Abstract
The severity of the 2017 – 2018 influenza season, combined with the low efficacy for some vaccine components, highlights the need to improve our current seasonal influenza vaccine. Thus, the National Institute of Allergy and Infectious Diseases (NIAID) recently announced a strategic plan to improve current influenza vaccines and eventually develop a “universal” influenza vaccine. This review will highlight the many different strategies being undertaken in pursuit of this goal and the exciting advances made by the influenza community. There is no doubt that an improved influenza vaccine is on the horizon.
Introduction
The 2017 – 2018 influenza season was a stark reminder that outbreaks of influenza virus are associated with significant morbidity and mortality worldwide. The Centers for Disease Control and Prevention reported over 30,000 laboratory-confirmed influenza-related hospitalizations and 171 confirmed pediatric deaths in the United States (1). Severe disease is most commonly seen in adults with underlying medical conditions including cardiovascular disease, metabolic disorders and obesity as examples. A severity assessment classified the 2017–18 season as high overall severity for each age group (children, adolescents, adults and older adults); something that hasn’t been observed since the 2003–04 season (1). The severity of this influenza season highlights the importance of measures to control and even prevent influenza virus infections.
Arguably, the most effective means to prevent influenza is through vaccination (https://www.cdc.gov/flu/consumer/prevention.htm). Yet, the hallmark of influenza viruses is the ability to undergo rapid antigenic variation due to the accumulation of mutations within the antibody-binding sites in the hemagglutinin (HA) and neuraminidase (NA) surface proteins, abrogating the binding of some antibodies (2). This antigenic drift requires that the World Health Organization (WHO) advisory group of experts meet biannually to analyze influenza surveillance data generated by the WHO Global Influenza Surveillance and Response System (GISRS) to determine if the influenza vaccine candidate viruses must be updated (http://www.who.int/influenza/vaccines/virus/recommendations/consultation201809/en/). Continual surveillance of circulating influenza viruses is crucial for the success of this process and timely production of our annual influenza vaccines (3, 4).
Currently, there are three main categories of annual vaccines approved by the Food and Drug Administration (FDA), the most common being the detergent-split inactivated influenza vaccine (IIV) (https://www.fda.gov/biologicsbloodvaccines/vaccines/approvedproducts/ucm093833.htm). IIV is composed of three or four candidate vaccine viruses (CVVs) including an influenza A H1N1 and H3N2 virus as well as influenza B viruses representing either one or both genetically distinct clades (Victoria or Yamagata). Some examples of the IIV include Fluzone®, Fluarix™, and Flucelvax®. CVVs used in the IIV can be grown in embryonated chicken eggs or Madin-Darby Canine Kidney (MDCK) cells, after which they are inactivated, purified and detergent-split (4). The antigen is primarily composed of the influenza HA protein, although trace amounts of NA protein may also be present. The vaccine is administered intramuscularly to elicit a protective antibody immune response. The second vaccine category is the recombinant influenza vaccine (RIV), known as Flublok®. The RIV is solely composed of the HA protein from the chosen CVVs for that particular year (5). Unlike IIV, RIV is produced and isolated solely in egg-free systems by expressing the HA in insect cells by baculovirus (6). Finally, a live attenuated influenza vaccine (LAIV) is available as FluMist®. Like the IIV, the LAIV is composed of three or four CVVs. However, these viruses have been engineered to grow at or below 33˚C, limiting replication to the upper respiratory tract (URT) (7, 8). Because they are attenuated live viruses, they elicit a more robust immune response including both B and T cell responses (9, 10).
While vaccines are our best line of defense against influenza, they can be improved. Driving antibody responses against the antigenic sites in the HA head is problematic given the constant drift in this region. It requires continually updating the vaccine to keep up with viral evolution. Growth of CVVs in eggs for vaccine production can also lead to mutations in the HA head region, reducing the efficacy of vaccine-generated antibody responses to circulating viruses (11, 12). This has been an issue for the H3N2 component of the vaccine for several recent influenza seasons (13) (https://www.scientificamerican.com/article/ldquo-the-problem-child-of-seasonal-flu-rdquo-beware-this-winter-rsquo-s-virus/). Finally, the IIV drives a strong humoral response to the HA component, which can be impacted by the immunization and infection history of the person (14–16).
In regards to the LAIV, which was initially licensed in the United States in 2003 for use in people ages 2 – 49 (17), low effectiveness against influenza A(H1N1)pdm09-like viruses circulating in the United States during the 2013–14 and 2015–16 seasons resulted in the Advisory Committee on Immunization Practices (ACIP) recommendation that LAIV not be used in the 2017–18 season (17). On February 21, 2018, ACIP recommended the LAIV as a vaccine option for the 2018–19 season (17). The reason for the low efficacy remains unclear but a study in the European version of the US vaccine showed substantial amounts of defective-interfering viral RNAs from both the influenza A and influenza B viruses (18). Given these challenges, the National Institute of Allergy and Infectious Diseases (NIAID) announced a strategic plan to improve current influenza vaccines and eventually lead to the development of a universal influenza vaccine (19). While a universal influenza vaccine would technically target all types of influenza viruses, for the purposes of the NIAID strategic plan “universal” refers to protection against both group 1 and 2 influenza A viruses (independent of influenza B protection). This review will highlight some advances in a few areas of the strategic plan.
HA-directed responses
The influenza HA and NA surface proteins are the major targets of immune responses elicited by vaccination, especially humoral responses (20). HA is the receptor binding protein in influenza. It consists of a globular head and a stem which is anchored to the viral envelope. The receptor binding portion of the head determines if the virus will bind to an α2,6 (mostly human), or α2,3 (mostly avian) linkage of the sialic acid. Once the virus gets internalized via endocytosis, an HA conformational change allows the virus to be released into the cytoplasm (21). Influenza A viruses are divided into two groups based on phylogenetic analysis of the HA protein (22). Group 1 contains H1 and H2 subtypes, which can sustain circulation in humans, as well as H5, H6, and H9, which can occasionally infect humans (22). The main subtype of group 2 is H3, but H7 and H10 can occasionally infect humans and be associated with severe and even fatal disease (22).
