Abstract
Dysfunction of microRNAs (miRs) has been implicated in the development and progression of various human cancers. Our previous study demonstrated that miR-101 inhibited bladder cancer cell proliferation and invasion through inhibition of c-FOS expression. As an miR generally has many targets, other targets of miR-101 may also serve important roles in bladder cancer progression. Reverse transcription-quantitative polymerase chain reaction and western blot analyses were used to examine mRNA and protein expression, respectively. Wound healing and Transwell assays were conducted to study cell migration and invasion, respectively. The luciferase reporter gene assay was performed to verify one of the targets of miR-101. The data in the present study indicate that the expression of miR-101 is significantly reduced in bladder cancer tissues compared with that in adjacent non-tumour tissues. In addition, miR-101 expression is also downregulated in bladder cancer cell lines compared with that in normal bladder epithelial cells. Furthermore, low expression of miR-101 was significantly associated with tumour metastasis, advanced clinical stage, and poor prognosis in bladder cancer. Frizzled class receptor 4 (FZD4) was identified as a novel target of miR-101 in bladder cancer cells. The expression of FZD4 was significantly upregulated in bladder cancer tissues and cell lines. Both miR-101 overexpression and FZD4 inhibition caused a significant reduction of the migration and invasion of bladder cancer cells, whereas overexpression of FZD4 reversed the suppressive effects of miR-101 on bladder cancer cell migration and invasion. In conclusion, it was demonstrated that miR-101 downregulation is associated with bladder cancer progression and that miR-101 can inhibit bladder cancer cell migration and invasion via directly targeting FZD4. The present study expands the understanding of the molecular mechanisms underlying bladder cancer progression.
Keywords: bladder cancer, microRNA, frizzled class receptor 4, migration, invasion
Introduction
Bladder cancer is one of the most common malignant tumours of the urinary system, and its incidence and mortality rate are increasing (1,2). Despite great advances in combined treatments, including surgical resection, chemotherapy and/or radiotherapy, the prognosis of patients is still unsatisfactory, mainly because of the high rate of recurrence and distant metastasis (3). Dysfunctions of certain oncogenes and tumour suppressors have been demonstrated to serve a key role during bladder cancer development and progression (4,5). Understanding the regulatory mechanism underlying the malignant progression of bladder cancer is important as it may benefit the development of effective strategies for the treatment of bladder cancer.
microRNAs (miRNAs or miRs), a class of non-coding RNAs that are ~22 nucleotides in length, are important gene regulators that bind to the 3′-untranslated region (UTR) of their target mRNAs, resulting in protein translation inhibition or mRNA degradation (6–8). A large number of miRNAs have been reported to regulate the expression of genes that are associated with physiological and pathological processes, including cell growth, proliferation, differentiation, apoptosis, migration, angiogenesis and tumourigenesis, among others (9–12). In recent years, a number of miRs, such as miR-124 (13), miR-21 (14), miR-5195 (15), miR-138 (16), and miR-576 (17) have been demonstrated to have key roles in bladder cancer.
Aberrant expression of miR-101 has previously been observed in various human cancers, such as gastric cancer (18), hepatocellular carcinoma (19), nasopharyngeal carcinoma (20), osteosarcoma (21), and breast cancer (22). Furthermore, miR-101 has been identified as a tumour suppressor in bladder cancer (23,24). Downregulation of miR-101 is associated with poor prognosis of patients with bladder transitional cell carcinoma (24). miR-101 directly targets vascular endothelial growth factor C, inhibits cell migration and invasion, and enhances cisplatin sensitivity by modulating the cyclooxygenase-2 pathway in bladder cancer cells (23,25). Our previous study illustrated that miR-101 has suppressive effects on bladder cancer cell proliferation and invasion via inhibiting the expression of its target c-FOS (26). As a miR generally has many target genes, other targets of miR-101 may also serve important roles during bladder cancer progression.
Therefore, the present study aimed to explore the regulatory mechanisms of miR-101 and frizzled class receptor 4 (FZD4), a putative target gene of miR-101, underlying the metastasis of bladder cancer in vitro.
Materials and methods
Clinical samples
The present study was approved by the Ethics Committee of Zhuzhou Central Hospital (Zhuzhou, China; ZZCH2017072). Bladder cancer tissue samples and matched adjacent non-tumour tissue samples 5 cm away were collected from 64 bladder cancer patients at Department of Urinary Surgery, Zhuzhou Central Hospital between May 2010 and May 2012. These patients included 44 males and 21 females 44–81 years old. Patients were excluded if they had received chemotherapy or radiotherapy prior to surgical resection. Written informed consent was obtained from all patients. The tissue samples were stored at −80°C prior to use.
Cell culture
The normal human bladder epithelial cell line SV-HUC-1 and four bladder cancer cell lines (HT-1376, T24 and 5637) were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% foetal bovine serum (FBS; Thermo Fisher Scientific, Inc.).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from the tissue samples and cell lines using TRIzol reagent, and the total RNA was then reverse transcribed into cDNA using a RevertAid First Strand cDNA Synthesis kit (both Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. RNA expression was measured by qPCR using a SYBR® Premix Ex Taq™ Tli RNaseH Plus PCR kit (Takara Bio, Inc., Otsu, Japan) on an ABI 7300 sequence detector (Thermo Fisher Scientific, Inc.). The reaction conditions were 95°C for 3 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 60 sec. GAPDH was used as the internal reference for mRNA, and U6 was used as the internal reference for miRNA. The relative expression was determined using the 2−ΔΔCq method (27). The primers for FZD4 were as follows: Forward, 5′-CCTCGGCTACAACGTGACC-3′ and reverse, 5′-TGCACATTGGCACATAAACAGA-3′. The primers for GAPDH were follows: Forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse, 5′-GCCATCACGCCACAGTTTC-3′. The primers for miR-101 (cat. no. HmiRQP0021) and U6 (cat. no. HmiRQP9001) were purchased from Guangzhou Fulengen Co., Ltd. (Guangzhou, China).
