Abstract
The aim of the present study was to define the role of Trx and Grx on metabolic thiol redox regulation and identify their protein and metabolite targets. The hepatocarcinoma-derived HepG2 cell line under both normal and oxidative/nitrosative conditions by overexpression of NO synthase (NOS3) was used as experimental model. Grx1 or Trx1 silencing caused conspicuous changes in the redox proteome reflected by significant changes in the reduced/oxidized ratios of specific Cys's including several glycolytic enzymes. Cys91 of peroxiredoxin-6 (PRDX6) and Cys153 of phosphoglycerate mutase-1 (PGAM1), that are known to be involved in progression of tumor growth, are reported here for the first time as specific targets of Grx1. A group of proteins increased their CysRED/CysOX ratio upon Trx1 and/or Grx1 silencing, including caspase-3 Cys163, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) Cys247 and triose-phosphate isomerase (TPI) Cys255 likely by enhancement of NOS3 auto-oxidation. The activities of several glycolytic enzymes were also significantly affected. Glycolysis metabolic flux increased upon Trx1 silencing, whereas silencing of Grx1 had the opposite effect. Diversion of metabolic fluxes toward synthesis of fatty acids and phospholipids was observed in siRNA-Grx1 treated cells, while siRNA-Trx1 treated cells showed elevated levels of various sphingomyelins and ceramides and signs of increased protein degradation. Glutathione synthesis was stimulated by both treatments. These data indicate that Trx and Grx have both, common and specific protein Cys redox targets and that down regulation of either redoxin has markedly different metabolic outcomes. They reflect the delicate sensitivity of redox equilibrium to changes in any of the elements involved and the difficulty of forecasting metabolic responses to redox environmental changes.
Keywords: Redox proteome, Thiol redox regulation, Glycolysis, S-nitrosation, NO synthase, Redoxins
Graphical abstract
Highlights
-
•
Trx1 and Grx1 Cys redox targets are abundant among Glycolytic enzymes.
-
•
PRDX6-Cys91 and PGAM-Cys153 are specific targets of Grx1.
-
•
Down regulation of thioredoxin and glutaredoxin have different metabolic outcomes.
-
•
Glutathione synthesis and membrane lipid composition are sensitive to Trx1 and Grx1 down regulation.
-
•
Redoxins down regulation also induce target Cys reductive changes under NOS3 overexpression.
1. Introduction
The importance of redox homeostasis for cancer cell survival in the context of energy metabolism is well established [2], [57], [83]. Sensitive cysteine residues on target proteins are at the center of regulatory mechanisms, whose redox state are controlled by two major cellular systems, the Thioredoxin (Trx)/Trx reductase (TrxR) and the Glutaredoxin (Grx)/glutathione (GSH) systems [30], [8].
Nitric oxide (NO) plays important roles in signal transduction [52]. It is synthesized in cells by NO synthase (NOS) isoenzymes whose expression may be regulated as part of cellular responses to several stimuli [1]. NO has contradictory effects on tumor cells, either pro-apoptotic or anti-apoptotic according to cell type, intracellular concentration range and subcellular site of its generation [23], [44]. Post-translational redox modifications (redox PTM) of target sensitive proteins and more specifically, S-nitrosation of sensitive cysteine residues in those proteins, is the basis of NO activity in signaling pathways, although other reversible oxidative modifications of cysteines are also to be expected since high levels of NO are accompanied by increased levels of reactive oxygen species (ROS) [85]. Reversible regulation of redox PTMs is necessary for this process to have regulatory and biological meaning. Intracellular levels of GSNO and S-nitrosated proteins are controlled by GSNO reductase, alcohol dehydrogenase class-3 (ADH III), which is a NADH-dependent enzyme that is conserved from bacteria to human and metabolizes GSNO to oxidized glutathione and NH3 [47]. GSNO reductase does not directly denitrosate SNO proteins, but as the GSNO pool is in equilibrium with protein thiols, reduction of GSNO by the GSNO reductase indirectly results in protein denitrosation [47].
Trx has been shown to catalyze denitrosation in vitro and to affect levels of cellular protein nitrosothiols. It has been shown to also catalyze the direct denitrosation of proteins in vivo through the action of its conserved active site dithiol, which in human Trx1 is Cys32 and Cys35 [4], [5]. NOS3 (or eNOS) itself is a target of Trx leading to activation of the enzyme as part of a regulatory mechanism of NOS3 by reversible auto-S-nitrosation [4], [15], [64], [67].
Mammalian Trx1 contains additional conserved Cys at positions 62, 69 and 73. Cys73 is particularly prone to S-nitrosation [26] and is implicated in the specific and reversible transfer of a nitrosothiol between Trx1 and caspase 3 with the resulting inhibition of apoptosis [60]. It has been shown that S-nitrosation of Cys73 is favored after formation of a disulfide between Cys32-Cys35, that attenuates Trx1 disulfide reductase and denitrosase activities [86]. Depending on the redox state of the cell, Trx1 catalyzes either trans-S-nitrosation or S-denitrosation so that upon inhibition of disulfide reduction or S-denitrosation activity, Cys73-SNO-Trx1 could catalyze trans-S-nitrosation of target proteins [26], [86]. Thus, as a master regulator of redox signaling, Trx1 would protect proteins via its reductase activity, but under highly oxidative environments, the Cys32/Cys35 oxidized disulfide form accumulates, Cys73 becomes S-nitrosated and Trx1 offers an alternative modality of protein regulation via transnitrosation [86].
Glutaredoxin is involved in post-translational modification of proteins by S-glutathionylation with a role mainly as a deglutathionylase [39], [54], [75]. Grx1 catalyzes reversible S-glutathionylation of protein targets involved in Glycolysis, energy sensing, calcium homeostasis, apoptosis and transcriptional regulation [59]. Grx1 has also been shown to counteract the proapoptotic action of NO in tumor cell lines [22], [35] and to regulate NOS3 activity by glutathionylation [9], an activity shared with Trx [78]. However, no direct evidence of Grx1 as denitrosase has been reported so far.
The thiol-based mechanism of NO multiple actions and the roles of Trx and Grx in the context of protein function regulation by reversible redox changes at target cysteines justify the study of cells under nitrosative stress. Exogenous addition of NO donors is a widely used experimental approach to this end, but endogenously produced RNS would better mimic a physiological situation. The levels of ROS and NO increase to 140% in NOS3 overexpressing HepG2 cells relative to control cells [21], which makes them a good experimental model to study the significance of Trx and Grx roles in the regulation of cellular physiology under nitrosative and oxidative conditions.
The Thiol Redox Proteome refers to the set of mapped protein cysteines showing reversible redox changes under given conditions or in response to stimuli [20]. Its detailed description may provide an insightful snapshot of the set of thiol redox switches [19], [38] in a cell under given conditions. Several qualitative and quantitative proteomic approaches have been devised to study the redox proteome [24], [27], [32], [43], [46], [50], [53], [55] and to define Trx target proteins either as reductase or transnitrosase [6], [46], [87] or target proteins of Grx as deglutathionylase [54]. The identification and modulation by reversible redox PTMs of key proteins that can control metabolic flow, would offer promising therapeutic candidates for a number of disease states such as cancer [80]. Moreover, redox signals work in tandem with other signals to control different cellular processes.
One common characteristic to all types of cancers is reprogramming of energy metabolism to generate ATP through intense glycolytic flux from glucose to lactate even when oxygen is present, a phenomenon known as “the Warburg effect”. Mitochondria remain functional and part of the glucose consumed is diverted into biosynthetic pathways upstream of pyruvate. Altered cell metabolism is a characteristic feature of many cancers resulting in changes to metabolite concentrations and eventually affecting cell signaling pathways and metabolic fluxes [79], [83]. There is a complex connection between metabolism and proliferation with many checkpoints still to be discovered.
We have previously assessed the role of NOS-3 overexpression on several metabolic checkpoints in HepG2 cells showing the prominence of the oxidative branch of the Pentose Phosphate Pathway (oxPPP) to direct the metabolic flux towards NADPH and the increase in Trx and Grx, but at the same time a decrease in nucleotide biosynthesis and proliferation [23]. Downregulation of Trx and Grx reduced cell proliferation and increased caspase-8 and caspase-3.
Here we have applied a quantitative proteomic and metabolomic approach to evaluate HepG2 cells under high levels of endogenous NO and ROS by NOS3 overexpression and ≈ 80% down-regulation of Grx1 or Trx1 with specific siRNA. Proteins undergoing significant redox changes have been identified, their sensitive cysteines have been mapped and their “reduced Cys/oxidized Cys” ratios have been determined. Known targets of redox PTMs have been confirmed and new targets have been discovered, mostly metabolic enzymes. Glycolytic metabolic flux, complex lipids and glutathione metabolism and protein degradation, among other pathways, were affected by Trx1 or Grx1 silencing, concomitant with redox changes at specific cysteines in glycolytic enzymes.
2. Material and methods
2.1. Materials and reagents
All reagents were of analytical grade and were purchased from Sigma, unless otherwise stated. HepG2 cell line used in this work was obtained from ATCC LGC Standards Company (Teddington, UK). Cell culture dishes and flasks were from TPP (Switzerland). Anti-Trx1 and anti-Grx1 were obtained from rabbit in our laboratory. Antibodies against PKM2, caspase-3, NOS-3 and β-actin were from Santa Cruz Biotechnology, (Dallas, TX, USA). Antibodies against PRDX6 were from Abcam (Cambridge, UK). ECL was from GE Healthcare (Wauwatosa, Wisconsin, USA). siRNA for Grx1 and Trx1 were from GE Healthcare Dharmacon (Wauwatosa, Wisconsin, USA). [14C-1]-Glucose, [14C-6]-Glucose and [3H-3]-Glucose were from Perkin Elmer (USA).
2.2. Cell growth conditions
Cells were transfected with the pcDNA/4TO (5100 bp; Invitrogen, Molecular Probes, Inc.) resulting in 4TO control cell line, as well as with the same expression vector containing NOS3 cDNA sequence (3462 bp; NCBI, ImaGenes, full length cDNA clone sequence BC063294) under the control of the cytomegalovirus promoter resulting in 4TO-NOS cell line. Cell lineages 4TO and 4TO-NOS were selected with zeocin (15 mg/L; Invitrogen) as was described by González et al. [21]. Cells were maintained in EMEM Medium (Eagle Minimum Essential Medium), pH 7.4, supplemented with 10% fetal bovine serum, 2.2 g/L NaHCO3, 1 mM sodium pyruvate, 100 U/L penicillin, 100 µg/mL streptomycin, 0.25 µg/mL amphotericin, and the corresponding selective zeocin antibiotic in 5% CO2 atmosphere at 37 °C.
2.3. Silencing of Trx1 and Grx1
Human Grx1 and Trx1 were down-regulated in non-transfected HepG2 cells (WT), 4TO and 4TO-NOS cells using specific siRNA in 6-well plates (20,000 cells/cm2) according to the manufacturer's recommendations (Dharmacon, GE Healthcare Life Sciences). Grx and Trx siRNA (25 nmol) were mixed with the transfection reagent DharmaFECT 1, previously pre-incubated with culture medium (antibiotic/antimycotic and serum free), and incubated for 20 min at room temperature. Afterwards, the interference solutions were added to cultured cells for 72 h in 2% culture medium in the absence of antibiotic/antimycotic solution [89]. Silencing of Trx 1 and Grx1 was always checked by Western blot and activity assay to confirm that their levels were reduced by ≈ 80% [22].
