Abstract
Glioma is a type of malignant tumor accounting for 80% of all brain cancer morbidity. The long non-coding RNA (lncRNA) PVT1 has been demonstrated to be an oncogenic lncRNA in other types of cancer. However, the role of PVT1 in glioma is still unknown. The aim of the present study was to investigate the role of PVT1 in glioma, and its potential association with microRNA (miR)-200a. miR-200a mimics and small interfering (si)RNA transfection were utilized to construct miR-200a overexpression and knockdown models to investigate the effect of miR-200a on glioma cells. Slow-virus infection was used to transfect cells. Western blotting and reverse transcription-quantitative polymerase chain reaction were applied for the quantitative analysis of mRNA and protein expression. Apoptosis of podocytes was detected by terminal deoxynucleotidyl-transferase-mediated dUTP nick end labelling staining. PVT1 expression in glioma was upregulated. In vitro, PVT1 silencing via transfection with si-PVT1 suppressed proliferation and invasion and induced G0/G1 phase arrest. Luciferase reporter assay revealed the association between miR-200a and the PVT1 3′-untranslated region. Furthermore, experiments examining both miR-200a and PVT1 indicated that miR-200a could reverse the effects of PVT1 on glioma cell phenotypes. The present study reveals the overexpression of PVT1 in glioma tissue and cells and the oncogenic role of PVT1 in gliomagenesis via sponging miR-200a, thus providing a potential biomarker for the early detection of glioma and prognosis prediction.
Keywords: long non-coding RNA, glioma, proliferation, invasion, microRNA-200a
Introduction
Glioma is the most common intracranial primary tumor and domestically accounts for 35–60% of brain tumors (1). As malignant glioma is an invasive growth, it is difficult to completely remove the tumor cells from within the surrounding brain tissue by surgery, which ultimately leads to poor prognoses (2). In addition, as the blood-brain barrier can limit the entry of anti-cancer drugs into the central nervous system, surgery combined with comprehensive chemotherapy or radiotherapy treatment is not sufficiently effective, and the median survival of patients with highly malignant glioblastoma remains <1 year (3). With the gradual elucidation of the effects of long-chain non-coding RNA (lncRNA) and microRNA (miRNA or miR) and the invasion mechanism of glioma, lncRNA and miRNA were demonstrated to serve a regulatory role in the glioma invasion process (4). Therefore, a novel method to clinically inhibit malignant glioma cell migration and invasion is required to identify lncRNA and miRNAs and explore the molecular mechanism by which they regulate glioma cell migration and invasion.
According to previous studies, lncRNA-PVT1 is highly expressed in breast cancer, prostate cancer, ovarian cancer, gastric cancer and lung cancer (4–7). However, the high expression of lncRNA-PVT1 in tumor cells remains unclear. In recent years, numerous studies have confirmed that miR-200 family expression disorders exist in multiple tumor tissues and participate in the regulation of biological characteristics such as tumor cell proliferation, migration and invasion (8–10). The miR-200 family inhibits epithelial-mesenchymal transition (EMT) by directly targeting the expression of zinc finger E-box binding homeobox 1/2 (ZEB1/ZEB2) and then inhibits the migration and invasion of tumor cells (11,12). miR-200a can inhibit the migration and invasion of cluster of differentiation 133/1+ ovarian cancer stem cells (13) and nasopharyngeal carcinoma cells (14) by downregulating ZEB2. In previous studies, miRNA arrays were used to screen miRNA expression profiles closely associated with their invasion and migration in high-grade and low-grade glioma cells (15,16). The miR-200 family was demonstrated to be more notably expressed in high-grade glioma cells than in low-grade glioma cells (16). The prominent feature of high-grade glioma cells compared with low-grade glioma cells is their high invasiveness (16). Therefore, it was speculate that the miR-200 family may participate in the regulation of glioma cell migration and invasion (17). In the present study, the association between lncRNA-PVT1 was investigated and miR-200a and their role in the migration and invasion of glioma cells was explored in order to reveal the key miRNAs and molecular mechanisms that regulate the migration of glioma cells and to identify novel methods for the clinical inhibition of glioma cell migration and invasion.
Materials and methods
Specimens and cell culture
Paired glioma samples (grade II–IV) and adjacent non-tumor tissues were obtained from 48 patients who had undergone gastrointestinal surgery between 2008 and 2013 at the First Hospital of Lanzhou University (Lanzhou, China). The patients' age range was from 30–70 years old. There were 23 female and 28 male patients recruited. All specimens were immediately frozen in liquid nitrogen and stored at 80°C until total RNA extraction. Written informed consent was obtained from all patients. No patient received chemotherapy or radiotherapy before surgery. The protocol was approved by the Research Ethics Committee of First Hospital of Lanzhou University (Gansu, China).