Western blotting
The cells were lysed in radio-immunoprecipitation assay (RIPA) lysis buffer containing protease inhibitors and phosphatase inhibitors (Thermo Fisher Scientific, Inc.), and the protein concentration was determined using a BCA protein assay kit (Thermo Fisher Scientific, Inc.). Proteins (50 µg per lane) were separated using 10% SDS-PAGE and then transferred onto polyvinylidene fluoride (PVDF) membranes (Thermo Fisher Scientific, Inc.). The membranes were then incubated with rabbit anti-FZD4 (cat. no. ab83042) and GAPDH primary antibodies (cat. no. ab9485; both 1:50; Abcam, Cambridge, MA, USA) at 4°C overnight and then with horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (cat. no. ab7090, 1:5,000; Abcam) at room temperature for 1 h. The immunofluorescence signals were detected with an enhanced chemiluminescence system (Thermo Fisher Scientific, Inc.), and a densitometric analysis was conducted using ImageJ software (version 1.47; National Institute of Health, Bethesda, MD, USA).
Cell transfection
In brief, 2×105 T24 cells/well seeded in a 6-well plate were transfected with 100 nM miR-101 mimic (cat. no. HmiR0009-MR04), negative control miR (miR-NC; cat. no. CmiR0001-MR04), miR-101 inhibitor (cat. no. HmiR-AN0021-SN-10), NC inhibitor (cat. no. CmiR-AN0001-SN; all Guangzhou Fulengen Co., Ltd.), FZD4 small interfering (si)RNA (cat. no. SR305441) or NC siRNA (cat. no. SR30004; both OriGene Technologies, Inc., Rockville, MD, USA), 100 nM miR-101 mimic + 1 mg blank vector (cat. no. AMS10002) or 100 nM miR-101 mimic + 1 mg pc-DNA3.1-FZD4 expression plasmid (cat. no. AMS10116; both Hunan Nanhua Aishi Pulin Biotechnology; NanHua Bio-medicine Co., Ltd., Changsha, China) using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.). At 48 h following cell transfection, the subsequent experiments were performed.
Bioinformatics analysis and dual luciferase reporter assay
Target Scan 7.1 software (www.targetscan.org) was applied to identify miR-101 target genes. The predicted miR-101 target sequence within the FZD4 3′-UTR or a mutant target sequence was cloned downstream of the luciferase gene driven by the cytomegalovirus (CMV) promoter, generating a wild type (WT) FZD4 or mutant type (MT) FZD4 luciferase reporter gene plasmid, respectively (Promega Corporation, Madison, WI, USA). According to the manufacturer's instruction, 293T cells were co-transfected with WT FZD4 or MT FZD4 luciferase reporter gene plasmid and miR-101 mimics or scramble miR mimics using Lipofectamine 2000. Following transfection for 48 h, the luciferase activity was detected using the Dual Luciferase Reporter Assay kit (Promega Corporation) on a LD400 luminometer (Beckman Coulter, Inc., Brea, CA, USA). Firefly luciferase activity was normalized to Renilla luciferase activity.
Cell migration assay
The transfected cells were cultured in 6-well plates for 24 h. Then, an artificial wound was created with a sterile 10-µl pipette tip on the confluent cell monolayer. Images were captured at 0 and 24 h following wound creation using an inverse microscope (magnification, ×40; Olympus Corporation, Tokyo, Japan).
Cell invasion assay
T24 cells (1×105 cells) were seeded into the upper chamber of a Transwell plate that was pre-coated with Matrigel (HyClone; GE Healthcare Life Sciences, Logan, UT, USA) containing 200 µl serum-free DMEM, and 500 µl DMEM supplemented with 10% FBS was added to the lower chamber. Following incubation at 37°C for 24 h, the cells in the upper chamber were removed, and the membrane was fixed at room temperature with 4% formaldehyde for 20 min and stained with Giemsa at room temperature for 10 min. The invading cells were counted under an inverse microscope (magnification, ×200; Olympus Corporation).
