Abstract
Two new series of heterocyclic derivatives with potential anticancer activity, in which a pyrrolo[1,2-b]pyridazine or a pyrrolo[2,1-a]phthalazine moiety was introduced in place of the 3′-hydroxy-4′-methoxyphenyl ring of phenstatin have been synthesised and their structure-activity relationship (SAR) was studied. Fourteen of the new compounds were evaluated for their in vitro cytotoxic activity by National Cancer Institute (NCI) against 60 human tumour cell lines panel. The best five compounds in terms of in vitro growth inhibition were screened in the second stage five dose-response studies, three of them showing a very good antiproliferative activity with GI50<100 nM on several cell lines including colon, ovarian, renal, prostate, brain and breast cancer, melanoma and leukemia. Docking experiments on the biologically active compounds showed a good compatibility with the colchicine binding site of tubulin.
Keywords: Anticancer; phenstatin; pyrrolo[1,2-b]pyridazine; pyrrolo[2,1-a]phthalazine; 3 + 2 dipolar cycloaddition; docking; N-heterocycles
Introduction
Considerable efforts have been focussed in the past decades, on the design and development of new antiproliferative drugs with improved efficiency, limited toxicity, cost-effectiveness, which are synchronously less prone to develop multidrug resistance1–3. Among the variety of targets used in this huge anticancer fight, tubulin targeting appears to be a key focus in cancer treatment, the research in this field remaining very active in past years4–7. After the success of Colchicine8, combretastatin A-49, vincristine or vinblastine10 as anticancer drugs acting by inhibiting tubulin polymerisation, research efforts focused on developing new colchicine binding site inhibitors with improved pharmacological profiles4,11.
One of the simplest known structures synthesised and tested as an anticancer agent in the past years is phenstatin12,13 which stand as one of the most potent tubulin polymerisation inhibitors by binding to the colchicine site of the tubulin and thus, interfering with the equilibrium dynamics associated with the cell division14,15. Because of its biological properties and structural simplicity, phenstatin continues to be a lead compound for rational design in anticancer therapy, the recent literature being plentiful of such phenstatin analogues16–18.
Pyrrolo-fused derivatives comprise a class of biologically active heterocyclic compounds which can serve as promising scaffolds for the development of anticancer, antimicrobial, antiviral, antimalarial, antitubercular, anti-inflammatory, and enzyme inhibiting drugs19. Among the fused pyrrolo-heterocyclic compounds, pyrrolo[1,2-b]pyridazines and its condensed pyrrolo[2,1-a]phthalazine system are compounds well known for their strong luminescence20,21 and photochromic properties22, and at the same time are promising in the field of drug design23,24, some derivatives being reported to have antimicrobial25,26, antifungal25 or anticancer effects27,28, or to act as acyl CoA:diacylglycerol acyltransferase (DGAT1) inhibitors24, JAK inhibitors29, HER-2 tyrosine kinase inhibitors30, IRAK4 inhibitors31, or MEK inhibitors32.
The replacement of one of the substituted phenyl ring of phenstatin with pyrrolo-fused heterocycles has been a major focus in rational drug design in the recent years, as there are several reported biological active phenstatin analogues containing an indole ring5,19, an indolizine ring33, or a pyrrolo[2,3-d]pyrimidine ring34. However, to our knowledge, there are no reported analogues of phenstatin with pyrrolo[1,2-b]pyridazine or pyrrolo[2,1-a]phthalazine scaffolds, respectively.
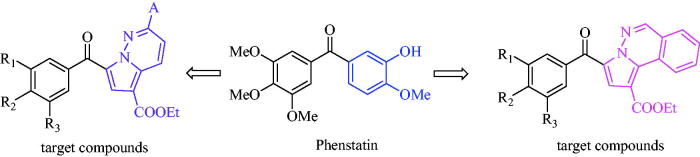
With the aim of exploring new potential antitumour scaffolds, the target compounds described in this paper possess a pyrrolo[1,2-b]pyridazin-7-yl or a pyrrolo[2,1-a]phthalazin-3-yl moiety in place of the 3′-hydroxy-4′-methoxyphenyl ring of phenstatin. In order to establish structure-activity relationships (SARs), we extended our structural modifications by introducing different substituents at position 2 of the pyrrolo[1,2-b]pyridazine unit, including methyl or 4-substituted phenyl rings (4-chlorophenyl, 4-bromophenyl or p-tolyl). At the same time, the trimethoxyphenyl ring of phenstatin was replaced either by 3,5-dihydroxyphenyl, 3,4-dimethoxyphenyl or 4-bromophenyl (Figure 1).
Figure 1.
The structures of phenstatin and the target compounds.
Materials and methods
Chemistry
All commercially available reagents and solvents employed were used without further purification. Melting points were recorded on an A. Krüss Optronic Melting Point Meter KSPI and are uncorrected. Proton and carbon nuclear magnetic resonance (δH, δC) spectra were recorded on a DRX-500 Bruker or a Bruker Avance 400 DRX spectrometers. The following abbreviations were used to designate chemical shift multiplicities: s: singlet, d: doublet, t: triplet, q: quartet, m: multiplet, bs: broad singlet, as: apparent singlet. All chemical shifts are quoted on the δ-scale in ppm. Coupling constants are given in Hz. IR spectra were recorded on a FTIR Shimadzu or Jasco 660 plus FTIR spectrophotometer. Analyses indicated by the symbols of the elements or functions were within ±0.4% of the theoretical values. Thin layer chromatography (TLC) was carried out on Merck silica gel 60F254 plates. Visualisation of the plates was achieved using a UV lamp (λmax = 254 or 365 nm).
Compounds 7d, 7h, 7l, 8d, 10a, and 10d were previously reported35–39.
General procedure for the synthesis of monoquaternary salts 7 and 10
1 mmol of heterocycle (pyridazine 1, 3-methylpyridazine 2, 3–(4-chlorophenyl)pyridazine) 3, 3–(4-bromophenyl)pyridazine 4, 3-(p-tolyl)pyridazine 5 or phthalazine 9 was dissolved in 7 ml acetone (for compounds 1–5) or acetonitrile for compound 9. Then 1.1 mmol of reactive halide (2-bromo-1–(3,4,5-trimethoxyphenyl)ethanone 6a, 2-bromo-1–(3,5-dimethoxyphenyl) ethanone 6 b, 2-bromo-1–(3,4-dimethoxyphenyl) ethanone 6c or 2-bromo-1–(4-bromophenyl) ethanone 6d) was added and the resulted mixture was stirred overnight at room temperature (to obtain compounds 7) or reflux (for synthesis of compounds 10). The reaction mixture was cooled and the formed precipitate was filtered and washed with diethyl ether to give the desired product which was used in the next reaction without any further purification. In case of salts 7e–g, the resulting salts have not crystallised; for these compounds, the solvent was removed under vacuum, and the resulting liquid was used in the next step.
