Abstract
Streptococcus pneumoniae sialidase SpNanB is an intramolecular trans-sialidase (IT-sialidase) and a virulence factor that is essential for streptococcal infection of upper and lower respiratory tract. SpNanB catalyzes the formation of 2,7-anhydro-N-acetylneuraminic acid (2,7-anhydro-Neu5Ac), a potential prebiotic that can be used as the sole carbon source of a common human gut commensal anaerobic bacterium. We report here the development of an efficient one-pot multienzyme (OPME) system for synthesizing 2,7-anhydro-Neu5Ac and its derivatives. Based on crystal structure analysis, an N-cyclohexyl derivative of 2,7-anhydro-neuraminic acid was designed, synthesized, and shown to be a selective inhibitor against SpNanB and another Streptococcus pneumoniae sialidase SpNanC. This study demonstrates a new strategy of synthesizing 2,7-anhydro-sialic acids in gram scale and the potential application of their derivatives as selective sialidase inhibitors.
Keywords: chemoenzymatic synthesis; 2,7-anhydro-sialic acids; sialidase; inhibitor; Streptococcus pneumoniae
TOC Graph
1. INTRODUCTION
Streptococcus pneumoniae is a common human pathogen that causes pneumonia, otitis media, septicemia, bacteremia, meningitis, and other serious diseases.1,2 It expresses up to three sialidases including SpNanA,3,4 SpNanB,5,6 and/or SpNanC.7,8 SpNanA is a hydrolytic sialidase catalyzing the hydrolysis of terminal α2–3-, α2–6-, and α2–8-linked sialic acid.4 SpNanB is an intramolecular trans-sialidase (IT-sialidase) which uses α2–3-linked sialosides as substrates to produce 2,7-anhydro-N-acetylneuraminic acid (2,7-anhydro-Neu5Ac, 1) (Figure 1).9,10 SpNanC catalyzes the selective cleavage of terminal α2–3-linked sialic acid to form 2,3-dehydro-2-deoxy-N-acetylneuraminic acid (Neu5Ac2en, DANA), a transition state analog of most hydrolytic sialidases.8
Fig. 1.
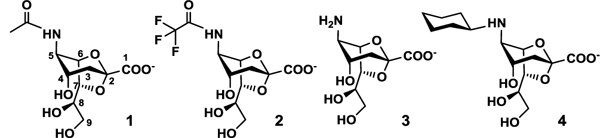
Structures of enzymatically synthesized 2,7-anhydro-Neu5Ac (1) and its chemoenzymatically synthesized derivatives 2–4.
SpNanB, along with SpNanA, provides sialic acid source and helps Streptococcus pneumoniae in biofilm formation, nutrition, colonization, and infection of the host.11–13 SpNanC plays a role of regulating the activity of hydrolytic sialidases including SpNanA as it catalyzes the production and the hydrolysis of hydrolytic sialidase inhibitor Neu5Ac2en.8 SpNanC has been identified as a Streptococcus pneumoniae marker for pneumococcal haemolytic uraemic syndrome in children.14
Sialidase inhibitors are proven successful anti-virus drugs,15 and are potential anti-bacteria reagents.16 Various types of inhibitors against SpNanA have been identified including Neu5Ac2en and derivatives,17,18 katsumadain A, artocarpin,19 and diazenylaryl sulfonic acids.20 However, effective inhibitors against SpNanB and/or SpNanC are not readily available. Neu5Ac2en inhibited SpNanB and SpNanC only weakly with an IC50 value falling in a submillimolar range.18 Oseltamivir21 did not show inhibitory activity against SpNanB nor SpNanC with a concentration up to 7.5 mM22 even though it inhibited Ruminococcus gnavus IT-sialidase RgNanH23 which shares a similar catalytic mechanism as SpNanB and produces the same 2,7-anhydro-Neu5Ac product. 2-N-Cyclohexylaminoethanesulfonic acid (CHES) was unexpectedly found to have weak inhibition against SpNanB.9 A family of β-amino-sulfonic acids was subsequently screened and the most potent inhibitor candidate identified had an IC50 value of 38.9 μM.24 Siastatin B25 also had a similar inhibitory activity against SpNanB and natural products katsumadain A and artocarpin were reported to be inhibitors against SpNanB.26 Recently, a natural product malabaricone C was reported to inhibit all three S. pneumoniae sialidases, with IC50 values in a submicromolar range.27 However, the control Neu5Ac2en was shown to have an IC50 value of 45.1 μM for SpNanB, which disagreed with other reports where Neu5Ac2en was a millimolar inhibitor against SpNanB.9,18,22,28
We hypothesized that the derivatives of SpNanB product, 2,7-anhydro-Neu5Ac (1), could be suitable selective inhibitors against SpNanB. 2,7-Anhydro-Neu5Ac (1) was initially characterized in 1982 as a sialic acid methanolysis byproduct,29 and later found in rat urine and human wet cerumen.30 It was shown to be a selective carbon source to support the growth of Ruminoccocus gnavus, a common human gut commensal anaerobic bacterium.31 Nevertheless, the roles and potential applications of 2,7-anhydro-Neu5Ac remain largely underexplored, partially due to the limited access to the 2,7-anhydro-sialic acids. There are only a few reports describing the chemical synthesis of 2,7-anhydro-sialic acids.32–34 Although the overall yield was improved and the synthetic route was shortened,34 the chemical synthetic methods required multistep protection and deprotection steps. Due to their high efficiency, excellent regio- and stereoselectivity, environmental friendly feature, as well as increasing accessibility, enzymes have been increasingly applied in organic synthesis. Especially, the use of enzymes in the synthesis of carbohydrates is growing rapidly.35,36 Recently, an enzymatic method was reported to produce 2,7-anhydro-Neu5Ac in milligram scales in a 33% overall yield by treating sialylglycoprotein fetuin with Ruminoccocus gnavus IT-sialidase RgNanH.37 An efficient method for large-scale synthesis of 2,7-anhydro-Neu5Ac and its derivatives is needed.
Herein, we report an efficient one-pot multienzyme (OPME) system for synthesizing 2,7-anhydro-Neu5Ac and its derivatives in gram-scale and preparative-scale with good overall yields. Moreover, we have demonstrated that it is possible to develop 2,7-anhydro-sialic acid derivatives as potential selective inhibitors against certain sialidases.
