SUMMARY
We integrated the genomic sequencing of 1,918 breast cancers, including 1,501 hormone receptor-positive tumors, with detailed clinical information and treatment outcomes. In 692 tumors previously exposed to hormonal therapy, we identified an increased number of alterations in genes involved in the mitogen-activated protein kinase (MAPK) pathway and in the estrogen receptor transcriptional machinery. Activating ERBB2 mutations and NF1 loss-of-function mutations were more than twice as common in endocrine resistant tumors. Alterations in other MAPK pathway genes (EGFR, KRAS, among others) and estrogen receptor transcriptional regulators (MYC, CTCF, FOXA1, and TBX3) were also enriched. Altogether, these alterations were present in 22% of tumors, mutually exclusive with ESR1 mutations, and associated with a shorter duration of response to subsequent hormonal therapies
INTRODUCTION
Breast cancers are categorized into molecularly distinct groups based on hormone receptor (HR) and HER2 status, which dictate different clinical outcomes and choice of therapies (Perou et al., 2000; Sorlie et al., 2001). Broad-based genomic characterization of breast cancer has largely established the landscape of inherited and somatic genomic alterations that typify each of these classes of primary disease (Banerji et al., 2012; Cancer Genome Atlas, 2012; Ciriello et al., 2015; Ellis et al., 2012; Nik-Zainal et al., 2016; Pereira et al., 2016; Shah et al., 2012; Stephens et al., 2012). In contrast to the abundance of genomic information from primary breast cancer, far less is known about the genomic alterations in metastatic tumors, the ultimate cause of death in most breast cancer patients. A detailed characterization of the genomic landscape of breast cancer metastasis could provide important insights including identifying (1) genomic drivers of metastatic disease progression, (2) the extent and clinical impact of tumoral heterogeneity, (3) the biologic determinants of variable response of individual patients to different therapies, and (4) additional potential therapeutic targets.
Recent tumor characterization efforts have begun to provide answers to some of these questions (Brastianos et al., 2015; De Mattos-Arruda et al., 2014; Ding et al., 2010; Juric et al., 2015; Savas et al., 2016; Yates et al., 2017). These studies have shown that while metastases are clonally related to their primary tumor, they nonetheless often acquire driver genomic alterations not present in the primary disease including mutations that inactivate the SWI-SNF and JAK2-STAT3 pathways (Yates et al., 2017). It is also clear that therapeutic pressures influence tumor evolution, which in turn, results in treatment resistance (Juric et al., 2015). These early findings reinforce the need for a comprehensive characterization of advanced breast cancers.
As part of a broad-based institutional effort, we have assembled a large genomic dataset of cancer patients with metastatic disease (Zehir et al., 2017) and, subsequently, have extracted detailed clinical annotation from each patient including the types of therapy administered over time and the observed clinical benefit. While encompassing all breast cancer subtypes, we have focused our efforts on HR+ tumors since they represent the largest subset and because they are frequently treated with defined lines of hormonal therapy, thereby enabling the study of resistance mechanisms. As with other therapies, intrinsic and acquired resistance to hormonal therapy leads to disease recurrence and limited clinical benefit (Ma et al., 2015). Recently, ligand-binding domain mutations in the estrogen receptor (ER) gene (ESR1) were shown to be present in 18% of endocrine-resistant HR+ breast cancers (Schiavon et al., 2015; Toy et al., 2013). While the discovery of ESR1 mutations offers valuable insights into the evolution of breast tumors under the selective pressure of therapy, only a fraction of breast cancer patients with endocrine resistant tumors harbor these mutations. In the remainder of patients, the mechanisms of endocrine resistance remain largely unexplained. In this study we aimed to perform a large clinico-genomic analysis to identify additional genomic alterations that might mediate resistance to hormonal therapy and provide a rationale for the development of therapeutic approaches to overcome resistance.
RESULTS
Genomic Features of Advanced, Post-Treatment Breast Cancer
We performed prospective targeted sequencing of 1,918 tumors from 1,756 breast cancer patients for whom detailed clinical annotation was available (Figure 1A). Tumors and matched normal DNA were analyzed using the Memorial Sloan Kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT) platform, which can identify somatic mutations, DNA copy number alterations, and select rearrangements in up to 468 cancer-associated genes, (Cheng et al., 2015; Zehir et al., 2017). While not designed for breast cancer gene discovery, sequencing was performed at high depth-of-coverage (771-fold average coverage) providing greater sensitivity than typical broader-scale sequencing approaches for the detection of subclonal mutational events (those present in only a subset of cancer cells) (Zehir et al., 2017).
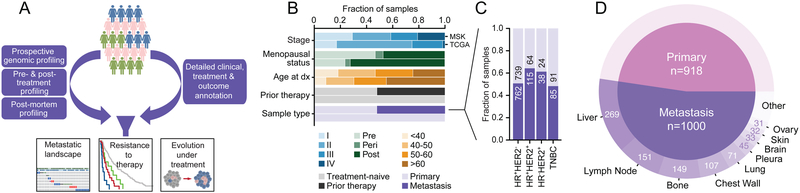
Figure 1: Clinical Characteristics of Prospectively Sequenced Advanced Breast Cancers.
(A) Study schema.
(B) Clinical features of this prospective cohort compared to a contemporary study of primary untreated breast cancers (TCGA).
(C) Composition and cohort size of primary and metastatic breast cancers by receptor subtype.
(D) Distribution of the biopsied metastatic disease sites and sample number in the study cohort.
In contrast to previous studies of untreated primary tumors including The Cancer Genome Atlas (TCGA) (Cancer Genome Atlas, 2012; Ciriello et al., 2015; Nik-Zainal et al., 2016; Stephens et al., 2012), this population was highly enriched for patients with high-risk clinical features at the time of diagnosis, including advanced stages, younger age, and premenopausal status (all p values < 10−4). Also, in contrast to the treatment-naïve TCGA dataset, 52% of tumors were collected after exposure to systemic anticancer therapy in the (neo)adjuvant (44%) and/or metastatic (27%) settings (median of three systemic treatments in the metastatic setting prior to sample collection, range 1-15, Figure 1B and Tables S1 and S2). Furthermore, 52% of biopsied tumors (n = 1,000) were from metastatic sites (Figure 1C and 1D). An important consideration is that these biopsies were performed relatively close in time to the initiation of the next line of therapy. Specifically, among the 912 tumor samples acquired prior to the start of any therapy, the tumors analyzed were collected a median of 40 days prior to therapy initiation (Figure S1A). The 782 specimens acquired after the completion of one or more lines of therapy were collected a median of 43 days after the cessation of the prior line of therapy and a median of 20 days prior to the start of the subsequent line of therapy (Figure S1B and S1C). The therapies that most commonly directly preceded or were administered directly after tumor collection were hormonal, either as a single agent or in combination with CDK4/6 inhibitors, MTOR inhibitors, or other agents, followed by chemotherapy-containing regimens (Figure S1D and S1E). Finally, 224 tumor samples were on-treatment biopsies (with 184 of these samples also exposed to prior lines of therapies), 67% of which were from patients treated with hormonal therapies (Figure S1F).
The prevalence and co-occurrence of genes previously shown to be recurrently altered in primary breast cancers were similar overall in this more aggressive and advanced cohort (Figure 2A; Table S3). Alterations in CDH1, TP53, GATA3, CCND1, PIK3CA, and PTEN arose in patterns consistent with their known associations with specific receptor types and histologies. Notably, the overall mutational burden of metastatic specimens was only modestly higher than primary tumors in HR+ disease, and there was no difference among HR− or triple-negative breast cancer patients (Figure S2A). While a subset of HER2+ tumors, as defined by clinical testing, lacked ERBB2 amplifications as inferred from the sequencing data (Figure 2A), the concordance between HER2 status as defined by immunohistochemistry or fluorescence in situ hybridization and ERBB2 amplification inferred from the sequencing data was 98% when performed on the same tumor sample (1778/1810 samples; Κ=0.9, 95% confidence interval: 0.87-0.94, STAR Methods).
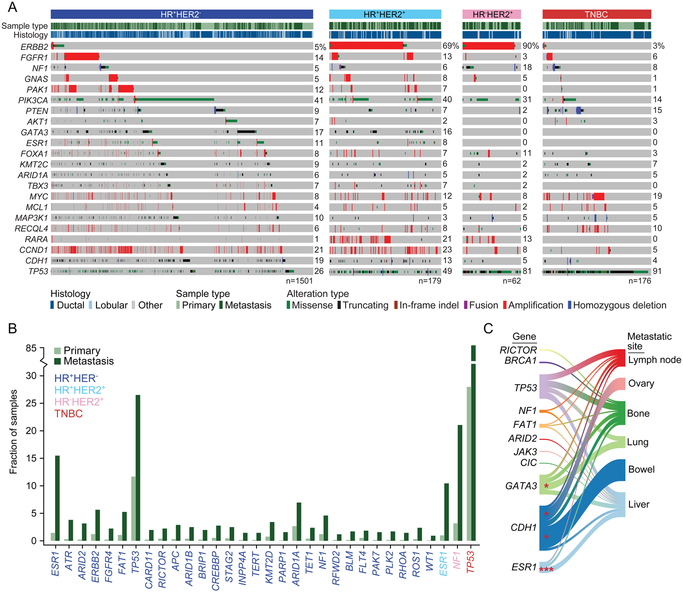
Figure 2: Genomic Characteristics of Prospectively Sequenced Advanced Breast Cancers.
(A) The pattern, frequency, and type of genomic alterations in key breast cancer genes by receptor type.
(B) The frequency difference among genes with a statistically significant increase (q < 0.05) of alterations in metastatic specimens as compared with primary tumors. The color of the gene symbol indicates statistical significance by receptor status.
(C) Recurrent genomic alterations (left) and their association with different organ sites of metastasis (right). Line thickness corresponds to the frequency of mutations arising in the indicated metastatic site. Shading identifies the relationships between genes and metastatic sites. Statistically significant associations are shown as asterisks (p < 0.05).
We decided next to identify mutations enriched in metastatic and/or therapy-resistant disease. While it would have been desirable to distinguish metastasis-specific from therapy-induced mutations, this has been an elusive goal in breast cancer for at least two reasons. First, a large proportion of breast cancer patients with early stage tumors receive postoperative adjuvant therapy with hormonal therapy, chemotherapy or both. Second, the occurrence of de novo metastatic disease is relatively rare. Our population is representative of this clinical reality as 87.5% of the biopsied metastatic tumors had been exposed to prior therapy in the adjuvant and/or metastatic settings. We nevertheless attempted an enrichment analysis in which we compared treatment-naïve primary tumors with the treatment-naïve metastatic specimens predominantly from de novo metastatic patients, but the analysis was insufficiently powered to identify robust differences as our cohort included only 125 treatment-naïve metastatic tumors (Figure S2B and S2C).