General procedure for preparation of compounds 8a–t and 11a–d
The cycloimmonium salt (7a–d or 10a–d) (1 mmol) and ethyl propiolate (1.1 mmol) were added to 10 ml of anhydrous acetone and the obtained suspension was stirred at room temperature. Then, a solution of triethylamine (TEA) (3 mmol, 3 equiv.) in anhydrous acetone (3 ml) was added drop-wise over 1 h (magnetic stirring) and the resulting mixture was then stirred overnight at room temperature. Water (10 ml) was added and the formed solid was collected by filtration to give a powder which was washed with 5 ml methanol. The product was crystallised from dichloromethane/methanol (1:1, v/v).
1–(2-Oxo-2–(3,4,5-trimethoxyphenyl)ethyl)pyridazin-1-ium bromide7a. Beige powder, Yield: 61%; m.p. 152–153 °C; IR (KBr, cm−1): 1672, 1583, 1416, 1319, 1165, 1128. 1H NMR (500 MHz DMSO-d6) δ 3.80 (s, 3H, OMe), 3.90 (s, 6H, 2 × OMe), 6.82 (s, 2H, H7), 7.39 (s, 2H, H10, H14), 8.76 (t, J = 6.0 Hz, 1H, H4), 8.90 (t, J = 6.0 Hz, 1H, H5), 9.74 (d, J = 3.5 Hz, 1H, H3), 9.74 (d, J = 5.0 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 56.4 (2 × OMe), 60.4 (OMe), 70.3 (C7), 106.2 (C10, C14), 136.0 (C5), 137.5 (C4), 143.3 (C12), 151.9 (C6), 153.1 (C11, C13), 154.7 (C3), 189.3 (C8). Anal. calcd. for C15H17BrN2O4: C, 48.80; H, 4.64; N, 7.59%. Found: C, 48.79; H, 4.55; N, 7.62%.
1–(2-Oxo-2–(3,5-dimethoxyphenyl)ethyl)pyridazin-1-ium bromide7b. Beige powder, Yield: 60%; m.p. 196–199 °C; IR (KBr, cm−1): 1684, 1586, 1438, 1334, 1209, 1157, 1058. 1H NMR (500 MHz DMSO-d6): δ 3.85 (s, 6H, 2 × OMe), 6.76 (s, 2H, H7), 6.94 (t, J = 2.0 Hz, 1H, H12), 7.21 (d, J = 2.5 Hz, 2H, H10, H14), 8.75 (dd, J = 8.0; 4.5 Hz, 1H, H4), 8.88 (m, 1H, H5), 9.74 (d, J = 5.5 Hz, 1H, H3), 9.90 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6): δ 55.8 (2 × OMe), 70.4 (C7), 106.3 (C10, C14), 106.5 (C12), 135.2 (C9), 136.0 (C5), 137.6 (C4), 151.9 (C6), 154.7 (C3), 160.9 (C11, C13), 190.2 (C8). Anal. calcd. for C14H15BrN2O3: C, 49.57; H, 4.46; N, 8.26%. Found: C, 49.59; H, 4.42; N, 8.32%.
1–(2-Oxo-2–(3,4-dimethoxyphenyl)ethyl)pyridazin-1-ium bromide7c. Beige solid, Yield: 60%; m.p. 123–124 °C; IR (KBr, cm−1): 2979, 1677, 1586, 1518, 1294, 1205, 1168. 1H NMR (500 MHz DMSO-d6): δ 3.85 (s, 3H, OMe), 3.90 (s, 3H, OMe), 6.79 (s, 2H, H7), 7.22 (d, J = 8.5 Hz, 1H, H13), 7.53 (d, J = 2.0 Hz, 1H, H10), 7.81 (dd, J = 8.5; 2.0 Hz, 1H, H14), 8.77 (m, 1H, H4), 8.91 (m, 1H, H5), 9.75 (dd, J = 5.0; 1.0 Hz, 1H, H3), 9.97 (d, J = 6.0 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6): δ 55.8 (OMe), 56.1 (OMe), 70.1 (C7), 110.4 (C10), 111.3 (C14), 123.7 (C13), 126.0 (C9), 136.0 (C5), 137.5 (C4), 151.9 (C6), 154.7 (C3), 148.9 (C11), 154.5 (C12), 188.7 (C8). Anal. calcd. for C14H15BrN2O3: C, 49.57; H, 4.46; N, 8.26%. Found: C, 49.55; H, 4.44; N, 8.30%.
1–(2-Oxo-2–(4-bromophenyl)ethyl)pyridazin-1-ium bromide7d. White solid, Yield: 66%; m.p. 235–237 °C; IR (KBr, cm−1): 3017, 2976, 1695, 1580, 1439, 1394, 1229, 976, 822. 1H NMR (400 MHz DMSO-d6): δ 6.75 (s, 2H, H7), 7.90 (d, J = 8.4 Hz, 2H, H11, H13), 8.03 (d, J = 8.4 Hz, 2H, H10, H14), 8.78 (m, 1H, H4), 8.92 (m, 1H, H5), 9.76 (dd, J = 6.0; 0.8 Hz, 1H, H3), 9.99 (d, J = 6.0 Hz, 1H, H6). 13 C NMR (100 MHz DMSO-d6): δ 70.2 (C7), 129.1 (C12), 130.4 (C10, C14), 132.2 (C11, C13), 135.4 (C9), 136.0 (C5), 137.5 (C4), 151.8 (C6), 154.7 (C3), 189.8 (C8). Anal. calcd. for C12H10Br2N2O: C, 40.26; H, 2.82; N, 7.82%. Found: C, 40.25; H, 2.79; N, 7.84%.
3-Methyl-1–(2-oxo-2–(3,4,5-trimethoxyphenyl)ethyl)pyridazin-1-ium bromide7e. Liquid, Yield: 50%; IR (cm−1): 3069, 2943, 1687, 1586, 1417, 1332, 1126, 1050, 921. 1H NMR (500 MHz DMSO-d6) δ 2.81 (s, 3H, Me), 3.80 (s, 3H, OMe), 3.89 (s, 6H, 2 × OMe), 6.76 (s, 2H, H7), 7.38 (s, 2H, H10, H14), 8.65 (d, J = 8.5 Hz, 1H, H4), 8.77 (dd, J = 8.5; 6.0 Hz, 1H, H5), 9.80 (d, J = 6.0 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6): δ 21.6 (Me), 56.4 (2 × OMe), 60.4 (OMe), 70.1 (C7), 106.2 (C10, C14), 128.5 (C9), 135.0 (C5), 138.4 (C4), 143.3 (C12), 149.7 (C6), 153.1 (2 x C11), 164.9 (C3), 189.3 (C8).
3-Methyl-1–(2-oxo-2–(3,5-dimethoxyphenyl)ethyl)pyridazin-1-ium bromide7f. Liquid, Yield: 50%; IR (cm−1): 3069, 2943, 1687, 1586, 1417, 1332, 1164, 1050, 921. 1H NMR (500 MHz DMSO-d6): δ 2.90 (s, 3H, Me), 3.84 (s, 6H, 2 × OMe), 6.63 (s, 2H, H7), 6.90 (t, J = 2.0 Hz, 1H, H12), 7.20 (d, J = 2.5 Hz, 2H, H10, H14), 8.67 (d, J = 8.5 Hz, 1H, H4), 8.78 (dd, J = 8.5; 5.5 Hz, 1H, H5), 9.87 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 21.6 (Me), 55.9 (2 × OMe), 70.3 (C7), 106.3 (C10, C14), 106.6 (C12); 135.0 (C9), 135.2 (C5), 138.4 (C4), 149.6 (C6), 160.8 (C11, C13), 164.8 (C3), 190.2 (C8).
