Abstract
The diversity of antigen specific adaptive receptors on the surface of B cells and in the population of secreted antibodies is enormous, but increasingly we are acquiring the technical capability to interrogate antibody repertoires in great detail. These antibody technologies have been especially pointed at understanding the complex issues of immunity to infection and disease caused by influenza virus, one of the most common and vexing medical problems in man. Influenza immunity is particularly interesting as a model system, because the antigenic diversity of influenza strains and proteins is high and constantly evolving. Discovery of canonical features in the subset of the influenza repertoire response that is broadly reactive for diverse influenza strains has spurred the recent optimism for creating universal influenza vaccines. Using new technologies for sequencing antibody repertoires at great depth is helping us to understand the central features of influenza immunity.
Keywords: Orthomyxoviridae Infections, Influenza A virus, Antibodies, Viral, Antibodies, Neutralizing, Repertoire
Introduction.
B cell repertoire diversity studies with influenza are interesting because of the wide extent of influenza variability. Influenza virus exhibits a significant challenge to the human immune system because of antigenic variability in field strains. The virus achieves genetic and antigenic diversity by two principal genetic mechanisms resulting in antigenic shift and antigenic drift. Influenza causes periodic human pandemics because the segmented viral genome allows creation of new viruses during coinfection of cells with viruses of two different antigenic subtypes. Genetic reassortment of the segments mixed during coinfection with avian, swine and human viruses allows complete changing of the surface proteins hemagglutinin (HA) and neuraminidase (NA) to subtypes never seen by humans) resulting in antigenic shifts. Such shifts or adaptation of avian viruses for human transmission were associated with large human pandemics cause by H1N1 in 1918, H2N2 in 1957, H3N2 in 1968, and a new H1N1 in 2009. The virus has a second genetic mechanism for diversity caused by the error-prone nature of the viral RNA-dependent RNA polymerase, which frequently introduces missense mutations. This occurrence is challenging because antigenic drift is thus caused by accumulating point mutations in the genes encoding HA and NA. Some of these mutations encode escape mutations for antibodies, and these viruses can be selected over time and enriched in the population. Gradual genetic drift in HA and NA genes causes the antigenic variation that reduces the protective effect of seasonal influenza vaccines. Genetic drift in influenza occurs in a direction over time (1), such that older individuals possess immunity to older strains, in patterns that can be recognized by the decade of birth. Human repertoire studies suggest that the potential diversity of the human antibody repertoire far exceeds that of the influenza antigen diversity, but the problem for vaccine prevention of new strains is a matter of timing. Our current influenza vaccine strategy is to make educated guesses about the likely dominant drifted strains based on molecular epidemiology studies, then to manufacture trivalent (with H1N1, H3N2 and B antigens) or quadrivalent (adding a second type B strain antigen) vaccines starting about 6 months before the season. In some cases, the dominant drifted strain can be mismatched antigenically, leading to suboptimal efficacy. Broader human antibody responses are desirable. Understanding the genetic and structural basis for broadly protective antibodies is a major current goal of the influenza immune repertoire field.
Concepts of repertoire.
The vocabulary surrounding the concept of repertoire is variously applied. In a broad sense, a repertoire is the collection of specificities that can comprise the variable receptors in the adaptive immune system. Vaccine scientists and infectious diseases investigators typically envision the adaptive immune repertoire as a functional system with diverse patterns of antigenic recognition. In this sense, the influenza repertoire is the collection of clones that recognize particular influenza HA or NA molecules, or collections of molecules, with varying patterns of epitope recognition, breadth and potency. This type of repertoire study is conducted mostly with proteins and viruses, using binding and virus inhibition assays and EM and crystallographic structural determinations. From a more fundamental immunological viewpoint, the adaptive immune receptor repertoire alternatively can be studied as a genetic system, comprising the recombined gene sequences encoding the receptors. This type of repertoire study is conducted with DNA sequencing technologies, especially deep sequencing with next generation sequencing (NGS) techniques. Here we will review both types of repertoire concepts, first the functional concepts, then second the genetic concepts.
Functional classes of antibodies revealed by recent influenza B cell studies.
Protective antibodies for influenza are directed to the two surface proteins HA and NA. There has been much more antibody and B cell work done on the response to the HA molecule than NA. The trimeric HA glycoprotein can be roughly thought of in antigenic terms as comprising two major domains, the membrane proximal stem domain and the membrane distal globular head domain. Neutralizing and protective antibodies have been isolated that recognize head or stem.