The intent of our analysis was, therefore, to survey the landscape of altered genes in advanced breast cancer, as a function of either metastasis or therapy, since they go hand in hand. Our gene-level enrichment analysis revealed that, overall, mutations in 32 genes were significantly more common in metastases compared with primary tumors (q < 0.05, Figure 2B). Twenty-nine of these genes (91%) arose in HR+HER2− disease, likely owing to the greater power of detection, as this was the largest subset analyzed (Figure 2B). While some mutant genes enriched in the metastatic context indicate their potential negative prognostic impact, others likely reflected the impact of prior therapy. In this regard, principal among the alterations were mutations in TP53, which were significantly more common in the metastatic samples of all receptor types except for HR−HER2+ disease (Figure S3A). As TP53 is a biomarker of poor prognosis (Ellis et al., 2012; Olivier et al., 2006), this finding likely reflects the advanced stage of many patients in this cohort. On the other hand, and reflecting the impact of prior therapy, ligand binding domain mutations in ESR1 were present in 18% of HR+HER2− patients whose sequenced tumor specimens were collected after endocrine therapy (n=128 of 692) (Chandarlapaty et al., 2016; Fribbens et al., 2016; Robinson et al., 2013; Toy et al., 2013). Other genes enriched in the HR+ metastatic context included epigenetic regulators (ARID1A, ARID2, and CREBBP) and members of the mitogen-activated protein kinase (MAPK) signaling cascade (ERBB2, NF1, and FGFR4). Some of these enrichments appeared to reflect organotropism in their pattern of metastasis (Figure 2C). For example, CDH1 mutations were associated with metastasis to the ovary, reflecting their high prevalence in lobular cancers and the preference of this subtype for ovarian metastasis (Harris et al., 1984). Conversely, GATA3 loss-of-function mutations were associated with lung metastasis, consistent with mouse model data demonstrating that loss of GATA3 expression is associated with increased pulmonary metastases (Dydensborg et al., 2009) and patient data showing that GATA3 expression is lower in those patients who develop lung metastases (McCleskey et al., 2015).
We next expanded our statistical power by performing a combined mutational hotspot analysis of this prospective cohort and retrospectively sequenced breast tumors (2,732 in total) (Cancer Genome Atlas, 2012). In total, we identified 313 statistically significant hotspot mutations in 72 genes (Figure S3B). Many of the mutational hotspots were in known breast cancer genes including PIK3CA, ERBB2, and others (Figure S3C). As an example, we identified 35 hotspot mutations in PIK3CA affecting 699 tumors (36.4%), many of which are known to induce constitutive phosphatidylinositol 3-kinase (PI3K) signaling and are being explored as predictive biomarkers of response to inhibitors of the pathway (Andre et al., 2016; Baselga et al., 2016; Juric et al., 2017; Mayer et al., 2016). Owing to the larger size of this cohort, we identified 12 previously unknown PIK3CA hotspots as significantly mutated in breast cancers. We expressed five of these variants of previously unknown significance in MCF10A cells and all induced greater levels of PI3K pathway activation than expression of the wild-type protein (Figure S3D).
We also identified hotspot mutations in genes not previously reported to be mutated in breast cancer. For example, SPOP E78K mutations were identified in three patients with HR+HER2− tumors collected following progression on hormonal therapy with aromatase inhibitors (AI, Figure S3E). Unlike the SPOP mutations described in prostate cancers (Barbieri et al., 2012), the E78K mutation affects a unique region of the MATH domain where rare loss-of-function mutations prevent SPOP-mediated degradation of ER in endometrial cancers (McCleskey et al., 2015). Additional hotspot mutations were identified in transcription factor genes including FOXA1, TBX3, and CTCF. Mutations in FOXA1 have previously been implicated in breast cancer pathogenesis (Ciriello et al., 2015), ER regulation (Augello et al., 2011), and were histology-specific. Lobular and ductal cancers possessed different FOXA1 mutant alleles positioned on either wing 2 or wing 1 of the Forkhead domain (Figure S3F). Hotspots in TBX3 (N297) and CTCF (H284 and Y226) were similarly histology-specific, arising exclusively in lobular and ductal cancers, respectively.
Discovering Mutations Associated with Hormonal Therapy Failure
To determine whether any of the alterations identified above were associated with resistance to hormonal therapy we collected detailed treatment histories for all sequenced patients and explored associations between genomic aberrations and response to therapy. Our cohort comprised a total of 1,501 HR+HER2− breast cancers that could be further divided into 809 hormonal therapy-naïve tumors and 692 post-hormonal therapy tumors (Figure 2A). In addition to the expected ESR1 hotspot mutations, ERBB2 and NF1 were the genes with the greatest difference in mutational frequency between pre- and post-hormonal therapy HR+HER2− breast cancers. In fact, functional hotspot mutations in ERBB2 and loss-of-function mutations in NF1 were more than twice as common in specimens acquired after endocrine therapy (p values = 0.00003 and 0.0004, respectively; Figure 3A). Interestingly, mutations in ESR1, ERRB2, and NF1 were mostly mutually exclusive.
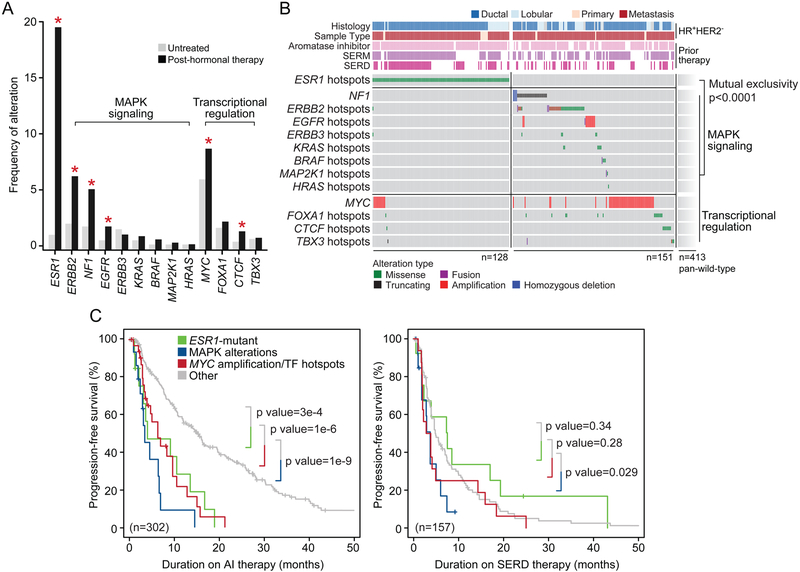
Figure 3: Genomic Landscape of Endocrine Resistance after Treatment with Hormonal Therapy.
(A) Frequency of recurrent functional genomic alterations as shown in (B) in tumors that were collected from patients that were either treatment-naïve or after treatment with hormonal therapy. Asterisks denote statistical significance between groups.
(B) Pattern and frequency of known or putative functional mutations and focal amplifications or deletions targeting effectors of MAPK signaling or aberrant transcriptional regulation in ESR1-mutant and ESR1-wild-type tumors after endocrine therapy (mutual exclusivity p value < 0.0001).
(C) Kaplan-Meier curves displaying progression-free survival of patients receiving aromatase inhibitor (AI) (left) or SERD (right) therapy based on genomic alterations. Tumors harboring hotspot mutations in ESR1 are displayed in green, those with known or putative functional alterations in effectors of MAPK signaling are shown in blue, and those with alterations in transcriptional regulators in red. Outcomes were compared with patients whose tumors were wild-type for such lesions (gray). All p values as indicated, log rank test.
See also Figure S4.
The mutual exclusivity between mutations in ESR1 and those in either ERBB2 or NF1 in specimens acquired after endocrine therapy (Figure 3B) implied that there may exist multiple non-overlapping genomic alterations that lead to a common molecular phenotype of therapeutic resistance. As both ERBB2 gain-of-function mutations and NF1 loss-of-function mutations induce RAS/RAF/MAPK pathway activity, we sought to explore whether other elements of this pathway might be associated with endocrine resistance. We performed a pathway-level analysis to identify additional RAS/RAF/MAPK signaling pathway alterations (henceforth referred to as MAPK pathway) present in tumors collected following hormonal therapy and explored their relationship with ESR1 alterations. We identified hotspot mutations in ERBB3, KRAS, HRAS, BRAF, and MAP2K1 (MEK1) that were present in both untreated and post-endocrine therapy samples. Notably, none of the post-treatment metastatic specimens with known or likely functional MAPK mutations harbored ESR1 mutations (Figure 3B). In addition, we identified 12 patients with focal amplifications of EGFR in metastases that had previously received the selective ER modulator (SERM) tamoxifen, with eight of these patients also having previously received AI. Here too, none of the post-endocrine therapy EGFR-amplified tumors harbored known ESR1 mutations that could explain treatment resistance. Moreover, these EGFR amplifications were not attributable to an elevated burden of DNA copy number alterations in metastatic breast cancers overall (Figure S4A), unlike the pattern seen in castration-resistant metastatic prostate cancers (Hieronymus et al., 2014; Taylor et al., 2010). Overall, 16% of all post-hormonal therapy HR+HER2− tumors lacking ESR1 mutations (n = 90) harbored known or presumed oncogenic mutations in one of multiple effectors of the MAPK pathway, a frequency of activating MAPK alterations significantly higher than that observed in untreated primary tumors (Figures 3A and 3B). Beyond alterations in effectors of MAPK, we also identified amplifications and hotspot mutations in MYC and CTCF respectively, both key transcriptional regulators, arising more commonly in post-hormonal therapy specimens. These transcription factor lesions, along with mutations in FOXA1 and TBX3, also largely arose in tumors that did not already possess either an ESR1 mutation or MAPK alteration (Figure 3B), indicating that they too may drive endocrine resistance. Taken together, the MAPK pathway and transcription factor lesions affected 27% of all ESR1-wild-type post-hormonal therapy HR+HER2− tumors (n = 564) and 22% of all post-hormonal therapy tumors (n = 692).