2. RESULTS AND DISCUSSION
Gram-Scale Enzymatic Synthesis of 2,7-Anhydro-Neu5Ac (1).
Similar to the function of leech IT-sialidase NanL,38 SpNanB was reported to be able to catalyze the formation of 2,7-anhydro-Neu5Ac directly from Neu5Ac.10 Nevertheless, our attempts to synthesize 2,7-anhydro-Neu5Ac (1) directly from N-acetylneuraminic acid (Neu5Ac) using SpNanB resulted in low yields (<20% by thin-layer chromatography analysis) despite the efforts in varying buffer, pH, SpNanB amount, and the concentration of Neu5Ac.
In order to obtain 2,7-anhydro-Neu5Ac (1) in gram-scale, a one-pot multienzyme (OPME) system containing four enzymes was developed. As shown in Scheme 1, Pasteurella multocida sialic acid aldolase (PmAldolase),39 Neisseria meningitidis CMP-sialic acid synthetase (NmCSS),40 and Pasteurella multocida sialyltransferase 1 M144D mutant (PmST1_M144D)41,42 were used for the in situ formation of α2–3-linked sialyllactose (3’-sialyllactose), which was the substrate of SpNanB for the production of 2,7-anhydro-Neu5Ac (1). PmAldolase was responsible for the formation of N-acetylneuraminic acid (Neu5Ac), the most common sialic acid form,43 from N-acetylmannosamine (ManNAc) and pyruvate. The Neu5Ac formed was activated by NmCSS to form cytidine 5’-monophosphate-N-acetylneuraminic acid (CMP-Neu5Ac) in the presence of cytidine 5’-triphosphate (CTP). PmST1_M144D was responsible for catalyzing the transfer of the Neu5Ac in CMP-Neu5Ac donor to lactose as the acceptor to form 3’-sialyllactose, which was used as the substrate of SpNanB for the formation of 2,7-anhydro-Neu5Ac as the target product.
Scheme 1.
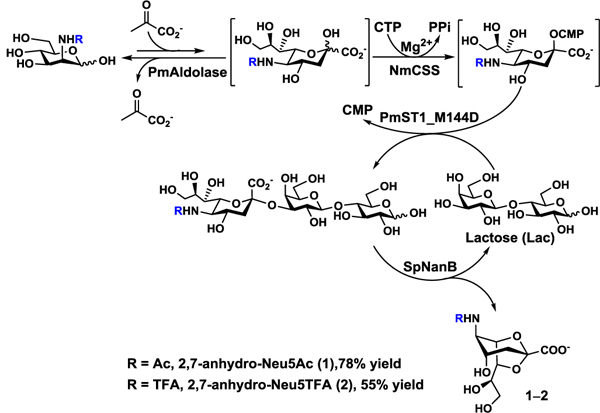
One-pot multienzyme (OPME) synthesis of 2,7-anhydro-Neu5Ac (1) and 2,7-anhydro-Neu5TFA (2).
Similar to the OPME synthesis of 2,3-dehydro-2-deoxy-sialic acids (Sia2ens),18 the optimal conditions for SpNanB-dependent OPME synthesis of 2,7-anhydro-Neu5Ac (1) were explored. Tris-HCl buffer at pH 7.5 was found to be a well suited condition to balance the activities of all four enzymes in the system. Lactose was chosen as the sialyltransferase acceptor due to its commercial availability and low cost. As lactose produced by the sialidase SpNanB-catalyzed reaction can be reused as the acceptor for sialyltransferase PmST1_M144D-catalyzed reaction (Scheme 1), it was used at 0.5 equivalent of the molar amount of the sialic acid precursor ManNAc in the system. The desired 2,7-anhydro-Neu5Ac (1) was obtained in 1.10 grams with a yield of 78% using this OPME method.
Pure 2,7-anhydro-Neu5Ac (1) was used to confirm a previous observation that similar to leech IT-sialidase,38 SpNanB can hydrolyze 2,7-anhydro-Neu5Ac to form Neu5Ac.9 For synthetic purpose, the 2,7-anhydro-Neu5Ac (1) hydrolysis activity of SpNanB was able to be minimized by controlling the reaction time and the amount of SpNanB.
Substrate Specificity Studies of SpNanB.
Substrate specificity studies of SpNanB confirmed that the enzyme was specific to α2–3-linked sialosides. A library of para-nitrophenol (pNP)-tagged α2–3-linked sialosides44 with derivatization at various positions of Neu5Ac showed that C5- and C9-modifications of Neu5Ac, including N-glycolylneuraminic acid (Neu5Gc), were well tolerated. However, sialosides containing 2-keto-3-deoxynonulsonic acid (Kdn) and its derivatives were not suitable substrates for SpNanB (Table S1 and Figure S1). The substrate promiscuities of SpNanB and the other three enzymes in the OPME system make it possible to synthesize 2,7-anhydro-sialic acids and derivatives with modifications at various positions.
It was interesting to notice that C7-modified sialic acids were also good substrates for SpNanB. These were unexpected as the hydroxyl group on C-7 was the nucleophile that attacked the anomeric carbon of sialic acid during the intramolecular trans-sialidase reaction.8 To confirm the observation from the colorimetric plate assay and identify the product, thin layer chromatography (TLC), mass spectrometry, and nuclear magnetic resonance (NMR) studies were carried out using Neu5Ac7N3α2–3GalβpNP, a sialoside with a C7-modified Neu5Ac, as the substrate for SpNanB. TLC analysis showed only two product spots throughout the reaction process, one of which representing GalβpNP. Mass spectrometry results showed a m/z value of 333.1051 (Figure S2), corresponding to 7-azido-Neu5Ac (7N3Neu5Ac) which was confirmed by NMR studies. These observations indicated that SpNanB can catalyze the hydrolysis of sialosides containing 7-modified sialic acids directly when the 7-hydroxyl group required for intramolecular trans-sialidase reaction is missing.
Design of a 2,7-Anhydro-Neu5Ac Derivative-Based SpNanB Inhibitor.
