Abstract
In their native environment, cells are immersed in a complex milieu of biochemical and biophysical cues. These cues may include growth factors, the extracellular matrix, cell–cell contacts, stiffness, and topography, and they are responsible for regulating cellular behaviors such as adhesion, proliferation, migration, apoptosis, and differentiation. The decision-making process used to convert these extracellular inputs into actions is highly complex and sensitive to changes both in the type of individual cue (e.g., growth factor dose/level, timing) and in how these individual cues are combined (e.g., homotypic/heterotypic combinations). In this review, we highlight recent advances in the development of engineering-based approaches to study the cellular decision-making process. Specifically, we discuss the use of biomaterial platforms that enable controlled and tailored delivery of individual and combined cues, as well as the application of computational modeling to analyses of the complex cellular decision-making networks.
Keywords: growth factors, extracellular matrix, cell–cell communication, intracellular signaling, microfluidics
1. INTRODUCTION
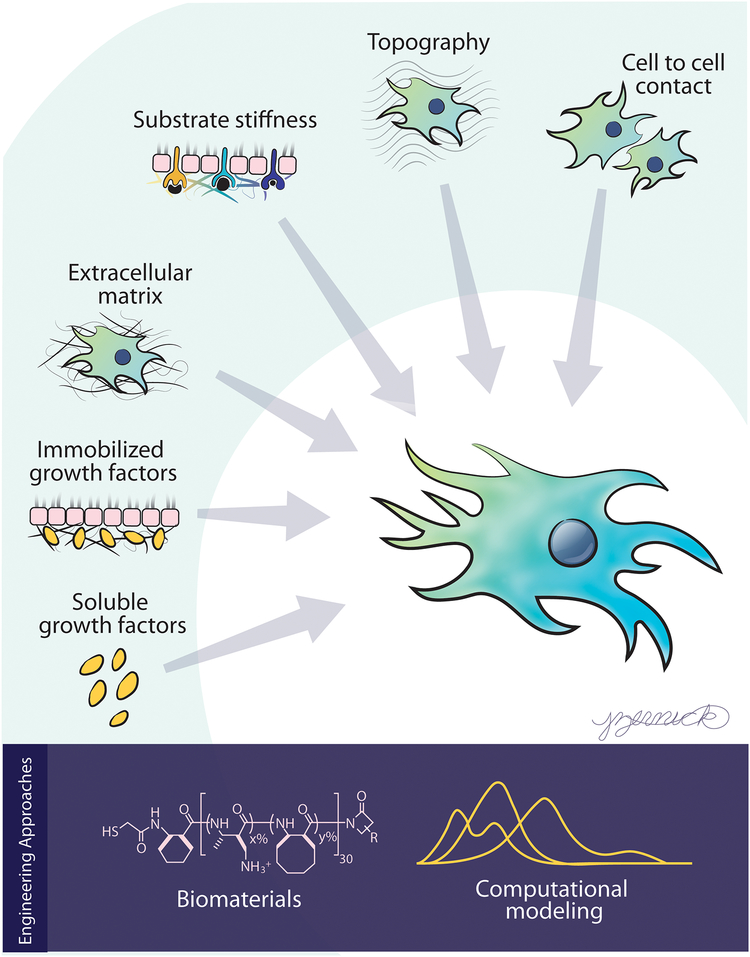
In vivo, cells are naturally exposed to a complex milieu of biochemical and biophysical cues that they constantly sample and then process to make decisions regarding their fate (1). These cues range from soluble growth factors to membrane-bound proteins on neighboring cells to tissue stiffness, and they change dynamically during tissue development, repair, and disease (Figure 1). Cells sense this extracellular information primarily through cell membrane-bound receptors, which initiate intracellular signaling cascades that integrate and ultimately convert this complex array of information into cellular responses such as adhesion, proliferation, migration, differentiation, and death. Understanding how cells interpret different cues and which signaling processes elicit different cellular decisions is critical to develop rational approaches to treat disease and engineer healthy tissues.
Figure 1.
Overview of cues that are received by cells and the engineering approaches covered in this review that can help decode the effects of these cues on cellular decisions.
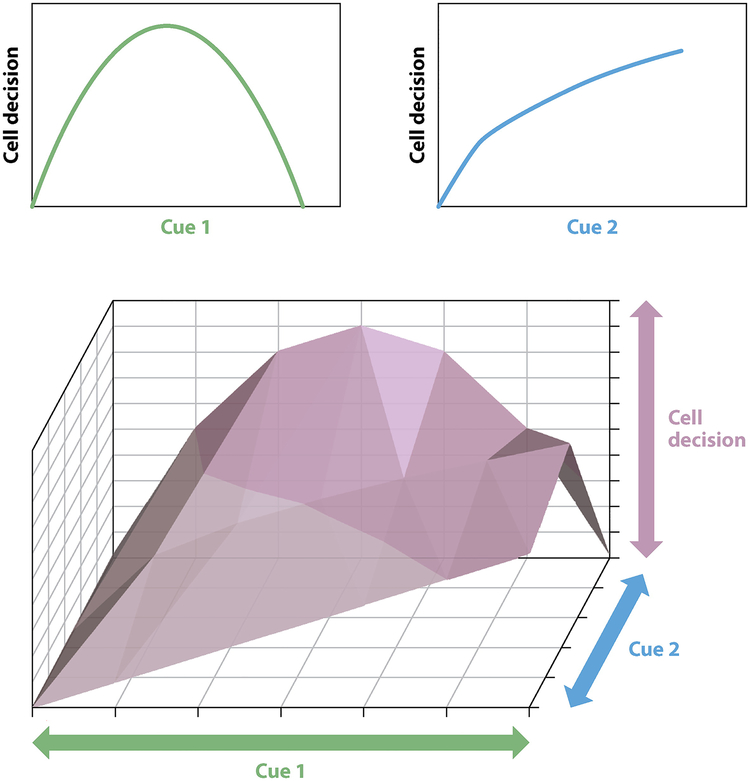
This review focuses on the development of engineering-based approaches to achieve controlled modulation of extracellular cues and their application to studies of cellular decision making. Specifically, we discuss platforms for perturbing the delivery of several types of biochemical cues (e.g., soluble factors, cell–cell interactions) and biophysical cues (e.g., substrate stiffness, topography), as well as advances in computational modeling to analyze the resulting multifactorial data sets. We first discuss approaches to the study of cellular decision making based on changing a single input. However, it is important to remember that although only a single input is varied in these studies, the cells are experiencing other cues. For example, with in vitro studies, researchers commonly make generalized statements such as “growth factor X increases the proliferation of cell type Y” when what they actually mean is “growth factor X increases the proliferation of cell type Y on tissue culture polystyrene (TCPS).” The omission of such qualifiers can be problematic, as the response of a cell to any single stimulus is dependent upon the properties of its environment. The stiffness of the culture substrate, the type of adhesive ligands available, and the density at which the cells are cultured are examples of features capable of dramatically altering the manner in which cells respond to a cue (Figure 2). Therefore, we also focus on recent advances where these engineering approaches have been used to study the effects of multiple cues in combination on cell fate.
Figure 2.
Cellular responses to variations in a single cue can vary depending on the cue. However, when the same cues are varied in concert, the resulting behavioral landscape can be complex and not obvious from the results of experiments that examined only individual variations.
2. ENGINEERING SINGLE CUES
2.1. Growth Factors
Cells exhibit distinct responses to different growth factors, and for an individual growth factor, there are numerous variables that influence the cellular decision-making process, including dose, timing, and presentation scheme. Recent studies have demonstrated that mathematical models, microfluidic platforms, or biomaterials-based methods can begin to decode the impact of these variables.
2.1.1. Effects of ligand dose.
It is widely understood that cells often show dose-dependent effects when treated with growth factors, and that this relationship varies for cell type and growth factor combinations. While the classic dose–response study in which cells are treated with a bolus of soluble growth factor is straightforward to implement, the results can vary depending on experimental conditions. For example, the level of Smad phosphorylation in response to transforming growth factor β (TGF-β) input depends on cell density (2). An experimental analysis led to the conclusion that this was due to increased ligand internalization and degradation as cell number increased, which resulted in decreased TGF-β receptor activation. Similarly, experiments showed that the response of ovarian cancer cells to insulin-like growth factor 1 (IGF1) depends on two cellular-mediated processes that decrease the level of free ligand available to activate IGF1 receptor (IGF1R): endocytosis/degradation and binding by insulin-like growth factor binding proteins (IGFBPs) (3). Through the use of a mass-action kinetic model, these processes could be simulated to predict the steady-state level of phosphorylated IGF1R, which showed a strong correlation to the extent of proliferation in response to IGF1. Similar findings have been reported for the epidermal growth factor (EGF) system, in which differences in ligand depletion rate were determined to be responsible for differences in mitogenic potency between EGF and transforming growth factor α (TGF-α) (4). While these three studies focused on receptor tyrosine kinases, a recent model demonstrated that ligand depletion is important for G protein–coupled receptors as well. Use of a multiscale model that incorporated cells positive for different receptors for CXCL12 and microfluidic source-sink experiments revealed that differences in ligand–receptor affinity induce different ligand gradients due to depletion kinetics, with ligand depletion-induced gradients that were short distance and steep most effective at promoting chemotactic migration (5).
An important consequence of these studies is that the cellular decision in response to changes in ligand dose is more properly described if ligand availability is given in terms of amount of growth factor/cell rather than traditional concentration units (e.g., nanograms per milliliter). The interpretation that concentration is not the best predictor of cell response may seem surprising, because in vitro assays of receptor–ligand binding equilibrium are governed by concentration-dependent kinetics. However, in intact cellular experiments, the actual concentration of ligand available for each receptor is dependent on multiple factors, such as cell number (which alters receptor number) and media volume (which affects the total amount of ligand, and therefore, ligand depletion kinetics). These insights should be considered when comparing experimental results across different scales. For example, the same concentration applied in a standard culture setup may deplete significantly faster in a microfluidic setting where cells are more concentrated relative to media volumes.