The predominant seasonal influenza vaccine-elicited immune response targets the globular head of HA for neutralization (23, 24). Antibodies bind to the HA to prevent binding to sialic acid or to prevent the conformational change that leads to fusion (25, 26). However, because of the previously mentioned mutability of the HA head, these responses only protect from identical strains of influenza. Emergence of vaccine escape variants leads to reduced protective immune responses (2, 27). To overcome this challenge, new strategies are being developed to target immune responses against the stem region of HA (28–30). The HA stem is highly conserved compared to the head making it a strong target for broadly protective immune responses (31). However, it is more difficult to mount immune responses to the stem due to the HA head’s immunodominance and steric hindrance of the stem (23, 24, 32). Thus, chimeric HA where the head of HA is changed and the stem is maintained have been developed (29, 33). Vaccination of mice with chimeric proteins of different group 1 virus heads (H1, H9, H6, or H5) with the same H1 stem could protect mice from challenge with a variety of group 1 viruses suggesting more broadly protective antibody responses. However, mice were not protected from challenge with H3N2 virus, an HA group 2 virus, demonstrating that protection did not extend to intergroup influenza virus strains (29). Similar results were shown in a ferret model (33).
Another strategy utilized for stalk-directed immune responses is a recombinant headless stem (34). While this strategy eliminates the immunodominance of the HA head, the resulting protein does not provide complete protection (34). Therefore, an initial dose of a vaccine that mimics a more natural infection will have to be utilized along with boosting of the primary immune response to elicit protective stem-directed antibody responses. Indeed, a recent study in ferrets demonstrated that a sequential immunization regimen can redirect the immune response towards conserved epitopes. Briefly, administration of a LAIV chimeric H8/1 (cH8/1) HA followed by a heterologous booster vaccination with a cH5/1N1 formalin inactivated non-adjuvanted whole virus resulted in low or undetectable titers in the URT after the A(H1N1)pdm09 virus challenge, supporting the further development of chimeric HA-based vaccination strategies (35). Boosting HA stem-directed antibodies can be done with either chimeric (29), or recombinant stable and correctly folded headless HA stem proteins (36, 37). One advantage of utilizing chimeric HA proteins is that people that have had natural influenza infection will already have some baseline stem-directed antibodies, which will benefit the boosting of the stem-directed antibody response (38–40). Of note, HA-stem directed antibodies are not necessarily neutralizing, and instead can employ different mechanisms, such as antibody-dependent cellular cytotoxicity (ADCC), to clear the virus after a permissive viral infection (41–43). Identifying approaches that can improve antibody responses to conserved regions of the HA head and stalk are an exciting development towards improved efficacy.
NA-directed responses
The second main surface protein of influenza virus, the NA protein, is a functional enzyme that cleaves sialic acid supporting the release of progeny virus during infection (21). Like HA, NA is divided into two groups based on phylogenetic analysis (22). The enzymatic activity of NA is the main target of antiviral drugs. While targeting this aspect of influenza replication is not necessarily sterilizing, it does limit viral replication and therefore decreases disease symptoms and viral spread (44–46). In fact, antiviral drugs against the enzymatic activity of NA are the only clinically relevant treatment for influenza infection, aside from supportive care. Therefore, targeting immune responses against the NA protein may be an important complement to HA-directed vaccines (47). A recent clinical study challenging young healthy adults with a pandemic H1N1 virus demonstrated better correlates of NA inhibition (NI) antibody titers with fewer, less severe, and less prolonged symptoms, as well as reduced viral shedding (46). In contrast, hemagglutination inhibition (HI) titers only correlated with a reduction in virus shedding (46). Similar results were seen with pre-existing anti-HA stalk antibodies as with HI titers (48). Another important aspect of NA-directed immune responses is that both inactivated and live-attenuated influenza vaccines induce increases in NI antibody titers. Additionally, NI antibody titers correlate with LAIV and IIV effectiveness (49, 50).
Despite these observations, some barriers must be overcome for the successful use of NA-directed immune responses by vaccination. First, like the HA stem, NA immunogenicity can be masked by the immunodominant HA head (51). However, because of the strong emphasis on HA quantification and standardization in influenza vaccines, low NA immunogenicity could be due to a lack of sufficient NA protein present in vaccines. While we know that vaccines contain some level of NA, given the increase in NI antibody titers after vaccination (52, 53), the content should be standardized. Yet before this can occur, we need better NA assays. The development of new and simplified techniques to determine NI antibody titers, including Enzyme Linked Lectin Assay (ELLA) and ELISA using NA in its native form, will help overcome these hurdles (54, 55). While these assays have good reproducibility amongst different laboratories (56), some caveats include steric hindrance or competition of HA-directed antibodies with NA-directed antibody activity (51). Therefore, ELISA using recombinant native form NA is probably the best option moving forward (57).
Once antigen content and NI assays have been standardized, the next hurdle to overcome will be the evolution or antigenic drift of NA. Although NA mutability is lower than HA, vaccine escapes and antiviral resistance strains are known to arise (58, 59). However, a positive aspect of influenza viruses is that HA subtypes and NA subtypes are not concordantly paired between group 1 and group 2 members. Driving vaccine responses against the conserved regions of both HA and NA may prove an exciting and important new approach to provide protective immunity and limit antigenic drift in the virus.
M2-directed responses
The third surface protein of influenza virus is the matrix protein 2 (M2). M2 functions in both viral entry and egress. During entry, M2 acts as a pH-dependent proton-selective ion channel that controls the internal acidity of virus particles in endosomes allowing for release of the nucleoprotein components (60). M2 also controls the pH of the Golgi lumen supporting viral assembly after replication (60–62). However, other functions of M2 are being uncovered (63, 64). In terms of vaccination, M2 serves as an interesting candidate given that it is highly conserved across multiple influenza virus strains (43, 64–66). In fact, avian influenza viruses also cross-react with human sera against the ectodomain of M2 (M2e) (67). Additionally, unlike HA and NA proteins, M2e mutations are non-existent for up to 11 passages in M2e-vaccinated mice (68), as well as rare and restricted in immunocompromised mice treated with anti-M2e antibodies (69). Despite the attractive nature of M2e as a vaccine antigen, its immunogenicity is low after natural infection (70).
To overcome this block, new strategies have been developed to induce M2e-directed antibody responses. The first of these fused M2e to the Hepatitis B virus core protein to form viral like particles (VLPs) with the M2e portion exposed on the surface (71). Those studies demonstrated that both intraperitoneal vaccination with adjuvants and intranasal vaccination without adjuvants protected against both group 1 and group 2 viruses (H1N1 and H3N2). Many other VLPs methods have now been employed with M2e (72–74). The protection elicited by antibodies to M2e are not sterilizing, and instead bind to the surface of virus-infected cells most likely acting through ADCC (75–78). The conservation and cross-protective properties of M2e are an exciting aspect of influenza vaccination improvement, and will most likely prove indispensable for the development of a universal influenza vaccine. Several recent reviews beautifully cover the state of knowledge of M2 and nucleoprotein (NP)-based vaccines (43, 66, 79).