Statistical analysis
The present data are expressed as mean ± standard deviations Statistical analysis was performed using SPSS 19.0 software (IBM Corp., Armonk, NY, USA). The differences between groups were analysed by one-way analysis of variance among three or more groups followed by a Tukey's post hoc test, or Student's t-test between two groups. In addition, a paired Student's t-test was used for analysing the differences of miR-101 and FZD4 expression between bladder cancer tissues and paired adjacent normal tissues. Pearson's χ2 test was also employed. Kaplan Meier analysis and a log-rank test were used to compare the survival rates of patients with bladder cancer grouped by low and high miR-101 expression. Furthermore, univariate and multivariate logistic regression analysis was used to confirm whether miR-101 expression was an independent prognostic factor for bladder cancer patients. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-101 is downregulated in bladder cancer tissues and cells
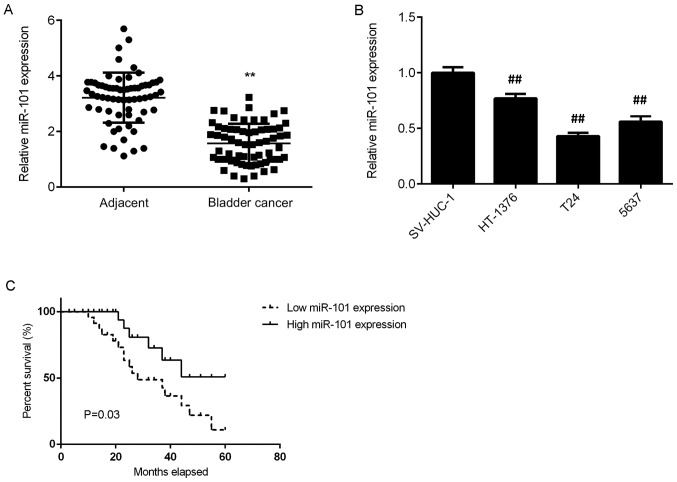
In the present study, RT-qPCR was conducted to examine the miR-101 expression in bladder cancer tissues. The data indicated that miR-101 was significantly downregulated in bladder cancer tissues compared with that in adjacent non-tumour tissues (Fig. 1A). Consistent with this observation, miR-101 was also significantly downregulated in bladder cancer cell lines (HT-1376, T24 and 5637) when compared with that in the normal bladder epithelial cell line SV-HUC-1 (Fig. 1B).
Figure 1.
(A and B) Reverse transcription-quantitative polymerase chain reaction was used to determine the miR-101 expression in bladder cancer tissues and cell lines. (C) Low miR-101 expression was associated with a shorter survival time of bladder cancer patients. **P<0.01 vs. Adjacent; ##P<0.01 vs. SV-HUC-1. miR, microRNA.
The clinical significance of miR-101 expression in bladder cancer was then studied. Bladder cancer patients were divided into two groups, a high miR-101 expression group and a low miR-101 expression group, according to the mean expression value (1.57) of miR-101 as the cut-off value. As presented in Table I, low miR-101 expression was significantly associated with lymph node metastasis and advanced clinical stage in bladder cancer. Further investigation demonstrated that low miR-101 expression was also significantly associated with shorter survival time of patients with bladder cancer (Fig. 1C). As presented in Table II, the result of multivariate Cox proportional hazard regression analysis indicated that miR-101 expression is an independent prognostic factor for bladder cancer patients. Taken together, these data suggest that the downregulation of miR-101 is associated with malignant progression and poor prognosis in patients with bladder cancer.
Table I.
Association between the microRNA-101 expression and clinicopathologic characteristics in bladder cancer.
| Variables | Total (n=65) | Low expression (n=32) | High expression (n=33) | P-value |
|---|---|---|---|---|
| Sex | 0.434 | |||
| Male | 44 | 20 | 24 | |
| Female | 21 | 12 | 9 | |
| Age (years) | 0.450 | |||
| ≤60 | 39 | 21 | 18 | |
| >60 | 26 | 11 | 15 | |
| Tumor size (cm) | 0.325 | |||
| ≤3 | 33 | 14 | 19 | |
| >3 | 32 | 18 | 14 | |
| Grade | 0.150 | |||
| Well and moderately | 50 | 22 | 28 | |
| Poor | 15 | 10 | 5 | |
| Clinical T stage | 0.012 | |||
| Ta+Tis+T1 | 27 | 8 | 19 | |
| T2-T4 | 38 | 24 | 14 | |
| Lymph node metastasis | 0.018 | |||
| Present | 21 | 15 | 6 | |
| Absent | 44 | 17 | 27 | |
| Distant metastasis | 0.053 | |||
| Present | 4 | 4 | 0 | |
| Absent | 61 | 28 | 33 |
Data are presented as n. miR, microRNA.
Table II.
Univariate and multivariate Cox hazard regression analysis.
| Characteristic | Univariate analysis Hazard Ratio (95% CI) | P-value | Multivariate analysis Hazard Ratio (95% CI) | P-value |
|---|---|---|---|---|
| miR-101 | 1.245 (0.602–2.554) | 0.544 | 0.257 (0.082–0.806) | 0.019 |
miR, microRNA; CI, confidence intervals.
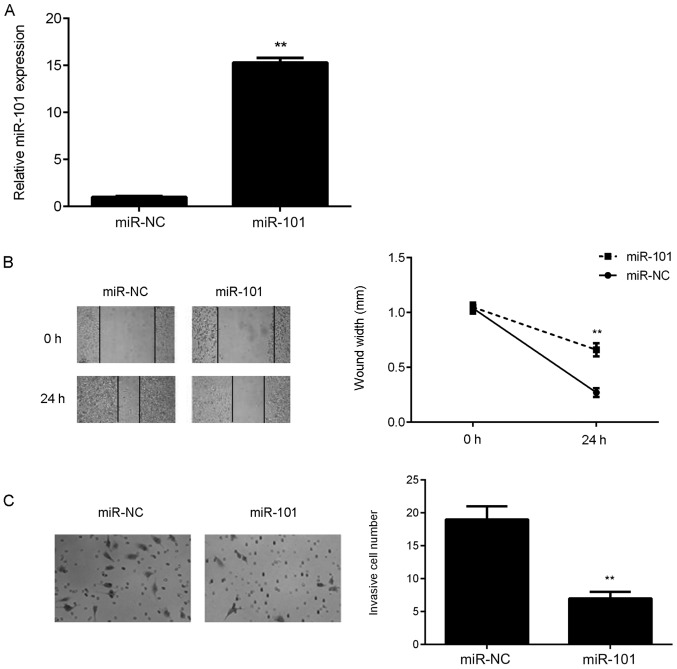
Overexpression of miR-101 inhibits the migration and invasion of bladder cancer cells
As the T24 cell line demonstrated the lowest expression of miR-101 among all bladder cancer cell lines, it was used the following in vitro experiments. As it had been demonstrated that the miR-101 levels were significantly reduced in bladder cancer tissues and cell lines, miR-101 mimics were used to transfect bladder cancer T24 cells to upregulate its expression. Following transfection, the miR-101 levels were significantly increased in the miR-101 group compared with those in the miR-NC group (Fig. 2A). A wound healing assay and Transwell assay were then conducted to study the effect of miR-101 overexpression on T24 cell migration and invasion. As presented in Fig. 2B and C, the overexpression of miR-101 led to a significant reduction in T24 cell migration and invasion.
