Abstract
Adenosine to inosine (A-to-I) editing is the most abundant form of RNA modification in mammalian cells, which is catalyzed by adenosine deaminase acting on the double-stranded RNA (ADAR) protein family. A-to-I editing is currently known to be involved in the regulation of the immune system, RNA splicing, protein recoding, microRNA biogenesis, and formation of heterochromatin. Editing occurs within regions of double-stranded RNA, particularly within inverted Alu repeats, and is associated with many diseases including cancer, neurological disorders, and metabolic syndromes. However, the significance of RNA editing in a large portion of the transcriptome remains unknown. Here, we review the current knowledge about the prevalence and function of A-to-I editing by the ADAR protein family, focusing on its role in the regulation of gene expression. Furthermore, RNA editing-independent regulation of cellular processes by ADAR and the putative role(s) of this process in gene regulation will be discussed.
Keywords: ADAR, RNA editing, non-coding RNA, Adenosine to inosine
Introduction
The extent and role of A-to-I RNA editing has only recently come to light with the advent of RNA-sequencing technologies. Given that inosine preferentially base pairs with cytosine, A-to-I editing has the potential to drastically alter the coding and structural properties of RNA. The first observation of RNA editing came from the characterization of an activity in Xenopus embryos causing differential migration of RNA:RNA duplexes on a gel [1]. Although this was thought to be a result of duplex unwinding, it was later determined to be caused by the destabilization of RNA duplexes through the C6 deamination of adenosine to inosine (A-to-I) by ADAR [2]. ADAR was later cloned in mammalian cells [3]. The ADAR family is highly conserved across metazoans, with the presence of ADAR members in the earliest-branching metazoan taxa, including sponges and ctenophores [4]. ADAR was later cloned in mammalian cells [3].
Humans have three ADAR genes (Figure 1): ADAR1, ADAR2 (also known as ADARB1 or RED1) and ADAR3 (also known as ADARB2 or RED2). ADAR1 has two major isoforms: an interferon inducible ADAR1 p150 that contains both the Zα and Zβ Z-DNA binding domains, and a constitutive ADAR1 p110 that lacks the N-terminal Zα Z-DNA binding domain [5, 6], while ADAR2 lacks any Z-DNA binding domains [7]. The Zβ domain lacks several conserved residues necessary for Z-DNA binding [8, 9]. Consequently, only the Zα domain has been shown to be able to bind Z-DNA [8], although its function is still unclear and debated. Although ADAR3 has not been shown to possess any catalytic activity, it is able to bind to both single (ssRNA) and double-stranded RNA (dsRNA) [7]. The significance of ADAR3 binding to ssRNA through its arginine-rich R-domain is unknown and this activity has only been demonstrated in vitro. The R-domain of ADAR3 has been shown to act as a nuclear localization sequence by interacting with importin subunit alpha-1 (KPNA2) [10]. The function of ADAR3 is still being explored, although evidence suggests that it may negatively regulate A-to-I editing in the brain and effect transcriptomic plasticity [7, 11]. In C. elegans, the catalytically inactive ADA-1 was found to regulate RNA editing by the catalytically active ADA-2 (Washburn 2014 Cell Reports), although it is unclear if a similar mechanism exists in humans.
Figure 1.
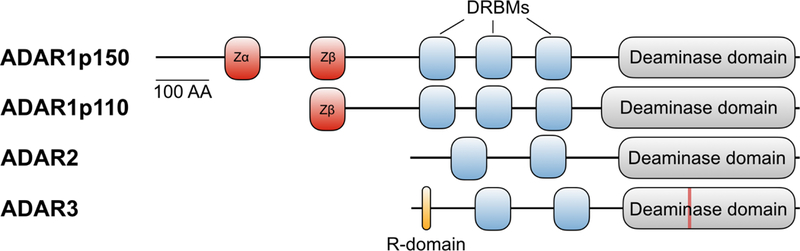
Human ADAR protein family. All ADAR family members contain double stranded RNA binding motifs (DRBMs) and a deaminase domain, which is mutated at a critical residue in ADAR 3 (red line). ADAR1 is expressed as a constitutive ADAR1p110 isoform, or as an interferon inducible ADAR1p110. AA, amino acids; Zα, Zα DNA binding motif; Zβ, Zβ DNA binding motif; R-domain, arginine-rich domain.
Both ADAR1 and ADAR2 are essential for normal growth and development. Loss of ADAR1 is embryonically lethal in mice by embryonic day E12.5 [12] due to impaired hematopoiesis and the development of severe type-I interferonopathy [13]. ADAR1 deficient hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs) show abolished capacity to repopulate an irradiated recipient, suggesting that ADAR1 has a role in both the differentiation and survival of these cells [14]. Furthermore, ADAR1 is essential for HSC and HPC maintenance in the fetal liver and bone marrow [13] and induced expression of a catalytically dead mutant of ADAR in HSCs results in mice developing severe hematopoietic defects [15]. Embryonic lethality of ADAR1 knockout mice can be rescued by subsequent deletion of MAVS [16], a protein that induces type-I interferon and NF-κB signaling, or the upstream effector MDA5 (also known as IFIH1) [17, 18] that is involved in the viral defense pathway by acting as a sensor for dsRNA [19]. This points to the role of ADAR editing in negatively regulating the MAVS-MDA5 pathway and preventing activation of an immune response by endogenous transcripts by destabilizing host-derived dsRNA structures. This destabilization prevents MDA5 from forming filaments and triggering an immune response as MDA5 is sensitive to bulges and mismatches in dsRNA [20] (see [21] for a more detailed overview). ADAR1 is also able to inhibit RIG-I activation by endogenous dsRNAs by directly competing with its binding for the substrate [22]. Both the editing and binding activities of ADAR are necessary to first of all suppress endogenous interferon responses and further prevent translational shutdown during such a response [23]. Mice harboring a genetic knockout of ADAR2 are born at normal mendelian ratios, suggesting that ADAR2 is dispensable for embryological development [24]. However, postnatal lethality is observed in ADAR2 null mice due to progressively worse brain seizures which result in death within 3 weeks of birth [24].
Both ADAR1 and ADAR2 have been reported to form homodimers and heterodimers with each other [25] presenting the possibility of multiple regulatory mechanisms controlling editing Recently, the editing and stability of CTN-RNA was found to be dependent upon the interaction between ADAR1 and ADAR2 in cells [26], suggesting that editing of select RNA substrates is dependent upon the cooperativity between these two ADAR members. It is however, unclear if this editing occurs through the heterodimerization of ADAR1 and ADAR2, or rather whether binding and editing of CTN-RNA by one ADAR isoform promotes the binding and editing activity of the other. Discrepancy between in vitro and in vivo heterodimerization data may suggest that post-translational modifications or other protein binding partners may be required to facilitate this interaction. The expression of ADAR3 however is restricted to certain brain regions and post-mitotic neurons [7, 27].
Role of A-to-I editing
Protein recoding
Given that A-to-I editing changes the base pairing properties of RNA, it is easy to envision its ability to alter the amino acid sequence of a protein. Although this is not the major role of ADAR editing, there are many notable examples where RNA editing changes protein function. ADAR2 editing of the AMPA type glutamate receptor 2 subunit (GRIA2) occurs at near 100% efficiency and results in an amino acid change of a glutamine to an arginine (Q/R) [28]. Editing of the Q/R site reduces Ca2+ permeability through the receptor by up to 30 times [24, 28]. This is essential for survival and genetic mutation of Q/R site rescues postnatal lethality of ADAR2 knockout mice [24]. Our understanding of why editing of the Q/R site occurs and is conserved [29] is incomplete, as a single point mutation could abrogate the need for any editing to occur. One postulated idea is that editing of GRIA2 may play a role in neural cell development and differentiation [30]. While earlier studies did not observe any obvious architectural differences in brain slices of GRIA2R/R mice [31], this idea cannot yet be discredited as changes may also occur at the microscopic or biochemical level. Recently, ADAR3 was found to be an important negative regulator of the editing of GRIA2 [11]. Notably, hypo-editing of GRIA2 in the setting of glioblastoma [32] is accompanied by an increase in ADAR3 protein expression [11]. The dsRNA binding domains of ADAR3 were found to be necessary for this activity, suggesting that ADAR3 is directly competing for editing site binding with other ADAR family members.