T cell-directed responses
In addition to HA-, NA-, and M2-directed antibody responses, some vaccine platforms, for example the LAIV, induce T cell responses to conserved antigens in influenza viruses (9, 43, 80). Unlike most antibody vaccine responses, T cell responses are not used to neutralize the virus and prevent infection, but limit viral spread (81). This is achieved by the quick elimination of infected cells by CD8+ T cells or by the concerted direction of the immune response by CD4+ T cells. Of these, CD8+ T cells have been the major focus of influenza-directed T cell responses to date. There are many influenza virus epitopes that are recognized by CD8+ T cells (20, 81). While those epitopes include HA and NA, other more highly conserved proteins like NP, matrix protein 1 (M1), and the polymerase proteins are of more interest to the universal influenza vaccination field (82, 83). The T cell epitopes against these proteins are very well conserved among different influenza virus strains (20, 81). It is no surprise, therefore, that there are many reports of CD8+ T cell responses correlating with high cross-protection against heterologous strains of influenza viruses in both mice and humans (84–88).
While CD4+ T cell responses to influenza infection have not received as much focus as CD8+ T cells, their role is still of importance (81, 89). Memory CD4+ T cells help direct a faster antibody response to mutated or immunologically-novel viral antigens, as well as the generation of new CD8+ T cell responses. In fact, a recent study using a novel platform of influenza vaccination in mice elucidated a major contribution of CD4+ T cells to the cross-protective anti-influenza immune response (79). In these experiments, a vaccinia virus encoding 5 proteins from an H5N1 viral strain was used for immunization and boosting of mice, followed by challenge with an H3N2 virus. This type of vaccination provided complete protection from heterologous virus challenge of mice. Previous studies using this vaccine platform showed the development of antibody responses capable of cross-reacting with different subtypes of viruses (90). However, serum transfer of vaccinated to naïve mice was unable to protect from challenge with a heterologous influenza strain (79). In contrast to the lack of protection from adoptive sera transfer, transfer of either CD4+ or CD8+ T cells protected mice from heterologous challenge. Likewise, depletion of CD4+ or CD8+ T cells, separately, prior to challenge did not alter the protection conferred by the vaccine (79). Of interest, when CD4+ T cells were depleted at time of vaccination, all protection was lost. Therefore, CD4+ and CD8+ T cells seem to be playing an equivalent protective role during challenge, and CD4+ T cells play a necessary role during vaccination.
This previous method of eliciting T cell responses through vector expression is not unique (43, 91), with some studies including mainly T cell antigens (92, 93), while other studies combine T cell and antibody antigens (94, 95). One study showed that intramuscular vaccination with a Modified Vaccinia virus Ankara (MVA) vector expressing the NP and M1 proteins greatly increased IFN-γ producing cells after ex vivo restimulation with peptides from the vaccine construct in humans (96). In subsequent studies, participants were vaccinated and challenged with influenza virus (97). When compared to unvaccinated controls, vaccinated subjects had less pronounced symptoms and lower shedding time during infection (97). Further, the MVA-NP+M1 vaccine boosted CD4+ and CD8+ T cell responses in subjects over 50 years of age (98) and can be used as an adjuvant to increase antibody responses towards IIV components without impacting T cell responses (99, 100). Another advantage of this adjuvant effect of vector vaccines is the possibility to combine with recombinant internal proteins, such as NP, to elicit the often-underappreciated role of non-neutralizing antibodies against those virus components (101).
While it is clear that T cells will be crucial for cross-protective immunity to diverse influenza virus strains, there are drawbacks that make it difficult to implement. First, as with most inflammatory responses, but especially with CD8+ T cell responses, the risk of immunopathology is high with such potent effector functions (102, 103). Therefore, it is important to balance a protective CD8+ T cell response to influenza with any associated tissue damage (81, 104). One possibility is to direct anti-influenza virus memory CD8+ T cell responses to the URT. Studies have shown that URT memory CD8+ T cells can prevent influenza virus dissemination to the lower respiratory tract (LRT) (105). Therefore, a strong CD8+ T cell response in the URT could attenuate the immune response necessary in the lungs and maximize non-damaging protection. This is indeed part of the objective when utilizing LAIV, which is given intranasally and is limited to URT replication. However, this leads to another barrier in T cell-directed immune responses to influenza; immune history.
Although immune history is not a problem specific to T cell-directed influenza vaccines, it is particularly apparent in this context. When introduced, the LAIV was of interest because of its live-attenuated nature (106). Therefore, such a vaccine should induce not only the antibodies necessary to target HA and NA, but also T cell responses to highly conserved antigens to confer cross-protection (9, 80). Although the calculated vaccine efficacy of the LAIV decreased as the years progressed, especially for pandemic H1N1 virus, the reasons behind the inefficacy were not investigated (107). Some studies show that LAIV induces T cell responses in children and does not in adults (14). Thus, on top of defective-interfering RNA, it is possible that the strength of antibody responses to the pandemic H1N1, or other antigenically similar viruses, neutralized the vaccine virus inhibiting the replication needed to elicit T cell responses (108). Such a limitation could be overcome with platforms such as vector vaccines expressing influenza proteins (79, 92–96). Overall, moving forward, it will be important to elicit strong B and T cell responses in our pursuit of improved influenza virus vaccines.
Challenges
Although beyond the scope of this review, it is important to note that a universal vaccine must also protect high-risk populations including the very young, aged adults, pregnant women, people with underlying health conditions, and overweight/obese individuals. We know that the IIV is less effective in high-risk populations (for example aged adults and overweight/obese individuals) due to underlying immune system complications (109–111). Age-related, and potentially weight-related changes in immune function likely contribute to a loss of influenza vaccine efficacy. These changes are likely to limit the applicability of a “universal vaccine” to high-risk populations, outside of the herd immune effects of vaccinating children and adults with a highly efficacious universal vaccine. One strategy for vaccine development in aged is moving toward the notion of “enhanced vaccines” to prevent the serious complications of influenza rather than a universal vaccine that is going to provide sterilizing immunity in this population. These are important consideration for the improvement of current influenza vaccines as well as in developing universal vaccines.