Figure 2.
T24 cells were transfected with an miR-101 mimic. Transfection with miR-NC was used as the control group. (A) Following transfection, the expression level of miR-101 was determined using reverse transcription-quantitative polymerase chain reaction. (B) Wound healing and (C) Transwell assays were used to examine the migration and invasion capacities of T24 cells, respectively. Magnification for wound healing assay, ×40. Magnification for Transwell assay, ×200. **P<0.01 vs. miR-NC. miR, microRNA; miR-NC, scrambled miR mimic.
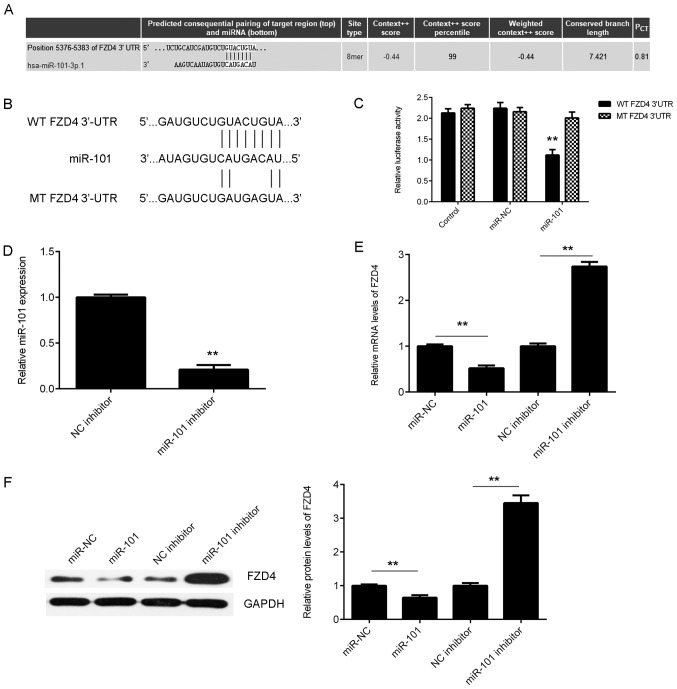
FZD4 is a target gene of miR-101 in bladder cancer cells
The putative targets of miR-101 in bladder cancer were then studied using bioinformatics analysis. A total of 796 genes have been predicted to be potential targets of miR-101 by TargetScan software (data not shown). Among these putative target genes, FZD4, which encodes a seven-transmembrane domain protein and is associated with β-catenin canonical signalling pathway, was selected to be studied (Fig. 3A), as a tumour suppressor miR-493 was reported to inhibit bladder cancer cell growth and migration ability by targeting FZD4 (27–29). To verify this predication, luciferase reporter gene plasmids were generated with either WT or MT FZD4 3′-UTR (Fig. 3B). Luciferase reporter gene assays were then conducted in 293T cells. As presented in Fig. 3C, the luciferase activity was significantly reduced in cells co-transfected with miR-101 mimics and WT FZD4 3′-UTR luciferase reporter gene plasmid compared with that in the control group, whereas cells co-transfected with miR-101 mimics and MT FZD4 3′-UTR luciferase reporter gene plasmid did not show increased luciferase activity.
Figure 3.
(A) TargetScan software predicted that FZD4 was a target gene of miR-101. (B) The binding sequences of miR-101 within the WT or MT 3′UTR of FZD4 are indicated. (C) Luciferase reporter gene assay was then conducted in 293T cells. Control, T24 cells transfected with WT or MT FZD4 plasmid, without miR-101 mimics; NC, T24 cells transfected with WT or MT FZD4 plasmid and scramble miR mimics. **P<0.01 vs. control. (D) RT-qPCR was performed to examine the expression of miR-101 in T24 cells transfected with miR-101 inhibitor or NC inhibitor. **P<0.01 vs. NC inhibitor. (E) RT-qPCR and (F) western blot analysis were conducted to examine the mRNA and protein expression levels of FZD4 in T24 cells transfected with miR-101 mimic, miR-NC, miR-101 inhibitor or NC inhibitor. **P<0.01. FZD4, frizzled class receptor 4; miR, microRNA; WT, wild type; MT, mutant; UTR, untranslated region; miR-NC, scramble miR mimics; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, negative control.