One of our research goals is to develop selective inhibitors against bacterial sialidases as chemical biological probes. We hypothesize that SpNanB product with substitutions that better fit the SpNanB substrate binding pocket would be selective inhibitors against SpNanB. Analysis of crystal structures of SpNanB complexed with various inhibitors indicated the presence of a hydrophobic pocket near the active site of SpNanB.9 In the structure (PDB ID: 4XHB) of SpNanB complexed with 2-N-cyclohexylaminoethansulfonic acid (CHES), the cyclohexyl group of CHES occupied the hydrophobic pocket, replacing the acetamido methyl group in 2,7-anhydro-Neu5Ac (1) bond to SpNanB in the structure complex.9,10 Further studies demonstrated that many functional groups could fit in this pocket with varying inhibitory activities against SpNanB.24 Superimposition of the bound 2,7-anhydro-Neu5Ac, CHES, and 2-[(3-chlorobenzyl)ammonio)ethanesulfonate revealed that the acetyl group in 2,7-anhydro-Neu5Ac occupies the same hydrophobic pocket in SpNanB as the cyclohexyl group and the 3-chlorobenzyl group of inhibitors (Figure 2). Therefore, we hypothesized that replacing the acetyl group in 2,7-anhydro-Neu5Ac (1) by a hydrophobic group such as cyclohexyl could result in a selective inhibitor against SpNanB.
Fig. 2.
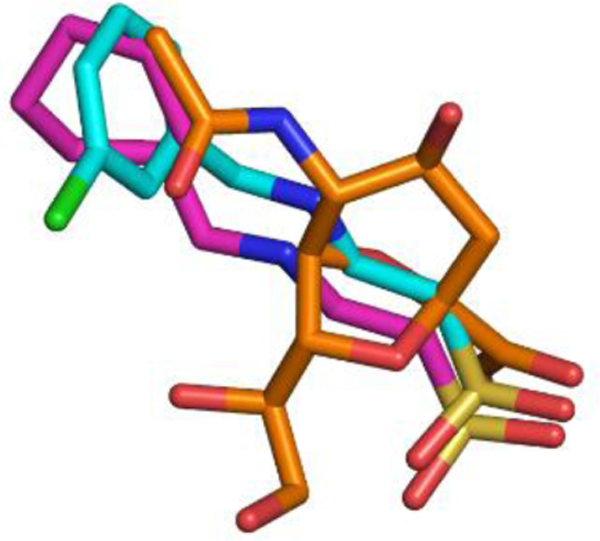
Superimposition of 2,7-anhydro-Neu5Ac (in brown sticks), CHES (in pink sticks), and 2-[(3-chlorobenzyl)ammonio)ethanesulfonate (in pale-blue sticks). The coordinates were extracted from Protein Data Bank (PDB IDs: 2WV1, 2VW2, and 4FPF).
Chemoenzymatic Synthesis and Derivatization of 2,7-Anhydro-Sialic Acids.
An efficient strategy to obtain N-substituted analogs of 2,7-anhydro-Neu5Ac (1) is to synthesize 2,7-anhydro-N-trifluoroacetylneuraminic acid (2,7-anhydro-Neu5TFA, 2) as a precursor for further chemical modification, since the N-trifluoroacetyl (N-TFA) group was shown previously to be a good mimic of N-acetyl group in enzyme-catalyzed reactions and can be easily removed and replace by other groups.45,46 Indeed, 2,7-anhydro-Neu5TFA (2) was successfully obtained from N-trifluoroacetylmannosamine (ManNTFA) as the starting material for the OPME system (Scheme 1). A lower yield (55%) obtained was most likely due to the lability of the N-trifluoroacetyl (N-TFA) group which could fall off during the reaction.46 The trifluoroacetyl group of the produced 2,7-anhydro-Neu5TFA (2) was readily removed under mild basic condition to form 2,7-anhydro-neuraminic acid (2,7-anhydro-Neu, 3) which was coupled with cyclohexanone by reductive amination to produce the desired 2,7-anhydro-N-cyclohexyl neuraminic acid (2,7-anhydro-Neu5Cyclohexyl, 4) in almost quantitative yield (Scheme 2).
Scheme 2.
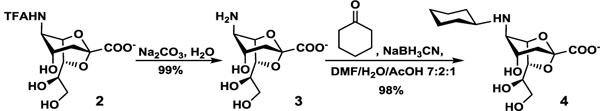
Synthesis of 2,7-anhydro-Neu5Cyclohexyl (4) from 2,7-anhydro-Neu5TFA (2).
Unlike 2,7-anhydro-Neu5Ac (1) which can be hydrolyzed by SpNanB, 2,7-anhydro-Neu5Cyclohexyl (4) was tested and was shown to be resistant to SpNanB hydrolysis.
Sialidase Inhibition Studies of 2,7-Anhydro-Sialic Acids.
The designed inhibitor 2,7-anhydro-Neu5Cyclohexyl (4), along with other 2,7-anhydro-sialic acids (1‒3), were tested for their inhibitory activities against several sialidases categorized in the glycoside hydrolase GH33 family in the Carbohydrate Active Enzyme (CAZy) database (www.cazy.org).47–49 Neu5Acα2–3GalβpNP was used as the substrate in a 384-well plate colorimetric assay.50,51 Each reaction mixture contained an inhibitor, the substrate, a sialidase of interest, and a β-galactosidase in an amount sufficient to cleave all GalβpNP generated from the sialidase-catalyzed reaction process to release para-nitrophenol (pNP), whose reading at A405 nm in a buffer of pH higher than 9.5 is correlated to the activity of the sialidase. The bacterial sialidases tested include SpNanA,52 SpNanB,52 and SpNanC,18 Pasteurella multocida sialyltransferase with α2‒3-sialidase activity (PmST1),41 Bifidobacterium infantis sialidase BiNanH2,53 as well as commercially available sialidases from Arthrobacter ureafaciens (AuSialidase), Clostridium perfringens (CpNanI), and Vibrio cholerae (VcSialidase). Recombinant human cytosolic sialidase hNEU254 was also tested.
At a concentration of 1 mM, no significant inhibition against any sialidases tested was observed for 2,7-anhydro-Neu5Ac (1) or 2,7-anhydro-Neu (3) (Table S2). In comparison, noticeable inhibitory activity was observed for 2,7-anhydro-Neu5Cyclohexyl (4) against SpNanA, SpNanB, and SpNanC. 2,7-Anhydro-Neu5TFA (2), the intermediate designed for the synthesis of 2,7-anhydro-Neu5Cyclohexyl (4), also showed some inhibitory activity against SpNanA, SpNanB, AuSialidase, and VcSialidase.
