Abstract
Background:
The GTPase of immunity-associated protein 5 is essential for lymphocyte homeostasis and survival. Recently, human GIMAP5 SNPs have been linked with an increased risk for asthma, while the loss of Gimap5 in mice has been associated with severe CD4+ T celldriven immune pathology.
Objective:
To identify the molecular and cellular mechanisms by which Gimap5-deficiency predisposes to allergic airway disease.
Methods:
CD4+ T cell polarization and the development of pathogenic CD4+ T cells were assessed in Gimap5-deficient mice and a human patient with a GIMAP5 loss-of-function (LOF) mutation. House dust mite (HDM)-induced airway inflammation was assessed using a complete Gimap5 LOF (Gimap5sph/sph) and conditional Gimap5fl/fl Cd4Cre/ert2 mice.
Results:
GIMAP5 LOF mutations in both mice and humans are associated with spontaneous polarization towards pathogenic TH17 and TH2 cells in vivo. Mechanistic studies in vitro reveal that impairment of Gimap5-deficient TH cell differentiation is associated with increased DNA damage, particularly during TH1 polarizing conditions. The DNA damage in Gimap5-deficient CD4+ T cells could be controlled by TGFβ, thereby promoting TH17 polarization. When challenged with HDM in vivo, Gimap5-deficient mice displayed an exacerbated asthma phenotype (inflammation and airway hyperresponsiveness) with increased development of TH2, TH17, and pathogenic TH17/ TH2 cells.
Conclusion:
Activation of Gimap5-deficient CD4+ T cells is associated with increased DNA damage and reduced survival that can be overcome by TGFβ. This leads to a selective survival of pathogenic TH17 cells, but also TH2 in humans and mice, ultimately promoting allergic airway disease.
Keywords: Allergic airway disease, house dust mite, Primary immune deficiency, GIMAP5, Th2/Th17 cells, TGFβ, DNA damage response, T cell polarization, immune homeostasis, lung disease
Capsule Summary
GIMAP5 LOF mutations in both mice and humans are associated with polarization of CD4+ T cells towards TH17 and TH2 cells in vivo. Gimap5-deficient mice develop more severe allergic airway disease upon HDM sensitization.
Introduction
Asthma is a chronic airway inflammatory disorder that develops through a variety of mechanisms, each leading to different clinical manifestations and responsiveness to treatment. While the majority of asthma cases can be controlled with standard treatments, a subset of asthmatics develop “severe” or “refractory” asthma.1 Recent work has shown that these cases are associated with the presence of TH2/TH17 CD4+ T cells and that these cells drive more severe disease than classical TH2 CD4+ T cells.2–4 Moreover, levels of IL-17A, or IL-17A producing CD4+ cells, in the lung, sputum, and blood of asthmatic patients correlate with disease severity.2–11However, both genetic and environmental factors that contribute to TH2 and TH17-associated asthma development remain unclear.
The GTPase of immunity-associated protein 5 (Gimap5) is critical for lymphocyte survival, with Gimap5-deficient CD4+ T cells demonstrating impaired survival and elevated DNA damage upon antigenic stimulation.12–14 Complete loss-of-function (LOF) mutations in GIMAP5, both in rodents and in humans, results in lymphopenia, loss of immunologic tolerance, and subsequent development of autoimmunity.12–21 In the context of Gimap5-deficient (Gimap5sph/sph) mice, this manifests as CD4+ T cells-driven colitis. Importantly, polymorphisms in GIMAP5 have been associated with increased risk for type 1 diabetes (T1D) and systemic lupus erythematosus (SLE), and a recent study revealed a SNP in the promoter region of GIMAP5 to be associated with asthma.22–25 It remains unclear, however, how polymorphisms in GIMAP5 predispose individuals to these inflammatory diseases.
In the current study, we show that Gimap5-deficiency drives the development of pathogenic CD4+ T cells with an exaggerated TH17 cell response and a more severe asthma phenotype.
Methods
Study design
For the proposed experiments involving the studies of immune responses in vivo, both male and female mice were used. All mice used were generated on a C57BL/6J background and the background purity was confirmed by whole genome SNP analysis. To minimize confounding secondary factors arising from lymphopenia and other late-stage pathologies that develop in Gimap5sph/sph mice, CD4+ T cells from 3–4 week-old mice were used unless otherwise noted. At this age, Gimap5sph/sph mice have relatively normal numbers of CD4+ T cells with a normal frequency of naïve T cells that exhibit a quiescent phenotype comparable to WT mice. Since we were interested in T cell proliferation/survival in WT vs Gimap5-deficient cells, we expected to find strong differences in measurement (e.g. normal vs absence of T cell survival/proliferation). In cases where the differences in the observed phenomena were clear and distinct, a group size of 6 mice was suitable to give statistically significant (i.e. P<0.05) data. For these studies, we estimated that, with a sample size of 6, we have a 99% power to detect at least 25% reduction in the % of proliferating cells in stimulated Gimap5sph/sph cells with a significant level (alpha) of 0.05 (two-tailed).
Mice and reagents
All experiments were performed according to US National Institutes of Health guidelines and were approved by the IACUC of The Cincinnati Children’s Hospital. C57BL/6J mice were obtained from Jackson. Deletion of Gimap5 in Gimap5flox/floxCd4cre-ert2 mice was induced by the administration of tamoxifen chow (40mg/kg body weight; Harlan Laboratories Teklad Diets). Gimap5sph/sph mice were generated as previously described (6) and bred in-house in the vivarium of Cincinnati Children’s Hospital. All mice were maintained under specific pathogen-free conditions.
All antibodies used for flow cytometry were purchased from eBioscience or Biolegend unless otherwise noted. Purified α-mouse-CD3 (17A2) and α-mouse-CD28 (37.51) antibodies (Biolegend) were used for mouse T cell activation. A fixable viability stain was purchased from eBioscience. 7-AAD was purchased from BD. PMA, ionomycin, and Brefeldin A were obtained from Sigma. αIFNγ (XMG1.2) and mIL-23 were obtained from eBioscience. TGFβ1, mIL-4, and hIL-2 were purchased from Miltenyi Biotech. αIL-4, IL-6, and mIL-1β, mIL-12, and TGFβ3 were obtained from PeproTech.
Generation of Gimap5flox/flox mice
CRISPR/Cas9 was used to insert LoxP sites flanking the second exon of Gimap5 (Fig. E3A). As the second exon contains the translation start site, deletion of this exon is expected to result into loss of Gimap5 protein (Fig. E5A). The accuracy of the insert was confirmed through sequencing (Fig. E3B). Presence of the LoxP sites was confirmed through PCR genotyping as shown in Fig. E3C. The PCR primers used are as follows: 5’ LoxP site PCR forward, 5’-GATC GTGCAACGACATGGG-3’, 5’ LoxP site PCR reverse, 5’-ACGACCTTGCCCGACTAGAG-3’, 3’ LoxP site PCR forward, 5’-CCACCCAAGGAGAGTTACCC-3’, 3’ LoxP site PCR reverse, 5’-GTCAAAGACAAGCTTCCACAGT-3’. Mice bearing the Gimap5fl/fl allele were then crossed those carrying the Cd4cre-ert2 and Foxp3cre transgenes to produce Gimap5fl/flCd4cre-ert2 or Gimap5fl/flFoxp3cre offspring. Deletion of Gimap5 was induced in Gimap5fl/flCd4cre-ert2 CD4+ T cells through the administration of tamoxifen chow (40mg/kg body weight; Harlan Laboratories Teklad Diets).
Flow cytometry and T cell analyses
T cell proliferation was quantified by incubating Mojo-purified (Biolegend) CD4+ T cells in 5 μM CTV in 0.2% FBS for 20 min. Cells were cultured in supplemented IMDM media containing 10% FBS, 2% penicillin/streptomycin, 1% L-glutamine, and 50 μM BME. Cells were either left unstimulated or stimulated with αCD3 (1 μg/ml)/αCD28 (2 μg/ml) in the presence of TGFβ1 (2 ng/mL). After 3 days incubation, proliferation was evaluated by analyzing CTV dilution by flow cytometry. Viability was determined by staining with a fixable viability dye. DNA damage was evaluated by γH2AX staining in conjunction with intracellular 7-AAD staining for cell cycle analysis. To evaluate lymphocyte populations ex vivo, lymphocytes from spleen, mesenteric lymph node (mLN), mediastinal lymph node (mdLN), and lung were isolated and stained with fluorochrome-conjugated antibodies for mouse CD3, CD4, CD19, CD44, γH2AX, Tbet, RORγt, Gata3, and Foxp3. To evaluate CD4+ T cell cytokine production capacity, lymphocytes from the spleen, mesenteric lymph node, mediastinal lymph node, and lung were restimulated with PMA (50 ng/mL) and ionomycin (500 ng/mL) in the presence of Brefeldin A for 6h before staining with fluorochrome-conjugated antibodies for mouse IFNγ, IL-13, and IL-17A. Except where noted in in vitro experiments, all data was generated from live cells as determined by fixable viability dye staining.
In vitro CD4+ T cell polarization
To test CD4+ T cell polarization, Mojo-purified (Biolegend) naïve CD4+ T cells were isolated from the spleen and peripheral lymph nodes (cervical, brachial, axillary, and inguinal) of 3–4week-old mice and labeled with 5 μM CTV in 0.2% FBS for 20 min. Cells were cultured supplemented RPMI media containing 10% FBS, 2% penicillin/streptomycin, and 50 μM BME. Cells were stimulated with αCD3 (1 μg/ml)/αCD28 (2 μg/ml) alone or in the presence of polarizing cytokines (Table E1).