As functional alterations targeting MAPK or various transcription factors arose in post-hormonal therapy breast cancers that preferentially lacked the known mechanism of endocrine resistance (ESR1 mutations) (mutual exclusivity p value < 0.0001), we assessed their association with response to hormonal therapies. Specifically, we examined patients with HR+HER2− metastatic disease for whom the sequenced biopsy was acquired prior to therapy with AI, SERM, or selective ER degrader (SERD) therapy. Patients with tumors harboring MAPK lesions had a significantly shorter progression-free survival (PFS) on AI compared with unaffected tumors (hereafter tumors lacking lesions in MAPK, ESR1, or transcription factors are referred to as pan-wild-type) with a median PFS of 3.5 versus 15.2 months, log-rank p value = 1.4×10−9. This outcome difference was similar among both ESR1-mutant patients and those patients with ESR1-wild-type tumors harboring transcription factor alterations (median PFS of 4.1 and 6.4 months respectively compared with 15.2 months in pan-wild-type patients; p values = 0.0003 and 1.3×10−6; Figure 3C). While there was no significant difference in PFS on SERM among patients with these mutational events (Figure S4B), this was not the case for SERD therapy response. Patients with ESR1 hotspot mutations or transcription factor mutations had a PFS similar to pan-wild-type patients on SERD, but MAPK-altered tumors had a shorter PFS on SERD than pan-wild-type patients (median PFS of 3.7 versus 4.8 months; p value = 0.029; Figure 3C).
The Timing of Candidate Mechanisms of Hormonal Therapy Resistance
To determine whether candidate genomic lesions associated with hormonal therapy resistance were present prior to therapy or whether such lesions arose under the selective pressure of therapy, we performed whole-exome sequencing (WES) in select patients who had adequate samples acquired prior to and after progression on hormonal therapy. We assembled an initial cohort of 30 breast cancer patients for whom we sequenced a treatment-naïve primary tumor, post-progression specimen, and a corresponding matched normal control (Table S4). As highlighted in three representative patients (Figure 4A and Figure S5), WES revealed clonally heterogeneous treatment-naïve primary tumors in which we did not detect the presence of the putative resistance MAPK mutation (sensitivity, ~4% of affected cancer cells) identified in the post-therapy, disease progression sample. In one of these representative patients, the pre-treatment and post-progression tumors shared clonal mutations (those present in all sequenced cancer cells) in PIK3CA and CDH1 as well as an FGFR1 focal amplification. While the pre-treatment tumor also had a subclone harboring a RHOA L22R hotspot mutation, this subclone was not detected post-progression, and was replaced by a KRAS G12R-mutant subclone as well as several additional lesions including a second PIK3CA mutation in cis (Figure 4A). Two other representative cases harbored loss-of-function NF1 mutations post-endocrine therapy. In both cases, neither NF1 mutation was detected in the pre-treatment primary tumor, with each demonstrating biallelic NF1 inactivation in the tumor collected following progression on hormonal therapy (Figure 4A). We then performed concomitant higher depth of coverage targeted sequencing (MSK-IMPACT) of the same matched pairs to increase our sensitivity for detecting subclonal mutations. This further sequencing provided sensitivity for robust mutation detection to ~1.3% of affected cancer cells, for which all results were concordant between sequencing modalities. We cannot exclude the possibility that key mutations pre-existed therapy but were present in a rarer subpopulation of cancers cells. Moreover, WES in these cases did not identify any other candidate lesion mediating endocrine therapy failure.
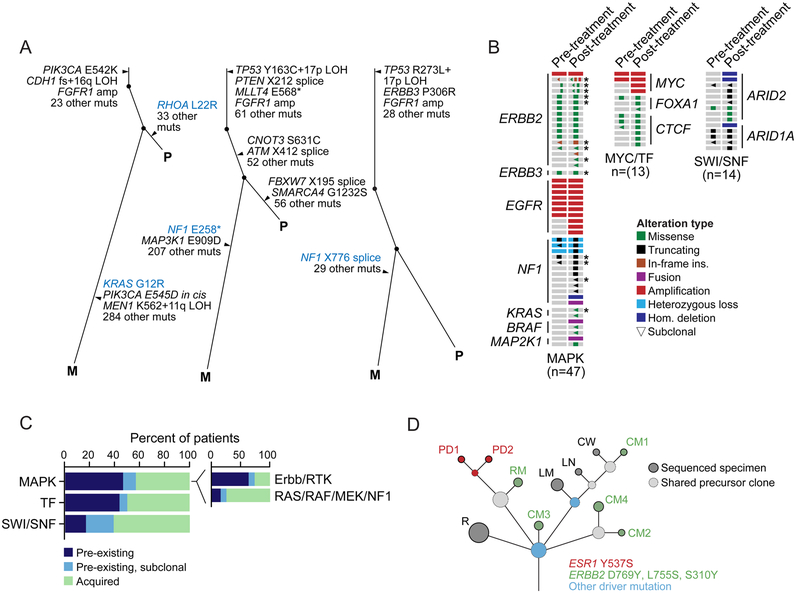
Figure 4: Tumor Evolution Under Endocrine Therapy.
(A) Evolutionary relationships among clones and somatic mutations inferred from whole-exome sequencing of three representative patients with paired treatment-naïve primary (P) and post-treatment metastatic (M) tumors indicating intra-tumoral heterogeneity and the acquisition of MAPK activating lesions after hormonal therapy (left, KRAS G12R; middle and right, two NF1 truncating mutations).
(B) Comparison of treatment-naïve primary tumors and their patient-matched post-endocrine therapy specimens in 74 patients with ESR1-wild-type HR+HER2− disease with diverse lesions in MAPK effectors, MYC and transcription factors (MYC/TF), or components of the SWI/SNF complex (as labeled) indicated pre-existing or acquired lesions.
(C) A summary of the rate of pre-existing and acquired lesions by pathway. Inset further subdivides MAPK alterations into receptor tyrosine kinase (Erbb/RTK) versus downstream RAS/RAF/MEK/NF1 lesions. Asterisks denote patients with multiple candidate resistance mutations.
(D) The clonal evolution of a HR+ metastatic breast cancer inferred from the sequencing of an initial recurrence (R) and ten metastatic sites collected at the time of autopsy. Circles are colored based on the presence of one or more breast cancer driver mutations (blue) or candidate mechanisms of endocrine resistance (red and green, as indicated). The size of the circle is proportional to the number of mutations defining the clone. Dashed circles represent shared precursor clones and solid circles represent the sequenced specimens. PD, pleural disease; RM, retroperitoneal mass; CM, cutaneous metastasis; LM, liver metastasis; LN, lymph node; CW, chest wall.
To broaden this analysis, we performed similar targeted sequencing on matched pre- and post-progression samples from 44 additional post-hormonal therapy patients (n = 74 total matched pairs, Table S4). Similar clonality analyses demonstrated that some putative resistance mechanisms identified in the prospective cohort existed prior to hormonal therapy in some patients whereas others appeared to be acquired or selected for under the selective pressure of treatment (Figures 4B and 4C; Table S4). For example, in the 27 patients for whom the post-progression specimen harbored either an EGFR amplification or ERBB2 mutation and we sequenced a treatment-naïve primary tumor, 60% and 82% were present prior to therapy, respectively (Figure 4B). Conversely, in the patients whose post-progression tumors harbored oncogenic mutations in downstream effectors of the MAPK signaling pathway such as NF1, KRAS, MAP2K1, and BRAF, these events were typically not detected in the treatment-naïve primary tumors and were either acquired or selected for under the selective pressure of endocrine therapy (in 74% of affected patients, Figure 4C). Among matched pre- and post-treatment tumors in patients with a candidate MYC amplification or transcription factor alteration, nearly half of the MYC amplifications or hotspot mutations in CTCF or FOXA1 were either subclonal and selected by, or were present prior to, hormonal therapy while the rest were acquired (Figures 4B). Finally, beyond these candidate mechanisms of endocrine resistance nominated by analysis of the prospective cohort, we also analyzed matched pair specimens from patients with no candidate resistant aberration with the goal of identifying other potential mechanisms of hormonal therapy failure. Although few were evident, loss-of-function lesions in ARID2 and ARID1A (encoding members of the SWI/SNF complex) were more often acquired, present more frequently in the post-treatment specimen (Figure 4C), a finding consistent with the enrichment of these lesions in the metastatic context (Figure 2A).
Patients with advanced breast cancer typically exhibit concurrent progression of several metastatic sites. These different metastatic locations are often clonally diverse and, as we have recently shown, therapeutic selective pressure may result in divergent or convergent evolution of different resistant clones that have the same resistant phenotype (Juric et al., 2015). To gain insights into the evolution of multiple progressing metastatic sites under the selective pressure of hormonal therapy, we performed a research autopsy on a patient with HR+ metastatic breast cancer and a treatment history that included multiple lines of hormonal therapy (Table S5). Multiple metastatic tissues were collected, ten of which were subjected to WES analysis. Assessing the evolutionary relationships among mutant clones and metastatic sites indicated that alterations in TP53, CDH1, FGFR1, and 40 additional mutations were clonal in the pre-mortem disease recurrence and all metastatic sites at autopsy (Figure S5B). However, it was notable that different metastatic lesions harbored different known or candidate mechanisms of endocrine resistance. The pleural disease harbored ESR1 Y537, which was not detected in either the pre-mortem cutaneous metastasis or any other metastatic sites at autopsy. In contrast, five additional spatially and organotypically distinct lesions possessed different ERBB2 driver mutations (S310, L755, and D769). None of these ERBB2 mutations were detected in the recurrence biopsy collected prior to hormonal therapy and none of the ERBB2-mutant metastatic lesions had evidence of ESR1 mutations (Figure 4D and S5B; Table S5).
To determine if these mutations existed in a fraction of cancer cells less than that detectable by our WES (≥80% power to detect somatic mutations in ~5% affected cancer cells per site), we performed droplet digital PCR (ddPCR) on all metastatic tumor specimens from the autopsy for each of the candidate mutations likely mediating endocrine resistance in this patient. These data confirmed that the metastatic sites positive for the ESR1 or ERBB2 mutations from WES were positive by ddPCR at similar predicted allele frequencies (Table S5). Moreover, all of the metastatic sites negative for these mutations from WES were similarly negative by ddPCR. Given that the empirical sensitivity of ddPCR was ~0.03% in these samples, these data indicate that the absence of these mutations in some metastatic sites was not due to false negatives from WES, although we cannot exclude the possibility that a subset of metastatic lesions harbored these mutations in <0.03% tumor cell subpopulations. These data indicate that at least some patients with advanced breast cancer have distinct and coexisting mechanisms of resistance to endocrine therapy in distinct tumor subclones that cannot be captured by a single biopsy of a metastatic site. While our data cannot establish the frequency with which such coexistent mechanisms of endocrine resistance emerge in treated breast cancer patients, recent studies of circulating tumor-derived cell-free DNA (cfDNA) from breast cancer patients indicate that the rate of ESR1 mutations in post-endocrine therapy HR+ breast cancers is higher than can be detected in tumor tissues, indicating that tumor biopsies alone are likely understating the frequency of these alterations due to tumor heterogeneity. Thus, while ERBB2 mutations may be mutually exclusive with ESR1 mutations in our broader prospectively sequenced cohort, subsequent cfDNA studies may show that these alterations are present in different subclones and thus co-occur in individual patients.