IC50 values were obtained for compounds which showed more than 50% inhibitory activity at 1 mM. As shown in Table 1, 2,7-anhydro-Neu5Cyclohexyl (4) was a micromolar inhibitor against SpNanB (IC50 = 180 ± 23 μM) and SpNanC (IC50 = 58.4 ± 2.4 μM). 2,7-Anhydro-Neu5TFA (2) was a high micromolar inhibitor against SpNanA (IC50 = 145 ± 16 μM) and AuSialidase (IC50 = 225 ± 34 μM).
Table 1.
IC50 Values of 2,7-anhydro-Neu5TFA (2) and 2,7-anhydro-Neu5Cyclohexyl (4) against bacterial sialidases.
| Sialidases | IC50 values of different inhibitors (μM) |
|
|---|---|---|
| (2) | (4) | |
| SpNanA | 145 ± 16 | 500 − 1000 |
| SpNanB | 250 − 500 | 180 ± 23 |
| SpNanC | > 1000 | 58.4 ± 2.4 |
| AuSialidase | 225 ± 34 | > 1000 |
| VcSialidase | 500 − 1000 | > 1000 |
The inhibitor 2,7-anhydro-Neu5Cyclohexyl (4) that we designed showed a noticeable improvement in inhibiting SpNanB and SpNanC compared to the non-modified SpNanB product 2,7-anhydro-Neu5Ac (1). Another improvement was that 2,7-anhydro-Neu5Cyclohexyl (4), but not 2,7-anhydro-Neu5Ac (1), was resistant to SpNanB hydrolysis. Although the IC50 values were still in a high micromolar range, 2,7-anhydro-Neu5Cyclohexyl (4) showed selectivity for the inhibition of all three Streptococcus pneumoniae sialidases among all sialidases tested. Therefore, we have demonstrated here that 2,7-anhydro-sialic acids with the potential for further improvement, could be a new type of scaffold for designing potential selective inhibitors against certain sialidases.
3. CONCLUSIONS
In conclusion, a novel one-pot multienzyme (OPME) strategy was developed for gram-scale and preparative synthesis of 2,7-anhydro-Neu5Ac (1) and 2,7-anhydro-Neu5TFA (2). The latter was further used to synthesize 2,7-anhydro-Neu (3) and 2,7-anhydro-Neu5Cyclohexyl (4), a designed sialidase inhibitor which showed improved inhibitory activity for SpNanA and more significantly for SpNanB and SpNanC, but not other sialidases tested. Both 2,7-anhydro-Neu5TFA (2) and 2,7-anhydro-Neu5Cyclohexyl (4) were shown to be high micromolar inhibitors selectively against certain bacterial sialidases. This study demonstrated an effective synthetic strategy for 2,7-anhydro-sialic acids and a new idea of exploring the family of 2,7-anhydro-sialic acids as potential selective sialidase inhibitors.
4. EXPERIMENTAL SECTION
Materials.
Recombinant sialidases were expressed and purified as reported previously for human cytosolic sialidase hNEU2,54 as well as bacterial sialidases from Streptococcus pneumoniae (SpNanA,52 SpNanB,52 and SpNanC18), Pasteurella multocida sialyltransferase 1 with α2–3-sialidase activity (PmST1),41 and Bifidobacterium infantis sialidase BiNanH2.53 Commercially available bacterial sialidases including those from Arthrobacter ureafaciens (Prozyme), Clostridium perfringens CpNanI (Sigma-Aldrich), and Vibrio cholerae were from Sigma-Aldrich. Aspergillus oryzae β-galactosidase was purchased from Sigma-Aldrich. Pasteurella multocida sialic acid aldolase (PmNanA),39 Neisseria meningitidis CMP-sialic acid synthetase (NmCSS),40 and Pasteurella multocida sialyltransferase 1 M144D mutant (PmST1_M144D)41,42 were expressed and purified as described previously. Siaα2–3GalβpNP44 used for substrate specificity studies were synthesized as described previously.
General methods.
Nuclear Magnetic Resonance (NMR) spectra were recorded in the NMR facility of the University of California, Davis on a Bruker Avance-400 NMR spectrometer (400 MHz for 1H, 100 MHz for 13C). Chemical shifts are reported in parts per million (ppm) on the δ scale. High resolution electrospray ionization (ESI) mass spectra were obtained using a Thermo Electron LTQ-Orbitrap Hybrid MS at the Mass Spectrometry Facility at the University of California, Davis. Specific rotation was recorded on a Rudolph Research Analytical Autopol IV automatic polarimeter. Column chromatography was performed using RediSep Rf silica columns or an ODS-SM (C18) column (51 g, 50 μm, 120 Å, Yamazen) on the CombiFlash® Rf 200i system. Thin layer chromatography (TLC) was performed on silica gel plates (Sorbent Technologies) using anisaldehyde sugar stain for detection. Gel filtration chromatography was performed with a column (100 cm × 2.5 cm) packed with Bio-Gel P-2 Fine resins (Bio-Rad). All reagents were at least of reagent grade and were used as supplied without further purification unless indicated.
One-pot multienzyme (OPME) synthesis of 2,7-anhydro-sialic acids.
ManNAc (1.0 g) or ManNTFA (300 mg), lactose (0.5 equiv.), sodium pyruvate (5 equiv.), CTP (1.5 equiv.) were dissolved in Tris-HCl buffer (100 mM, pH 7.5, 100 or 20 mL) containing 20 mM of MgCl2. The pH of the solution was further adjusted to 7.5 with 4 M of NaOH. PmAldolase (8.0 or 4.0 mg), NmCSS (4.0 or 2.0 mg), PmST1_M144D (8.0 or 4.0 mg), SpNanB (4.0 or 2.0 mg) were added and the reaction was incubated in an isotherm incubator for 16 h at 37 °C with agitation at 100 rpm. The reaction was quenched by adding the same volume of ice-cold ethanol and incubating at 4 °C for 1 h. The formed precipitates were removed by centrifugation and the supernatant was concentrated. The residue was purified gradually by passing it through a BioGel P-2 gel filtration column, a silica column (EtOAc:MeOH:H2O = 4:2:0.2, by volume), followed by a C18 column (100% H2O) to produce the pure compound.
5-Acetamido-2,7-anhydro-3,5-dideoxy-α-D-glycero-D-galacto-non-2-ulopyranosonic acid (2,7-anhydro-Neu5Ac, 1).
