ABSTRACT
The biological activities and the structural arrangement of adevonin, a novel antimicrobial peptide, were investigated. The trypsin inhibitor ApTI, isolated from Adenanthera pavonina seeds, was used as a template for screening 18-amino acid peptides with predicted antimicrobial activity. Adevonin presented antimicrobial activity and minimum inhibitory concentrations (MIC) ranging from 1.86 to 7.35 µM against both Gram-positive and – negative bacterial strains. Moreover, adevonin exerted time-kill effects within 10 min and both susceptible and drug-resistant bacterial strains were affected by the peptide. In vitro and in vivo assays showed that, at MIC concentration, adevonin did not affect human fibroblasts (MRC-5) viability or Galleria mellonella survival, respectively. Hemolytic activity was observed only at high peptide concentrations. Additionally, nucleic acid efflux assays, gentian violet uptake and time-kill kinetics indicate that the antimicrobial activity of adevonin may be mediated by bacterial membrane damage. Furthermore, molecular dynamic simulation in the presence of SDS micelles and anionic membrane bilayers showed that adevonin acquired a stable α-helix secondary structure. Further studies are encouraged to better understand the mechanism of action of adevonin, as well as to investigate the anti-infective activity of this peptide.
KEYWORDS: Adenanthera pavonina, antimicrobial peptide, rational design, α-helical
1. Introduction
The emergence and spreading of multidrug-resistant bacteria strains have increased in recent decades. Known as ‘superbugs’ [1,2], these bacteria strains display resistance against different antibiotic classes and their appearance has spread at alarming rates. High bacterial resistance levels present a potential public health risk difficulting infectious diseases control that further aggravating clinical conditions. In order to address this global threat, research and development of new antimicrobial drugs with mechanisms of action that differ from traditional antibiotics are of great importance [3]. In this context, antimicrobial peptides (AMPs) represent a potential class of molecules to be used in the prevention and control of bacterial infections.
AMPs are a class of antimicrobials with broad-spectrum activities, usually composed of 10 to 50 amino acids, also presenting an overall positive charge and large hydrophobic patches [4–6]. They are classified into 4 major structural categories, according to the secondary structure adopted during the interaction with the bacterial membrane, including α-helix, β-sheet, loop coil, and extended structural arrangements [7]. In addition to the AMPs obtained from natural sources, numerous AMPs have been designed based on natural peptides [8]. As a result of knowledge derived from natural peptides and database screening, shorter and more specific AMPs have been designed. The rational design and chemical synthesis of AMPs may overcome issues related to stability, bioavailability and their resistance to proteolysis [9]. For AMPs to become a therapeutic option several challenges must be overcome, including antibacterial effectiveness in vitro and in vivo, along with low toxicity rates at the therapeutic dose [10]. The use of innovative rational and computational design strategies have the potential to increase the discovery of novel therapeutic peptides at a lower cost, with enhanced effectiveness and broad-spectrum activities [10,11].
Here we report the rational design of a new AMP derived from the Adenanthera pavonina trypsin inhibitor (ApTI) using prediction tools, including Collection of Anti-Microbial Peptides (CAMPR3) [12] and The Antimicrobial Peptide Database (APD3) [13]. ApTI belongs to the Kunitz inhibitor family, and is composed of two polypeptide chains; a large chain of 16 kDa and a smaller one (5 kDa), which are linked by a disulfide bridge [14]. Since trypsin inhibitors can have several biological activities, including antifungal and antimicrobial, we investigated the bactericidal properties of ApTI. Analyzing the ApTI amino acid sequence we found peptide sequences with predicted antimicrobial activity, homologous to AMPs with α-helical secondary structure. From the ApTI amino acid sequence, AMPs were predicted selecting peptides containing 18 amino acid residues in length with the intention of forming a complete 5-turn α-helix. The first predicted AMP comprised the amino acid residues 8–25 from ApTI. This peptide was further synthesized and antimicrobial assays were performed. As a result, no antimicrobial activity was observed and, for this fact, we carried out amino acid substitutions to increase the net positive charge and hydrophobicity features of the peptide, thus obtaining a rationalized AMP denominated adevonin. We, therefore, describe the design, antimicrobial and cytotoxic activities, as well as structural prediction of adevonin. Moreover, the in vivo effect of adevonin on Galleria mellonella larvae was evaluated. Furthermore, we investigated whether the mechanism of action of adevonin involves damage of bacterial membranes.
2. Material and methods
2.1. Rational design
The protein sequence of ApTI (A. pavonina trypsin inhibitor) was used for the prediction of peptides containing 18 amino acid residues. The prediction was made through the CAMPR3 database [12]. The sequence located between residues 8–25 was selected based on SVM, RF, ANN and DA algorithms. Modifications of residues and their locations were made with the objective of forming a peptide that acquired a net positive charge and a hydrophobic portion to determine its antimicrobial activity. Helical wheel diagrams were built to guide the rearrange of the amino acid residues within the adevonin sequence. The APD3 [13] was used to obtain parameters of hydrophobic ratio, the net charge of the molecule and the Boman index. The peptide generated was denominated adevonin.
2.2. Peptide synthesis
The adevonin peptide and the original template peptide were purchased from Genone (Rio de Janeiro, RJ, Brazil), which determined the intact mass as well as the degree of purity of the peptide. The synthesis was performed using the solid-phase methodology.