Concluding remarks
Influenza virus is a moving target. Rapid evolution allows escape from the protective immune responses generated during natural infection or vaccination to seasonal viruses. Therefore, to provide broad and long-lasting immunity, we need to consider an “all-inclusive” approach to vaccination (Figure 1). This would include a concerted immune response to conserved regions of different influenza virus proteins driving both B and T cell responses. Many groups are evaluating vaccines to different proteins utilizing distinct platforms for administrations, the impact of immune history or imprinting on vaccine responsiveness, as well as evaluating new methods for preparing vaccine viruses independent of eggs or even cell culture. The results of these studies will lead to improved vaccines against seasonal influenza viruses. They can serve as the template for generation of broadly protective long-lasting immunity that may protect against emerging influenza strains. Yet we must keep in mind that even the best vaccines may have less than ideal efficacy in high-risk populations; a hurdle that might only be overcome by “enhanced vaccines” to prevent serious complications in these populations, or by herd immunity, to ensure population-wide protection against influenza viruses.
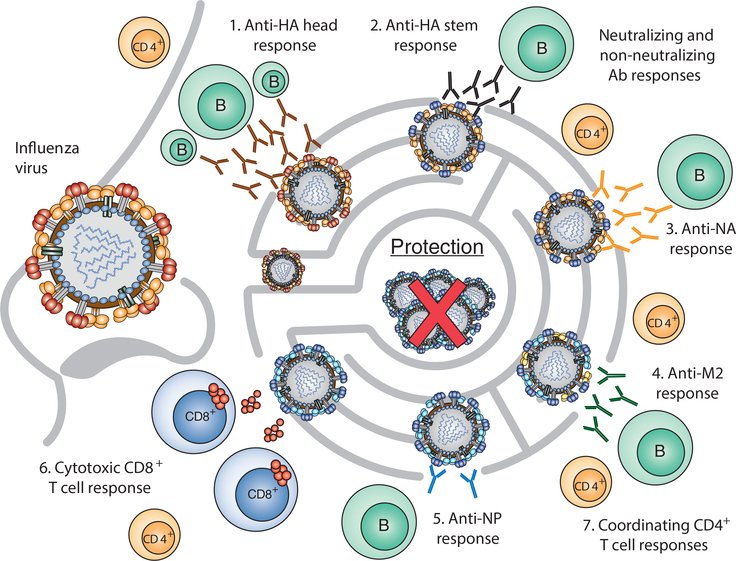
Figure 1. A wholistic approach to vaccination.
Different immune responses are elicited upon influenza infection or vaccination: (1) the immunodominant anti-HA globular head responses apply pressure to select mutations in antigenic sites (drift variants); (2) anti-HA stem responses are more broadly cross-protective because of higher conservation of stem antigens; (3) the anti-NA response targets the enzymatic site to prevent virus entry and spread; (4) M2 antibody targets provide a better cross-protective response due to conservation across influenza viruses; (5) less immunodominant anti-NP antibody responses can be targeted to provide better protection using conserved internal proteins; (6) cytotoxic CD8+ T cell responses target highly conserved internal proteins that provide higher cross-protection to drift variants and heterosubtypic viruses; (7) CD4+ T cell responses help to coordinate both cytotoxic CD8+ T cells and antibody responses to antigens from viral infections and vaccinations. Eliciting all of these pressures against the virus will most likely reduce vaccine escapes and lead to a better universal vaccine.
Acknowledgments
L.D.E. is supported by T32AI106700–02. S.S.C is supported by ALSAC and HHSN272201400006C.
References
- 1.Garten R, Blanton L, Elal AIA, Alabi N, Barnes J, Biggerstaff M, Brammer L, Budd AP, Burns E, Cummings CN, Davis T, Garg S, Gubareva L, Jang Y, Kniss K, Kramer N, Lindstrom S, Mustaquim D, O’Halloran A, Sessions W, Taylor C, Xu X, Dugan VG, Fry AM, Wentworth DE, Katz J, and Jernigan D 2018. Update: Influenza Activity in the United States During the 2017–18 Season and Composition of the 2018–19 Influenza Vaccine. MMWR Morb Mortal Wkly Rep 67: 634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrova VN, and Russell CA 2018. The evolution of seasonal influenza viruses. Nat Rev Microbiol 16: 47–60. [DOI] [PubMed] [Google Scholar]
- 3.Soema PC, Kompier R, Amorij JP, and Kersten GF 2015. Current and next generation influenza vaccines: Formulation and production strategies. Eur J Pharm Biopharm 94: 251–263. [DOI] [PubMed] [Google Scholar]
- 4.Wong SS, and Webby RJ 2013. Traditional and new influenza vaccines. Clin Microbiol Rev 26: 476–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox MM, Izikson R, Post P, and Dunkle L 2015. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther Adv Vaccines 3: 97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cox MM, Patriarca PA, and Treanor J 2008. FluBlok, a recombinant hemagglutinin influenza vaccine. Influenza Other Respir Viruses 2: 211–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin H, Lu B, Zhou H, Ma C, Zhao J, Yang CF, Kemble G, and Greenberg H 2003. Multiple amino acid residues confer temperature sensitivity to human influenza virus vaccine strains (FluMist) derived from cold-adapted A/Ann Arbor/6/60. Virology 306: 18–24. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann E, Mahmood K, Chen Z, Yang CF, Spaete J, Greenberg HB, Herlocher ML, Jin H, and Kemble G 2005. Multiple gene segments control the temperature sensitivity and attenuation phenotypes of ca B/Ann Arbor/1/66. J Virol 79: 11014–11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoft DF, Babusis E, Worku S, Spencer CT, Lottenbach K, Truscott SM, Abate G, Sakala IG, Edwards KM, Creech CB, Gerber MA, Bernstein DI, Newman F, Graham I, Anderson EL, and Belshe RB 2011. Live and inactivated influenza vaccines induce similar humoral responses, but only live vaccines induce diverse T-cell responses in young children. J Infect Dis 204: 845–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohn KG, Bredholt G, Brokstad KA, Pathirana RD, Aarstad HJ, Tondel C, and Cox RJ 2015. Longevity of B-cell and T-cell responses after live attenuated influenza vaccination in children. J Infect Dis 211: 1541–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katz JM, Naeve CW, and Webster RG 1987. Host cell-mediated variation in H3N2 influenza viruses. Virology 156: 386–395. [DOI] [PubMed] [Google Scholar]
- 12.Wang ML, Katz JM, and Webster RG 1989. Extensive heterogeneity in the hemagglutinin of egg-grown influenza viruses from different patients. Virology 171: 275–279. [DOI] [PubMed] [Google Scholar]