Mouse asthma model
Because Gimap5sph/sph mice show progressive changes in differentiated CD4+ T cells with age,12,14 IT injections started at 4 weeks of age, when a similar frequency of naïve CD4+ T cells can be observed compared to WT littermates.12,14 WT C57BL/6 and Gimap5sph/sph mice were administered nine, 25 μl intratracheal injections of 25 μg HDM or saline over 3 weeks. For older (6–8 wk) WT, Gimap5fl/flCd4cre-ert2, and Gimap5fl/flFoxp3cre mice, doses of 50 μg HDM in 50 μl saline were used. WT and Gimap5fl/flCd4cre-ert2 mice were placed on tamoxifen chow (40mg/kg body weight; Harlan Laboratories Teklad Diets) 7 days prior to the first I.T. injection in relevant experiments. In HDM experiments, mice were sacrificed 24h after the last exposure.
Airway hyperresponsiveness (AHR)
Mice were anesthetized with Ketamine, Xylazine, and Acepromazine (100, 20, and 10 mg/mL respectively mixed at a ratio of 4:1:1). Invasive measurements of airway responsiveness were made using the flexiVent apparatus (Scireq, Montreal, Canada). Mouse tracheas were cannulated with a 19-gauge blunt needle; mice were ventilated at 150 breaths/min, 3.0 cm water positive end-expiratory pressure. Two total lung capacity perturbations were then performed for airway recruitment before baseline measurement and subsequent methacholine challenges were performed. Dynamic resistance (R) was assessed following exposure to increasing concentrations of aerosolized methacholine (0, 12.5, 25, 50, and 100 mg/mL). The average of the 3 highest R values with a coefficient of determination of 0.9 or greater (as determined by flexiVent software) was used to determine the dose-response curve.
BALF cell collection and analysis
Bronchoalveolar lavage was performed by cannulation of the trachea. Lungs were lavaged with 1 mL PBS. After counting the number of cells with a hemocytometer, cells were spun onto slides and stained with the HEMA3 stain set (Fisher Scientific). After de-identifying samples, approximately 200 cells were counted and the number of macrophages, neutrophils, and eosinophils were calculated.
Isolation and characterization of lung-infiltrating cells
After lavage, lungs were removed and the upper right lobe was minced and digested in 2 ml RMPI + 0.5 mg/ml Liberase DL (Roche Diagnostics) and 0.5 mg/ml DNAse I (Sigma) at 37ºC for 30 min. Cells were then passed over a 70 μm cell strainer. Cells were washed with PBS and counted. ~1×106 cells were stimulated with 50 ng/mL PMA + 500 ng/mL ionomycin for 6h. After 2 hours, Brefeldin A was added. CD4+ T cells were stained with a viability dye (eBioscience Live/Dead stain) and antibodies for CD3, CD4, CD44, and γδTCR; intracellular staining for IL-13, IL-17A, and IFNγ was performed after fixation and permeabilization according to manufacturer’s instructions (eBioscience). Foxp3 staining was performed on separate cells that had not been restimulated ex vivo.
Histology
Lung tissue was collected and immediately fixed in 10% buffered formalin solution, followed by routine paraffin embedding. Periodic acid-Schiff staining (PAS) and hematoxylin/eosin (H&E) staining was performed on 4 μm sections from the paraffin-embedded tissue blocks for conventional light microscopy analysis.
Analysis of human T cells
For all studies concerning human cells, informed consent was obtained and studies were approved by the CCHMC institutional IRB. PBMCs were isolated from whole blood of a GIMAP5-deficient patient, a healthy parent (heterozygous for GIMAP5 rs72650695), and unrelated healthy controls by Ficoll-Paque Plus density gradient centrifugation. Isolated PBMCs were cultured in RPMI, 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 2mM Lglutamine, and 10 mM HEPES and were stimulated with PMA (50 ng/mL) and ionomycin (500 ng/mL) in the presence of Brefeldin A and analyzed for IFNγ, IL-17A, and IL-13 production by flow cytometry.
Immunoblotting
Protein lysates from Mojo-isolated (BioLegend) CD4+ T cells were prepared according to standard methods from resting cells. Lysates were separated using 10% Bis-Tris Gels, transferred onto nitrocellulose, and immunoblotted with primary antibodies to Gimap5 (MAC421)12 and βactin (CST).
Analysis and Graphics Software
Flow cytometry data were analyzed using FlowJo. All data were analyzed for statistical significance using GraphPad Prism4® software (GraphPad Software, San Diego, CA). Graphical abstract was created with BioRender. Figures components were generated in FlowJo or GraphPad Prism and assembled in Adobe Illustrator.
Statistical analysis
Data are expressed as mean ± SDs except for where otherwise noted. For studies comparing T cells from C57BL/6J, Gimap5sph/sph; and/or Gimap5fl/flCd4cre-ert2 mice, Student’s two-tailed test or ANOVA followed by Sidak’s multiple comparisons test for three or more groups were used. Data were considered statistically significant if P values were <0.5.
Results
GIMAP5 LOF mutations in mice and human promote development of TH17/TH2 CD4+ T cells in vivo
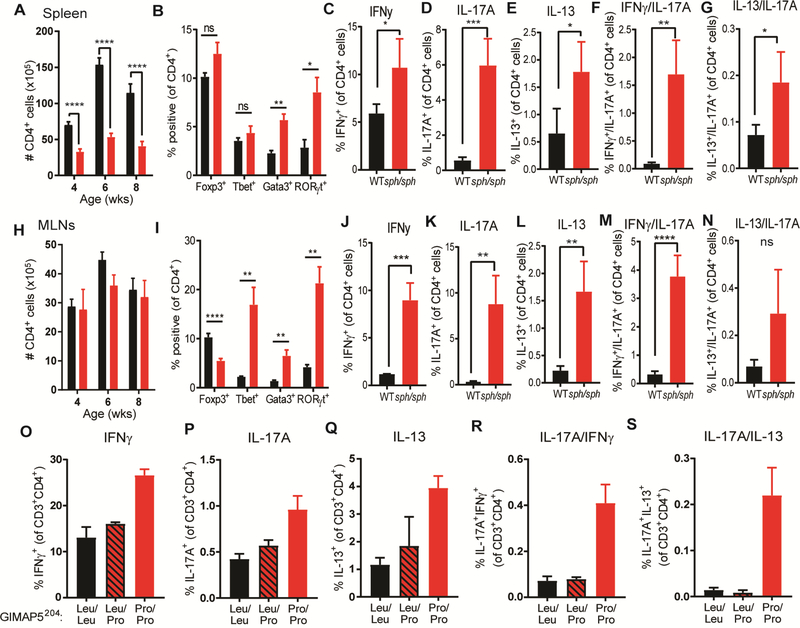
Given that polymorphisms in GIMAP5 have been associated with increased risk for type 1 diabetes (T1D), systemic lupus erythematosus (SLE), and asthma.22–25, and our previous studies revealed the development of CD4-dependent colitis in Gimap5-deficient mice,14,15 we first investigated whether baseline differences in CD4+ T cell polarization exist in vivo. To assess this, we examined the CD4+ T cell compartment in the spleen and mesenteric lymph nodes (mLN) of 8-week old WT and Gimap5sph/sph mice. As previously described, CD4+ T cells were reduced in the spleen of Gimap5sph/sph mice (Fig. 1A). Importantly, however, an increased proportion of the remaining Gimap5sph/sph CD4+ T cells were polarized to TH2 cells (Gata3+CD4+) and TH17 cells (RORγt+CD4+), but not TH1 (Tbet+, CD4+) or Treg cells (Foxp3+, CD4+) compared to WT mice (Fig. 1B). Furthermore, Gimap5sph/sph splenic CD4+ T cells more frequently produced IFNγ, IL-17A, and/or IL-13 compared to WT CD4+ T cells (Fig. 1C-E). Notably, Gimap5sph/sph mice had expanded frequencies of CD4+ T cells that produced both IFNγ+IL-17A+ or IL-13+IL-17A+ (Fig. 1F,G). These cells have been associated with pathology in several autoimmune diseases and asthma, respectively.2,3,26–31
FIG 1.
Enhanced In vivo polarization of Gimap5-deficent mouse and human CD4+ T cells. (A) Number of CD4+ T cells in the spleen of 8wk WT (black bars) and Gimap5sph/sph (red bars) mice. (B) frequency of splenic CD4+ T cells expressing Foxp3, Tbet, Gata3, or RORγt (n=7). (C-G) Expression of IFNγ, IL-17A, and/or IL-13 in splenic CD4+ T cells after PMA/ionomycin stimulation (n=4). (H) Number of CD4+ T cells in the mesenteric lymph node of 8wk mice. (I) frequency of CD4+ T cells expressing Foxp3, Tbet, Gata3, or RORγt in the mesenteric lymph node (n=7). (J-N) Expression of IFNγ, IL-17A, and/or IL-13 in CD4+ T cells from the mesenteric lymph node after PMA/ionomycin stimulation (n=4). Bars represent mean values ± SD. Statistical significance is determined by Student’s two-tailed test. (O-S) Frequency of CD4+ T cells producing IFNγ, IL-17A, and/or IL-13 upon PMA/ionomycin stimulation. T cells isolated from the PBMCs of a GIMAP5 patient (GIMAP5204-Pro/Pro), his heterozygous parent, and unrelated controls (n=2, 2, and 7, respectively). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
Interestingly, the mLNs of Gimap5sph/sph mice contained similar numbers of total CD4+ T cells compared to WT littermates, but a higher frequency expressed Tbet, Gata3, or RORγt compared to WT littermates (Fig. 1H,I). Similar to the spleen, an increased proportion of CD4+ T cells in the mLN were IFNγ+, IL-17A+, IL-13+, and the rarer IFNγ+IL-17A+ double producers (Fig. 1J-N). Together these observations suggest that loss of Gimap5 is associated with an increased polarization of effector CD4+ T cells, including the development of pathogenic CD4+ T cells that may drive immunopathology.