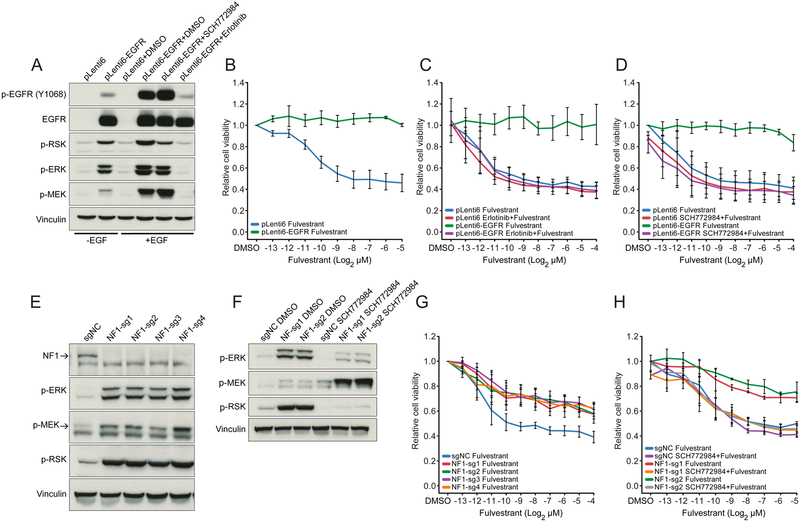
We next sought to validate the role of several of the candidate genomic aberrations identified here in mediating resistance to hormonal therapy. As ERBB2-amplified tumors are known to be resistant to hormonal therapy (Johnston et al., 2009; Kaufman et al., 2009; Marcom et al., 2007; Schwartzberg et al., 2010), we decided to model EGFR amplification and NF1 loss, the next two most common alterations in our cohort known to activate the MAPK signaling pathway. We reasoned that the observed focal amplifications of EGFR, a closely related tyrosine kinase receptor with ERBB2, were likely to also result in hormonal therapy resistance. We thus generated isogenic ER+ MCF7 cells with or without EGFR overexpression (Figure 5A). Notably, fulvestrant reduced the viability of MCF7 cells stably transfected with the empty pLenti6 vector, but not those overexpressing EGFR (Figure 5B), a resistance phenotype that could be reversed upon co-treatment with a combination of fulvestrant and the EGFR kinase inhibitors erlotinib or gefitinib (Figure 5C and S6A). In addition, selective targeting of MAPK signaling with one of multiple structurally distinct ERK inhibitors (Figure 5D and S6B-C) re-sensitized MCF7-EGFR cells to fulvestrant.
Figure 5: Functional Validation of MAPK Activation as Mechanism of Resistance to Endocrine Therapy.
(A) Western blot showing expression and phosphorylation of EGFR and activation of MAPK pathway effectors in MCF7 cells transduced with pLenti6 mock control and pLenti6-EGFR vector.
(B and D) Cell viability of MCF7 cells stably transfected with pLenti6 mock control or pLenti6-EGFR treated with fulvestrant (B), fulvestrant + 5 mM erlotinib (C), or fulvestrant + 500 nM SCH772984 (D).
(E) Western blot showing activation of MAPK pathway effectors in four independent pools of NF1-knockout and control (scrambled-sgNC) MCF7 cells.
(F) Change in expression of phosphorylated ERK, MEK and RSK in cells treated with 500 nM SCH772984 versus control (DMSO). Results are shown for two independent MCF7 pools expressing NF1-targeted guide RNAs and a control (scrambled-sgNC) guide RNA.
(G and H) Cell viability following treatment with fulvestrant (G), or fulvestrant + 500 nM SCH772984 (H). Results are shown for cells in which NF1 was knocked out using four different NF1-targeted guide RNAs. Error bars represent standard deviations.
See also Figure S6.
We similarly investigated the association between endocrine resistance and NF1 loss. We established four independent pools of NF1-knockout MCF7 cells using CRISPR/Cas9 editing with four different NF1-targeted guide RNAs and confirmed loss of NF1 expression by western blot (Figure 5E). Loss of NF1 expression in these breast cancer cells was associated with MAPK pathway activation as measured by increased levels of phosphorylated ERK, MEK and RSK (Figures 5E and 5F). Furthermore, loss of NF1 expression was associated with reduced fulvestrant sensitivity in these cells (Figure 5G). Treatment with the ERK inhibitor SCH772984 suppressed MAPK signaling and reversed the resistance phenotype (Figures 5F and 5H), as did treatment with other ERK inhibitors (Figures S6D and S6E). Collectively, these data indicate that alterations that result in loss of NF1 expression in ER+ breast cancer cells are sufficient to induce MAPK signaling and confer resistance to hormonal therapy.
DISCUSSION
This clinico-genomic study of a large cohort of clinically phenotyped patients with advanced breast cancer provides insights into the genomic alterations present in metastatic disease. Overall, mutations in 32 genes were significantly more common in metastases compared with primary tumors. Our study was enriched for patients with HR+ disease and, in this subgroup alone, 29 genes were more commonly mutated in metastatic versus primary tumors. Our data suggest that some of these alterations were a consequence of selective therapeutic pressure and mechanisms of systemic therapy resistance. Specifically, 22% of post-hormonal therapy HR+HER2− tumors had largely non-overlapping lesions in one of the multiple effectors of MAPK signaling or in MYC or other transcription factors. Importantly, such mutations were mutually exclusive with hotspot mutations in ESR1.
Increased MAPK pathway signaling in vitro can induce an ER-negative phenotype in breast cancer (Creighton et al., 2006) and thus the finding of frequent MAPK alterations in hormonal therapy resistant metastatic tumors is analogous to the lineage plasticity that mediates androgen receptor independence in resistant prostate tumors (Mu et al., 2017; Watson et al., 2015). Patients with MAPK alterations had poor responses to hormonal therapy, suggesting that ER dependence is attenuated in these patients. Indeed, we could replicate in a breast cancer model system that EFGR overexpression or loss of NF1 expression induces a state of endocrine resistance that is reversed by ERK inhibitors, in both cases suggesting a potential therapeutic strategy.
While an exploratory clinical finding, it was also notable that patients having HR+HER2− breast cancer with mutations in one of several transcription factors had, unlike MAPK-altered tumors, a shorter duration of response to AI but not SERD therapy. Pioneering factors such as FOXA1 play a role in chromatin remodeling and alter transcription binding site accessibility (Zaret and Carroll, 2011) and previous work has shown cooperativity among FOXA1 and ER to facilitate estrogen-induced gene expression (Carroll et al., 2005; Toska et al., 2017). The clinical response data here suggest that reduced ER activity but retained ER protein levels may suffice to support this cooperation and result in poorer outcomes on AI treatment whereas the SERD-induced loss of ER expression may abolish FOXA1-ER cooperation resulting in no difference in therapeutic outcomes. While suggestive, such potential differences require further exploration as part of subsequent mechanistic studies in appropriate breast cancer models.
Finally, while MAPK and ESR1 mutations were mutually exclusive at the level of individual patients in the prospective sequencing cohort, they were found to coexist (ERBB2 and ESR1) in separate metastatic lesions from a patient from which multiple tumors were available for analysis. These data suggest that ERBB2 and ESR1 mutations are biologically distinct mechanisms of resistance to hormonal therapy that may coexist in individual patients, but when both are present, are likely to exist in distinct tumor subclones. This finding broadens the data in ESR1-mutant tumors that indicate multiple distinct activating mutations in ESR1 can coexist in different drug-resistant cell populations, all subclonally, in post-endocrine treated patients (Spoerke et al., 2016). The data thus establish a strong rationale for the rapid validation and adoption of technologies to sensitively profile circulating tumor-derived cfDNA to capture the molecular heterogeneity of treatment resistance as a guide to therapy selection. Moreover, data from this patient indicated that convergent evolution exists with multiple independently arising ERBB2 mutations in different metastatic sites, implying that a limited number of therapeutically vulnerable pathways are activated in response to hormonal therapy. Nevertheless, the presence of multiple coexistent mutations in individual patients with very advanced disease resulting in the same phenotype would make it challenging to develop combination therapies that would eradicate all resistance clones, and hence, the need to treat the disease with more effective therapies earlier in the course of its evolution.
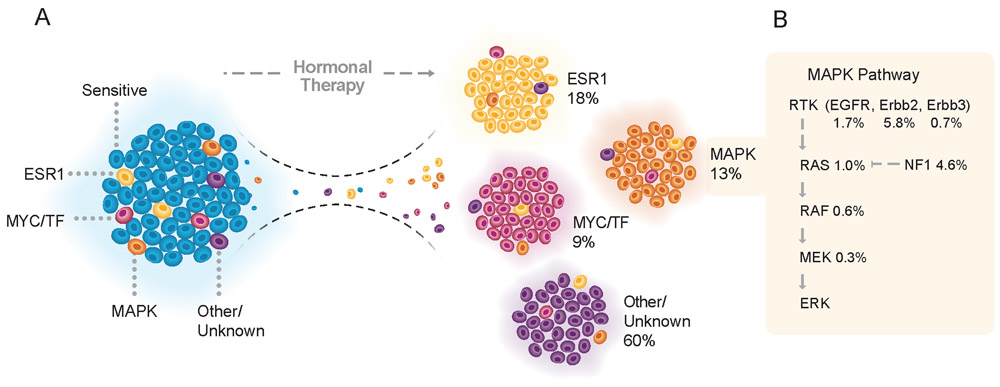
Our findings suggest an emerging taxonomy of endocrine resistant breast cancer that categorizes resistant tumors into four groups: (1) tumors harboring mutations of ESR1, (2) tumors bearing functional lesions in the MAPK pathway, (3) tumors with alterations in the machinery of transcriptional regulation, and (4) pan-wild-type tumors with a still occult mechanism of drug resistance (Figure 6A). The consequences of these findings could influence clinical practice. While implying that tumors harboring ESR1 mutations, MAPK alterations, or alterations in the ER transcriptional machinery are unlikely to benefit from AI and should be considered for alternative treatments, these findings could also inform potential approaches to revert resistance. For example, there are several opportunities for therapeutic intervention that could be tested in combination with hormonal therapy (Figure 6B). HER2 kinase inhibitors are active in breast tumors harboring ERBB2 mutations (Hyman et al., 2018). Similarly, EGFR inhibitors have been shown to be active in patients with breast cancer, although our data suggest that these agents were likely not previously studied in the correct clinical context (Baselga et al., 2013; Osborne et al., 2011). Moreover, MAPK pathway inhibitors including selective MEK and ERK inhibitors are also under active clinical evaluation in patients with breast and other cancer types (Flaherty et al., 2012; Morris et al., 2013). We anticipate that a more comprehensive and multi-modality analysis of pan-wild-type tumors will further refine this classification. For example, recent studies have identified mutations in non-coding regions of transcription factors (Rheinbay et al., 2017), so it is likely that broader molecular profiling methods may assist in the identification of yet unknown drivers of therapy resistance.