Many known examples of protein recoding by RNA editing require the formation of specific dsRNA structures (see [33] for a more comprehensive overview) and occur at regions where the underlying DNA sequence is evolutionarily conserved, although overall conservation between all identified ADAR targets is very low [34]. Recently, cephalopods have been found to be able to undergo extensive A-to-I editing in order to diversity their proteome, especially in neural cells in which recoding leads to alterations in neural excitability and morphology [35]. However, this appears to be an event specific to cephalopods and occurs at the expense of genomic diversity, as all recoding sites are highly conserved within the taxon [35].
RNA export, splicing and stability
ADAR1 can shuttle in and out of the nucleus through interactions with the nuclear import receptor transportin-1 [36] and the nuclear export receptor exportin-5 [36, 37]. While nuclear import of ADAR is inhibited by its binding of dsRNA substrates, nuclear export is not [36]. As such, it is believed that ADAR1 may mediate the export of specific RNAs out of the nucleus (Figure 2). A-to-I editing itself mediates nuclear retention of hyper-edited dsRNAs through selective binding of inosine-rich RNAs to NONO (Figure 2) [38]. One function of nuclear retention is to permit those RNAs that are not required in the cytoplasm under normal conditions to be quickly exported immediately after some form of cellular stress. The classical example of nuclear retention involves modulation of the levels of mCAT2, a protein encoding for an arginine transporter and involved in the nitric oxide pathway [39]. CTN-RNA is transcribed from the mCAT2 gene using an alternate upstream promoter and contains an extended 3’UTR sequence [40]. The 3’UTR of CTN-RNA forms a dsRNA structure that is extensively bound and edited by ADAR2, promoting its stabilization and half-life [26, 40, 41]. Upon cellular stress, the 3’UTR of CTN-RNA is cleaved which allows for its subsequent nuclear export into the cytoplasm where it results in increased production of the mCAT2 protein [40]. A similar mechanism has been found for the Nicolin 1 gene [42]. It is feasible that A-to-I hyper-edited sites exist in a subset of RNAs that are retained in the nucleus to be released under specific stimuli. As a large proportion of A-to-I editing occurs within intronic sequences [43], an obvious function of A-to-I editing mediated nuclear retention is to retain incorrectly spliced or un-spliced transcripts in the nucleus. Interestingly, as paraskeckles are not found in human embryonic stem cells (ESCs) due to the lack of the non-coding RNA NEAT1 and only appear upon ESC differentiation, there is no nuclear retention of edited RNAs [44] despite the high level of ADAR editing in ESCs [45]. Although there is ongoing debate as to whether paraspeckles are essential for cellular function, as mice lacking NEAT1 are viable and display no obvious phenotypic changes under laboratory condition [46], many argued that they may only be necessary during times of stress [47]. This idea is in line with the functional retention in edited RNAs for quick export to the cytoplasm in the event of a stimulus. For instance, NEAT1 is dramatically upregulated following viral infection and exerts antiviral effects [48] and NEAT1 knockout mice fail to become pregnant [49], showing clear phenotypic effects. It would be interesting to see the effects of challenging NEAT1 knockout mice with infection or a tumor on mouse survival.
Figure 2.
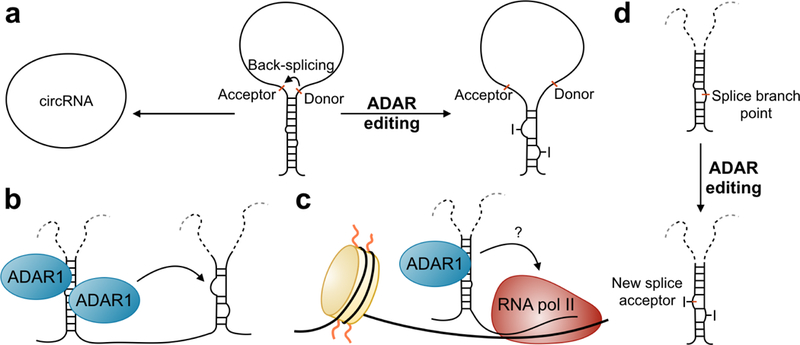
ADAR editing at inverted Alu repeat regions. Editing of Alu repeats can antagonize circRNA formation (a), induce editing of regions in-cis by bringing ADAR in proximity to a less-favorable binding site (b), effect transcription (c), and create or destroy splice sites (d).
A-to-I editing and N6-methyladenosine (m6A) RNA modifications are generally mutually exclusive [50]. A-to-I edited RNA residues can no longer be methylated by the m6A complex and 6mA modified RNA is a poor substrate for A-to-I editing [51]. Indeed, there is a negative correlation between m6A and levels of A-to-I editing in cells [50] suggesting that these two types of RNA modifications can directly antagonize the functions of one another.
A-to-I editing has been heavily implicated in the regulation of RNA splicing by introducing a splice donor or acceptor site, or abolishing a splice branch site. ADAR2 self-regulates its expression and catalytic activity by modulating its own alternative splicing [52]. A-to-I editing by ADAR2 can convert an intronic AA dinucleotide into an AI dinucleotide that can function like the canonical AG 3’ splice site acceptor [52]. The ability of editing events to alter splicing has been documented for several other genes [53–56]. As many such editing events occur in the context of inverted Alu repeats, it has been argued that Alu expansion in the human genome helped to promote transcript diversity through alternative splicing and exonization (reviewed in [57]). The binding of ADAR to RNA has also been proposed to interfere with spliceosomal assembly [58], which could in part explain the role of ADAR in controlling aberrant exonization of Alu elements in the human genome [59].
ADAR is directly involved in regulating the stability of certain RNA species inside the cell. The binding of ADAR2 was found to antagonize the binding of the RNA decay-promoting protein PARN, an RNA deadenylase, to CTN-RNA in an editing independent manner [41]. Likewise, ADAR1 binding to inverted Alu repeats in the 3’UTR of multiple transcripts is believed to directly inhibit the binding of the RNA-decay protein and stress-response gene Stuaufen-1 [60]. This function of ADAR1 acts to protect cells from stress-induced apoptosis in an editing independent manner [60]. Alternatively, ADAR1 interaction with HuR increases transcript stability [61]. HuR belongs to the embryonic lethal abnormal vision (ELAV), an RNA-binding protein family that preferentially binds to single stranded AU-rich RNA sequences [62, 63]. AU-rich elements (AREs) are found in the 3’UTR of up to 8% of all mRNAs, including cytokines and cell cycle genes [64, 65], and are one of the best characterized elements involved in RNA instability. The binding of ADAR1 and HuR to such RNAs is cooperative to increase transcript stability as knockdown of ADAR1 abolishes HuR binding [61].
RNAi
Given that miRNA biogenesis requires the formation of dsRNA intermediates, it is easy to envision the role of ADAR in this process. ADAR can antagonize miRNA biogenesis through editing dependent or independent means. Editing of pri-miRNA or pre-miRNAs can result in many outcomes that depend upon the location and extend of A-to-I editing. RNA editing has large and global effects on RNA secondary structure [66]. Editing of a particular pri-miRNA can suppress its subsequent processing by Drosha [67] or DICER [68]. Alternatively, if editing occurs in the “seed” region of the miRNA it has the potential to alter its target binding sites and change the function of the miRNA as it will now silence a completely different set of genes, as has been demonstrated for miR-376 [69] and a miRNA encoded by human herpesvirus [70]. ADAR protein binding can also suppress miRNA biogenesis and the efficacy of siRNA by directly competing with RNAi processing machinery for the binding of the pre-miRNA in an editing-independent fashion [71, 72]. More recently, ADAR1 has been shown to heterodimerize with DICER to stimulate miRNA biogenesis [73–75], although what determines the formation of this interaction between DICER and ADAR to facilitate either a stimulatory or inhibitory effect remains to be determined. Furthermore, ADAR binding to RNA can antagonize miRNA by reducing the accessibility go the AGO2-miRNA complex to its target site [76].