Previously, we described a patient with a LOF mutation in GIMAP5 (rs72650695; a missense variant leading to a Leu204→Pro amino acid change and a complete loss of protein expression)14 resulting in an immunodeficiency with features of autoimmunity. Functional characterization of CD4+ T cells from this patient revealed strikingly similar defects to Gimap5-deficient mouse CD4+ T cells.14 We therefore asked if the patient has similarly skewed CD4+ T cell compartment. Strikingly, a higher proportion of the patient’s circulating CD4+ T cells expressed IL-17A, IL-13, or IFNγ compared to the patient’s heterozygous mother or unrelated controls upon stimulation with PMA and ionomycin (Fig. 1O-Q). Furthermore, an increased frequency of CD4+ T cells co-expressed IL-17A+IL-13+ or IL-17A+IFNγ+ (Fig. 1R,S). While we were only able to study a single patient with GIMAP5-deficiency, the increased development of pathogenic peripheral TH2 and TH17 cells in the GIMAP5-defcient patient is consistent with our mouse data.
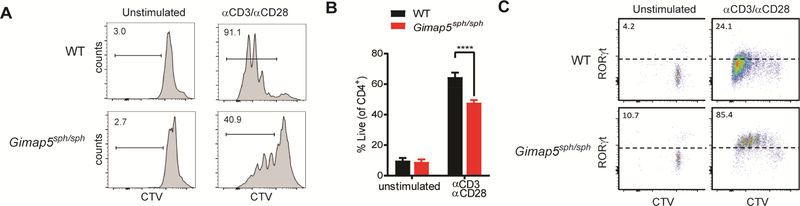
Notably, this predilection towards TH17 polarization can be observed in in vitro culture systems. In previous studies, we observed that bulk Gimap5sph/sph CD4+ T cells have an impaired proliferation and survival upon stimulation with αCD3/αCD28 (Fig. 2A-B) and only a small subset is able to survive and proliferate.12,14 A more in-depth evaluation of surviving cells demonstrates expression of high levels of RORγt, indicating either a selective survival of TH17polarized cells or a predisposition of surviving cells to adopt a TH17 (Fig. 2C). Together, these data suggest that GIMAP5-deficiency in both humans and mice causes CD4+ T cells to preferentially adopt a TH17 phenotype in vitro and in vivo.
FIG 2.
Preferential expression of RORγt by surviving Gimap5sph/sph CD4+ T cells upon TCR stimulation. (A) Proliferation of WT and Gimap5sph/sph CD4+ T cells after 3 days stimulation with αCD3 (1 μg/ml) + αCD28 (2 μg/ml). (B) Survival CD4+ T cells after 3d stimulation. (C) Expression of RORγt in live CD4+ T cells after 3d stimulation with αCD3/αCD28. Plots depict live cells (C), while bar graphs (B) represent mean values ± SD (n=4). All experiments were performed at least three times. Statistical significance is determined by Student’s two-tailed test. ****P<0.0001
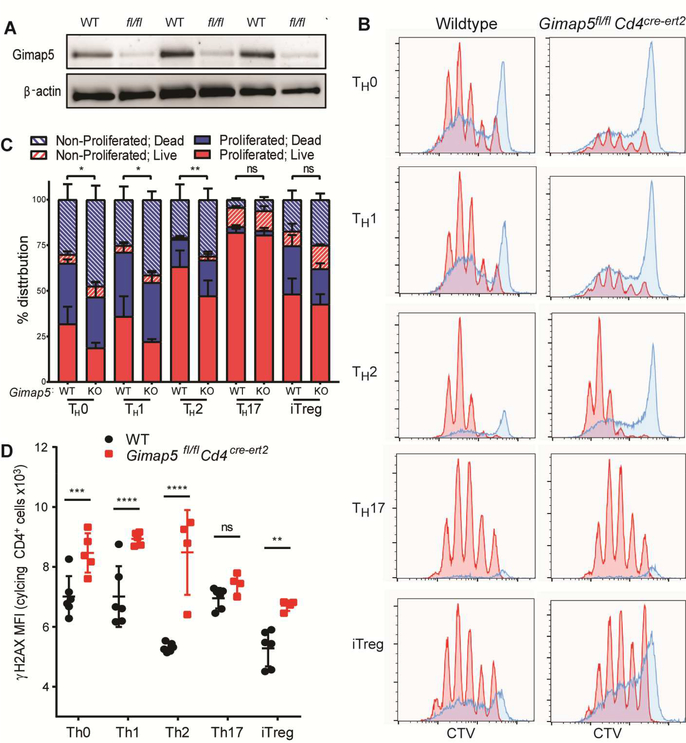
Selective survival of Gimap5-deficient CD4+ T cells under different polarizing conditions
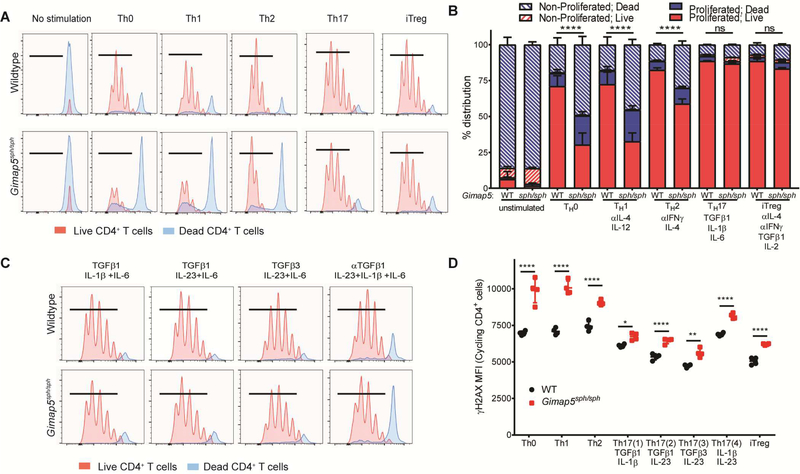
Both Gimap5-deficient rodents and humans develop lymphopenia12–19 and it is thus unclear whether the increased prevalence of TH17 cells in vivo is due to a skewing of polarization or selective survival of TH17 cells. Similarly, the expression of RORγt by almost all surviving Gimap5-deficient CD4+ T cells could be due to the survival of cells, or merely as the consequence of surviving cells polarizing to TH17 cells in vitro. To differentiate between these possibilities, naïve CD4+ T cells were isolated from 3–4-week-old wildtype and Gimap5sph/sph mice and stimulated with αCD3/αCD28 in TH0, TH1, TH2, TH17, or iTreg polarizing conditions for 3 days (detailed in Table E1) before evaluating survival and proliferation. As with total CD4+ T cells, naïve Gimap5sph/sph CD4+ T cells exhibited reduced survival and proliferation when stimulated with αCD3/αCD28 (TH0 conditions) compared to WT CD4+ T cells (Fig. 3A,B). A similar defect was observed in TH1 polarizing conditions. This defect was primarily characterized by Gimap5sph/sph cells unable to complete cell cycle, although a considerable proportion of cells died after cycling. Stimulation in TH2 polarizing conditions resulted in a more bifurcated response, with a subset of cells exhibiting strong proliferation while a significant number of cells died without completing cell cycle. Strikingly, compared to WT, naïve Gimap5sph/sph CD4+ T cell survival and proliferation was completely normal in TH17 and iTreg polarizing conditions. Notably, both TH17 and iTreg polarizing conditions include TGFβ1; we asked if TGFβ signaling promoted Gimap5sph/sph CD4+ T cell survival and proliferation during Th17 polarization. Previously, Lee et al. identified different TH17 polarizing conditions using combinations of TGFβ1, TGFβ3, IL-6, IL-23, and IL-1β (Table E1).32 To test the contribution of TGFβ and the TH17 transcriptional program to Gimap5sph/sph CD4+ T cell survival, we stimulated naïve CD4+ T cells in TH17 polarizing conditions containing TGFβ1, TGFβ3, or anti-TGFβ1. While Gimap5sph/sph CD4+ T cell survival and proliferation were improved in TH17 polarizing conditions lacking TGFβ signaling over TH0 conditions, survival was significantly reduced compared to TH17 polarizing conditions containing TGFβ1 or TGFβ3 (Fig. 3C). These data suggest that TGFβ is a critical mediator of Gimap5-deficient CD4+ T cell survival ultimately promoting the development of pathogenic TH17 cells.
FIG 3.