2.1.2. Time-dependent effects.
Importantly, the most common experimental setup—treatment with a bolus of growth factor at a single time point—does not accurately recapitulate many physiological systems in which growth factors are present for short periods of time or are replenished by transport from the blood stream. Through microfluidic approaches or simply the addition or removal of stimuli-containing media at set times, it has become apparent that stimulation dynamics can affect cellular decision making. For example, Gaudet and colleagues (6) showed that treatment with tumor necrosis factor α (TNF-α) for as little as 10 s was sufficient to induce activation of the nuclear factor κB (NF-κB) pathway. Interestingly, cell apoptosis for TNF-α exposure for 1 min was similar to that for continuous exposure, and significantly higher than exposure for 1 h, possibly due to changes in cross talk between downstream pathways. As another example, exposure of cells to a short pulse (30 s) of TGF-β resulted in transient activation of Smad phosphorylation, but repeated short pulses led to sustained Smad phosphorylation (7). While the response to single pulses was graded with respect to TGF-β dose, multiple pulses led to switch-like behavior. These differential responses may allow cells to respond to a larger range of doses present for short time periods, but filter out constant low-level stimuli.
2.1.3. Effects of ligand presentation scheme.
While treating cells with growth factors in solution is easy to implement experimentally, there are numerous physiological examples where growth factors are found physically tethered to residues in the extracellular matrix (ECM), effectively immobilizing them (8). The impact of growth factors present in these different forms is an area of intense interest in the development of biomaterials to direct cellular decisions. In a seminal study, Kuhl & Griffith (9) developed a method to tether murine EGF (which has a single primary amine) onto poly(ethylene oxide)-functionalized surface using amine-reactive chemistry. In its tethered form, EGF prevents EGF receptor (EGFR) endocytosis, resulting in sustained phosphorylation of extracellular signal–regulated kinase (ERK) and reduced Fas ligand–induced apoptosis of mesenchymal stem cells (MSCs) (10). Importantly, this sustained activation of ERK did not interfere with differentiation toward osteogenic or adipogenic lineage (11). More recently, this group adapted its immobilization scheme for tricalcium phosphate scaffolds and showed that tethered EGF increased MSC survival nearly fourfold when injected into immunocompetent mice (12). The amine-reactive chemistry employed to immobilize EGF is not amenable to all growth factors, as many have additional lysine residues that are critical for biological function. As an alternative approach, photoinitiated thiol–acrylate chemistry has been used to covalently tether growth factors such as TGF-β onto poly(ethylene glycol) (PEG) hydrogels (13). The tethered TGF-β promoted chondrogenesis of MSCs at similar levels as TGF-β in its soluble form, indicating that the covalent binding did not affect the bioactivity of the ligand. Meanwhile, a novel platform to immobilize vascular endothelial growth factor (VEGF) relies on a two-step process in which VEGF is first immobilized through its heparin-binding domain to dictate orientation and then covalently “locked” in place through a photoreactive group (14).
Cells are capable of responding to ligands in both their soluble and immobilized forms, raising the question of how the presentation scheme affects cellular decisions. However, comparing the two methods directly is challenging. For example, in soluble growth factor experiments the cells are already in culture prior to stimulation, whereas in immobilized growth studies cells undergo the process of attachment and spreading in the presence of the stimuli. To control for this, studies have utilized inhibitors against the growth factor receptor during the process of initial seeding in order to control the onset of signaling (15). Alternatively, one can temporally control the contact time between cells and ligand. For example, cells cultured on a polydimethylsiloxane (PDMS) surface were inverted and placed on top of a surface where growth factor was immobilized (16). The time in contact can be regulated to reflect the short timescales (minutes) of soluble factor–based experiments. Through the use of this system, VEGF receptor 2 and p38 phosphorylation was extended compared with soluble delivery and occurred independently of ligand internalization (16). Altered signaling was also observed for keratinocytes cultured on immobilized EGF versus those treated with soluble EGF; immobilized EGF led to lower ERK and Akt phosphorylation but increased collective migration, whereas soluble EGF induced high ERK and Akt phosphorylation with increased cell proliferation (17).
An additional complication that arises when comparing soluble to immobilized conditions is determining how to match the doses appropriately. Through quantification of immobilization efficiency and the use of concentration in terms of molecules of growth factor per cell (based on insights outlined in Section 2.1.2), we recently treated keratinocytes with dose-matched soluble and immobilized EGF. Under these conditions, phospholipase C γ1 (PLCγ1) was specifically activated by immobilized EGF as a result of differences in EGFR internalization, and activation of PLCγ1 resulted in increased single-cell migrational persistence and faster wound closure (15).
2.2. Insoluble Extracellular Matrix Proteins
In addition to polypeptide growth factors, the insoluble ECM surrounding cells plays a critical role in modulating cellular signaling to influence cellular decisions such as proliferation, differentiation, and migration. While differences in biochemical cues (e.g., binding and degradation sites) between ECM molecules affect cell behavior, we focus on recent advances demonstrating how ECM physical cues, such as mechanical stiffness and topography, affect cellular decisions and can be explored through biomaterials-based approaches.
2.2.1. Effects of extracellular matrix stiffness.
The elastic moduli of different tissues in the body vary from <1 kPa in the brain to >1 GPa for calcified bone (18, 19), although the modulus of most noncalcified tissues is generally below 50 kPa, with much of the mechanical characteristics of a tissue resulting from the ECM. The first observation that mechanical properties alone affect cellular spreading and motility was nearly 20 years ago (20). Such findings have been observed in numerous cell types since, although the effects are commonly found to depend on cell type (21). While the effect of stiffness has been examined in natural materials such as collagen, examining a range of stiffnesses without impacting other properties such as ligand density is difficult in natural materials. Therefore, most studies have relied on the use of synthetic systems such as polyacrylamide (PAA), PDMS, PEG, or polysaccharide-based biomaterials such as alginate or hyaluronic acid (HA), which are rendered cell adhesive by incorporation of ECM molecules or adhesive peptide sequences.
Using PAA gels modified with collagen, Discher and colleagues (22) reported that MSCs showed differentiation patterns that were dependent on the substrate elasticity. Most notably, this report observed differentiation that matched the mechanical properties of native tissue—that is, cells on the softest materials expressed neural markers, and those on stiffer substrates showed osteogenic markers. While these results were initially attributed solely to the difference in stiffness between the substrates, later reports have suggested that this observation may result from intrinsic variations in collagen conjugation (23). Briefly, as PAA gel formulations are changed to achieve different stiffnesses, the pore size of the surface is changed, which alters spacing between collagen conjugation points and potentially affects how the collagen transmits the mechanics of the substrate to the cells. This interpretation was supported by evidence that MSCs cultured on collagen-modified PDMS substrates were not affected by stiffness and that changes in crosslinker concentration influenced the spacing (but not amount) of collagen on PAA, where MSCs demonstrated stiffness sensitivity (23). However, the relationship between collagen linkage site spacing, stiffness, and stem cell differentiation remains unresolved, as subsequent studies have employed alternative approaches to isolate these variables and have reached different conclusions (24). Taken together, these studies demonstrate that decoupling the impact of stiffness from other variables is complex, even in relatively simple two-dimensional (2D) cultures. Yet, this challenge of accounting for multiple aspects of the microenvironment also serves as a source of inspiration for continual retooling of biomaterials-based approaches to enable isolation of specific single cues within a complex multi-cue system.
Of course, many cells experience the ECM in three dimensions in vivo. To examine the impact of stiffness in such environments, it is necessary to use materials that are compatible with cell encapsulation. A study using HA modified with RGD peptides found that MSCs show differences in cell shape and spreading that depend on both the stiffness of the material and the dimensionality of the matrix (25). For example, cells spread more on stiffer 2D substrates but showed a biphasic response as stiffness was varied in three-dimensional (3D) gels. Intriguingly, the Yes-associated protein/transcriptional coactivator with PDZ-binding motif (YAP/TAZ) nuclear/cytoplasmic ratio increased with stiffness in 2D but decreased with stiffness in 3D gels, demonstrating that investigations of downstream signaling may help clarify differences in cellular decisions in these different contexts (25).
One limitation of many hydrogel systems is that the adhesion is incorporated through relatively short adhesive peptides, which do not transmit force in the same manner as intact ECM fibers. For example, fibrin has a higher stiffness in tension than under compression due to the buckling of fibers under compression. This nonlinearity has been examined through finite element models, which predict that fiber microbuckling leads to a decrease in local compression stiffness and provides a mechanism by which cells can sense changes at greater distances in native ECM than is possible in a linear polymer (26). To examine how changes in fiber stiffness impact cells, Chen and colleagues (27) employed a method in which methacrylated dextran functionalized with RGD is electrospun into fibers of varied diameter, density, and anisotropy, and cross-linked for differing lengths of time to modulate stiffness. Cells on fibers with lower stiffness were able to reorganize the fibers, resulting in a local increase in the density of RGD, increased focal adhesion formation, and ultimately increased proliferation. These studies demonstrate that cellular behavior is influenced by the unique mechanical properties of fibrous matrices, and suggests that cellular responses in native ECM will be much more heterogeneous than observed in model systems.
Importantly, stiffness within a tissue is not constant over time and may undergo significant changes during the pathogenesis of conditions such as fibrosis and cancer (28). To examine how cells respond to dynamic changes in stiffness, investigators have developed numerous materials-based approaches that enable cells to be cultured at one stiffness for a period of time before the material is either softened or hardened. For example, the use of methacrylated HA allows two possible cross-linking reactions to be exploited: a Michael-type addition with subsaturating levels of DTT and free-radical polymerization of the remaining available sites (29). MSCs cultured on these gels show increased area and differentiation to an osteogenic lineage with dynamic stiffening. As an alternative, gels made of PEG–norbornene have been dynamically stiffened by swelling with additional PEG–norbornene/PEG–thiol and a second photocrosslinking step (30). Valvular interstitial cells (VICs) encapsulated in soft gels have a myofibroblastic morphology, whereas cells in stiff gels are rounder; when VICs were allowed to spread in soft gels prior to stiffening, the myofibroblastic morphology was retained. Intriguingly, markers for the VIC activated phenotype were partially reversed with stiffening, suggesting that mechanics can regulate differentiation independently of morphology (30). Additional approaches have been developed to examine the impact of softening materials. For example, VICs cultured on stiff 2D PEG–alkyne gels cross-linked with a photodegradable azide cross-linker showed that the activated phenotype was lost with softening (31). Note that this is the opposite trend observed in 3D gels (30), further demonstrating the complicated effect of mechanotransduction on cellular decisions.