Figure 6: A Taxonomy of Mechanisms of Resistance to Endocrine Therapy.
(A) Tumors after hormonal therapy failure may be divided into four categories, depending on the genomic aberrations present in the refractory lesions. Tumors harboring ESR1 mutations (yellow) were responsible for 18% of the tumors relapsing after endocrine therapy. Functional lesions in the MAPK pathway (orange) and mutations in the machinery of transcriptional regulation (MYC/TF, red) were responsible for 13% and 9% of the resistant cases, respectively. Pan-wild-type tumors with a still occult mechanism of resistance to hormonal therapy (purple) accounted for the remaining 60% of cases. These alterations may preexist in the pre-treatment tumors and expand or be acquired under the selective pressure of endocrine therapy.
(B) Frequency of alterations in the MAPK signaling pathway in tumors following hormonal therapy.
Our analysis of genomic associations with hormonal therapy revealed genomic correlates of endocrine resistance at smaller sample sizes than would be possible if the signal of association was diluted by passenger (non-functional) mutations. Nevertheless, our study represents only a single institutional dataset that while carefully clinically annotated, reflects retrospective clinical data collection that may limit the interpretation of our findings and thus requires independent confirmation. Future efforts to nominate increasingly rare genomic mechanisms of endocrine resistance will require even greater samples sizes that would be facilitated by multi-institutional, cooperative data sharing efforts.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by Dr. José Baselga at baselgaj@mskcc.org
EXPERIMENTAL MODEL AND SUBJECT DETAIL
Study Population and Clinical Annotation
A total of 1918 breast tumor specimens from 1756 patients underwent prospective genomic profiling with return of results to patients and their physicians between April 2014 and March 2017. This study was approved by the Memorial Sloan Kettering Cancer Center Institutional review board and all patients provided written informed consent for tumor sequencing and review of patient medical records for detailed demographic, pathologic, and treatment information (NCT01775072). The characteristics of the patients are provided in Table S1. Detailed treatment history data was obtained for each patient and included all lines of systemic therapy from time of diagnosis of invasive carcinoma to the study data lock in April 2017 (Table S2 and S3). The exact regimens as well as the dates of start and stop of therapy were recorded for each treatment line. For each treatment line, the time of biopsy collection for MSK-IMPACT testing was compared with the treatment start and stop dates. The MSK-IMPACT biopsy was therefore categorized as: 1) pre-treatment, 2) on-treatment, and 3) post-treatment corresponding to the acquisition of the MSK-IMPACT sample prior to the start, during receipt, or after completion of therapy.
Cell Lines
MCF7 cells were obtained from ATCC (HTB-22) and were cultured in DMEM/F-12 (Corning) and supplemented with 10% FBS, MEM non-essential amino acids (Corning), 50 U/ml penicillin, and 50 ng/ml streptomycin under normal oxygen conditions (5% CO2, 37 °C). MCF10A cells were obtained from ATCC (CRL-10317) and were similarly cultured in DMEM/F-12 (Corning) and supplemented with 10% FBS, Cholera toxin (100 ng/ml, Sigma C8052), EGF (20 ng/ml, Sigma E9644), 50 U/ml penicillin, and 50 ng/ml streptomycin under normal oxygen conditions (5% CO2, 37 °C). 293T cells were obtained from ATCC (CRL-3216) and amphotropic cells were kindly provided by the laboratory of N. Rosen. Both 293T and amphotropic cells were cultured in DMEM (Corning) supplemented with 10% FBS under normal oxygen conditions (5% CO2, 37 °C). pLenti6-V5 was obtained from Invitrogen and pLenti6-EGFR was kindly provided by C. Costa (Massachusetts General Hospital). pBABE puro HA PIK3CA, pBABE puro HA PIK3CA E545K, and pBABE puro HA PIK3CA H1047R were gifts from J. Zhao (Addgene plasmid #12522, Addgene plasmid #12525, Addgene plasmid #12524) (Zhao et al., 2005). Site-directed mutagenesis (Stratagene) was performed on the wild-type HA PIK3CA plasmid. Plasmids were Sanger-sequenced to verify the mutation or deletion. Mutagenesis primers are documented in Table S6.
METHOD DETAILS
Pathology and Receptor Subtype Classification
The histologic subtypes of breast cancer were determined based on clinical pathology reports of the primary tumors and metastatic sites. Pathology subtypes were classified as either invasive ductal carcinoma (IDC), invasive lobular carcinoma (ILC), mixed ductal and lobular carcinoma (mixed IDC/ILC), and several additional rare histologies (Table S1). The TNM classification and overall tumor stage at diagnosis was defined per the American Joint Committee on Cancer (AJCC) 7th edition staging system. Patients defined as having metastatic disease were confirmed to have had a biopsy of a metastatic site that identified breast cancer. Tumor grade was defined based on the Nottingham combined histologic grade of the primary breast tumor (Elston-Ellis modification of Scarff-Bloom-Richardson grading system) (Bloom and Richardson, 1957; Elston and Ellis, 1991). The primary tumors with total tumor score of 3-5 were classified as G1 (well differentiated); 6-7: as G2 (moderately differentiated), and 8-9: as G3 (poorly differentiated). Patients were classified into breast cancer subtypes based on ER and PR IHC results and the HER2 IHC and/or FISH results rendered at the time of diagnosis in accordance with the American Society of Clinical Oncology (ASCO) and College of American Pathology (CAP) guideline recommendations (Hammond et al., 2010; Wolff et al., 2014).
Prospective Sequencing and Analysis
For all 1918 patients, tumor and patient-matched normal DNA was extracted, respectively, from formalin-fixed paraffin embedded (FFPE) tumor biopsy samples and mononuclear cells from peripheral blood. All specimens underwent next-generation sequencing in our Institutional CLIA-certified laboratory using MSK-IMPACT, a hybridization capture-based next-generation sequencing assay, which analyzes all protein-coding exons of between 341 and 468 cancer-associated genes (Table S2), all as previously described (Cheng et al., 2015; Zehir et al., 2017). Samples were sequenced using either the 341-gene version 1 panel (n=432); the 410-gene version 2 panel (n=968); or the 468-gene version 3 panel (n=518). Average sequencing coverage across all tumors was 771-fold. Somatic mutations, DNA copy number alterations, and structural rearrangements were identified as previously described (Cheng et al., 2015) and all mutations were manually reviewed (Table S3).
Mutational and DNA Copy Number Analysis
In addition to the prospectively sequenced patients described here, we also included mutational data from the whole-exome sequencing of 1298 patients with primary untreated breast cancer from previously published studies (Cancer Genome Atlas, 2012; Stephens et al., 2012). For the purposes of cross-comparison between cohorts, all mutational data independent of platform were uniformly re-annotated to GRCh37 gene transcripts in Ensembl release 75 (Gencode 19) and a single canonical transcript for each gene was used to annotate mutations. Mutation effect is reported using Variant Effect Predictor (VEP) ver. 81 and vcf2maf ver. 1.6.3. To identify significant hotspot mutations, we ran an established algorithm sensitive for detecting rare hotspot mutations (Chang et al., 2016). In addition to the gene-level amplification and deletion calls generated by the clinical laboratory pipeline, genome-wide total and allele-specific DNA copy number was determined using the FACETS algorithm (Shen and Seshan, 2016) for prospectively sequenced patients. Purity, average ploidy, and allele-specific integer-copy number for each segment were then determined by maximum likelihood.
ERBB2 Amplification Status by Prospective Sequencing
The concordance between traditional methods of HER2 amplification detection (IHC and/or FISH) and ERBB2 amplification detection by the MSK-IMPACT assay was assessed. Details of the bioinformatics pipeline to detect copy number alterations and the validation of ERBB2 amplification detection by MSK-IMPACT have been previously published (Ross et al., 2017). Tumors with IHC/FISH and prospective sequencing on the same tumor sample were included in this analysis. The concordance between IHC/FISH and MSK-IMPACT sequencing is summarized in the table below. The overall concordance was 98% (1,778 of 1,810 samples). There was excellent agreement between the two testing methods (Kappa: 0.9, 95% confidence interval: 0.87-0.94).
| IHC/FISH | |||
|---|---|---|---|
| Amplified | Non-amplified | ||
| MSK-IMPACT | Amplified | 169 | 5 |
| Non-amplified | 27 | 1609 | |
However, among those cases HER2+ by IHC and/or FISH, 28 of 196 total (14%) did not meet criteria for amplification by MSK-IMPACT sequencing. Reasons for this discordance included low tumor content (14 of 27), a relatively low level of amplification by FISH (9 of 27), and intratumoral heterogeneity with only a discrete population of tumor cells showing amplification (3 of 27). One additional discordant sample and one of the discordant samples that showed a low level of amplification by FISH also had an ERBB2 mutation. Among cases non-amplified by IHC and/or FISH, five cases (<1% of 1614) did meet criteria for copy number gain/borderline amplification by MSK-IMPACT. Two of the five cases showed a low-level copy number gain by MSK-IMPACT and one of these discordant samples also had an ERBB2 mutation. Two of the five cases showed amplification by MSK-IMPACT in the setting of high background noise in the copy number profile, requiring confirmation of amplification by an alternate method. The final discordant case showed extensive high-grade ductal carcinoma in situ (DCIS) that was HER2+ and contributed to the copy number gain by MSK-IMPACT, however, the invasive carcinoma was HER2−.