Subsequently, experiments were conducted to clarify whether miR-101 could negatively regulate FZD4 expression in bladder cancer cells. T24 cells were transfected with miR-101 mimics or an miR-101 inhibitor. The data indicated that transfection with miR-101 mimics significantly increased the miR-101 levels (Fig. 2A) and that transfection with miR-101 inhibitor significantly reduced the miR-101 level in T24 cells (Fig. 3D). The mRNA and protein expression levels of FZD4 were then studied. As presented in Fig. 3E and F, miR-101 overexpression significantly inhibited the mRNA and protein levels of FZD4, whereas inhibition of miR-101 significantly increased the mRNA and protein levels of FZD4 in T24 cells. Accordingly, these findings demonstrated that FZD4 is a novel target gene of miR-101 in bladder cancer cells.
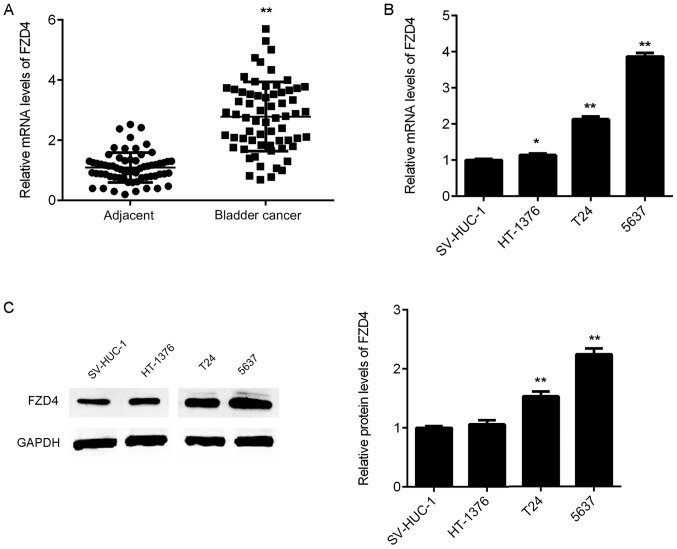
Upregulation of FZD4 in bladder cancer
RT-qPCR was then used to examine the expression of FZD4 in bladder cancer tissues. As indicated in Fig. 4A, the mRNA expression of FZD4 was significantly upregulated in bladder cancer tissue samples compared with that in adjacent non-tumour tissue samples. In addition, the mRNA and protein expression levels of FZD4 were also increased in bladder cancer cell lines compared with those in SV-HUC-1 cells (Fig. 4B and C). Therefore, FZD4 is upregulated in bladder cancer.
Figure 4.
(A) The mRNA expression of FZD4 in bladder cancer tissues and adjacent non-tumour tissues was determined using RT-qPCR. **P<0.01 vs. adjacent. (B) RT-qPCR and (C) western blot analysis were used to examine the FZD4 mRNA and protein expression levels in bladder cancer cell lines and normal bladder epithelial SV-HUC-1 cells. *P<0.05, **P<0.01 vs. SV-HUC-1. FZD4, frizzled class receptor 4; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Inhibition of FZD4 inhibits the migration and invasion of bladder cancer cells
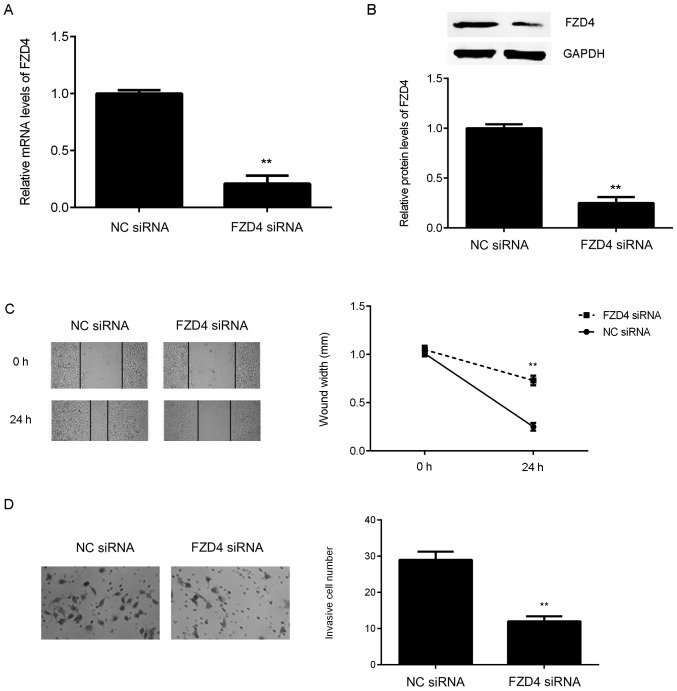
As FZD4 is significantly upregulated in bladder cancer cell lines, FZD4 siRNA was used to transfect bladder cancer T24 cells in order to downregulate its expression. Following transfection with FZD4 siRNA, the mRNA and protein expression levels of FZD4 were significantly reduced in T24 cells compared with those in the NC siRNA group (Fig. 5A and B). A wound healing assay and a Transwell assay were then conducted to examine the effect of FZD4 inhibition on the migration and invasion of T24 cells. As presented in Fig. 5C and D, knockdown of FZD4 significantly inhibited T24 cell migration and invasion.
Figure 5.
T24 cells were transfected with NC siRNA or FZD4 siRNA. Following transfection, (A) Reverse transcription-quantitative polymerase chain reaction and (B) western blot analysis were used to examine the FZD4 mRNA and protein expression levels. (C) Wound healing and (D) Transwell assays were used to examine the migration and invasion capacities of T24 cells. Magnification for wound healing assay, ×40. Magnification for Transwell assay, ×200. **P<0.01 vs. NC siRNA. NC, negative control, siRNA, small interfering RNA; FZD4, frizzled class receptor 4.