The human glioma cell lines U-87MG, U373MG, U251, SHG44, T98G and CHG-5, and human astrocyte normal cells HEB were purchased from the Cellular Resource Center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). Cells were cultured at 37°C in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (both Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and were dissociated with 0.25% trypsin. In addition, the U-87 cell line used is derived from a glioblastoma of unknown origin and has been authenticated as the U-87MG strain by STR profiling. Furthermore, the U-373-MG cell line used is known to be a derivative of the human U-251-MG astrocytoma cell line, and has been authenticated as the U-373-MG strain by STR profiling.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA from glioma tissue and cells was extracted using TRIzol reagent (Life Technologies; Thermo Fisher Scientific, Inc.) and the RNA was measured using a NanoDrop-2000 spectrophotometer. The cDNA was then synthesized using PrimeScript RT Master Mix System (Takara Biotechnology Co., Ltd., Dalian, China) following the manufacturer's protocol. The PCR solution was prepared on ice as follows: 1 µl cDNA, 1 µl primers and 10 µl of SYBR Green qRT-PCR Master Mix (5 µl; Invitrogen; Thermo Fisher Scientific, Inc.). The final volume was then adjusted to 20 µl using RNase-free water. All reactions were carried out in an ABI FAST 7500 system. The relative expression level of each gene was calculated using the 2−ΔΔCq method (9). Each reaction was repeated three times. Primers for PCR amplification were as follows: PVT1 forward, 5′-AAAACGGCAGCAGGAAATGT-3′ and reverse, 5′-ATTCCCATAGAAGGGGCAGG-3′; miR-200a forward, 5′-CGTAACACTGTCTGGTAACGATGT-3′; and U6 forward, 5′-CTCGCTTCGGCAGCACA-3′. The reverse primers for miR-200a and the internal reference U6 are universal primers from the EXPRESS SYBR® GreenER™ miRNA qRT-PCR kit (Invitrogen; Thermo Fisher Scientific, Inc.). GAPDH (forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse, 5-AGCCTTCTCCATGGTGGTGAAGAC-3) was used as a reference protein. The PCR conditions included an initial denaturation step of 94°C for 2 min, followed by 30 cycles of 94°C for 30 sec, 59°C for 30 sec and 72°C for 2 min, and a final elongation step at 72°C for 10 min.
LncRNA profiling
RNA purity and integrity were analyzed using an Agilent Bioanalyzer 2100 for lncRNA microarray. Qualified RNA was purified using RNase-free DNase and RNeasy mini kit (Qiagen, Inc., Valencia, CA, USA). Total RNA was then amplified and labeled using a Low Input Quick Amp Labeling kit, One-Color (Agilent Technologies, Inc., Santa Clara, CA, USA) following the manufacturer's protocol. The RNeasy mini kit was then used to purify labeled complementary (c)RNA. Each slide was hybridized at 65°C with 600 ng Cy3-labeled cRNA using a Gene Expression Hybridization kit (Agilent Technologies, Inc.) in a hybridization oven (Agilent Technologies, Inc.). Following 17 h hybridization, slides were washed in staining dishes (Thermo Fisher Scientific, Inc.) with a Gene Expression Wash Buffer kit (Agilent Technologies, Inc.). Slides were scanned with an Agilent microarray scanner (Agilent Technologies, Inc.) using default settings and dye channel. Finally, data were extracted with Feature Extraction Software 10.7 (Agilent Technologies, Inc.). Raw data were normalized using a quantile algorithm with Gene Spring software 11.0 (Agilent Technologies, Inc.).
Subcellular fractionation
The nuclear and cytosolic fractions of U87 cells were separated with a PARIS kit (Life Technologies; Thermo Fisher Scientific, Inc.). RNA was isolated immediately from the fractions using TRI reagent (Molecular Research Center, Inc., Cincinnati, OH, USA). qPCR was conducted using SYBR Green Master mix system (Invitrogen; Thermo Fisher Scientific, Inc.) in a final reaction volume of 20 µl in a Mx3000 Stratagene PCR amplifier (Agilent Technologies, Inc.). The primers were used at a final concentration of 0.5 µM. The sequences for the primers used for qPCR amplification were as follows: PVT1 forward, 5′-AAAACGGCAGCAGGAAATGT-3′ and reverse, 5′-ATTCCCATAGAAGGGGCAGG-3′. After the reactions, the cycle threshold (CT) values were determined using fixed threshold settings. The miRNA expression in the cytosol and nuclei was normalized to U6 snRNA and mRNA expression in the cells was normalized to GAPDH. Primer sequences, thermocycling conditions and the quantification method used as the same as previously described.
Cell transfection
The short hairpin RNA (shRNA) sequence targeting lncRNA-PVT1 was ligated into the pLKO.1-Puro vector (Invitrogen; Thermo Fisher Scientific, Inc.). The lentivirus was packaged in 293 cells (Cellular Resource Center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences), and lentiviruses were collected from the supernatant. U87 cells (1×105 cells/well) were seeded on 6-well plates and incubated at 37°C for a further 24 h prior to transfection. The cells were then transduced lentiviruses using Lipofectamine 3000 (Thermo Fisher Scientific, Inc.) at 37°C for 24 h. The medium was then replaced with 100 µl complete culture medium at 37°C. Following 48 h of transfection, the cells were used for subsequent experiments. A total of 100 nM small interfering (si)RNA duplexes (si-PVT#1, si-PVT#2 and si-PVT#3), miR-200a mimics, a miR-200a inhibitor and negative control (NC) RNA duplexes was synthesized by Shanghai Jieli Biotechnology Co., Ltd., (Shanghai, China) for use in transient transfection assays. siRNA sequences were as follows: si-PVT#1 5′-GUGAUUUACCAGUCAGUGAAU-3′, si-PVT#2 5′-GUGAUUAACAGUCCCUGAAU-3′ and si-PVT#3 5′-GUGAAAUGCCAGUAUAUGAAU-3′. miRNA sequences were as follows: miR-200a mimics forward, 5′-UGGCAGUCUCUUAGAUGGUGG-3′ and reverse, 5′-GCCAUGCAAGACACUGCCAGG-3′; miR-200a inhibitor forward, 5′-GAGGGCAGAATCATCACGAAGT-3′ and reverse, 5′-TGAGAGATCTGGTTCCCGAAAC-3′ negative control forward, 5′-GCTCGTGGCTTAGGAGATTG-3′ and reverse, 5′-CTGGCAAAGCATGAGGAACT-3′.