1.10 g, 78%, white solid. (c 0.5, H2O); 1H NMR (400 MHz, D2O) δ 4.51 (bs, 1H, H-6), 4.40 (dd, J = 7.8, 0.8 Hz, 1H, H-7), 3.95–3.90 (m, 1H, H-4), 3.89 (bs, 1H, H-5), 3.72 (dd, J = 11.8, 2.8 Hz, 1H, H-9), 3.56 (dd, J = 11.8, 6.0 Hz, 1H, H-9’), 3.50 (ddd, J = 7.8, 6.0, 2.8 Hz, 1H, H-8), 2.14 (dd, J = 15.2, 5.6 Hz, 1H, H-3ax), 2.00 (s, 3H, COCH3), 2.00–1.92 (m, 1H, H-3eq); 13C NMR (100 MHz, D2O) δ 174.0, 173.5, 105.5, 77.1, 76.6, 71.9, 66.8, 62.2, 51.9, 35.2, 21.8; HRMS (ESI) Anal. Calcd for C11H16NO8 [M-H]-: 290.0881, Found: 290.0886.
2,7-Anhydro-3,5-dideoxy-5-trifluoroacetamido-α-D-glycero-D-galacto-non-2-ulopyranosonic acid (2,7-anhydro-Neu5TFA, 2).
219.8 mg, 55%, white solid. (c 0.6, H2O); 1H NMR (400 MHz, D2O) δ 4.65 (bs, 1H, H-6), 4.48 (dd, J = 7.8, 0.8 Hz, 1H, H-7), 4.07–4.04 (m, 2H, H-4, H-5), 3.78 (dd, J = 11.6, 2.8 Hz, 1H, H-9), 3.62 (dd, J = 11.6, 6.2 Hz, 1H, H-9’), 3.57 (ddd, J = 7.8, 6.2, 2.8 Hz, 1H, H-8), 2.24 (dd, J = 15.2, 5.4 Hz, 1H, H-3ax), 2.05 (d, J = 15.2 Hz, 1H, H-3eq); 13C NMR (100 MHz, D2O) δ 173.9, 158.5 (q, J = 38.0 Hz), 115.7 (q, J = 284.0 Hz), 105.6, 76.8, 76.5, 71.9, 66.1, 62.2, 53.0, 35.4; HRMS (ESI) Anal. Calcd for C11H13NO8F3 [M-H]-: 344.0599, Found: 344.0591.
Chemical derivatization at C-5 of 2,7-anhydro-sialic acids.
5-Amino-2,7-anhydro-3,5-dideoxy-α-D-glycero-D-galacto-non-2-ulopyranosonic acid (2,7-anhydro-Neu, 3).
2,7-Anhydro-Neu5TFA (30.0 mg, 0.0817 mmol) was dissolved in Na2CO3 aqueous solution (2 mL, pH = 9) and the reaction was stirred for 16 h. Without neutralization, the solution was directly passed through a BioGel P-2 gel filtration column to produce 2,7-anhydro-Neu (20.1 mg, 99%) as a white solid. (c 0.25, H2O); 1H NMR (400 MHz, D2O) δ 4.60 (bs, 1H, H-6), 4.42 (dd, 1H, J = 7.8, 0.6 Hz, H-7), 4.09–4.02 (m, 1H, H-4), 3.77 (dd, J = 11.6, 2.8 Hz, 1H, H-9), 3.61 (dd, J = 11.6, 6.0 Hz, 1H, H-9’), 3.56 (ddd, J = 7.8, 6.0, 2.8 Hz, 1H, H-8), 3.16 (bs, 1H, H-5), 2.26 (dd, J = 15.4, 5.6 Hz, 1H, H-3ax), 2.04 (d, J = 15.4 Hz, 1H, H-3eq); 13C NMR (100 MHz, D2O) δ 173.9, 105.7, 77.6, 76.6, 71.9, 67.3, 62.3, 52.8, 34.8; HRMS (ESI) Anal. Calcd for C9H14NO7 [M-H]-: 248.0776, Found: 248.0771.
2,7-Anhydro-5-cyclohexylamido-3,5-dideoxy-α-D-glycero-D-galacto-non-2-ulopyranosonic acid (2,7-anhydro-Neu5Cyclohexyl, 4).
To a stirred solution of 2,7-anhydro-Neu (15.0 mg, 0.0602 mmol) in a mixed solvent of DMF/H2O/AcOH (7:2:1, 2 mL), cyclohexanone (62 μL, 0.60 mmol) and NaBH3CN (38 mg, 0.60 mmol) were added and the reaction was stirred for 30 min. The solvent was concentrated in vacuo. The crude product was re-dissolved in water (1 mL) and was passed through a BioGel P-2 gel filtration column to produce 2,7-anhydro-Neu5Cyclohexyl (20.8 mg, 98%) as a white solid. (c 1.1, H2O); 1H NMR (400 MHz, D2O) δ 4.77 (bs, 1H, H-6), 4.45 (d, J = 7.6 Hz, 1H, H-7), 4.23–4.16 (m, 1H, H-4), 3.77 (dd, J = 11.8, 2.8 Hz, 1H, H-9), 3.62 (dd, J = 11.8, 5.8 Hz, 1H, H-9’), 3.57 (ddd, J = 7.6, 5.8, 2.8 Hz, 1H, H-8), 3.36 (bs, 1H, H-5), 3.21 (bs, 1H, H-1’ hexyl), 2.29 (dd, J = 15.4, 5.8 Hz, 1H, H-3ax), 2.17–1.98 (m, 3H, H-3eq, H hexyl), 1.93–1.77 (m, 2H, H hexyl), 1.75–1.61 (m, 1H, H hexyl), 1.45–1.24 (m, 4H, H hexyl), 1.25–1.09 (m, 1H, H hexyl); 13C NMR (100 MHz, D2O) δ 173.5, 105.6, 76.7, 75.3, 71.8, 64.4, 62.2, 56.0, 55.5, 35.3, 30.0, 29.7, 24.7, 24.19, 24.15; HRMS (ESI) Anal. Calcd for C15H24NO7 [M-H]-: 330.1558, Found: 330.1559.
SpNanB-catalyzed hydrolysis assays of 2,7-anhydro-Neu5Ac (1) and 2,7-anhydro-Neu5Cyclohexyl (4).