2.4. Sample preparation for redox proteomics
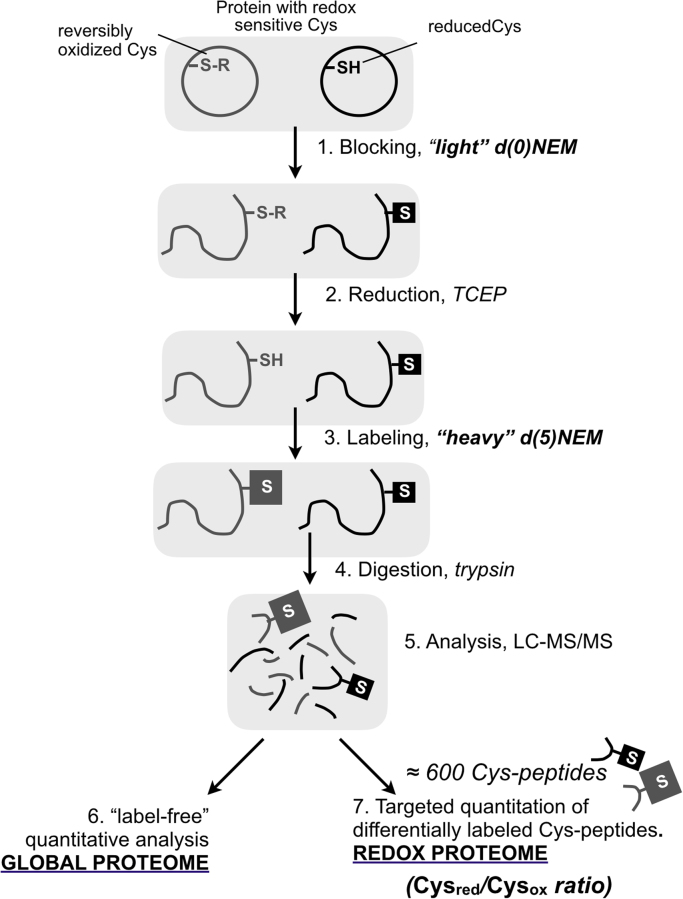
The experiments were routinely carried out at 100,000 cells/cm2 and the cells were washed with PBS. The methodological strategy is summarized in Fig. 1 as described before [55]. Reversibly oxidized Cys were labeled with “heavy” NEM in which 5 hydrogen atoms had been substituted by deuterium atoms here named “d(5)NEM” and reduced cysteines were blocked with “light” NEM, here named “d(0)NEM”. Cell extracts were obtained with lysis solution (50 mM ammonium bicarbonate, 50 mM d(0)NEM, 0.5% CHAPS, 1 mM PMSF); samples were centrifuged at 15,000g for 5 min at 4 °C, excess d(0)NEM was removed using Zeba spin desalting columns (Thermo Scientific). 100 µg of protein were diluted up to 160 µl with 25 mM ammonium bicarbonate, incubated with denaturing reagent by addition of 10 µl of 1% w/v RapiGest (Waters) in 25 mM ammonium bicarbonate, incubated at 80 °C for 10 min and vortexed. 10 µl of a 100 mM solution of TCEP was added followed by incubation at 60 °C for 10 min to reduce the reversibly oxidized cysteines that were subsequently alkylated by adding 10 µl of 200 mM d(5)NEM and incubated at room temperature for 30 min. An aliquot was taken at this point to check the procedure by SDS-PAGE.
Fig. 1.
Proteomics experimental strategy. The procedure follows the already classical three-step approach. In this case, the thiol blocking agent was NEM, the cysteine reductant was TCEP and the newly formed thiols were labeled with “heavy” d(5)-NEM in which 5 hydrogen atoms had been substituted by deuterium atoms. LC-MS/MS data were analyzed for global protein changes with MaxQuant software for “label-free” quantitation [12]. Redox protein changes were analyzed from the set of Cys-peptides identified by targeted quantification using Skyline [48] and calculating the “light”(reduced)/“heavy”(oxidized) Cys ratio. See M&M section for a detailed description.
Proteolytic digestion was performed by addition of 10 µl 12.5 ng/µl of trypsin (Promega) in 25 mM ammonium bicarbonate and incubated at 37 °C temperature overnight. Protein digestion was stopped by addition of 3 µl trifluoroacetic acid (1.5% final concentration). Digested samples were dialyzed through detergent removal column (Pierce) to eliminate any possible rest of CHAPS and dried in speedvac.
2.5. LC–MS/MS
Protein analyses were performed at the Proteomics Facility (SCAI) at the University of Córdoba. Peptides were scanned and fragmented with the LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nano-UHPLC Ultimate 3000 (Dionex-Thermo Scientifics). Chromatography conditions were: mobile phase solution A: 0.1% formic acid in ultrapure water; mobile phase solution B: 80% acetonitrile, 0.1% formic acid. A chromatography gradient was performed in C18 nano-capillary column (Acclaim PepMap C18, 75 µm internal diameter, 3 µm particle size, Dionex-Thermo Scientifics) as follows: 5 min, 4% solution B; 60 min, 4–35% solution B; 10 min, 35–80% B; 10 min, 80% B; 10 min 4% B. The nano-electrospray voltage was set to 1300 V and the capillary voltage to 50 V at 190 °C. The LTQ Orbitrap XL was operated in parallel mode, allowing for the accurate measurement of the precursor survey scan (400–1500 m/z) in the Orbitrap selection, a 30,000 full-width at half-maximum (FWHM) resolution at m/z 400 concurrent with the acquisition of top five CID Data-Dependent MS/MS scans in the LIT for peptide sequence. Singly charged ions were excluded. The normalized collision energies used were 35% for CID. The maximum injection times for MS and MS/MS were set to 500 ms and 50 ms, respectively. The precursor isolation width was 3 amu and the exclusion mass width was set to 5 ppm. Monoisotopic precursor selection was allowed and singly charged species were excluded. The minimum intensity threshold for MS/MS was 500 counts for the linear ion trap and 1000 counts for the Orbitrap. MS2 spectra were searched with SEQUEST engine against a database of Uniprot_Human_Nov2014 (www.uniprot.org). Peptides were generated from a tryptic digestion with up to one missed cleavage, including NEM and (d5)NEM modification in Cys and methionine oxidation as dynamic modifications. Statistical data were calculated with percolator tool against decoy database using 1% FDR as threshold for significance.
2.6. Label-free MS protein quantification
The analysis of MS raw data of the two studies was performed using the MaxQuant (v1.5.7.0) [12] and Perseus (v1.5.6.0) [82] software. Three RAW data files per sample from 3 separate experiments were analyzed. Proteins were identified by searching raw data against the human UniprotKB/Swiss-Prot protein database (February 2018 version). The modifications NEM and d(5)NEM and methionine oxidation was set as variable modifications for the second study. Cleavage specificity was by trypsin, allowing for a maximum of two missed cleavages, a mass tolerance of 10 ppm for precursors and 0.01 Da for fragment ions. The false discovery (FDR) cut-off for protein identification was 1%. Enabling the “match between runs” option allowed for identification transfer between samples. Similar proteins were grouped, and only unique peptides were used for quantification. Identified from reverse database or contaminants hits proteins were removed prior to further analysis. Finally, the resultant list was analyzed according to the instructions of the software developers [82]. The criteria for considering a differentially expressed protein were that it was identified and quantified using at least two unique peptides; has a fold change of at least 1.5 and had a P ≤ 0.05 value.
2.7. Targeted analysis of differentially labeled Cys residues
The method devised has been described before [55]. Briefly, Cys-containing peptides detected with identical amino acid sequences and both d(0) and d(5) NEM modifications independently with a confident individual peptide ion score were considered redox peptides. Redox peptides detected from Proteome Discoverer analyses of RAW files were selected for targeted analysis using m/z data and retention times with the open software Skyline [48]. Targeted analysis applying m/z, retention times, and fragmentation spectra for peptide selection allowed the calculation of the reduced/oxidized ratio (or d(0)/d(5) NEM) of the Cys residues using the individual parent ion intensities. The individual reduced/oxidized ratio for redox Cys peptides in each sample was used to calculate an average ratio of reduced/oxidized calculated for the specific Cys residues.
2.8. Metabolomic analysis
Metabolomic analyses were performed at Metabolon, NC USA and the samples were prepared following the specific guidelines. Global biochemical profiles were determined in 2 × 106 HepG2 cells collected from different treatment groups (wild type, 4TO and 4TO-NOS cells treated with specific siRNA for Trx and Grx or with non-target siRNA). Each experiment was done four times and the dry cell pellets were immediately frozen in liquid nitrogen and stored at −80 °C until shipment. Samples were prepared using the automated MicroLab STAR® system from Hamilton Company. To remove protein, dissociate small molecules bound to protein or trapped in the precipitated protein matrix, and to recover chemically diverse metabolites, proteins were precipitated with methanol under vigorous shaking for 2 min (Glen Mills GenoGrinder 2000) followed by centrifugation. The resulting extract was divided into five fractions: two for analysis by two separate reverse phase (RP)/UPLC-MS/MS methods with positive ion mode electrospray ionization (ESI), one for analysis by RP/UPLC-MS/MS with negative ion mode ESI, one for analysis by HILIC/UPLC-MS/MS with negative ion mode ESI, and one sample was reserved for backup. Raw data was extracted, peak-identified and QC processed using Metabolon's hardware and software. Peaks were quantified using area-under-the-curve and normalized.
2.9. Biotin switch technique
All operations were performed in darkness and following the protocol described by Martínez-Ruiz et al. [51] with some modifications. Treated cells were resuspended in lysis buffer (50 mM Tris-HCl, pH 7.4, 300 mM NaCl, 5 mM EDTA, 0.1 mM neocuproine, 1% Triton X-100, 1 mM PMSF, 1 µg/mL aprotinin and 2 µg/mL leupeptin) and incubated on ice for 15 min. Samples were centrifuged at 10,000g, 4 °C for 15 min and supernatants were collected. Extracts were adjusted to 1.0 mg/mL of protein and were blocked with 4 volumes of blocking buffer (225 mM HEPES, pH 7.2, 0.9 mM EDTA, 90 µM neocuproine, 2.5% SDS and 50 mM NEM) at 37 °C for 30 min with stirring. After blocking, samples were precipitated with cold acetone and pellets were resuspended in HENS buffer (250 mM HEPES, pH 7.2, 1 mM EDTA, 0.1 mM neocuproine, 1% SDS) with 100 mM ascorbate and 1 mM biotin-HPDP and incubated for 1 h at room temperature. Finally, samples were passed through Zeba spin desalting columns (Thermo Scientific Pierce) to eliminate ascorbate and biotinylated proteins were detected by western blot with “anti-biotin”. In addition, the biotinylated proteins were captured with neutravidin-agarose (Pierce) and recovered using 100 mM ß-mercaptoethanol for subsequent Western blot analysis of specific target proteins.
2.10. Rate of glycolysis and Pentose Phosphate Pathway (PPP)
These were measured following the protocol described by Requejo-Aguilar et al. [68] with some modifications. The resuspended cells (1.5 ×105 cells) were incubated in “Elliot” buffer (122 mM NaCl, 11 mM Na2HPO4, 0.4 mM KH2PO2, 1.2 mM SO4Mg, 3.1 mM KCl, 1.3 mM CaCl2, pH7.4) in the presence of 5 μCi of D-[3-3H]glucose,(0.5 μCi mL−1of D-[1–14C]glucose or 1 μCi mL−1 of [6–14C]glucose and 5 mM D-glucose in sealed vials. The glycolytic flux was measured by assaying the rate of 3H2O production from [3-3H]glucose, and the PPP flux as the difference between [1–14C]glucose and [6–14C]glucose incorporated into 14CO2. Glucose concentrations in Elliot buffer were measured by enzymatic analysis spectrophotometrically.
2.11. Measurement of enzymatic activities, lactate and protein
All glycolytic enzymatic activities were measured in fresh cell lysates obtained in the absence of reducing agents. Cell lysates were prepared with lysis buffer containing 50 mM HEPES pH 7.2, 2 mM EDTA, 100 mM NaCl, 1% Triton X-100, 1 mM PMSF. Commercial preparations of pure enzymes were used to calibrate the assays and to determine the linear dependence activity range. Pyruvate kinase (PK) and triose phosphate isomerase (TPI) activities were measured according to the method described by Fielek and Mohrenweiser [16] without DTT in the assay mixture. Glyceraldehyde-3-phosphate dehydrogenase (G3PDH) activity was measured as an increase in absorption at 340 nm resulting from reduction of NAD+ at 25 °C. The reaction mixture contained 130 mM Tris-HCl, pH 8.0, 0.25 mM NAD+, and 5 mM DL-glyceraldehyde-3-phosphate. Three technical replicates were routinely done for each independent experiment. Lactate concentration in the culture medium after 72 h of treatment with the siRNA Grx1 or siRNA Trx1 was determined by an enzymatic colorimetric assay (505 nm) using the kit Labkit (Chemelex, S.A). Protein concentration was determined by the Bradford method (Bio-Rad) using BSA as standard.
2.12. SDS-PAGE and Western blotting
SDS-PAGE was performed with homogeneous 10% non-reducing gels for detection of biotinylated proteins and 12% Criterium XT Precast Gel (Bio-Rad) for detection of specific proteins (Trx1, PRDX6, PKM2, caspase-3, NOS3). After electrophoresis, proteins were transferred to a nitrocellulose membrane with a semi-dry electrophoretic transfer system (Bio-Rad). The membranes were incubated overnight at 4 °C with the corresponding dilutions of primary antibodies: 1:4000 against biotin, 1:1000 against PRDX6, 1:500 against PKM2, caspase-3 and NOS3, 1:500 against Trx1 and Grx1. Then washed and incubated with the corresponding secondary antibodies conjugated to peroxidase (anti-rabbit, anti-goat or anti-mouse) used at 1:8000 dilution and the chemiluminescent signal was induced by ECL reagent (Thermo-Fisher). Ponceau staining was used as cell protein-loading control and actin was used as reference for quantitative densitometric normalization.