Heterochromatin formation and gene regulation
Vigilin is an RNA binding protein that contains 14 K-homology (KH) domains and has been implicated in both heterochromatin formation and chromosomal segregation in Drosophila and humans [77, 78]. The C-terminal domain of Vigilin has been shown to interact with SUV39H1 [78], a histone methyltransferase involved in depositing the heterochromatic H3K9me3 mark [79]. Vigilin has been shown to bind to hyper-edited RNAs (predominantly inverted Alu repeats) (Figure 3) and interact with both ADAR and the Ku70/86 complex that is involved in non-homologous end joining (NHEJ) [79], suggesting that ADAR editing may play a role in both the formation of heterochromatin and the DNA repair pathway. However, a direct role of ADAR in either of these processes has never been shown. Of note, the DNA damage response (DDR) results in an increase in type I interferon signaling [80]. In certain cell types including macrophages, this interferon response may promote the DDR pathway [81]. Given that ADAR edited RNAs have been implicated in NHEJ and type I interferon signaling promotes expression of ADAR1p150, it would be interesting to see if this protein has any role in the DDR.
Figure 3.
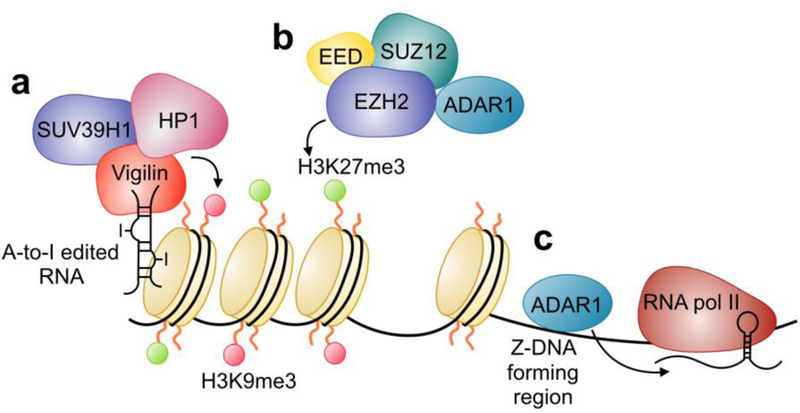
ADAR control of heterochromatin and gene transcription. A-to-I edited RNA binds to Vigilin to influence heterochromatin deposition (a). ADAR interaction with the PRC2 complex may regulate the deposition of H3K27me3 and gene silencing (b). ADAR binding to Z-DNA in transcribed regions could aid in recruiting the enzyme to newly formed transcripts.
Several recent mass spectrometry screens have identified ADAR1 enrichment in heterochromatin [82] with the H3K9me3 mark [83, 84], as well as in association with the Polycomb Repressive Complex (PRC) components EED and PRC1 [85], suggesting that ADAR may directly play a role in the formation of heterochromatin (Figure 3) in an editing-dependent or editing-independent way. The Zα domain has been shown to be involved in protein localization to cytoplasmic stress granules [86, 87] and viral defense [88]. Z-DNA is a left-helical form of DNA [89] that may be transiently formed in alternating purine-pyrimidine DNA stretches in response to torsional stress [90, 91] such as negative supercoiling that arises in the region behind a moving polymerase [92]. Expression of the Zα domain of human ADAR1 fused to the Gal4 activation domain in yeast enhanced transcription from a reporter containing a poly[d(CG)] Z-DNA forming region in its promoter [93], suggesting that ADAR1 p150 may also have an editing independent role in transcriptional regulation (Figure 3), though this has yet to be experimentally determined. As ADAR1 p150 is an interferon inducible transcript that may have the capability of binding to Z-DNA it is tempting to speculate that ADAR1 p150 may regulate gene expression during an immune response.
Regulation of A-to-I editing
Editing-site selectivity
Early studies suggested that inosine was present at levels up to 1 in every 17,000 nucleotides in rat tissue [94], making it one of the most abundant mammalian RNA modifications. Moreover, RNA editing is at least 100 times more prevalent in primates compared to rodents [95], likely due to the high abundance of Alu elements in our genomes which may contribute to transcriptomic complexity [96, 97]. However, due to the difficulties of detecting A-to-I editing events, we have only recently characterized a substantial proportion of the editing sites in human tissues. ADAR editing requires the formation of double-stranded RNA [2]. Interestingly, both dsRNA and DNA:RNA duplexes have structural similarities as they both form an A-form helix [98, 99]. While, in vitro studies showed the ability of both ADAR1 and ADAR2 to modify the DNA strand of a DNA-RNA duplex [100], no significant DNA editing was observed above background levels when human ADAR was fused to Cas9 which would put ADAR in the context of such a duplex [101]. Interestingly, deamination of the RNA strand of DNA:RNA duplexes was shown in vitro [100], presenting the possibility of RNA editing in the absence of double-stranded RNA formation, although this is yet to be shown in vivo. However, EMSA data has also shown that binding of the catalytic domain of ADAR2 to a target substrate is sensitive to 2’ deoxy substitutions opposite the editing site [102], although this study was never carried out with the full-length enzymes.
The mechanism guiding editing site selectivity are poorly understood. While ADAR1 and ADAR2 can edit some of the same substrates, most substrates can only be edited by one of the two enzymes. Prediction of ADAR editing sites remains poor [103]. ADAR editing involves a base-flipping mechanism in which the reactive base is flipped out of the RNA duplex to be accessed by the active site of the enzyme [104]. Due to the structural constraints of such a feat, there is a nearest neighboring preference for A or U on the 5’ side and G on the 3’ side of the A to be edited [104, 105]. Catalytic activity is further stimulated if there is a C mismatch opposite the base to be edited [106]. Selectivity of editing sites is contributed to by both the catalytic domain and the dsRBDs [103, 107].
To date, over 400,000 individual editing events have been reported in the human transcriptome [108]. However, earlier studies estimated that up to 100 million editing sites may exist in the human genome, with many editing events occurring at levels of under 1% [109]. This discrepancy is due to the computational difficulty of identifying bona fide editing sites within sequencing data, given that editing sites must be discriminated from single nucleotide polymorphisms (SNPs) and the difficulty of identifying editing sites in repetitive regions of the genome where they are most abundant and hyper editing tends to occur. Moreover, it is unclear if such low editing frequencies are functionally relevant. The majority (97.7%) of editing sites occur within repetitive sites, particularly in the context of inverted Alu repeats [108].
A-to-I editing events occur in either a site specific or promiscuous manner. Site-specific editing occurs at sites that are more conserved in the genome and form imperfect dsRNA structures and includes editing of coding regions and miRNA sites [34, 95, 110]. Promiscuous editing however occurs in the context of long dsRNA structures that have few bulges and mismatches [111], making inverted Alu repeats perfect candidates for this type of editing. Alu elements are ~300 nucleotide primate-specific short interspersed nuclear elements (SINEs) that comprise 10% of the human genome. Given that Alu elements frequently occur in clusters and are over-represented in gene-rich regions of the genome [112], this presents the intriguing notion that Alu elements may have been co-opted by the ADAR machinery to diversify the human transcriptome. Indeed, editing of inverted Alu repeats has been shown to be involved in many processes including circular RNA biogenesis, induction of editing in cis, transcriptional elongation and splicing (Figure 4).
Figure 4.
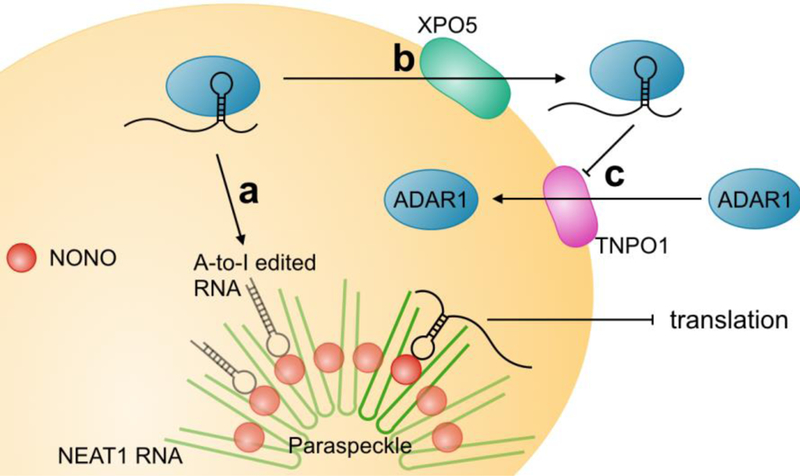
ADAR control of RNA export. Hyper-editing of RNA by ADAR causes RNA retention through binding of inosine-rich RNA to the RNA-binding protein NONO in paraspeckles (a). While RNA-bound ADAR can be effectively exported from the nucleus (b), nuclear import is inhibited. Only free ADAR is able to shuttle back into the nucleus (c).