Cell growth assay
Cell growth was measured using MTT (5 mg/ml; Sigma-Aldrich; Merck KGaA). Cells were transfected in 96-well plates with siRNA or vector and incubated at 37°C for 48 h, followed by the addition of 20 µl Cell Titer 96 AQueous One solution (Thermo Fisher Scientific, Inc.) and further incubation at 37°C for 1–4 h. Absorbance was recorded at 490 nm. Six replicates were conducted in the assay.
Wound healing assay
Cells were cultured in 6-well plates with DMEM supplemented with 10% FBS. When the cells in the six-well plate reached 80% confluence, the cell layer was scratched with a sterile 10-ml pipette tip, washed with medium, and incubated at 37°C with DMEM containing 1% FBS for 48 h. To prevent cell proliferation, the cells were preincubated with mitomycin C (10 µg/ml; Adooq Bioscience LLC, Irvine, CA, USA) for 1 h at 37°C. Images were captured at different time points (0 and 48 h) using a light microscope at a magnification of ×200.
Transwell assay
U87 cells were adjusted to a concentration of 2.0×105 with serum-free medium following routine digestion by Trypsin-EDTA solution (Invitrogen; Thermo Fisher Scientific, Inc.). Then, 200 µl cell suspension was transferred to the upper chamber of a Transwell plate. The chamber was implanted in 24-well plates. Then, 500 µl complete medium containing 10% FBS was added to the lower chamber, and the plates were incubated at 37°C for 24 h. Following removing the Transwell chamber, the remaining cells were removed from the upper chambers with cotton swabs. Then, the cells were fixed at 10 min at room temperature with 3.7% formaldehyde and stained at room temperature for 1 min with crystal violet. The migrated cells were counted under using a light microscope (magnification, ×200) and the mean number of migrated cells per field of view (of five random fields) were counted, which was considered an indicator of cell invasion ability.
Cell cycle distribution and apoptosis analysis
A flow cytometry assay was performed in order to detect the effect of downregulating lncRNA-PVT1 on cell cycle distribution and apoptosis. For the cell cycle distribution analysis, U87 cells were transfected with si-PVT1, collected 72 h later, subjected to trypsinization, and fixed at 4°C for 18 h with ice-cold 70% ethanol. The fixed cells were stained at room temperature for 30 min with with 500 µl propidium iodide (PI)/RNase buffer (BD Biosciences, San Jose, CA, USA) and analyzed using a flow cytometer. For cell apoptosis analysis, U87 cells transfected with si-PVT1 were collected 72 h later, stained at room temperature for 30 min with Annexin V-fluorescein isothiocycanate (FITC)/propidium iodide (PI) apoptosis detection kit (BD Biosciences). The cells were analyzed using a flow cytometer and Cell Quest acquisition software (version 2.9; BD Biosciences). Three replicates were performed.
Immunohistochemistry assays
The tumors from the xenograpft mice were fixed using 10% formaldehyde for 2 h at room temperature, embedded in paraffin and processed into 5-µm-thick sections. Antigen retrieval was performed on the tumor sections using Lab Vision™ Tris-HCl buffer for heat-induced epitope retrieval (cat no. AP-9005-050; Thermo Fisher Scientific, Inc.) for 15 min at 65°C. Following cooling at room temperature, the sections were washed with distilled water and blocked in 10% normal goat serum (Thermo Fisher Scientific, Inc.) at room temperature for 30 min. Paraffin sections were stained with hematoxylin at room temperature for 5 min and eosin at room temperature for 3 min. Ki-67 were conducted by using the streptavidin-peroxidase method (Zhongshan Golden Bridge Biotech Co., Ltd., Beijing, China). The tissue sections were deparaffinized with xylene and rehydrated with decreasing concentrations of ethanol. The sections were incubated overnight using a primary rabbit polyclonal antibody against Ki-67 (1:500; cat. no. 16667; Abcam, Cambridge, UK) in a humidified container at 4°C. Subsequently, proteins were incubated with rabbit horseradish-peroxidase-labeled immunoglobulin G (1:5,000; cat. no. ab6728; Abcam) for 12 h at 4°C. Images were acquired using a Leica IX71 light microscope (Olympus, Japan) at a magnification of ×200.
Luciferase assay
To examine interactions between PVT1 and miR-200a, wild-type PVT and mutant PVT were combined with pmirGLO-promotor vector (Shanghai GenePharma Co., Ltd., Shanghai, China) according to the manufacturer's protocol. 293 cells were plated at a density of 8×103 cells/well in 96-well plates and cultured at 37°C with 5% CO2 overnight. Cells were then co-transfected with 100 ng wild-type or mutant luciferase reporter plasmids, and 400 ng miR-200a mimic or NC miRNAs using Lipofectamine 3000. After 48 h, luciferase activity was measured using the Bright-Glo™ Luciferase assay system (Promega Corporation, Madison, WI, USA) and normalized to that of Renilla luciferase activity.
Western blot analysis
Protein samples from U87 cells were exrtracted and homogenized using radioimmunoprecipitation assay lysis buffer (Invitrogen; Thermo Fisher Scientific, Inc.) and quantified via the standard BCA method. Equal amounts of protein from each sample (50 µg/lane) were separated by SDS-PAGE on a 10% gel and electroblotted onto nitrocellulose membranes. Following blocking with TBST supplemented with 5% skimmed milk at 4°C overnight, the membranes were incubated with primary antibodies at 4°C overnight. The primary antibodies used were as follows: anti-NCAM1 (1:1,000; cat. no. 9272), anti-PTEN (1:1,000; cat. no. 79156) and anti-GAPDH (1:2,000; cat. no. 8245; Abcam). GAPDH was used as the internal control. The membranes were then incubated with horseradish peroxidase-conjugated secondary anti-primary IgG antibodies (1:5,000; cat. no. 7097; Abcam) for 1 h at room temperature. Following a wash with TBS, immunoreactive bands on the membrane were visualized using SuperSignal West Pico Chemiluminescent Substrate Trial kit (Thermo Fisher Scientific, Inc.), the results analyzed with Quantity One software (version v4.6; Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) was used for statistical analysis. Data are presented as the mean ± standard deviation. Statistical evaluation was performed using a paired Student's t-test for lncRNA-PVT1 levels in glioma tissues, and unpaired data were compared using unpaired Student's t-tests. Comparisons of data between multiple groups were analyzed using one way analysis of variance followed by Dunnett's test. P<0.05 was considered to indicate a statistically significant difference. Each experiment was repeated in triplicate.