2.13. Statistics
Where appropriate, results are expressed as mean ± SEM of at least three independent experiments. Data were compared using ANOVA with the least significant difference test post hoc multiple comparison analysis. The threshold for statistically significant differences was set at p ≤ 0.05. In the case of Global and Redox Proteome, data were analyzed using Student's T-test comparing control vs siRNA treatment. The threshold for this statistically significant differences was set at q ≤ 0.05 according to the method of Storey [77] using the R-package. In the case of the metabolomic analysis following normalization to Bradford protein concentration and log transformation a mixed model ANOVA was used to identify biochemicals that differed significantly between experimental groups; statistical significance was set at p ≤ 0.05.
3. Results and discussion
Overexpression of NOS3 in HepG2 cells caused a ≈ 25% increase in ROS, NO and NOS activity compared with the same cells transformed with the empty vector [21] and constituted a physiological model for endogenously oxidative/nitrosative stressed cells to study the role of Trx1 and Grx1. Treatment with Trx1 or Grx1 specific siRNA down-regulated the levels and activities of either redoxin by ≈ 80% (Ref. Fig. 5 in [22]). These cells were collected and lysed under conditions that allowed blocking of reduced protein thiols with “light” d(0)NEM followed by reduction of reversibly oxidized Cys and labelling with “heavy” d(5)NEM (Fig. 1). Samples were prepared and analyzed by a proteomics LC-MS/MS approach allowing for global “label-free” quantitation and for targeted analysis of differentially labeled Cys-peptides [55], [71].
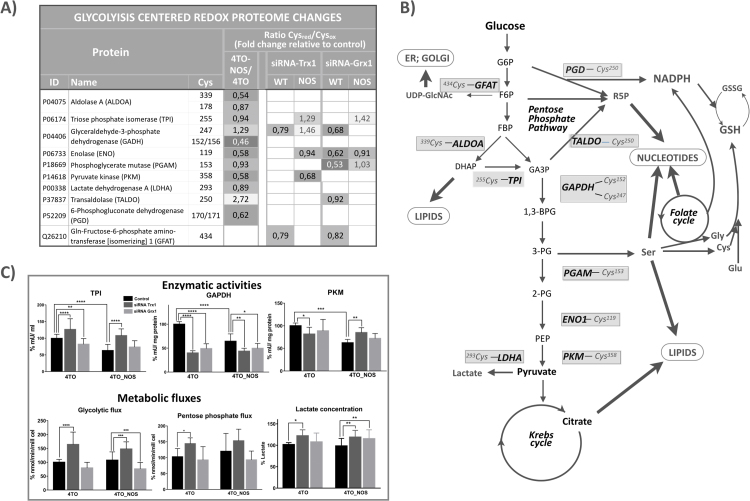
Fig. 5.
Silencing Trx1 or Grx1 affects glycolytic enzymes redox state and activities and glycolytic flux. A) Enzymes of Glycolysis and related pathways undergoing significant (q≤0.1) cysteine redox changes on NOS3 overexpression and upon Trx1 or Grx1 down-regulation have been selected. The precise cysteine residues affected and the changes in the values of their reduced/oxidized cysteine ratios relative to the control (4TO-NOS vs 4TO; siRNA treated vs untreated) are shown. The values for siRNA treated 4TO-NOS cells have been weighted by subtracting the changes observed in siRNA treated 4TO control cells; B) Glycolysis centered metabolic network showing the main glycolytic stream and off-shooting pathways; the final destinations of metabolic fluxes are indicated with thick arrows; the enzymes with redox altered cysteines and their residue position are highlighted. C) Enzymatic activities of triosephosphate isomerase, glyceraldehyde-3-phosphate dehydrogenase and pyruvate kinase, Glycolysis and Pentose Phosphate Pathway fluxes and lactate concentration in siRNA Trx1 and siRNAGrx1 treated 4TO_NOS and 4TO cells are shown relative to untreated 4TO control cells. Between 4 and 9 independent experiments were done for each parameter and a Student t-test was calculated for statistical significance with values p ≤ 0.001, 0.001 >p ≤ 0.01 and 0.01 >p ≤ 0.05 marked with *** , * * and * , respectively.
3.1. Global proteome
Quantitative differences between the proteomes of WT, 4TO and 4TO-NOS, treated or not with siRNA-Trx1 and siRNA-Grx1, were determined by “label-free” analysis. The significant differential proteomes were determined from roughly 600 unique proteins identified in each study. Only 14 proteins varied significantly between 4TO-NOS and 4TO HepG2 cells and a smaller number changed upon either siRNA treatment in 4TO or 4TO-NOS cells, as shown in Suppl. File 1.
Overexpression of NOS3 and down-regulation of Trx1 or Grx1 in HepG2 cells has marked phenotypical consequences like slow-down of proliferation and apoptosis exacerbation as previously reported [22]. To investigate whether these phenotypical changes might have been triggered by redox postranslational modification of key proteins, we analyzed the redox proteome.
3.2. Redox proteome
Trx1 and Grx1 main action is to reverse oxidative changes in protein thiols, not only those involved in regulatory postranslational modifications of metabolic and signaling proteins, but also those associated with thiol-dependent ROS scavenging enzymes, like peroxiredoxins. Any change in the levels of either redoxin should have consequences on the redox state of sensitive proteins which could reveal detailed knowledge on their functions. Silencing with specific siRNA provides a gentler experimental approach to mimic physiological down-regulation events compared to a full knockout, although detection of the expected subtle changes in molecular indicators requires precise analytical methods to study the redox proteome.
3.2.1. NOS3 overexpression alters the Redox Proteome
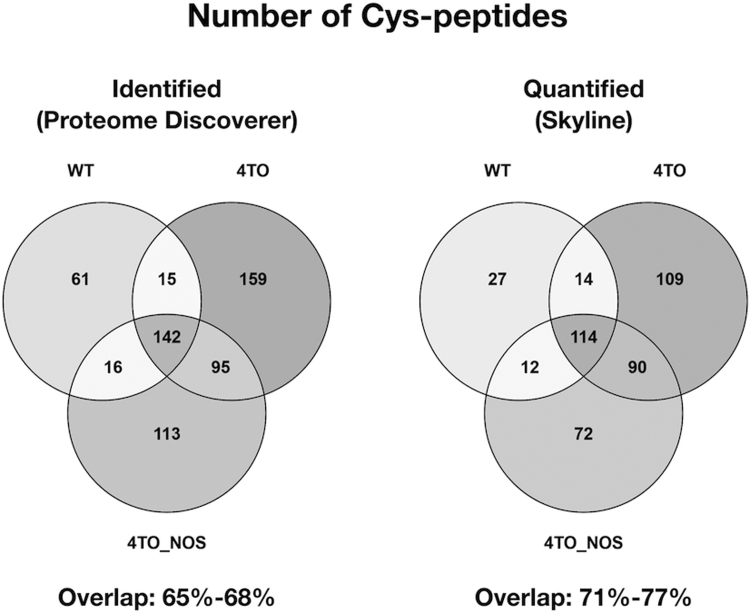
The levels of ROS and NO had been measured in 4TO-NOS cells by standard general methods (DCFH2-DA fluorescence and colorimetric nitrite determination, respectively) showing an increase of 40% in the cells overexpressing NOS3 [21]. Now, the Cysred/Cysox ratio was determined for every Cys peptide identified/quantified in WT (234/168 peptides), 4TO (411/328 peptides) and 4TO-NOS (366/289 peptides) HepG2 cells. Overlapping of Cys-peptides between samples was 65–68% for identified peptides and 71–77% for quantified peptides (Suppl. Fig. 2). The majority of quantified peptides were in the Cys-SH state with Cysred/Cysox ratios ranging from nearly 40 to around 2. Ninety-four Cys-peptides underwent significant changes in their Cysred/Cysox ratios when the cells overexpressed NOS3 compared to cells transfected with empty vector (Fig. 2; Suppl. File 2). Changes were predominantly oxidative, but 13 proteins were ≥ 1.5 fold reduced in the oxidative/nitrosative environment of NOS3 overexpressing cells. Their reductive change could be direct or indirect consequence of the observed marked induction of Trx1 and Grx1 [22], but other perturbations in metabolic and signaling pathways induced by NOS3 overexpression could also be responsible.
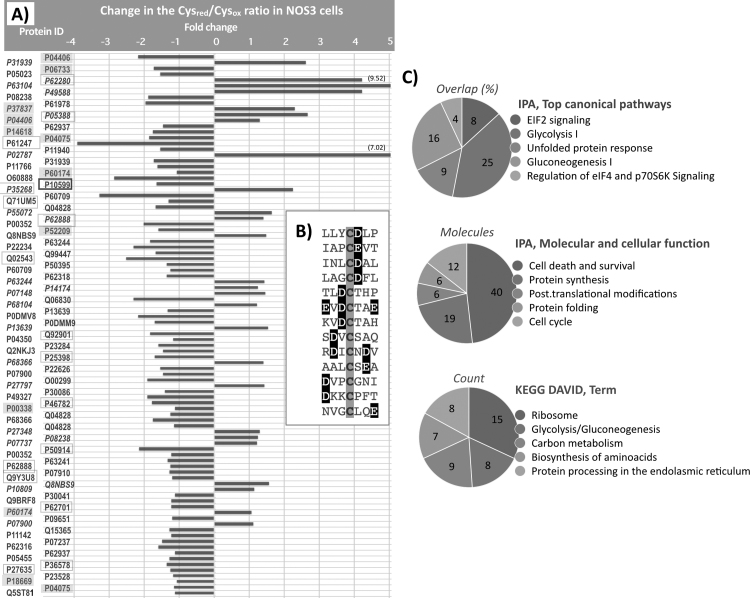
Fig. 2.
NOS3 over expression alters the Reduced/Oxidized ratio of specific Cys on target proteins. A) The UniProt ID of the protein is shown in the vertical axis and the relative change in the Cysred/Cysox ratio in 4TO_NOS cells is indicated in the horizontal axis as fold change relative to the ratio in control 4TO cells transformed with the empty vector. Proteins are ordered according to their statistical q score (Storey & Tibshirani) from 0.0000516 (top) to 0.0497383 (bottom). Proteins with negative values were more oxidized after NOS3 overexpression; those with positive values (IDs in italic) were more reduced. Two proteins showed reductive changes higher than fivefold as indicated. P10599, square boxed with thick line, is Trx1(C73); ID of enzymes of Glycolysis and its branching pathways are highlighted with grey squares and ribosomal proteins are square boxed with thin line. B) sequence context around the cysteine residue of 13 peptides whose reduced/oxidized ratio increased ≥ 1.5 on NOS3 overexpression, showing the presence of acidic residues (white letters highlighted in black) close to the affected cysteine (grey shadowed). Protein names and Cys mapping are detailed in sheet “NOS3 Vo” of the Excel file shown in Suppl Fig. 2. C) Systems Biology analysis of the whole set of redox modified proteins using Ingenuity Pathway Analysis (Qiagen) and DAVID [Huang et al. Nature Protoc. 2009;4(1):44–57]. The five most significant terms from each analysis are shown with p values ranging from 9.18E−19 to 9.75E−04.
The presence of acidic residues around the modified cysteine in these redox target proteins is particularly apparent (Fig. 2B). 77% (10/13) of the proteins showing an increase in their reduced/oxidized Cys ratio had at least one Glu or Asp between positions + 2 and −2, centered on the target cysteine, whereas this percentage was lower (46%; 16/35) for the proteins undergoing an oxidative change. The accumulated content of Asp and Glu in the average protein from the UniProtKB TrEMBL is 11.65%, therefore, the frequency of at least one acidic residue at these positions is markedly higher than expected at random and could be related to the sensitivity of these Cys residues to nitrosative stress induced redox changes.
Taken together, these changes show that NOS3 overexpression exerts oxidative pressure on a number of protein cysteine residues with potential regulatory effects mainly at the level of Glycolysis/gluconeogenesis, protein synthesis and folding, endoplasmic reticulum processes, and cell death and survival (Fig. 2C). To check whether Trx1 or Grx1 are involved in this redox disturbance, both redoxins were down-regulated with specific siRNA and the redox state of individual Cys residues was measured again.
3.2.2. Specific protein cysteines underwent oxidative and reductive changes upon Trx1 or Grx1 silencing depending on NOS3 overexpression
Silencing of Trx1 or Grx1 to ≈ 20% their normal levels in WT and NOS3-overexpressing HepG2 cells did result in significant redox changes of specific protein cysteines, compared with their respective controls. We had previously reported that the activities of both redoxins decreased to the same extent as Trx1 and Grx1 protein levels in their respective siRNA treated cells, excluding the induction of other active isoforms [22]. Moreover, analysis of the differential proteome of these cells in the present study did not show compensatory induction of isoforms.