Preferential survival of Gimap5sph/sph CD4+ T cells in TH17 and iTreg polarizing conditions is associated with TGFβ. (A-B) Proliferation and survival of naïve CD4+ T cells from WT and Gimap5sph/sph mice stimulated 3d in non-polarizing conditions (TH0) or TH1, TH2, TH17, or iTreg polarizing conditions. Blue histograms represent dead cells; red histograms show live cells. Hatched bars represent non-proliferated cells; solid bars represent proliferated cells. (C) Proliferation of live/dead CD4+ T cells after 3d stimulation in different TH17 polarizing conditions. (D) γH2AX expression (DNA damage) in live CD4+ T cells in cycle after 3d stimulation in different polarizing conditions. Data represent mean values ± SD (n=4). All experiments were performed at least three times. Statistical significance is determined by ANOVA followed by Sidak’s multiple comparisons test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
Activation of Gimap5sph/sph CD4+ T cells results in increased DNA damage that is mitigated by TGFβ1
We have previously reported that poor survival of Gimap5sph/sph CD4+ T cells upon αCD3/αCD28 stimulation correlated with exacerbated DNA damage.14 We therefore asked whether the reduced survival in different polarizing conditions was associated with exacerbated DNA damage. Specifically, we assessed the level of phosphorylated histone H2AX (γH2AX) in live, dividing CD4+ T cells as a marker for double-stranded DNA breaks. Interestingly, high levels of γH2AX were observed even in WT CD4+ T cells when stimulated under TH0, TH1, or TH2 conditions, while TH17 and iTreg conditions were associated with comparatively modest DNA damage after 3 days of polarizing conditions (Fig. 3D). Importantly, Gimap5sph/sph CD4+ T cells consistently showed elevated DNA damage in TH0, TH1 or TH2 conditions compared to WT (Fig. 3D), while γH2AX levels in both WT and Gimap5sph/sph CD4+ T cells were reduced in polarizing conditions containing either TGFβ1 or TGFβ3. Notably, elevated DNA damage was also observed in Gimap5sph/sph TH1 (Tbet+) cells directly ex vivo, indicating that this phenotype remains relevant in vivo (Fig E1). Together, these data indicate that Gimap5sph/sph CD4+ T cells are unable to control DNA damage that occurs in TH0, TH1 and TH2 polarizing conditions.
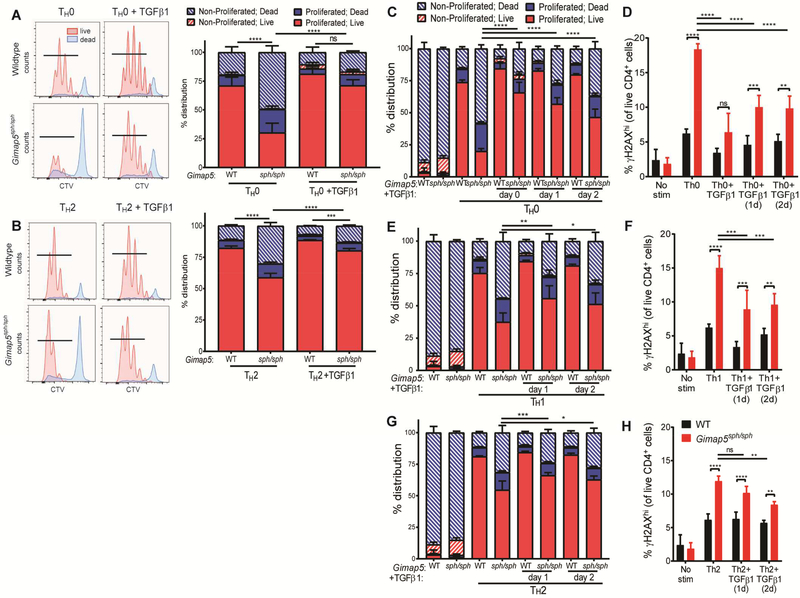
Given the preferential survival of Gimap5-deficient CD4+ T cells stimulated in TGFβ-containing- TH17 and iTreg polarizing conditions, we next asked if TGFβ1 signaling was sufficient to rescue CD4+ T cells cultured in polarizing conditions associated with poor survival. Strikingly, proliferation and survival of Gimap5sph/sph CD4+ T cells were almost completely rescued with the addition of TGFβ1 in these polarizing conditions (Fig. 4A,B).
FIG 4.
TGFβ1 rescues survival and proliferation of naïve Gimap5sph/sph CD4+ T cells. Proliferation and survival of naïve WT and Gimap5sph/sph CD4+ T cells after 3d stimulation in (A) TH0 or (B) TH2 polarizing conditions ± 2 ng/mL TGFβ1. Blue histograms represent dead cells; red histograms show live cells. Survival, proliferation, and γH2AX expression of naïve CD4+ T cells stimulated in (C-D) TH0, (E-F) TH1, (G-H) or TH2 polarizing conditions. At 1 or 2 days after initial stimulation, 2 ng/ml TGFβ1 was added to the indicated culture. Data represent mean values ± SD (n=4). Statistical significance is determined by ANOVA followed by Sidak’s multiple comparisons test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
Notably, TGFβ is cleaved to its active form in the lung (and other tissues) after tissue injury,33–36 resulting in a TGFβ-rich environment. As naïve T cells are first activated in lymph nodes before migrating to the affected tissue, we asked if exposure to TGFβ1 during the cycling phase when DNA damage is most pronounced was sufficient to promote survival of Gimap5sph/sph CD4+ T cells. Indeed, the addition of TGFβ1 after 1 or 2 days of stimulation/polarization robustly improved Gimap5sph/sph CD4+ T cell survival, proliferation, and control of DNA damage in TH0, TH1, and TH2 polarizing conditions, although it was insufficient to completely abrogate the effects of Gimap5-deficiency (Fig. 4C-H). These data demonstrate that, independent of TH17 or iTreg polarization, TGFβ1 is sufficient to rescue the survival of Gimap5sph/sph CD4+ T cells. Moreover, TGFβ1 signaling is not required at the time of initial αCD3/αCD28 stimulation. These studies thus identify TGFβ as an important mediator during CD4+ T cell differentiation limiting DNA damage and improving survival and proliferation of activated Gimap5sph/sph CD4+ T cells and promoting the selective development of pathogenic TH17 cells.
Gimap5-deficiency exacerbates allergic airway pathology
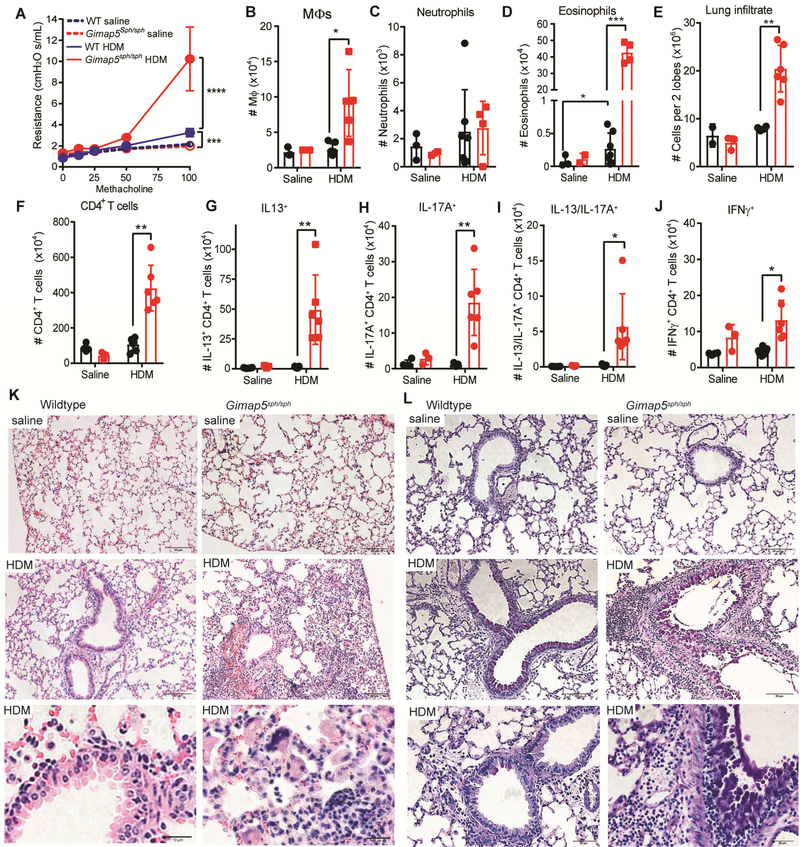
A recent study identifies GIMAP5 as a modifier gene in asthma.25 Specifically, a 5’ SNP (rs6965571) in the promoter region of GIMAP5 is associated with ~3-fold increased risk of asthma in a prospective Swedish birth cohort25, although the mechanisms responsible for increased asthma risk are unclear. Given our findings that both GIMAP5-deficient mice and human display a high frequency of TH2 and TH2/TH17 pathogenic CD4+ T cells in vivo, and that TH2/TH17 allergic responses exacerbate the asthma phenotype, we assessed the role of Gimap5 in allergic airway disease. Based on the changes in T cell polarization described above, we predicted that loss of Gimap5 would increase sensitivity to allergic airway disease and we therefore used a mild model of experimentally-induced allergic airway disease in C57BL/6J mice.37,38 Specifically, we sensitized Gimap5sph/sph and control C57BL/6 wildtype (WT) mice with nine intratracheal (IT) injections of 25 μg house dust mite (HDM) during a 3-week period as previously described,39 after which immune pathology was assessed. One day following the last exposure, mice were evaluated for airway hyperresponsiveness (AHR) and BAL inflammatory profile. Strikingly, HDM-sensitized Gimap5sph/sph mice exhibited significantly higher AHR compared to WT mice and saline-treated mice (Fig. 5A). The observed increase in AHR corresponded with a significantly increased infiltration of eosinophils and macrophages in the bronchoalveolar lavage fluid (BALF) in HDM-sensitized Gimap5sph/sph mice (Fig. 5B-D).