HA stem domain antibodies.
The recognition of human antibodies to the stem region is relatively new. When the avian H5N1 virus crossed species from birds to humans and revealed it high pathogenicity nature, many vaccinologists and immunologists committed to finding prevention and treatment approaches for H5N1 viruses. Remarkably, some humans with prior exposure to seasonal influenza viruses but not avian viruses exhibited seropositivity to H5 antigens, which was unexpected. Soon thereafter, stem-reactive human monoclonal antibodies (mAbs) were reported that exhibited cross-reactivity for H5 and H1 HA molecules, or broader (2–5). The discovery of the stem epitope that is the target for broadly protective antibodies has excited the influenza research community because of the potential to achieve broadly protective or universal immunity with a stem antigen vaccine. The stem region is relatively conserved across subtypes, and also the stem domain evolves more slowly under immune pressure than the head domain (6). The first cross-reactive stem antibodies to be described in 2009, F10 and CR6261 have been well characterized, and even broader stem antibodies are continually reported. A very broad stem antibody that recognizes both antigenic Group 1 and 2 influenza type A viruses, FI6, has been described (7). Many stem antibodies appear to mediate protective effects in animal models using ADCC activity, as the potency in virus neutralization tests of stem antibodies as a class tends to be lower than that of human antibodies to the head domain especially to the receptor binding site (RBS). As a proof-of-principle for stem vaccine design, some human mAbs are in clinical testing in therapeutic challenge studies, including the Crucell stem antibodies CR6261 and CR8020. Stem vaccines have been developed on these principles of immunity, using HA molecules that are chimerized using head domains from virus subtypes that do not circulate in humans with the stem domain from H1 subtype, “headless “HA constructs and other stem focusing strategies (8–13). If the antibodies being tested in clinical trials mediate a significant viral reduction, they also could be considered for development as prophylactic or therapeutic biological drugs. Similarly, Baker and colleagues used Rosetta software enabled structure-based protein design to develop small protein molecules that mimic the structure and inhibitory function of the stem antibody CR6261 (14, 15).
HA head domain antibodies.
The most common human antibody responses to influenza HA are directed to the head domain, especially the RBS. Many head domain specific mAbs with excellent neutralizing potency have been isolated. The challenge for head domain-based immunity is that the helices and loops surrounding the RBS are hypervariable. Much of the head domain surface is very mutable, without disturbing the attachment and fusion functions mediated by HA. As above, the antigenic drift is directional in both a genetic and an antigenic manner, and the effect of small numbers of amino acid changes can have outsized effects within and antigenic cluster of residues if they facilitate evasion of a dominant immune response (16). Some antibodies bind to the RBS directly and block attachment to sialic acid bearing receptors (an effect that can be mimicked in the hemagglutination inhibition assay using red blood cells which are rich in surface sialic acid). Changes in HA receptor specificity (from the α2,3 sialic acid binding preference of avian strains to the α2,6 sialic acid preference in human transmissible strains) drives some of the more dramatic changes in the HA head domains, especially in the RBS. When a major shift in receptor specificity occurs, only a small subset of broadly neutralizing antibodies is likely to bind in such a way as to accommodate these changes (17). The driving forces for maintenance or shift of preferred sialic acid receptor binding and the pressure on the HA RBS region to evade neutralizing antibody interact, exhibiting complex network effects (18).