A selection of target proteins with ≥ 1.5 or ≤ 0.67 fold redox changes is presented in Fig. 3A. Three groups can be distinguished in this table. i) Proteins in the upper part of the table showed sensitive cysteines which underwent an oxidative shift in WT cells, but a reductive shift in NOS3 overexpressing cells, upon silencing of either redoxin. ii) A second group in the middle part of the table shows cysteines that were more oxidized only in 4TO-NOS cells upon either redoxin silencing. iii) Finally, a third group was only sensitive towards Trx1 silencing almost exclusively in NOS3-overexpressing cells. Six proteins: Cofilin-1, 40 S ribosomal protein S5 and isoforms Gamma, Theta/delta and Beta/alpha of the 14–3-3 family of proteins, showed high sensitivity to both redoxins but their redox changes, though conspicuous, did not display clear trends and have been omitted.
Fig. 3.
Changes in the reduced/oxidized ratio of specific Cys on target proteins upon Trx1 and Grx1 silencing. A) The UniProt ID and name of the protein, the sequence of the Cys-peptide and the position of the Cys residue affected are shown together with the redox change observed expressed as fold change of the Cysred/Cysox ratio relative to the non-silenced cells. Oxidative changes are colored red and reductive changes are green. Only proteins showing fold changes ≥ 1.5 or ≤ 0.66 are included. All the values are statistically significant according to their q value (Storey & Tibshirani). Cells colored yellow with a value of 1.00 represent non-significant changes; ND, peptide not detected. B) Putative consensus sequences of Trx1 and Grx1 related sensitive Cys-peptides; the sequences of the peptides showing significant redox changes upon Trx1 or Grx1 silencing in WT and NOS3-overexpressing HepG2 cells were subjected to Motif-X ([10]; http://motif-x.med.harvard.edu) with the following parameters: central character “C”, width “13”, occurrences “20”, significance “0.0001”, background “ipi. HUMAN.fasta”; motif scores for logos 1, 2 and 3 were 8.57, 6.30 and 4.86, respectively. C) Two Cys peptides specifically sensitive towards Grx1 but not Trx1 belonging to Phosphoglyceromutase-1(Cys153) and Peroxiredoxin-6(Cys91) were highly conserved as shown by pairwise alignment with a 17 residue window around the sensitive Cys, matrix EBLOSUM62, gap penalty 2; a very high conservation score was found with 2 identities (11.8%) and 6 similarities (35.3%).
These changes could be the consequence of direct interaction of either redoxin with the target proteins or could be due to indirect effects. They were almost exclusively oxidative in WT cells as one would expect from the canonical antioxidant functions of Grx1 and Trx1. However, in NOS3 overexpressing cells, the effect of silencing the redoxins, especially Trx1, also resulted in reductive changes in several protein cysteines. The reason for this apparently contradictory result could rely on the fact that NOS3 itself is sensitive to redox modification and can be activated by Trx1 and Grx1 through denitrosation and deglutathionylation [4], [78]. Hence, decreasing the levels of these redoxins by specific siRNA should result in higher levels of redox modified NOS3, lower NOS3 activity and lower protein S-nitrosation levels followed by higher reduced/oxidized ratio of sensitive cysteines in target proteins. We had previously determined the level of Tyr nitration, a good indication of the degree of nitrosative/oxidative stress, in siRNA-Trx1 and siRNA-Grx1 treated WT and NOS3 overexpressing HepG2 cells [22]. Nitro-tyrosine levels were markedly higher in WT HepG2 cells but decreased to 50% in NOS3 overexpressing HepG2 cells which is a further evidence of NOS3 inactivation upon Trx1 and Grx1 down-regulation.
3.2.3. Redoxins-sensitive cysteines show specific sequence motifs
Defining sequence signatures around redox sensitive cysteines would help predict and find redox regulated target proteins. A search for short linear motifs in the sequences around the redox modified Cys's shown in Fig. 3A using the motif-x algorithm [10] produced three motifs with high enrichment score among the input set of proteins compared with the background occurrence in the whole human proteome (Fig. 3B). There is a Lys at position + 1 or + 4 in motifs 1 and 2 respectively and a threonine at + 4 in motif 3. It is well known that nucleophilic reactivity of Cys's is dependent on ionization to thiolate favored by low pKa. Computational and experimental results indicate that the surrounding charged side chains can contribute, but do not primarily control, the thiolate pKa in the Trx superfamily and other proteins with reactive Cys [69], [70]. However, a search for denitrosation sites in proteins found two potential motifs with lysine at + 6 and + 7 [87] and a structural analysis of cysteine S-nitrosation sites found lysine at position + 3 and threonine at + 5 with moderate frequency [49]. The presence of a lysine residue near the target Cys is also common to human proteins S-nitrosation motifs [42]. A study that combined sequence, structure, and electrostatic approaches predicted that Thr, its hydroxyl group, and hydrogen-bonding capabilities play an important role in Cys deprotonation and reactivity [70]. Hence, our results with Lys or Thr upstream of Cys fit in with the current structural landscape of redox sensitive Cys's, where some signatures have been put forward but evidences for a strong consensus sequence motif has not yet been achieved.
Two proteins were specifically sensitive to Grx1 silencing, PGAM-1(Cys153) and PRDX6(Cys91). Alignment of both Cys-peptides showed a striking conservation score around the affected Cys stressing their assignment as Grx1 targets (Fig. 3C). These Cys's are positioned in a coil in the protein structure with 3.78% and 4.00% solvent accessibility, respectively, and are enriched in acidic residues around the central cysteine. They fit in the Homo sapiens HC05 group glutathionylation motif as catalogued at dbGSH (http://csb.cse.yzu.edu.tw/dbGSH). PRDX6 is 1-Cys peroxiredoxin with prominent features as it depends on GSH and GST-Pi for peroxidase activity and also has phospholipase A2 (PLA2) activity, which plays a role for activation of NADPH oxidase 1 and 2 (NOX1, NOX2) [17], [41]. The Grx-dependent redox modified Cys at position 91 is not the canonical catalytic (“peroxidatic”) cysteine that lies at position 47 of PRDX6. Cys91 has not been given attention so far and has been routinely substituted by Ser in the recombinant protein, supposedly to avoid unwanted thiol oxidation and “shuffling” and to facilitate handling. Further experiments have been undertaken to find out whether the Grx-specific redox sensitivity of PRDX6 Cys91 plays a role on any of the protein functions.
3.2.4. Part of the observed cysteine redox changes were due to direct S-nitrosation
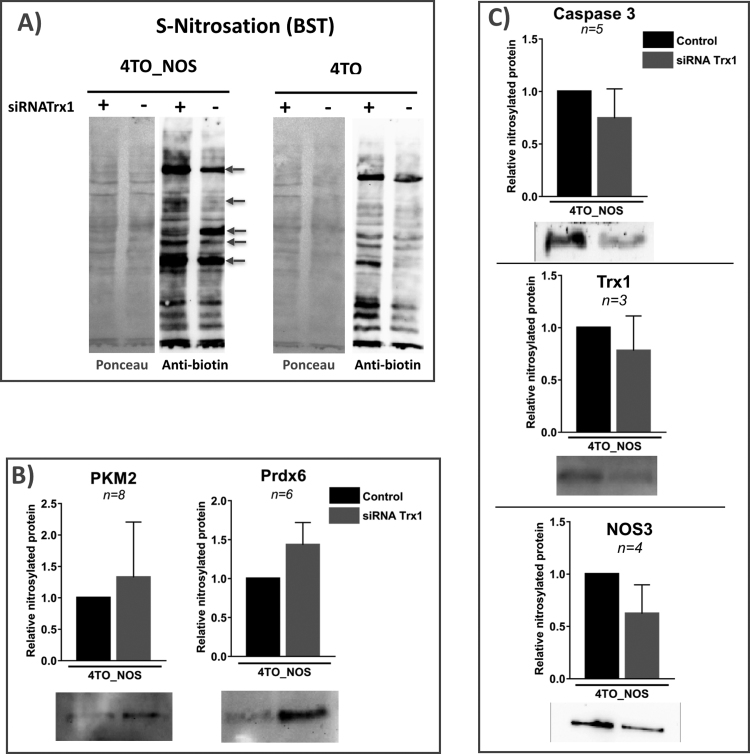
As determined previously [21], NOS3 overexpression not only increases the levels of NO but those of other ROS as well, so reversible oxidation of Cys residues could be due to S-nitrosation or other types of oxidative modifications induced by ROS. To ascertain whether S-nitrosation was actually taking place, we have carried out the Biotin Switch Technique (BST). We have limited this analysis to siRNA-Trx1 treated cells, since Trx1 is well-known to display denitrosase activity.
Analysis of total biotinylated proteins by Western blotting showed clear differences between control and NOS3 overexpressing HepG2 cells as expected. When we compared Trx1 silenced cells with their control, no significant differences were detected at the level of total protein, but interestingly clear differences were found in band patterns indicating changes in S-nitrosated Cys's at the level of individual proteins: some proteins increased while others decreased upon Trx1 silencing (Fig. 4A). These results are in agreement with the finding that several proteins underwent a significant reductive shift on their key Cys's (Fig. 3).
Fig. 4.
Protein S-nitrosation in HepG2 cells over expressing NOS3 and treated with specific siRNA-Trx1. A) NOS3 over expressing HepG2 cells, 4TO-NOS, and control 4TO cells transformed with empty vector were silenced or not, for Trx1 and subject to BST; biotinylated proteins constituting the “S-nitrosome” were separated on SDS-PAGE, transferred to Nitrocellulose membrane and revealed with avidin-peroxidase conjugate. Membranes stained with Ponceau reagent before transfer are presented in the left panels to show the protein load. Arrows point to bands with increased or diminished intensity upon Trx1 silencing. B) and C) biotinylated proteins from 4TO-NOS HepG2 cells were captured on avidin-agarose, eluted with ß-ME, subjected to Western blot and revealed with specific antibodies to 5 selected proteins as indicated. The intensity of the bands was quantified by image analysis as described in M&M; relative abundances of each protein in the fraction of biotinylated proteins compared to non-silenced control cells are shown, together with the number of replicas and the bars showing standard deviation; trimmed images from membranes showing representative band patterns are included below each graph.
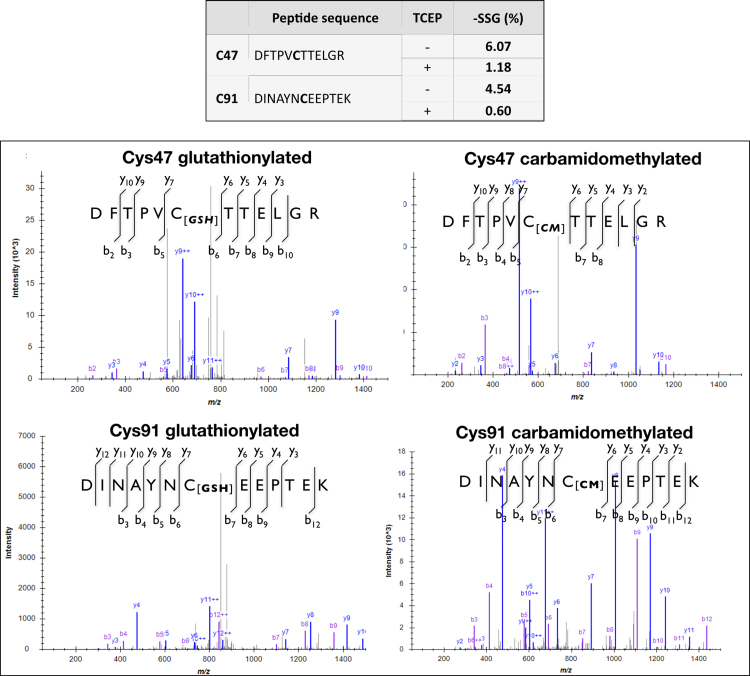
To test individual proteins, PRDX6, PKM2, caspase-3, NOS3 and Trx1 were chosen to analyze their nitrosation state on Trx1 silencing by the BST as shown in Fig. 4B and C. All five proteins were captured with the BST, indicating that they are targets of S-nitrosation. Despite the high variability of the technique, a clear trend was observed in 4TO-NOS cells showing higher amounts of S-nitrosated PRDX6 and PKM in Trx1-silenced cells than in control cells. This trend was also observed in 4TO and in normal HepG2 cells (not shown). PRDX6 and PKM are likely substrates of Trx1 denitrosase activity, although the involved cysteines cannot be mapped with this technique and we cannot tell whether the S-nitrosated Cys are Cys91 or Cys47 in PRDX6 and Cys358 or another Cys in PKM. The peptide containing peroxidatic Cys47 of PRDX6 was not detected in any of our samples. We have previous experience on the difficulty of detecting this tryptic peptide [65], but it was detected using different technical tools in another study where Cys47 was actually more oxidized in sciatic nerve of old compared to young mice [56]. Since we have found that the “extra” Cys91 of PRDX6 is not sensitive to Trx1 but to Grx1 silencing (Fig. 3), which has no denitrosase activity, it is tempting to conclude that Cys47 might be target of S-nitrosation in a Trx1-dependent manner while Cys91 might be a Grx1-dependent target of glutathionylation. A preliminary mass spectrometric analysis of recombinant PRDX6 showed that both Cys47 and Cys91 are liable to spontaneous reversible glutahionylation (Suppl. Fig. 1). These data open the door to interesting insights into PRDX6 molecular properties and functions.