- 13.Skowronski DM, Janjua NZ, De Serres G, Sabaiduc S, Eshaghi A, Dickinson JA, Fonseca K, Winter AL, Gubbay JB, Krajden M, Petric M, Charest H, Bastien N, Kwindt TL, Mahmud SM, Van Caeseele P, and Li Y 2014. Low 2012–13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS One 9: e92153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He XS, Holmes TH, Zhang C, Mahmood K, Kemble GW, Lewis DB, Dekker CL, Greenberg HB, and Arvin AM 2006. Cellular immune responses in children and adults receiving inactivated or live attenuated influenza vaccines. J Virol 80: 11756–11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Myers JL, Bostick DL, Sullivan CB, Madara J, Linderman SL, Liu Q, Carter DM, Wrammert J, Esposito S, Principi N, Plotkin JB, Ross TM, Ahmed R, Wilson PC, and Hensley SE 2013. Immune history shapes specificity of pandemic H1N1 influenza antibody responses. J Exp Med 210: 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cobey S, and Hensley SE 2017. Immune history and influenza virus susceptibility. Curr Opin Virol 22: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grohskopf LA, Sokolow LZ, Fry AM, Walter EB, and Jernigan DB 2018. Update: ACIP Recommendations for the Use of Quadrivalent Live Attenuated Influenza Vaccine (LAIV4) - United States, 2018–19 Influenza Season. MMWR Morb Mortal Wkly Rep 67: 643–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gould PS, Easton AJ, and Dimmock NJ 2017. Live Attenuated Influenza Vaccine contains Substantial and Unexpected Amounts of Defective Viral Genomic RNA. Viruses 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erbelding EJ, Post DJ, Stemmy EJ, Roberts PC, Augustine AD, Ferguson S, Paules CI, Graham BS, and Fauci AS 2018. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis 218: 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bui HH, Peters B, Assarsson E, Mbawuike I, and Sette A 2007. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. Proc Natl Acad Sci U S A 104: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fields BN, Knipe DM, Howley PM 2013. Orthomyxoviridae. Wolters Kluwer Health/Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- 22.Krammer F 2015. Emerging influenza viruses and the prospect of a universal influenza virus vaccine. Biotechnol J 10: 690–701. [DOI] [PubMed] [Google Scholar]
- 23.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr., and Wilson IA 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krammer F, Garcia-Sastre A, and Palese P 2018. Is It Possible to Develop a “Universal” Influenza Virus Vaccine? Potential Target Antigens and Critical Aspects for a Universal Influenza Vaccine. Cold Spring Harb Perspect Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knossow M, Gaudier M, Douglas A, Barrere B, Bizebard T, Barbey C, Gigant B, and Skehel JJ 2002. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology 302: 294–298. [DOI] [PubMed] [Google Scholar]
- 26.Reading SA, and Dimmock NJ 2007. Neutralization of animal virus infectivity by antibody. Arch Virol 152: 1047–1059. [DOI] [PubMed] [Google Scholar]
- 27.Krammer F, and Palese P 2015. Advances in the development of influenza virus vaccines. Nat Rev Drug Discov 14: 167–182. [DOI] [PubMed] [Google Scholar]
- 28.Krammer F, and Palese P 2013. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3: 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krammer F, Pica N, Hai R, Margine I, and Palese P 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87: 6542–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, and Palese P 2010. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog 6: e1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrews SF, and McDermott AB 2018. Shaping a universally broad antibody response to influenza amidst a variable immunoglobulin landscape. Curr Opin Immunol 53: 96–101. [DOI] [PubMed] [Google Scholar]
- 32.Vareckova E, Mucha V, Ciampor F, Betakova T, and Russ G 1993. Monoclonal antibodies demonstrate accessible HA2 epitopes in minor subpopulation of native influenza virus haemagglutinin molecules. Arch Virol 130: 45–56. [DOI] [PubMed] [Google Scholar]
- 33.Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, and Palese P 2012. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol 86: 5774–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, Garcia-Sastre A, and Palese P 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nachbagauer R, Krammer F, and Albrecht RA 2018. A Live-Attenuated Prime, Inactivated Boost Vaccination Strategy with Chimeric Hemagglutinin-Based Universal Influenza Virus Vaccines Provides Protection in Ferrets: A Confirmatory Study. Vaccines (Basel) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding Z, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu W, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, and Radosevic K 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349: 1301–1306. [DOI] [PubMed] [Google Scholar]
- 37.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, and Graham BS 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21: 1065–1070. [DOI] [PubMed] [Google Scholar]
- 38.Moody MA, Zhang R, Walter EB, Woods CW, Ginsburg GS, McClain MT, Denny TN, Chen X, Munshaw S, Marshall DJ, Whitesides JF, Drinker MS, Amos JD, Gurley TC, Eudailey JA, Foulger A, DeRosa KR, Parks R, Meyerhoff RR, Yu JS, Kozink DM, Barefoot BE, Ramsburg EA, Khurana S, Golding H, Vandergrift NA, Alam SM, Tomaras GD, Kepler TB, Kelsoe G, Liao HX, and Haynes BF 2011. H3N2 influenza infection elicits more cross-reactive and less clonally expanded anti-hemagglutinin antibodies than influenza vaccination. PLoS One 6: e25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrammert J, Koutsonanos D, Li GM, Edupuganti S, Sui J, Morrissey M, McCausland M, Skountzou I, Hornig M, Lipkin WI, Mehta A, Razavi B, Del Rio C, Zheng NY, Lee JH, Huang M, Ali Z, Kaur K, Andrews S, Amara RR, Wang Y, Das SR, O’Donnell CD, Yewdell JW, Subbarao K, Marasco WA, Mulligan MJ, Compans R, Ahmed R, and Wilson PC 2011. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med 208: 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margine I, Hai R, Albrecht RA, Obermoser G, Harrod AC, Banchereau J, Palucka K, Garcia-Sastre A, Palese P, Treanor JJ, and Krammer F 2013. H3N2 influenza virus infection induces broadly reactive hemagglutinin stalk antibodies in humans and mice. J Virol 87: 4728–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jegaskanda S, Job ER, Kramski M, Laurie K, Isitman G, de Rose R, Winnall WR, Stratov I, Brooks AG, Reading PC, and Kent SJ 2013. Cross-reactive influenza-specific antibody-dependent cellular cytotoxicity antibodies in the absence of neutralizing antibodies. J Immunol 190: 1837–1848. [DOI] [PubMed] [Google Scholar]