3-Methyl-1–(2-oxo-2–(3,4-dimethoxyphenyl)ethyl)pyridazin-1-ium bromide7g. Liquid; Yield: 51%; IR (cm−1): 2969, 1682, 1586, 1517, 1270, 1019, 806. 1H NMR (DMSO-d6, 500 MHz) δ 2.80 (s, 3H, Me), 3.84 (s, 3H, OMe), 3.84 (s, 3H, OMe), 6.78 (s, 2H, H7), 7.18 (d, J = 8.5 Hz, 1H, H12), 7.51 (d, J = 2.0 Hz, 1H, H10), 7.79 (dd, J = 8.5; 2.0 Hz, 1H, H14), 8.68 (1H, d, J = 8.5 Hz, H4), 8.80 (1H, dd, J = 8.5; 5.5 Hz, H5), 9.93 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6): δ = 21.7 (Me), 55.8 (OMe), 56.0 (OMe), 70.0 (C7), 110.5 (C10), 111.4 (C14), 123.8 (C13), 126.1 (C9), 135.0 (C5), 138.3 (C4), 149.7 (C6), 148.9 (C11), 154.5 (C12), 164.8 (C3), 188.7 (C8).
3-Methyl-1–(2-oxo-2–(4-bromophenyl)ethyl)pyridazin-1-ium bromide7h. Beige solid, Yield: 70%; m.p. 216–218 °C; IR (KBr, cm−1): 3013, 2976, 1692, 1586, 1468, 1231, 1072, 988, 828. 1H NMR (500 MHz DMSO-d6) δ 2.81 (s, 3H, Me), 6.74 (s, 2H, H7), 7.89 (d, J = 8.5 Hz, 2H, H11, H13), 8.02 (d, J = 8.5 Hz, 2H, H10, H14), 8.66 (d, J = 8.5 Hz, 1H, H4), 8.78 (dd, J = 8.5; 6.0 Hz, 1H, H5), 9.82 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 21.6 (Me), 70.0 (C7), 129.1 (C12); 130.4 (C10, C14), 132.3 (C11, C13), 132.4 (C9), 135.0 (C5), 138.4 (C4), 149.7 (C6), 164.8 (C3), 189.8 (C8). Anal. calcd. for C13H12Br2N2O: C, 41.97; H, 3.25; N, 7.53%. Found: C, 42.00; H, 3.18; N, 7.55%.
3–(4-Chlorophenyl)-1–(2-oxo-2–(3,4,5-trimethoxyphenyl)ethyl)pyridazin-1-ium bromide7i. Beige solid, Yield 74%; m.p. 186–188 °C; IR (KBr, cm−1): 3059, 2932, 1682, 1599, 1450, 1343, 1132. 1H NMR (500 MHz DMSO-d6) δ 3.81 (s, 3H, OMe), 3.90 (s, 6H, 2 × OMe), 6.91 (s, 2H, H7), 7.42 (s, 2H, H10, H14), 7.76 (d, J = 7.5 Hz, 2H, H17, H19), 8.25 (d, J = 8.0 Hz, 2H, H16, H20), 8.96 (dd, J = 9.0; 4.5 Hz, 1H, H5), 9.33 (d, J = 9.0 Hz, 1H, H4), 9.93 (d, J = 4.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 56.3 (2 × OMe), 60.3 (OMe), 70.5 (C7), 106.2 (C10, C14), 128.4 (C9), 129.6 (C16, C20, C17, C19), 130.5 (C15), 134.7 (C4), 136.3 (C5), 137.7 (C18), 143.2 (C12), 150.0 (C6), 153.0 (C11, C13), 159.9 (C3), 189.1 (C8). Anal. calcd. for C21H20BrClN2O4: C, 52.57; H, 4.20; N, 5.84%. Found: C, 52.55; H, 4.18; N, 5.87%.
3–(4-Chlorophenyl)-1–(2-oxo-2–(3,5-dimethoxyphenyl)ethyl)pyridazin-1-ium bromide7j. Beige solid, Yield: 75%; m.p. 220–222 °C; IR (KBr, cm−1): 3034, 2980, 1697, 1599, 1456, 1356, 1204, 1153, 841. 1H NMR (500 MHz DMSO-d6) δ 3.86 (s, 6H, 2 × OMe), 6.83 (s, 2H, H7), 6.95 (as, 1H, H12), 7.24 (as, 2H, H10, H14), 7.77 (d, J = 8.5 Hz, 2H, H17, H19), 8.25 (d, J = 8.5 Hz, 2H, H16, H20), 8.95 (dd, J = 9.0; 5.5 Hz, 1H, H5), 9.31 (d, J = 9.0 Hz, 1H, H4), 9.87 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 55.8 (2 × OMe), 70.7 (C7), 106.3 (C10, C14), 106.6 (C12); 129.7 (C16, C20), 129.8 (C17, C19), 130.6 (C15), 134.8 (C4), 135.2 (C9), 136.4 (C5), 137.8 (C18), 150.1 (C6), 160.2 (C3), 160.9 (C11, C13); 190.0 (C8). Anal. calcd. for C20H18BrClN2O3: C, 53.41; H, 4.03; N, 6.23%. Found: C, 53.45; H, 4.00; N, 6.27%.
3–(4-Chlorophenyl)-1–(2-oxo-2–(3,4-dimethoxyphenyl)ethyl)pyridazin-1-ium bromide7k. Beige solid, Yield 77%; m.p. 144–146 °C; IR (KBr, cm−1): 3055, 2936, 1680, 1597, 1518, 1452, 1333, 1271, 1157, 1092, 1015. 1H NMR (500 MHz DMSO-d6) δ 3.86 (s, 3H, OMe), 3.91 (s, 3H, OMe), 6.79 (s, 2H, H7), 7.23 (d, J = 8.0 Hz, 1H, H13), 7.55 (s, 1H, H10), 7.76 (d, J = 7.5 Hz, 2H, H17, H19), 7.82 (d, J = 8.0 Hz, 1H, H14), 8.24 (d, J = 7.5 Hz, 2H, H16, H20), 8.94 (dd, J = 8.5; 5.5 Hz, 1H, H5), 9.30 (d, J = 8.5 Hz, 1H, H4), 9.87 (d, J = 4.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 55.8 (OMe), 56.0 (OMe), 70.3 (C7), 110.5 (C10), 111.3 (C13), 123.7 (C14), 126.0 (C9), 129.7 (C16, C20, C17, C19), 130.6 (C15), 134.7 (C4), 136.3 (C5), 137.8 (C18), 148.9 (C11), 150.1 (C6), 154.6 (C12); 160.1 (C3), 188.4 (C8). Anal. calcd. for C20H18BrClN2O3: C, 53.41; H, 4.03; N, 6.23%. Found: C, 53.44; H, 3.99; N, 6.27%.