In order to study the impact of dynamic changes in mechanics, it would be ideal to utilize materials that can be softened or stiffened without changing the core polymer chemistry. In one such approach, alginate gels were developed with embedded liposomes containing gold nanoparticles and either CaCl2 or a chelator (32). When exposed to light, the heat from the nanoparticles released the lipsosome cargo and caused an increase or decrease in material stiffness; when fibroblasts were cultured in gels that stiffened, they were observed to go from an amoeboid morphology to rounded. One limitation of this system is that a single liposome formulation is used to both soften and stiffen the gel, preventing reversible patterns from being tested. As an alternative, PEG cross-linked with azobenzene containing cross-linking peptides can be softened or stiffened by the use of light to convert azobenzene between cis and trans configurations (33). Development of these systems may lead to an improved understanding of how changes in tissue mechanics influence cellular decisions during the dynamic processes of development and disease progression.
2.2.2. Effects of extracellular matrix topography.
Another property of the ECM that can influence cellular behavior is its topography. Numerous studies have demonstrated that cells respond to microscale patterns such as ridges by altering their shape to conform to the pattern (reviewed in Reference 34). To date, most of these studies have used static substrates in which the pattern is present as the cells are added to the material and remains unchanged throughout the experiment. In contrast, the topography of the native ECM is dynamic throughout development and disease. In order to determine whether cells respond to dynamic changes in topography, PDMS substrates were subjected to stretch and exposure to UV and ozone (35). When the stretch was released, the substrate presented lamellar wrinkles that could be reversed by restretching the material. MSCs responded to this material with changes in cell shape and alignment when the patterns changed, although there appeared to be a delay of approximately 1 day (35). This approach can also present more complex patterns, suggesting that it could be used to mimic changes observed in development or disease.
More recently, focus has moved from microscale effects to the impacts of the topography of the ECM fibers with which cells interact. Electrospinning of an elastin-like protein with fiber diameters ranging from 0.8 to 2.0 μm revealed differences in topography as characterized by arc/chord ratio and z-dimension variation (36). These altered substrate characteristics influence cell adhesions; on 0.8-μm fibers endothelial cells have similar z-dimensional morphology as on flat substrates, whereas on wider fibers the cells conform to the surface of the fibers. On these wider fibers, the cells have disrupted VE-cadherin and increased speed and proliferation. Cells on these wider fibers also had increased YAP nuclear localization; experimental tests with small interfering RNA (siRNA) against YAP and inhibitors of the YAP/TEAD interaction suggest that this was necessary for topography-mediated cellular decisions (36). Although the fiber orientation in most studies is random, analyses of intact tissue have revealed patterns in the ECM fibers that change in diseases (37, 38). These complex patterns can be reconstructed using multiphoton microscopy to “read” the pattern and multiphoton excited photochemistry to “write” it (39). Such techniques will enable much finer control to determine the impact of different parameters of ECM topography.
Of course, cells in vivo experience fiber topography in three dimensions. Seminal research from the Keely lab found that the radial alignment of collagen fibers around tumors accompanies progression in breast cancer (40) and predicts poor outcomes (41). To examine the impact of collagen I fiber alignment in three dimensions, Reinhart-King and colleagues (42) utilized magnetic particles embedded in collagen to induce fiber alignment during gelation. Because cells can remodel these fibers, these authors focused on the initial event of breast cancer cell spreading and found that cells elongated faster, ultimately resulting in a higher percentage of motile cells. In aligned matrices, cellular filipodia demonstrated clear anisotropy with preferential elongation along the direction of the fibers, and the length and lifetime of these protrusions were regulated by Rac1 and FAK (42). These results suggest that the aligned matrices associated with poor outcomes in breast cancer may exert their effect by directing cellular decisions that enable a faster and more efficient invasion process. It will be interesting to examine the effect of fiber alignment on other migratory mechanisms (i.e., collective, amoeboid) and in more complex 3D geometries, such as those that can be generated by two-photon laser scanning lithography (43).
2.3. Cell–Cell Contacts
Cell interactions with their neighbors in vivo influence cellular decisions. Although these interactions include the exchange of soluble factors and force transmission through the ECM, there are additional mechanisms that rely on direct cell–cell contact such as juxtracrine signaling, desmosomes, adherens and tight junctions, and gap junctions (44). At the simplest level, cell–cell contacts have been studied using cells seeded at different densities (45) or as spheroids (46). In order to study the impact of cell contacts in a more controlled manner, microcontact printing has been utilized to print ECM islands/stripes with varying dimensions to accommodate defined numbers of cells (47). Recently, this method was extended to develop small aggregates (<10 cells) that are then released from the surface by brief exposure to collagenase and encapsulated in 3D scaffolds (48). Quantification of albumin production suggested that cell–cell contacts supported hepatocyte differentiation for significantly longer times, suggesting that pre-engineering cell–cell contacts may be useful for the development of microtissues.
With these approaches it is difficult to separate the impact of soluble interactions from the effects of juxtracrine signaling or other cell–cell contacts. A key example of juxtracrine signaling is that of ephrin/Eph, which enable bidirectional communication between cells during developmental processes such as angiogenesis (49). To directly assay the impact of ephrin-A1 (ligand for EphA2), PEG hydrogels were modified with Fc–ephrin-A1; endothelial cells cultured on 2D gels of this material demonstrated increased proliferation and tubule formation with the addition of ephrin-A1 (50). Incorporation of ephrin-A5/EphA5 into PEG hydrogels increases the viability of dissociated primary mouse β-islet cells (51). Intriguingly, ephrin-A5/EphA5 were effective at improving survival at nanomolar concentrations, whereas the RGD peptide sequence was effective only at micromolar levels, suggesting that engineering cell–cell signaling may be more critical than cell–ECM interactions to regulate cellular survival for some cell types.
Another critical juxtacrine interaction in development is the delta/notch system (52). To examine the impact of the notch ligand Jagged-1 on stem cell proliferation and differentiation, investigators indirectly immobilized histidine-tagged Jagged-1 to surfaces through adsorbed anti-poly-histidine (53). Jagged-1 modification increased differentiation of human embryonic stem cells to an ectoderm phenotype and cardiovascular progenitor cells to cardiomyocytes. More recently, an agent-based model was utilized to simulate the effects of altered delta/notch signaling on intestinal stem cell differentiation in colonic crypts (54). The computational model predicted, and experiments in a mouse model of notch hyperactivation supported, that lateral inhibition resulting from delta/notch signaling in the transit-amplifying zone of the crypt was essential to control the ratio of differentiated stem cell fates. Taken together, these studies suggest that future efforts to regulate cellular decisions may benefit by expanding beyond soluble growth factors to examine juxtacrine-mediated signaling.
In a similar vein, preliminary studies isolating the effects of cellular junctions suggest that incorporation of adherens junctions will provide an additional mechanism to instruct cell behavior. The use of nonlinear diffusion modeling predicted that cell–cell adhesion would support collective migration by enabling cells to pull their neighbors, rather than hindering migration by increasing drag (55). This prediction was experimentally confirmed by siRNA knockdown of α-catenin in keratinocytes, which resulted in significantly less wound closure. In an alternate approach to analyze the impact of cell–cell junctions, HA gels that incorporated the HAV motif from N-cadherin were observed to increase initial differentiation of MSCs to chondrocytes both in vitro and in vivo (56). Follow-up research by the same group determined that cells in HAV-modified gels were rounder, with increased nuclear β-catenin (57). Interestingly, these authors observed a similar increase in nuclear β-catenin in cells in small clusters in control gels, suggest that varying cell cluster size or N-cadherin peptide levels might enable regulation of β-catenin localization to influence downstream decisions.
3. ENGINEERING MULTIPLE CUES
It may seem overwhelming to consider that many well-accepted cell responses may hold true only in a narrow range of environments, or to postulate exactly how many qualifiers must be given for an experimental finding in order to accurately summarize its context. However, it is unlikely that all cues exert equivalent levels of influence on cell behavior. The design of novel engineered scaffold environments provides an avenue to achieve independent and combined variation of individual types of cues, from which we may start to identify a hierarchy of cue influences for a given cellular response. Computational models may also be applied to further enrich analysis of the complex data sets emerging from such multifactorial investigations.
3.1. Homotypic Cue Combinations
In their native environments, cells are immersed in a complex mixture containing thousands of soluble factors. Some of these factors provide complementary or synergistic cues, whereas others force the cell to resolve conflicting information. The delivery of growth factors and growth factor combinations has been studied extensively in two dimensions on TCPS, but the development of more advanced biomaterial platforms has enabled greater control over the delivery of these soluble cues while also providing the cells with a more physiologically relevant microenvironment. A canonical example of balancing growth factor combinations to yield results not possible with either factor alone is the stimulation of angiogenesis with VEGF and platelet-derived growth factor (PDGF) (58). A study using biomaterial scaffolds to achieve controlled delivery of VEGF and PDGF found that a combination of these factors increased vessel size and functionality compared with either factor alone (59), an effect that could be further optimized by sequential delivery of VEGF followed by PDGF-BB (60).
Many other growth factor combinations have been delivered from biomaterial scaffolds or controlled-release particles to yield similar effects in the context of osteogenesis (61), vascularization (62), neurogenesis (63), and other cellular outcomes (64). When faced with any of these combinations, cells must interpret the different soluble cues to ultimately reach a decision that integrates the separate inputs and yields a specific cellular response; engineering approaches have also been implemented to understand the mechanisms by which cells make these decisions. Specifically, large-scale computational techniques have been applied to analyze how intracellular signaling networks coordinate to translate multi–growth factor exposure to changes in cell behavior. Research by Lauffenburger and colleagues (65) described a data-driven principal components–based model capable of predicting cell response from the unique patterns of intracellular signal mediators induced by multiple soluble cues. This study analyzed the cellular response of 9 pairwise combinations of 3 different soluble cues (TNF-α, EGF, and insulin) by measuring levels of 19 different intracellular signaling mediators and 4 behavior outputs linked to apoptosis at several distinct time points. The resulting model accurately predicted the multiple time-dependent apoptotic responses that result from various combinations of prodeath TNF-α and prosurvival EGF and insulin. Furthermore, this model found previously undiscovered feedback loops that play a significant role in the signaling pathways leading to apoptosis (65). Understanding the ways in which cells integrate information from multiple soluble cues could ultimately enable greater optimization and control over these events, which may be translated to inform approaches for tissue regeneration or design targeted inhibitors to halt dominant pathways in disease processes.