Results
lncRNA-PVT1 is upregulated in glioma tissues and cells
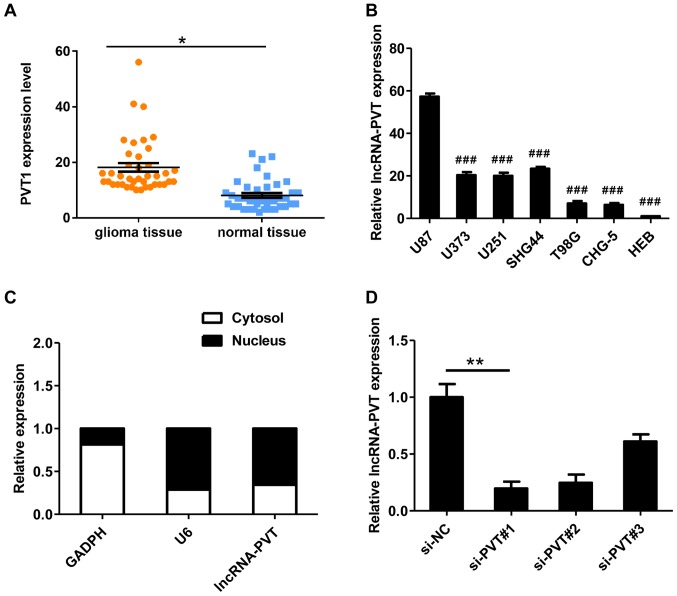
RT-qPCR was used to detect lncRNA-PVT1 levels in glioma tissues from 48 patients. The expression of lncRNA-PVT in glioma tissues was significantly higher compared with normal tissues (Fig. 1A). It was also demonstrated that lncRNA-PVT was upregulated in glioma cell lines compared with HEB cells (Fig. 1B) and preferentially located in the nucleus (Fig. 1C). In summary, these data suggest that lncRNA-PVT is highly expressed in glioma.
Figure 1.
The expression of lncRNA-PVT is associated with glioma progression. (A) lncRNA-PVT levels in glioma tissues of 48 independent patients. The adjacent non-tumor tissues were used for comparison. (B) Abundance of lncRNA-PVT in glioma cell lines compared with that in the neuron cell line, HEB. (C) Localization of lncRNA-PVT in glioma cells. U6 and GAPDH were used as markers of nuclear and cytoplasmic localization, respectively. (D) Expression levels of lncRNA-PVT were inhibited by small interfering RNAs in glioma cells. Data are presented as the mean ± standard deviation of triplicate experiments. *P<0.05, **P<0.01 as indicated. ###P<0.001 vs. HEB. lncRNA, long non-coding RNA; si, small interfering RNA; NC, negative control.
To assess the possible role of lncRNA-PVT in glioma, three different siRNAs targeting lncRNA-PVT1 were designed (named si-PVT#1-3) for transfection into U87 cells. These three siRNAs were effective at reducing the level of endogenous lncRNA-PVT (Fig. 1D). To avoid the possibility of an off-target, only si-PVT#1 was used for the follow-up experiments.
Knockdown of lncRNA-PVT inhibits glioma cell growth and invasion
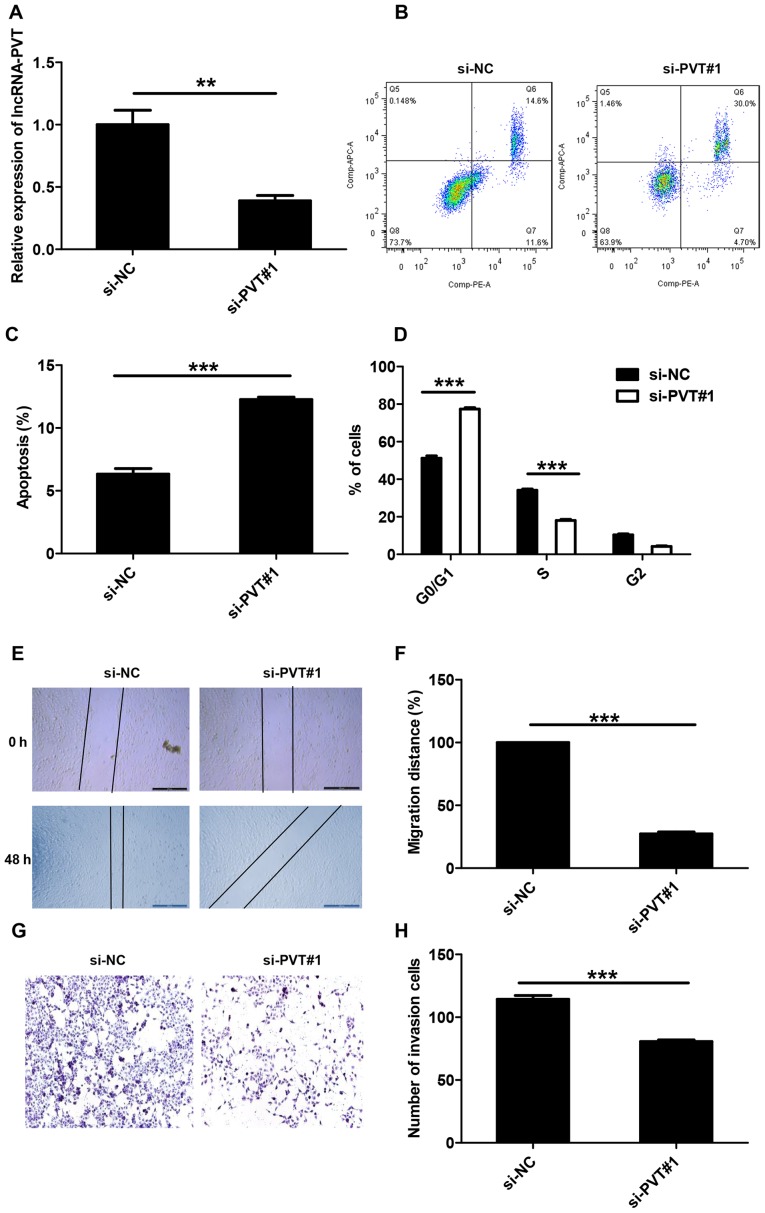
RT-qPCR was used to detect lncRNA-PVT1 levels and the results demonstrated that lncRNA-PVT1 levels in the si-NC group was 2.4 times higher compared with the si-PVT#1 group; therefore the transfection was successful (Fig. 2A). To explore the potential mechanism by which lncRNA-PVT1 promotes the proliferation of glioma cells in vitro, flow cytometry was used to differentiate the apoptotic population and cell cycle distribution of lncRNA-PVT and control glioma cells. Compared with the control group, the si-PVT#1 group exhibited a significant increase in the percentage of early apoptotic cells (Fig. 2B and C). At the same time, significant G0/G1 phase arrest was observed in cells transfected with si-PVT#1 (Fig. 2D).