2.3. Antimicrobial activities
The antimicrobial activities of adevonin, the template peptide, and ApTI were evaluated by the broth microdilution method [15]. The bacterial inoculum was prepared using the direct growth method. The followed bacteria strains were assayed: Escherichia coli (ATCC 35218), Klebsiella oxytoca (ATCC 13182), Klebsiella pneumoniae (ATCC 70603), Serratia marcesens (ATCC 13880), Klebsiella pneumoniae KpC+ (001825971 isolated) and Staphylococcus aureus (ATCC 80958). Isolated colonies on Mueller Hinton agar (MHA) were placed in sterile 0.9% NaCl solution until it reached turbidity equal to 0.5 on the MacFarland scale; absorbance was read at 625 nm. The suspension was diluted 1:20, to provide 5 × 106 colony forming units (CFU) per mL−1. A stock aliquot of the peptides was prepared in sterile 0.9% NaCl solution. From this solution, serial dilutions were made. Eighty μl of Mueller Hinton broth (MHB), 10 µL of the peptide solution and 10 μl of the bacterial inoculum were added to each well of the microplate. As a positive control, 80 µL MH broth, 10 µL chloramphenicol at 125 µM and 10 µL of the bacterial inoculum were added to the well. As a negative control, 80 µl of MH broth and 10 µL of sterile 0.9% NaCl solution and 10 µL of the bacterial inoculum were added to the well. The sterility control contained 100 µL of MH broth. Samples and controls were performed in triplicate. The microplate was incubated at 37°C with stirring. Absorbance readings were performed at 595 nm at 30-minute intervals on a Multiskan GO microplate reader. The percentage of inhibition of bacterial growth was calculated using the last reading of the exponential phase of bacterial growth: % inhibition of growth = [1 − (Sample/Acn)] × 100, where Sample is the absorbance of the sample and Acn is the absorbance of the negative control. The minimal inhibitory concentration (MIC) was determined as the lowest concentration at which bacterial growth was inhibited by 100%, using adevonin in concentrations of 29.6 to 1.84 µM. All experiments were performed in triplicate.
2.4. Hemolytic assays
Type O human erythrocytes were washed three times with 0.15 M PBS, followed by two centrifugations for 8 min at 1,381 × g and a centrifugation for 8 min at 2,455 × g. After washing, the erythrocytes were resuspended to 0.5% (v/v) in 0.15 M phosphate buffered saline (PBS). The erythrocyte suspension (750 µL) was added to microtubes along with the solution containing the adevonin peptide (750 µL). The peptide was diluted serial dilutions ranging from 350 to 2.73 μM. The microtubes were incubated at 37°C for 1 h. After incubation, the microtubes were centrifuged for 10 min at 2,455 × g. The supernatants were transferred to a 96-well polystyrene microplate and absorbance read at 414 nm. Triton X-100 (0.06%) was used as a positive control (100% hemolysis) and PBS was used as a negative control [16].
2.5. Cytotoxic activity analysis
The human fibroblast line from lung tissue MRC-5 was kindly provided by Dr. Emerson S. Lima (Federal University of Amazonas, Manaus, AM, Brazil). Cells were cultured in RPMI-1640 medium and DMEM (Gibco, Brazil), respectively supplemented with 10% fetal bovine serum (FBS), 100 U.mL−1 of penicillin and 100 μg.mL−1 of streptomycin (Gibco, Brazil) at 37°C in a 5% CO2 incubator. Cytotoxicity was evaluated according to Mosman [17], based on the enzymatic reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) (Sigma) to form crystals of formazan by mitochondrial and cellular dehydrogenase enzymes. Cells (6x103 cells/well) were grown in 96-well tissue culture plates and exposed to various concentrations of adevonin (10–0.3 μM) for 24, 48 and 72 h. After the incubation period, the cells were washed with PBS, 100 μL of MTT solution (1 mg.mL−1 diluted in culture medium) were added. After 4 h of incubation, the formazan crystals were resuspended with 100 μL of dimethylsulfoxide (DMSO) and read at 630 nm on the ThermoPlate TP-READER reader. Three independent experiments were performed in triplicate. Cell viability was calculated from the following formula: Cell viability (%) = (Abs sample/Abs negative control) x100.
2.6. G. mellonella model studies
The whole cycle of G. mellonella was maintained at 28°C. Insects were fed with an artificial diet consisting of honey and several flours. Larvae weighing 220–260 mg were randomly selected to comprise groups of ten larvae. Adevonin was diluted in sterile PBS to the concentration of 100 mg.kg−1 of larvae. Sterile PBS was used as control. Sample (10 µL) was inoculated by injection into the hemocoel via the last right proleg, using a Hamilton syringe (Sigma-Aldrich). Petri dishes were maintained at 37°C and were observed daily during 120 h; larvae were considered as dead when they did not respond to touch [18].
2.7. Time-dependent bactericidal activity evaluation
A bacterial culture of K. oxytoca (ATCC 13182) and S. aureus (ATCC 80958) were incubated overnight in a MHA plate. Isolated colonies were diluted to match 0.5 McFarland standards and treated with the MIC concentration of adevonin for K. oxytoca (1.84 µM) and S. aureus (7.35 µM), respectively. The control used was a bacterial suspension with no treatment. Aliquots were withdrawn from each tube and transferred to MHA plates at time intervals of 0, 5, 10, 15, 20, 25, 30, 60, 90 and 120 min. A control of assay was carried out with chloramphenicol (4 ug.mL−1) at the same time intervals. The plates were incubated overnight a 37°C and counts of CFU were performed [19].
2.8. Efflux of UV-absorbing materials
About 5 ml of bacterial culture of K. oxytoca (ATCC 13182) and S. aureus (ATCC 80958) were incubated overnight in Brain heart infusion (BHI) with the cell density adjusted with PBS (pH 7.4) to an OD620 nm of 2.0. The suspension was centrifuged at 400 × g for 14 min, the supernatant was discarded and the pellet was washed twice and resuspended in PBS (pH 7.4). One-hundred-and-eighty µl of the bacterial suspension were added to 20 µL adevonin peptide solution (1.84 µM and 7.35 µM for K. oxytoca and S. aureus, respectively). The untreated bacterial suspension was used as a control. After incubation at 37°C for 60 min, bacteria were centrifuged at 13,400 × g for 15 min, the supernatant was collected and the absorbance read at 260 nm [20].