Circular RNAs (circRNAs) are a class of non-coding RNAs (ncRNAs) that have a variety of cellular functions including acting as micro RNA (miRNA) sponges [113], regulating RNA polymerase II associated transcription [114], and interacting with RNA-binding proteins to perform diverse functions such as controlling cell cycle progression [115]. One of the major methods of circRNA biogenesis involves the presence of reverse complementary sequences such as Alu repeats flanking the RNA and promoting back-splicing and circularization [116]. ADAR editing negatively regulates circRNA biogenesis by editing and destabilizing the flanking Alu repeat sequences, making circRNA production less favorable [117]. Alu editing has also been shown to act as cis-acting editing inducer elements (EIEs) for distant sites on the same RNA molecule and allow for the editing of short double-stranded sites that could not otherwise be edited efficiently by ADAR [118]. This has been shown for a number of transcripts including the I/M editing site in the Gabra transcript [118] and the Q/R editing site of the GluA2 transcript [119]. Transcriptome wide analyses have shown that adjacent Alu sequences inserted in the same orientation are overrepresented in the human genome, compared to those inserted in an inverted orientation [120–122]. The presence of inverted Alu elements within a gene appeared to interfere with RNA Pol II activity as polymerase occupancy was decreased downstream of a transcribed inverted Alu region, indicating that the double-stranded structure formed by the RNA influences polymerase activity and gene expression [122]. This secondary structure formation by inverted repeat Alus appears to influence the kinetics of RNA polymerase, enabling for a global mechanism to control for gene expression rate. Indeed, transcripts containing inverted Alu elements are more lowly expressed compared to those that do not [57, 122]. Others have found that the presence of inverted Alu sequences in the 3’UTR has the potential to alter translation efficiency [123]. Whether ADAR editing of these inverted repeats has implications for both transcription and translation by possibly destabilizing and alternating the double-stranded structures or recruiting a different repertoire of RNA binding proteins is an unanswered question in the field.
The expression of ADAR1 and ADAR2 has been reported to be differentially regulated by the DNA-methyltransferase (DNMT) inhibitor 5’-azacytidine and the global histone deacetylase (HDAC) inhibitor trichostatin A [124], suggesting an interplay between global gene expression and ADAR activity. DNMT inhibitors are known to induce interferon signaling through the upregulation of endogenous retroviruses (ERVs) [125]. Consequently, differential regulation of ADAR activity in the setting of DNMT and HDAC inhibitors could be part of an endogenous protective mechanism against ERVs.
A-to-I editing in disease
Mutations in ADAR1 are known to contribute to or cause several inflammatory disorders including Aicardi-Goutières syndrome (AGS) [126] and systemic lupus erythematosus (SLE) [127]. While homozygous mutations in ADAR that abolish its catalytic activity are embryonically lethal, heterozygous mutations cause dyschomatosis symmetrica hereditaria (DSH) [128]. DSH often manifests simply as a skin disease causing hyper- or hypo-pigmentation of the hands and feet and rarely results in neurological deterioration [129] although the pathogenesis of this disorder is unknown. Altered levels of A-to-I editing has also been reported for many cognitive and psychiatric disorders including Alzheimers [130, 131], depression [132] and schizophrenia [133, 134]. Interestingly, editing of the serotonin 2C receptor (5-HT2C) is altered in depressed suicide patients [135, 136]. Treatment of mice with fluoxetine (Prozac), a drug commonly used to treat depression, resulted in editing changes to 5-HT2C that were exactly opposite to those seen in depressed suicide patients [135], suggesting that one mechanism through which this drug works is by targeting and altering ADAR editing in neural tissue. ADAR2 may also play a role in the pathogenesis of type 2 diabetes (T2D), as knockdown of ADAR2 in rat insulinoma cells and primary pancreatic islets reduced glucose-stimulated insulin secretion [137]. Furthermore, ADAR2 is metabolically regulated in islet cells [138].
Finally, ADAR activity has been heavily implicated in the pathogenesis and progression of many cancer types including lung, breast and blood cancers [139–143]. For example, editing of focal adhesion kinase (FAK) RNA by ADAR promotes transcript stability and results in increased FAK protein expression that enhances cancer cell invasion and metastasis [139]. As another example, JAK/STAT signaling was shown to increase the expression of ADAR1 in leukemic stem cells, allowing for the editing and impairment of let-7 miRNA biogenesis [142]. Let-7 has known tumor suppressor functions [144] and its expression must be suppressed in stem cells to allow for self-renewal [145]. Genome-wide studies have identified many editing sites that are hypo- or hyper-edited in the setting of cancer [146]. Many of these sites are found in non-coding regions, however a subset of these editing sites were identified to result in non-synonymous RNA mutations. For example, editing of antizyme inhibitor 1 (AZIN1) by ADAR1 in the context of liver cancer confers increased cell proliferation by promoting AZIN1 protein stability and increasing polyamine production [147]. For a more detailed review on the role of ADAR enzymes in cancer, see [148, 149].
Conclusions
In conclusion, we show the pervasive role of ADAR in the regulation of many aspects of RNA function, including regulating the biogenesis of circRNA, miRNA, RNA export and splicing. The advent of sequencing technologies has allowed us to better understand the breadth of RNA editing and its contribution to transcriptomic diversity in a variety of cell, tissue, and disease settings. However, many outstanding question remain in the field including: (1) how ADAR activity is regulated in cells by nucleo-cytoplasmic shuttling, or by post-translational modifications and other protein-protein interactions, (2) the role of ADAR in gene silencing by heterochromatin, (3) whether ADAR activity influenced the evolution and exaptation of Alu elements in the human genome, (4) the prevalence and function of nuclear retained hyper-edited RNAs, (5) the interplay between A-to-I editing and 6mA base modification and how these modifications influence target gene expression, as well as (6) the role of the RNA editing enzymes in cancer progression and autoimmune disorders.
Acknowledgements
This project was supported by NIAID PO1 AI099783–01, AI111139–01 and NIDDK DK104681–01 and NIMH R01 113407–01 to KVM.
Footnotes
The authors declare no conflict of interest.