FIG 5.
Gimap5sph/sph mice exhibit increased AHR, CD4+ T cell polarization, and airway pathology upon HDM sensitization. C57BL/6 WT (black bars) and Gimap5sph/sph (red bars) mice were administered 25 μg HDM 9 times IT over three weeks. (A) Airway hyper-responsiveness (AHR) in response to methacholine challenge 1 day after the last HDM challenge (saline: n=3; HDM: n=5–6). (B-D) Number of macrophages, neutrophils, and eosinophils in the BAL fluid (saline: n=3; HDM: n=4–7). (E-F). Number of (E) total lung-infiltrating cells and (F) CD4+ T cells (in 2 lobes). (G-J) Percentage of lung-infiltrating CD4+CD3+ T cells expressing IL-13, IL-17A, IL-13 and IL-17A, or IFNγ upon restimulation with PMA/ionomycin for 6h (saline: n=3; HDM: n=6–7). (K-L) Representative (K) H&E and (L) PAS images of lung tissue from WT and Gimap5sph/sph mice given saline or HDM. Data represents mean values ± SD. Statistical significance is determined by ANOVA followed by Sidak’s multiple comparisons test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
To determine whether exacerbated AHR and eosinophil/macrophage infiltration correlated with increased development of pathogenic CD4+ T cells, total lung infiltrating cells from the upper right lobe for each mouse were quantified, restimulated with PMA and ionomycin, and evaluated for cytokine production. The lungs from HDM-sensitized Gimap5sph/sph mice had significantly more inflammatory infiltrate (Fig. 5E) and, importantly, exhibited a four-fold increase in infiltrating CD4+ T cells compared to WT mice (Fig. 5F). The latter involved a marked increase in CD4+ T cells producing IL-13 or IL-17 (Fig. 5G,H) and importantly also included a significant increase in pathogenic IL-13+IL-17A+ CD4+ T cells (Fig. 5I) that have been associated with severe immune pathology/disease. Similarly, lung-draining mediastinal lymph nodes of Gimap5sph/sph mice displayed an increase in IL-13+, IL-17A+, and IL-13+IL-17A+ CD4+ T cells (Fig. E2). Furthermore, HDM-sensitization resulted in distortion of normal lung architecture, increased airway mucus production, marked edema, and lymphocytic infiltration in the interstitial space, representative of perivascular inflammation (Fig. 5K-L). Together, these data exhibit that loss-of-function mutations in Gimap5 exacerbate the asthma phenotype which is associated with a significant expansion of pathogenic TH2/TH17 cells.
Polarization defects in Gimap5-deficient CD4+ T cells are cell-intrinsic
Besides CD4+ T cells, Gimap5 is expressed in innate lymphoid cells (ILCs) and our previous studies indicate NK cell development/survival is markedly affected in Gimap5-deficient mice.12,19 Given the critical role for ILC2 and ILC3s in CD4+ T cell differentiation and asthma,40–45 we next asked if the observed CD4+ T cells defects were cell intrinsic or whether other cell types might contribute to the impaired polarization of CD4+ T cells. To do so, we generated the Gimap5fl/fl mouse using CRISPR/Cas9 on a C57BL/6 background. Loxp sites were placed flanking Exon 2 of Gimap5 (Fig. E3) and subsequent Gimap5fl/fl mice were crossed to Cd4cre-ert2 mice, allowing the tamoxifen-inducible deletion of Gimap5 in CD4+ T cells (cKO). Effective ablation of Gimap5 expression in CD4+ T cells was achieved with the administration of tamoxifen in chow (Fig. 6A).
FIG 6.
Impaired survival and increased DNA damage associated with Gimap5-deficiency is CD4+ T cell intrinsic. (A) Expression of Gimap5 in CD4+ T cells isolated from WT and Gimap5fl/flCd4cre-ert2 mice after tamoxifen-induced deletion of Gimap5. Immunoblot shows three independent cKO mice. (B-C) Proliferation and survival of naïve CD4+ T cells from WT and Gimap5fl/flCd4cre-ert2 mice stimulated 3d in nonpolarizing conditions (TH0) or TH1, TH2, TH17, or iTreg polarizing conditions. Blue histograms represent dead cells; red histograms show live cells. (D) γH2AX expression in live, cycling CD4+ T cells after 3d stimulation in TH0, TH1, TH2, TH17, or iTreg polarizing conditions. Data represent mean values ± SD (n=4–5). Statistical significance is determined by ANOVA followed by Sidak’s multiple comparisons test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
Similar to CD4+ T cells isolated from the Gimap5sph/sph mouse, Gimap5fl/flCd4cre-ert2 naïve CD4+ T cells demonstrated impaired survival and proliferation in TH0, TH1, and TH2 polarizing conditions compared to WT controls. Furthermore, no significant difference in survival and proliferation between WT and cKO CD4+ T cells was observed in TH17 polarizing conditions (Fig. 6B-C). Together, these results correlated with DNA damage observed in cycling CD4+ T cells with cKO CD4+ T cells displaying elevated γH2AX compared to WT CD4+ T cells stimulated in TH0, TH1, TH2, and iTreg polarizing conditions. Similar to Gimap5sph/sph CD4+ T cells, no difference in survival/proliferation was observed in TH17 polarizing conditions. These data confirm that the impaired survival and increased DNA damage in stimulated Gimap5-deficient CD4+ T cells are cell intrinsic.
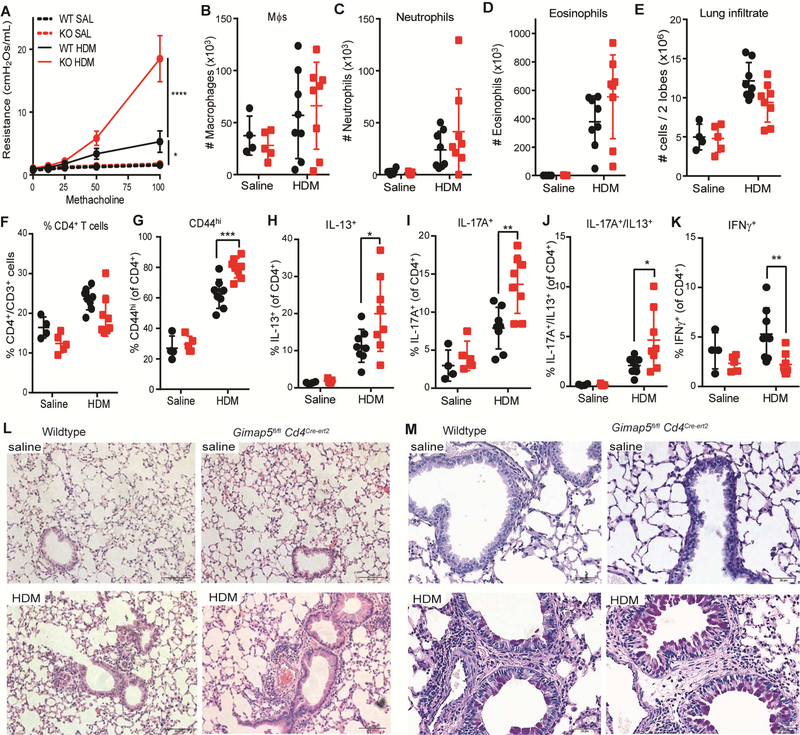
Loss of Gimap5 in CD4+ T cells is sufficient to drive exacerbated AHR and pathology
As pathogenic CD4+ T cells are one of the primary drivers of HDM-induced lung pathology, we asked if Gimap5-deficiency in CD4+ T cells alone was sufficient to drive more severe disease in the HDM model of allergic asthma. To do so, one week after starting the tamoxifen chow, allergic airway disease was induced WT and Gimap5fl/flCd4cre-ert2 mice as above. Comparable to Gimap5sph/sph mice, conditional deletion of Gimap5 in CD4+ T cells was sufficient to drive increased AHR in HDM-sensitized mice (Fig 7A). While exacerbated AHR was not specifically associated with changes in the numbers of macrophages and neutrophils in the BALF, the number of infiltrating eosinophils trended upwards in HDM-sensitized Gimap5fl/flCd4cre-ert2 mice compared to WT mice (Fig. 7B-D). Interestingly, while the number of lung-infiltrating CD4+ T cells in HDM-sensitized Gimap5fl/flCd4cre-ert2 mice was similar, or even slightly reduced, compared to WT mice (Fig. 7E-F), a higher proportion expressed an activated phenotype (CD44hi) (Fig. 7G). Furthermore, they exhibited an increased frequency of CD4+ T cells producing IL-13, IL-17A, or both IL-17A and IL-13 together (Fig. 7H-J), while the frequency of IFNγ+ CD4+ T cells was reduced compared to WT mice (Fig.7 K). Notably, similar levels of regulatory CD4+ T cells were observed in the lungs WT and Gimap5fl/flCd4cre-ert2 mice, indicating that the development of iTreg cells and/or recruitment is not affected in this model (Fig. E4A). B cells were likewise unaffected (Fig. E4B). Similar to the Gimap5sph/sph mice, HDM sensitization resulted in edema with perivascular inflammation and increased mucus production in the lungs of cKO mice (Fig. 7L-M). These data reveal that the conditional loss of Gimap5 in CD4+ T cells is sufficient to drive the development of pathogenic CD4+ T cells and exacerbate allergy-like lung disease upon HDM-sensitization.