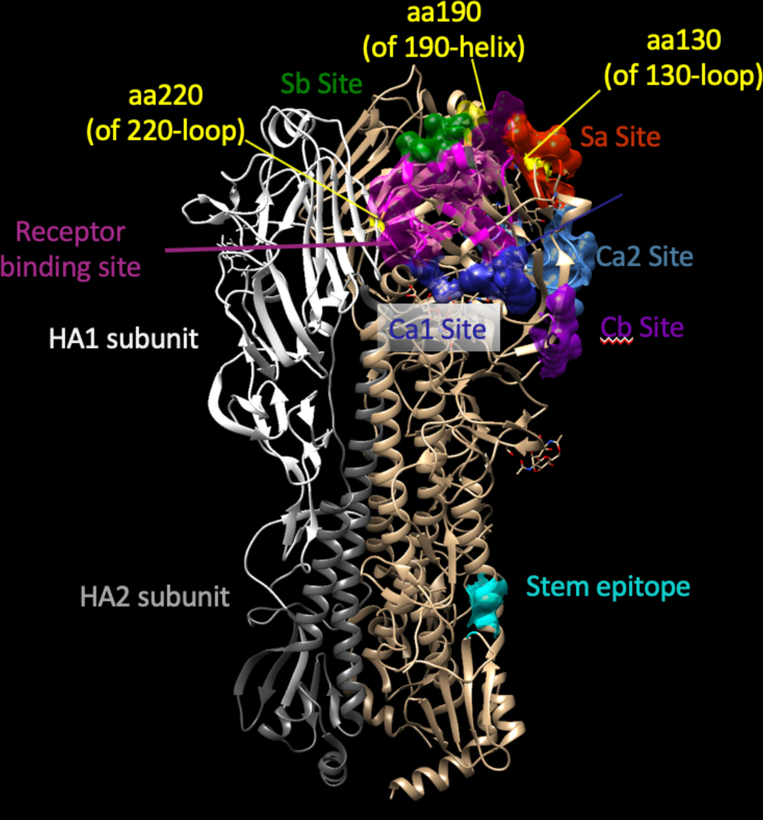
Major antigenic sites on the HA head have been designated based principally on early murine mAb epitope mapping. The legacy nomenclatures for antibody recognition of these sites on HA uses slightly different numbering of amino acids for H1 and H3. The designations for antigenic sites also differ. For H1, the sites were named during mapping of murine mAbs on the HA of the early prototype isolate H1N1 A/Puerto Rico/8/34 (A/PR/8/34) and the sites are designated Sa, Sb, Ca1, Ca2 and Cb (19). For H3N2 strains the HA head domain principal antigenic sites for murine mAbs were designated A, B, C, D, and E (20, 21). The site designations oversimplify the modes of recognition to some extent, since most human neutralizing mAbs recognize broad footprints that contact 2 or more of these sites. Our 1918 influenza mAb 1F1 contacts residues in the Sa, Sb and Ca2 sites, and contacts the RBS (22). From those same studies of 1918 virus survivors, the HA-specific antibody 2D1 that neutralizes 1918 virus contacts residues in diverse sites including sites Sb and Ca1 (23). Some HA antibodies recognize quaternary structures that are formed by a surface formed only by two HA protomers in the HA trimer (24). As the number of antibody epitopes and antigen-antibody complexes has proliferated, the use of structure-based descriptions has become a common practice. The immunodominant structural features around the RBS called the 130-loop, the 150-loop, the 190-helix and the 220-loop incorporate the number of a residue in that structural element to designate the antigenic site. Representative epitope designations with antigenic and structural nomenclatures forH1 are shown in Figure 1.
Figure 1. Structural features and antigenic sites on influenza hemagglutinin protein.
Features are mapped onto the hemagglutinin crystal structure of a 2009 H1N1 influenza virus (PDB 3LZG). HA is a trimer; the subunits of one protomer are shown on the leftmost protomer in white (HA1) or grey (HA2). Conformational antigenic sites Sa (red), Sb (green), Ca1 (dark blue), Ca2 (light blue) and Cb (purple) are mapped onto an adjacent protomer. The receptor binding site is indicated in pink. The position of amino acids in the 130-loop, 190-helix and 220-loop are indicated in yellow. Contact residues for a typical stem antibody are indicated in cyan.
Canonical genetic features of influenza virus-specific repertoires.