Literature Cited
- 1.Bass BL & Weintraub H (1987) A developmentally regulated activity that unwinds RNA duplexes, Cell 48, 607–613. [DOI] [PubMed] [Google Scholar]
- 2.Bass BL & Weintraub H (1988) An unwinding activity that covalently modifies its double-stranded RNA substrate, Cell 55, 1089–98. [DOI] [PubMed] [Google Scholar]
- 3.O’Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A & Keller W (1995) Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase, Molecular and cellular biology 15, 1389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grice LF & Degnan BM (2015) The origin of the ADAR gene family and animal RNA editing, BMC Evolutionary Biology 15, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George CX & Samuel CE (1999) Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible, Proceedings of the National Academy of Sciences 96, 4621–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herbert A, Alfken J, Kim Y-G, Mian IS, Nishikura K & Rich A (1997) A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase, Proceedings of the National Academy of Sciences 94, 8421–8426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CX, Cho DS, Wang Q, Lai F, Carter KC & Nishikura K (2000) A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains, RNA (New York, NY) 6, 755–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YG, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL & Rich A (2003) A role for Z-DNA binding in vaccinia virus pathogenesis, Proceedings of the National Academy of Sciences of the United States of America 100, 6974–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Athanasiadis A, Placido D, Maas S, Brown BA 2nd, Lowenhaupt K & Rich A (2005) The crystal structure of the Zbeta domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains, Journal of molecular biology 351, 496–507. [DOI] [PubMed] [Google Scholar]
- 10.Maas S & Gommans WM (2009) Identification of a selective nuclear import signal in adenosine deaminases acting on RNA, Nucleic Acids Research 37, 5822–5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oakes E, Anderson A, Cohen-Gadol A & Hundley HA (2017) Adenosine Deaminase That Acts on RNA 3 (ADAR3) Binding to Glutamate Receptor Subunit B Pre-mRNA Inhibits RNA Editing in Glioblastoma, Journal of Biological Chemistry 292, 4326–4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mannion Niamh M., Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting Chris P., McLaughlin Paul J., Jantsch Michael F., Dorin J, Adams Ian R., Scadden ADJ, Öhman M., Keegan Liam P. & O’Connell Mary A. (2014) The RNA-Editing Enzyme ADAR1 Controls Innate Immune Responses to RNA, Cell Reports 9, 1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartner JC, Walkley CR, Lu J & Orkin SH (2009) ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling, Nature immunology 10, 109–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.XuFeng R, Boyer MJ, Shen H, Li Y, Yu H, Gao Y, Yang Q, Wang Q & Cheng T (2009) ADAR1 is required for hematopoietic progenitor cell survival via RNA editing, Proceedings of the National Academy of Sciences 106, 17763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piskol R, Chalk A, Higuchi M, Seeburg P, Li JB, Hartner J & Walkley CR (2013) A-To-I RNA Editing By ADAR1 Is Essential For Hematopoiesis, Blood 122, 1199. [Google Scholar]
- 16.Seth RB, Sun L, Ea CK & Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3, Cell 122, 669–82. [DOI] [PubMed] [Google Scholar]
- 17.Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM & Stetson DB (2015) Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development, Immunity 43, 933–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH & Walkley CR (2015) RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself, Science (New York, NY) 349, 1115–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T & Akira S (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses, Nature 441, 101–5. [DOI] [PubMed] [Google Scholar]
- 20.Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, Zhang C-Z & Hur S (2018) Breaching Self-Tolerance to Alu Duplex RNA Underlies MDA5-Mediated Inflammation, Cell 172, 797–810.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liddicoat BJ, Chalk AM & Walkley CR (2016) ADAR1, inosine and the immune sensing system: distinguishing self from non-self, Wiley Interdisciplinary Reviews: RNA 7, 157–172. [DOI] [PubMed] [Google Scholar]
- 22.Yang S, Deng P, Zhu Z, Zhu J, Wang G, Zhang L, Chen AF, Wang T, Sarkar SN, Billiar TR & Wang Q (2014) ADAR1 Limits RIG-I RNA Detection and Suppresses IFN Production Responding to Viral and Endogenous RNAs, Journal of immunology (Baltimore, Md : 1950) 193, 3436–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung H, Calis JJA, Wu X, Sun T, Yu Y, Sarbanes SL, Dao Thi VL, Shilvock AR, Hoffmann HH, Rosenberg BR & Rice CM (2018) Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown, Cell 172, 811–824.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R & Seeburg PH (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2, Nature 406, 78–81. [DOI] [PubMed] [Google Scholar]
- 25.Chilibeck KA, Wu T, Liang C, Schellenberg MJ, Gesner EM, Lynch JM & MacMillan AM (2006) FRET analysis of in vivo dimerization by RNA-editing enzymes, The Journal of biological chemistry 281, 16530–5. [DOI] [PubMed] [Google Scholar]
- 26.Anantharaman A, Gholamalamdari O, Khan A, Yoon J-H, Jantsch MF, Hartner JC, Gorospe M, Prasanth SG & Prasanth KV (2017) RNA-editing enzymes ADAR1 and ADAR2 coordinately regulate the editing and expression of Ctn RNA, FEBS Letters 591, 2890–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F & Uhlen M (2014) Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics, Molecular & cellular proteomics : MCP 13, 397–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R & Seeburg PH (1993) RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency, Cell 75, 1361–70. [DOI] [PubMed] [Google Scholar]
- 29.Li IC, Chen Y-C, Wang Y-Y, Tzeng B-W, Ou C-W, Lau Y-Y, Wu K-M, Chan T-M, Lin W-H, Hwang S-PL & Chow W-Y (2014) Zebrafish Adar2 Edits the Q/R Site of AMPA Receptor Subunit gria2α Transcript to Ensure Normal Development of Nervous System and Cranial Neural Crest Cells, PLOS ONE 9, e97133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Whitney NP, Peng H, Erdmann NB, Tian C, Monaghan DT & Zheng JC (2008) Calcium-permeable AMPA receptors containing Q/R-unedited GluR2 direct human neural progenitor cell differentiation to neurons, The FASEB Journal 22, 2888–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kask K, Zamanillo D, Rozov A, Burnashev N, Sprengel R & Seeburg PH (1998) The AMPA receptor subunit GluR-B in its Q/R site-unedited form is not essential for brain development and function, Proceedings of the National Academy of Sciences of the United States of America 95, 13777–13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maas S, Patt S, Schrey M & Rich A (2001) Underediting of glutamate receptor GluR-B mRNA in malignant gliomas, Proceedings of the National Academy of Sciences 98, 14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishikura K (2016) A-to-I editing of coding and non-coding RNAs by ADARs, Nature reviews Molecular cell biology 17, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinto Y, Cohen HY & Levanon EY (2014) Mammalian conserved ADAR targets comprise only a small fragment of the human editosome, Genome Biology 15, R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liscovitch-Brauer N, Alon S, Porath HT, Elstein B, Unger R, Ziv T, Admon A, Levanon EY, Rosenthal JJC & Eisenberg E Trade-off between Transcriptome Plasticity and Genome Evolution in Cephalopods, Cell 169, 191–202.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fritz J, Strehblow A, Taschner A, Schopoff S, Pasierbek P & Jantsch MF (2009) RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1, Molecular and cellular biology 29, 1487–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulsen H, Nilsson J, Damgaard CK, Egebjerg J & Kjems J (2001) CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain, Molecular and cellular biology 21, 7862–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z & Carmichael GG (2001) The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs, Cell 106, 465–75. [DOI] [PubMed] [Google Scholar]
- 39.MacLeod CL (1996) Regulation of cationic amino acid transporter (CAT) gene expression, Biochemical Society transactions 24, 846–52. [DOI] [PubMed] [Google Scholar]
- 40.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ & Spector DL (2005) Regulating gene expression through RNA nuclear retention, Cell 123, 249–63. [DOI] [PubMed] [Google Scholar]