Redox sensitivity of PKM2 Cys358 has been reported before [2], [53], however the identity of its redox reversible modification was not determined. Our data show higher S-nitrosation levels of PKM2 in Trx1 silenced cells in parallel with reversible oxidative modification of Cys358 suggesting PKM2 as target of Trx1 denitrosase activity. On the other hand, caspase-3, Trx1 and NOS3 itself, were S-nitrosated in NOS3 overexpressing cells, apparently to a lesser extent when Trx1 was silenced (Fig. 3C).
These results confirm that S-nitrosation contributes to redox changes taking place on target Cys's in HepG2 cells depending on the overexpression of NOS3 and affected by the prevailing levels of Trx1.
3.2.5. Proteins with redoxin-sensitive cysteines are abundant in Glycolysis
Clustering of redox sensitive proteins with DAVID [33], [34] revealed a significant enrichment in Glycolysis in either normal or NOS3 overexpressing HepG2 cells for both redoxins, but with higher score for Grx1 silencing. Enzymes belonging to Glycolysis or its branching pathways affected by thiol redox changes upon Tx1 or Grx1 silencing or upon overexpression of NOS3 are listed in Fig. 5A and are shown in context schematically in Fig. 5B.
3.2.5.1. Glyceraldehyde-3-phosphate dehydrogenase
The peptide containing Cys152/Cys156 of GAPDH was markedly more oxidized in NOS3 overexpressing cells. Cys152 is part of the active site and its oxidation renders the enzyme inactive [25], [61]. We confirmed that GAPDH activity decreased in NOS3 overexpressing HepG2 cells (Fig. 5C). Silencing of Trx1 or Grx1 in normal HepG2 cells did result in diminished levels of GAPDH activity as expected, but this loss correlated with oxidation of another Cys residue, Cys247, and not with the redox state of Cys152/156, that did not change (Fig. 5A). GAPDH Cys247 was indeed more oxidized in Trx1 and Grx1 down-regulated WT cells, but more reduced in NOS3 overexpressing cells when Trx1 was silenced. Reversible S-thiolation of GAPDH was earlier reported [73] and since then Cys247 or its equivalent has been mapped and found to be S-glutathionylated, S-nitrosated and S-acetylated [24] even in plants [3], as well as a predicted target of Trx trans-nitrosation [86].
It would seem that GAPDH-C247 is prone to oxidative modification by nitrosation or glutathionylation under the prevailing redox cellular environment in normal HepG2 cells and to its reversal by either Trx or Grx, respectively. When this fragile equilibrium is displaced by down regulation of either redoxin, the oxidative state of Cys247 increases. However, as discussed in Section 2b, decreasing the levels of Trx1 by specific siRNA in NOS3 overexpressing cells should result in higher levels of redox modified NOS3, lower NOS3 activity and lower GAPDH-C247 nitrosation levels. The levels of nitro-Tyr, an indication of ROS and RNS, did actually decrease down to 50% on either redoxin silencing [22]. This would explain the reductive shift in GAPDH-C247.
Besides its conventional metabolic role, a number of studies have identified the participation of GAPDH in diverse cellular functions and these oxidative changes not only affect its glycolytic function but also stimulate the participation of GAPDH in cell death [11], [25]. Our findings point to Cys247 as a sensitive target of the antioxidant activity of Trx1 and Grx1 in WT cells with consequences on its enzymatic activity.
The rational to explain the decrease in activity of GAPDH would be that it is rewarding for the cells to slow down the glycolytic flux at this point to facilitate upstream diversion of glucose catabolism toward the Pentose Phosphate Pathway (PPP) and to increase NADPH production in this prevalent oxidative environment. However, it has yet to be demonstrated whether inactivation of GAPDH is reflected directly in slowing down of glycolytic flux. Actually, we have observed that inactivation of GAPDH in siRNA-Trx1 treated cells is paralleled by an increase in glycolytic flux (Fig. 5C). Alterations in glycolytic enzymes can regulate autophagy e.g. the moonlighting behaviour of GAPDH enables its direct interaction with mTOR [58] or translocate to the nucleus and upregulate Atg12 [11], both mechanisms being related to the activation of autophagic process. siTrx1 and siGrx1 increased caspase-3 and TUNEL in 4TO and 4TO-NOS3 cells [22] and increased the oxidative status of the heat shock 70 kDa protein 4 (HspA4) Cys270 (Fig. 3A), an interactor with HspA8 (heat shock cognate 71 protein), which drives chaperone-mediated autophagy [13]. Redox changes at target cysteines in GAPDH and HspA4 suggest that Trx1 and Grx1 signaling promoted a shift from autophagic survival to apoptotic pathway.
3.2.5.2. Phosphoglyceromutase-1
(PGAM1) Cys153 was more oxidized in WT cells when Grx1 was down-regulated. PGAM1 is commonly upregulated in human cancers and regulates anabolic biosynthesis by controlling intracellular levels of 3-phosphoglycerate (3-PG) and 2-phosphoglycerate (2-PG) to promote tumor growth [28]. Both 3-PG and 2-PG are allosteric regulators of glycolytic branching pathways: 3-PG inhibits 6-phosphogluconate dehydrogenase (6PGD) of PPP, whereas 2-PG activates phosphoglycerate dehydrogenase (PHGDH) of glycine and serine synthesis pathway. It is worth noting that activation of PGAM1 by phosphorylation at Tyr26, common in human cancer cells, promotes cell proliferation and tumor growth [29]. Cys153 is not close to the active site but it has been noted that proteomes of Cys PTMs have localized these modifications primarily in non-catalytic regions [24]. Moreover, Cys153 is in the same region of the protein as Tyr26 and Lys100. It has been demonstrated that increased levels of ROS stimulate PGAM Lys100 deacetylation and activity by promoting its interaction with SIRT2 [88]; its modification by oxidation could equally affect its activity or even the phosphorylation and acetylation state of Tyr26 and Lys100 located nearby.
Cellular response to oxidative stress are mediated by the HIF-1α, which is required for the upregulation of mRNAs encoding glucose transporters and glycolytic enzymes, with the notable exception of PGAM [37], indicating a possible postranscriptional regulatory mechanism. Sensitivity of PGAM1 activity to thiol specific reagents was initially reported for the rabbit enzyme [66], but to our knowledge, this is the first time that a Grx1-dependent redox change is observed and mapped to Cys153 suggesting a novel form of regulation that could contribute to modulation of glycolytic flux for biosynthetic and antioxidant purposes in response to the redox environment.
3.2.5.3. Pyruvate Kinase
PKM2 (P14618-1) behaves similarly: its Cys358 is close to the allosteric activator fructose-1,6-bisphosphate (FBP) binding site and is sensitive to oxidative stress [2], [53]. It has been shown that ROS-dependent inhibition of PKM2 is needed to maintain the availability of glucose-6-phosphate (G-6-P) for flux into the PPP and to sustain cell survival when endogenous ROS accumulate. Regulation of PKM2 via oxidation of Cys358 is critical under these conditions for optimal tumor growth [2]. Coherent with this, we show here that NOS3 overexpression oxidizes Cys358 accompanied by a significant decrease in activity (Fig. 5C). Cys358 is further oxidized when Trx1 is down-regulated, but surprisingly, PKM activity is significantly recovered to the same level as in siRNA-Trx1 treated control cells (Fig. 5C). An increase in the PKM1/PKM2 isoenzyme ratio induced by redox regulation of splicing events as reported in hepatoma cells [84] would explain this result. These data demonstrate the complexity of Trx1 involvement in maintaining the redox state of this Cys358 and the activity of this critical enzyme.
3.2.5.4. Enolase
Cys119 of Alpha-enolase is sensitive to oxidative conditions in NOS3 cells and is a target of Grx1 antioxidant activity in WT cells (Fig. 5A). This cysteine was found to be reactive toward mercury resin specific for Cys-SNO [24] and its glutathionylation in SH-SY5Y neuroblastoma cells resulted in loss of enzymatic activity [36]. Further studies will confirm the regulatory role of this Grx1 dependent redox change of α-Enolase Cys119.
3.2.5.5. Triose phosphate isomerase
TPI Cys255 was more reduced when either Trx1 or Grx1 were silenced in NOS3 cells (Fig. 5A). TPI from several organisms has been shown to be regulated by redox changes involving glutathione at a cysteine equivalent to human Cys255. The equivalent Cys in P. falciparum TPI, Cys217, is located at the interphase of a complex formed between TPI and Trx [74], whereas A. thaliana TPI is inactivated by glutathionylation at Cys218 in a manner reverted by Grx [14]. Our results agree with these showing a significant decrease in TPI activity when Grx1 is silenced in control cells. However, silencing of either Trx1 or Grx1 in NOS3 overexpressing cells increased the activity of TPI in parallel with reduction of Cys255, a likely consequence of NOS3 inactivation, as already discussed above. It is tempting to speculate that TPI Cys255 could act as a thiol redox switch to help divert the glycolytic flow from DHAP towards lipid synthesis as part of a metabolic response to changes in the redox environment [45].
Several enzymes from Glycolysis off-shooting pathways showed significant redox changes: Lactate dehydrogenase A (LDHA) Cys293 and 6-phosphogluconate dehydrogenase (PGD) Cys170/171 were slightly more oxidized in NOS3 cells; Glutamine-Fructose-6-phosphate amino-transferase [isomerizing] 1 (GFAT) Cys434 was sensitive to both Trx1 and Grx1 down-regulation; and Transaldolase (TALDO) Cys250 was markedly more reduced in NOS3 cells
Altogether, these data show widespread redox sensitivity of key cysteines in glycolytic enzymes eventually affecting their activities. The prominent role played by Trx1 and Grx1 on these thiol redox switches could be meaningful as part of a pleiotropic redoxin reductive action as a kind of “redox regulon” [20] to coordinate an integrated response to ROS that would balance pyruvate and NADPH production and biosynthetic flux diversion (Fig. 5B). However, the precise consequences of these redoxin-dependent redox changes on metabolic fluxes cannot be predicted from the changes of individual enzymatic activities [81]. Knowledge of Flux Control Coefficients of each enzyme would be necessary and the fluxes through glycolysis branched pathways should also be taken into account. As a first approach we have measured glycolytic metabolic flux and metabolites levels, as described below.
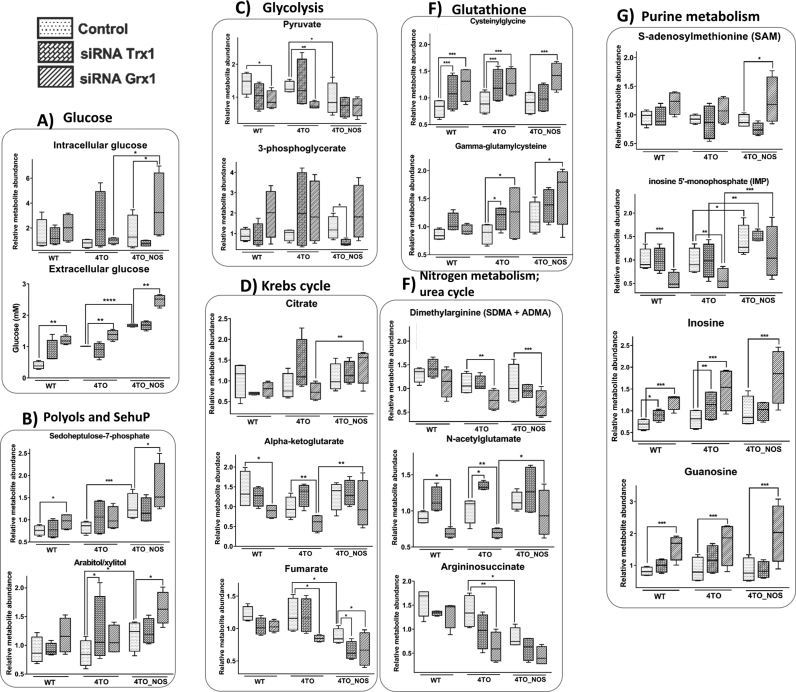
3.3. Glycolytic flux and metabolite profiles were differentially affected by Trx1 and Grx1 silencing
Glycolytic and Pentose Phosphate Pathway fluxes were determined using radiolabelled glucose and measuring the formation of [3H]H2O from [3-3H]glucose during the reaction catalysed by enolase and that of [14C]CO2 from [1–14C]glucose by 6-phophogluconate dehydrogenase, respectively. A metabolomic analysis was also undertaken. As shown in Fig. 5C, Trx1 silencing had a stimulating effect on both pathways in either NOS3 overexpressing cells and control cells, whereas Grx1 silencing had the opposite effect in all cases. These effects could be a reflection of the differential action of both redoxins on the redox state of glycolytic enzymes (see Fig. 5A).