3–(4-Chlorophenyl)-1–(2-oxo-2–(4-bromophenyl)ethyl)pyridazin-1-ium bromide7l. Yellow solid, Yield: 75%; m.p. 200–202 °C; IR (KBr, cm−1): 3055, 2992, 1690, 1587, 1447, 1395, 1333, 1233, 1096, 986, 824. 1H NMR (500 MHz DMSO-d6) δ 6.86 (s, 2H, H7), 7.76 (d, J = 6.5 Hz, 2H, H17, H19), 7.90 (d, J = 6.0 Hz, 2H, H11, H13), 8.04 (d, J = 6.0 Hz, 2H, H10, H14), 8.25 (d, J = 6.5 Hz, 2H, H16, H20), 8.96 (bs, 1H, H5), 9.33 (d, J = 7.5 Hz, 1H, H4), 9.93 (bs, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 70.5 (C7), 129.2 (C12), 129.7 (C16, C20), 129.8 (C17, C19), 130.4 (C11, C13), 130.6 (C15), 132.3 (C10, C14), 132.4 (C9), 134.8 (C4), 136.4 (C5), 137.8 (C18), 150.1 (C6), 160.1 (C3), 189.7 (C8). Anal. calcd. for C18H13Br2ClN2O: C, 46.14; H, 2.80; N, 5.98%. Found: C, 46.14; H, 2.77; N, 6.01%.
3–(4-Bromophenyl)-1–(2-oxo-2–(3,4,5-trimethoxyphenyl)ethyl)pyridazin-1-ium bromide7m. Brown solid, Yield: 75%; m.p. 185–187 °C; IR (KBr, cm−1): 2930, 1680, 1585, 1456, 1340, 1132. 1H NMR (500 MHz DMSO-d6) δ 3.82 (s, 3H, OMe), 3.92 (s, 6H, 2 × OMe), 6.90 (s, 2H, H7), 7.43 (s, 2H, H10, H14), 7.91 (d, J = 8.0 Hz, 2H, H17, H19), 8.18 (d, J = 8.0 Hz, 2H, H16, H20), 8.97 (dd, J = 9.0; 4.5 Hz, 1H, H5), 9.33 (d, J = 9.0 Hz, 1H, H4), 9.92 (d, J = 4.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 56.4 (2 × OMe), 60.4 (OMe), 70.6 (C7), 106.2 (C10, C14), 126.8 (C18), 128.5 (C9), 129.9 (C16, C20), 130.9 (C15), 132.6 (C17, C19), 134.7 (C4), 136.4 (C5), 143.3 (C12), 150.1 (C6), 153.0 (C11, C13), 160.3 (C3), 189.2 (C8). Anal. calcd. for C21H20Br2N2O4: C, 48.12; H, 3.85; N, 5.34%. Found: C, 48.15; H, 3.79; N, 5.37%.
3–(4-Bromophenyl)-1–(2-oxo-2–(3,5-dimethoxyphenyl)ethyl)pyridazin-1-ium bromide7n. Brown solid, Yield: 73%; m.p. 217–220 °C; IR (KBr, cm−1): 3078, 2926, 1695, 1591, 1452, 1385, 1103, 1074. 1H NMR (500 MHz DMSO-d6) δ 3.85 (s, 6H, 2 × OMe), 6.82 (s, 2H, H7), 7.23 (s, 2H, H10, H14), 7.91 (d, J = 8.0 Hz, 2H, H17, H19), 8.17 (d, J = 8.0 Hz, 2H, H16, H20), 8.95 (dd, J = 9.0; 4.5 Hz, 1H, H5), 9.30 (d, J = 9.0 Hz, 1H, H4), 9.86 (d, J = 4.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 56.7 (2 × OMe), 70.7 (C7), 106.6 (C12), 107.1 (C10, C14), 127.7 (C18), 128.9 (C16, C20), 130.8 (C15), 132.7 (C17, C19), 134.8 (C4), 135.2 (C9), 136.3 (C5), 150.8 (C6), 160.3 (C3), 160.9 (C11, C13), 189.9 (C8). Anal. calcd. for C20H18Br2N2O3: C, 48.61; H, 3.67; N, 5.67%. Found: C, 48.65; H, 3.66; N, 5.67%.
3–(4-Bromophenyl)-1–(2-oxo-2–(3,4-dimethoxyphenyl)ethyl)pyridazin-1-ium bromide7o. Brown solid, Yield: 72%; m.p. 210–212 °C; IR (KBr, cm−1): 3049, 2975, 1697, 1596, 1453, 1358, 1261, 1207. 1H NMR (500 MHz DMSO-d6) δ 3.86 (s, 3H, 2 OMe), 3.91 (s, 3H, 2 OMe), 6.80 (s, 2H, H7), 7.23 (d, J = 8.5 Hz, 1H, H13), 7.55 (d, J = 2.0 Hz, 1H, H10), 7.83 (dd, J = 8.5; 2.0 Hz, 1H, H14), 7.91 (d, J = 8.5 Hz, 2H, H17, H19), 8.17 (d, J = 8.5 Hz, 2H, H16, H20), 8.95 (dd, J = 9.0; 5.5 Hz, 1H, H5), 9.31 (d, J = 9.0 Hz, 1H, H4), 9.87 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 55.8 (OMe), 56.1 (OMe), 70.4 (C7), 110.4 (C10), 111.3 (C13), 123.7 (C14), 126.0 (C9), 126.9 (C18), 129.9 (C16, C20), 131.0 (C15), 132.7 (C17, C19), 134.7 (C4), 136.4 (C5), 148.9 (C11), 150.1 (C6), 154.6 (C12); 160.3 (C3), 188.5 (C8). Anal. calcd. for C20H18Br2N2O3: C, 48.61; H, 3.67; N, 5.67%. Found: C, 48.62; H, 3.64; N, 5.68%.
3–(4-Bromophenyl)-1–(2-oxo-2–(4-bromophenyl)ethyl)pyridazin-1-ium bromide7p. Brown solid, Yield: 80%; m.p. 206–208 °C; IR (KBr, cm−1): 3055, 1690, 1587, 1447, 1387, 1333, 1233, 1076, 986, 827. 1H NMR (500 MHz DMSO-d6) δ 6.84 (s, 2H, H7), 7.90 (d, J = 8.5 Hz, 2H, H11, H13), 7.91 (d, J = 8.0 Hz, 2H, H17, H19), 8.04 (d, J = 8.0 Hz, 2H, H16, H20), 8.17 (d, J = 8.5 Hz, 2H, H10, H14), 8.95 (dd, J = 8.0; 5.5 Hz, 1H, H5), 9.32 (d, J = 8.0 Hz, 1H, H4), 9.89 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 70.5 (C7), 126.9 (C18), 129.2 (C12), 129.9 (C10, C14), 130.4 (C16, C20), 131.0 (C15), 132.2 (C11, C13), 132.4 (C9), 132.6 (C17, C19), 134.8 (C4), 136.4 (C5), 150.1 (C6), 160.3 (C3), 189.6 (C8). Anal. calcd. for C18H13Br3N2O: C, 42.14; H, 2.55; N, 5.46%. Found: C, 42.12; H, 2.54; N, 5.48%.