FIG 7.
Gimap5-deficiency in CD4+ T cells is sufficient to drive increased AHR, CD4+ T cell polarization, and airway pathology upon HDM sensitization. C57BL/6 WT (black data points) and Gimap5fl/flCd4cre-ert2 (red data points) mice were administered 50 μg HDM 9 times IT over three weeks. (A) Airway hyper-responsiveness (AHR) in response to methacholine challenge 1 day after the last HDM challenge (saline: n=4–5; HDM: n=7–8). (B-D) Number of macrophages, neutrophils, and eosinophils in the BAL fluid (saline: n=4–5; HDM: n=8). (E-F). Number of (E) total lung-infiltrating cells (in 2 lobes) and (F) frequency of CD4+CD3+ T cells therein. (G) Frequency of CD4+CD3+ T cells expressing an activated (CD44hi) phenotype. (H-K) Percentage of lung-infiltrating CD4+CD3+ T cells expressing IL-13, IL-17A, IL-13 and IL-17A, or IFNγ upon restimulation with PMA/ionomycin for 6h (saline: n=4–5; HDM: n=8). (L-M) Representative (L) H&E and (M) PAS images of lung tissue from WT and Gimap5fl/flCd4cre-ert2 mice given saline or HDM. Data represents mean values ± SD. Statistical significance is determined by ANOVA followed by Sidak’s multiple comparisons test. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
As we have previously shown that Gimap5-deficient Tregs have impaired suppressive capacity, we next asked defective Tregs were responsible for the development of more severe airway disease. Interestingly, selective ablation of Gimap5 in Tregs was insufficient to drive exacerbated airway disease and development of pathogenic CD4+ T cells, indicating that loss of Gimap5 intrinsically promotes the polarization of pathogenic CD4+ T cells (Fig E5).
Discussion
Herein we describe a key role for Gimap5 in the prevention of pathogenic CD4+ T cell development and show that loss of Gimap5 is associated with an increased risk for allergic lung disease. Specifically, HDM sensitization of Gimap5-deficient mice results in severe airway hyperresponsiveness, eosinophil recruitment to the lung, and an exacerbated TH2/TH17 response. The increased airway inflammation was also observed in conditional loss of Gimap5 in CD4+ T cells demonstrating that these cells are the primary driver of HDM-induced lung pathology. Moreover, even in the absence of HDM sensitization, Gimap5sph/sph CD4+ T cells progressively adopt an inflammatory phenotype, with TH17 cells being particularly prominent. Importantly, the abnormal CD4+ T cell polarization profile is also observed in a GIMAP5-deficient patient that exhibited an increased development of pathogenic CD4+ T cells in the periphery. Thus, despite the observed lymphopenia in both Gimap5-deficient mice and humans, the selective survival of pathogenic TH17 cells may be a key driver of immune pathology in allergic airway disease.
Our current findings that loss of GIMAP5 leads to pathogenic CD4+ T cell development may help to understand how GIMAP5 polymorphism rs6965571 in the promoter region of GIMAP5 predisposes to asthma development.25 The rs6965571 SNP is adjacent to a Foxo1 binding site (upstream) and an NFATc1 binding motif (downstream) and could affect the binding of these transcription factors (TFs) to regulate GIMAP5 expression. Interestingly, our previous studies identified an impaired Foxo1 expression15 and NFATc1 activity upon stimulation of Gimap5-deficient CD4+ T cells.14 Thus, a potential feed-forward loop exists in which Gimap5 expression is required for the expression and activity of TFs that in turn regulate the transcriptional expression of GIMAP5. Thus the SNP may cause a progressive loss of GIMAP5 expression following T cell activation leading to an increased development of TH2/TH17 cells that promote immune pathology in the lung. Previously, the presence of TH2/ TH17 cells has been associated with more severe disease, and adoptive transfer experiments in mice have demonstrated that these cells are more pathogenic than classic TH2 cells.2–4 Furthermore, elevated IL-17A, either from these double producers or classic TH17 cells, has been linked to more severe disease and steroid resistance in human patients.5–11 This elevated IL-17A can drive increased pathology directly through recruitment of neutrophils or, as shown by Hall et al., or indirectly by modulating TH2 pathogenicity by enhancing IL-13/STAT6 signaling.46,47 Thus a critical future question to address is to identify the effect of the rs6965571 on GIMAP5 expression upon T cell activation and to establish whether patients with this SNP have an increased number of pathogenic (TH17) CD4+ T cells.
While DNA damage occurs normally upon mitogenic stimulation in CD4+ T cells, a key finding is the exacerbated DNA damage in cycling Gimap5-deficient CD4+ T cells that is associated with reduced T cell survival that is independent of Fas-signaling or intrinsic apoptotic cascades.14 Importantly, our previous studies identified a key role for Gimap5 in regulating GSK3 activity and subsequent control of the DNA damage response during CD4+ T cell proliferation.14 Here we show that DNA damage was particularly associated with TH1 and TH2 polarizing conditions. Interestingly, the presence of TGFβ could limit the extent of DNA damage even in “TH1” and “TH2” polarizing conditions. Previous studies have shown that TGFβ signaling can promote DNA damage repair and cell viability by inducing activation of ataxia-telangiectasia-mutated (ATM),48,49 suggesting that TGFβ signaling can overcome the exacerbated DNA damage observed in activated Gimap5-deficient CD4+ T cells. Moreover, we show that initiation of TGFβ signaling even 2 days after initial TCR stimulation is sufficient to reduce cell death and DNA damage in vitro in Gimap5-deficient CD4+ T cells. In vivo, this would translate towards improved survival of newly activated CD4+ T cells encountering TGFβ in the lung or other TGFβ-rich tissues. We posit that the ability of TGFβ to overcome the survival defects associated with Gimap5-deficiency could affect the CD4+ T cell compartment by promoting the selective survival of TH17 cells and other pathogenic CD4+ T cells infiltrating the TGFβ-rich asthmatic lung, where they drive further immune pathology.
Somewhat paradoxically, Gimap5-deficient mice fail to induce iTregs in vivo and retained Tregs have reduced suppressive capacity.14,15 It remains unclear why TGFβ rescues TH17 CD4+ T cell development, but not iTreg polarization and function, although our previous data suggest that the uncontrolled activity of GSK3β in the absence of Gimap5 may be critical to this process. Several independent studies demonstrate that GSK3β activity promotes TH17 polarization while impairing iTreg activity, providing one possible mechanism by which TH17 polarization is favored in the absence of Gimap5.50–56 Furthermore, Foxo family members, which are poorly expressed in Gimap5-deficient CD4+ T cells,15 are key TFs in Treg development and function.57,58 Impaired expression of these TFs is predicted to be a critical factor in the inability of Gimap5sph/sph mice to develop iTregs. This dysbalance between pathogenic CD4+ T cells and functional regulatory T cells is one of the primary driving factors behind the immune pathologies associated with Gimap5 deficiency. Unfortunately, the inability of TGFβ to rescue iTreg function, its promotion of the development of pathogenic TH17 CD4+ T cells, and its critical role in the resolution of inflammation makes it an impractical target in clinical applications. However, therapeutic targeting of other links in the GIMAP5-pathway, such as GSK3β, may offer more promise in clinical options for patients with GIMAP5-associated asthma.
Supplementary Material
Key Messages.
GIMAP5 LOF mutations in mice and humans result in increased TH2/TH17 polarization in vivo.
Gimap5-deficienct CD4+ T cells exhibit increased DNA damage and reduced survival upon activation that can be overcome by TGFβ.
Gimap5-deficient mice develop more severe allergic airway disease upon HDM sensitization.
Acknowledgements:
We thank G. Butcher for graciously providing the MAC421 αGimap5 antibody.
Funding: The research was funded by the Crohn’s & Colitis Foundation of America (#3793) and the NIH, PHS Grant P30 DK078392 (Integrative Morphology Core of the Cincinnati Digestive Disease Research Core Center) and R21# R21AI131050.