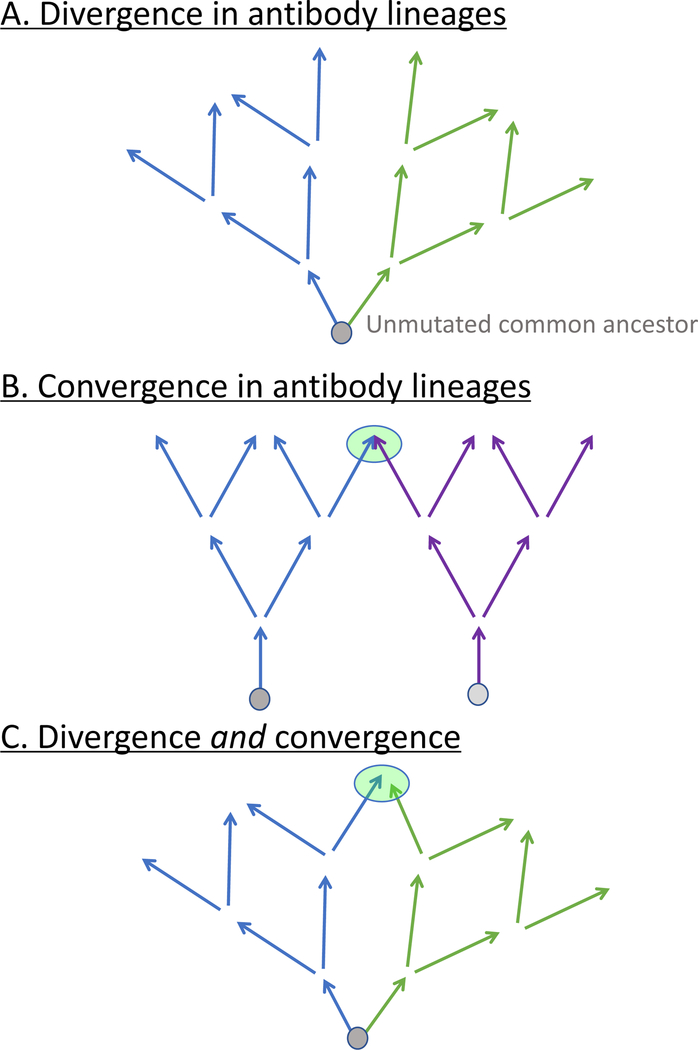
Expressed antibody genes are complex in nature because they incorporate 2 (light chain; variable [V] or joining [J]) or 3 (heavy chain; V, diversity [D], or J) germline genes during recombination at the genomic DNA level. The junctions between V-D (non-templated 1 or N1 region) and D-J (non-templated 2 or N2 region) joins are hypervariable because isoforms of the enzyme terminal deoxynucleotidyl transferase can remove some 3′ end V, 5′ or 3′ end D, or 5′ end J gene segment nucleotides or add on nucleotides to V, D, or J gene segment ends that are non-templated (N additions) or copied as palindromic sequences to the ends of the V (derived from the ends of V, called P additions). The collection of features encoded by the choice of V, D and J segments and the non-templated N1 and N2 regions could generate combinatorial and junctional diversity on the order of ~ 1011 antibody heavy chains alone. Antibody genes also undergo somatic hypermutation, especially during memory responses in the germinal centers following secondary exposure to antigens, resulting in point mutations or insertions/deletions (indels) that encode somatic variants of the original recombination. Clearly, shared (or “public”) antibody variable gene usage is important in some responses that are commonly observed, but also somatic hypermutation plays an important role in achieving antigen-specific responses. Interestingly, somatic mutations cause genetic divergence within one clonal lineage, but also mutations can create similar antibody gene sequences in independent clonal lineages through convergent evolution (25) (Figure 2). Subjects vaccinated against influenza virus show convergent antibody rearrangements (26). This type of sequence convergence from diverse clonotypes into common sequence motifs in sequence from individuals who have received the same antigen exposure also has been reported for other antigens. Interestingly, each of these genetic features (common V or D gene usage and convergent amino acid motifs, and indels) has been described to contribute to common features of influenza specific repertoires, as discussed below.
Figure 2. Schematic representation of genetic repertoire patterns typical in clonal lineages.
A) Clonal divergence from the unmutated common ancestor sequence from the original recombination occurs by the addition of somatic mutations and indels. B) Convergence occurs in clonal lineages when two different clonotypes (which differ in V, junction, and/or J genes at the original recombination) achieve a similar sequence or structure by introduction of somatic mutations. C) Both lineage divergence and subsequent convergence can occur within a single clonotype.
A variable (V) heavy chain gene that is associated with influenza responses.
When frequent use of an antibody heavy chain V gene is associated with antigen specificity, this observation typically suggests that the germline-encoded HCDR2 loop is inherently fit for interacting with the target antigen. VH1-69 is an unusual antibody heavy chain gene segment, since some alleles of this gene encode for a loop that is unusually hydrophobic for a loop project into solute, and thus inherently capable of interacting with recessed hydrophobic pockets on antigens. VH1-69-encoded antibodies are identified frequently in virus-specific repertoires for diverse viruses because of this property. The influenza HA stem region possesses a shallow hydrophobic pocket, and the prototypic stem antibodies that launched the current ‘universal flu’ movement were encoded by VH1-69 with an allele encoding a Phe residue at the tip of CDR2. Use of the VH1-69 gene segment is not sufficient to identify HA stem antibodies as additional residues are required in other regions of the antibody to interact with the stem. Also, now broad stem antibodies encoded by other VH genes have been reported, so also it can be said that VH1–69 is not necessary to make a stem antibody. Nevertheless, the use of this gene is a very common feature in many stem antibodies.