1–(2-Oxo-2–(3,4,5-trimethoxyphenyl)ethyl)-3-(p-tolyl)pyridazin-1-ium bromide7q. Beige solid, Yield: 78%; m.p. 208–210 °C; IR (KBr, cm−1): 3040, 2990, 2928, 1688, 1587, 1454, 1343, 1132, 990. 1H NMR (500 MHz DMSO-d6) δ 2.43 (s, 3H, Me), 3.81 (s, 3H, OMe), 3.91 (s, 6H, 2 × OMe), 6.87 (s, 2H, H7), 7.42 (s, 2H, H10, H14), 7.48 (d, J = 8.0 Hz, 2H, H17, H19), 8.13 (d, J = 8.0 Hz, 2H, H16, H20), 8.92 (dd, J = 9.0; 5.5 Hz, 1H, H5), 9.28 (d, J = 9.0 Hz, 1H, H4), 9.86 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 21.1 (Me), 56.4 (2 × OMe), 60.4 (OMe), 70.6 (C7), 106.2 (C10, C14), 127.8 (C16, C20), 128.5 (C9), 128.9 (C15), 130.2 (C17, C19), 134.3 (C4), 136.2 (C5), 143.2 (C18), 143.3 (C12), 149.6 (C6), 153.1 (C11, C13), 161.1 (C3), 189.3 (C8). Anal. calcd. for C22H23BrN2O4: C, 57.53; H, 5.05; N, 6.10%. Found: C, 57.55; H, 5.01; N, 6.13%.
1–(2-Oxo-2–(3,5-dimethoxyphenyl)ethyl)-3-(p-tolyl)pyridazin-1-ium bromide7r. Beige solid, Yield: 75%; m.p. 180–182 °C; IR (KBr, cm−1): 3042, 2974, 1695, 1591, 1456, 1202, 1155, 808. 1H NMR (500 MHz DMSO-d6) δ 2.43 (s, 3H, Me), 3.86 (s, 6H, 2 × OMe), 6.82 (s, 2H, H7), 6.94 (t, J = 2.5 Hz, 1H, H12), 7.24 (d, J = 2.5 Hz, 2H, H10, H14), 7.48 (d, J = 8.5 Hz, 2H, H17, H19), 8.13 (d, J = 8.5 Hz, 2H, H16, H20), 8.90 (dd, J = 9.0; 5.5 Hz, 1H, H5), 9.27 (d, J = 9.0 Hz, 1H, H4), 9.84 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 21.1 (Me), 55.8 (2 × OMe), 70.7 (C7), 106.3 (C10, C14), 106.6 (C12); 127.8 (C16, C20), 128.9 (C15), 130.2 (C17, C19), 134.3 (C4), 135.2 (C9), 136.1 (C5), 143.2 (C18), 149.6 (C6), 160.9 (C11, C13); 161.1 (C3), 190.1 (C8). Anal. calcd. for C21H21BrN2O3: C, 58.75; H, 4.93; N, 6.53%. Found: C, 58.77; H, 4.89; N, 6.55%.
1–(2-Oxo-2–(3,4-dimethoxyphenyl)ethyl)-3-(p-tolyl)pyridazin-1-ium bromide7s. Beige solid, Yield: 75%; m.p. 150–151 °C; IR (KBr, cm−1): 3009, 2962, 1679, 1590, 1518, 1455, 1268, 1160, 1024. 1H NMR (500 MHz DMSO-d6) δ 2.42 (s, 3H, Me), 3.86 (s, 3H, OMe), 3.91 (s, 3H, OMe), 6.79 (s, 2H, H7), 6.94 (d, J = 8.5 Hz, 1H, H13), 7.55 (d, J = 1.5 Hz, 2H, H10), 7.48 (d, J = 8.0 Hz, 2H, H17, H19), 7.83 (dd, J = 8.5; 1.5 Hz, 1H, H14), 8.13 (d, J = 8.0 Hz, 2H, H16, H20), 8.90 (dd, J = 8.5; 5.5 Hz, 1H, H5), 9.27 (d, J = 9.0 Hz, 1H, H4), 9.83 (d, J = 5.5 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 21.1 (Me), 55.8 (OMe), 56.1 (OMe), 70.3 (C7), 110.4 (C10), 111.3 (C13); 123.7 (C14), 126.1 (C9), 127.9 (C16, C20), 128.9 (C15), 130.3 (C17, C19), 134.3 (C4), 136.1 (C5), 143.3 (C18), 148.9 (C11), 149.7 (C6), 154.5 (C12), 161.1 (C3), 188.6 (C8). Anal. calcd. for C21H21BrN2O3: C, 58.75; H, 4.93; N, 6.53%. Found: C, 58.78; H, 4.90; N, 6.56%.
1–(2-Oxo-2–(4-bromophenyl)ethyl)-3-(p-tolyl)pyridazin-1-ium bromide7t. Yellow solid, Yield: 77%; m.p. 208–210 °C; IR (KBr, cm−1): 3022, 2988, 1694, 1584, 1452, 1219, 984. 1H NMR (500 MHz DMSO-d6) δ 2.42 (s, 3H, Me), 6.84 (s, 2H, H7), 7.47 (d, J = 8.0 Hz, 2H, H11, H13), 7.91 (d, J = 8.5 Hz, 2H, H17, H19), 8.05 (d, J = 8.5 Hz, 2H, H16, H20), 8.13 (d, J = 8.0 Hz, 2H, H10, H14), 8.91 (dd, J = 9.0; 6.0 Hz, 1H, H5), 9.28 (d, J = 9.0 Hz, 1H, H4), 9.86 (d, J = 6.0 Hz, 1H, H6). 13 C NMR (125 MHz DMSO-d6) δ 21.1 (Me), 70.5 (C7), 127.9 (C16, C20), 128.9 (C15), 129.2 (C12), 130.2 (C17, C19), 130.4 (C11, C13), 132.3 (C10, C14), 132.5 (C9), 134.3 (C4), 136.1 (C5), 143.2 (C18), 149.7 (C6), 161.0 (C3), 189.7 (C8). Anal. calcd. for C19H16Br2N2O: C, 50.92; H, 3.60; N, 6.25%. Found: C, 50.93; H, 3.56; N, 6.26%.