- 42.DiLillo DJ, Tan GS, Palese P, and Ravetch JV 2014. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcgammaR interactions for protection against influenza virus in vivo. Nat Med 20: 143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epstein SL 2018. Universal Influenza Vaccines: Progress in Achieving Broad Cross-Protection In Vivo. Am J Epidemiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murphy BR, Kasel JA, and Chanock RM 1972. Association of serum anti-neuraminidase antibody with resistance to influenza in man. N Engl J Med 286: 1329–1332. [DOI] [PubMed] [Google Scholar]
- 45.Couch RB, Atmar RL, Franco LM, Quarles JM, Wells J, Arden N, Nino D, and Belmont JW 2013. Antibody correlates and predictors of immunity to naturally occurring influenza in humans and the importance of antibody to the neuraminidase. J Infect Dis 207: 974–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Memoli MJ, Shaw PA, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Fargis S, Risos K, Powers JH, Davey RT Jr., and Taubenberger JK 2016. Evaluation of Antihemagglutinin and Antineuraminidase Antibodies as Correlates of Protection in an Influenza A/H1N1 Virus Healthy Human Challenge Model. MBio 7: e00417–00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eichelberger MC, Morens DM, and Taubenberger JK 2018. Neuraminidase as an influenza vaccine antigen: a low hanging fruit, ready for picking to improve vaccine effectiveness. Curr Opin Immunol 53: 38–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park JK, Han A, Czajkowski L, Reed S, Athota R, Bristol T, Rosas LA, Cervantes-Medina A, Taubenberger JK, and Memoli MJ 2018. Evaluation of Preexisting Anti-Hemagglutinin Stalk Antibody as a Correlate of Protection in a Healthy Volunteer Challenge with Influenza A/H1N1pdm Virus. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clements ML, Betts RF, Tierney EL, and Murphy BR 1986. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J Clin Microbiol 24: 157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, and Ohmit SE 2015. Antibody to Influenza Virus Neuraminidase: An Independent Correlate of Protection. J Infect Dis 212: 1191–1199. [DOI] [PubMed] [Google Scholar]
- 51.Kosik I, and Yewdell JW 2017. Influenza A virus hemagglutinin specific antibodies interfere with virion neuraminidase activity via two distinct mechanisms. Virology 500: 178–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cate TR, Rayford Y, Nino D, Winokur P, Brady R, Belshe R, Chen W, Atmar RL, and Couch RB 2010. A high dosage influenza vaccine induced significantly more neuraminidase antibody than standard vaccine among elderly subjects. Vaccine 28: 2076–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hassantoufighi A, Zhang H, Sandbulte M, Gao J, Manischewitz J, King L, Golding H, Straight TM, and Eichelberger MC 2010. A practical influenza neutralization assay to simultaneously quantify hemagglutinin and neuraminidase-inhibiting antibody responses. Vaccine 28: 790–797. [DOI] [PubMed] [Google Scholar]
- 54.Couzens L, Gao J, Westgeest K, Sandbulte M, Lugovtsev V, Fouchier R, and Eichelberger M 2014. An optimized enzyme-linked lectin assay to measure influenza A virus neuraminidase inhibition antibody titers in human sera. J Virol Methods 210: 7–14. [DOI] [PubMed] [Google Scholar]
- 55.Rajendran M, Nachbagauer R, Ermler ME, Bunduc P, Amanat F, Izikson R, Cox M, Palese P, Eichelberger M, and Krammer F 2017. Analysis of Anti-Influenza Virus Neuraminidase Antibodies in Children, Adults, and the Elderly by ELISA and Enzyme Inhibition: Evidence for Original Antigenic Sin. MBio 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eichelberger MC, Couzens L, Gao Y, Levine M, Katz J, Wagner R, Thompson CI, Hoschler K, Laurie K, Bai T, Engelhardt OG, participants E. s., and Wood J 2016. Comparability of neuraminidase inhibition antibody titers measured by enzyme-linked lectin assay (ELLA) for the analysis of influenza vaccine immunogenicity. Vaccine 34: 458–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prevato M, Ferlenghi I, Bonci A, Uematsu Y, Anselmi G, Giusti F, Bertholet S, Legay F, Telford JL, Settembre EC, Maione D, and Cozzi R 2015. Expression and Characterization of Recombinant, Tetrameric and Enzymatically Active Influenza Neuraminidase for the Setup of an Enzyme-Linked Lectin-Based Assay. PLoS One 10: e0135474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kilbourne ED, Johansson BE, and Grajower B 1990. Independent and disparate evolution in nature of influenza A virus hemagglutinin and neuraminidase glycoproteins. Proc Natl Acad Sci U S A 87: 786–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sandbulte MR, Westgeest KB, Gao J, Xu X, Klimov AI, Russell CA, Burke DF, Smith DJ, Fouchier RA, and Eichelberger MC 2011. Discordant antigenic drift of neuraminidase and hemagglutinin in H1N1 and H3N2 influenza viruses. Proc Natl Acad Sci U S A 108: 20748–20753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schnell JR, and Chou JJ 2008. Structure and mechanism of the M2 proton channel of influenza A virus. Nature 451: 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ciampor F, Thompson CA, Grambas S, and Hay AJ 1992. Regulation of pH by the M2 protein of influenza A viruses. Virus Res 22: 247–258. [DOI] [PubMed] [Google Scholar]
- 62.Sakaguchi T, Leser GP, and Lamb RA 1996. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J Cell Biol 133: 733–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen BJ, Leser GP, Jackson D, and Lamb RA 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol 82: 10059–10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wohlgemuth N, Ye Y, Fenstermacher KJ, Liu H, Lane AP, and Pekosz A 2017. The M2 protein of live, attenuated influenza vaccine encodes a mutation that reduces replication in human nasal epithelial cells. Vaccine 35: 6691–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schotsaert M, De Filette M, Fiers W, and Saelens X 2009. Universal M2 ectodomain-based influenza A vaccines: preclinical and clinical developments. Expert Rev Vaccines 8: 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolpe A, Schepens B, Fiers W, and Saelens X 2017. M2-based influenza vaccines: recent advances and clinical potential. Expert Rev Vaccines 16: 123–136. [DOI] [PubMed] [Google Scholar]