A diversity (D) heavy chain gene that is associated with influenza responses.
The central loop structure of the heavy chain CDR3 is encoded by the D gene. In many memory B cells, the sequence of the D gene from the original recombination is so mutated that a D gene is no longer recognizable. Therefore, HCDR3 amino acid motifs (discussed below) are more recognizable than a common D gene. Still, there are likely to be a number of D germline genes that encode sequences that enable initial low affinity binding in the germline configuration to HA or NA. One example that has been recognized is the use of the D3–9 gene in broadly neutralizing antibodies that recognize the HA stem (27). Much of the interaction surface of some stem antibodies is encoded by D3–9, including in antibodies that bind the dominant stem epitope in differing orientations and binding poses. Remarkably, some stem antibodies that incorporate D3–9 use the gene segment in differing reading frames in the recombined sequence.
HCDR3 amino acid motifs that are associated with influenza responses.
In some respects, the antibody fold that forms the framework and even the CDR loops serve simply as scaffolds to deliver a very small number of amino acids that function as the effector molecules for virus binding and inhibition. Patterns of preferred amino acids in the antigen binding surface of antibodies most often has been identified in antibodies that insert a CDR loop into the RBS. Contact residues in the RBS exhibit a high level of conservation because this region must preserve the capacity to bind sialic acid bearing receptors. Since the RBS has a limited diversity of residues in positions that contact sialic acid, it stands to reason that only a few selected amino acids on the antibody loops can mediate the interaction with this area of the RBS, and the antibody loops should in some way mimic the chemical or structural features of sialic acid. Indeed, several specific amino acid patterns have been identified in antibody CDRs as comprising canonical motifs for interaction with the influenza HA RBS. One of these canonical modes of interaction is the display of an aspartate residue on a CDR that provides a favorable charge interaction with amino acids in the RBS. The backbone atoms of aspartic acid can mimic the acetamido groups of the RBS and a carboxylic acid mimics the carboxylate of sialic acid. Many mAbs the interact with the RBS possess an aspartate or dipeptide with an aspartic acid-hydrophobic motif positioned properly to interact with the RBS position (28). A number of H1 HA-specific neutralizing antibodies have been described with this feature, including the CH65 (29, 30) and 5J8 (31, 32) antibodies, which are especially interesting as a pair since the binding pose of each on the HA determines the breadth of molecular recognition for diverse H1 HA molecules. The complementary breadth of these two antibodies determined by the angle of approach to the RBS likely would cover most H1 strains. (33). The presence of aspartate residue on a CDR inserting into the RBS is not sufficient to mediate the interaction, as structure of the H5.3 antibody with H5 HA shows that this CDR orients the aspartate away from the RBS even though it is near the tip of the RBS (34, 35).
A second canonical mode of interaction with the RBS is mediated by display of a Phe or Tyr aromatic residue on an antibody loop, which can form pi-pi interactions between the HA and mAb CDR (36). This interaction also uses a CDR to mimic sialic acid binding to HA. The antibody protein remarkably mimics the interaction of the carbohydrate sialic acid by placing its amino acid structure into the RBS with a backbone carbonyl group. One antibody, F045–092, was reported that uses both the Phe aromatic residue interaction and carboxylate side chain of an Asp residue to interact with the RBS (37, 38).
One way to achieve breadth of interaction with the HA is to reduce the contact region of the antibody with HA. The antibody C05 is an antibody that accomplishes this feat using an especially long HCDR3 loop that interacts with the conserved residues in the base of the RBS but not the variable amino acids around the rim of the RBS (39).
The occurrence of insertions/deletions in clonal lineages.