2,7-Anhydro-Neu5Ac (1) or 2,7-anhydro-Neu5Cyclohexyl (4) (10 mM final concentration) was treated with SpNanB (1 mg/mL) in NaOAc buffer (100 mM, pH = 6.0) or Tris-HCl buffer (100 mM, pH = 7.0) for 24 h. Thin layer chromatography (TLC) and mass spectrometry showed that most 2,7-anhydro-Neu5Ac (1) was converted to Neu5Ac, whereas 2,7-anhydro-Neu5Cyclohexyl (4) remained intact.
Substrate specificity studies of SpNanB.
Substrate specificity assays were carried out in duplicates in 384-well plates in a final volume of 20 μL in NaOAc buffer (200 mM, pH 5.5) containing a sialoside selected from Siaα2–3GalβpNP or Siaα2–6GalβpNP (0.3 mM), β-galactosidase (12 μg), and SpNanB (0.4 μg). The reactions were incubated for 30 min at 37 °C, and were stopped by adding 40 μL of 0.5 M CAPS buffer (N-cyclohexyl-3-aminopropane sulfonic acid, pH 10.5) to each well. The amount of the para-nitrophenolate formed was determined by measuring the A405 nm of the reaction mixtures using a microplate reader.
Inhibition assays.
Inhibition assays were carried out in duplicates in 384-well plates in a final volume of 20 μL containing Neu5Acα2–3GalβpNP (0.3 mM) and β-galactosidase (12 μg) with or without inhibitors. The assay conditions varied for different sialidases as described below: SpNanA (0.0015 μg), NaOAc buffer (100 mM, pH 6.0); SpNanB (0.003 μg), NaOAc buffer (100 mM, pH 6.0); SpNanC (0.01 μg), MES buffer (100 mM, pH 6.5); AuSialidase (1.0 mU), NaOAc buffer (100 mM, pH 5.5); CpNanI (1.3 mU), MES buffer (100 mM, pH 5.0); VcSialidase (0.57 mU), NaCl (150 mM), CaCl2 (10 mM), NaOAc buffer (100 mM, pH 5.5); PmST1 (0.4 μg), CMP (0.4 mM), NaOAc buffer (100 mM, pH 5.5); BiNanH2 (0.029 μg), NaOAc buffer (100 mM, pH 5.0); hNEU2 (1.2 μg), MES buffer (100 mM, pH 5.0). The reactions were incubated for 30 min at 37 °C, and were stopped by adding 40 μL of 0.5 M CAPS buffer (N-cyclohexyl-3-aminopropane sulfonic acid, pH 10.5) to each well. The amount of the para-nitrophenolate formed was determined by measuring the A405 nm of the reaction mixtures using a microplate reader.
The percentage inhibition was determined using a concentration of 1 mM of each inhibitor. The reaction without any inhibitors was used as a control. IC50 values were obtained by varying the concentrations of inhibitors from 0 to 1000 μM to obtain concentration-response plots of the inhibitors. The values of IC50 were calculated by the software Grafit 5.0.
Supplementary Material
ACKNOWLEDGMENT
This work was partially supported by the National Institutes of Health (NIH) grant R01AI130684.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Tables S1 and S2, Figure S1 and S2, and NMR spectra of products (PDF).
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Fine MJ; Smith MA; Carson CA; Mutha SS; Sankey SS; Weissfeld LA; Kapoor WN Prognosis And Outcomes of Patients with Community-Acquired Pneumonia: A Meta-Analysis. JAMA 1996, 275, 134–141. [PubMed] [Google Scholar]
- (2).Reichler MR; Allphin AA; Breiman RF; Schreiber JR; Arnold JE; McDougal LK; Facklam RR; Boxerbaum B; May D; Walton RO The Spread of Multiply Resistant Streptococcus pneumoniae at a Day Care Center in Ohio. J. Infect. Dis. 1992, 166, 1346–1353. [DOI] [PubMed] [Google Scholar]
- (3).Camara M; Boulnois G; Andrew P; Mitchell T A Neuraminidase from Streptococcus pneumoniae has the Features of a Surface Protein. Infect. Immun. 1994, 62, 3688–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Xu G; Li X; Andrew PW; Taylor GL Structure of the Catalytic Domain of Streptococcus pneumoniae Sialidase NanA. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2008, 64, 772–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Camara M; Mitchell TJ; Andrew PW; Boulnois GJ Streptococcus pneumoniae Produces at least Two Distinct Enzymes with Neuraminidase Activity: Cloning and Expression of a Second Neuraminidase Gene in Escherichia coli. Infect. Immun. 1991, 59, 2856–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Berry AM; Lock RA; Paton JC Cloning and Characterization of NanB, a Second Streptococcus pneumoniae Neuraminidase Gene, and Purification of the NanB Enzyme from Recombinant Escherichia coli. J. Bacteriol. 1996, 178, 4854–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Pettigrew MM; Fennie KP; York MP; Daniels J; Ghaffar F Variation in the Presence of Neuraminidase Genes among Streptococcus pneumoniae Isolates with Identical Sequence Types. Infect. Immun. 2006, 74, 3360–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Xu G; Kiefel MJ; Wilson JC; Andrew PW; Oggioni MR; Taylor GL Three Streptococcus pneumoniae Sialidases: Three Different Products. J. Am. Chem. Soc. 2011, 133, 1718–1721. [DOI] [PubMed] [Google Scholar]
- (9).Xu G; Potter JA; Russell RJ; Oggioni MR; Andrew PW; Taylor GL Crystal Structure of the NanB Sialidase from Streptococcus pneumoniae. J. Mol. Biol. 2008, 384, 436–449. [DOI] [PubMed] [Google Scholar]
- (10).Gut H; King SJ; Walsh MA Structural and Functional Studies of Streptococcus pneumoniae Neuraminidase B: An Intramolecular Trans-sialidase. FEBS Lett. 2008, 582, 3348–3352. [DOI] [PubMed] [Google Scholar]