- 67.Grandea AG 3rd, Olsen OA, Cox TC, Renshaw M, Hammond PW, Chan-Hui PY, Mitcham JL, Cieplak W, Stewart SM, Grantham ML, Pekosz A, Kiso M, Shinya K, Hatta M, Kawaoka Y, and Moyle M 2010. Human antibodies reveal a protective epitope that is highly conserved among human and nonhuman influenza A viruses. Proc Natl Acad Sci U S A 107: 12658–12663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerhard W, Mozdzanowska K, and Zharikova D 2006. Prospects for universal influenza virus vaccine. Emerg Infect Dis 12: 569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zharikova D, Mozdzanowska K, Feng J, Zhang M, and Gerhard W 2005. Influenza type A virus escape mutants emerge in vivo in the presence of antibodies to the ectodomain of matrix protein 2. J Virol 79: 6644–6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Black RA, Rota PA, Gorodkova N, Klenk HD, and Kendal AP 1993. Antibody response to the M2 protein of influenza A virus expressed in insect cells. J Gen Virol 74 ( Pt 1): 143–146. [DOI] [PubMed] [Google Scholar]
- 71.Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, and Fiers W 1999. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nat Med 5: 1157–1163. [DOI] [PubMed] [Google Scholar]
- 72.Ionescu RM, Przysiecki CT, Liang X, Garsky VM, Fan J, Wang B, Troutman R, Rippeon Y, Flanagan E, Shiver J, and Shi L 2006. Pharmaceutical and immunological evaluation of human papillomavirus viruslike particle as an antigen carrier. J Pharm Sci 95: 70–79. [DOI] [PubMed] [Google Scholar]
- 73.Bessa J, Schmitz N, Hinton HJ, Schwarz K, Jegerlehner A, and Bachmann MF 2008. Efficient induction of mucosal and systemic immune responses by virus-like particles administered intranasally: implications for vaccine design. Eur J Immunol 38: 114–126. [DOI] [PubMed] [Google Scholar]
- 74.Denis J, Acosta-Ramirez E, Zhao Y, Hamelin ME, Koukavica I, Baz M, Abed Y, Savard C, Pare C, Lopez Macias C, Boivin G, and Leclerc D 2008. Development of a universal influenza A vaccine based on the M2e peptide fused to the papaya mosaic virus (PapMV) vaccine platform. Vaccine 26: 3395–3403. [DOI] [PubMed] [Google Scholar]
- 75.Simhadri VR, Dimitrova M, Mariano JL, Zenarruzabeitia O, Zhong W, Ozawa T, Muraguchi A, Kishi H, Eichelberger MC, and Borrego F 2015. A Human Anti-M2 Antibody Mediates Antibody-Dependent Cell-Mediated Cytotoxicity (ADCC) and Cytokine Secretion by Resting and Cytokine-Preactivated Natural Killer (NK) Cells. PLoS One 10: e0124677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jegerlehner A, Schmitz N, Storni T, and Bachmann MF 2004. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol 172: 5598–5605. [DOI] [PubMed] [Google Scholar]
- 77.Wang R, Song A, Levin J, Dennis D, Zhang NJ, Yoshida H, Koriazova L, Madura L, Shapiro L, Matsumoto A, Yoshida H, Mikayama T, Kubo RT, Sarawar S, Cheroutre H, and Kato S 2008. Therapeutic potential of a fully human monoclonal antibody against influenza A virus M2 protein. Antiviral Res 80: 168–177. [DOI] [PubMed] [Google Scholar]
- 78.El Bakkouri K, Descamps F, De Filette M, Smet A, Festjens E, Birkett A, Van Rooijen N, Verbeek S, Fiers W, and Saelens X 2011. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J Immunol 186: 1022–1031. [DOI] [PubMed] [Google Scholar]
- 79.Valkenburg SA, Li OTW, Li A, Bull M, Waldmann TA, Perera LP, Peiris M, and Poon LLM 2018. Protection by universal influenza vaccine is mediated by memory CD4 T cells. Vaccine 36: 4198–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Powell TJ, Strutt T, Reome J, Hollenbaugh JA, Roberts AD, Woodland DL, Swain SL, and Dutton RW 2007. Priming with cold-adapted influenza A does not prevent infection but elicits long-lived protection against supralethal challenge with heterosubtypic virus. J Immunol 178: 1030–1038. [DOI] [PubMed] [Google Scholar]
- 81.Grant EJ, Chen L, Quinones-Parra S, Pang K, Kedzierska K, and Chen W 2014. T-cell immunity to influenza A viruses. Crit Rev Immunol 34: 15–39. [DOI] [PubMed] [Google Scholar]
- 82.Grebe KM, Yewdell JW, and Bennink JR 2008. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect 10: 1024–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nachbagauer R, and Krammer F 2017. Universal influenza virus vaccines and therapeutic antibodies. Clin Microbiol Infect 23: 222–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Epstein SL, Kong WP, Misplon JA, Lo CY, Tumpey TM, Xu L, and Nabel GJ 2005. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine 23: 5404–5410. [DOI] [PubMed] [Google Scholar]
- 85.McMichael AJ, Gotch FM, Noble GR, and Beare PA 1983. Cytotoxic T-cell immunity to influenza. N Engl J Med 309: 13–17. [DOI] [PubMed] [Google Scholar]
- 86.Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, Bean T, Barclay W, Deeks JJ, and Lalvani A 2013. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med 19: 1305–1312. [DOI] [PubMed] [Google Scholar]
- 87.Wang Z, Wan Y, Qiu C, Quinones-Parra S, Zhu Z, Loh L, Tian D, Ren Y, Hu Y, Zhang X, Thomas PG, Inouye M, Doherty PC, Kedzierska K, and Xu J 2015. Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8(+) T cells. Nat Commun 6: 6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hayward AC, Wang L, Goonetilleke N, Fragaszy EB, Bermingham A, Copas A, Dukes O, Millett ER, Nazareth I, Nguyen-Van-Tam JS, Watson JM, Zambon M, Flu Watch G, Johnson AM, and McMichael AJ 2015. Natural T Cell-mediated Protection against Seasonal and Pandemic Influenza. Results of the Flu Watch Cohort Study. Am J Respir Crit Care Med 191: 1422–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC, Lambkin-Williams R, Gilbert A, Oxford J, Nicholas B, Staples KJ, Dong T, Douek DC, McMichael AJ, and Xu XN 2012. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 18: 274–280. [DOI] [PubMed] [Google Scholar]