2.9. Uptake of gentian violet
About 5 mL of K. oxytoca (ATCC 13182) and S. aureus (ATCC 80958) bacterial culture was incubated overnight in BHI. The bacterial suspension was centrifuged at 4,500 × g for 5 min at 4°C. The supernatant was discarded and the pellet washed twice and resuspended in PBS (pH 7.4). The bacterial suspension was adjusted to 0.5 on the McFarland scale, determined with a turbidimeter. One-hundred-and-eighty µl of the bacterial suspension was added to the wells of the microplate together with 20 µL of the adevonin peptide solution (1.84 µM and 7.35 µM for K. oxytoca and S. aureus, respectively). The control contained 180 µL of the bacterial suspension and 20 µL of PBS (pH 7.4). After incubation at 37°C for 30 min, the bacteria were collected by centrifugation at 9,300 × g for 5 min and resuspended in PBS solution containing 10µL.mL−1 of crystal violet. The sample and control were incubated at 37°C for 10 min. After this time, the suspension was centrifuged at 13,400 × g for 15 min. The pellet was discarded, the supernatant was collected and the absorbance read at 570 nm [21]. The absorbance of the violet crystal solution was also measured. For the calculation of the capture percentage, the crystal violet solution was considered as 100%. The value of violet crystal uptake by the sample and the control was calculated using the following formula: Capture of crystal violet = (Abs sample/Abs crystal violet solution) × 100.
2.10. Molecular modeling
Initially, the primary sequence of peptide adevonin was submitted to Blastp in order to find the best template structure for comparative modeling simulations. However, no template structures were found, taking into account parameters such as the sequences’ identity, coverage, score and e-value. Thus, the tridimensional theoretical models for the adevonin peptide were built on the I-TASSER server [22] using a hierarchical approach for peptide structure prediction. The lowest free-energy model was then selected for validation procedures. Firstly, PROCHECK [23] evaluated the peptide’s geometry, stereochemistry, and energy distribution, also calculating its average score for dihedral angles, allied to the backbone covalent forces. ProSA-web [24] was also used to evaluate the folding quality of the selected model. Structure visualization was performed in Pymol [25].
2.11. Molecular dynamics in solution and SDS
Molecular dynamic simulations for adevonin were carried out in water, using the single point charge (SPC) water model. The simulations were performed using the GROMOS96 43A1 force field from the GROMACS v.5.0.4 computational package [26]. The validated tridimensional theoretical model for adevonin was used as the initial structure in the simulations and further immersed in 5,094 water molecules in a cubic box. Chloride ions were added to neutralize the system’s charge. The simulations were done under ionic strength of 0.2 M NaCl. MD simulations in sodium dodecyl sulfate (SDS) were carried out in dodecahedron boxes, where the adevonin peptide was placed into contact with a SDS micelle constituted of 100 detergents. SDS micelles were built, and their topologies generated using the CHARMM-GUI server [27]. The geometry of water molecules was constrained using the SETTLE algorithm [26]. Moreover, the LINCS algorithm was used to link all the atom-bond lengths. Particle Mesh Ewald (PME) was used for electrostatic corrections with a radius cut-off of 1.4 nm to minimize the computational simulation time. The same radius cut-off was used for van der Waals interactions. The list of the neighbors of each atom was updated every 10 simulation steps of 2 fs each. The steepest descent algorithm (50,000 steps) was applied for energy minimization. The system underwent a normalization of temperature and pressure to 300 K and 1 bar using the velocity rescaling thermostat (NVT) and the Parrinello-Rahman barostat (NPT), respectively, for 100 ps. The system with minimized energy and balanced temperature and pressure was submitted to molecular dynamics simulation during 100 ns.
2.12. Molecular dynamics in POPC/POPG bilayers
Molecular dynamic studies were also performed in a membrane environment using a 50 Å2 1-palmito-yl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC)/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylglycerol (POPG) bilayer, in a molar ratio of 1:1. The CHARMM-GUI server [27] was used to place adevonin into the bilayer using the insertion method. The topology files for POPC and POPG were obtained from the CHARMM-GUI server [27], and the assembled system was simulated using the GROMOS96 53a6 force field. Similarly to the simulation in water, the peptide/bilayer system underwent a normalization of temperature and pressure to 300 K and 1 bar using the velocity rescaling thermostat (NVT) and the Parrinello-Rahman barostat (NPT), respectively, for 100 ps. The system with minimized energy and balanced temperature and pressure was submitted to molecular dynamics simulation during 100 ns, using the leap-frog algorithm.
3. Results
3.1. Rational design
AMPs containing 18 amino acid residues were selected from CAMPR3 database [12] using the sliding window approach from the entire ApTI sequence. The first predicted AMP was located between residues 8–25 (Figure 1(a)). The sequence GNFLRNGGSYYIVPAFRG was chosen as it was predicted as an AMP by Support Vector Machine (SVM – 0.549), Random Forest (RF – 0.873), Artificial Neural Network (ANN – AMP) and Discriminant Analysis (DA – 0.856) algorithms. Data were reported with decimal numbers and the closer the number to 1.0 the higher the probability of AMP activity. The original peptide contained residues commonly found in AMPs, including Tyr, Phe, and Arg, and displayed a + 2 net charge and 33% hydrophobicity. The Boman Index for the original peptide was calculated as 1.18 kcal.mol−1, suggesting a moderate potential for binding to biological membranes. The alignment of the original peptide with AMPs from the APD [13] revealed homology with α-helical AMPs from amphibians, including parkerin [28] and temporin [29]. These AMPs show a similar mechanism of action, involving bacteria membrane disruption. Even sharing a homologous amino acid sequence, we investigated whether the original peptide could also assume amphipathic α-helical conformation through the helical wheel diagram, based on the distribution of hydrophobic and hydrophilic amino acids (Figure 1). The distribution of hydrophobic amino acid residues occurs in three different regions along the diagram, suggesting that the original peptide might not assume this conformation.
Figure 1.