Figure 2.
Knockdown lncRNA-PVT inhibits glioma migration and invasion, and arrests the cell cycle at G0/G1 phase. (A) lncRNA-PVT1 levels were detected by reverse transcription-quantitative polymerase chain reaction to demonstrate transfection success. (B) lncRNA-PVT knockdown induces apoptosis and G0/G1 arrest of glioma cells and inhibits the invasion of glioma cells. (C) The effect of lncRNA-PVT knockdown on the apoptosis of U87 cells was determined by measuring the percentage of Annexin V-stained cells using flow cytometry. (D) Effects of lncRNA-PVT knockdown on cell cycle arrest. (E and F) lncRNA-PVT silencing decreased the migration rate of U87 cells. (G and H) Knockdown of lncRNA-PVT inhibited glioma cell invasion (magnification, ×200). Data are presented as the mean ± standard deviation of triplicate experiments. **P<0.01, ***P<0.001 as indicated. lncRNA, long non-coding RNA; si, small interfering RNA; NC, negative control.
To determine whether the expression level of lncRNA-PVT is associated with glioma, wound-healing experiments and Transwell experiments were used to analyze the effect of lncRNA-PVT knockdown on U87 cell invasion. Compared with control treatment, knockdown of lncRNA-PVT inhibited cell migration (Fig. 2E and F) and significantly reduced cell invasion (Fig. 2G and H).
lncRNA-PVT is negatively associated with the expression of miR-200a
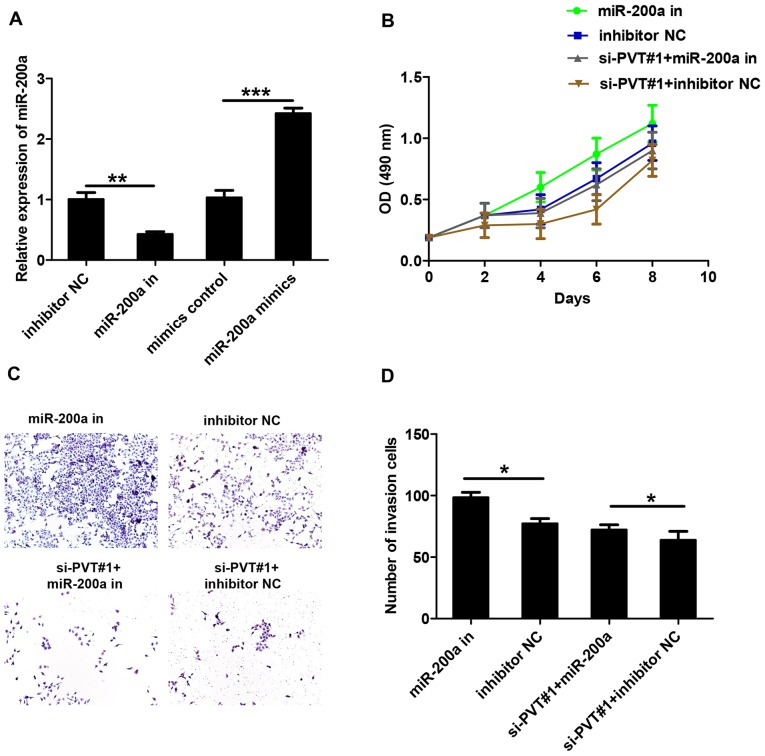
RT-qPCR was used to detect miR-200a levels, and the results demonstrated transfection success (Fig. 3A). It has been reported previously that miRNAs interact with lncRNAs to regulate their expression levels (10,11). To verify the effect of miR-200a on lncRNA-PVT, U87 cells were transfected with miR-200a inhibitors and si-PVT#1. MTS proliferation assays demonstrated that miR-200a inhibitors were able to eliminate the effects of si-PVT#1 on cell viability (Fig. 3B). A Transwell invasion assay demonstrated that miR-200a inhibitors enhanced the invasion of U87 cells compared with the inhibitor NC group (Fig. 3C and D). In addition, the co-transfection of si-PVT#1 with miRNA-200a inhibitors eliminated the inhibitory effect of si-PVT#1 on glioma cell invasion when compared with the si-PVT#1 + inhibitor NC group. These data indicate that miR-200a can inhibit the function of lncRNA-PVT.