3.3.1. Grx1 silencing slows down Glycolysis and stimulates lipogenesis
The relationships between cysteine redox state, enzymatic activities and metabolic fluxes are not straightforward, the metabolic profiles in siRNA-Grx1 treated cells were coherent with lower glycolytic flux showing increased levels of extracellular and intracellular glucose (Fig. 6A). Sedoheptulose-7-phosphate (Su7P) accumulated in WT and NOS3 overexpressing HepG2 cells treated with siRNA-Grx1 (Fig. 6B). This was accompanied by increased levels of several polyols including arabitol, xylitol, ribitol, mannitol and sorbitol, indicative of transaldolase (TALDO) malfunctioning [76], and decreased levels of ribose. These metabolic changes indicate slowing down of glucose processing through the non-oxidative part of PPP as a consequence of Grx1 silencing. Redox changes detected in TALDO-Cys250, 6-phosphogluconate dehydrogenase (PGD) Cys170/171 and TPI-Cys255 could be involved in the underlying regulatory mechanisms.
Fig. 6.
Metabolites concentrations in normal and NOS3 overexpressing HepG2 under Trx1 and Grx1 down-regulation. A metabolomics analysis was performed in normal (WT), 4TO and 4TO_NOS cells treated with siRNA-Trx1, siRNA-Grx1 or control siRNA-nonTarget as described in Materials & Methods. A selection of metabolites representing the most conspicuous changing trends when cells are treated with siRNA-Trx1 or siRNA-Grx1 is shown. The results presented in this figure are part of a larger metabolic study. Extracellular glucose concentration was determined in culture media by a standard method independently of the metabolomic analysis.
Another effect of Grx1 silencing was an increase in 3-phosphoglycerate (3PG) and lower levels of pyruvate (Fig. 6C) coincident with PGAM-Cys153 oxidation and likely deviation of 3PG toward serine pathway. Moreover, unchanged or increased levels of citrate and lower levels of α-ketoglutarate and fumarate (Fig. 6D), might indicate slowing down of Krebs cycle but deviation of citrate to fatty acid biosynthesis. Accordingly, elevated levels of acyl-carnitine (Fig. 6H) and de novo synthesis of phospholipids, was evidenced in siRNA-Grx1 treated cells, represented by high levels of phosphatidylcholine and the family of 2-arachidonoyl phospholipids with the exception of phosphatidylserine (Figs. 6L and 6N). The increase in 2-arachidonoyl phospholipids runs parallel with oxidation of PRDX6 Cys91 and it is worth noting the specificity of cytosolic Ca2+-independent PLA2 activity for sn-2-arachidonic acid glycerophospholipids [63], a precursor of eicosanoids, which represent a class of lipid mediators. Moreover, arachidonic acid has been shown to mediate the known proliferative action of PRDX6 [72]. These changes speak of active membrane remodeling and/or lipid signaling events in Grx1 down-regulated cells.
Levels of several intermediary metabolites of purine pathway were markedly elevated in Grx1 silenced cells with the exception of IMP but including S-adenosylmethionine (SAM) (Fig. 6G). Elevated levels of SAM should indicate intensification of the methionine/folate pathway connected to glutathione synthesis and supplier of NADPH as an alternative to PPP (Fig. 5B). The synthesis of glutathione was actually exacerbated as indicated by increased levels of gamma-glutamylcysteine and cysteinylglycine (Fig. 6F).
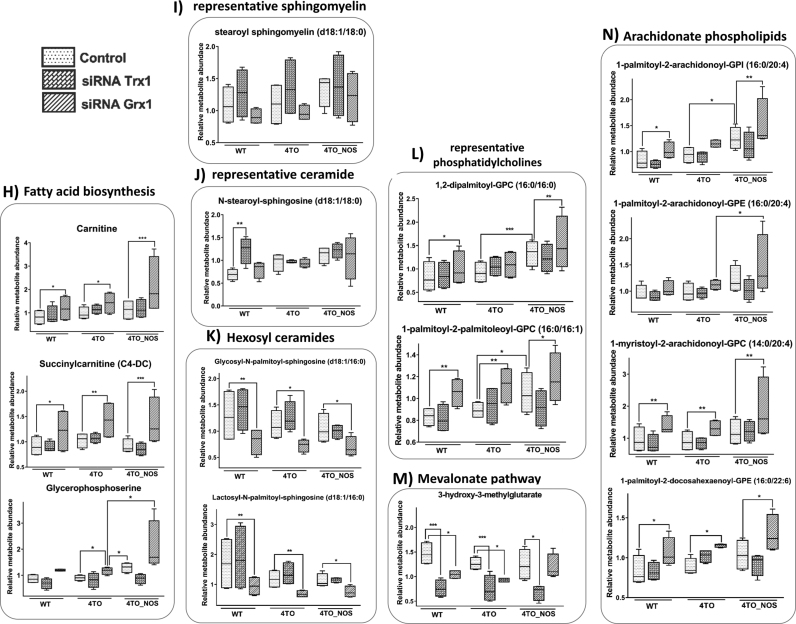
3.3.2. Trx1 silencing induces membrane remodeling
There were similarities and differences between the effects of Grx1 and Trx1 silencing in metabolites’ levels. Increased glutathione metabolism (Fig. 6F) and a marked decrease in 3-hydroxy-3-methylglutarate (Fig. 6M) of mevalonate pathway are among the coincidences, but the levels of gamma-glutamylaminoacids is higher in siRNA-Grx1 (see Suppl. File 3) and siRNA-Trx1 did not affect Su7P and glucose levels (Fig. 6B). Trx silencing elicited trending and significant increases in dipeptides (see Suppl. File 3) that may reflect increased proteasome-mediated elimination of oxidized proteins in the setting of oxidative stress. Increases in protein degradation could suggest Keap1-Nrf2 signaling and increased autophagy. Notably, many of these changes were not observed in NOS3 overexpressing 4TO_NOS cells.
Trx1 silencing also showed signs of lipid remodeling but differed from Grx1 in promoting a trend of increasing sphingomyelins and ceramides (Figs. 6I and 6J). Ceramide is involved in apoptotic processes and in the formation of membrane raft redox signaling platforms (MRRSP) that participate in the assembly and activation of NOX complexes [62]. These changes are consistent with the pro-apoptotic effect of Trx and Grx silencing in these cells as we had already reported [22]. The sensitivity of sphingomyelinases to redox changes is a key to the cross-talk between sphingolipids and redox signaling through the regulation of NADPH oxidase, mitochondrial integrity, NOS, and antioxidant enzymes [7]. The trend of sphingomyelins’ pool increase on Trx1 silencing could have an influence on mitochondrial lipid composition and integrity. However, cardiolipin, a typical and prominently functional mitochondrial phospholipid, was not detected. Hence, involvement of mitochondria cannot be excluded and is worth of further attention.
4. Conclusions
In the cell there is a delicate balance between effective redox signaling mechanisms and potentially damaging oxidative damage. Oxidative/nitrosative induced PTMs are critical for the accumulation of redox modified proteins, induction of autophagy, and further cellular apoptosis in unsolved pathophysiological intracellular responses while redox signaling through the transient oxidation/reduction of key Cys residues in regulatory proteins plays a role in many of the hallmarks of cancer [31].
The data presented herein show that Trx and/or Grx are involved in redox modifications of targeted cysteines of several glycolytic enzymes affecting their activity, although changes in the activity of one single enzyme cannot be extrapolated to equivalent changes in overall metabolic flux. These changes are part of a widespread adaptive mechanism aimed at redistributing metabolic fluxes between Glycolysis and its off-shooting pathways to respond to subtle changes in the cellular redox environment. Trx and Grx share a number of protein Cys redox targets but down regulation of either redoxin has markedly different metabolic outcomes: silencing of Trx1 stimulates glycolytic flux while silencing of Grx1 decelerates it.
Besides its canonical antioxidant action, Trx1 also contributes to oxidative modifications of protein thiols, likely by activation of NOS3, reflecting the delicate sensitivity of redox equilibrium to changes in any of the elements involved and the difficulty of forecasting metabolic responses to redox environmental changes.
A correlation can be put forward between the reversible oxidation of Cys91 of PRDX6 upon Grx1 silencing that is accompanied by a significative increase of sn-2-arachidonoyl containing phospholipids and Cys91 sensitivity to glutathionylation, considering the well-known deglutathionylase activity of Grx.
To our knowledge, this is the first demonstration of a role for Grx1 and Trx1 in the crosstalk between lipids metabolism and redox signaling. Further and deeper insights into the underlying mechanisms, i.e. possible involvement of PRDX6 and Nrf2 signaling [40], CD95 ligand or TNFα induced activation of sphingomyelinases and ceramide induced MRRSP activation of NOX [18], etc., are expected to be discovered with ongoing focused experiments.
Acknowledgements
This research has been financed by grants from the Spanish Ministry of Economy and Competitiveness (BFU2016‐80006-P), the Andalusian Government (Consejería de Economía, Innovación, Ciencia y Empleo, BIO‐0216 and CTS‐6264 and Consejería de Igualdad, Salud y Políticas Sociales, PI-00025-2013 and PI-0198-2016), Institute of Health Carlos III, Spain (PI13/00021, PI16/00090, CIBEREHD and CIBERNED) co-financed by European Development Regional Fund, Europe. We are indebted to Beatriz Carmona (EJ17-BIO216, Programa de Empleo Joven, FEDER/Junta de Andalucía) for her excellent technical assistance in the Biochemistry and Molecular Biology lab.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.11.007.
Appendix A. Supplementary material
Fig. S1.
Redox status analysis of Cys47 and Cys91 in native and TCEP treated recombinant Prdx6. Recombinant human PRDX6 overproduced in E. coli with His-tag at the C-terminus was incubated with 1 mM TCEP (+) or buffer alone (-) for 30 min at room temperature. It was then incubated with 10 mM IAM, digested with trypsin and analyzed by LC-MS/MS. Peptides containing Cys47 and Cys91, either carbamidomethylated (-SH) or glutathionylated (-SSG) were quantified using the free software Skyline. Peptide intensity data were normalized to the average intensity of 3 constant, miss-cleavage resistant, non-cystine peptides (MPGGLLLGDVAPNFEANTTVGR [0−22]; LPFPIIDDR [97−105]; LSILYPATTGR [144−154]). The percentage of glutathionylated Cys is shown in the last column of the table. Fragmentation MS/MS spectra of the peptides containing Cys47 and Cys91 in the original state and after reduction with TCEP are shown; the different ionic y and b series are sketched on the sequence of each peptide highlighting the cysteine modification, either carbamidomethylated (reduced) or glutathionylated. Mass spectrometry analysis was done on a Thermo Orbitrap Fusion (Q-OT-qIT, Thermo Scientific) mass spectrometer operated in positive mode. Protein identification was performed using Proteome Discoverer (version 2.1.0.81, Thermo Scientific). MS2 spectra were searched with SEQUEST engine against a database of Uniprot_Homo sapiens and Escherichia coli_ Feb2017 (www.uniprot.org). Peptides were generated by theoretical tryptic digestion allowing up to one missed cleavage, carbamidomethylation, dioxidation, trioxidation and glutathionylation of cysteine and oxidation of methionine as variable modifications. Precursor mass tolerance was 10 ppm and product ions were searched at 0.1 Da tolerance. Peptide spectral matches (PSM) were validated using percolator based on q-values at 1% FDR. Quantification of peptides was done using the free software Skyline.
Supplementary material Suppl. File 1. Quantitative analysis of proteins relative abundancies in HepG2 cells after several treatments. 4TO_NOS HepG2 cells overexpressing NOS3 and cells transformed with the empty vector (4TO), treated or not with siRNA-Trx1 or siRNA-Grx1 were quantified relative to untreated controls, including 4TO-NOS versus 4TO, using MacxQuant software as described in Materials & Methods. Only proteins with p values ≤ 0.05 and analyzed with at least 2 unique peptides are shown.