Insertions and deletions in antibody gene sequences occur in a subset of sequences in antibody genes from memory B cells. These sequence alterations result from DNA duplication events following DNA strand breaks that occur during the somatic hypermutation process. Many insertions occur at mutational hot spots for error-prone DNA polymerases. In some cases, the insertion enhances the affinity of binding of an influenza antibody. For example, we isolated the influenza H1 HA-specific mAb 2D1 from a circulating B cell in the peripheral blood of a 1918 influenza pandemic survivor (40). In addition to point mutations, the antibody gene has a 9 base pair insertion in the framework 3 of the heavy chain, adjacent to the HCDR2. We showed that the structure of 2D1 in complex with the HA of the 1918 pandemic H1N1 influenza virus exhibited an unusual conformation and relative orientation of the HCDR1 and HCDR2 loops in this neutralizing MAb (23). The insertion causes displacement of the HCDR1, resolving a steric conflict in the parental antibody-antigen interaction by moving a CDR away from the region of interaction, rather than creating a new interaction in the antibody-antigen interface (41).
The coming wave of influenza-specific genetic repertoires based on NGS studies.
Bulk antibody variable gene repertoire sequencing is interesting for comparing individuals with healthy vs disease states, but also now we are deploying approaches for using antibody NGS repertoire technologies to study antigen-specific responses, such as those to influenza virus vaccination or infection. NGS data from B cells in the circulating peripheral blood samples of recently vaccinated subjects show that certain antibody sequences are over-represented in circulation at those time points, and thus these clonotypes are likely to be specific for the recent exposure. Plasmablasts circulate briefly for a few days during a period about a week after influenza vaccination, and these cells have a very high copy number of antibody mRNA per cell, and thus these sequences are over-represented in bulk sequence repertoires at those timepoints. Analysis of NGS libraries made from serial blood samples taken at timepoints before and soon after vaccination are especially helpful for this type of analysis, since the presence of clonally-expanded B cells can be inferred by the presence of the same antibody gene rearrangements in libraries from sequential timepoints (26, 42). So, over-representation (unexpectedly high frequency) is a principal tool for identifying putative antigen specific clones in such repertoire studies. A second tool is the identification of sequence similarity or ‘convergence’ of antibody gene rearrangements in repertoires from different individuals with the same exposure, such as two individuals who have received the same seasonal influenza vaccine. If two heavy chain antibody variable gene sequences are encoded by the same inferred germline VH and JH gene segments and have identical length CDR3s with identical or very similar CDR3 sequences, these sequences can be considered members of a single clonotype. Each somatic variant of the clonotype may be considered a different clone. Lineages of clones that are assigned to a single clonotype can be constructed by alignments, which may suggest these clones were derived from a single B cell with an unmutated sequence (the computationally inferred sequence is sometimes termed the reverted unmutated ancestor [RUA] or unmutated common ancestor [UCA]). When sequences from two different individuals with a common exposure share the same inferred germline VH and JH gene segments and have identical length CDR3s with identical or very similar CDR3 sequences, we can term these sequences to be members of a ‘public clonotype’ (43). We believe such sequences are of particular interest, because they can reveal canonical features of common responses in populations, and influenza-specific public clonotypes could be interesting to target with structure-based reverse vaccine design programs for universal influenza vaccines that would be effective in a broad distribution of subjects. Proof-of-principle has been established for this reverse vaccinology approach in previous studies of another respiratory pathogen, respiratory syncytial virus (44).
Effect of synthetic gene technologies on validation of repertoire studies.
If one uses a longer read NGS technique to obtain full-length variable region sequences, it is possible to synthesize cDNAs encoding the variable region, clone those cDNA into a full-length immunoglobulin expression encoding an Fc region, and express a corresponding recombinant human IgG. Recent improvements in the throughput of synthetic DNA synthesis technologies allow individual investigators to synthesize dozens of genes at a time rapidly on the bench with on-instrument Gibson cloning to rapidly generate expression vectors, for instance using a BioXP instrument (Synthetic Genomics). Even larger scale synthesis is possible now with chip-based synthesis methods developed by Twist Biosciences that enable synthesis of thousands of antibodies at a time. We have used this approach recently to rapidly express over 1,000 individual antibodies obtained from individual plasmablast cells in a single blood sample from an influenza-infected individual. Making the antibody proteins encoded by such putative influenza-specific B cells allows downstream validation of the specificity, structure and function of antibodies encoded by the sequences. This type of experiment is already revealing findings about the enormous diversity of clonotypes made in the response to influenza, and the specificity of the response. For example, early studies suggest that antibodies to the HA protein, which is the only correlate of vaccine-induced immunity currently accepted by regulatory authorities, comprise a minority of the B cell response to infection.
Conclusion.