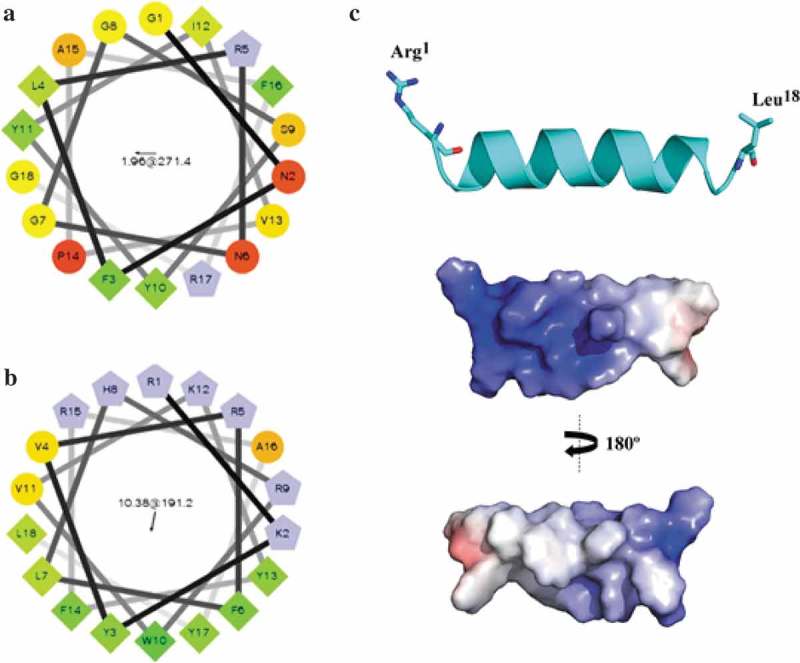
The arrangement of adevonin. Helical wheel diagrams for the original peptide and adevonin. The original AMP obtained from ApTI (a) and the rationalized AMP, adevonin (b), are shown. Hydrophilic residues were shown as circles; hydrophobic residues as diamonds; potentially negatively-charged residues are shown as triangles and positively-charged residues are shown as pentagons. The most hydrophobic residue is green and the amount of green decreases proportionally with hydrophobicity, with the yellow color indicating zero hydrophobicity. The hydrophilic residues are encoded in red, with pure red representing the greatest hydrophilicity (uncharged) and the shade of red decreasing in proportion to hydrophilicity. Potentially charged residues are in light blue. The arrows inside the diagram represent the hydrophobic moment of the peptides. Diagrams were built using the server Helical Wheel Projection (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi). (c) Lowest free energy theoretical model for the adevonin peptide. Adaptive Poisson-Boltzmann Solver (APBS) electrostatic potential of peptide adevonin; potential ranges from – 5 kT/e (red) to + 5 kT/e (blue).
In order to obtain a more cationic and amphipathic peptide, we carried out changes in the amino acid composition of the original peptide. This rationa-lized AMP, adevonin (RKYVRFLHRWVKYFRAYL), had a higher number of Arg, Lys, and hydrophobic amino acid residues (Tyr and Trp), increasing both the net positive charge (+6) and the hydrophobic ratio (44%). These modifications increased the Boman Index by two-fold (2.65 kcal.mol−1). Furthermore, to create an amphipathic α-helical structure and improve the propensity of adevonin to form α-helix, we performed a helical wheel diagram-guided design to change the positions of the amino acid residues in adevonin, thus favoring amphipathicity (Figure 1(b)). Once the design of adevonin was concluded, this peptide was synthesized with 96% purity, had its molecular weight of 2,502.04 Da determined by MALDI-TOF/MS (Supplementary material S1) and was submitted to antimicrobial, cytotoxic and hemolytic assays.
3.2. Antibacterial activities
The original peptide derived form ApTI was synthesized and tested against five bacterial strains. The antimicrobial activity of ApTI, the original peptide and adevonin were evaluated for the determination of MIC values. All the molecules were used in the assays from the same initial concentration (30 µM), followed by serial dilution. The MICs for all molecules is summarized in Table 1. ApTI and the original peptide were not active against any of the bacterial strains tested; whereas adevonin presented MICs ranging from <2 µM to 7.35 µM against susceptible bacterial strains. The antimicrobial activity of adevonin was further assessed against a carbapenem-ase-producing Klebsiella pneumoniae strain (KpC+ 001825971 isolate). The MIC of adevonin against K. pneumoniae KpC+ was 25.6 µM; in contrast, the commercial antibiotic, Imipenem, was unable to control bacterial growth at the concentration of 250 µM. Combined, these results demonstrated that the modifications carried out in the original peptide contributed to the antibacterial activity observed in adevonin.
Table 1.
Antimicrobial activity of adevonin, the original peptide, and ApTI.
| MIC (µM) |
|||
|---|---|---|---|
| Microorganism | adevonin | Original AMP | ApTI |
| Gram-negative | |||
| Escherichia coli ATCC 35218 | 7.35 | >30.0 | >30.0 |
| Klebsiella oxytoca ATCC 13182 | 1.84 | >30.0 | >30.0 |
| Klebsiella pneumoniae ATCC 70603 | 3.67 | >30.0 | >30.0 |
| Serratia marcesens ATCC 13880 | 1.84 | >30.0 | >30.0 |
| Klebsiella pneumoniae KpC+ 001825971 | 25.6 | n.d. | n.d. |
| Gram-positive | |||
| Staphylococcus aureus ATCC 80958 | 7.35 | >30.0 | >30.0 |
aMIC determined by broth microdilution method [15]. n.d.: not determined.
3.3. Hemolytic activity
We studied its hemolytic properties using human type O erythrocytes. At MIC concentration, adevonin did not exert hemolytic effects. Increasing the adevonin concentration we determined that hemolysis occurred at 175 µM (Supplementary material S3), a concentration 24-fold higher than MIC.