FZD4 overexpression abolished the effects of miR-101 on bladder cancer cells
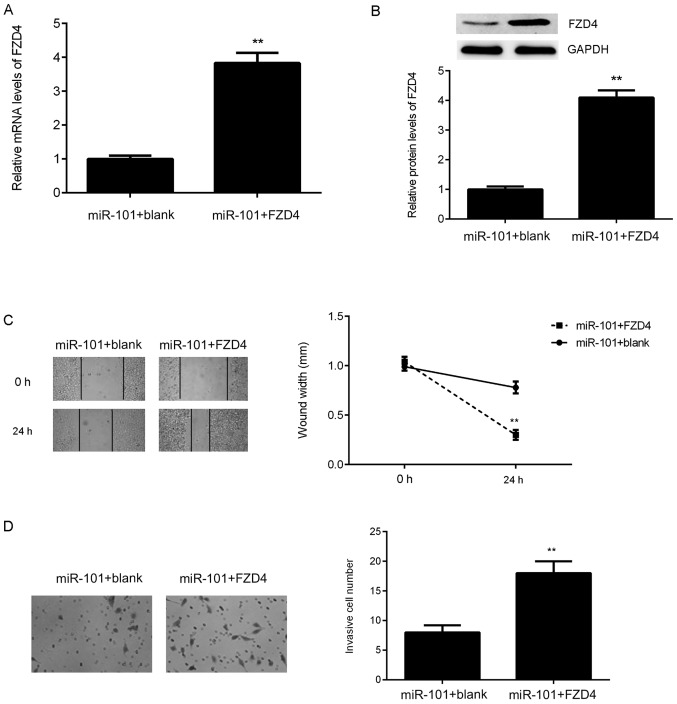
As FZD4 was negatively regulated by miR-101 in bladder cancer cells, it was speculated that FZD4 may be associated with miR-101-mediated T24 cell migration and invasion. To verify this hypothesis, miR-101-overexpressing cells were then transfected with an FZD4 expression plasmid. As presented in Fig. 6A and B, the expression levels of FZD4 were significantly upregulated in the miR-101+FZD4 group compared with those in the miR-101+blank group. It was then demonstrated that FZD4 overexpression significantly abolished the suppressive effects of miR-101 on T24 cell migration and invasion (Fig. 6C and D). These findings suggest that miR-101 inhibits the migration and invasion of bladder cancer cells via directly targeting FZD4.
Figure 6.
miR-101-overexpressing T24 cells were transfected with blank vector or FZD4 expression plasmid. Following transfection, (A) reverse transcription-quantitative polymerase chain reaction and (B) western blot analysis were used to examine the FZD4 mRNA and protein expression levels. (C) Wound healing and (D) Transwell assays were used to examine the migration and invasion capacities of T24 cells. Magnification for wound healing assay, ×40. Magnification for Transwell assay, ×200. **P<0.01 vs. miR-101+blank. miR, microRNA; FZD4, frizzled class receptor 4.
Discussion
The regulatory mechanism of miR-101 in the metastasis of bladder cancer remains unclear. In the present study it was demonstrated that miR-101 was downregulated in bladder cancer and that its reduced expression was significantly associated with tumour metastasis, advanced clinical stage, and poor prognosis. FZD4 was identified as a novel target of miR-101. The expression of FZD4 was significantly upregulated in bladder cancer and negatively regulated by miR-101 in bladder cancer cells. Both miR-101 overexpression and FZD4 inhibition caused a significant reduction in the migration and invasion of bladder cancer cells, while overexpression of FZD4 reversed the suppressive effects of miR-101 on bladder cancer cell migration and invasion.
miR-101 has been observed to be frequently downregulated in some common cancers and is thought to serve a tumour suppressive role (30,31). For instance, miR-101 is downregulated in cholangiocarcinoma, and overexpression of miR-101 significantly inhibits cholangiocarcinoma cell proliferation and tumour growth (31). In addition, miR-101-3p can inhibit the proliferation and migration of hepatocellular carcinoma cells by inhibition of Rab5a expression (30). Recently, it has also been suggested that miR-101 may function as a tumour suppressor in bladder cancer. Zhang et al previously demonstrated that miR-101 is downregulated in bladder transitional cell carcinoma, and low expression of miR-101 has been associated with a shorter survival time of patients (24). In the present study, it was demonstrated that miR-101 is downregulated in bladder cancer, and its reduced expression is significantly associated with lymph node metastasis, advanced clinical stage and poor prognosis of patients.
Furthermore, several target genes of miR-101 have previously been identified in bladder cancer. For example, Hu et al demonstrated that miR-101 suppresses the motility of bladder cancer cells via targeting c-Met (32). Our previous study demonstrated that miR-101 serves an inhibitory role in the regulation of the proliferation and invasion of bladder cancer cells by directly targeting c-FOS (26). Additionally, miR-101 has been implicated in chemotherapy resistance in bladder cancer (25). Bu et al reported that miR-101 enhances cisplatin sensitivity in human bladder cancer cells by modulating the cyclooxygenase-2 pathway (25). In addition, it has been demonstrated that miR-101 is regulated by long non-coding RNA (lncRNA) in bladder cancer cells. Liu et al reported that lncRNA SPRY4-intronic transcript 1 sponged miR-101-3p to promote the proliferation and metastasis of bladder cancer cells through increasing the expression of enhancer of zeste 2 polycomb repressive complex 2 subunit (33).