Protecting against influenza infection and disease by vaccination is challenging because of the constant antigenic drift and periodic antigenic shift that occurs in field strains. The human antibody repertoire clearly has the capacity to recognize and neutralize virtually any influenza strain, but the challenge is to use a limited number of immunogens to induce the most broadly protective or ‘universal’ antibody responses prior to infection. Recent studies of the functional antibody repertoires in humans, and the use of NGS to define genetic repertoires, have revealed that the human immune system can generate very broad and potent antibodies. Repertoire and human monoclonal antibody studies now enable researchers to isolate tens of thousands of naturally occurring human antibodies that inhibit influenza. By searching through these repertoires, and using new technologies in synthetic genomics, we are increasingly able to express and characterize thousands of antibodies to identify broad and potent antibodies that could be used for prophylaxis or therapy against a broad array of influenzas. Furthermore, computational design of novel antigens, based on the plethora of antigen-antibody complexes that are being reported, allows us to recapitulate the conformational of minimal protective epitopes with synthetic vaccine constructe. Such findings point the way toward development and testing of broader and more potent experimental influenza vaccines.
Acknowledgments
Funding statement. This work was supported by U.S. N.I.H. grants U19 AI117905 and HHS contract HHSN272201400024C and by a grant from the Human Vaccines Project.
References
- 1.Volz EM, Koelle K, and Bedford T. 2013. Viral Phylodynamics. Plos Comput Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, and Goudsmit J. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3: e3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, and Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16: 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, and Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okuno Y, Isegawa Y, Sasao F, and Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67: 2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkpatrick E, Qiu X, Wilson PC, Bahl J, and Krammer F. 2018. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci Rep 8: 10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, and Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333: 850–856. [DOI] [PubMed] [Google Scholar]
- 8.Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, Garcia-Sastre A, and Palese P. 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. MBio 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu XY, Hoffman RMB, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, de Man M, Ding ZQ, Apetri A, Kukrer B, Sneekes-Vriese E, Tomkiewicz D, Laursen NS, Lee PS, Zakrzewska A, Dekking L, Tolboom J, Tettero L, van Meerten S, Yu WL, Koudstaal W, Goudsmit J, Ward AB, Meijberg W, Wilson IA, and Radosevic K. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349: 1301–1306. [DOI] [PubMed] [Google Scholar]
- 10.Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang LS, Zhang Y, Joyce MG, Lingwood D, Moin SM, Andersen H, Okuno Y, Rao SS, Harris AK, Kwong PD, Mascola JR, Nabel GJ, and Graham BS. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nature Medicine 21: 1065-+. [DOI] [PubMed] [Google Scholar]
- 11.Valkenburg SA, Mallajosyula VVA, Li OTW, Chin AWH, Carnell G, Temperton N, Varadarajan R, and Poon LLM. 2016. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci Rep-Uk 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hai R, Krammer F, Tan GS, Pica N, Eggink D, Maamary J, Margine I, Albrecht RA, and Palese P. 2012. Influenza Viruses Expressing Chimeric Hemagglutinins: Globular Head and Stalk Domains Derived from Different Subtypes. Journal of Virology 86: 5774–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krammer F, Pica N, Hai R, Margine I, and Palese P. 2013. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87: 6542–6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, Myers CA, Kamisetty H, Blair P, Wilson IA, and Baker D. 2012. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol 30: 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, and Baker D. 2011. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 332: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus ADME, and Fouchier RAM. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305: 371–376. [DOI] [PubMed] [Google Scholar]
- 17.Wu NC, Grande G, Turner HL, Ward AB, Xie J, Lerner RA, and Wilson IA. 2017. In vitro evolution of an influenza broadly neutralizing antibody is modulated by hemagglutinin receptor specificity. Nature Communications 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu NC, Thompson AJ, Xie J, Lin CW, Nycholat CM, Zhu XY, Lerner RA, Paulson JC, and Wilson IA. 2018. A complex epistatic network limits the mutational reversibility in the influenza hemagglutinin receptor-binding site. Nature Communications 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerhard W, Yewdell J, Frankel ME, and Webster R. 1981. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290: 713–717. [DOI] [PubMed] [Google Scholar]
- 20.Wilson IA, Skehel JJ, and Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289: 366–373. [DOI] [PubMed] [Google Scholar]