The presence of many different ECM molecules within the cellular microenvironment may also be described as a type of homotypic cue combination. ECM molecules are capable of binding to cells via different integrin and nonintegrin receptors, and changes in ECM composition or in the fragmentation of a single type of ECM molecule can signal important information about tissue health and integrity (28). The use of scaffolds containing multiple ECM components [e.g., fibrin plus collagen (66) or Matrigel itself (67)] is common in tissue engineering, although these matrices are less typically used in the context of cellular decision making, which requires the blended scaffolds to be compared with scaffolds composed of individual ECM components. One method to facilitate the production of multiple ECM combinations and their controls, originally described by Bhatia and colleagues (68), involves spotting microarrays of individual and combined ECM proteins atop thin PAA gels and analyzing the cellular responses. This technique has been applied to analyze large numbers of ECM combinations in the context of hepatic differentiation and hepatocyte function (68) and is amenable to combination with soluble factor delivery (69). Although performed in 2D experiments, this approach could provide high-throughput and inexpensive screening of ECM conditions to then translate to 3D experiments. Through the use of a PEG-based scaffold, combinations of RGD, collagen I, and HA have been examined in the context of chondrogenesis (70). For several chondrogenic outcomes, a combination of RGD with either collagen I or HA did not lead to a change in cellular behavior relative to RGD alone; however, the combination of all three insoluble cues stimulated a synergistic increase in markers such as collagen type I production (70).
Reductionist approaches, which use discrete peptide sequences to mimic elements of native ECM proteins, may also be used to create multi-cue ECM environments. ECM-derived peptides are less unwieldy to combine in scaffold synthesis than whole proteins, and peptide-modified matrices have had widespread use in tissue engineering (71). One ECM feature that can be mimicked using multipeptide materials is the presentation of synergistic or cryptic sites within an ECM protein, sites that are not captured by simple presentation of a single peptide. For example, several groups have synthesized difunctional peptide chains that contain RGD, its synergism site PHSRN, and an inert spacer in between to mimic the spacing of these two sequences in native fibronectin (72, 73). This divalent sequence synergistically increased cell adhesion compared with either peptide alone (72, 73). A multifactorial combination of different adhesive peptides has also revealed synergistic and opposing actions of peptide pairs (74). A sequence allowing self-assembly was coupled to each peptide, and hydrogels were formed in a modular manner using varied combinations of the peptides across a range of concentrations. Using endothelial cell proliferation as the desired outcome, this study (74) found several instances in which the level of effect of one peptide depended upon the presence of a second peptide at a specific concentration. Additionally, this research demonstrated how factorial design can identify interactions between cues and Design of Experiment approaches such as response surface methodology can then identify optimized combinations for a particular cellular decision (74).
3.2. Heterotypic Cue Combinations
In addition to homotypic combinations, cells are exposed to diverse classes of stimuli within their microenvironment, giving rise to the presentation of heterotypic cue combinations. Construction of in vitro environments that enable the study of cellular decision making in response to heterotypic cues has required the development of engineered platforms that permit controlled presentation and independent variation of these cues.
3.2.1. Stiffness and soluble cues.
Most studies that examine the cellular response to growth factors or other soluble cues employ TCPS as the cell culture substrate. However, because different cell types in the body sit in different mechanical milieus, a one-size-fits-all approach to providing a culture substrate is not appropriate to elucidate physiologically relevant behaviors. As described in Section 2.2.1, numerous cell decisions, such as differentiation, migration, and proliferation, can be strongly influenced by substrate stiffness. The signaling cascades and cytoskeletal restructuring initiated by these mechanotransduction events have consequences for the manner in which cells respond to soluble factors. Additionally, even within an individual tissue, the mechanical environment undergoes changes with development, repair, and disease stage, thus motivating the investigation of how these biophysical changes influence cellular decision making when faced with other surrounding cues.
2D culture of cells on PAA gels has remained the most common approach for investigating the impact of stiffness on the cellular response to additional cues. An early demonstration of interplay between stiffness and soluble cues was presented in 2006 by Engler et al. (22) (also see Section 2.2.1, above). Specifically, the cell culture media supplements that are commonly used to direct lineage-specific differentiation were found to be most effective when paired with the substrate stiffness that corresponded to the target tissue type. For example, muscle-induction medium stimulated the greatest expression of myoblast markers when the MSCs were cultured on PAA substrates exhibiting a muscle-like elastic modulus (11 kPa). Similar results were obtained for the combination of osteogenic medium and stiffness, indicating a synergy between media- and stiffness-based induction of cellular differentiation. These findings were also dependent on the extent of lineage commitment at the time of soluble cue delivery; the addition of differentiation media that opposed the stiffness-induced differentiation lineage (e.g., treatment of cells on a soft, neurogenic substrate with osteogenic medium) was effective at trans-differentiating the cells only at early time points. Thus, for more committed cells, the influence of substrate stiffness appeared to be dominant compared with the soluble differentiation stimuli.
Other groups have similarly achieved synergistic induction of cell differentiation by using a combination of mechanical and soluble cues on a PAA-based platform (75, 76). Moreover, the stiffness at which a cell is most responsive to a growth factor can vary with growth factor identity. Such an effect was observed for chondrocyte differentiation, in which the greatest increases in cellular stiffness and traction forces were achieved through a combination of TGF-β1 and the stiffest substrates examined (90 kPa), but the converse was true for IL-1β, which induced maximal effects on cellular stiffness and traction force when applied to cells on soft (1 kPa) materials (75). This stiffness-dependent sensitization to soluble differentiation cues has been observed across many other cell types; the range of mechanics and type of soluble stimulus depends on the type of tissue being examined (18, 22, 77, 78).
The interplay between stiffness and soluble cues has also been investigated in the context of tissue repair and disease. For example, keratinocyte functions related to dermal wound healing are modulated by a combination of stiffness and EGF, wherein cells were unresponsive to EGF on low-stiffness materials (1 kPa) but exhibited significantly enhanced sensitivity to EGF with respect to proliferation, migration, and hypertrophy when cultured on stiffer substrates (30 or 100 kPa) (79). Even though EGF was ineffective on soft substrates, there was still robust EGFR phosphorylation in response to EGF delivery, highlighting that receptor activation cannot always be used as a proxy for the extent of growth factor–induced signaling. Additionally, because tissue stiffness changes during repair, these findings have implications for timing the delivery of soluble factors to the appropriate healing stage. Increased stiffness also led to increased EGF sensitivity in epithelial MDCK cells (80). In this case, the amount of EGF needed to overcome contact-inhibited cell proliferation was reduced 100-fold by increasing substrate stiffness by only 4.5-fold (80). These responses may be due in part to disruption of cell–cell contacts on stiffer substrates (80); together with the increased sensitivity to EGF, these findings may be relevant to understanding the role of microenvironmental stiffness in tumor progression, and identifying the underlying signaling could yield potential treatment targets.
Although 2Dculture on PAA gels offers advantages with respect to its ease of use, reproducibility, and long history as a culture substrate, it is limited in its ability to provide more biologically complex milieus or examine cell behaviors (e.g., invasion) that can occur only in three dimensions. The use of more sophisticated biomaterial platforms to investigate the effect of microenvironment stiffness on the cellular response to soluble cues has generally yielded findings consistent with those obtained on PAA gels. However, unlike PAA, these scaffold platforms have incorporated varied cross-linking or modification methods intended to achieve improved control over other scaffold properties (e.g., pore size, ECM presentation) (81–84). In matrices ranging from silk fibroin (81) to modified gelatin (82), scaffold stiffness modulates the cellular response to soluble factors in a manner that depends strongly on both the specific cell type and specific cue. While stiffness is often named as the dominant cue in guiding cellular decision making in multi-cue systems (85–87), it is important to recognize that such a statement also requires qualifiers to be accurate. Even within a single cell type, identification of a dominant cue depends strongly on both the specific soluble factor being delivered and the specific cellular response being measured (77).
Although most studies examining the combination of biophysical and soluble cues have focused on the impact of native molecules on outcomes such as cellular differentiation, there is an emerging appreciation that microenvironmental stiffness may also be an important modulator of the cellular response to nonnative molecules (e.g., drugs). Increased matrix stiffness increases tumor cell malignancy (88), but it has been unclear how such changes in stiffness may influence the sensitivity of the tumor cells to antitumorigenic agents. Not surprisingly, the findings in these studies have been dependent on the cell type being investigated (84, 89). However, in cases where the tumor cells were responsive to the cytotoxic agent, the drug-induced stimulation of apoptosis was most effective on the softest substrates. This finding was consistent across 2D and 3D experiments using materials ranging from PAA (89, 90) to multiple types of PEG derivatives (83, 84). Interestingly, however, there were cellular outcomes for which the drug efficacy was greatest on stiff substrates. Specifically, colony formation by hepatocellular carcinoma cells was reduced by cisplatin or fluorouracil treatment on 12-kPa, but not 1 kPa, substrates (90); this result was attributed to culture on soft substrates causing the selective enrichment of a colony-forming cell phenotype. Thus, although an antineoplastic agent may be more successful at reducing overall cell number in stiffer tumors, this could come at the cost of increasing the proportion of cancer stem cells in the tumor, illustrating the complexities involved in designing therapeutic strategies when faced with opposing cellular decisions. However, such challenges may also inspire new cotreatment approaches that are tailored to the specific tumor microenvironment. Additionally, these results highlight the limitations and potential for false-discovery rates using traditional in vitro drug screening on TCPS, which is far stiffer than even the stiffest tumors.