- 90.Poon LL, Leung YH, Nicholls JM, Perera PY, Lichy JH, Yamamoto M, Waldmann TA, Peiris JS, and Perera LP 2009. Vaccinia virus-based multivalent H5N1 avian influenza vaccines adjuvanted with IL-15 confer sterile cross-clade protection in mice. J Immunol 182: 3063–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Valkenburg SA, Leung NHL, Bull MB, Yan LM, Li APY, Poon LLM, and Cowling BJ 2018. The Hurdles From Bench to Bedside in the Realization and Implementation of a Universal Influenza Vaccine. Front Immunol 9: 1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lambe T, Carey JB, Li Y, Spencer AJ, van Laarhoven A, Mullarkey CE, Vrdoljak A, Moore AC, and Gilbert SC 2013. Immunity against heterosubtypic influenza virus induced by adenovirus and MVA expressing nucleoprotein and matrix protein-1. Sci Rep 3: 1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vitelli A, Quirion MR, Lo CY, Misplon JA, Grabowska AK, Pierantoni A, Ammendola V, Price GE, Soboleski MR, Cortese R, Colloca S, Nicosia A, and Epstein SL 2013. Vaccination to conserved influenza antigens in mice using a novel Simian adenovirus vector, PanAd3, derived from the bonobo Pan paniscus. PLoS One 8: e55435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Price GE, Soboleski MR, Lo CY, Misplon JA, Pappas C, Houser KV, Tumpey TM, and Epstein SL 2009. Vaccination focusing immunity on conserved antigens protects mice and ferrets against virulent H1N1 and H5N1 influenza A viruses. Vaccine 27: 6512–6521. [DOI] [PubMed] [Google Scholar]
- 95.Price GE, Soboleski MR, Lo CY, Misplon JA, Quirion MR, Houser KV, Pearce MB, Pappas C, Tumpey TM, and Epstein SL 2010. Single-dose mucosal immunization with a candidate universal influenza vaccine provides rapid protection from virulent H5N1, H3N2 and H1N1 viruses. PLoS One 5: e13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Berthoud TK, Hamill M, Lillie PJ, Hwenda L, Collins KA, Ewer KJ, Milicic A, Poyntz HC, Lambe T, Fletcher HA, Hill AV, and Gilbert SC 2011. Potent CD8+ T-cell immunogenicity in humans of a novel heterosubtypic influenza A vaccine, MVA-NP+M1. Clin Infect Dis 52: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lillie PJ, Berthoud TK, Powell TJ, Lambe T, Mullarkey C, Spencer AJ, Hamill M, Peng Y, Blais ME, Duncan CJ, Sheehy SH, Havelock T, Faust SN, Williams RL, Gilbert A, Oxford J, Dong T, Hill AV, and Gilbert SC 2012. Preliminary assessment of the efficacy of a T-cell-based influenza vaccine, MVA-NP+M1, in humans. Clin Infect Dis 55: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Antrobus RD, Lillie PJ, Berthoud TK, Spencer AJ, McLaren JE, Ladell K, Lambe T, Milicic A, Price DA, Hill AV, and Gilbert SC 2012. A T cell-inducing influenza vaccine for the elderly: safety and immunogenicity of MVA-NP+M1 in adults aged over 50 years. PLoS One 7: e48322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mullarkey CE, Boyd A, van Laarhoven A, Lefevre EA, Veronica Carr B, Baratelli M, Molesti E, Temperton NJ, Butter C, Charleston B, Lambe T, and Gilbert SC 2013. Improved adjuvanting of seasonal influenza vaccines: preclinical studies of MVA-NP+M1 coadministration with inactivated influenza vaccine. Eur J Immunol 43: 1940–1952. [DOI] [PubMed] [Google Scholar]
- 100.Antrobus RD, Berthoud TK, Mullarkey CE, Hoschler K, Coughlan L, Zambon M, Hill AV, and Gilbert SC 2014. Coadministration of seasonal influenza vaccine and MVA-NP+M1 simultaneously achieves potent humoral and cell-mediated responses. Mol Ther 22: 233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Carragher DM, Kaminski DA, Moquin A, Hartson L, and Randall TD 2008. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J Immunol 181: 4168–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Enelow RI, Mohammed AZ, Stoler MH, Liu AN, Young JS, Lou YH, and Braciale TJ 1998. Structural and functional consequences of alveolar cell recognition by CD8(+) T lymphocytes in experimental lung disease. J Clin Invest 102: 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhao Y, Zhang YH, Denney L, Young D, Powell TJ, Peng YC, Li N, Yan HP, Wang DY, Shu YL, Kendrick Y, McMichael AJ, Ho LP, and Dong T 2012. High levels of virus-specific CD4+ T cells predict severe pandemic influenza A virus infection. Am J Respir Crit Care Med 186: 1292–1297. [DOI] [PubMed] [Google Scholar]
- 104.Duan S, and Thomas PG 2016. Balancing Immune Protection and Immune Pathology by CD8(+) T-Cell Responses to Influenza Infection. Front Immunol 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pizzolla A, Nguyen THO, Smith JM, Brooks AG, Kedzieska K, Heath WR, Reading PC, and Wakim LM 2017. Resident memory CD8(+) T cells in the upper respiratory tract prevent pulmonary influenza virus infection. Sci Immunol 2. [DOI] [PubMed] [Google Scholar]
- 106.Belshe RB, Edwards KM, Vesikari T, Black SV, Walker RE, Hultquist M, Kemble G, Connor EM, and C.-T. C. E. S. Group. 2007. Live attenuated versus inactivated influenza vaccine in infants and young children. N Engl J Med 356: 685–696. [DOI] [PubMed] [Google Scholar]
- 107.Singanayagam A, Zambon M, Lalvani A, and Barclay W 2018. Urgent challenges in implementing live attenuated influenza vaccine. Lancet Infect Dis 18: e25–e32. [DOI] [PubMed] [Google Scholar]
- 108.Lewnard JA, and Cobey S 2018. Immune History and Influenza Vaccine Effectiveness. Vaccines (Basel) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Paich HA, Sheridan PA, Handy J, Karlsson EA, Schultz-Cherry S, Hudgens MG, Noah TL, Weir SS, and Beck MA 2013. Overweight and obese adult humans have a defective cellular immune response to pandemic H1N1 influenza A virus. Obesity (Silver Spring) 21: 2377–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Karlsson EA, Hertz T, Johnson C, Mehle A, Krammer F, and Schultz-Cherry S 2016. Obesity Outweighs Protection Conferred by Adjuvanted Influenza Vaccination. MBio 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kosaraju R, Guesdon W, Crouch MJ, Teague HL, Sullivan EM, Karlsson EA, Schultz-Cherry S, Gowdy K, Bridges LC, Reese LR, Neufer PD, Armstrong M, Reisdorph N, Milner JJ, Beck M, and Shaikh SR 2017. B Cell Activity Is Impaired in Human and Mouse Obesity and Is Responsive to an Essential Fatty Acid upon Murine Influenza Infection. J Immunol 198: 4738–4752. [DOI] [PMC free article] [PubMed] [Google Scholar]