- 21.Wilson IA, and Cox NJ. 1990. Structural basis of immune recognition of influenza virus hemagglutinin. Annu Rev Immunol 8: 737–771. [DOI] [PubMed] [Google Scholar]
- 22.Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE Jr., Wilson IA, and Basler CF. 2012. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog 8: e1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr., and Wilson IA. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knossow M, Gaudier M, Douglas A, Barrere B, Bizebard T, Barbey C, Gigant B, and Skehel JJ. 2002. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology 302: 294–298. [DOI] [PubMed] [Google Scholar]
- 25.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Briney BS, Smith SA, Basler CF, and Crowe JE Jr. 2011. Epitope-specific human influenza antibody repertoires diversify by B cell intraclonal sequence divergence and interclonal convergence. J Immunol 187: 3704–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson KJ, Liu Y, Roskin KM, Glanville J, Hoh RA, Seo K, Marshall EL, Gurley TC, Moody MA, Haynes BF, Walter EB, Liao HX, Albrecht RA, Garcia-Sastre A, Chaparro-Riggers J, Rajpal A, Pons J, Simen BB, Hanczaruk B, Dekker CL, Laserson J, Koller D, Davis MM, Fire AZ, and Boyd SD. 2014. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe 16: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu NC, Yamayoshi S, Ito M, Uraki R, Kawaoka Y, and Wilson IA. 2018. Recurring and Adaptable Binding Motifs in Broadly Neutralizing Antibodies to Influenza Virus Are Encoded on the D3–9 Segment of the Ig Gene. Cell Host Microbe 24: 569–578 e564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee PS, and Wilson IA. 2015. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol 386: 323–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, and Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108: 14216–14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, and Harrison SC. 2015. Viral receptor-binding site antibodies with diverse germline origins. Cell 161: 1026–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE Jr., Ward AB, and Wilson IA. 2013. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol 87: 12471–12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, and Crowe JE Jr. 2011. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol 85: 10905–10908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crowe JE Jr. 2017. Principles of Broad and Potent Antiviral Human Antibodies: Insights for Vaccine Design. Cell Host Microbe 22: 193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thornburg NJ, Nannemann DP, Blum DL, Belser JA, Tumpey TM, Deshpande S, Fritz GA, Sapparapu G, Krause JC, Lee JH, Ward AB, Lee DE, Li S, Winarski KL, Spiller BW, Meiler J, and Crowe JE Jr. 2013. Human antibodies that neutralize respiratory droplet transmissible H5N1 influenza viruses. J Clin Invest 123: 4405–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winarski KL, Thornburg NJ, Yu Y, Sapparapu G, Crowe JE Jr., and Spiller BW. 2015. Vaccine-elicited antibody that neutralizes H5N1 influenza and variants binds the receptor site and polymorphic sites. Proc Natl Acad Sci U S A 112: 9346–9351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr., and Wilson IA. 2013. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20: 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, and Kurosawa Y. 2011. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol 85: 11048–11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, and Wilson IA. 2014. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5: 3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O’Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, and Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489: 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, Stevens J, Wilson IA, Aguilar PV, Altschuler EL, Basler CF, and Crowe JE Jr. 2008. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455: 532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krause JC, Ekiert DC, Tumpey TM, Smith PB, Wilson IA, and Crowe JE Jr. 2011. An insertion mutation that distorts antibody binding site architecture enhances function of a human antibody. MBio 2: e00345–00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boyd SD, Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, Simen BB, Hanczaruk B, Nguyen KD, Nadeau KC, Egholm M, Miklos DB, Zehnder JL, and Fire AZ. 2009. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med 1: 12ra23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Setliff I, McDonnell WJ, Raju N, Bombardi RG, Murji AA, Scheepers C, Ziki R, Mynhardt C, Shepherd BE, Mamchak AA, Garrett N, Karim SA, Mallal SA, Crowe JE Jr., Morris L, and Georgiev IS. 2018. Multi-Donor Longitudinal Antibody Repertoire Sequencing Reveals the Existence of Public Antibody Clonotypes in HIV-1 Infection. Cell Host Microbe 23: 845–854 e846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, Connell MJ, Stevens E, Schroeter A, Chen M, Macpherson S, Serra AM, Adachi Y, Holmes MA, Li Y, Klevit RE, Graham BS, Wyatt RT, Baker D, Strong RK, Crowe JE Jr., Johnson PR, and Schief WR. 2014. Proof of principle for epitope-focused vaccine design. Nature 507: 201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]