Abbreviations used
- AHR
Airway hyper responsiveness
- BALF
Bronchoalveolar lavage fluid
- cKO
conditional knockout
- GIMAP5
GTPase of Immunity-associated protein 5
- HDM
House dust mite
- IT
Intratracheal
- iTreg
induced regulatory T cell
- LOF
Loss-of-function
- γH2AX
phosphorylated histone H2AX
- mLN
mesenteric lymph node
- mdLN
mediastinal lymph node
- PMA
Phorbol 12-myristate-13-acetate
- SLE
Systemic lupus erythematosus
- SNPs
Single nucleotide polymorphisms
- T1D
Type 1 diabetes
- WT
Wildtype
Footnotes
Disclosure of conflict of interest: The authors declare that they have no relevant conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hekking PPW, Wener RR, Amelink M, Zwinderman AH, Bouvy ML, Bel EH. The prevalence of severe refractory asthma. J Allergy Clin Immunol [Internet]. 2015;135:896–902. Available from: 10.1016/j.jaci.2014.08.042 [DOI] [PubMed] [Google Scholar]
- 2.Irvin C, Zafar I, Good J, Rollins D, Christianson C, Gorska MM, et al. Increased frequency of dual-positive TH2/TH17 cells in bronchoalveolar lavage fluid characterizes a population of patients with severe asthma. J Allergy Clin Immunol [Internet]. 2014;134:1175–1186.e7. Available from: 10.1016/j.jaci.2014.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y-H, Voo KS, Liu B, Chen C-Y, Uygungil B, Spoede W, et al. A novel subset of CD4 + T H 2 memory/effector cells that produce inflammatory IL-17 cytokine and promote the exacerbation of chronic allergic asthma. J Exp Med [Internet]. 2010;207:2479–91. Available from: http://www.jem.org/lookup/doi/10.1084/jem.20101376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosmi L, Maggi L, Santarlasci V, Capone M, Cardilicchia E, Frosali F, et al. Identification of a novel subset of human circulating memory CD4+ T cells that produce both IL-17A and IL-4. J Allergy Clin Immunol [Internet]. 2010;125:222–230.e4. Available from: 10.1016/j.jaci.2009.10.012 [DOI] [PubMed] [Google Scholar]
- 5.Molet S, Hamid Q, Davoine F, Nutku E, Taha R, Pagé N, et al. IL-17 is increased in asthmatic airways and induces human bronchial fibroblasts to produce cytokines. J Allergy Clin Immunol. 2001;108:430–8. [DOI] [PubMed] [Google Scholar]
- 6.Pene J, Chevalier S, Preisser L, Venereau E, Guilleux M-H, Ghannam S, et al. Chronically Inflamed Human Tissues Are Infiltrated by Highly Differentiated Th17 Lymphocytes. J Immunol [Internet]. 2008;180:7423–30. Available from: http://www.jimmunol.org/cgi/doi/10.4049/jimmunol.180.11.7423 [DOI] [PubMed] [Google Scholar]
- 7.Chien JW, Lin CY, Yang KD, Lin CH, Kao JK, Tsai YG. Increased IL-17A secreting CD4+ T cells, serum IL-17 levels and exhaled nitric oxide are correlated with childhood asthma severity. Clin Exp Allergy. 2013;43:1018–26. [DOI] [PubMed] [Google Scholar]
- 8.Bullens DMA, Truyen E, Coteur L, Dilissen E, Hellings PW, Dupont LJ, et al. IL-17 mRNA in sputum of asthmatic patients: Linking T cell driven inflammation and granulocytic influx? Respir Res. 2006;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Ramli W, Préfontaine D, Chouiali F, Martin JG, Olivenstein R, Lemière C, et al. TH17-associated cytokines (IL-17A and IL-17F) in severe asthma. J Allergy Clin Immunol. 2009;123:1185–7. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Yang J, Gao Y, Guo W. Th17 Immunity in Patients with Allergic Asthma. Int Arch Allergy Immunol [Internet]. 2010;151:297–307. Available from: https://www.karger.com/?doi=10.1159/000250438 [DOI] [PubMed] [Google Scholar]
- 11.Nanzer AM, Chambers ES, Ryanna K, Richards DF, Black C, Timms PM, et al. Enhanced production of IL-17A in patients with severe asthma is inhibited by 1α,25-dihydroxyvitamin D3 in a glucocorticoid-independent fashion. J Allergy Clin Immunol [Internet]. 2013;132:297–304.e3. Available from: 10.1016/j.jaci.2013.03.037 [DOI] [PubMed] [Google Scholar]
- 12.Barnes MJ, Aksoylar H, Krebs P, Bourdeau T, Arnold CN, Xia Y, et al. Loss of T Cell and B Cell Quiescence Precedes the Onset of Microbial Flora-Dependent Wasting Disease and Intestinal Inflammation in Gimap5-Deficient Mice. J Immunol [Internet]. 2010;184:3743 Available from: http://uc.summon.serialssolutions.com/2.0.0/link/0/eLvHCXMw3V1Lj9MwELa6CNBKCMHy2PCQfAAuqy6xk9jNgcN2BSzPy3bPkZM4IqKvbRKV_Sn8W2ZsN0mrInHmlrZOmna-zMMz8w0hlBWjGLz-INPwpCkJPnnm85RnIWd5LEyDWG-Uz2DD5Ne99z9I-SvYOPT3Jie4A28yAWN7eN2Uhqkpw04onekcuRwAE1g9bbL_s9JwMGED [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X-L, Serrano D, Ghobadi F, Mayhue M, Hoebe K, Ilangumaran S, et al. TCR and IL-7 Signaling Are Altered in the Absence of Functional GTPase of the Immune Associated Nucleotide Binding Protein 5 (GIMAP5). PLoS One [Internet]. 2016;11:e0151837 Available from: http://dx.plos.org/10.1371/journal.pone.0151837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patterson AR, Endale M, Lampe K, Aksoylar HI, Flagg A, Woodgett JR, et al. Gimap5-dependent inactivation of GSK3β is required for CD4+ T cell homeostasis and prevention of immune pathology. Nat Commun [Internet]. 2018;9:430 Available from: http://www.nature.com/articles/s41467-018-02897-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aksoylar HI, Lampe K, Barnes MJ, Plas DR, Hoebe K. Loss of immunological tolerance in Gimap5-deficient mice is associated with loss of Foxo in CD4+ T cells. J Immunol [Internet]. 2012;188:146–54. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3258489&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer HJ, Witte a.-K, Walter L, Gro ne H-J, van den Brandt J, Reichardt HM Distinct roles of T-cell lymphopenia and the microbial flora for gastrointestinal and CNS autoimmunity. FASEB J [Internet]. 2016;1–9. Available from: http://www.fasebj.org/cgi/doi/10.1096/fj.15-277384 [DOI] [PubMed]
- 17.van den Brandt J, Fischer HJ, Walter L, Hunig T, Kloting I, Reichardt HM. Type 1 Diabetes in BioBreeding Rats Is Critically Linked to an Imbalance between Th17 and Regulatory T Cells and an Altered TCR Repertoire. J Immunol [Internet]. 2010;185:2285–94. Available from: http://www.jimmunol.org/cgi/doi/10.4049/jimmunol.1000462 [DOI] [PubMed] [Google Scholar]
- 18.Poussier P, Ning T, Murphy T, Dabrowski D, Ramanathan S. Impaired post-thymic development of regulatory CD4+25+ T cells contributes to diabetes pathogenesis in BB rats. J Immunol [Internet]. 2005;174:4081–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15778366http://www.jimmunol.org/content/174/7/4081.full.pdf%5Cnhttp://www.ncbi.nlm.nih.gov/pubmed/15778366 [DOI] [PubMed] [Google Scholar]
- 19.Schulteis RD, Chu H, Dai X, Chen Y, Edwards B, Haribhai D, et al. Impaired survival of peripheral T cells, disrupted NK/NKT cell development, and liver failure in mice lacking Gimap5. Blood [Internet]. 2008;112:4905 Available from: http://uc.summon.serialssolutions.com/2.0.0/link/0/eLvHCXMwlV1Za9wwEB7S0gtKaTdt4h6ghx4vMVldtkWf0qXblNA8lO2z0NoSMSTuEtuQ_vuO5CPepYX2zcfICGbGM580-gaAUJcpzPp5btHTTIo5eT5na5YLRguVhANik1Y-I0jc2b3HxPe4K97utuh5TD3Xl7yDSZDy9vv9dDnuFwjOul4FiI-FSml_SO5v39gOQgMx8CQI [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nitta T, Nasreen M, Seike T, Goji A, Ohigashi I, Miyazaki T, et al. IAN family critically regulates survival and development of T lymphocytes. PLoS Biol. 2006;4:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cousins L, Graham M, Tooze R, Carter C, Miller JR, Powrie FM, et al. Eosinophilic Bowel Disease Controlled by the BB Rat-Derived Lymphopenia/Gimap5 Gene. Gastroenterology. 2006;131:1475–85. [DOI] [PubMed] [Google Scholar]
- 22.Hellquist A, Skoog T, Vendelin J, Cunninghame-Graham DS, Vyse TJ, Kere J, et al. The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. J Med Genet [Internet]. 2007;44:314 Available from: http://uc.summon.serialssolutions.com/2.0.0/link/0/eLvHCXMwnV1Nb9QwEB1VCBBShUr5ChTkA1RcWhI7sZNDD1VFgUMlDnC2HMeBrXY3q00idfvrO2N7V7sVB8RxY2cTZ2Y8M_bzGwCWtWWFUb-wDi3NKIzJbcprbnOeNZX0B8S2SvnAGjJGiModXOLpfPLHYyvjR1z64mqnLSWJdKk5w9hdVWV1PHTd9Gwx8_e52-PQ0bMOmZpW [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lim MK, Sheen DH, Kim SA, Won SK, Lee SS, Chae SC, et al. IAN5 polymorphisms are associated with systemic lupus erythematosus. Lupus [Internet]. 2009;18:1045 Available from: http://search.ebscohost.com/login.aspx?direct=true&db=mnh&AN=19762377&site=ehost-live&scope=site [DOI] [PubMed] [Google Scholar]
- 24.Shin JH, Janer M, McNeney B, Blay S, Deutsch K, Sanjeevi CB, et al. IA-2 autoantibodies in incident type I diabetes patients are associated with a polyadenylation signal polymorphism in GIMAP5. Genes Immun [Internet]. 2007;8:503 Available from: http://uc.summon.serialssolutions.com/2.0.0/link/0/eLvHCXMw3V1bq9QwEA6LKAgi3l31QB70ICyFNEmb9sGH5XhbUBFdfQ1pmi4re3Zl24IL_nhnmt52UTjPvnbStHS-JDPTmW8IoWGRpGD1C-tgpRkFNrllPONW8jBP46ZAbNTKZ9LRCQ3X_gctL-YBn5m62sEHWmc7TAnECAYG0LEC10dYF0OEtaVRLWeY8GVa1XQZ6AZbNhwM [DOI] [PubMed] [Google Scholar]