Ethyl 7–(3,4,5-trimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8a. Beige powder, Yield: 55%; m.p. 188–190 °C; IR (KBr, cm−1): 1703, 1635, 1584, 1474, 1323, 1217, 1128, 1051; 1H NMR (500 MHz CDCl3) δ 1.41 (t, J = 7.0 Hz, 3H, CH3), 3.90 (s, 6H, 2 × OMe), 3.96 (s, 3H, OMe), 4.40 (q, J = 7.0 Hz, 2H, CH2), 7.15 (dd, J = 9.0; 4.5 Hz, 1H, H3), 7.17 (s, 2H, H12, H16), 7.77 (s, 1H, H6), 8.53 (as, 1H, H4), 8.67 (d, J = 9.0 Hz, 1H,H2). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 55.5 (2 × OMe), 60.6 (CH2), 61.2 (OMe), 105.5 (C5), 107.4 (C12, C16), 117.6 (C3), 124.4 (C6), 126.8 (C7), 128.2 (C2), 133.5 (C8), 134.2 (C14), 142.2 (C11), 144.3 (C4), 153.2 (C13, C15), 163.7 (COO), 183.6 (C10). Anal. calcd. for C20H20N2O6: C, 62.49; H, 5.24; N, 7.29%. Found: C, 64.59; H, 5.15; N, 7.32%.
Ethyl 7–(3,5-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8b. Yellow powder, Yield: 57%; m.p. 178–180 °C; IR (KBr, cm−1): 1685, 1631, 1587, 1479, 1358, 1225, 1047 (C-O); 1H NMR (500 MHz CDCl3) δ 1.40 (t, J = 7.0 Hz, 3H, CH3), 3.85 (s, 6H, 2 × OMe), 4.39 (q, J = 7.0 Hz, 2H, CH2), 6.68 (s, 1H, H14), 7.01 (d, J = 1.5 Hz, 2H, H12, H16), 7.15 (dd, J = 9.5; 4.5 Hz, 1H, H3), 7.78 (s, 1H, H6), 8.54 (d, J = 3.0 Hz, 1H, H4), 8.67 (d, J = 9.0 Hz, 1H, H2). 13 C NMR (125 MHz CDCl3) δ 14.6 (CH3), 55.8 (2 × OMe), 60.6 (CH2), 104.7 (C14), 105.5 (C5), 107.6 (C12, C16), 117.8 (C3), 125.2 (C6), 126.7 (C7), 128.2 (C2), 133.7 (C8), 141.1 (C11), 144.4 (C4), 160.8 (C13, C15), 163.7 (COO), 184.1 (C10). Anal. calcd. for C19H18N2O5: C, 64.40; H, 5.12; N, 7.91%. Found: C, 64.52; H, 5.05; N, 7.99%.
Ethyl 7–(3,4-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8c. Beige solid, Yield: 50%; m.p. 123–124 °C; IR (KBr, cm−1): 1704, 1629, 1580, 1371, 1132, 1023; 1H NMR (500 MHz CDCl3) δ 1.41 (t, J = 7.0 Hz, 3H, CH3), 3.96 (s, 3H, OMe), 3.98 (s, 3H, OMe), 4.39 (q, J = 7.0 Hz, 2H, CH2), 6.95 (d, 1H, J = 8.0 Hz, H15), 7.12 (dd, J = 9.5; 4.5 Hz, 1H, H3), 7.55 (d, J = 1.5 Hz, 1H, H12), 7.56 (dd, J = 8.0; 1.5 Hz, 1H, H16), 7.75 (s, 1H, H6), 8.50 (dd, J = 4.0; 2.0 Hz, 1H, H4), 8.65 (dd, J = 9.0; 2.0 Hz, 1H, H2). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 56.2 (OMe), 56.3 (OMe), 60.5 (CH2), 105.2 (C5), 110.1 (C12), 112.0 (C15), 117.3 (C3), 123.9 (C16), 124.8 (C6), 126.9 (C7), 128.2 (C2), 131.7 (C11), 133.2 (C8), 144.2 (C4), 149.2 (C13), 153.2 (C14), 163.8 (COO), 184.4 (C10). Anal. calcd. for C19H18N2O5: C, 64.40; H, 5.12; N, 7.91%. Found: C, 64.44; H, 5.07; N, 7.97%.
Ethyl 7–(4-bromobenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8d. Yellow solid, Yield: 57%; m.p. 133–135 °C; IR (KBr, cm−1): 1707, 1638, 1468, 1235, 1190, 1096; 1H NMR (500 MHz CDCl3) δ 1.40 (t, J = 7.0 Hz, 3H, CH3), 4.38 (q, J = 7.0 Hz, 2H, CH2), 7.17 (dd, J = 9.0; 4.5 Hz, 1H, H3), 7.65 (d, J = 8.0 Hz, 2H, H13, H15), 7.72 (s, 1H, H6), 7.77 (d, J = 8.0 Hz, 2H, H12, H16), 8.54 (dd, J = 4.0; 1.5 Hz, 1H, H4), 8.67 (dd, J = 9.5; 4.0 Hz, 1H, H2). 13 C NMR (125 MHz CDCl3) δ 14.6 (CH3), 60.7 (CH2), 105.7 (C5), 118.0 (C3), 125.0 (C6), 126.4 (C7), 127.4 (C14), 128.2 (C2), 131.2 (C12, C16), 131.9 (C13, C15), 133.8 (C8), 137.9 (C11), 144.5 (C4), 163.6 (COO), 183.3 (C10). Anal. calcd. for C17H13BrN2O3: C, 54.71; H, 3.51; N, 7.51%. Found: C, 54.74; H, 3.47; N, 7.57%.
Ethyl 2-methyl-7–(3,4,5-trimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8e. Beige solid; Yield: 47%; m.p. 91–93 °C; IR (KBr, cm−1): 2978,1695, 1656, 1586, 1470, 1368, 1233, 1124, 754; 1H NMR (500 MHz CDCl3) δ 1.40 (t, J = 7.0 Hz, 3H, CH3), 2.64 (s, 3H,Me), 3.90 (s, 6H, 2 × OMe), 3.95 (s, 3H, OMe), 4.38 (q, J = 7.0 Hz, 2H, CH2), 7.02 (d, J = 9.0 Hz, 1H, H3), 7.18 (s, 2H, H12, H16), 7.68 (s, 1H, H6), 8.52 (d, J = 9.0 Hz, 1H, H4). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 22.4 (Me), 56.5 (2 × OMe), 60.5 (CH2), 61.2 (OMe), 105.1 (C5), 107.5 (C12, C16), 119.7 (C3), 123.8 (C6), 126.6 (C7), 127.6 (C11), 132.3 (C8), 134.3 (C14), 142.2 (C4), 153.1 (C13, C15), 153.4 (C2), 163.9 (COO), 183.5 (C10). Anal. calcd. for C21H22N2O6: C, 63.31; H, 5.57; N, 7.03%. Found: C, 63.29; H, 5.55; N, 7.00%.