3.2.2. Stiffness and extracellular matrix cues.
Due to the challenges of designing materials with independently varied biophysical and biochemical properties, studies of the combination of increased stiffness and increased adhesive ligand density have often been somewhat inadvertent. Investigators have frequently employed type I collagen gels, which are easily constructed (i.e., without the need for experience in materials chemistry), allow 3D cell culture, and can be varied in stiffness via an increase in collagen concentration (91–93). However, this approach simultaneously increases the density of ECM ligands, which can also influence cellular decisions. Although concurrent increases in stiffness and ECM density are physiologically relevant to many conditions, such as breast cancer (94) and fibrosis (95), the ability to decouple these variables is important to both accurately control their values and study their contributions to tissue function. The dissection of these variables can be simplified via the use of 2D platforms based on PAA or PEG, which allow independent modulation of stiffness and ECM density to be accomplished in a rather straightforward manner (86, 87). For example, using separate sets of substrates, Barcus et al. (87) sought to understand the contributions of stiffness versus collagen density in regulating prolactin signaling by breast cancer cells, as their earlier research had discovered increased prolactin signaling in traditional collagen gels where stiffness and density were simultaneously increased (92). Separating these variables, these authors found that stiffness was the dominant regulator of the cellular response to prolactin and elucidated pathways involved in this response, thereby identifying a potential novel target for inhibiting these tumorigenic signals (87).
Efforts to synthesize alternative, 3D biomaterial platforms that enable independent variation of stiffness and ECM cues (including both ligand density and topography) have been wide ranging, with a recent push to include more of the native ECM structure. Independent variation of scaffold stiffness and peptide density by use of PEG- or alginate-based hydrogels and covalently tethered ECM-derived peptides has been possible for more than 20 years (96, 97). However, this reductionist approach can omit information provided by the full ECM molecule that is essential for addressing some biological questions. Thus, more recently developed approaches have combined various PEG derivatives or alginate with whole ECM proteins (98–101), methacrylated gelatin alone or modified with whole proteins (102, 103), or HA with other ECM components (104–106). Alternatively, other groups have devised methods to increase the elastic modulus of collagen gels without changing collagen concentration via the introduction of cross-linking molecules (107, 108). Using these platforms, various cell types have been found to respond to changes in material stiffness, as well as changes in ECM identity, in a manner that is dependent upon the cell type and measured outcome. Several of these studies have also illustrated the importance of ECM features that are not typically captured in reductionist approaches, such as the necessity of a fibrous collagen structure in promoting breast cancer cell invasion (102).
However, despite the creation of various 3D biomaterial environments that enable independent tuning of mechanical and ECM properties, relatively few studies have intentionally merged these features to examine their combined effects. Rather, many investigations employing scaffolds with decoupled variables have evaluated ECM variations at a single stiffness, or vice versa. The evaluation of scaffold properties independently of one another has drawbacks, as it can be difficult to predict the cellular response to a combination of cues from their response to the cues in isolation. Substrate stiffness affects the kinetics of interactions between cells and ECM ligands, modulating the lifetime of the integrin-ligand bond, which in turn affects cytoskeletal assembly and cellular differentiation (109). Individual ECM proteins also differ in their ability to transmit or shield cells from mechanical forces due to the dependence of ligand accessibility on mechanical tension in the ECM (110). An increase in ligand density on a soft substrate may yield no effect, but a simultaneous increase in stiffness and ligand density can result in synergistic increases in a cell differentiation marker, as was demonstrated for osteogenic differentiation of MSCs in PEG gels (111). Using a PEG-based system that included collagen I and varied amounts of fibronectin and laminin, Yang and colleagues (98) demonstrated an interdependence of mechanical and biochemical properties in regulating osteogenesis of adipose-derived stem cells, wherein the optimal stiffness for achieving osteogenesis depended on the type and concentration of ECM cues present. These findings highlight the importance of pursuing a combinatorial approach to material design, rather than performing sequential optimization of biomaterial properties.
3.2.3. Adhesion and soluble cues.
Communication between growth factor receptors and adhesion receptors (both cell–ECM and cell–cell) has been extensively documented in many types of cells (112). The cross talk and physical interactions between these classes of receptors can yield an adhesion-dependent response to growth factors. These effects can be observed in simplified approaches that deliver a soluble cue to cells cultured on ECM-coated TCPS. For example, the responsiveness of mammary epithelial cells to insulin or EGF is dictated by the type of ECM present, with basement membrane promoting the response to insulin and collagen I promoting the response to EGF (113). ECM coatings can also differentially regulate cellular responsiveness to pathological soluble stimuli, as demonstrated by the finding that fibrin provides a supportive environment for TGF-β1-induced nodule formation by VICs while the same cells were largely resistant to the influence of TGF-β1 when cultured on collagen I (114). A similar dependence of soluble cue signaling on ECM identity has been demonstrated in 3D matrices. For example, endometriotic epithelial cells were responsive to macrophage-conditioned media when cultured on basement membrane, but not on collagen I or TCPS (115). These findings demonstrate the inability of traditional culture platforms (e.g., TCPS) to capture important cellular behaviors and responses.
Combinations of ECM ligands and soluble cues have also been widely explored in more tailored biomaterial environments. Because PEG or alginate alone is nonadhesive to cells, matrices fabricated from these elements commonly include an adhesive peptide, and experiments using these matrices often involve the application of a soluble stimulus. The most typical example of this approach is the application of differentiation medium to cells cultured in scaffolds with a range of adhesive ligand densities (116, 117). In these environments, cell–ECM adhesion often acts as a permissive switch, enabling the cells to respond to the delivered soluble cues (118). However, this effect also depends on the type of ECM ligand presented, as different peptides bind to different integrin receptors, affecting integrin-specific interactions with growth factor receptors (112). In many of these studies, cell–ECM interactions are the dominant cue in cellular decision making. In other circumstances, by contrast, soluble cues are the primary driver of cellular decision making, with ECM identity serving to fine-tune the cellular response (69). Overall, these observations raise important considerations for designing the in vitro environment, where the absence of cellular response to a soluble cue could be a physiologically inaccurate response caused by absence of the requisite ECM input in that specific in vitro setup.
Cell–cell contact represents another type of adhesion that is able to regulate cellular responsiveness to soluble cues. Cell–cell contact and mitogenic growth factors often work in opposition to each other, as proliferation is attenuated by contact inhibition but stimulated by many growth factors. In vivo, contact inhibition generally overrides the influence of soluble growth factors (119), preventing uncontrolled cell growth. However, using a combination of micropatterned substrates to control cell placement and gene delivery to modulate E-cadherin levels, Asthagiri and colleagues (45) demonstrated that contact inhibition controls the proliferative capacity of mammary epithelial cells only below a certain threshold level of EGF. Moreover, the growth factor threshold does not remain at a set level, but instead varies with the extent of cell–cell contact. Cells must also resolve conflicting cues provided by cell–cell contact and soluble growth factors to make decisions in the context of cell migration. A controlled system was engineered using a microfluidic approach to compare the effects of growth factor and contact-inhibition cues at the single-cell level (120). Similar to the above-mentioned findings for proliferation, a threshold of EGF existed over which the promigration stimulus provided by this cue overruled the contact inhibition of locomotion in rat mammary tumor cells. Understanding the context dependence and hierarchy of factors that modulate events such as cell proliferation and migration can thus inform the development of approaches to inhibit tumor metastasis and growth.
3.2.4. Other cue combinations.
It is not possible to address all possible cue combinations within a single review. Therefore, the preceding sections have focused on a few of the more prevalent cue combinations that are being investigated using engineering-based approaches. However, robust investigations of cellular decision making in response to many other multi-cue systems are under way. For example, variations in substrate topography can sensitize cells to various growth factors or ECM cues (121–124). Also, there have been several reports of cell–cell interactions modulating the cellular response to changes in scaffold stiffness (125–127). A common theme in all of these studies is that they were enabled by the construction of novel culture environments that allow independent modulation of individual cues.
The exhaustive number of potential cue combinations has also motivated the development of platforms that enable high throughout in executing multi-cue experiments. Most studies have focused upon rather limited combinations of cues, in part because of the unwieldiness of generating and analyzing larger numbers of conditions. In an effort to address this issue, several elegant studies have described the generation of arrayed platforms (85, 128–131). Microwell arrays containing thin PEG hydrogel films have been used to enable combinatorial screening of substrate stiffness, cell adhesion proteins, chemokines, and signaling proteins on cellular behavior (85). Specifically, Lutolf and colleagues found that adipogenic differentiation of MSCs was affected by both biochemical and biophysical cues. To decouple the relative effects of these inputs, these authors developed generalized linear models to find the strength of individual effects as well as cases of synergy/antagonism. Through this analysis, Lutolf and colleagues concluded that stiffness played the dominant role in regulating this behavior, with biochemical cues fine-tuning the cellular response. In separate research, robotic liquid-dispensing technology was employed to pattern more than 1,000 unique microenvironments, constructed using PEG derivatives modified to vary stiffness, degradability, ECM type, cell–cell interaction components, and soluble factors (129). Interestingly, an examination of mouse embryonic stem cell proliferation and self-renewal across these conditions revealed soluble factors to be the dominant cues and the strongest predictor of cell outcomes. Other groups have used micropatterning methods to examine combinations of cell–cell contact and soluble cues (130) or ECM composition and soluble cues (128), or have employed photolithography to pattern simultaneous gradients of stiffness and ligand density by using HA hydrogels (131). Each of these studies enabled investigation of a larger number of conditions than traditional, discrete methods. The development of such higher-throughput methods also creates the need for more systems-level computational techniques to analyze the resulting data (129).
4. FUTURE DIRECTIONS
Most of the studies we have discussed have looked at cells at a population-averaged level under the assumption of homogeneous inputs, but it is not the population of cells that chooses to live or die—it is the individual cell. Methods to analyze and characterize cell behavior at the single-cell level include single-cell -omic measures such as RNA-Seq (132), flow/mass cytometry (133, 134), and imaging-based approaches (135). Of these, only imaging is able to track the same cell in time to monitor both signaling events and downstream effects. Numerous tools have been developed to monitor individual signaling pathways in real time, including fluorescent reporters to monitor protein localization, kinase activity (136, 137), hydrogen peroxide localization (138), and Ca2+ (139). Importantly, many of these readouts can be multiplexed with single-cell endpoint analysis such as single-molecule fluorescent in situ hybridization for gene expression (140) or microwells for the analysis of secreted proteins (141). Additionally, imaging modalities such as second harmonic generation can characterize the variation in fiber topography (142) and quantify forces from fiber displacement (143). Combining these signaling and microenvironment readouts may enable a deeper understanding of how the ECM influences cell behavior.