Treatment-Naïve Primary and Post-Treatment Metastatic Matched Pairs
To assess whether specific molecular alterations associated with endocrine treatment failure pre-existed therapy, we acquired and sequenced matched treatment-naïve primary tumor specimens from a subset of affected patients. Pathology review, FFPE tissue sectioning, and DNA extraction were performed as described previously (Al-Ahmadie et al., 2016; Cheng et al., 2015; Zehir et al., 2017). After excluding samples with insufficient tumor tissue, insufficient data quality due to low total DNA quantity and purity, or that failed library preparation, a total of 74 patients had matched treatment-naïve primary, post-endocrine therapy tumor, and matched normal control specimens available for analysis and were included in further either MSK-IMPACT (as described above) or whole-exome sequencing analysis. Of these, 30 patients had sufficient tumor tissue for both whole-exome and higher depth of coverage MSK-IMPACT sequencing. The remaining 44 patients had tumor tissue sufficient for only MSK-IMPACT (Table S4).
For these 74 patients, we sequenced a total of 164 tumor specimens. In total, 95 tumor samples were sequenced as part of our prospective clinical sequencing cohort while the remaining 69 treatment-naïve primary tumor samples were acquired retrospectively and sequenced in the research setting. For the samples subjected to MSK-IMPACT, median sequencing coverage across all tumors was 577-fold with 98% of all targeted exons were covered at 100x or more (Table S4). The prospectively sequenced samples were analyzed for somatic mutations and DNA copy number alterations in the clinical setting using analytical methods including manual review as previously described (Cheng et al., 2015; Zehir et al., 2017). Retrospectively sequenced samples were also analyzed in the research setting using somatic mutation and DNA copy number alteration detection pipelines identical to those in the clinical laboratory. For somatic mutations detected in only the post-treatment metastatic sample(s) of the patient, we genotyped that mutant allele for sub-detection read support in the corresponding treatment-naïve primary tumor using custom processing tools (https://github.com/mskcc/GetBaseCountsMultiSample). For a mutation to be considered detected using this approach, we required at least two properly paired supporting reads that were neither PCR duplicates nor QC failed and having minimum mapping and base quality scores of 20. Otherwise, all principle somatic mutation calling was performed with previously described methods (Zehir et al., 2017). Finally, all index lesions called in prospective sequencing data of the post-treatment metastatic sample for patients for whom a matched treatment-naïve primary tumor was sequenced were manually reviewed if present in the primary tumor with low read support. To determine the clonality of each mutation, we performed allele-specific DNA copy number inference using FACETS (Shen and Seshan, 2016) and the fraction of mutated cancer cells for all retained somatic mutations (cancer cell fraction, CCF) was calculated as previously described (McGranahan et al., 2015). Clonal mutations were those with a CCF (assuming the number of mutant copies was equal to the number of copies of the more frequent allele, major copy number) greater than 0.8 or the upper bound of the CCF confidence interval was >0.85. Mutations with CCFs not meeting this condition were defined a subclonal.
DNA copy number alterations were detected as described previously (Cheng et al., 2015). We used the following criteria for calling amplifications and heterozygous/homozygous deletions. Genes with mean segment fold-change (ratio of normalized sequencing depth in tumor to normal) of >1.8 with adjusted p value < 0.05 were called amplified. Similarly, for homozygous deletions we required a mean whole-gene fold-change < −2 or at least one exon within the affected gene having a fold-change < −2, with both criteria requiring adjusted statistical significance of p value < 0.05. Briefly, statistical significance (also described in Cheng et al., 2015) is determined by evaluating how extreme the segment is from the observed background distribution of segments that are clustered around fold-change of 1. In addition, to increase the sensitivity to detect amplifications in the matched treatment-naïve primary tumors, we performed copy number analysis using FACETS and called amplifications where the total copy number determined by FACETS was greater than or equal to 6 independent of genome doubling. To determine heterozygous losses, we performed FACETS analysis on all the matched pairs of tumors and identified genes with a copy number state of 1 and 0 (total copy number and local copy number, see Shen and Seshan, 2016). For gene fusions, we manually inspected the sites of the detected in the post-treatment metastatic sample for fusion supporting reads in the treatment-naïve primary tumor using IGV (http://software.broadinstitute.org/software/igv/).
Index lesions are annotated as absent (not detected given the sensitivity of detection), subclonal, or clonal for somatic mutations and absent or present for copy number alterations. For 13 patients with multiple candidate index lesions, the selected alteration for frequency comparisons (Figure 4B) was chosen based on the descending frequency of alterations within the affected genes in post-endocrine treated ESR1-wild-type (Figure 3A). Fifteen of the 74 patients for whom we sequenced matched pairs of treatment-naïve primary and post-treatment metastatic tumors had multiple metastatic tumors sequenced prospectively. If a candidate index lesion was detected in only a subset of the metastatic tumors, the lesion was designated as subclonal irrespective of whether it arose clonally in one or more metastases.
For whole-exome sequencing, 250 ng of DNA was used to prepare Illumina Hiseq libraries with the Kapa DNA library preparation chemistry (Kapa Biosystems) and barcoded adaptors (IDT). 500 ng of library was captured by hybridization using the SureSelect XT HumanAllExon V4 (Agilent). PCR amplification of the libraries was carried out for 8 cycles in the pre-capture step and for 7 cycles post capture. For samples of very limited quantities (50 ng or less starting material), post-capture PCR was performed up to 15 cycles. Samples were run on a HiSeq 4000 in a 125 bp/125 bp paired end run, using the SBS chemistry (Illumina). In total, 65 tumor specimens corresponding to 30 treatment-naïve primary and 35 post-treatment samples were sequenced to a median depth of 358-fold coverage. The processing and analysis of whole-exome sequencing data was performed as previously described (Al-Ahmadie et al., 2016). To remove likely FFPE artifacts, we excluded somatic mutations that (1) were not present in the matched normal specimens, but were previously reported as germline variants in ExAC, (2) were called mutant alleles in an extended panel of unmatched normal specimens, and (3) possessed mutant allele frequencies <3% in the called tumor sample. Clonality estimates were determined as described above for matched pairs sequenced with MSK-IMPACT. CCF density plots in Figure S5 show hotspot mutations and index lesions identified in the matched pairs analysis. For fusions and indels identified by the expanded non-exonic region coverage of MSK-IMPACT that was not captured by the WES design, we manually reviewed each event in sequencing data and included the events in Figure S5 if detected by MSK-IMPACT sequencing of the same sample.
Autopsy Specimen Acquisition, Sequencing, and Analysis
One patient witnessed by her legally authorized representative provided premortem written informed consent for postmortem tissue collection and sequencing that was approved by the Memorial Sloan Kettering Cancer Center Institutional Review Board. A total of 49 distinct tumor samples were collected from a diverse number of anatomic sites including skin, lung, lymph nodes, liver and pleura. Each tumor sample was trimmed of normal tissue before half was snap frozen in liquid nitrogen and the other half placed in formalin for overnight fixation and paraffin embedding. Formalin fixed histologic sections of each piece of tissue were screened for tumor cellularity and quality, and frozen sections cut from the matching snap frozen tissue for macrodissection. Ultimately, 12 metastatic tumor tissues were selected for whole-exome sequencing. Two specimens were excluded from subsequent analysis owing to low tumor purity as estimated by FACETS analysis. The remaining WES sequence analysis was performed as described above. Tumor specimens were sequenced to a median coverage of 296-fold (Table S5). Purity, ploidy, genome doubling, and cancer cell fractions for all mutations in all specimens were inferred. Tumor purity ranged from 38 to 63% tumor cell content as estimated from sequencing data. Based on the depth of coverage, estimated tumor purity, and both the observed allele fractions and cancer cell fractions of the mutations of interest, we inferred having greater than or equal to 80% power to detect somatic mutations down to 5% CCF across nearly all tissues in the autopsy. Evolutionary relationships among clones and metastatic sites were inferred using the union of somatic mutations called in any of the 10 metastatic tissues. Somatic variant sites included were those 1) covered at 20-fold or greater in all tumor and matched normal specimens, 2) supported by greater than 3 reads in the tumor, 3) present in zero reads in the matched normal, and 4) with a mutant allele fraction in any affected tumor of greater than 3%.
Digital droplet PCR was performed on ESR1 Y537S, ERBB2 D769Y, ERBB2 L755S, and ERBB2 S310Y in all metastatic specimens that underwent whole-exome sequencing to confirm their presence (Table S5). Assays were designed and ordered through Bio-Rad (Table S6)
Cycling conditions were tested to ensure optimal annealing/extension temperature as well as optimal separation of positive from empty droplets. All reactions were performed on a QX200 ddPCR system (Bio-Rad). Each sample was evaluated in technical duplicates. PCR reactions contained primers and probes, DNA and digital PCR Supermix for probes (no dUTP). Reactions were partitioned into a median of ~16,000 droplets per well using the QX200 droplet generator. Emulsified PCRs were run on a 96-well thermal cycler using cycling conditions identified during the optimization step (95°C 10’;40 cycles of 94°C 30’’ 55°C 1’, 98°C 10’, 4°C hold). Plates were read and analyzed with the QuantaSoft software to assess the number of droplets positive for mutant DNA, wild-type DNA, both, or neither. Empirical sensitivity of 0.03% was assessed from total droplet counts across all metastatic sites and assessed mutations and we required that greater than 3 mutant droplets existed before a site was confirmed mutated.
Experimental Validation
For lentivirus transduction, 293T cells were seeded into 10cm dishes 16 hr before transfection. 1.5 μg pMD2.G envelope vector, 3 μg packaging vector pCMV-dR8.2, and 4.5 μg pLenti6-EGFR or pLenti6-V5 were added to 1 ml jetPRIME buffer (Polyplus) and then 18 μl jetPRIME transfection reagent was added for 10 min incubation before adding to the cells. EGFR plasmid was kindly provided by Anthony Faber from VCU Massey Cancer Center. Medium was refreshed 6 hr post-transfection and the supernatant of 293T cells containing lentivirus was collected 48 hr post-transfection to infect MCF7 cells with polybrene (8 μg/ml) for 24 hr and then positive transduced cells were selected with 10 μg/ml blasticidin (GIBCO) for 3 days. For retrovirus transduction, amphotropic cells were seeded into 6-well plates 16 hr before transfection. 1.8 μg plasmids of interest and 0.2 μg pMD2.G envelope vector were added to 200 μl jetPRIME buffer (Polyplus) and then 4 μl jetPRIME transfection reagent was added for 10 minutes incubation before adding to the cells. Medium was refreshed 6 hr post-transfection and the supernatant of amphotropic cells containing retrovirus was collected 48 hr post-transfection to infect MCF10A cells with polybrene (8 μg/ml) for 24 hr and then positive transduced cells were selected with 2 μg/ml puromycin (GIBCO) for 3 days.