3.4. Cell cytotoxicity
Cellular cytotoxicity was assessed by exposing fibroblasts to adevonin at different concentrations (0.3, 0.6, 1.25, 2.5, 5 and 10 µM) and time (24, 48 and 72 h). A significant reduction of cell viability, of 25% on average, was observed in each time interval in the highest peptide concentration (Supplementary material S4).
3.5. Effect of adevonin on G. mellonella survival
Finally, adevonin was administered by intra hemocoel injection in G. mellonella larvae and the deaths of individual larvae determined during 120 h. At this time, 100% of larvae were alive, similarly to observations in the control group, which was injected with PBS.
3.6. Time-dependent bactericidal activity
AMPs that act at plasma membrane level usually kill bacteria during the first minutes of contact, differently of molecules that act on intracellular targets [6]. To determine whether the interaction of adevonin with the bacterial membrane participates in its mechanism of action, we incubated K. oxytoca (ATCC 13182) and S. aureus (ATCC 80958) with adevonin and, aliquots of suspension were transferred to MHA plates at different intervals. As seen in Table 2, the number of CFU drastically decreased within 10 min for K. oxytoca and 25 min for S. aureus, while during longer exposures no viable CFU were observed. The antibiotic chloramphenicol (4 µg.mL−1) exhibited a moderated inhibition of bacterial growth within 90 min, showing bacterial growth after 120 min of incubation. In the control cultures, all intervals produced a high number of CFU, demonstrating that exposure to adevonin was responsible for the bacterial death. Adevonin compromised the viability of K. oxytoca and S. aureus, strains belonging to Gram-negative and -positive bacteria, respectively, therefore demonstrating a bactericidal effect at MIC concentration, in contrast to chloramphenicol, that demonstrated a bacteriostatic mechanism of action.
Table 2.
Time-kill adevonin kinetics against K. oxytoca ATCC 13182 and S. aureus ATCC 80958. Control corresponds to bacteria suspensions incubated with NaCl 0.9%. Adevonin was incubated at MIC concentration and the respective treatments. The antibiotic chloramphenicol was used at 4 µg.mL−1. The results are showed as CFU/mL−1.
| Time (min) | K. oxytoca control | K. oxytoca + adevonin | K. oxytoca + antibiotic | S. aureus + adevonin | S. aureus control | S. aureus + antibiotic |
|---|---|---|---|---|---|---|
| 0 | >105 | >105 | >105 | >105 | >105 | >105 |
| 5 | >105 | >105 | >105 | >105 | >105 | >105 |
| 10 | >105 | 2 | >105 | >105 | >105 | >105 |
| 15 | >105 | 0 | >105 | >105 | >105 | >105 |
| 20 | >105 | 0 | >105 | 102 | >105 | >105 |
| 25 | >105 | 0 | >105 | 8 | >105 | 102 |
| 30 | >105 | 0 | >105 | 0 | >105 | 102 |
| 60 | >105 | 0 | 103 | 0 | >105 | 35 |
| 90 | >105 | 0 | 52 | 0 | >105 | 30 |
| 120 | >105 | 0 | 102 | 0 | >105 | 103 |
3.7. Efflux of UV-absorbing materials and uptake of gentian violet
To substantiate our finding that adevonin induces damages on the bacterial membrane, we determined the efflux of UV-absorbing materials by K. oxytoca (ATCC 13182) and S. aureus (ATCC 80958) when exposed to adevonin at the MIC concentration, as depicted in Figure 2(a). Adevonin prompted an increased absorbance at 260 nm, indicating a 5-fold increase in nucleic acid and protein release after incubation with adevonin. This increase is compatible with the release of intracellular material due to the loss of the integrity of the plasma membrane [6]. Additionally, the uptake of gentian violet by bacteria further demonstrated the ability of the adevonin to compromise membrane integrity, as adevonin-incubated bacteria presented an increase of 30% in the capture of gentian violet (Figure 2(b)). Taken together, these results support the ability of adevonin to induce damage on bacterial membranes.
Figure 2.
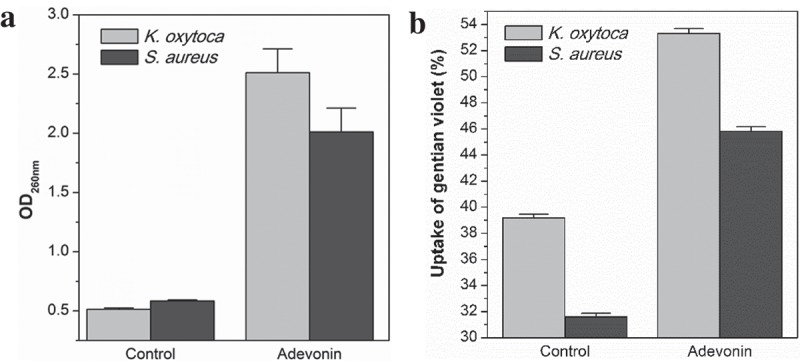
(a) Efflux of UV-absorbing materials. (b) Uptake of gentian violet. Control corresponds to bacteria incubated without peptide. Vertical bars correspond to the standard deviation.
3.8. Molecular modeling
In order to obtain insights into the structural adevonin behavior in different environments, molecular modeling and dynamics simulations were carried out. Initially, adevonin was modeled and the lowest free-energy model submitted to validation procedures regarding its fold quality and stereochemistry. The Ramachandran Plot for the lowest energy theoretical structure of adevonin showed that 87.5% (n = 14) of residues were located in the most favorable regions and 12.5% (n = 4) were in the region that had been additionally permitted, indicating no steric hindrance. Adevonin presented a z-score value of 0.02, showing that the structure was within the range of scores found for peptides of similar size deposited in the Protein Data Bank (PDB) [30] and determined by nuclear magnetic resonance (NMR). The PROCHECK server [23] showed that adevonin had an average score for the dihedral angles along with covalent forces of the main chain within the expected range for a reliable structure (>−0.05). Molecular modeling suggested that adevonin might adopt an amphipathic, helical structure (Figure 1(c)). These findings were also supported by the calculated electrostatic potential for adevonin, revealing amphipathic properties.