- 41.Anantharaman A, Tripathi V, Khan A, Yoon J-H, Singh DK, Gholamalamdari O, Guang S, Ohlson J, Wahlstedt H, Öhman M, Jantsch MF, Conrad NK, Ma J, Gorospe M, Prasanth SG & Prasanth KV (2017) ADAR2 regulates RNA stability by modifying access of decay-promoting RNA-binding proteins, Nucleic Acids Research 45, 4189–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen LL, DeCerbo JN & Carmichael GG (2008) Alu element-mediated gene silencing, EMBO J 27, 1694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Athanasiadis A, Rich A & Maas S (2004) Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome, PLoS biology 2, e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen L-L & Carmichael GG (2009) Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA, Molecular cell 35, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osenberg S, Paz Yaacov N, Safran M, Moshkovitz S, Shtrichman R, Sherf O, Jacob-Hirsch J, Keshet G, Amariglio N, Itskovitz-Eldor J & Rechavi G (2010) Alu Sequences in Undifferentiated Human Embryonic Stem Cells Display High Levels of A-to-I RNA Editing, PLoS ONE 5, e11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakagawa S, Naganuma T, Shioi G & Hirose T (2011) Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice, The Journal of Cell Biology 193, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leslie M (2011) Paraspeckles may provide stress relief, The Journal of Cell Biology 193, 2–2. [Google Scholar]
- 48.Ma H, Han P, Ye W, Chen H, Zheng X, Cheng L, Zhang L, Yu L, Wu X, Xu Z, Lei Y & Zhang F (2017) The Long Noncoding RNA NEAT1 Exerts Antihantaviral Effects by Acting as Positive Feedback for RIG-I Signaling, Journal of virology 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC & Hirose T (2014) The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice, Development (Cambridge, England) 141, 4618–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiang J-F, Yang Q, Liu C-X, Wu M, Chen L-L & Yang L (2018) N6-Methtladenosines Modulate A-to-I RNA Editing, Molecular Cell 69, 126–135.e6. [DOI] [PubMed] [Google Scholar]
- 51.Véliz EA, Easterwood LM & Beal PA (2003) Substrate Analogues for an RNA-Editing Adenosine Deaminase: Mechanistic Investigation and Inhibitor Design, Journal of the American Chemical Society 125, 10867–10876. [DOI] [PubMed] [Google Scholar]
- 52.Rueter SM, Dawson TR & Emeson RB (1999) Regulation of alternative splicing by RNA editing, Nature 399, 75–80. [DOI] [PubMed] [Google Scholar]
- 53.Flomen R, Knight J, Sham P, Kerwin R & Makoff A (2004) Evidence that RNA editing modulates splice site selection in the 5-HT2C receptor gene, Nucleic Acids Res 32, 2113–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E & Rechavi G (2010) Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates, Proceedings of the National Academy of Sciences of the United States of America 107, 12174–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A & Ast G (2007) Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome, Genome Biology 8, R127.-R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E & Ast G (2007) RNA-editing-mediated exon evolution, Genome Biol 8, R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tajaddod M, Jantsch MF & Licht K (2016) The dynamic epitranscriptome: A to I editing modulates genetic information, Chromosoma 125, 51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Licht K, Kapoor U, Mayrhofer E & Jantsch MF (2016) Adenosine to Inosine editing frequency controlled by splicing efficiency, Nucleic Acids Res 44, 6398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samuel CE (2011) Adenosine Deaminases Acting on RNA (ADARs) are both Antiviral and Proviral Dependent upon the Virus, Virology 411, 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakurai M, Shiromoto Y, Ota H, Song C, Kossenkov AV, Wickramasinghe J, Showe LC, Skordalakes E, Tang HY, Speicher DW & Nishikura K (2017) ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay, Nature structural & molecular biology 24, 534–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang IX, So E, Devlin JL, Zhao Y, Wu M & Cheung VG (2013) ADAR regulates RNA editing, transcript stability, and gene expression, Cell Rep 5, 849–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES & Gorospe M (2003) Role of the RNA-binding protein HuR in colon carcinogenesis, Oncogene 22, 7146–54. [DOI] [PubMed] [Google Scholar]
- 63.Meisner NC, Hackermuller J, Uhl V, Aszodi A, Jaritz M & Auer M (2004) mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure, Chembiochem : a European journal of chemical biology 5, 1432–47. [DOI] [PubMed] [Google Scholar]
- 64.Shaw G & Kamen R (1986) A conserved AU sequence from the 3’ untranslated region of GM-CSF mRNA mediates selective mRNA degradation, Cell 46, 659–67. [DOI] [PubMed] [Google Scholar]
- 65.Bakheet T, Williams BRG & Khabar KSA (2006) ARED 3.0: the large and diverse AU-rich transcriptome, Nucleic Acids Research 34, D111–D114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Solomon O, Di Segni A, Cesarkas K, Porath HT, Marcu-Malina V, Mizrahi O, Stern-Ginossar N, Kol N, Farage-Barhom S, Glick-Saar E, Lerenthal Y, Levanon EY, Amariglio N, Unger R, Goldstein I, Eyal E & Rechavi G (2017) RNA editing by ADAR1 leads to context-dependent transcriptome-wide changes in RNA secondary structure, Nature communications 8, 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R & Nishikura K (2006) Modulation of microRNA processing and expression through RNA editing by ADAR deaminases, Nature structural & molecular biology 13, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R & Nishikura K (2007) RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex, EMBO reports 8, 763–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG & Nishikura K (2007) Redirection of silencing targets by adenosine-to-inosine editing of miRNAs, Science (New York, NY) 315, 1137–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gandy SZ, Linnstaedt SD, Muralidhar S, Cashman KA, Rosenthal LJ & Casey JL (2007) RNA editing of the human herpesvirus 8 kaposin transcript eliminates its transforming activity and is induced during lytic replication, Journal of virology 81, 13544–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Heale BS, Keegan LP & O’Connell MA (2011) The Effect of RNA Editing and ADARs on miRNA Biogenesis and Function, Advances in experimental medicine and biology 700, 76–84. [DOI] [PubMed] [Google Scholar]
- 72.Yang W, Wang Q, Howell KL, Lee JT, Cho D-SC, Murray JM & Nishikura K (2005) ADAR1 RNA Deaminase Limits Short Interfering RNA Efficacy in Mammalian Cells, The Journal of biological chemistry 280, 3946–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ota H, Sakurai M, Gupta R, Valente L, Wulff BE, Ariyoshi K, Iizasa H, Davuluri RV & Nishikura K (2013) ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing, Cell 153, 575–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishikura K, Sakurai M, Ariyoshi K & Ota H (2013) Antagonistic and stimulative roles of ADAR1 in RNA silencing, RNA biology 10, 1240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qi L, Song Y, Chan THM, Yang H, Lin CH, Tay DJT, Hong H, Tang SJ, Tan KT, Huang XX, Lin JS, Ng VHE, Maury JJP, Tenen DG & Chen L (2017) An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer, Nucleic Acids Res 45, 10436–10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brümmer A, Yang Y, Chan TW & Xiao X (2017) Structure-mediated modulation of mRNA abundance by A-to-I editing, Nature communications 8, 1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huertas D, Cortes A, Casanova J & Azorin F (2004) Drosophila DDP1, a multi-KH-domain protein, contributes to centromeric silencing and chromosome segregation, Current biology : CB 14, 1611–20. [DOI] [PubMed] [Google Scholar]
- 78.Zhou J, Wang Q, Chen L-L & Carmichael GG (2008) On the mechanism of induction of heterochromatin by the RNA-binding protein vigilin, RNA (New York, NY) 14, 1773–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD & Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases, Nature 406, 593–9. [DOI] [PubMed] [Google Scholar]