Fig. S2.
Distribution of identified and quantified Cys-peptides between the different samples. Venn diagrams have been drawn with the lists of identified peptides (those unambiguously ascribed by the Proteome Discoverer search engine) and quantified peptides (those selected by the Skyline software for quantitative analysis) in the three different cell types studied. The range of overlapping percentage is also indicated.
Supplementary material Suppl. File 2. Redox Proteome of HepG2 cells. The values of reduced/oxidized ratios of Cys peptides identified and quantified in samples of HepG2 cells from 3 independent experiments treated with siRNA-Grx1 and siRNA-Trx1 and calculated statistical parameters are shown; Data from WT, 4TO (Vo) and 4TO_NOS (NOS3) cells are shown in separate sheets.
Supplementary material Suppl. File 3 Metabolome analysis. Data from WT, 4TO (V) and 4TO_NOS (N3) cells treated with siRNA-Trx1 (T), siRNA-Grx1 (G) or with unspecific control siRNA (C) with 4 replicas each. Four sheets are presented displaying an alphabetical list of the metabolites detected, a pathway heat map and box-plots of each metabolite ordered alphabetically or clustered by pathways.
References
- 1.Alderton W.K., Cooper C.E., Knowles R.G. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anastasiou D., Poulogiannis G., Asara J.M., Boxer M.B., Jiang J.-K., Shen M., Bellinger G., Sasaki A.T., Locasale J.W., Auld D.S., Thomas C.J., Vander Heiden M.G., Cantley L.C. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bedhomme M., Adamo M., Marchand C.H., Couturier J., Rouhier N., Lemaire S.D., Zaffagnini M., Trost P. Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem J. 2012;445:337–347. doi: 10.1042/BJ20120505. [DOI] [PubMed] [Google Scholar]
- 4.Benhar M. Nitric oxide and the thioredoxin system: a complex interplay in redox regulation. Biochim. Biophys. Acta. 2015;1850:2476–2484. doi: 10.1016/j.bbagen.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Benhar M., Forrester M.T., Hess D.T., Stamler J.S. Regulated Protein Denitrosylation by Cytosolic and Mitochondrial Thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benhar M., Thompson J.W., Moseley M.A., Stamler J.S. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat O.M., Yuan X., Li G., Lee R., Li P.-L. Sphingolipids and Redox Signaling in Renal Regulation and Chronic Kidney Diseases. Antioxid Redox Signal, ars. 2018;2017:7129–7154. doi: 10.1089/ars.2017.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchanan B.B., Balmer Y. Redox regulation: a broadening horizon. Annu Rev Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- 9.Chen C.-A., De Pascali F., Basye A., Hemann C., Zweier J.L. Redox modulation of endothelial nitric oxide synthase by glutaredoxin-1 through reversible oxidative post-translational modification. Biochemistry. 2013;52:6712–6723. doi: 10.1021/bi400404s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou M.F., Schwartz D. Biological sequence motif discovery using motif-x. Curr Protoc Bioinformatics. 2011:15–24. doi: 10.1002/0471250953.bi1315s35. Chapter 13, Unit13. [DOI] [PubMed] [Google Scholar]
- 11.Colell A., Green D.R., Ricci J.-E. Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 2009;16:1573–1581. doi: 10.1038/cdd.2009.137. [DOI] [PubMed] [Google Scholar]
- 12.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 13.Dodson M., Darley-Usmar V., Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumont S., Bykova N.V., Pelletier G., Dorion S., Rivoal J. Cytosolic Triosephosphate Isomerase from Arabidopsis thaliana Is Reversibly Modified by Glutathione on Cysteines 127 and 218. Front Plant Sci. 2016;7:1942. doi: 10.3389/fpls.2016.01942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erwin P.A., Lin A.J., Golan D.E., Michel T. Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J Biol Chem. 2005;280:19888–19894. doi: 10.1074/jbc.M413058200. [DOI] [PubMed] [Google Scholar]
- 16.Fielek S., Mohrenweiser H.W. Erythrocyte Enzyme Deficiencies Assessed with a Miniature Centrifugal Analyzer. Clin. Chem. 1979;25:384–388. [PubMed] [Google Scholar]
- 17.Fisher A.B. Peroxiredoxin 6 in the repair of peroxidized cell membranes and cell signaling. Arch Biochem Biophys. 2017;617:68–83. doi: 10.1016/j.abb.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gault C.R., Obeid L.M., Hannun Y.A. An Overview of Sphingolipid Metabolism: From Synthesis to Breakdown. Adv. Exp. Med. Biol. 2010;688:1–23. doi: 10.1007/978-1-4419-6741-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Go Y.-M., Jones D.P. Thiol/disulfide redox states in signaling and sensing. Critical Reviews in Biochemistry and Molecular Biology. 2013:1–19. doi: 10.3109/10409238.2013.764840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Go Y.M., Jones D.P. The Redox Proteome. Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.R113.464131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González R., Ferrín G., Aguilar-Melero P., Ranchal I., Linares C.I., Bello R.I., la Mata de M., Gogvadze V., Bárcena J.A., Alamo J.M., Orrenius S., Padillo F.J., Zhivotovsky B., Muntané J. Targeting hepatoma using nitric oxide donor strategies. Antioxid Redox Signal. 2013;18:491–506. doi: 10.1089/ars.2011.4476. [DOI] [PubMed] [Google Scholar]
- 22.González R., López-Grueso M.J., Muntané J., Bárcena J.A., Padilla C.A. Redox regulation of metabolic and signaling pathways by thioredoxin and glutaredoxin in NOS-3 overexpressing hepatoblastoma cells. Redox Biology. 2015;6:122–134. doi: 10.1016/j.redox.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González R., Molina-Ruiz F.J., Bárcena J.A., Padilla C.A., Muntané J. Regulation of Cell Survival, Apoptosis, and Epithelial-to-Mesenchymal Transition by Nitric Oxide-Dependent Post-Translational Modifications. Antioxid Redox Signal. 2018;29:1312–1332. doi: 10.1089/ars.2017.7072. [DOI] [PubMed] [Google Scholar]
- 24.Gould N.S., Evans P., Martínez-Acedo P., Marino S.M., Gladyshev V.N., Carroll K.S., Ischiropoulos H. Site-Specific Proteomic Mapping Identifies Selectively Modified Regulatory Cysteine Residues in Functionally Distinct Protein Networks. Chemistry & Biology. 2015;22:965–975. doi: 10.1016/j.chembiol.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hara M.R., Agrawal N., Kim S.F., Cascio M.B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J.H., Tankou S.K., Hester L.D., Ferris C.D., Hayward S.D., Snyder S.H., Sawa A. S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 2005;7:665–674. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 26.Hashemy S.I., Holmgren A. Regulation of the catalytic activity and structure of human thioredoxin 1 via oxidation and S-nitrosylation of cysteine residues. J Biol Chem. 2008;283:21890–21898. doi: 10.1074/jbc.M801047200. [DOI] [PubMed] [Google Scholar]
- 27.Held J.M., Danielson S.R., Behring J.B., Atsriku C., Britton D.J., Puckett R.L., Schilling B., Campisi J., Benz C.C., Gibson B.W. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Molecular & Cellular Proteomics. 2010;9:1400–1410. doi: 10.1074/mcp.M900643-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hitosugi T., Zhou L., Elf S., Fan J., Kang H.-B., Seo J.H., Shan C., Dai Q., Zhang L., Xie J., Gu T.-L., Jin P., Alečković M., LeRoy G., Kang Y., Sudderth J.A., DeBerardinis R.J., Luan C.-H., Chen G.Z., Muller S., Shin D.M., Owonikoko T.K., Lonial S., Arellano M.L., Khoury H.J., Khuri F.R., Lee B.H., Ye K., Boggon T.J., Kang S., He C., Chen J. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hitosugi T., Zhou L., Fan J., Elf S., Zhang L., Xie J., Wang Y., Gu T.-L., Alečković M., LeRoy G., Kang Y., Kang H.-B., Seo J.H., Shan C., Jin P., Gong W., Lonial S., Arellano M.L., Khoury H.J., Chen G.Z., Shin D.M., Khuri F.R., Boggon T.J., Kang S., He C., Chen J. Tyr26 phosphorylation of PGAM1 provides a metabolic advantage to tumours by stabilizing the active conformation. Nat Commun. 2013;4:1790. doi: 10.1038/ncomms2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmgren A., Johansson C., Berndt C., Lönn M.E., Hudemann C., Lillig C.H. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 31.Hornsveld M., Dansen T.B. The Hallmarks of Cancer from a Redox Perspective. Antioxid Redox Signal. 2016;25:300–325. doi: 10.1089/ars.2015.6580. [DOI] [PubMed] [Google Scholar]
- 32.Hu W., Tedesco S., McDonagh B., Bárcena J.A., Keane C., Sheehan D. Selection of thiol- and disulfide-containing proteins of Escherichia coli on activated thiol-Sepharose. Analytical Biochemistry. 2010;398:245–253. doi: 10.1016/j.ab.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 35.Inadomi C., Murata H., Ihara Y., Goto S., Urata Y., Yodoi J., Kondo T., Sumikawa K. Overexpression of glutaredoxin protects cardiomyocytes against nitric oxide-induced apoptosis with suppressing the S-nitrosylation of proteins and nuclear translocation of GAPDH. Biochemical and Biophysical Research Communications. 2012:1–6. doi: 10.1016/j.bbrc.2012.07.118. [DOI] [PubMed] [Google Scholar]
- 36.Ishii T., Uchida K. Induction of reversible cysteine-targeted protein oxidation by an endogenous electrophile 15-deoxy-delta12,14-prostaglandin J2. Chem Res Toxicol. 2004;17:1313–1322. doi: 10.1021/tx049860+. [DOI] [PubMed] [Google Scholar]
- 37.Iyer N.V., Kotch L.E., Agani F., Leung S.W., Laughner E., Wenger R.H., Gassmann M., Gearhart J.D., Lawler A.M., Yu A.Y., Semenza G.L. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karplus P.A., Poole L.B. Cysteine-based redox switches in enzymes. Antioxid Redox Signal. 2011;14:1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klatt P., Lamas S. Regulation of protein function by S‐glutathiolation in response to oxidative and nitrosative stress. FEBS J. 2000;267:4928–4944. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 40.Kuda O., Brezinova M., Silhavy J., Landa V., Zidek V., Dodia C., Kreuchwig F., Vrbacky M., Balas L., Durand T., Hübner N., Fisher A.B., Kopecky J., Pravenec M. Nrf2-mediated Antioxidant Defense and Peroxiredoxin 6 are Linked to Biosynthesis of Palmitic Acid Ester of 9-Hydroxystearic Acid. Diabetes. 2018 doi: 10.2337/db17-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon J., Wang A., Burke D.J., Boudreau H.E., Lekstrom K.J., Korzeniowska A., Sugamata R., Kim Y.-S., Yi L., Ersoy I., Jaeger S., Palaniappan K., Ambruso D.R., Jackson S.H., Leto T.L. Peroxiredoxin 6 (Prdx6) supports NADPHoxidase1 (Nox1)-based superoxide generation and cell migration. Free Radic Biol Med. 2016 doi: 10.1016/j.freeradbiomed.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee T.-Y., Chen Y.-J., Lu T.-C., Huang H.-D., Chen Y.-J. SNOSite: exploiting maximal dependence decomposition to identify cysteine S-nitrosylation with substrate site specificity. PLoS ONE. 2011;6:e21849. doi: 10.1371/journal.pone.0021849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leichert L.I., Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C.-Q., Wogan G.N. Nitric oxide as a modulator of apoptosis. Cancer Lett. 2005;226:1–15. doi: 10.1016/j.canlet.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 45.Li P.-L., Gulbins E. Bioactive Lipids and Redox Signaling: Molecular Mechanism and Disease Pathogenesis. Antioxid Redox Signal. 2018 doi: 10.1089/ars.2017.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lindahl M., Mata-Cabana A., Kieselbach T. The disulfide proteome and other reactive cysteine proteomes: analysis and functional significance. Antioxid Redox Signal. 2011;14:2581–2642. doi: 10.1089/ars.2010.3551. [DOI] [PubMed] [Google Scholar]