In the present study, FZD4 was identified as a novel target gene of miR-101, and it was demonstrated that the expression of FZD4 is negatively mediated by miR-101 in bladder cancer cells. FZD4, a member of the frizzled gene family, encodes a seven-transmembrane domain protein (28). The members of this family are receptors for the Wingless type MMTV integration site family of signalling proteins and the majority are coupled to the β-catenin canonical signalling pathway (28). Previously, miR-493, a tumour suppressor, was reported to inhibit bladder cancer cell growth and migration ability by inhibiting the expression of FZD4 (29). In the present study, it was demonstrated that knockdown of FZD4 caused a significant reduction in the migration and invasion of bladder cancer cells, similarly to the effects of miR-101 overexpression. Based on the above findings, it was hypothesised that FZD4 may be associated with the miR-101-mediated migration and invasion of bladder cancer cells. Further investigation demonstrated that FZD4 overexpression abolished the suppressive effects of miR-101 on the migration and invasion of bladder cancer cells, which supports the present hypothesis.
In conclusion, the present study demonstrated that miR-101 is downregulated in bladder cancer and can inhibit bladder cancer cell migration and invasion via directly targeting FZD4. The present study expands the understanding of molecular mechanisms underlying bladder cancer progression.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors' contributions
LC collected clinical tissues, examined the expression in tissues, and wrote the manuscript. YL designed the study and revised the manuscript. ZH, ZY, WL, FY, TL, LS and YZ performed the in vitro experiments.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Zhuzhou Central Hospital (Zhuzhou, China) and written informed consent was obtained from all patients.
Patient consent for publication
Written informed consent was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Skeldon SC, Larry Goldenberg S. Bladder cancer: A portal into mens health. Urol Oncol. 2015;33:40–44. doi: 10.1016/j.urolonc.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 4.Ghafouri-Fard S, Nekoohesh L, Motevaseli E. Bladder cancer biomarkers: Review and update. Asian Pac J Cancer Prev. 2014;15:2395–2403. doi: 10.7314/APJCP.2014.15.6.2395. [DOI] [PubMed] [Google Scholar]
- 5.Du C, Gao Y, Xu S, Jia J, Huang Z, Fan J, Wang X, He D, Guo P. KLF5 promotes cell migration by up-regulating FYN in bladder cancer cells. FEBS Lett. 2016;590:408–418. doi: 10.1002/1873-3468.12069. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Y, Yang C, Wang K, Liu X, Liu Q. MicroRNA-33b inhibits the proliferation and migration of osteosarcoma cells via targeting hypoxia-inducible factor-1α. Oncol Res. 2017;25:397–405. doi: 10.3727/096504016X14743337535446. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Zhang Y, Dai Q, Zeng F, Liu H. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the Rac1/JNK pathway via targeting MiR-509. Oncol Res. 2018 doi: 10.3727/096504017X14957939026111. [DOI] [PubMed] [Google Scholar]
- 8.Yang M, Zhai X, Ge T, Yang C, Lou G. MiR-181a-5p promotes proliferation and invasion, and inhibits apoptosis of cervical cancer cells via regulating inositol Polyphosphate-5-Phosphatase A (INPP5A) Oncol Res. 2017;26:703–712. doi: 10.3727/096504017X14982569377511. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Wang Y, Li J, Xu C, Zhang X. MicroRNA-139-5p inhibit cell proliferation and invasion by targeting RHO-associated coiled-coil containing protein kinase 2 in ovarian cancer. Oncol Res. 2018;26:411–420. doi: 10.3727/096504017X14974343584989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S, Hui Y, Li X, Jia Q. Silencing of lncRNA-CCDC26 restrains the growth and migration of glioma cells in vitro and in vivo via targeting miR-203. Oncol Res. 2018;26:1143–1154. doi: 10.3727/096504017X14965095236521. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Wang C, Zhou B, Liu M, Liu Y, Gao R. miR-126-5p restoration promotes cell apoptosis in cervical cancer by targeting Bcl2l2. Oncol Res. 2017;25:463–470. doi: 10.3727/096504016X14685034103879. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Luo C, Qiu J. MiR-181a inhibits cervical cancer development via downregulating GRP78. Oncol Res. 2017;25:1341–1348. doi: 10.3727/096504017X14867268787969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Wu Q, Xu B, Wang P, Fan W, Cai Y, Gu X, Meng F. MiR-124 exerts tumor suppressive functions on the cell proliferation, motility and angiogenesis of bladder cancer by fine-tuning UHRF1. FEBS J. 2015;282:4376–4388. doi: 10.1111/febs.13502. [DOI] [PubMed] [Google Scholar]
- 14.Yang X, Cheng Y, Li P, Tao J, Deng X, Zhang X, Gu M, Lu Q, Yin C. A lentiviral sponge for miRNA-21 diminishes aerobic glycolysis in bladder cancer T24 cells via the PTEN/PI3K/AKT/mTOR axis. Tumour Biol. 2015;36:383–391. doi: 10.1007/s13277-014-2617-2. [DOI] [PubMed] [Google Scholar]