For the Ribonucleoproteins (RNP) mediated knockout of NF1, gRNAs targeting the NF1 gene were designed using Benchling (http://www.benchling.com) and ordered as crRNAs together with negative control crRNAs, tracrRNA and Cas9 proteins from IDT. RNP were assembled and nucleofected into cells following the manufacturer’s instruction. Briefly, for each reaction, 2 μl of 200 μM crRNAs and 200 μM tracrRNA were mixed and heated at 95 °C for 5 min, and cool to room temperature gradually. 3.36 μl of crRNAs:tracrRNA complex, 4.76 μl of Cas9 protein (61 μM) and 1.88 μl PBS were incubated at room temperature for 30 min to form RNP. 1 × 106 MCF7 cells were nucleofected with 10 μl of RNP complex and 2.9 μl of Cas9 electroporation enhancer (IDT) by program P-20 using nucleofection solution V (Lonza). Independent pools of cells were selected and characterized for further experiments. Knockout efficiency was analyzed by western blot 5 days post nucleofection. The negative control crRNA (NC#1) was ordered from IDT (Catalog #: 1072544) and the crRNAs targeting NF1 were as follows: sgNF1-1: CGGTTACCTGCTCGTCGAAG, sgNF1-2: CTCGTCGAAGCGGCTGACCA, sgNF1-3: AGTCAGTACTGAGCACAACA and sgNF1-4: GTTGTGCTCAGTACTGACTT.
Western blot was performed as previously described (Castel et al., 2016). Briefly, cells were washed with ice-cold PBS and lysed on ice for 30 min with RIPA lysis buffer supplemented with protease inhibitor (Roche) and phosphatase inhibitor (Thermo Scientific). Protein concentration was determined by the BCA assay (Pierce) according to the manufacturer’s protocol. Samples were prepared for loading by adding 4x sample buffer (Invitrogen) and heating at 100 °C for 10 min. Total proteins were separated by SDS-PAGE on 4–12% Bis-Tris gradient gels (Invitrogen). Proteins were electrophoretically transferred to NC membrane (Bio-Rad), which was blocked in 5% BSA with TBST (Boston BioProducts). Membranes were incubated with primary antibodies in 5% BSA/TBST overnight at 4°C. HRP-conjugated secondary antibody incubation was performed for 1 hr at room temperature in 2% BSA/TBST and signals were visualized by ECL (Super Signal West Femto, Thermo Scientific). Primary antibodies used in this study were: rabbit anti-EGFR (1:2,000; CST 4267), rabbit anti-pEGFR (Y1068) (1:1,000; CST 3777), rabbit anti-AKT (1:2,000; CST 9272), rabbit anti-pAKT (T308) (1:1,000, CST 2965), rabbit anti-pAKT (S473) (1:1,000; CST 4060), rabbit anti-p110α (1:1,000; CST 4249), rabbit anti-HA (1:2,000; CST 3724), rabbit anti-Vinculin (1:2000; CST 13901), rabbit anti-p-MEK (1:1000; CST 9154), rabbit anti-p-ERK (1:2000; CST 4370), rabbit anti-p-RSK (1:2000; Abcam ab32413), mouse anti-NF1 (1:500; Santa Cruz sc-376886), and rabbit anti-ACTB (1:5,000; CST 4970).
Cell viability was performed using the CellTiter-Glo assay (Promega) per the manufacturer’s protocol. For the EGFR experiments, MCF7 cells were seeded in 96 well plates and the next day EGF (100 ng/ml, GIBCO PHG0311) and the indicated treatments were added for 4 days. For the NF1 experiments, MCF7 cells were seeded in 96 well plates and the next day the indicated treatments were added for 4 days. Luminescence was determined by microplate reader (Biotek) and cell viability of each well was normalized by the DMSO treated groups. Compounds used in this study were: gefitinib (Selleck, S1025), erlotinib (Selleck, S1023), SCH772984 (Selleck, S7101), BVD-523 (Selleck, S7854), VX-11e (Selleck, S7709) and fulvestrant (Sigma, I4409).
QUANTIFICATION AND STATISTICAL ANALYSIS
To identify genes with mutations enriched in metastatic patients, we compared metastatic MSK patient samples to a cohort of primary breast cancers that combined both the TCGA cohort as well as MSK patients with sequenced primary samples. A given gene must have had mutations present in at least 5 patients to be considered. Statistical significance was determined by Fisher exact test that compared the number of primary samples that possessed mutations in the given gene to those identified in metastatic samples. Resulting p values were corrected for multiple hypothesis testing with Benjamini and Hochberg method. We determined the association between genomic alterations and progression-free survival (PFS) with disease progression on therapy or patient death. Due to sample size considerations, we restricted this analysis to 661 patients with metastatic HR+HER2− breast cancer who received hormonal therapy. We categorized hormonal therapies into six major classes of therapy including: 1) aromatase inhibitors including letrozole, exemestane, or anastrozole; 2) selective estrogen receptor modulators (SERM) including tamoxifen, raloxifene, or toremifene; 3) selective estrogen receptor degraders (SERD) including fulvestrant; 4) mTORC1 inhibitors such as everolimus in combination with aromatase inhibitors; and CDK4/CDK6 inhibitors such as palbociclib; 5) in combination with aromatase inhibitors; or 6) in combination with SERDs such as fulvestrant. We used univariate Cox proportional hazard models to determine the association between genomic alterations in each gene or set of genes and PFS. The analysis was restricted to patients for whom the sequenced biopsy was acquired directly prior to the first line of the indicated treatment. For patients with multiple lines of the same class of therapy, only the first treatment line from that class that was started after the MSK-IMPACT biopsy was included in the analysis. We tested the proportionality assumption of the Cox regression model through time-dependency analysis of selected genetic alterations (cox.zph function of the R package survival). We rejected the null hypotheses with a two-sided α = 0.05. We corrected the analysis for multiple comparisons using the Benjamini-Hochberg method whenever appropriate. Recurrent hotspot mutations were identified as previously described (Chakravarty et al., 2017; Chang et al., 2016). Statistically significant hotspots were those with a q value<0.01.
For the cell viability studies, the Luminescence was determined by microplate reader (Biotek) and cell viability of each well was normalized by the DMSO treated groups. The mean and standard deviation (SD) were calculated for each group.
DATA AND SOFTWARE AVAILABILITY
The assembled prospective somatic mutational and clinical data have been deposited for visualization and download in the cBioPortal for Cancer Genomics (http://www.cbioportal.org/study?id=breast_msk_2018) and provided as supplemental tables.
The accession number for the deposited whole-exome sequencing data reported in this paper is dbGaP: phs001674.v1.p1.
Supplementary Material
SIGNIFICANCE.
The genomic evolution of breast cancers exposed to systemic therapy and its effects on clinical outcome have not been broadly characterized. Collectively, our findings suggest an emerging taxonomy of hormone-resistant breast cancer that includes, in addition to already known ESR1 mutations, functional lesions in the MAPK pathway and in the estrogen receptor transcriptional regulation machinery. Our findings could influence clinical practice, as patients with tumors harboring ESR1 mutations, or alterations in the MAPK pathway and in the estrogen receptor transcriptional machinery, are likely to have limited benefit from aromatase inhibitors. Furthermore, some of these alterations are therapeutically targetable including ERBB2-activating mutations, NF1 loss, and EGFR amplification, and thus the findings could lead to strategies to revert or delay drug resistance.
ACKNOWLEDGEMENTS
We thank members of the Marie-Josée and Henry R. Kravis Center for Molecular Oncology for useful discussions. This work was supported by National Institutes of Health awards P30 CA008748, U54 OD202355, R01 CA190642 (to J.B.), R01 CA207244 (to D.M.H. and B.S.T.), and R01 CA204749 (to B.S.T.); the American Cancer Society (127350-RSG-15-067-01-TBG), Breast Cancer Alliance Young Investigator Award (to P.R.), the Sontag Foundation, the Josie Robertson Foundation, Stand up to Cancer, and the Breast Cancer Research Foundation.
Footnotes
DECLARATION OF INTERESTS
J.B. has received in the past honoria from Roche and Novartis. He serves on the board of Varian Medical Systems, Bristol-Myers Squibb, and Foghron, and on the scientific advisory board of Grail, PMV Pharma, Apogen and Northern Biologicals, and Tango, and is a founder of Venthera.