- (11).Manco S; Hernon F; Yesilkaya H; Paton JC; Andrew PW; Kadioglu A Pneumococcal neuraminidases A and B Both Have Essential Roles During Infection of the Respiratory Tract and Sepsis. Infect. Immun. 2006, 74, 4014–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Burnaugh AM; Frantz LJ; King SJ Growth of Streptococcus pneumoniae on Human Glycoconjugates is Dependent upon the Sequential Activity of Bacterial Exoglycosidases. J. Bacteriol. 2008, 190, 221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Trappetti C; Kadioglu A; Carter M; Hayre J; Iannelli F; Pozzi G; Andrew PW; Oggioni MR Sialic Acid: A Preventable Signal for Pneumococcal Biofilm Formation, Colonization, and Invasion of the Host. J. Infect. Dis. 2009, 199, 1497–1505. [DOI] [PubMed] [Google Scholar]
- (14).Janapatla RP; Hsu MH; Hsieh YC; Lee HY; Lin TY; Chiu CH Necrotizing Pneumonia Caused by NanC-Carrying Serotypes is Associated with Pneumococcal Haemolytic Uraemic Syndrome in Children. Clin. Microbiol. Infect. 2013, 19, 480–486. [DOI] [PubMed] [Google Scholar]
- (15).von Itzstein M The War Against Influenza: Discovery And Development of Sialidase Inhibitors. Nat. Rev. Drug Discov. 2007, 6, 967–974. [DOI] [PubMed] [Google Scholar]
- (16).Chen GY; Chen X; King S; Cavassani KA; Cheng J; Zheng X; Cao H; Yu H; Qu J; Fang D; Wu W; Bai XF; Liu JQ; Woodiga SA; Chen C; Sun L; Hogaboam CM; Kunkel SL; Zheng P; Liu Y Amelioration of Sepsis by Inhibiting Sialidase-Mediated Disruption of the CD24-SiglecG Interaction. Nat. Biotechnol. 2011, 29, 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gut H; Xu G; Taylor GL; Walsh MA Structural Basis for Streptococcus pneumoniae NanA inhibition by Influenza Antivirals Zanamivir And Oseltamivir Carboxylate. J. Mol. Biol. 2011, 409, 496–503. [DOI] [PubMed] [Google Scholar]
- (18).Xiao A; Li Y; Li X; Santra A; Yu H; Li W; Chen X Sialidase-Catalyzed One-Pot Multienzyme (OPME) Synthesis of Sialidase Transition-State Analogue Inhibitors. ACS Catal. 2018, 8, 43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Walther E; Richter M; Xu Z; Kramer C; von Grafenstein S; Kirchmair J; Grienke U; Rollinger JM; Liedl KR; Slevogt H; Sauerbrei A; Saluz HP; Pfister W; Schmidtke M Antipneumococcal Activity of Neuraminidase Inhibiting Artocarpin. Int. J. Med. Microbiol. 2015, 305, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hoffmann A; Richter M; von Grafenstein S; Walther E; Xu ZL; Schumann L; Grienke U; Mair CE; Kramer C; Rollinger JM; Liedl KR; Schmidtke M; Kirchmair J Discovery and Characterization of Diazenylaryl Sulfonic Acids as Inhibitors of Viral and Bacterial Neuraminidases. Front. Microbiol. 2017, 8, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kim CU; Lew W; Williams MA; Liu H; Zhang L; Swaminathan S; Bischofberger N; Chen MS; Mendel DB; Tai CY; Laver WG; Stevens RC Influenza Neuraminidase Inhibitors Possessing a Novel Hydrophobic Interaction in the Enzyme Active Site: Design, Synthesis, and Structural Analysis of Carbocyclic Sialic Acid Analogues with Potent Anti-Influenza Activity. J. Am. Chem. Soc. 1997, 119, 681–690. [DOI] [PubMed] [Google Scholar]
- (22).Hayre JK; Xu G; Borgianni L; Taylor GL; Andrew PW; Docquier JD; Oggioni MR Optimization of a Direct Spectrophotometric Method to Investigate the Kinetics and Inhibition of Sialidases. BMC Biochem. 2012, 13, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Tailford LE; Owen CD; Walshaw J; Crost EH; Hardy-Goddard J; Le Gall G; de Vos WM; Taylor GL; Juge N Discovery of Intramolecular Trans-Sialidases in Human Gut Microbiota Suggests Novel Mechanisms of Mucosal Adaptation. Nat. Commun. 2015, 6, 7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Brear P; Telford J; Taylor GL; Westwood NJ Synthesis and Structural Characterisation of Selective Non-Carbohydrate-Based Inhibitors of Bacterial Sialidases. ChemBioChem 2012, 13, 2374–2383. [DOI] [PubMed] [Google Scholar]
- (25).Umezawa H; Aoyagi T; Komiyama T; Morishima H; Hamada M Purification and Characterization of a Sialidase Inhibitor, Siastatin, Produced by Streptomyces . J. Antibiot. 1974, 27, 963–969. [DOI] [PubMed] [Google Scholar]
- (26).Grienke U; Richter M; Walther E; Hoffmann A; Kirchmair J; Makarov V; Nietzsche S; Schmidtke M; Rollinger JM Discovery of Prenylated Flavonoids with Dual Activity Against Influenza Virus and Streptococcus pneumoniae. Sci. Rep. 2016, 6, 27156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Park JY; Hwan Lim S; Ram Kim B; Jae Jeong H; Kwon HJ; Song GY; Bae Ryu Y; Song Lee W Sialidase Inhibitory Activity of Diarylnonanoid and Neolignan Compounds Extracted from the Seeds of Myristica fragrans. Bioorg. Med. Chem. Lett. 2017, 27, 3060–3064. [DOI] [PubMed] [Google Scholar]
- (28).Walther E; Xu ZL; Richter M; Kirchmair J; Grienke U; Rollinger JM; Krumbholz A; Saluz HP; Pfister W; Sauerbrei A; Schmidtke M Dual Acting Neuraminidase Inhibitors Open New Opportunities to Disrupt the Lethal Synergism between Streptococcus pneumoniae and Influenza Virus. Front. Microbiol. 2016, 7, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Lifely MR; Cottee FH Formation and Identification of Two Novel Anhydro Compounds Obtained by Methanolysis of N-Acetylneuraminic Acid and Carboxyl-Reduced, Meningococcal Polysaccharide. Carbohydr. Res. 1982, 107, 187–197. [Google Scholar]
- (30).Suzuki M; Suzuki A; Yamakawa T; Matsunaga E Characterization of 2,7-Anhydro-N-acetylneuraminic Acid in Human Wet Cerumen. J. Biochem. 1985, 97, 509–515. [DOI] [PubMed] [Google Scholar]