Figure 3.
miR-200a inhibits the function of lncRNA-PVT1. (A) Reverse transcription-quantitative polymerase chain reaction to detect the expression of miR-200a. (B) The cell viability of U87 cells transfected with inhibitor NC or si-PVT#1 and miR-200a inhibitors was measured. (C and D) The invasive ability of glioma cells co-transfected with inhibitor NC or si-PVT#1 and miR-200a inhibitors was determined (magnification, ×200). Data are presented as the mean ± standard deviation of triplicate experiments. *P<0.05, **P<0.01, ***P<0.001 as indicated. miR, microRNA; lncRNA, long non-coding RNA; NC, negative control; si, small interfering RNA; in, inhibitor.
Interaction between lncRNA-PVT1 and miR-200a
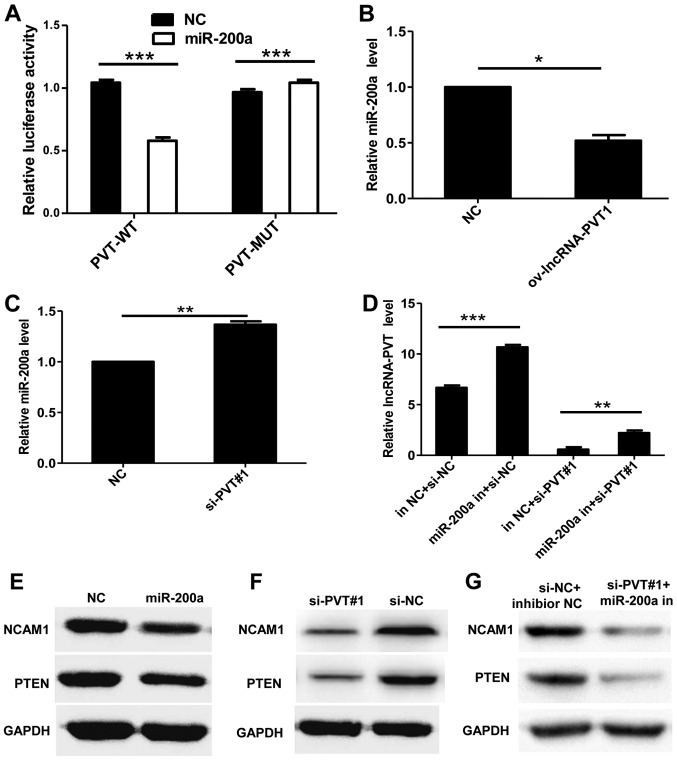
To evaluate the association between lncRNA-PVT1 and miR-200a, lncRNA-PVT1 or lncRNA-PVT1 containing a miR-200a binding site mutation were ligated into the psiCHECK vector and named PVT-WT and PVT-MUT, respectively. Following transfection with miR-200a mimic and PVT-WT or PVT-MUT, expression vectors were co-transfected into cells, and the luciferase activity was measured by a dual-luciferase assay, and the results demonstrated a significant decrease in enzyme activity, compared with controls (Fig. 4A). In addition, overexpression of lncRNA-PVT1 significantly inhibited the expression of miR-200a, whereas the silencing of lncRNA-PVT1 increased the expression of miR-200a (Fig. 4B and C). However, inhibition of miR-200a expression enhanced the expression of lincRNA-PVT1. The expression of lncRNA-PVT1 was attenuated following the co-transfection of miR-200a inhibitor with si-PVT#1 into cells compared with transfection with the NC control (Fig. 4D). Previous studies have reported that miR-200a can regulate the expression of phosphatase and tensin homolog and neural cell adhesion molecule 1 (18). Western blot analysis demonstrated that overexpression of miR-200a inhibited the expression of these genes (Fig. 4E). The inhibition of the expression of these genes was also observed in U87 cells transfected with si-PVT#1 (Fig. 4F). In addition, the expression of these genes was inhibited when the miR-200a inhibitor and si-PVT#1 were co-transfected into U87 cells (Fig. 4G). These results suggest that miR-200a is targeted by lncRNA PVT1.
Figure 4.
Interaction between lncRNA-PVT1 and miR-200a. (A) Luciferase activity of PVT1 mRNA 3′-untranslated region (WT or MUT) in the presence or the absence of miR-200a by dual-luciferase reporter system. (B) Overexpression of lncRNA-PVT1 significantly inhibited the expression of miR-200a, (C) whereas silencing lncRNA-PVT1 significantly increased the expression of miR-200a. (D) The expression of lncRNA-PVT1 was attenuated following co-transfection of miR-200a inhibitors with si-PVT#1 into cells compared with the transfection of the NC. (E) Western blot analysis demonstrated that overexpression of miR-200a inhibited the expression of these genes. (F) The inhibition of gene expression was also observed in U87 cells transfected with si-PVT#1. (G) The expression of these genes was also inhibited when the miR-200a inhibitor and si-PVT#1 were co-transfected into U87 cells. Data are presented as the mean ± standard deviation of triplicate experiments. *P<0.05, **P<0.01, ***P<0.001 as indicated. lncRNA, long non-coding RNA; miR, microRNA; WT, wild-type; MUT, mutant; si, small interfering RNA; NC, negative control; in, inhibitor; NCAM1, neural cell adhesion molecule 1; PTEN, phosphatase and tensin homolog.