3.9. Molecular dynamics
Since the results pointed bacterial membrane as a potential target of adevonin, we further investigated the secondary structure of adevonin in different environments, including water (ionic strength: 0.2 M NaCl), SDS micelles and an anionic lipid bilayer composed of POPC/POPG at a ratio of 1:1. The simulations were carried out through 100 ns of MD simulation. In water, adevonin presented a small, central ordered region with both amino and carboxy-terminal domains exhibiting considerable disorder from 30 ns (Figure 3(a)). The unstructured molecule reflects high amplitude of mean square deviation (RMSD), root mean square fluctuation (RMSF), and radius of gyration (Rg) (Figure 3(d)). These deviations accompanied the structural changes (destabilization) of the molecule, supporting the unfolded state in solution. The MD simulation in SDS and POPC/POPG demonstrated that adevonin presents high stability in these environments (Figure 3(b,c)), forming an amphipathic helix with low deviation in the RMSD, RMSF, and Rg during the entire simulation (Figure 3(e,f)). The MD simulation corroborated the results previously obtained, supporting the relationship between antibacterial activities and damage on the bacterial membrane with its conformational stability in environments that mimics a Gram-negative bacteria membrane (Figure 3(c)).
Figure 3.
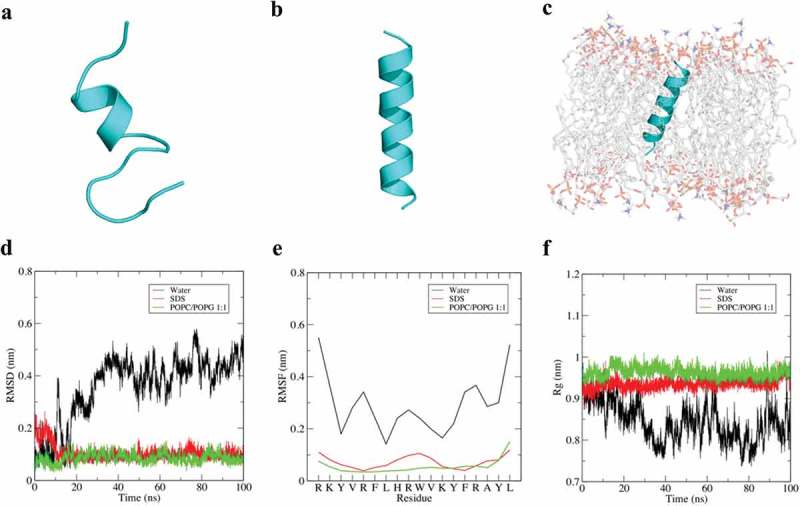
Dynamic simulation of adevonin in different environments. Structural snapshots of adevonin in water (a), SDS (b) and POPC/POPG 1:1 (c) after 100 ns of MD simulation. The MD analyzes, including root mean square deviation (RMSD) (d), root mean square fluctuation (RMSF) (e) and radius of gyration (Rg) are also shown. N-terminus is always at the top.
4. Discussion
Since the introduction of antibiotics in the late 40s of the last century, an array of treatment options have been available for the bacterial infections treatment [1,2,31]. Currently, an impressive bacterial infections number have become resistant to the antibiotic treatment of choice, particularly bacteria in the six high-priority group of pathogens called the ‘ESKAPE’ group (E. faecium, S. aureus, K. pneumoniae, A. baumanii, P. aeruginosa, and Enterobacter species) [10,31]. The decline in the development of new antibacterial molecular entities since the 1980s has made an important contribution to bacterial resistance [31], spurring investigations to find new alternatives to control bacterial infections, such as the use of AMPs.
The isolation and characterization of new AMPs are time-consuming and labor-intensive. As such, the prediction or design of AMPs based on natural protein or peptide templates represents a strategy to expand the AMP repository [8]. Some biochemical characteristics are determinant for AMP activity and specificities, including size, primary sequence, net charge, secondary structure, hydrophobicity and amphipathicity [32]. Based on these features, prediction tools can be used for database-aided design. The original peptide predicted from the ApTI sequence was thought to display antibacterial activity; however, in vitro assays revealed that it had no activity against bacteria. Further investigations suggested that the original AMP structure might not assume any secondary conformation. Many AMPs adopt a secondary structure only upon interaction with cell membranes [31], where this folding is essential to the peptide’s bactericidal activity.
The AMPs parkerin and temporin, homologous to the original peptide, are characterized by showing α-helical secondary conformation followed interaction with the bacterial membrane. From this interaction, these AMPs promote the lysis of bacterial membranes [28,29]. Based on that we carried out a helical wheel diagram-guided design to change the positions of the amino acid residues in adevonin to favor amphipathicity. The addition of positive residues in the adevonin peptide was though to increase the initial attraction of adevonin to the bacterial membrane through electrostatic interaction [4]. The mixed cationic and hydrophobic composition of AMPs makes them well suited for interacting with and perturbing microbial cytoplasmic membranes, typically presenting anionic lipid content [6]. Adevonin presented activity against Gram-negative and Gram-positive bacterial strains, with MICs ranging from 1.84 to 7.35 µM, similar to other recently reported AMPs [33], including melitin (9–18 µM), magainin-2-amide (40–50 µM), indolicin (2–16 µM), and cecropin P1 (0.3–17 µM) [34–38]. For K. pneumoniae KpC+ 001825971, the MIC value obtained was 25.6 µM. Assays using the same K. pneumoniae KpC+ 001825971 strain and carried out by Cardoso et al. [33], reported a MIC value for Pa-MAP 1.9 of 96 µM, a concentration almost 4 times higher compared to adevonin. Interestingly, in that study, Pa-MAP 1.9 was able to inhibit the formation and eradicate K. pneumoniae KpC+ 001825971 biofilms at 1.1 µM, demonstrating that the bactericidal and anti-biofilm activities involve different and complex mechanisms. Importantly, the susceptibility of K. pneumoniae KpC+ to adevonin does not correlate with preexisting resistance to carbapenems. Therefore, additional studies of adevonin activity on inhibition of the eradication of bacterial biofilms are encouraged.