- 25.Heinonen MT, Laine A-P, Söderhäll C, Gruzieva O, Rautio S, Melén E, et al. GIMAP GTPase Family Genes: Potential Modifiers in Autoimmune Diabetes, Asthma, and Allergy. J Immunol [Internet]. 2015;194:5885 Available from: http://www.jimmunol.org/content/194/12/5885.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahern PP, Schiering C, Buonocore S, McGeachy MJ, Cua DJ, Maloy KJ, et al. Interleukin-23 Drives Intestinal Inflammation through Direct Activity on T Cells. Immunity [Internet]. 2010;33:279–88. Available from: 10.1016/j.immuni.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, et al. Late Developmental Plasticity in the T Helper 17 Lineage. Immunity [Internet]. 2009;30:92–107. Available from: 10.1016/j.immuni.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Annunziato F, Cosmi L, Santarlasci V, Maggi L, Liotta F, Mazzinghi B, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kebir H, Ifergan I, Alvarez JI, Bernard M, Poirier J, Arbour N, et al. Preferential recruitment of interferon-y-expressing TH17 cells in multiple sclerosis. Ann Neurol. 2009;66:390–402. [DOI] [PubMed] [Google Scholar]
- 30.Harbour SN, Maynard CL, Zindl CL, Schoeb TR, Weaver CT. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc Natl Acad Sci [Internet]. 2015;112:201415675 Available from: http://www.pnas.org.remote.library.osaka-u.ac.jp/content/112/22/7061.long [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Biagini Myers JM, Brandt EB, Ryan PH, Lindsey M, Mintz-Cole RA, et al. β-Glucan exacerbates allergic asthma independent of fungal sensitization and promotes steroid-resistant TH2/TH17 responses. J Allergy Clin Immunol [Internet]. 2017;139:54–65.e8. Available from: 10.1016/j.jaci.2016.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee Y, Awasthi A, Yosef N, Quintana FJ, Xiao S, Peters A, et al. Induction and molecular signature of pathogenic TH17 cells. Nat Immunol. 2012;13:991–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tatler AL, Jenkins G. TGF- b Activation and Lung Fibrosis. [DOI] [PubMed]
- 34.Jenkins RG, Su X, Su G, Scotton CJ, Camerer E, Laurent GJ, et al. Ligation of protease-activated receptor 1 enhances α v β 6 integrin – dependent TGF-β activation and promotes acute lung injury. J Clin incestigation. 2006;116:1606–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kitamura H, Cambier S, Somanath S, Barker T, Minagawa S, Markovics J, et al. Mouse and human lung fibroblasts regulate dendritic cell trafficking, airway inflammation, and fibrosis through integrin α v β 8 – mediated activation of TGF- β. J Clincal Investig. 2011;121:40–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindstedt K a, Wang Y, Shiota N, Saarinen J, Hyytiäinen M, Kokkonen JO, et al. Activation of paracrine TGF-beta1 signaling upon stimulation and degranulation of rat serosal mast cells: a novel function for chymase. FASEB J [Internet]. 2001;15:1377–88. Available from: http://www.ncbi.nlm.nih.gov/pubmed/11387235 [DOI] [PubMed] [Google Scholar]
- 37.Herz U, Renz H, Wiedermann U. Animal models of type I allergy using recombinant allergens. Methods. 2004;32:271–80. [DOI] [PubMed] [Google Scholar]
- 38.Gueders MM, Paulissen G, Crahay C, Quesada-Calvo F, Hacha J, Van Hove C, et al. Mouse models of asthma: A comparison between C57BL/6 and BALB/c strains regarding bronchial responsiveness, inflammation, and cytokine production. Inflamm Res. 2009;58:845–54. [DOI] [PubMed] [Google Scholar]
- 39.Brandt EB, Kovacic MB, Lee GB, Gibson AM, Acciani TH, Le Cras TD, et al. Diesel exhaust particle induction of IL-17A contributes to severe asthma. J Allergy Clin Immunol [Internet]. 2013;132:1194–1204.e2. Available from: 10.1016/j.jaci.2013.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HY, Lee HJ, Chang YJ, Pichavant M, Shore SA, Fitzgerald KA, et al. Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat Med [Internet]. 2014;20:54–61. Available from: 10.1038/nm.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hekking PP, Loza MJ, Pavlidis S, de Meulder B, Lefaudeux D, Baribaud F, et al. Pathway discovery using transcriptomic profiles in adult-onset severe asthma. J Allergy Clin Immunol. 2017;1–11. [DOI] [PubMed] [Google Scholar]
- 42.Wallrapp A, Riesenfeld SJ, Burkett PR, Abdulnour REE, Nyman J, Dionne D, et al. The neuropeptide NMU amplifies ILC2-driven allergic lung inflammation. Nature [Internet]. 2017;549:351–6. Available from: 10.1038/nature24029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Motomura Y, Morita H, Moro K, Nakae S, Artis D, Endo TA, et al. Basophil-derived interleukin-4 controls the function of natural helper cells, a member of ILC2s, in lung inflammation. Immunity [Internet]. 2014;40:758–71. Available from: 10.1016/j.immuni.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 44.Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, et al. A role for IL-25 and IL-33–driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med [Internet]. 2013;210:2939–50. Available from: http://www.jem.org/lookup/doi/10.1084/jem.20130351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Halim TYF, Steer CA, Mathä L, Gold MJ, Martinez-Gonzalez I, McNagny KM, et al. Group 2 innate lymphoid cells are critical for the initiation of adaptive T helper 2 cell-mediated allergic lung inflammation. Immunity. 2014;40:425–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hall SL, Baker T, Lajoie S, Richgels PK, Yang Y, McAlees JW, et al. IL-17A enhances IL-13 activity by enhancing IL-13–induced signal transducer and activator of transcription 6 activation. J Allergy Clin Immunol [Internet]. 2017;139:462–471.e14. Available from: 10.1016/j.jaci.2016.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Busse WW, Holgate S, Kerwin E, Chon Y, Feng JY, Lin J, et al. Randomized, double-blind, placebo-controlled study of brodalumab, a human anti-IL-17 receptor monoclonal antibody, in moderate to severe asthma. Am J Respir Crit Care Med. 2013;188:1294–302. [DOI] [PubMed] [Google Scholar]
- 48.Hardee ME, Marciscano AE, Medina-Ramirez CM, Zagzag D, Narayana A, Lonning SM, et al. Resistance of glioblastoma-initiating cells to radiation mediated by the tumor microenvironment can be abolished by inhibiting transforming growth factor-β. Cancer Res. 2012;72:4119–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu C Te Hsieh CC, Yen TC Chen WC, Chen MF. TGF-β1 mediates the radiation response of prostate cancer. J Mol Med. 2014;93:73–82. [DOI] [PubMed] [Google Scholar]
- 50.Beurel E, Yeh W-I, Michalek SM, Harrington LE, Jope RS. Glycogen synthase kinase-3 is an early determinant in the differentiation of pathogenic Th17 cells. J Immunol. 2011;186:1391–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan M, Winslow MM, Chen L, Kuo A, Felsher D, Crabtree GR. Enhanced NFATc1 nuclear occupancy causes T cell activation independent of CD28 costimulation. J Immunol. 2007;178:4315–21. [DOI] [PubMed] [Google Scholar]
- 52.Graham JA, Fray M, De Haseth S, Lee KM, Lian MM, Chase CM, et al. Suppressive regulatory T cell activity is potentiated by glycogen synthase kinase 3β inhibition. J Biol Chem. 2010;285:32852–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welsh GI, Miyamoto S, Priced NT, Safer B, Proud CG. T-cell activation leads to rapid stimulation of translation initiation factor eIF2B and inactivation of glycogen synthase kinase-3. J Biol Chem. 1996;271:11410–3. [DOI] [PubMed] [Google Scholar]
- 54.Ohteki T, Parsons M, Zakarian a, Jones RG, Nguyen LT, Woodgett JR, et al. Negative regulation of T cell proliferation and interleukin 2 production by the serine threonine kinase GSK-3. J Exp Med. 2000;192:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beurel E, Michalek SM, Jope RS. Innate and adaptive immune responses regulated by glycogen synthae kinase-3 (GSK3). Tremds Immunol. 2010;31:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Beurel E, Kaidanovich-Beilin O, Yeh W-I, Song L, Palomo V, Michalek SM, et al. Regulation of Th1 Cells and Experimental Autoimmune Encephalomyelitis by Glycogen Synthase Kinase-3. J Immunol [Internet]. 2013;190:5000–11. Available from: http://www.jimmunol.org/cgi/doi/10.4049/jimmunol.1203057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kerdiles YM, Stone EL, Beisner DL, McGargill MA, Ch’en IL, Stockmann C, et al. Foxo Transcription Factors Control Regulatory T Cell Development and Function. Immunity [Internet]. 2010;33:890–904. Available from: 10.1016/j.immuni.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouyang W, Beckett O, Ma Q, Paik JH, Depinho RA, Li MO. Foxo proteins cooperatively control the differentiation of Foxp3 + regulatory T cells. Nat Immunol [Internet]. 2010;11:618–27. Available from: 10.1038/ni.1884 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.