Discussion
Although thousands of lncRNAs have been studied, the functional studies of lncRNA have just begun. Previous functional studies have demonstrated that lncRNA may function as an oncogene in the development and progression of human cancer (2,3).
lncRNAs form a class of non-coding RNAs with a transcript length of >200 nt (19). As lncRNAs lack a coding sequence region, they cannot encode a protein, and they work only in the form of RNA (19). Compared with miRNA, lncRNA is longer, its structure is more complex, and its mechanisms of action are more diverse (19). The transcripts produced by 4–9% of the mammalian genome sequence are lncRNA (the corresponding ratio of protein-coding RNA is 1%) (19). Therefore, lncRNA regulation of life processes may be more extensive (20). Previous studies have demonstrated that lncRNA is associated with important physiological processes such as chromatin silencing and genomic imprinting (21,22). In recent years, lncRNA has been demonstrated to be closely associated with the development of various tumors (19). Certain lncRNAs serve an important role in the development of tumors (23). Certain abnormal expression patterns of lncRNAs in tumors have an important role in the cancer diagnosis, treatment and prognosis (24). lncRNA is gradually receiving greater focus in tumor research. For example, lncRNA-HOTAIR can promote breast cancer metastasis by interacting with the polycomb repressive complex (25). Yang et al (26,27) have also demonstrated that the lncRNAs highly expressed in hepatocellular carcinoma and low expression in tumor have important effects on the development of hepatocellular carcinoma.
The PVT1 gene is often heterotopic in the tumor and is expressed with other genes, as Nagoshi demonstrated in the expression of fusion genes of PVT1-neurobeachin and WW domain containing oxidoreductase in multiple myeloma (28). According to the published literature, lncRNA-PVT1 is highly expressed in breast cancer, prostate cancer, ovarian cancer, gastric cancer and lung cancer (6,7,28). However, the role of high lncRNA-PVT1 expression in tumor cells is not very clear. In the present study, it was demonstrated that lncRNA-PVT1 promotes the proliferation and invasion of tumor cells.
Previous studies have demonstrated that exogenous overexpression of miR-200a can promote the invasion of U87 cells and that inhibition of miR-200a expression can inhibit the invasion of U87 cells (16,29). Thus, it was speculated that miR-200a serves an important role in glioma invasion and may be one of the key miRNAs associated with the regulation of glioma cell migration.
In the present study, it was demonstrated that lncRNA-PVT1 is frequently overexpressed in glioma tissue, indicating the carcinogenic activity of lncRNA-PVT1. The discovery of the function of lncRNA-PVT1 was also demonstrated by inhibiting the expression of lncRNA-PVT1 in glioma cells. In recent years, a number of studies have confirmed miR-200 family expression disorders in a variety of tumor tissues, wherein they participate in tumor cell proliferation, migration, and invasion and the regulation of biological processes (8–10,29). Although miR-200a has been demonstrated to target many protein-coding genes, the present study suggested that miR-200a also targets lncRNA-PVT1. It was demonstrated that the expression of lncRNA-PVT1 and miR-200a in clinical glioma tissue was negatively associated. This suggests that the expression of miR-200a in glioma tissue can inhibit the expression of lncRNA-PVT1.
In conclusion, the present data suggest that lncRNA-PVT1 is upregulated in human glioma tissue and can be used as a negative prognostic factor in patients with glioma. Decreasing the expression of lncRNA-PVT1 inhibits the proliferation and invasion of glioma cells and induces apoptosis. lncRNA-PVT1 acts as an oncogene by upregulating miR-200a. The present study reveals the overexpression of PVT1 in glioma tissue and cells and the oncogenic role of PVT1 in gliomagenesis via sponging of miR-200a, thus providing a potential biomarker for the early detection and prognostic prediction of glioma. It has been reported that miR-200a can directly target the mRNA of β-catenin (CTNNB1 transcription). The downregulation of miR-200a may affect the tumor development by activating the wnt pathway and causing epithelial-mesenchymal transition. In further research, we will continue to explore the possible mRNA and biologic process associated with glioma genesis.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and Technology Project of Lanzhou City (grant no. 2015-2-67) and the Health Industry Research Plan of Gansu Province (grant no. GWGL2014-49).
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YZ and GY designed the present study and analyzed the data. YL and GY performed the experiments.