A concern about the use of AMPs is with regards to their toxicity against mammalian cells. The hemolytic activity observed for AMPs varies widely [33,39]. Hemolytic activity was observed when the adevonin concentration was 24 times higher than the MIC, suggesting that the peptide is potentially safe for administration. Assays with human MRC-5 fibroblasts with adevonin at low concentrations demonstrated a low cytotoxicity of the peptide. Additionally, when adevonin was injected in G. mellonella it had no adverse effects on larvae survival. Furthermore, the safety of concentrations could be improved with a further modification of the adevonin amino acid sequence. In addition to the decrease in hemolytic activity and cytotoxicity, changes in the adevonin structure may contribute to increase the bactericidal activity. The most common modifications are the addition of amino-terminal acetylation, carboxy-terminal amidation, the introduction of non-standard amino acids, the synthesis of all-D-enantiomer and peptide cyclization [8,40–42].
The short period necessary to eliminate bacterial growth, associated with the increase in cytoplasmic content after adevonin incubation suggests that the membrane is the primary target of the peptide. To support our MD simulation results that suggested that adevonin adopts a stable secondary structure in anionic and membrane-like environments, we determined the exposure time necessary to kill bacteria, as well as the extravasation of cytoplasmic contents. The recruitment of anionic lipids to a region of the bacterial membrane results in the concentration of negative charge in a domain at which cationic peptides may congregate, possibly causing the membrane damages [6,8]. As a result of damages on the bacterial membrane is possible occurs slow leakage of contents and/or depolarization of the membrane or yet a complete membrane lysis [8].
The outer membrane of Gram-negative bacteria executes the crucial role of providing an extra layer of protection against bacteria, representing a selective barrier. These permeability properties affect the susceptibility of the microorganism to current antibiotics, which essentially target intracellular processes. Small hydrophilic drugs, such as β-lactams, use the pore-forming porins to access to the cell interior, while hydrophobic drugs diffuse across the lipid bilayer. Modifications in the lipid or protein composition modulate antibiotic sensitivity. For this reason, the identification of molecules able to kill bacteria by different mechanisms of action is crucial [43]. Adevonin was able to eliminate bacterial growth of both Gram-negative and – positive groups, including antibiotic-resistant strains (K. pneumoniae KpC+), demonstrating the potential of AMPs to become a new class of broad-spectrum antibiotics. Since AMPs act directly on the bacterial membrane, initially by electrostatic interaction, it is unlikely that the composition of the outer membrane can change to such an extent that the physico-chemical properties of the target of AMPs are altered.
In conclusion, we describe herein the design and biological activity of adevonin, a rationalized AMP designed from ApTI. Adevonin is highly active against both sensitive and drug-resistant bacteria strains, including Gram-negative strains from the ESKAPE group. Adevonin showed that its secondary structure varies according to the environment, acquiring α-helical conformation in presence of bacterial membrane, which in turn is associated with antimicrobial activity. Based on these results we propose to further investigate the anti-infective activity of this peptide in vivo in cutaneous and/or systemic infection models. Nevertheless, this molecule could serve as a template for the design of further AMPs, increasing the repertoire of molecules that may provide effective bacterial infection control.
Funding Statement
This work was supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico [407127/2013-5] and Fundação de Apoio ao Desenvolvimento do Ensino, Ciência e Tecnologia do Estado de Mato Grosso do Sul [009/2015].
Acknowledgments
The authors thank the Brazilian development agencies CNPq, CAPES, FUNDECT, FAPDF, and FINEP, for the financial support.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data can be accessed here.
References
- [1].Alanis AJ.Resistance to antibiotics: are we in the post-antibiotic era? Arch Med Res. 2005;36(6):697–705. [DOI] [PubMed] [Google Scholar]
- [2].Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529(7586):336. [DOI] [PubMed] [Google Scholar]
- [3].Organization WH Global action plan on antimicrobial resistance. 2015. 2017. [Google Scholar]
- [4].Ye H. Molecular design of antimicrobial peptides based on hemagglutinin fusion domain to combat antibiotic resistance in bacterial infection. J Pept Sci. 2018;24(3):e3068. [DOI] [PubMed] [Google Scholar]
- [5].Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: key components of the innate immune system. Crit Rev Biotechnol. 2012;32(2):143–171. [DOI] [PubMed] [Google Scholar]
- [6].Wimley WC. Describing the mechanism of antimicrobial peptide action with the interfacial activity model. ACS Chem Biol. 2010;5(10):905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Takahashi D, Shukla SK, Prakash O, et al. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie. 2010;92(9):1236–1241. [DOI] [PubMed] [Google Scholar]
- [8].Wang G. Antimicrobial peptides: discovery, design and novel therapeutic strategies. Wallingford, Oxforshire, (UK): CABI; 2017. [Google Scholar]
- [9].McGrath DM, Barbu EM, Driessen WH, et al. Mechanism of action and initial evaluation of a membrane active all-D-enantiomer antimicrobial peptidomimetic. Proc Natl Acad Sci USA. 2013;110(9):3477–3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Fjell CD, Hiss JA, Hancock RE, et al. Designing antimicrobial peptides: form follows function. Nat Rev Drug Discov. 2012;11(1):37. [DOI] [PubMed] [Google Scholar]