- (31).Crost EH; Tailford LE; Monestier M; Swarbreck D; Henrissat B; Crossman LC; Juge N The mucin-degradation strategy of Ruminococcus gnavus: The importance of intramolecular trans-sialidases. Gut Microbes 2016, 7, 302–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Furuhata K; Takeda K; Ogura H Studies on Sialic Acids XXIV. Synthesis of 2,7-Anhydro-N-acetylneuraminic Acid. Chem. Pharm. Bull. (Tokyo) 1991, 39, 817–819. [Google Scholar]
- (33).Furuhata K; Ogura H Studies on Sialic Acids. XXX. Synthesis of 2,7-Anhydrosialic Acid. Chem. Pharm. Bull. (Tokyo) 1992, 40, 3197–3200. [Google Scholar]
- (34).Asressu KH; Wang CC Concise Synthesis of 2,7-Anhydrosialic Acid Derivatives and Its Application. Carbohydr. Res. 2017, 453–454, 44–53. [DOI] [PubMed] [Google Scholar]
- (35).Gijsen HJ; Qiao L; Fitz W; Wong CH Recent Advances in the Chemoenzymatic Synthesis of Carbohydrates and Carbohydrate Mimetics. Chem. Rev. 1996, 96, 443–474. [DOI] [PubMed] [Google Scholar]
- (36).Koeller KM; Wong CH Synthesis of Complex Carbohydrates and Glycoconjugates: Enzyme-Based and Programmable One-Pot Strategies. Chem. Rev. 2000, 100, 4465–4494. [DOI] [PubMed] [Google Scholar]
- (37).Monestier M; Latousakis D; Bell A; Tribolo S; Tailford LE; Colquhoun IJ; Le Gall G; Yu H; Chen X; Rejzek M; Dedola S; Field RA; Juge N Membrane-Enclosed Multienzyme (MEME) Synthesis of 2,7-Anhydro-Sialic Acid Derivatives. Carbohydr. Res. 2017, 451, 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Luo Y; Li SC; Li YT; Luo M The 1.8 A Structures of Leech Intramolecular Trans-sialidase Complexes: Evidence of Its Enzymatic Mechanism. J. Mol. Biol. 1999, 285, 323–332. [DOI] [PubMed] [Google Scholar]
- (39).Li Y; Yu H; Cao H; Lau K; Muthana S; Tiwari VK; Son B; Chen X Pasteurella multocida Sialic Acid Aldolase: A Promising Biocatalyst. Appl. Microbiol. Biotechnol. 2008, 79, 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Yu H; Yu H; Karpel R; Chen X Chemoenzymatic Synthesis of CMP-Sialic Acid Derivatives by a One-Pot Two-Enzyme System: Comparison of Substrate Flexibility of Three Microbial CMP-Sialic Acid Synthetases. Bioorg. Med. Chem. 2004, 12, 6427–6435. [DOI] [PubMed] [Google Scholar]
- (41).Yu H; Chokhawala H; Karpel R; Yu H; Wu B; Zhang J; Zhang Y; Jia Q; Chen X A Multifunctional Pasteurella multocida Sialyltransferase: A Powerful Tool for the Synthesis of Sialoside Libraries. J. Am. Chem. Soc. 2005, 127, 17618–17619. [DOI] [PubMed] [Google Scholar]
- (42).Sugiarto G; Lau K; Qu J; Li Y; Lim S; Mu S; Ames JB; Fisher AJ; Chen X A Sialyltransferase Mutant with Decreased Donor Hydrolysis and Reduced Sialidase Activities for Directly Sialylating LewisX. ACS Chem. Biol. 2012, 7, 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Angata T; Varki A Chemical Diversity in the Sialic Acids and Related α-Keto Acids: An Evolutionary Perspective. Chem. Rev. 2002, 102, 439–470. [DOI] [PubMed] [Google Scholar]
- (44).Chokhawala HA; Yu H; Chen X High-Throughput Substrate Specificity Studies of Sialidases by Using Chemoenzymatically Synthesized Sialoside Libraries. ChemBioChem 2007, 8, 194–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Liu R; Xu Y; Chen M; Weiwer M; Zhou X; Bridges AS; DeAngelis PL; Zhang Q; Linhardt RJ; Liu J Chemoenzymatic Design of Heparan Sulfate Oligosaccharides. J. Biol. Chem. 2010, 285, 34240–34249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Chen Y; Thon V; Li Y; Yu H; Ding L; Lau K; Qu J; Hie L; Chen X One-Pot three-Enzyme Synthesis of UDP-GlcNAc Derivatives. Chem. Commun. 2011, 47, 10815–10817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Henrissat B A Classification of Glycosyl Hydrolases Based on Amino Acid Sequence Similarities. Biochem. J. 1991, 280, 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Henrissat B; Bairoch A New Families in the Classification of Glycosyl Hydrolases Based on Amino Acid Sequence Similarities. Biochem. J. 1993, 293, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Henrissat B; Bairoch A Updating the Sequence-Based Classification of Glycosyl Hydrolases. Biochem. J. 1996, 316, 695–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Eschenfelder V; Brossmer R Synthesis of p-Nitrophenyl 5-Acetamido-3,5-Dideoxy-α-D-Glycero-D-Galacto-2-Nonulopyranosidonic Acid, a Chromogenic Substrate for Sialidases. Carbohydr. Res. 1987, 162, 294–297. [DOI] [PubMed] [Google Scholar]
- (51).Kodama H; Baum LG; Paulson JC Synthesis of Linkage-Specific Sialoside Substrates for Colorimetric Assay of Neuraminidases. Carbohydr. Res. 1991, 218, 111–119. [DOI] [PubMed] [Google Scholar]
- (52).Tasnima N; Yu H; Li Y; Santra A; Chen X Chemoenzymatic Synthesis of para-Nitrophenol (pNP)-Tagged α2–8-Sialosides and High-Throughput Substrate Specificity Studies of α2–8-Sialidases. Org. Biomol. Chem. 2017, 15, 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Sela DA; Li Y; Lerno L; Wu S; Marcobal AM; German JB; Chen X; Lebrilla CB; Mills DA An Infant-Associated Bacterial Commensal Utilizes Breast Milk Sialyloligosaccharides. J. Biol. Chem. 2011, 286, 11909–11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Li Y; Cao H; Yu H; Chen Y; Lau K; Qu J; Thon V; Sugiarto G; Chen X Identifying Selective Inhibitors Against the Human Cytosolic Sialidase NEU2 by Substrate Specificity Studies. Mol. BioSyst. 2011, 7, 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