Ethics approval and consent to participate
Written informed consent was obtained from all patients. The protocol was approved by the Research Ethics Committee of First Hospital of Lanzhou University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Tsuzuki T, Izumoto S, Ohnishi T, Hiraga S, Arita N, Hayakawa T. Neural cell adhesion molecule L1 in gliomas: Correlation with TGF-beta and p53. J Clin Pathol. 1998;51:13–17. doi: 10.1136/jcp.51.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleber S, Sancho-Martinez I, Wiestler B, Beisel A, Gieffers C, Hill O, Thiemann M, Mueller W, Sykora J, Kuhn A, et al. Yes and PI3K bind CD95 to signal invasion of glioblastoma. Cancer Cell. 2008;13:235–248. doi: 10.1016/j.ccr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro Oncol. 2012;14(Suppl 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, Nishida K, Okuda K, Sakamoto N, Kobayashi S, Yamamoto-Sugitani M, et al. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res. 2012;72:4954–4962. doi: 10.1158/0008-5472.CAN-12-0213. [DOI] [PubMed] [Google Scholar]
- 5.Yu C, Wang Y, Li G, She L, Zhang D, Chen X, Zhang X, Qin Z, Cao H, Liu Y. LncRNA PVT1 promotes malignant progression in squamous cell carcinoma of the head and neck. J Cancer. 2018;9:3593–3602. doi: 10.7150/jca.26465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan B, Wu HY, Lv DJ, Zhou XM, Zhong LR, Lei B, Zhang SB, Mao XM. Downregulation of lncRNA PVT1 expression inhibits proliferation and migration by regulating p38 expression in prostate cancer. Oncol Lett. 2018;16:5160–5166. doi: 10.3892/ol.2018.9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y, Du H, Bao L, Liu W. LncRNA PVT1 promotes ovarian cancer progression by silencing miR-214. Cancer Biol Med. 2018;15:238–250. doi: 10.20892/j.issn.2095-3941.2017.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, Wang X, Zhu Y, Zhu J, Lai Q. miR-200b-3p inhibits proliferation and induces apoptosis in colorectal cancer by targeting Wnt1. Mol Med Rep. 2018;18:2571–2580. doi: 10.3892/mmr.2018.9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren W, Gao L, Qiang C, Li S, Zheng J, Wang Q, Zhi Y, Cai G, Kong X, Zhou M, et al. Kindlin-2-mediated upregulation of ZEB2 facilitates migration and invasion of oral squamous cell carcinoma in a miR-200b-dependent manner. Am J Transl Res. 2018;10:2529–2541. [PMC free article] [PubMed] [Google Scholar]
- 10.Pillman KA, Phillips CA, Roslan S, Toubia J, Dredge BK, Bert AG, Lumb R, Neumann DP, Li X, Conn SJ, et al. miR-200/375 control epithelial plasticity-associated alternative splicing by repressing the RNA-binding protein Quaking. EMBO J. 2018;37(pii):e99016. doi: 10.15252/embj.201899016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng YB, He X, Huang YF, Wu QN, Zhou YC, Hao DJ. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol Res. 2017;25:1207–1214. doi: 10.3727/096504017X14886679715637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia H, Ng SS, Jiang S, Cheung WK, Sze J, Bian XW, Kung HF, Lin MC. miR-200a-mediated downregulation of ZEB2 and CTNNB1 differentially inhibits nasopharyngeal carcinoma cell growth, migration and invasion. Biochem Biophys Res Commun. 2010;391:535–541. doi: 10.1016/j.bbrc.2009.11.093. [DOI] [PubMed] [Google Scholar]
- 13.Wang C, Kang L, Wang X, Liu Y, Zhao X. Expression of miR-200a and chemotherapeutic treatment efficacy of glioma. Oncol Lett. 2018;15:5767–5771. doi: 10.3892/ol.2018.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Chiyomaru T, Fukuhara S, Saini S, Majid S, Deng G, Shahryari V, Chang I, Tanaka Y, Enokida H, Nakagawa M, et al. Long non-coding RNA HOTAIR is targeted and regulated by miR-141 in human cancer cells. J Biol Chem. 2014;289:12550–12565. doi: 10.1074/jbc.M113.488593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu J, Rodova M, Nanta R, Meeker D, Van Veldhuizen PJ, Srivastava RK, Shankar S. NPV-LDE-225 (Erismodegib) inhibits epithelial mesenchymal transition and self-renewal of glioblastoma initiating cells by regulating miR-21, miR-128 and miR-200. Neuro Oncol. 2013;15:691–706. doi: 10.1093/neuonc/not011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y, Chen N, Sun F, Fan Q. CREB up-regulates long non-coding RNA HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. doi: 10.1093/nar/gkq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palazzo AF, Lee ES. Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front Genet. 2018;9:440. doi: 10.3389/fgene.2018.00440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 20.Serviss JT, Johnsson P, Grander D. An emerging role for long non-coding RNAs in cancer metastasis. Front Genet. 2014;5:234. doi: 10.3389/fgene.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–772. doi: 10.1016/j.tibs.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhang L, Peng D, Sood AK, Dang CV, Zhong X. Shedding light on the dark cancer genomes: Long noncoding RNAs as novel biomarkers and potential therapeutic targets for cancer. Mol Cancer Ther. 2018;17:1816–1823. doi: 10.1158/1535-7163.MCT-18-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helsmoortel H, Everaert C, Lumen N, Ost P, Vandesompele J. Detecting long non-coding RNA biomarkers in prostate cancer liquid biopsies: Hype or hope? Noncoding RNA Res. 2018;3:64–74. doi: 10.1016/j.ncrna.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: A new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chisholm KM, Wan Y, Li R, Montgomery KD, Chang HY, West RB. Detection of long non-coding RNA in archival tissue: Correlation with polycomb protein expression in primary and metastatic breast carcinoma. PLoS One. 2012;7:e47998. doi: 10.1371/journal.pone.0047998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang F, Huo XS, Yuan SX, Zhang L, Zhou WP, Wang F, Sun SH. Repression of the long noncoding RNA-LET by histone deacetylase 3 contributes to hypoxia-mediated metastasis. Mol Cell. 2013;49:1083–1096. doi: 10.1016/j.molcel.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Yang F, Zhang L, Huo XS, Yuan JH, Xu D, Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al. Long noncoding RNA high expression in hepatocellular carcinoma facilitates tumor growth through enhancer of zeste homolog 2 in humans. Hepatology. 2011;54:1679–1689. doi: 10.1002/hep.24563. [DOI] [PubMed] [Google Scholar]
- 28.Nagoshi H, Taki T, Hanamura I, Nitta M, Otsuki T, Nishida K, Okuda K, Sakamoto N, Kobayashi S, Yamamoto-Sugitani M, et al. PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res. 2012;72:4954–4962. doi: 10.1158/0008-5472.CAN-12-0213. [DOI] [PubMed] [Google Scholar]
- 29.Yu F, Zheng Y, Hong W, Chen B, Dong P, Zheng J. MicroRNA-200a suppresses epithelial-to-mesenchymal transition in rat hepatic stellate cells via GLI family zinc finger 2. Mol Med Rep. 2015;12:8121–8128. doi: 10.3892/mmr.2015.4452. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.