- 47.Liu L., Hausladen A., Zeng M., Que L., Heitman J., Stamler J.S. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 48.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marino S.M., Gladyshev V.N. Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of trans-nitrosylation. J Mol Biol. 2010;395:844–859. doi: 10.1016/j.jmb.2009.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martínez-Acedo P., Núñez E., Gómez F.J.S., Moreno M., Ramos E., Izquierdo-Álvarez A., Miro-Casas E., Mesa R., Rodriguez P., Martínez-Ruiz A., Dorado D.G., Lamas S., Vázquez J. A novel strategy for global analysis of the dynamic thiol redox proteome. Mol Cell Proteomics. 2012;11:800–813. doi: 10.1074/mcp.M111.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martínez-Ruiz A., Lamas S. Detection and proteomic identification of S-nitrosylated proteins in endothelial cells. Arch Biochem Biophys. 2004;423:192–199. doi: 10.1016/j.abb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 52.Martínez-Ruiz A., Araújo I.M., Izquierdo-Álvarez A., Hernansanz-Agustín P., Lamas S., Serrador J.M. Specificity in S-nitrosylation: a short-range mechanism for NO signaling? Antioxid Redox Signal. 2013;19:1220–1235. doi: 10.1089/ars.2012.5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDonagh B., Ogueta S., Lasarte G., Padilla C.A., Bárcena J.A. Shotgun redox proteomics identifies specifically modified cysteines in key metabolic enzymes under oxidative stress in Saccharomyces cerevisiae. J Proteomics. 2009;72:677–689. doi: 10.1016/j.jprot.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 54.McDonagh B., Pedrajas J.R., Padilla C.A., Bárcena J.A. Thiol redox sensitivity of two key enzymes of heme biosynthesis and pentose phosphate pathways: uroporphyrinogen decarboxylase and transketolase. Oxid Med Cell Longev. 2013;2013 doi: 10.1155/2013/932472. 932472–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDonagh B., Sakellariou G.K., Smith N.T., Brownridge P., Jackson M.J. Differential cysteine labeling and global label-free proteomics reveals an altered metabolic state in skeletal muscle aging. J Proteome Res. 2014;13:5008–5021. doi: 10.1021/pr5006394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McDonagh B., Scullion S.M., Vasilaki A., Pollock N., McArdle A., Jackson M.J. Ageing-induced changes in the redox status of peripheral motor nerves imply an effect on redox signalling rather than oxidative damage. Free Radic Biol Med. 2016;94:27–35. doi: 10.1016/j.freeradbiomed.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Metallo C.M., Vander Heiden M.G. Understanding metabolic regulation and its influence on cell physiology. Molecular Cell. 2013;49:388–398. doi: 10.1016/j.molcel.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mi N.L., Sang H.H., Kim J., Koh A., Chang S.L., Jung H.K., Jeon H., Kim D.-H., Suh P.-G., Sung H.R. Glycolytic flux signals to mTOR through glyceraldehyde-3-phosphate dehydrogenase-mediated regulation of Rheb. Mol Cell Biol. 2009;29:3991–4001. doi: 10.1128/MCB.00165-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mieyal J.J., Gallogly M.M., Qanungo S., Sabens E.A., Shelton M.D. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10:1941–1988. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mitchell D.A., Marletta M.A. Thioredoxin catalyzes the S-nitrosation of the caspase-3 active site cysteine. Nature Chemical Biology. 2005;1:154–158. doi: 10.1038/nchembio720. [DOI] [PubMed] [Google Scholar]
- 61.Mohr S., Stamler J.S., Brüne B. Posttranslational modification of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosylation and subsequent NADH attachment. J Biol Chem. 1996;271:4209–4214. doi: 10.1074/jbc.271.8.4209. [DOI] [PubMed] [Google Scholar]
- 62.Morgan M.J., Kim Y.-S., Liu Z. Lipid rafts and oxidative stress-induced cell death. Antioxid Redox Signal. 2007;9:1471–1483. doi: 10.1089/ars.2007.1658. [DOI] [PubMed] [Google Scholar]
- 63.Murakami M., Kudo I. Phospholipase A2. J. Biochem. 2002;131:285–292. doi: 10.1093/oxfordjournals.jbchem.a003101. [DOI] [PubMed] [Google Scholar]
- 64.Patel J.M., Zhang J., Block E.R. Nitric oxide-induced inhibition of lung endothelial cell nitric oxide synthase via interaction with allosteric thiols: role of thioredoxin in regulation of catalytic activity. Am. J. Respir. Cell Mol. Biol. 1996;15:410–419. doi: 10.1165/ajrcmb.15.3.8810647. [DOI] [PubMed] [Google Scholar]
- 65.Pedrajas J.R., McDonagh B., Hernández-Torres F., Miranda-Vizuete A., González-Ojeda R., Martínez-Galisteo E., Padilla C.A., Bárcena J.A. Glutathione Is the Resolving Thiol for Thioredoxin Peroxidase Activity of 1-Cys Peroxiredoxin Without Being Consumed During the Catalytic Cycle. Antioxid Redox Signal. 2016;24:115–128. doi: 10.1089/ars.2015.6366. [DOI] [PubMed] [Google Scholar]
- 66.Prehu M.O., Prehu C., Calvin M.C., Rosa R. Rabbit M type phosphoglyceromutase: comparative effects of two thiol reagents antibody reaction and hybridization studies. Comp. Biochem. Physiol., B. 1988;89:257–262. doi: 10.1016/0305-0491(88)90220-9. [DOI] [PubMed] [Google Scholar]
- 67.Ravi K., Brennan L.A., Levic S., Ross P.A., Black S.M. S-nitrosylation of endothelial nitric oxide synthase is associated with monomerization and decreased enzyme activity. Proc Natl Acad Sci USA. 2004;101:2619–2624. doi: 10.1073/pnas.0300464101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Requejo-Aguilar R., Lopez-Fabuel I., Jimenez-Blasco D., Fernandez E., Almeida A., Bolaños J.P. DJ1 represses glycolysis and cell proliferation by transcriptionally up-regulating Pink1. Biochem J. 2015;467:303–310. doi: 10.1042/BJ20141025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Roos G., Foloppe N., Messens J. Understanding the pK(a) of redox cysteines: the key role of hydrogen bonding. Antioxid Redox Signal. 2013;18:94–127. doi: 10.1089/ars.2012.4521. [DOI] [PubMed] [Google Scholar]
- 70.Salsbury F.R., Knutson S.T., Poole L.B., Fetrow J.S. Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schilling B., Rardin M.J., MacLean B.X., Zawadzka A.M., Frewen B.E., Cusack M.P., Sorensen D.J., Bereman M.S., Jing E., Wu C.C., Verdin E., Kahn C.R., MacCoss M.J., Gibson B.W. Platform-independent and Label-free Quantitation of Proteomic Data Using MS1 Extracted Ion Chromatograms in Skyline: APPLICATION TO PROTEIN ACETYLATION AND PHOSPHORYLATION. Molecular & Cellular Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmitt A., Schmitz W., Hufnagel A., Schartl M., Meierjohann S. Peroxiredoxin 6 triggers melanoma cell growth by increasing arachidonic acid-dependent lipid signalling. Biochem J. 2015;471:267–279. doi: 10.1042/BJ20141204. [DOI] [PubMed] [Google Scholar]
- 73.Schuppe-Koistinen I., Moldéus P., Bergman T., Cotgreave I.A. S-thiolation of human endothelial cell glyceraldehyde-3-phosphate dehydrogenase after hydrogen peroxide treatment. Eur J Biochem. 1994;221:1033–1037. doi: 10.1111/j.1432-1033.1994.tb18821.x. [DOI] [PubMed] [Google Scholar]
- 74.Shahul H.M.S., Sarma S.P. The structure of the thioredoxin-triosephosphate isomerase complex provides insights into the reversible glutathione-mediated regulation of triosephosphate isomerase. Biochemistry. 2012;51:533–544. doi: 10.1021/bi201224s. [DOI] [PubMed] [Google Scholar]
- 75.Shelton M.D., Chock P.B., Mieyal J.J. Glutaredoxin: role in reversible protein s-glutathionylation and regulation of redox signal transduction and protein translocation. Antioxid Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 76.Stincone A., Prigione A., Cramer T., Wamelink M.M.C., Campbell K., Cheung E., Olin-Sandoval V., Grüning N.-M., Krüger A., Tauqeer Alam M., Keller M.A., Breitenbach M., Brindle K.M., Rabinowitz J.D., Ralser M. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev. 2014;90:927–963. doi: 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Subramani J., Kundumani-Sridharan V., Hilgers R.H.P., Owens C., Das K.C. Thioredoxin Uses a GSH-independent Route to Deglutathionylate Endothelial Nitric-oxide Synthase and Protect against Myocardial Infarction. J Biol Chem. 2016;291:23374–23389. doi: 10.1074/jbc.M116.745034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sullivan L.B., Gui D.Y., Heiden M.G.V. Altered metabolite levels in cancer: implications for tumour biology and cancer therapy. Nat. Rev. Cancer. 2016;16:680–693. doi: 10.1038/nrc.2016.85. [DOI] [PubMed] [Google Scholar]
- 80.Tennant D.A., Durán R.V., Gottlieb E. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 81.Teusink B., Passarge J., Reijenga C.A., Esgalhado E., van der Weijden C.C., Schepper M., Walsh M.C., Bakker B.M., van Dam K., Westerhoff H.V., Snoep J.L. Can yeast glycolysis be understood in terms of in vitro kinetics of the constituent enzymes? Testing biochemistry. Eur J Biochem. 2000;267:5313–5329. doi: 10.1046/j.1432-1327.2000.01527.x. [DOI] [PubMed] [Google Scholar]
- 82.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry–based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 83.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei L., Dai Y., Zhou Y., He Z., Yao J., Zhao L., Guo Q., Yang L. Oroxylin A activates PKM1/HNF4 alpha to induce hepatoma differentiation and block cancer progression. Cell Death and Disease. 2017;8:e2944. doi: 10.1038/cddis.2017.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wei T., Chen C., Hou J., Xin W., Mori A. Nitric oxide induces oxidative stress and apoptosis in neuronal cells. Biochim. Biophys. Acta. 2000;1498:72–79. doi: 10.1016/s0167-4889(00)00078-1. [DOI] [PubMed] [Google Scholar]
- 86.Wu C., Liu T., Chen W., Oka S.-I., Fu C., Jain M.R., Parrott A.M., Baykal A.T., Sadoshima J., Li H. Redox regulatory mechanism of transnitrosylation by thioredoxin. Molecular & Cellular Proteomics. 2010;9:2262–2275. doi: 10.1074/mcp.M110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wu C., Parrott A.M., Fu C., Liu T., Marino S.M., Gladyshev V.N., Jain M.R., Baykal A.T., Li Q., Oka S., Sadoshima J., Beuve A., Simmons W.J., Li H. Thioredoxin 1-mediated post-translational modifications: reduction, transnitrosylation, denitrosylation, and related proteomics methodologies. Antioxid Redox Signal. 2011;15:2565–2604. doi: 10.1089/ars.2010.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu Y., Li F., Lv L., Li T., Zhou X., Deng C.-X., Guan K.-L., Lei Q.-Y., Xiong Y. Oxidative stress activates SIRT2 to deacetylate and stimulate phosphoglycerate mutase. Cancer Res. 2014;74:3630–3642. doi: 10.1158/0008-5472.CAN-13-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang L., Wu D., Wang X., Cederbaum A.I. Depletion of cytosolic or mitochondrial thioredoxin increases CYP2E1-induced oxidative stress via an ASK-1-JNK1 pathway in HepG2 cells. Free Radic Biol Med. 2011;51:185–196. doi: 10.1016/j.freeradbiomed.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Suppl. File 1. Quantitative analysis of proteins relative abundancies in HepG2 cells after several treatments. 4TO_NOS HepG2 cells overexpressing NOS3 and cells transformed with the empty vector (4TO), treated or not with siRNA-Trx1 or siRNA-Grx1 were quantified relative to untreated controls, including 4TO-NOS versus 4TO, using MacxQuant software as described in Materials & Methods. Only proteins with p values ≤ 0.05 and analyzed with at least 2 unique peptides are shown.
Supplementary material Suppl. File 2. Redox Proteome of HepG2 cells. The values of reduced/oxidized ratios of Cys peptides identified and quantified in samples of HepG2 cells from 3 independent experiments treated with siRNA-Grx1 and siRNA-Trx1 and calculated statistical parameters are shown; Data from WT, 4TO (Vo) and 4TO_NOS (NOS3) cells are shown in separate sheets.
Supplementary material Suppl. File 3 Metabolome analysis. Data from WT, 4TO (V) and 4TO_NOS (N3) cells treated with siRNA-Trx1 (T), siRNA-Grx1 (G) or with unspecific control siRNA (C) with 4 replicas each. Four sheets are presented displaying an alphabetical list of the metabolites detected, a pathway heat map and box-plots of each metabolite ordered alphabetically or clustered by pathways.