REFERENCES
- Al-Ahmadie HA, Iyer G, Lee BH, Scott SN, Mehra R, Bagrodia A, Jordan EJ, Gao SP, Ramirez R, Cha EK, et al. (2016). Frequent somatic CDH1 loss-of-function mutations in plasmacytoid variant bladder cancer. Nat Genet 48, 356–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre F, Campone M, Ciruelos EM, Iwata H, Liobl S, Rugo HS, Wilke C, Mills D, Chol M, Longin A, et al. (2016). SOLAR-1: A phase III study of alpelisib + fulvestrant in men and postmenopausal women with HR+/HER2− advanced breast cancer (BC) progressing on or after prior aromatase inhibitor therapy. J Clin Oncol 34, TPS618–TPS618. [Google Scholar]
- Augello MA, Hickey TE, and Knudsen KE (2011). FOXA1: master of steroid receptor function in cancer. EMBO J 30, 3885–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. (2012). Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486, 405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. (2012). Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44, 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baselga J, Cortes J, De Laurentiis M, Dieras V, Harbeck N, Hsu JY, Ng V, Schimmoller F, Wilson TR, Im Y, et al. (2016). SANDPIPER: Phase III study of the PI3-kinase (PI3K) inhibitor taselisib (GDC-0032) plus fulvestrant in patients (pts) with estrogen receptor (ER)-positive, HER2-negative locally advanced or metastatic breast cancer (BC) enriched for pts with PIK3CA-mutant tumors. J Clin Oncol 34, TPS617–TPS617. [Google Scholar]
- Baselga J, Gomez P, Greil R, Braga S, Climent MA, Wardley AM, Kaufman B, Stemmer SM, Pego A, Chan A, et al. (2013). Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J Clin Oncol 31, 2586–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom HJ, and Richardson WW (1957). Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer 11, 359–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, Van Allen EM, Lawrence MS, Horowitz PM, Cibulskis K, et al. (2015). Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discov 5, 1164–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas N (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. (2005). Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122, 33–43. [DOI] [PubMed] [Google Scholar]
- Castel P, Ellis H, Bago R, Toska E, Razavi P, Carmona FJ, Kannan S, Verma CS, Dickler M, Chandarlapaty S, et al. (2016). PDK1-SGK1 Signaling Sustains AKT-Independent mTORC1 Activation and Confers Resistance to PI3Kalpha Inhibition. Cancer Cell 30, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, et al. (2017). OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandarlapaty S, Chen D, He W, Sung P, Samoila A, You D, Bhatt T, Patel P, Voi M, Gnant M, et al. (2016). Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol 2, 1310–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, Gao J, Socci ND, Solit DB, Olshen AB, et al. (2016). Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 34, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, et al. (2015). Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn 17, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. (2015). Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 163, 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, and El-Ashry D (2006). Activation of mitogen-activated protein kinase in estrogen receptor alpha-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor alpha-negative human breast tumors. Cancer Res 66, 3903–3911. [DOI] [PubMed] [Google Scholar]
- De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CK, Nuciforo P, Bidard FC, Aura C, Saura C, Peg V, et al. (2014). Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol 25, 1729–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. (2010). Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dydensborg AB, Rose AA, Wilson BJ, Grote D, Paquet M, Giguere V, Siegel PM, and Bouchard M (2009). GATA3 inhibits breast cancer growth and pulmonary breast cancer metastasis. Oncogene 28, 2634–2642. [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. (2012). Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston CW, and Ellis IO (1991). Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19, 403–410. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, Demidov LV, Hassel JC, Rutkowski P, Mohr P, et al. (2012). Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 367, 107–114. [DOI] [PubMed] [Google Scholar]
- Fribbens C, O'Leary B, Kilburn L, Hrebien S, Garcia-Murillas I, Beaney M, Cristofanilli M, Andre F, Loi S, Loibl S, et al. (2016). Plasma ESR1 Mutations and the Treatment of Estrogen Receptor-Positive Advanced Breast Cancer. J Clin Oncol 34, 2961–2968. [DOI] [PubMed] [Google Scholar]
- Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. (2010). American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med 134, e48–72. [DOI] [PubMed] [Google Scholar]
- Harris M, Howell A, Chrissohou M, Swindell RI, Hudson M, and Sellwood RA (1984). A comparison of the metastatic pattern of infiltrating lobular carcinoma and infiltrating duct carcinoma of the breast. Br J Cancer 50, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Schultz N, Gopalan A, Carver BS, Chang MT, Xiao Y, Heguy A, Huberman K, Bernstein M, Assel M, et al. (2014). Copy number alteration burden predicts prostate cancer relapse. Proc Natl Acad Sci U S A 111, 11139–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman DM, Piha-Paul SA, Won H, Rodon J, Saura C, Shapiro GI, Juric D, Quinn DI, Moreno V, Doger B, et al. (2018). HER kinase inhibition in patients with HER2- and HER3-mutant cancers. Nature 554, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S, Pippen J Jr., Pivot X, Lichinitser M, Sadeghi S, Dieras V, Gomez HL, Romieu G, Manikhas A, Kennedy MJ, et al. (2009). Lapatinib combined with letrozole versus letrozole and placebo as first-line therapy for postmenopausal hormone receptor-positive metastatic breast cancer. J Clin Oncol 27, 5538–5546. [DOI] [PubMed] [Google Scholar]
- Juric D, Castel P, Griffith M, Griffith OL, Won HH, Ellis H, Ebbesen SH, Ainscough BJ, Ramu A, Iyer G, et al. (2015). Convergent loss of PTEN leads to clinical resistance to a PI(3)K[agr] inhibitor. Nature 518, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric D, Krop I, Ramanathan RK, Wilson TR, Ware JA, Sanabria Bohorquez SM, Savage HM, Sampath D, Salphati L, Lin RS, et al. (2017). Phase I Dose-Escalation Study of Taselisib, an Oral PI3K Inhibitor, in Patients with Advanced Solid Tumors. Cancer Discov 7, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman B, Mackey JR, Clemens MR, Bapsy PP, Vaid A, Wardley A, Tjulandin S, Jahn M, Lehle M, Feyereislova A, et al. (2009). Trastuzumab plus anastrozole versus anastrozole alone for the treatment of postmenopausal women with human epidermal growth factor receptor 2-positive, hormone receptor-positive metastatic breast cancer: results from the randomized phase III TAnDEM study. J Clin Oncol 27, 5529–5537. [DOI] [PubMed] [Google Scholar]
- Ma CX, Reinert T, Chmielewska I, and Ellis MJ (2015). Mechanisms of aromatase inhibitor resistance. Nat Rev Cancer 15, 261–275. [DOI] [PubMed] [Google Scholar]
- Marcom PK, Isaacs C, Harris L, Wong ZW, Kommarreddy A, Novielli N, Mann G, Tao Y, and Ellis MJ (2007). The combination of letrozole and trastuzumab as first or second-line biological therapy produces durable responses in a subset of HER2 positive and ER positive advanced breast cancers. Breast Cancer Res Treat 102, 43–49. [DOI] [PubMed] [Google Scholar]
- Mayer IA, Abramson V, Formisano L, Balko JM, Estrada MV, Sanders M, Juric D, Solit D, Berger MF, Won H, et al. (2016). A Phase Ib Study of Alpelisib (BYL719), a PI3Kalpha-specific Inhibitor, with Letrozole in ER+/HER2-Negative Metastatic Breast Cancer. Clin Cancer Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleskey BC, Penedo TL, Zhang K, Hameed O, Siegal GP, and Wei S (2015). GATA3 expression in advanced breast cancer: prognostic value and organ-specific relapse. Am J Clin Pathol 144, 756–763. [DOI] [PubMed] [Google Scholar]
- McGranahan N, Favero F, de Bruin EC, Birkbak NJ, Szallasi Z, and Swanton C (2015). Clonal status of actionable driver events and the timing of mutational processes in cancer evolution. Sci Transl Med 7, 283ra254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris EJ, Jha S, Restaino CR, Dayananth P, Zhu H, Cooper A, Carr D, Deng Y, Jin W, Black S, et al. (2013). Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov 3, 742–750. [DOI] [PubMed] [Google Scholar]
- Mu P, Zhang Z, Benelli M, Karthaus WR, Hoover E, Chen CC, Wongvipat J, Ku SY, Gao D, Cao Z, et al. (2017). SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, et al. (2016). Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, et al. (2006). The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12, 1157–1167. [DOI] [PubMed] [Google Scholar]
- Osborne CK, Neven P, Dirix LY, Mackey JR, Robert J, Underhill C, Schiff R, Gutierrez C, Migliaccio I, Anagnostou VK, et al. (2011). Gefitinib or placebo in combination with tamoxifen in patients with hormone receptor-positive metastatic breast cancer: a randomized phase II study. Clin Cancer Res 17, 1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, Pugh M, Jones L, Russell R, Sammut SJ, et al. (2016). The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 7, 11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. (2000). Molecular portraits of human breast tumours. Nature 406, 747–752. [DOI] [PubMed] [Google Scholar]
- Rheinbay E, Parasuraman P, Grimsby J, Tiao G, Engreitz JM, Kim J, Lawrence MS, Taylor-Weiner A, Rodriguez-Cuevas S, Rosenberg M, et al. (2017). Recurrent and functional regulatory mutations in breast cancer. Nature 547, 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DR, Wu YM, Vats P, Su F, Lonigro RJ, Cao X, Kalyana-Sundaram S, Wang R, Ning Y, Hodges L, et al. (2013). Activating ESR1 mutations in hormone-resistant metastatic breast cancer. Nat Genet 45, 1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross DS, Zehir A, Cheng DT, Benayed R, Nafa K, Hechtman JF, Janjigian YY, Weigelt B, Razavi P, Hyman DM, et al. (2017). Next-Generation Assessment of Human Epidermal Growth Factor Receptor 2 (ERBB2) Amplification Status: Clinical Validation in the Context of a Hybrid Capture-Based, Comprehensive Solid Tumor Genomic Profiling Assay. J Mol Diagn 19, 244–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas P, Teo ZL, Lefevre C, Flensburg C, Caramia F, Alsop K, Mansour M, Francis PA, Thorne HA, Silva MJ, et al. (2016). The Subclonal Architecture of Metastatic Breast Cancer: Results from a Prospective Community-Based Rapid Autopsy Program "CASCADE". PLoS Med 13, e1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, Fenwick K, Kozarewa I, Lopez-Knowles E, Ribas R, et al. (2015). Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Sci Transl Med 7, 313ra182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzberg LS, Franco SX, Florance A, O'Rourke L, Maltzman J, and Johnston S (2010). Lapatinib plus letrozole as first-line therapy for HER-2+ hormone receptor-positive metastatic breast cancer. Oncologist 15, 122–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. (2012). The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R, and Seshan VE (2016). FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res 44, e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. (2001). Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98, 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoerke JM, Gendreau S, Walter K, Qiu J, Wilson TR, Savage H, Aimi J, Derynck MK, Chen M, Chan IT, et al. (2016). Heterogeneity and clinical significance of ESR1 mutations in ER-positive metastatic breast cancer patients receiving fulvestrant. Nat Commun 7, 11579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC, Nik-Zainal S, Martin S, Varela I, Bignell GR, et al. (2012). The landscape of cancer genes and mutational processes in breast cancer. Nature 486, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. (2010). Integrative genomic profiling of human prostate cancer. Cancer Cell 18, 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toska E, Osmanbeyoglu HU, Castel P, Chan C, Hendrickson RC, Elkabets M, Dickler MN, Scaltriti M, Leslie CS, Armstrong SA, et al. (2017). PI3K pathway regulates ER-dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science (New York, N Y) 355, 1324–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toy W, Shen Y, Won H, Green B, Sakr RA, Will M, Li Z, Gala K, Fanning S, King TA, et al. (2013). ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat Genet 45, 1439–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson PA, Arora VK, and Sawyers CL (2015). Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 15, 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, Allred DC, Bartlett JM, Bilous M, Fitzgibbons P, et al. (2014). Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 138, 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Knappskog S, Wedge D, Farmery JHR, Gonzalez S, Martincorena I, Alexandrov LB, Van Loo P, Haugland HK, Lilleng PK, et al. (2017). Genomic Evolution of Breast Cancer Metastasis and Relapse. Cancer Cell 32, 169–184 e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret KS, and Carroll JS (2011). Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25, 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, and Roberts TM (2005). The oncogenic properties of mutant p110alpha and p110beta phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci U S A 102, 18443–18448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The assembled prospective somatic mutational and clinical data have been deposited for visualization and download in the cBioPortal for Cancer Genomics (http://www.cbioportal.org/study?id=breast_msk_2018) and provided as supplemental tables.
The accession number for the deposited whole-exome sequencing data reported in this paper is dbGaP: phs001674.v1.p1.