- 15.Jiang Z, Zhang Y, Cao R, Li L, Zhong K, Chen Q, Xiao J. MiR-5195-3p inhibits proliferation and invasion of human bladder cancer cells by directly targeting oncogene KLF5. Oncol Res. 2017;25:1081–1087. doi: 10.3727/096504016X14831120463349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun DK, Wang JM, Zhang P, Wang YQ. MicroRNA-138 regulates metastatic potential of bladder cancer through ZEB2. Cell Physiol Biochem. 2015;37:2366–2374. doi: 10.1159/000438590. [DOI] [PubMed] [Google Scholar]
- 17.Liang Z, Li S, Xu X, Xu X, Wang X, Wu J, Zhu Y, Hu Z, Lin Y, Mao Y, et al. MicroRNA-576-3p inhibits proliferation in bladder cancer cells by targeting cyclin D1. Mol Cells. 2015;38:130–137. doi: 10.14348/molcells.2015.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riquelme I, Tapia O, Leal P, Sandoval A, Varga MG, Letelier P, Buchegger K, Bizama C, Espinoza JA, Peek RM, et al. miR-101-2, miR-125b-2 and miR-451a act as potential tumor suppressors in gastric cancer through regulation of the PI3K/AKT/mTOR pathway. Cell Oncol (Dordr) 2016;39:23–33. doi: 10.1007/s13402-015-0247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng F, Liao YJ, Cai MY, Liu TH, Chen SP, Wu PH, Wu L, Bian XW, Guan XY, Zeng YX, et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet. 2015;11:e1004873. doi: 10.1371/journal.pgen.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Liu T, Zhang T, Du S, Xie GX, Lin X, Chen L, Yuan Y. MiR-101 sensitizes human nasopharyngeal carcinoma cells to radiation by targeting stathmin 1. Mol Med Rep. 2015;11:3330–3336. doi: 10.3892/mmr.2015.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang K, Zhang Y, Ren K, Zhao G, Yan K, Ma B. MicroRNA-101 inhibits the metastasis of osteosarcoma cells by downregulation of EZH2 expression. Oncol Rep. 2014;32:2143–2149. doi: 10.3892/or.2014.3459. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Li L, Guo R, Li X, Lu Y, Guan X, Gitau SC, Wang L, Xu C, Yang B, Shan H. miR-101 promotes breast cancer cell apoptosis by targeting Janus kinase 2. Cell Physiol Biochem. 2014;34:413–422. doi: 10.1159/000363010. [DOI] [PubMed] [Google Scholar]
- 23.Lei Y, Li B, Tong S, Qi L, Hu X, Cui Y, Li Z, He W, Zu X, Wang Z, Chen M. miR-101 suppresses vascular endothelial growth factor C that inhibits migration and invasion and enhances cisplatin chemosensitivity of bladder cancer cells. PLoS One. 2015;10:e0117809. doi: 10.1371/journal.pone.0117809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Qi F, Cao Y, Chen M, Zu X. Down-regulated microRNA-101 in bladder transitional cell carcinoma is associated with poor prognosis. Med Sci Monit. 2014;20:812–817. doi: 10.12659/MSM.890300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bu Q, Fang Y, Cao Y, Chen Q, Liu Y. Enforced expression of miR-101 enhances cisplatin sensitivity in human bladder cancer cells by modulating the cyclooxygenase-2 pathway. Mol Med Rep. 2014;10:2203–2209. doi: 10.3892/mmr.2014.2455. [DOI] [PubMed] [Google Scholar]
- 26.Long Y, Wu Z, Yang X, Chen L, Han Z, Zhang Y, Liu J, Liu W, Liu X. MicroRNA-101 inhibits the proliferation and invasion of bladder cancer cells via targeting c-FOS. Mol Med Rep. 2016;14:2651–2656. doi: 10.3892/mmr.2016.5534. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Chang H, Rattner A, Nathans J. Frizzled receptors in development and disease. Curr Top Dev Biol. 2016;117:113–139. doi: 10.1016/bs.ctdb.2015.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueno K, Hirata H, Majid S, Yamamura S, Shahryari V, Tabatabai ZL, Hinoda Y, Dahiya R. Tumor suppressor microRNA-493 decreases cell motility and migration ability in human bladder cancer cells by downregulating RhoC and FZD4. Mol Cancer Ther. 2012;11:244–253. doi: 10.1158/1535-7163.MCT-11-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheng Y, Li J, Zou C, Wang S, Cao Y, Zhang J, Huang A, Tang H. Downregulation of miR-101-3p by hepatitis B virus promotes proliferation and migration of hepatocellular carcinoma cells by targeting Rab5a. Arch Virol. 2014;159:2397–2410. doi: 10.1007/s00705-014-2084-5. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Han C, Zhu H, Song K, Wu T. miR-101 inhibits cholangiocarcinoma angiogenesis through targeting vascular endothelial growth factor (VEGF) Am J Pathol. 2013;182:1629–1639. doi: 10.1016/j.ajpath.2013.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z, Lin Y, Chen H, Mao Y, Wu J, Zhu Y, Xu X, Xu X, Li S, Zheng X, Xie L. MicroRNA-101 suppresses motility of bladder cancer cells by targeting c-Met. Biochem Biophys Res Commun. 2013;435:82–87. doi: 10.1016/j.bbrc.2013.04.042. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X, Wang M, Huang C, Wang L, Zeng F, Jiang G. LncRNA SPRY4-IT1 sponges miR-101-3p to promote proliferation and metastasis of bladder cancer cells through up-regulating EZH2. Cancer Lett. 2017;388:281–291. doi: 10.1016/j.canlet.2016.12.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.