- [11].Farris M, Steinberg A. Mitrecin A, an endolysin‐like bacteriolytic enzyme from a newly isolated soil streptomycete. Lett Appl Microbiol. 2014;58(5):493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Waghu FH, Barai RS, Gurung P, et al. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2015;44(D1):D1094–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44(D1):D1087–D1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Richardson M, Campos FAP, Xavier-Filho J, et al. The amino acid sequence and reactive (inhibitory) site of the major trypsin isoinhibitor (DE5) isolated from seeds of the Brazilian Carolina tree (Adenanthera pavonina L.). Biochim Biophys Acta. 1986;872(1–2):134–140. [Google Scholar]
- [15].CLSI. Performance standards for antimicrobial susceptibility testing. Wayne (PA): Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- [16].Uggerhøj LE, Poulsen TJ, Munk JK, et al. Rational design of alpha-helical antimicrobial peptides: do’s and don’ts. ChemBioChem. 2015;16(2):242–253. [DOI] [PubMed] [Google Scholar]
- [17].Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65(1–2):55–63. [DOI] [PubMed] [Google Scholar]
- [18].Maguire R, Duggan O, Kavanagh K. Evaluation of Galleria mellonella larvae as an in vivo model for assessing the relative toxicity of food preservative agents. Cell Biol Toxicol. 2016;32(3):209–216. [DOI] [PubMed] [Google Scholar]
- [19].Mitić-Ćulafić D, Vuković-Gačić BS, Knežević-Vukčević JB, et al. Comparative study on the antibacterial activity of volatiles from sage (Salvia officinalis L.). Arch Biol Sci. 2005;57(3):173–178. [Google Scholar]
- [20].Zhou K, Zhou W, Li P, et al. Mode of action of pentocin 31-1: an antilisteria bacteriocin produced by Lactobacillus pentosus from Chinese traditional ham. Food Control. 2008;19(8):817–822. [Google Scholar]
- [21].Vaara M, Vaara T. Outer membrane permeability barrier disruption by polymyxin in polymyxin-susceptible and-resistant Salmonella typhimurium. Antimicrob Agents Chemother. 1981;19(4):578–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yang J, Yan R, Roy A, et al. The I-TASSER suite: protein structure and function prediction. Nat Methods. 2015;12(1):7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Laskowski RA, MacArthur MW, Moss DS, et al. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283–291. [Google Scholar]
- [24].Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(suppl_2):W407–W410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].DeLano WL. The PyMOL molecular graphics system version 2.0. San Carlos (CA); 2002. [Google Scholar]
- [26].Abraham MJ, Murtola T, Schulz R, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- [27].Wu EL, Cheng X, Jo S, et al. CHARMM‐GUI membrane builder toward realistic biological membrane simulations. J Comput Chem. 2014;35(27):1997–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lu Z, Zhai L, Wang H, et al. Novel families of antimicrobial peptides with multiple functions from skin of Xizang plateau frog, Nanorana parkeri. Biochimie. 2010;92(5):475–481. [DOI] [PubMed] [Google Scholar]
- [29].Wang H, Lu Y, Zhang X, et al. The novel antimicrobial peptides from skin of Chinese broad-folded frog, Hylarana latouchii (Anura: ranidae). Peptides. 2009;30(2):273–282. [DOI] [PubMed] [Google Scholar]
- [30].Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lohner K. Membrane-active antimicrobial peptides as template structures for novel antibiotic agents. Curr Top Med Chem. 2017;17(5):508–519. [PubMed] [Google Scholar]
- [32].Shagaghi N, Palombo EA, Clayton AH, et al. Antimicrobial peptides: biochemical determinants of activity and biophysical techniques of elucidating their functionality. World J Microbiol Biotechnol. 2018;34(4):62. [DOI] [PubMed] [Google Scholar]
- [33].Cardoso MH, Ribeiro SM, Nolasco DO, et al. A polyalanine peptide derived from polar fish with anti-infectious activities. Sci Rep. 2016;6:21385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Beschiaschvili G, Seelig J. Melittin binding to mixed phosphatidylglycerol/phosphatidylcholine membranes. Biochemistry. 1990;29(1):52–58. [DOI] [PubMed] [Google Scholar]
- [35].Maloy WL, Kari UP. Structure–activity studies on magainins and other host defense peptides. Biopolymers. 1995;37(2):105–122. [DOI] [PubMed] [Google Scholar]
- [36].Subbalakshmi C, Krishnakumari V, Sitaram N, et al. Interaction of indolicidin, a 13-residue peptide rich in tryptophan and proline and its analogues with model membranes. J Biosci (Bangalore). 1998;23(1):9–13. [Google Scholar]
- [37].Gazit E, Boman A, Boman HG, et al. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry. 1995;34(36):11479–11488. [DOI] [PubMed] [Google Scholar]
- [38].Melo MN, Ferre R, Castanho MA. Antimicrobial peptides: linking partition, activity and high membrane-bound concentrations. Nature Rev Microbiol. 2009;7(3):245. [DOI] [PubMed] [Google Scholar]
- [39].Park IY, Park CB, Kim MS, et al. Parasin I, an antimicrobial peptide derived from histone H2A in the catfish, Parasilurus asotus. FEBS Lett. 1998;437(3):258–262. [DOI] [PubMed] [Google Scholar]
- [40].Rodriguez-Granillo A, Annavarapu S, Zhang L, et al. Computational design of thermostabilizing D-amino acid substitutions. J Am Chem Soc. 2011;133(46):18750–18759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hirt H, Hall JW, Larson E, et al. A D-enantiomer of the antimicrobial peptide GL13K evades antimicrobial resistance in the Gram positive bacteria Enterococcus faecalis and Streptococcus gordonii. PLoS One. 2018;13(3):e0194900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Li W, Sun Z, O’Brien-Simpson NM, et al. The Nα-Methyl effect arginine of selective substitution D-or on the activity of the proline-rich antimicrobial peptide, Chex1-Arg20. Antimicrob Anticancer Pept. 2018;5(1):1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Delcour AH. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta. 2009November27;1794(5):808–816. PubMed PMID: PMC2696358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