The goal for many biomedical engineers is to utilize our understanding of cellular decision making to regulate these decisions toward some desired outcome, such as wound healing or tissue regeneration. As single-cell experiments have demonstrated that, even for uniform stimuli, there is considerable heterogeneity with respect to an individual cellular decision, and that cells can execute multiple decisions in response to one cue, achieving a desired outcome requires determining how to best tip the balance of cellular decisions toward the ultimate goal. For example, in a recent study our groups analyzed the collective response of reepithelialization in response to changes in both stiffness and EGF dose, both of which could be incorporated into a wound healing approach (79). We determined that both stiffness and EGF influenced cell proliferation, migration, and spreading. Using partial least squares regression modeling, we determined that cell persistence and cell area were the most strongly correlated to wound closure, suggesting that approaches that increase cell spreading (in particular, stiffness) and cell persistence [e.g., immobilization of EGF (15)] should be combined. This approach—to deconstruct the collective response, decode the contributions of individual cellular behaviors, and then identify methods to regulate them—could be broadly adapted to aid in the design of therapeutic strategies.
ACKNOWLEDGMENTS
The writing of this review was supported by grants from the National Institutes of Health (1DP2CA195766 to P.K.K.; R01GM099031 to K.S.M.; R21CA202040 to P.K.K. and K.S.M.; R21EY026222 to K.S.M.; R01HL093281 to K.S.M.; and R21EB019508 to K.S.M.), the National Science Foundation (CBET-1401584 to K.S.M. and P.K.K.), and the American Cancer Society (RSG-13–026–01-CSM to P.K.K.).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Downward J 2001. The ins and outs of signalling. Nature 411:759–62 [DOI] [PubMed] [Google Scholar]
- 2.Clarke DC, Brown ML, Erickson RA, Shi Y, Liu X. 2009. Transforming growth factor β depletion is the primary determinant of Smad signaling kinetics. Mol. Cell. Biol 29:2443–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tian D, Kreeger PK. 2014. Analysis of the quantitative balance between insulin-like growth factor (IGF)-1 ligand, receptor, and binding protein levels to predict cell sensitivity and therapeutic efficacy. BMC Syst. Biol 8:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reddy CC, Wells A, Lauffenburger DA. 1996. Receptor-mediated effects on ligand availability influence relative mitogenic potencies of epidermal growth factor and transforming growth factor alpha. J. Cell Physiol 166:512–22 [DOI] [PubMed] [Google Scholar]
- 5.Chang SL, Cavnar SP, Takayama S, Luker GD, Linderman JJ. 2015. Cell, isoform, and environment factors shape gradients and modulate chemotaxis. PLOS ONE 10:e0123450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee RE, Qasaimeh MA, Xia X, Juncker D, Gaudet S. 2016. NF-κB signalling and cell fate decisions in response to a short pulse of tumour necrosis factor. Sci. Rep 6:39519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zi Z, Feng Z, Chapnick DA, Dahl M, Deng D, et al. 2011. Quantitative analysis of transient and sustained transforming growth factor β signaling dynamics. Mol. Syst. Biol 7:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinbergs ID, Brown L, Edelman ER. 1996. Cellular response to transforming growth factor-beta1 and basic fibroblast growth factor depends on release kinetics and extracellular matrix interactions. J. Biol. Chem 271:29822–29 [DOI] [PubMed] [Google Scholar]
- 9.Kuhl PR, Griffith-Cima LG. 1996. Tethered epidermal growth factor as a paradigm for growth factor–induced stimulation from the solid phase. Nat. Med 2:1022–27 [DOI] [PubMed] [Google Scholar]
- 10.Fan VH, Tamama K, Au A, Littrell R, Richardson LB, et al. 2007. Tethered epidermal growth factor provides a survival advantage to mesenchymal stem cells. Stem Cells 25:1241–51 [DOI] [PubMed] [Google Scholar]
- 11.Rodrigues M, Blair H, Stockdale L, Griffith L, Wells A. 2013. Surface tethered epidermal growth factor protects proliferating and differentiating multipotential stromal cells from FasL-induced apoptosis. Stem Cells 31:104–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nuschke A, Rodrigues M, Rivera J, Yates C, Whaley D, et al. 2016. Epidermal growth factor tethered to β-tricalcium phosphate bone scaffolds via a high-affinity binding peptide enhances survival of human mesenchymal stem cells/multipotent stromal cells in an immune-competent parafascial implantation assay in mice. Stem Cells Transl. Med 5:1580–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCall JD, Anseth KS. 2012. Thiol–ene photopolymerizations provide a facile method to encapsulate proteins and maintain their bioactivity. Biomacromolecules 13:2410–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson SM, Chen TT, Iruela-Arispe ML, Segura T. 2009. The phosphorylation of vascular endothelial growth factor receptor 2 (VEGFR-2) by engineered surfaces with electrostatically or covalently immobilized VEGF. Biomaterials 30:4618–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim CS, Mitchell IP, Desotell AW, Kreeger PK, Masters KS. 2016. Immobilized epidermal growth factor stimulates persistent, directed keratinocyte migration via activation of PLCγ1. FASEB J 30:2580–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson SM, Shergill B, Barry ZT, Manousiouthakis E, Chen TT, et al. 2011. VEGF internalization is not required for VEGFR-2 phosphorylation in bioengineered surfaces with covalently linked VEGF. Integr. Biol 3:887–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Puccinelli TJ, Bertics PJ, Masters KS. 2010. Regulation of keratinocyte signaling and function via changes in epidermal growth factor presentation. Acta Biomater 6:3415–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Discher DE, Mooney DJ, Zandstra PW. 2009. Growth factors, matrices, and forces combine and control stem cells. Science 324:1673–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mente PL, Lewis JL. 1994. Elastic modulus of calcified cartilage is an order of magnitude less than that of subchondral bone. J. Orthop. Res 12:637–47 [DOI] [PubMed] [Google Scholar]
- 20.Pelham RJ Jr., Wang Y. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility. PNAS 94:13661–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeung T, Georges PC, Flanagan LA, Marg B, Ortiz M, et al. 2005. Effects of substrate stiffness on cell morphology, cytoskeletal structure, and adhesion. Cell Motil. Cytoskelet 60:24–34 [DOI] [PubMed] [Google Scholar]
- 22.Engler AJ, Sen S, Sweeney HL, Discher DE. 2006. Matrix elasticity directs stem cell lineage specification. Cell 126:677–89 [DOI] [PubMed] [Google Scholar]
- 23.Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, et al. 2012. Extracellular-matrix tethering regulates stem-cell fate. Nat. Mater 11:642–49 [DOI] [PubMed] [Google Scholar]
- 24.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, et al. 2014. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat. Mater 13:979–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caliari SR, Vega SL, Kwon M, Soulas EM, Burdick JA. 2016. Dimensionality and spreading influence MSC YAP/TAZ signaling in hydrogel environments. Biomaterials 103:314–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notbohm J, Lesman A, Rosakis P, Tirrell DA, Ravichandran G. 2015. Microbuckling of fibrin provides a mechanism for cell mechanosensing. J. R. Soc. Interface 12:20150320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baker BM, Trappmann B, Wang WY, Sakar MS, Kim IL, et al. 2015. Cell-mediated fibre recruitment drives extracellular matrix mechanosensing in engineered fibrillar microenvironments. Nat. Mater 14:1262–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonnans C, Chou J, Werb Z. 2014. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol 15:786–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guvendiren M, Burdick JA. 2012. Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun 3:792. [DOI] [PubMed] [Google Scholar]
- 30.Mabry KM, Lawrence RL, Anseth KS. 2015. Dynamic stiffening of poly(ethylene glycol)-based hydrogels to direct valvular interstitial cell phenotype in a three-dimensional environment. Biomaterials 49:47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirschner CM, Alge DL, Gould ST, Anseth KS. 2014. Clickable, photodegradable hydrogels to dynamically modulate valvular interstitial cell phenotype. Adv. Healthc. Mater 3:649–57 [DOI] [PubMed] [Google Scholar]
- 32.Stowers RS, Allen SC, Suggs LJ. 2015. Dynamic phototuning of 3D hydrogel stiffness. PNAS 112:1953–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosales AM, Mabry KM, Nehls EM, Anseth KS. 2015. Photoresponsive elastic properties of azobenzene-containing poly(ethylene-glycol)-based hydrogels. Biomacromolecules 16:798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gasiorowski JZ, Murphy CJ, Nealey PF. 2013. Biophysical cues and cell behavior: the big impact of little things. Annu. Rev. Biomed. Eng 15:155–76 [DOI] [PubMed] [Google Scholar]
- 35.Guvendiren M, Burdick JA. 2013. Stem cell response to spatially and temporally displayed and reversible surface topography. Adv. Healthc. Mater 2:155–64 [DOI] [PubMed] [Google Scholar]
- 36.Mascharak S, Benitez PL, Proctor AC, Madl CM, Hu KH, et al. 2017. YAP-dependent mechanotransduction is required for proliferation and migration on native-like substrate topography. Biomaterials 115:155–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutson HN, Marohl T, Anderson M, Eliceiri K, Campagnola P, Masters KS. 2016. Calcific aortic valve disease is associated with layer-specific alterations in collagen architecture. PLOS ONE 11:e0163858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen B, Campbell KR, Tilbury K, Nadiarnykh O, Brewer MA, et al. 2016. 3D texture analysis for classification of second harmonic generation images of human ovarian cancer. Sci. Rep 6:35734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao L, Kupfer ME, Jung JP, Yang L, Zhang P, et al. 2017. Myocardial tissue engineering with cells derived from human-induced pluripotent stem cells and a native-like, high-resolution, 3-dimensionally printed scaffold. Circ. Res 120:1318–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, Keely PJ. 2006. Collagen reorganization at the tumor–stromal interface facilitates local invasion. BMC Med 4:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conklin MW, Eickhoff JC, Riching KM, Pehlke CA, Eliceiri KW, et al. 2011. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol 178:1221–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carey SP, Goldblatt ZE, Martin KE, Romero B, Williams RM, Reinhart-King CA. 2016. Local extracellular matrix alignment directs cellular protrusion dynamics and migration through Rac1 and FAK. Integr. Biol 8:821–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Culver JC, Hoffmann JC, Poche RA, Slater JH, West JL, Dickinson ME. 2012. Three-dimensional biomimetic patterning in hydrogels to guide cellular organization. Adv. Mater 24:2344–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brucher BL, Jamall IS. 2014. Cell–cell communication in the tumor microenvironment, carcinogenesis, and anticancer treatment. Cell Physiol. Biochem 34:213–43 [DOI] [PubMed] [Google Scholar]