- 80.Brzostek-Racine S, Gordon C, Van Scoy S & Reich NC (2011) The DNA Damage Response Induces Interferon, Journal of immunology (Baltimore, Md : 1950) 187, 5336–5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morales AJ, Carrero JA, Hung PJ, Tubbs AT, Andrews JM, Edelson BT, Calderon B, Innes CL, Paules RS, Payton JE & Sleckman BP (2017) A type I IFN-dependent DNA damage response regulates the genetic program and inflammasome activation in macrophages, eLife 6, e24655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Becker JS, McCarthy RL, Sidoli S, Donahue G, Kaeding KE, He Z, Lin S, Garcia BA & Zaret KS (2017) Genomic and Proteomic Resolution of Heterochromatin and Its Restriction of Alternate Fate Genes, Mol Cell 68, 1023–1037 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ji X, Dadon DB, Abraham BJ, Lee TI, Jaenisch R, Bradner JE & Young RA (2015) Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions, Proceedings of the National Academy of Sciences of the United States of America 112, 3841–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soldi M & Bonaldi T (2013) The Proteomic Investigation of Chromatin Functional Domains Reveals Novel Synergisms among Distinct Heterochromatin Components, Molecular & Cellular Proteomics 12, 764–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cao Q, Wang X, Zhao M, Yang R, Malik R, Qiao Y, Poliakov A, Yocum AK, Li Y, Chen W, Cao X, Jiang X, Dahiya A, Harris C, Feng FY, Kalantry S, Qin ZS, Dhanasekaran SM & Chinnaiyan AM (2014) The Central Role of EED in the Orchestration of Polycomb Group Complexes, Nature communications 5, 3127.-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ng SK, Weissbach R, Ronson GE & Scadden ADJ (2013) Proteins that contain a functional Z-DNA-binding domain localize to cytoplasmic stress granules, Nucleic Acids Research 41, 9786–9799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.George CX, Ramaswami G, Li JB & Samuel CE (2016) Editing of cellular self RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses, Journal of Biological Chemistry [DOI] [PMC free article] [PubMed]
- 88.Ward SV, George CX, Welch MJ, Liou L-Y, Hahm B, Lewicki H, de la Torre JC, Samuel CE & Oldstone MB (2011) RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis, Proceedings of the National Academy of Sciences of the United States of America 108, 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G & Rich A (1979) Molecular structure of a left-handed double helical DNA fragment at atomic resolution, Nature 282, 680–6. [DOI] [PubMed] [Google Scholar]
- 90.Klysik J, Stirdivant SM, Larson JE, Hart PA & Wells RD (1981) Left-handed DNA in restriction fragments and a recombinant plasmid, Nature 290, 672–7. [DOI] [PubMed] [Google Scholar]
- 91.Haniford DB & Pulleyblank DE (1983) Facile transition of poly[d(TG) x d(CA)] into a left-handed helix in physiological conditions, Nature 302, 632–4. [DOI] [PubMed] [Google Scholar]
- 92.Peck LJ & Wang JC (1985) Transcriptional block caused by a negative supercoiling induced structural change in an alternating CG sequence, Cell 40, 129–137. [DOI] [PubMed] [Google Scholar]
- 93.Oh D-B, Kim Y-G & Rich A (2002) Z-DNA-binding proteins can act as potent effectors of gene expression in vivo, Proceedings of the National Academy of Sciences 99, 16666–16671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Paul MS & Bass BL (1998) Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA, The EMBO Journal 17, 1120–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ramaswami G & Li JB (2014) RADAR: a rigorously annotated database of A-to-I RNA editing, Nucleic Acids Res 42, D109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mattick JS (2010) RNA as the substrate for epigenome-environment interactions: RNA guidance of epigenetic processes and the expansion of RNA editing in animals underpins development, phenotypic plasticity, learning, and cognition, BioEssays : news and reviews in molecular, cellular and developmental biology 32, 548–52. [DOI] [PubMed] [Google Scholar]
- 97.Mattick JS & Mehler MF (2008) RNA editing, DNA recoding and the evolution of human cognition, Trends in neurosciences 31, 227–33. [DOI] [PubMed] [Google Scholar]
- 98.Lipfert J, Skinner GM, Keegstra JM, Hensgens T, Jager T, Dulin D, Köber M, Yu Z, Donkers SP, Chou F-C, Das R & Dekker NH (2014) Double-stranded RNA under force and torque: Similarities to and striking differences from double-stranded DNA, Proceedings of the National Academy of Sciences of the United States of America 111, 15408–15413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Broyde S & Hingerty B (1978) ‘A’ forms of RNAs in single strands, duplexes and RNA-DNA hybrids, Nucleic Acids Research 5, 2729–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zheng Y, Lorenzo C & Beal PA (2017) DNA editing in DNA/RNA hybrids by adenosine deaminases that act on RNA, Nucleic Acids Res 45, 3369–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI & Liu DR (2017) Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage, Nature 551, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Phelps KJ, Tran K, Eifler T, Erickson AI, Fisher AJ & Beal PA (2015) Recognition of duplex RNA by the deaminase domain of the RNA editing enzyme ADAR2, Nucleic Acids Research 43, 1123–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eggington JM, Greene T & Bass BL (2011) Predicting sites of ADAR editing in double-stranded RNA, Nature communications 2, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Matthews MM, Thomas JM, Zheng Y, Tran K, Phelps KJ, Scott AI, Havel J, Fisher AJ & Beal PA (2016) Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity, Nature structural & molecular biology 23, 426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Polson AG & Bass BL (1994) Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase, The EMBO Journal 13, 5701–5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wong SK, Sato S & Lazinski DW (2001) Substrate recognition by ADAR1 and ADAR2, RNA (New York, NY) 7, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stephens OM, Haudenschild BL & Beal PA (2004) The Binding Selectivity of ADAR2’s dsRBMs Contributes to RNA-Editing Selectivity, Chemistry & Biology 11, 1239–1250. [DOI] [PubMed] [Google Scholar]
- 108.Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, Liu KI, Zhang R, Ramaswami G, Ariyoshi K, Gupte A, Keegan LP, George CX, Ramu A, Huang N, Pollina EA, Leeman DS, Rustighi A, Goh YPS, Chawla A, Del Sal G, Peltz G, Brunet A, Conrad DF, Samuel CE, O’Connell MA, Walkley CR, Nishikura K & Li JB (2017) Dynamic landscape and regulation of RNA editing in mammals, Nature 550, 249–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E & Levanon EY (2014) A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes, Genome research 24, 365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Warnefors M, Liechti A, Halbert J, Valloton D & Kaessmann H (2014) Conserved microRNA editing in mammalian evolution, development and disease, Genome Biology 15, R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Carmi S, Church GM & Levanon EY (2011) Large-scale DNA editing of retrotransposons accelerates mammalian genome evolution, Nature communications 2, 519. [DOI] [PubMed] [Google Scholar]
- 112.Lander ES,Linton LM,Birren B,Nusbaum C,Zody MC,Baldwin J,Devon K,Dewar K,Doyle M,FitzHugh W,Funke R,Gage D,Harris K,Heaford A,Howland J,Kann L,Lehoczky J,LeVine R,McEwan P,McKernan K,Meldrim J,Mesirov JP,Miranda C,Morris W,Naylor J,Raymond C,Rosetti M,Santos R,Sheridan A,Sougnez C,Stange-Thomann Y,Stojanovic N,Subramanian A,Wyman D,Rogers J,Sulston J,Ainscough R,Beck S,Bentley D,Burton J,Clee C,Carter N,Coulson A,Deadman R,Deloukas P,Dunham A,Dunham I,Durbin R,French L,Grafham D,Gregory S,Hubbard T,Humphray S,Hunt A,Jones M,Lloyd C,McMurray A,Matthews L,Mercer S,Milne S,Mullikin JC,Mungall A,Plumb R,Ross M,Shownkeen R,Sims S,Waterston RH,Wilson RK,Hillier LW,McPherson JD,Marra MA,Mardis ER,Fulton LA,Chinwalla AT,Pepin KH,Gish WR,Chissoe SL,Wendl MC,Delehaunty KD,Miner TL,Delehaunty A,Kramer JB,Cook LL,Fulton RS,Johnson DL,Minx PJ,Clifton SW,Hawkins T,Branscomb E,Predki P,Richardson P,Wenning S,Slezak T,Doggett N,Cheng JF,Olsen A,Lucas S,Elkin C,Uberbacher E,Frazier M, et al. (2001) Initial sequencing and analysis of the human genome, Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 113.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK & Kjems J (2013) Natural RNA circles function as efficient microRNA sponges, Nature 495, 384–8. [DOI] [PubMed] [Google Scholar]
- 114.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H & Shan G (2015) Exon-intron circular RNAs regulate transcription in the nucleus, Nature structural & molecular biology 22, 256–64. [DOI] [PubMed] [Google Scholar]
- 115.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P & Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2, Nucleic Acids Res 44, 2846–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liang D & Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production, Genes & development 28, 2233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C & Rajewsky N (2015) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals, Cell Rep 10, 170–7. [DOI] [PubMed] [Google Scholar]