Ethyl 2-methyl-7–(3,5-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8f. Beige solid, Yield: 45%; m.p. 96–98 °C; IR (KBr, cm−1): 2937, 1681, 1654, 1592, 1436, 1361, 1232, 1158, 754; 1H NMR (500 MHz CDCl3) δ 1.39 (t, J = 7.0 Hz, 3H, CH3), 2.65 (s, 3H,Me), 3.84 (s, 6H, 2 × OMe), 4.37 (q, J = 7.0 Hz, 2H, CH2), 6.68 (s, 1H, H14), 7.02 (overlapped signals, 3H, H3, H12, H16), 7.69 (s, 1H, H6), 8.52 (d, J = 9.5 Hz, 1H, H4). 13 C NMR (125 MHz CDCl3) δ 14.6 (CH3), 22.4 (Me), 55.8 (2 × OMe), 60.5 (CH2), 104.6 (C14), 105.2 (C5), 107.6 (C12, C16), 119.9 (C3), 124.6 (C6), 126.6 (C7), 127.6 (C11), 132.5 (C8), 141.2 (C4), 153.5 (C2), 160.7 (C13, C15), 163.8 (COO), 184.1 (C10). Anal. calcd. for C20H20N2O5: C, 65.21; H, 5.47; N, 7.60%. Found: C, 65.18; H, 5.45; N, 7.63%.
Ethyl 2-methyl-7–(4-bromobenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8h. Beige solid, Yield: 42%; m.p. 145–147 ˚C; IR (KBr, cm−1): 2990, 1701, 1645, 1547, 1462, 1362, 1261, 1229, 1188, 1098, 754; 1H NMR (500 MHz CDCl3) δ 1.40 (t, J = 7.0 Hz, 3H, CH3), 2.65 (s, 3H, Me), 4.38 (q, J = 7.0 Hz, 2H, CH2), 7.04 (d, J = 9.5 Hz, 1H, H3), 7.63 (s, 1H, H6), 7.65 (d, J = 8.0 Hz, 2H, H13, H15), 7.77 (d, J = 8.0 Hz, 2H, H12, H16), 8.54 (d, J = 9.0 Hz, 1H, H4). 13 C NMR (125 MHz CDCl3) δ 14.6 (CH3), 22.4 (Me), 60.6 (CH2), 105.4 (C5), 120.1 (C3), 124.4 (C6), 126.3 (C7), 127.3 (C14), 127.6 (C4), 131.2 (C12, C16), 131.8 (C13, C15), 132.6 (C8), 138.1 (C11), 153.7 (C2), 163.7 (COO), 183.3 (C10). Anal. calcd. for C18H15BrN2O3: C, 55.83; H, 3.90; N, 7.23%. Found: C, 55.85; H, 3.87; N, 7.26%.
Ethyl 2–(4-chlorophenyl)-7–(3,4,5-trimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8i. Beige solid, Yield: 41%; m.p. 230–232 ˚C; IR (KBr, cm−1): 2984, 1697, 1657, 1583, 1503, 1460, 1314, 1234, 1169, 1130, 808; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.90 (s, 6H, 2 × OMe), 3.97 (s, 3H, OMe), 4.42 (q, J = 7.0 Hz, 2H, CH2), 7.20 (s, 2H, H12, H16), 7.47 (d, J = 8.5 Hz, 2H, H19, H21), 7.47 (d, J = 9.0 Hz, 1H, H4), 7.79 (s, 1H, H6), 8.01 (d, J = 8.5 Hz, 2H, H18, H22), 8.69 (d, J = 9.0 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 56.5 (2 × OMe), 60.6 (CH2), 61.2 (OMe), 105.6 (C5), 107.3 (C12, C16), 115.8 (C4), 124.4 (C6), 127.1 (C7), 128.4 (C3, C18, C22), 129.5 (C19, C21), 132.2 (C11), 133.6 (C17), 134.2 (C8), 136.9 (C20), 142.3 (C14), 151.1 (C2), 153.2 (C13, C15), 163.7 (COO), 183.6 (C10). Anal. calcd. for C26H23ClN2O6: C, 63.10; H, 4.68; N, 5.66%. Found: C, 63.15; H, 4.67; N, 5.69%.
Ethyl 2–(4-chlorophenyl)-7–(3,5-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8j. Beige solid, Yield: 41%; m.p. 193–195 ˚C; IR (KBr, cm−1): 2949, 1694, 1659, 1599, 1458, 1298, 1094, 810; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.85 (s, 6H, 2 × OMe), 4.40 (q, J = 7.0 Hz, 2H, CH2), 6.71 (bs, 1H, H14), 7.05 (d, J = 2.0 Hz, 2H, H12, H16), 7.47 (d, J = 8.5 Hz, 2H, H19, H21), 7.59 (d, J = 9.5 Hz, 1H4), 7.81 (s, 1H, H6), 8.01 (d, J = 8.5 Hz, 2H, H18, H22), 8.70 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 55.8 (2 × OMe), 60.6 (CH2), 104.8 (C14), 105.7 (C5), 107.5 (C12, C16), 115.9 (C4), 125.0 (C6), 127.1 (C7), 128.4 (C18, C22), 128.5 (C3), 129.5 (C19, C21), 132.4 (C8), 133.7 (C17), 136.8 (C20), 141.1 (C11), 151.2 (C2), 160.9 (C13, C15), 163.7 (COO), 184.2 (C10). Anal.calcd. for C25H21ClN2O5: C, 64.59; H, 4.55; N, 6.03%. Found: C, 64.64; H, 4.50; N, 6.10%.
Ethyl 2–(4-chlorophenyl)-7–(3,4-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8k. Beige solid, Yield: 42%; m.p. 161–163 ˚C; IR (KBr, cm−1): 2982, 1696, 1649, 1595, 1462, 1269, 1234, 1090, 806; 1H NMR (500 MHz CDCl3) δ 1.43 (t, J = 7.0 Hz, 3H, CH3), 3.97 (s, 3H, OMe), 3.99 (s, 3H, OMe), 4.41 (q, J = 7.0 Hz, 2H, CH2), 6.95 (d, J = 8.0 Hz, 1H, H15), 7.46 (d, J = 8.5 Hz, 2H, H19, H21), 7.56 (m, 3H, H12, H16, H4), 7.76 (s, 1H, H6), 8.00 (d, J = 8.5 Hz, 2H, H18, H22), 8.68 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.8 (CH3), 56.3 (OMe), 56.4 (OMe), 60.7 (CH2), 105.4 (C5), 110.2 (C15), 112.0 (C12), 115.7 (C4), 124.2 (C6), 124.9 (C16), 127.4 (C7), 128.6 (C3, C18, C22), 129.5 (C19, C21), 131.9 (C11), 132.1 (C17), 133.8 (C8), 136.9 (C20), 151.1 (C2), 149.4 (C13), 153.4 (C14), 163.9 (COO), 183.5 (C10). Anal. calcd. for C25H21ClN2O5: C, 64.59; H, 4.55; N, 6.03%. Found: C, 64.60; H, 4.49; N, 6.07%.