- 45.Kim JH, Kushiro K, Graham NA, Asthagiri AR. 2009. Tunable interplay between epidermal growth factor and cell–cell contact governs the spatial dynamics of epithelial growth. PNAS 106:11149–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Wever O, Hendrix A, De Boeck A, Eertmans F, Westbroek W, et al. 2014. Single cell and spheroid collagen type I invasion assay. Methods Mol. Biol 1070:13–35 [DOI] [PubMed] [Google Scholar]
- 47.Mrksich M, Dike LE, Tien J, Ingber DE, Whitesides GM. 1997. Using microcontact printing to pattern the attachment of mammalian cells to self-assembled monolayers of alkanethiolates on transparent films of gold and silver. Exp. Cell Res 235:305–13 [DOI] [PubMed] [Google Scholar]
- 48.Li CY, Stevens KR, Schwartz RE, Alejandro BS, Huang JH, Bhatia SN. 2014. Micropatterned cell–cell interactions enable functional encapsulation of primary hepatocytes in hydrogel microtissues. Tissue Eng. A 20:2200–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pasquale EB. 2008. Eph–ephrin bidirectional signaling in physiology and disease. Cell 133:38–52 [DOI] [PubMed] [Google Scholar]
- 50.Moon JJ, Lee SH, West JL. 2007. Synthetic biomimetic hydrogels incorporated with ephrin-A1 for therapeutic angiogenesis. Biomacromolecules 8:42–49 [DOI] [PubMed] [Google Scholar]
- 51.Lin CC, Anseth KS. 2011. Cell-cell communication mimicry with poly(ethylene glycol) hydrogels for enhancing β-cell function. PNAS 108:6380–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bray SJ. 2006. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol 7:678–89 [DOI] [PubMed] [Google Scholar]
- 53.Tung JC, Paige SL, Ratner BD, Murry CE, Giachelli CM. 2014. Engineered biomaterials control differentiation and proliferation of human-embryonic-stem-cell-derived cardiomyocytes via timed Notch activation. Stem Cell Rep 2:271–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toth B, Ben-Moshe S, Gavish A, Barkai N, Itzkovitz S. 2017. Early commitment and robust differentiation in colonic crypts. Mol. Syst. Biol 13:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nardini JT, Chapnick DA, Liu X, Bortz DM. 2016. Modeling keratinocyte wound healing dynamics: Cell–cell adhesion promotes sustained collective migration. J. Theor. Biol 400:103–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bian L, Guvendiren M, Mauck RL, Burdick JA. 2013. Hydrogels that mimic developmentally relevant matrix and N-cadherin interactions enhance MSC chondrogenesis. PNAS 110:10117–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vega SL, Kwon M, Mauck RL, Burdick JA. 2016. Single cell imaging to probe mesenchymal stem cell N-cadherin mediated signaling within hydrogels. Ann. Biomed. Eng 44:1921–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. 2000. Vascular-specific growth factors and blood vessel formation. Nature 407:242–48 [DOI] [PubMed] [Google Scholar]
- 59.Chen RR, Silva EA, Yuen WW, Mooney DJ. 2007. Spatio-temporal VEGF and PDGF delivery patterns blood vessel formation and maturation. Pharm. Res 24:258–64 [DOI] [PubMed] [Google Scholar]
- 60.Hao X, Silva EA, Månsson-Broberg A, Grinnemo K-H, Siddiqui AJ, et al. 2007. Angiogenic effects of sequential release of VEGF-A165 and PDGF-BB with alginate hydrogels after myocardial infarction. Cardiovasc. Res 75:178–85 [DOI] [PubMed] [Google Scholar]
- 61.Vo TN, Kasper FK, Mikos AG. 2012. Strategies for controlled delivery of growth factors and cells for bone regeneration. Adv. Drug Deliv. Rev 64:1292–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martino MM, Brkic S, Bovo E, Burger M, Schaefer DJ, et al. 2015. Extracellular matrix and growth factor engineering for controlled angiogenesis in regenerative medicine. Front. Bioeng Biotechnol 3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willerth SM. 2017. Biomimetic strategies for replicating the neural stem cell niche. Curr. Opin. Chem. Eng 15:8–14 [Google Scholar]
- 64.Lee K, Silva EA, Mooney DJ. 2011. Growth factor delivery–based tissue engineering: general approaches and a review of recent developments. J. R. Soc. Interface 8:153–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. 2005. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 310:1646. [DOI] [PubMed] [Google Scholar]
- 66.Walters BD, Stegemann JP. 2014. Strategies for directing the structure and function of three-dimensional collagen biomaterials across length scales. Acta Biomater 10:1488–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Regier MC, Montanez-Sauri SI, Schwartz MP, Murphy WL, Beebe DJ, Sung KE. 2017. The influence of biomaterials on cytokine production in 3D cultures. Biomacromolecules 18:709–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flaim CJ, Chien S, Bhatia SN. 2005. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods 2:119–25 [DOI] [PubMed] [Google Scholar]
- 69.Kaylan KB, Ermilova V, Yada RC, Underhill GH. 2016. Combinatorial microenvironmental regulation of liver progenitor differentiation by Notch ligands, TGFβ, and extracellular matrix. Sci. Rep 6:23490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carrion B, Souzanchi MF, Wang VT, Tiruchinapally G, Shikanov A, et al. 2016. The synergistic effects of matrix stiffness and composition on the response of chondroprogenitor cells in a 3D precondensation microenvironment. Adv. Healthc. Mater 5:1192–202 [DOI] [PubMed] [Google Scholar]
- 71.Rahmany MB, Van Dyke M. 2013. Biomimetic approaches to modulate cellular adhesion in biomaterials: a review. Acta Biomater 9:5431–37 [DOI] [PubMed] [Google Scholar]
- 72.Benoit DS, Anseth KS. 2005. The effect on osteoblast function of colocalized RGD and PHSRN epitopes on PEG surfaces. Biomaterials 26:5209–20 [DOI] [PubMed] [Google Scholar]
- 73.Ochsenhirt SE, Kokkoli E, McCarthy JB, Tirrell M. 2006. Effect of RGD secondary structure and the synergy site PHSRN on cell adhesion, spreading and specific integrin engagement. Biomaterials 27:3863–74 [DOI] [PubMed] [Google Scholar]
- 74.Jung JP, Moyano JV, Collier JH. 2011. Multifactorial optimization of endothelial cell growth using modular synthetic extracellular matrices. Integr. Biol 3:185–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen C, Xie J, Deng L, Yang L. 2014. Substrate stiffness together with soluble factors affects chondrocyte mechanoresponses. ACS Appl. Mater. Interfaces 6:16106–16 [DOI] [PubMed] [Google Scholar]
- 76.Allen JL, Cooke ME, Alliston T. 2012. ECM stiffness primes the TGFβ pathway to promote chondrocyte differentiation. Mol. Biol. Cell 23:3731–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Banks JM, Mozdzen LC, Harley BA, Bailey RC. 2014. The combined effects of matrix stiffness and growth factor immobilization on the bioactivity and differentiation capabilities of adipose-derived stem cells. Biomaterials 35:8951–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park JS, Chu JS, Tsou AD, Diop R, Tang Z, et al. 2011. The effect of matrix stiffness on the differentiation of mesenchymal stem cells in response to TGF-β. Biomaterials 32:3921–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wickert LE, Pomerenke S, Mitchell I, Masters KS, Kreeger PK. 2016. Hierarchy of cellular decisions in collective behavior: implications for wound healing. Sci. Rep 6:20139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim J-H, Asthagiri AR. 2011. Matrix stiffening sensitizes epithelial cells to EGF and enables the loss of contact inhibition of proliferation. J. Cell Sci 124:1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Floren M, Bonani W, Dharmarajan A, Motta A, Migliaresi C, Tan W. 2016. Human mesenchymal stem cells cultured on silk hydrogels with variable stiffness and growth factor differentiate into mature smooth muscle cell phenotype. Acta Biomater 31:156–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tan S, Fang JY, Yang Z, Nimni ME, Han B. 2014. The synergetic effect of hydrogel stiffness and growth factor on osteogenic differentiation. Biomaterials 35:5294–306 [DOI] [PubMed] [Google Scholar]
- 83.Chang FC, Tsao CT, Lin A, Zhang M, Levengood SL, Zhang M. 2016. PEG-chitosan hydrogel with tunable stiffness for study of drug response of breast cancer cells. Polymers 8:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tokuda EY, Leight JL, Anseth KS. 2014. Modulation of matrix elasticity with PEG hydrogels to study melanoma drug responsiveness. Biomaterials 35:4310–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gobaa S, Hoehnel S, Lutolf MP. 2015. Substrate elasticity modulates the responsiveness of mesenchymal stem cells to commitment cues. Integr. Biol 7:1135–42 [DOI] [PubMed] [Google Scholar]
- 86.Hansen TD, Koepsel JT, Le NN, Nguyen EH, Zorn S, et al. 2014. Biomaterial arrays with defined adhesion ligand densities and matrix stiffness identify distinct phenotypes for tumorigenic and nontumorigenic human mesenchymal cell types. Biomater. Sci 2:745–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Barcus CE, Keely PJ, Eliceiri KW, Schuler LA. 2016. Prolactin signaling through focal adhesion complexes is amplified by stiff extracellular matrices in breast cancer cells. Oncotarget 7:48093–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumar S, Weaver VM. 2009. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev 28:113–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zustiak S, Nossal R, Sackett DL. 2014. Multiwell stiffness assay for the study of cell responsiveness to cytotoxic drugs. Biotechnol. Bioeng 111:396–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schrader J, Gordon-Walker TT, Aucott RL, van Deemter M, Quaas A, et al. 2011. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 53:1192–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wozniak MA, Desai R, Solski PA, Der CJ, Keely PJ. 2003. ROCK-generated contractility regulates breast epithelial cell differentiation in response to the physical properties of a three-dimensional collagen matrix. J. Cell Biol 163:583–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barcus CE, Keely PJ, Eliceiri KW, Schuler LA. 2013. Stiff collagen matrices increase tumorigenic prolactin signaling in breast cancer cells. J. Biol. Chem 288:12722–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Willits RK, Skornia SL. 2004. Effect of collagen gel stiffness on neurite extension. J. Biomater. Sci. Polym. Ed 15:1521–31 [DOI] [PubMed] [Google Scholar]
