswaran R, McNeish IA, Wang Y, Lemoine NR. Expression of the IAP protein family is dysregulated in pancreatic cancer cells and is important for resistance to chemotherapy. Int J Cancer. 2007;120(11):2344–2352. doi: 10.1002/ijc.22554. [DOI] [PubMed] [Google Scholar]
- 35.Fulda S. Promises and challenges of Smac mimetics as Cancer therapeutics. Clin Cancer Res. 2015;21(22):5030–5036. doi: 10.1158/1078-0432.CCR-15-0365. [DOI] [PubMed] [Google Scholar]
- 36.Duckett CS. IAP proteins: sticking it to Smac. Biochem J. 2005;385(1):e1–e2. doi: 10.1042/BJ20041800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abreu MT, Peek RM., Jr Gastrointestinal malignancy and the microbiome. Gastroenterology. 2014;146(6):1534–1546. doi: 10.1053/j.gastro.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Wang X, Evers BM. Induction of cIAP-2 in human colon cancer cells through PKC delta/NF-kappa B. J Biol Chem. 2003;278(51):51091–51099. doi: 10.1074/jbc.M306541200. [DOI] [PubMed] [Google Scholar]
- 39.Chu ZL, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW. Suppression of tumor necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-kappaB control. Proc Natl Acad Sci U S A. 1997;94(19):10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3(5):330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 42.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 43.Sheikh R, Walsh N, Clynes M, O'Connor R, McDermott R. Challenges of drug resistance in the management of pancreatic cancer. Expert Rev Anticancer Ther. 2010;10(10):1647–1661. doi: 10.1586/era.10.148. [DOI] [PubMed] [Google Scholar]
- 44.Niero EL, Rocha-Sales B, Lauand C, Cortez BA, de Souza MM, Rezende-Teixeira P, Urabayashi MS, Martens AA, Neves JH, Machado-Santelli GM. The multiple facets of drug resistance: one history, different approaches. J Exp Clin Cancer Res. 2014;33:37. doi: 10.1186/1756-9966-33-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 2010;28(20):3219–3226. doi: 10.1200/JCO.2009.27.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baguley BC. Multiple drug resistance mechanisms in cancer. Mol Biotechnol. 2010;46(3):308–316. doi: 10.1007/s12033-010-9321-2. [DOI] [PubMed] [Google Scholar]
- 48.Midgley R, Kerr D. Colorectal cancer. Lancet. 1999;353(9150):391–399. doi: 10.1016/S0140-6736(98)07127-X. [DOI] [PubMed] [Google Scholar]
- 49.Miura K, Karasawa H, Sasaki I. cIAP2 as a therapeutic target in colorectal cancer and other malignancies. Expert Opin Ther Targets. 2009;13(11):1333–1345. doi: 10.1517/14728220903277256. [DOI] [PubMed] [Google Scholar]
- 50.Krajewska M, Kim H, Kim C, Kang H, Welsh K, Matsuzawa S, Tsukamoto M, Thomas RG, Assa-Munt N, Piao Z, et al. Analysis of apoptosis protein expression in early-stage colorectal cancer suggests opportunities for new prognostic biomarkers. Clin Cancer Res. 2005;11(15):5451–5461. doi: 10.1158/1078-0432.CCR-05-0094. [DOI] [PubMed] [Google Scholar]
- 51.Mendoza-Rodriguez M, Arevalo Romero H, Fuentes-Panana EM, Ayala-Sumuano JT, Meza I. IL-1beta induces up-regulation of BIRC3, a gene involved in chemoresistance to doxorubicin in breast cancer cells. Cancer Lett. 2017;390:39–44. doi: 10.1016/j.canlet.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. Fusobacterium nucleatum promotes Chemoresistance to colorectal Cancer by modulating autophagy. Cell. 2017;170(3):548–563. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, et al. Analysis of fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017;358(6369):1443–1448. doi: 10.1126/science.aal5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primer sequences of qPCR and ChIP-qPCR and siRNA sequences used in the study. (XLSX 10 kb)
Table S2. Information of antibody used in the study. (XLSX 9 kb)
Figure S1. The peak map of DNA sequencing about the constructions of wild-type and mutant BIRC3 promoters. (PDF 712 kb)
Figure S2. Distribution of Fn abundance in CRC patients (n = 94). The relative abundance of Fn in FFPE CRC tissues was determined by 2-ΔCt. The patients were ranked according to abundance of Fn. A cut-off value of 0.1 was set to distinguish the high Fn abundance group (n = 21) from the low/negative Fn abundance group(n = 73). (TIF 508 kb)
Data Availability Statement
All data are included in the manuscript and the additional files.