- 118.Daniel C, Silberberg G, Behm M & Ohman M (2014) Alu elements shape the primate transcriptome by cis-regulation of RNA editing, Genome Biol 15, R28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Daniel C, Widmark A, Rigardt D & Ohman M (2017) Editing inducer elements increases A-to-I editing efficiency in the mammalian transcriptome, Genome Biol 18, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stenger JE, Lobachev KS, Gordenin D, Darden TA, Jurka J & Resnick MA (2001) Biased distribution of inverted and direct Alus in the human genome: implications for insertion, exclusion, and genome stability, Genome research 11, 12–27. [DOI] [PubMed] [Google Scholar]
- 121.Lobachev KS, Stenger JE, Kozyreva OG, Jurka J, Gordenin DA & Resnick MA (2000) Inverted Alu repeats unstable in yeast are excluded from the human genome, EMBO J 19, 3822–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tajaddod M, Tanzer A, Licht K, Wolfinger MT, Badelt S, Huber F, Pusch O, Schopoff S, Janisiw M, Hofacker I & Jantsch MF (2016) Transcriptome-wide effects of inverted SINEs on gene expression and their impact on RNA polymerase II activity, Genome Biology 17, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Capshew CR, Dusenbury KL & Hundley HA (2012) Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing, Nucleic Acids Res 40, 8637–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Uchida H & Ito S (2015) Differential regulation of expression of RNA-editing enzymes, ADAR1 and ADAR2, by 5-aza-2’-deoxycytidine and trichostatin A in human neuronal SH-SY5Y cells, Neuroreport 26, 1089–94. [DOI] [PubMed] [Google Scholar]
- 125.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Buhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Mergoub T, Chan TA, Baylin SB & Strick R (2015) Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses, Cell 162, 974–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rice GI, Kasher PR, Forte GMA, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, Jenkinson EM, Bacino CA, Battini R, Bertini E, Brogan PA, Brueton LA, Carpanelli M, Laet CD, de Lonlay P, del Toro M, Desguerre I, Fazzi E, Garcia-Cazorla À, Heiberg A, Kawaguchi M, Kumar R, Lin J-PSM, Lourenco CM, Male AM, Marques W, Mignot C, Olivieri I, Orcesi S, Prabhakar P, Rasmussen M, Robinson RA, Rozenberg F, Schmidt JL, Steindl K, Tan TY, van der Merwe WG, Vanderver A, Vassallo G, Wakeling EL, Wassmer E, Whittaker E, Livingston JH, Lebon P, Suzuki T, McLaughlin PJ, Keegan LP, O’Connell MA, Lovell SC & Crow YJ (2012) Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature, Nature genetics 44, 1243–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Laxminarayana D, Khan IU & Kammer G (2002) Transcript mutations of the alpha regulatory subunit of protein kinase A and up-regulation of the RNA-editing gene transcript in lupus T lymphocytes, Lancet (London, England) 360, 842–9. [DOI] [PubMed] [Google Scholar]
- 128.Xing Q, Wang M, Chen X, Qian X, Qin W, Gao J, Wu S, Gao R, Feng G & He L (2005) Identification of a novel ADAR mutation in a Chinese family with dyschromatosis symmetrica hereditaria (DSH), Archives of dermatological research 297, 139–42. [DOI] [PubMed] [Google Scholar]
- 129.Kondo T, Suzuki T, Ito S, Kono M, Negoro T & Tomita Y (2008) Dyschromatosis symmetrica hereditaria associated with neurological disorders, The Journal of dermatology 35, 662–6. [DOI] [PubMed] [Google Scholar]
- 130.Khermesh K, D’Erchia AM, Barak M, Annese A, Wachtel C, Levanon EY, Picardi E & Eisenberg E (2016) Reduced levels of protein recoding by A-to-I RNA editing in Alzheimer’s disease, RNA (New York, NY) 22, 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gaisler-Salomon I, Kravitz E, Feiler Y, Safran M, Biegon A, Amariglio N & Rechavi G (2014) Hippocampus-specific deficiency in RNA editing of GluA2 in Alzheimer’s disease, Neurobiology of aging 35, 1785–91. [DOI] [PubMed] [Google Scholar]
- 132.Weissmann D, van der Laan S, Underwood MD, Salvetat N, Cavarec L, Vincent L, Molina F, Mann JJ, Arango V & Pujol JF (2016) Region-specific alterations of A-to-I RNA editing of serotonin 2c receptor in the cortex of suicides with major depression, Translational Psychiatry 6, e878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kubota-Sakashita M, Iwamoto K, Bundo M & Kato T (2014) A role of ADAR2 and RNA editing of glutamate receptors in mood disorders and schizophrenia, Molecular Brain 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Silberberg G, Lundin D, Navon R & Ohman M (2012) Deregulation of the A-to-I RNA editing mechanism in psychiatric disorders, Human molecular genetics 21, 311–21. [DOI] [PubMed] [Google Scholar]
- 135.Gurevich I, Englander MT, Adlersberg M, Siegal NB & Schmauss C (2002) Modulation of serotonin 2C receptor editing by sustained changes in serotonergic neurotransmission, The Journal of neuroscience : the official journal of the Society for Neuroscience 22, 10529–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Niswender CM, Herrick-Davis K, Dilley GE, Meltzer HY, Overholser JC, Stockmeier CA, Emeson RB & Sanders-Bush E (2001) RNA editing of the human serotonin 5-HT2C receptor. alterations in suicide and implications for serotonergic pharmacotherapy, Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 24, 478–91. [DOI] [PubMed] [Google Scholar]
- 137.Yang L, Zhao L, Gan Z, He Z, Xu J, Gao X, Wang X, Han W, Chen L, Xu T, Li W & Liu Y (2010) Deficiency in RNA editing enzyme ADAR2 impairs regulated exocytosis, FASEB journal : official publication of the Federation of American Societies for Experimental Biology 24, 3720–32. [DOI] [PubMed] [Google Scholar]
- 138.Gan Z, Zhao L, Yang L, Huang P, Zhao F, Li W & Liu Y (2006) RNA editing by ADAR2 is metabolically regulated in pancreatic islets and beta-cells, The Journal of biological chemistry 281, 33386–94. [DOI] [PubMed] [Google Scholar]
- 139.Amin EM, Liu Y, Deng S, Tan KS, Chudgar N, Mayo MW, Sanchez-Vega F, Adusumilli PS, Schultz N & Jones DR (2017) The RNA-editing enzyme ADAR promotes lung adenocarcinoma migration and invasion by stabilizing FAK, Science Signaling 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chan TH, Qamra A, Tan KT, Guo J, Yang H, Qi L, Lin JS, Ng VH, Song Y, Hong H, Tay ST, Liu Y, Lee J, Rha SY, Zhu F, So JB, Teh BT, Yeoh KG, Rozen S, Tenen DG, Tan P & Chen L (2016) ADAR-Mediated RNA Editing Predicts Progression and Prognosis of Gastric Cancer, Gastroenterology 151, 637–650 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fumagalli D, Gacquer D, Rothe F, Lefort A, Libert F, Brown D, Kheddoumi N, Shlien A, Konopka T, Salgado R, Larsimont D, Polyak K, Willard-Gallo K, Desmedt C, Piccart M, Abramowicz M, Campbell PJ, Sotiriou C & Detours V (2015) Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome, Cell Rep 13, 277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zipeto MA, Court AC, Sadarangani A, Delos Santos NP, Balaian L, Chun HJ, Pineda G, Morris SR, Mason CN, Geron I, Barrett C, Goff DJ, Wall R, Pellecchia M, Minden M, Frazer KA, Marra MA, Crews LA, Jiang Q & Jamieson CHM (2016) ADAR1 Activation Drives Leukemia Stem Cell Self-Renewal by Impairing Let-7 Biogenesis, Cell stem cell 19, 177–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Rossetti C, Picardi E, Ye M, Camilli G, D’Erchia AM, Cucina L, Locatelli F, Fianchi L, Teofili L, Pesole G, Gallo A & Sorrentino R (2017) RNA editing signature during myeloid leukemia cell differentiation, Leukemia 31, 2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA & Jacks T (2008) Suppression of non-small cell lung tumor development by the let-7 microRNA family, Proceedings of the National Academy of Sciences of the United States of America 105, 3903–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Copley MR, Babovic S, Benz C, Knapp DJ, Beer PA, Kent DG, Wohrer S, Treloar DQ, Day C, Rowe K, Mader H, Kuchenbauer F, Humphries RK & Eaves CJ (2013) The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells, Nature cell biology 15, 916–25. [DOI] [PubMed] [Google Scholar]
- 146.Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner Henrica M. J., Eterovic AK, Yuan Y, Li J, Nair N, Minelli R, Tsang Yiu H., Cheung Lydia W. T., Jeong Kang J., Roszik J, Ju Z, Woodman Scott E., Lu Y, Scott Kenneth L., Li Jin B., Mills Gordon B. & Liang H (2015) The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers, Cancer Cell 28, 515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen L, Li Y, Lin CH, Chan THM, Chow RKK, Song Y, Liu M, Yuan Y-F, Fu L, Kong KL, Qi L, Li Y, Zhang N, Tong AHY, Kwong DL-W, Man K, Lo CM, Lok S, Tenen DG & Guan X-Y (2013) Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma, Nature Medicine 19, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wang C, Zou J, Ma X, Wang E & Peng G (2017) Mechanisms and implications of ADAR-mediated RNA editing in cancer, Cancer letters 411, 27–34. [DOI] [PubMed] [Google Scholar]
- 149.Fritzell K, Xu LD, Lagergren J & Ohman M (2017) ADARs and editing: The role of A-to-I RNA modification in cancer progression, Seminars in cell & developmental biology [DOI] [PubMed]