- 94.Lu P, Weaver VM, Werb Z. 2012. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol 196:395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wells RG. 2013. Tissue mechanics and fibrosis. Biochim. Biophys. Acta 1832:884–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhu J 2010. Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering. Biomaterials 31:4639–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee KY, Mooney DJ. 2012. Alginate: properties and biomedical applications. Prog. Polym. Sci 37:106–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nii M, Lai JH, Keeney M, Han LH, Behn A, et al. 2013. The effects of interactive mechanical and biochemical niche signaling on osteogenic differentiation of adipose-derived stem cells using combinatorial hydrogels. Acta Biomater 9:5475–83 [DOI] [PubMed] [Google Scholar]
- 99.Chaudhuri O, Koshy ST, Branco da Cunha C, Shin JW, Verbeke CS, et al. 2014. Extracellular matrix stiffness and composition jointly regulate the induction of malignant phenotypes in mammary epithelium. Nat. Mater 13:970–78 [DOI] [PubMed] [Google Scholar]
- 100.Hwang NS, Varghese S, Li H, Elisseeff J. 2011. Regulation of osteogenic and chondrogenic differentiation of mesenchymal stem cells in PEG-ECM hydrogels. Cell Tissue Res 344:499–509 [DOI] [PubMed] [Google Scholar]
- 101.Jung JP, Sprangers AJ, Byce JR, Su J, Squirrell JM, et al. 2013. ECM-incorporated hydrogels cross-linked via native chemical ligation to engineer stem cell microenvironments. Biomacromolecules 14:3102–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Berger AJ, Linsmeier KM, Kreeger PK, Masters KS. 2017. Decoupling the effects of stiffness and fiber density on cellular behaviors via an interpenetrating network of gelatin-methacrylate and collagen. Biomaterials 141:125–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hutson CB, Nichol JW, Aubin H, Bae H, Yamanlar S, et al. 2011. Synthesis and characterization of tunable poly(ethylene glycol): gelatin methacrylate composite hydrogels. Tissue Eng. A 17:1713–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suri S, Schmidt CE. 2010. Cell-laden hydrogel constructs of hyaluronic acid, collagen, and laminin for neural tissue engineering. Tissue Eng. A 16:1703–16 [DOI] [PubMed] [Google Scholar]
- 105.Tarus D, Hamard L, Caraguel F, Wion D, Szarpak-Jankowska A, et al. 2016. Design of Hyaluronic Acid Hydrogels to Promote Neurite Outgrowth in Three Dimensions. ACS Appl. Mater. Interfaces 8:25051–59 [DOI] [PubMed] [Google Scholar]
- 106.Wei Z, Lewis DM, Xu Y, Gerecht S. 2017. Dual cross-linked biofunctional and self-healing networks to generate user-defined modular gradient hydrogel constructs. Adv. Healthc. Mater 6:1700523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mason BN, Starchenko A, Williams RM, Bonassar LJ, Reinhart-King CA. 2013. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater 9:4635–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Francis-Sedlak ME, Uriel S, Larson JC, Greisler HP, Venerus DC, Brey EM. 2009. Characterization of type I collagen gels modified by glycation. Biomaterials 30:1851–56 [DOI] [PubMed] [Google Scholar]
- 109.Jiang L, Sun Z, Chen X, Li J, Xu Y, et al. 2016. Cells sensing mechanical cues: stiffness influences the lifetime of cell–extracellular matrix interactions by affecting the loading rate. ACS Nano 10:207–17 [DOI] [PubMed] [Google Scholar]
- 110.Seong J, Tajik A, Sun J, Guan JL, Humphries MJ, et al. 2013. Distinct biophysical mechanisms of focal adhesion kinase mechanoactivation by different extracellular matrix proteins. PNAS 110:19372–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gandavarapu NR, Alge DL, Anseth KS. 2014. Osteogenic differentiation of human mesenchymal stem cells on α5 integrin binding peptide hydrogels is dependent on substrate elasticity. Biomater. Sci 2:352–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ross RS. 2004. Molecular and mechanical synergy: cross-talk between integrins and growth factor receptors. Cardiovasc. Res 63:381–90 [DOI] [PubMed] [Google Scholar]
- 113.Lee YJ, Streuli CH. 1999. Extracellular matrix selectively modulates the response of mammary epithelial cells to different soluble signaling ligands. J. Biol. Chem 274:22401–8 [DOI] [PubMed] [Google Scholar]
- 114.Rodriguez KJ, Masters KS. 2009. Regulation of valvular interstitial cell calcification by components of the extracellular matrix. J. Biomed. Mater. Res. A 90:1043–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pollock K, Jaraczewski TJ, Carroll MJ, Lebovic DI, Kreeger PK. 2014. Endometriotic epithelial cell response to macrophage-secreted factors is dependent on extracellular matrix context. Cell Mol. Bioeng 7:409–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chai C, Leong KW. 2007. Biomaterials approach to expand and direct differentiation of stem cells. Mol. Ther 15:467–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kilian KA, Mrksich M. 2012. Directing stem cell fate by controlling the affinity and density of ligand–receptor interactions at the biomaterials interface. Angew. Chem. Int. Ed. Engl 51:4891–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zieris A, Prokoph S, Levental KR, Welzel PB, Grimmer M, et al. 2010. FGF-2 and VEGF functionalization of starPEG-heparin hydrogels to modulate biomolecular and physical cues of angiogenesis. Biomaterials 31:7985–94 [DOI] [PubMed] [Google Scholar]
- 119.Hanahan D, Weinberg RA. 2000. The hallmarks of cancer. Cell 100:57–70 [DOI] [PubMed] [Google Scholar]
- 120.Lin B, Yin T, Wu YI, Inoue T, Levchenko A. 2015. Interplay between chemotaxis and contact inhibition of locomotion determines exploratory cell migration. Nat. Commun 6:6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Foley JD, Grunwald EW, Nealey PF, Murphy CJ. 2005. Cooperative modulation of neuritogenesis by PC12 cells by topography and nerve growth factor. Biomaterials 26:3639–44 [DOI] [PubMed] [Google Scholar]
- 122.Raghunathan V, McKee C, Cheung W, Naik R, Nealey PF, et al. 2013. Influence of extracellular matrix proteins and substratum topography on corneal epithelial cell alignment and migration. Tissue Eng. A 19:1713–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tocce EJ, Liliensiek SJ, Broderick AH, Jiang Y, Murphy KC, et al. 2013. The influence of biomimetic topographical features and the extracellular matrix peptide RGD on human corneal epithelial contact guidance. Acta Biomater 9:5040–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kim DH, Provenzano PP, Smith CL, Levchenko A. 2012. Matrix nanotopography as a regulator of cell function. J. Cell Biol 197:351–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cosgrove BD, Mui KL, Driscoll TP, Caliari SR, Mehta KD, et al. 2016. N-Cadherin adhesive interactions modulate matrix mechanosensing and fate commitment of mesenchymal stem cells. Nat. Mater 15:1297–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ye K, Cao L, Li S, Yu L, Ding J. 2016. Interplay of matrix stiffness and cell–cell contact in regulating differentiation of stem cells. ACS Appl. Mater. Interfaces 8:21903–13 [DOI] [PubMed] [Google Scholar]
- 127.Mao AS, Shin JW, Mooney DJ. 2016. Effects of substrate stiffness and cell–cell contact on mesenchymal stem cell differentiation. Biomaterials 98:184–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Flaim CJ, Teng D, Chien S, Bhatia SN. 2008. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev 17:29–39 [DOI] [PubMed] [Google Scholar]
- 129.Ranga A, Gobaa S, Okawa Y, Mosiewicz K, Negro A, Lutolf MP. 2014. 3D niche microarrays for systems-level analyses of cell fate. Nat. Commun 5:4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chen S, Bremer AW, Scheideler OJ, Na YS, Todhunter ME, et al. 2016. Interrogating cellular fate decisions with high-throughput arrays of multiplexed cellular communities. Nat. Commun 7:10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Rape AD, Zibinsky M, Murthy N, Kumar S. 2015. A synthetic hydrogel for the high-throughput study of cell–ECM interactions. Nat. Commun 6:8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, et al. 2009. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6:377–82 [DOI] [PubMed] [Google Scholar]
- 133.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, et al. 2011. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332:687–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, et al. 2004. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell 118:217–28 [DOI] [PubMed] [Google Scholar]
- 135.Gaudet S, Miller-Jensen K. 2016. Redefining signaling pathways with an expanding single-cell toolbox. Trends Biotechnol 34:458–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Regot S, Hughey JJ, Bajar BT, Carrasco S, Covert MW. 2014. High-sensitivity measurements of multiple kinase activities in live single cells. Cell 157:1724–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Albeck JG, Mills GB, Brugge JS. 2013. Frequency-modulated pulses of ERK activity transmit quantitative proliferation signals. Mol. Cell 49:249–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Warren EA, Netterfield TS, Sarkar S, Kemp ML, Payne CK. 2015. Spatially-resolved intracellular sensing of hydrogen peroxide in living cells. Sci. Rep 5:16929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kniss-James AS, Rivet CA, Chingozha L, Lu H, Kemp ML. 2017. Single-cell resolution of intracellular T cell Ca2+ dynamics in response to frequency-based H2O2 stimulation. Integr. Biol 9:238–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee RE, Walker SR, Savery K, Frank DA, Gaudet S. 2014. Fold change of nuclear NF-κB determines TNF-induced transcription in single cells. Mol. Cell 53:867–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lu Y, Xue Q, Eisele MR, Sulistijo ES, Brower K, et al. 2015. Highly multiplexed profiling of single-cell effector functions reveals deep functional heterogeneity in response to pathogenic ligands. PNAS 112:E607–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Campagnola PJ, Loew LM. 2003. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat. Biotechnol 21:1356–60 [DOI] [PubMed] [Google Scholar]
- 143.Notbohm J, Lesman A, Tirrell DA, Ravichandran G. 2015. Quantifying cell-induced matrix deformation in three dimensions based on imaging matrix fibers. Integr. Biol 7:1186–95 [DOI] [PubMed] [Google Scholar]