Ethyl 2–(4-chlorophenyl)-7–(4-bromobenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8l. White solid; Yield: 45%; m.p. 163–165 ˚C; IR (KBr, cm−1) 3051, 2988, 1694, 1458, 1242, 1207, 1090, 808; 1H N,R (500 MHz CDCl3) δ 1.43 (t, J = 7.0 Hz, 3H, CH3), 4.41 (q, J = 7.0 Hz, 2H, CH2), 7.47 (d, J = 8.5 Hz, 2H, H19, H21), 7.59 (d, J = 9.5 Hz, H4), 7.67 (d, J = 8.0 Hz, 2H, H12, H16), 7.76 (s, 1H, H6), 7.78 (d, J = 8.0 Hz, 2H, H13, H15), 7.96 (d, J = 8.5 Hz, 2H, H18, H22), 8.70 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 60.7 (CH2), 105.9 (C5), 116.1 (C4), 124.8 (C6), 126.9 (C14), 127.4 (C7), 128.4 (C18, C22), 128.5 (C3), 129.5 (C19, C21), 132.4 (C8), 133.5 (C17), 131.1 (C13, C15), 131.9 (C12, C16), 137.0 (C20), 138.0 (C11), 151.3 (C2), 163.6 (COO), 183.5 (C10). Anal. calcd. for C23H16BrClN2O3: C, 57.11; H, 3.33; N, 5.79%. Found: C, 57.10; H, 3.29; N, 5.85%.
Ethyl 2–(4-bromophenyl)-7–(3,4,5-trimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8m. Beige solid, Yield: 50%; m.p. 238–240 ˚C; IR (KBr, cm−1): 2984, 2930, 1697, 1657, 1586, 1503, 1458, 1314, 1234, 1169, 1128, 808, 752; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.90 (s, 6H, 2 × OMe), 3.97 (s, 3H, OMe), 4.42 (q, J = 7.0 Hz, 2H, CH2), 7.20 (s, 2H, H12, H16), 7.58 (d, J = 9.0 Hz, H4), 7.63 (d, J = 8.5 Hz, 2H, H19, H21), 7.80 (s, 1H, H6), 7.94 (d, J = 8.5 Hz, 2H, H18, H22), 8.69 (d, J = 9.0 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 56.5 (2 × OMe), 60.6 (CH2), 61.2 (OMe), 105.6 (C5), 107.4 (C12, C16), 115.7 (C4), 124.4 (C6), 125.3 (C20), 127.1 (C7), 128.5 (C3), 128.7 (C18, C22), 132.5 (C19, C21), 132.2 (C11), 134.1 (C17), 134.2 (C8), 142.3 (C14), 151.2 (C2), 153.2 (C13, C15), 163.7 (COO), 183.6 (C10). Anal. calcd. for C26H23BrN2O6: C, 57.90; H, 4.30; N, 5.19%. Found: C, 57.95; H, 4.27; N, 5.24%.
Ethyl 2–(4-bromophenyl)-7–(3,5-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8n. Beige solid, Yield: 51%; m.p. 193–195 ˚C; IR (KBr, cm−1): 2982, 1695, 1659, 1591, 1458, 1298, 1155, 1096, 808; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.85 (s, 6H, 2 × OMe), 4.40 (q, J = 7.0 Hz, 2H, CH2), 6.71 (t, J = 2.5 Hz, 1H, H14), 7.05 (d, J = 2.5 Hz, 2H, H12, H16), 7.58 (d, J = 9.5 Hz, H4), 7.63 (d, J = 8.5 Hz, 2H, H19, H21), 7.81 (s, 1H, H6), 7.94 (d, J = 8.5 Hz, 2H, H18, H22), 8.70 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 55.8 (2 × OMe), 60.6 (CH2), 104.8 (C14), 105.7 (C5), 107.5 (C12, C16), 115.9 (C4), 125.0 (C6), 125.2 (C20), 127.1 (C7), 128.4 (C3), 128.7 (C18, C22), 132.4 (C19, C21, C8), 134.1 (C17), 141.1 (C11), 151.2 (C2), 160.9 (C13, C15), 163.7 (COO), 184.2 (C10). Anal. calcd. for C25H21BrN2O5: C, 58.95; H, 4.16; N, 5.50%. Found: C, 58.94; H, 4.09; N, 5.55%.
Ethyl 2–(4-bromophenyl)-7–(3,4-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8o. Beige solid, Yield: 52%; m.p. 166–167 ˚C; IR (KBr, cm−1): 2975, 2929, 1719, 1680, 1592, 1458, 1269, 1236, 1147, 1087; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.97 (s, 3H, OMe), 3.99 (s, 3H, OMe), 4.41 (q, J = 7.0 Hz, 2H, CH2), 6.95 (d, J = 7.5 Hz, 1H, H15), 7.57–7.62 (m, 5H, H12, H16, H4, H19, H21), 7.76 (s, 1H, H6), 7.93 (d, J = 8.0 Hz, 2H, H18, H22), 8.67 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 56.2 (OMe), 56.3 (OMe), 60.6 (CH2), 105.4 (C5), 110.1 (C15), 111.8 (C12), 115.5 (C4), 124.1 (C6), 124.8 (C16), 125.1 (C20), 127.3 (C7), 128.4 (C3), 128.7 (C18, C22), 131.7 (C11), 132.4 (C19, C21), 132.0 (C8), 134.2 (C17), 149.3 (C13), 151.1 (C2), 153.2 (C14), 163.8 (COO), 183.4 (C10). Anal. calcd. for C25H21BrN2O5: C, 58.95; H, 4.16; N, 5.50%. Found: C, 58.97; H, 4.10; N, 5.53%.
Ethyl 2–(4-bromophenyl)-7–(4-bromobenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8p. White solid, Yield: 45%; m.p. 171–173 ˚C; IR (KBr, cm−1): 3051, 2988, 1694, 1657, 1458, 1242, 1209, 1094, 1072, 806, 748; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 4.41 (q, J = 7.0 Hz, 2H, CH2), 7.63 (d, J = 8.5 Hz, 2H, H19, H21), 7.59 (d, J = 9.5 Hz, H4), 7.67 (d, J = 8.0 Hz, 2H, H12, H16), 7.76 (s, 1H, H6), 7.77 (d, J = 8.0 Hz, 2H, H13, H15), 7.88 (d, J = 8.5 Hz, 2H, H18, H22), 8.70 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.6 (CH3), 60.7 (CH2), 105.9 (C5), 116.0 (C4), 124.8 (C6), 125.3 (C20), 126.9 (C14), 127.4 (C7), 128.5 (C3), 128.7 (C18, C22), 131.1 (C19, C21), 131.9 (C13, C15), 132.4 (C8), 132.5 (C12, C16), 133.9 (C17), 138.0 (C11), 151.4 (C2), 163.6 (COO), 183.5 (C10). Anal. calcd. for C23H16Br2N2O3: C, 52.30; H, 3.05; N, 5.30%. Found: C, 52.30; H, 3.00; N, 5.32%.
Ethyl 2-(p-tolyl)-7–(3,4,5-trimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8q. White solid, Yield: 40%; m.p. 198–200 ˚C; IR (KBr, cm−1): 3020, 2978, 2943, 1697, 1657, 1586, 1503, 1460, 1333, 1234, 1130, 1130, 806, 750; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 2.41 (s, 3H, Me), 3.89 (s, 6H, 2 × OMe), 3.97 (s, 3H, OMe), 4.41 (q, J = 7.0 Hz, 2H, CH2), 7.20 (s, 2H, H12, H16), 7.29 (d, J = 8.5 Hz, 2H, H19, H21), 7.60 (d, J = 9.5 Hz, 1H4), 7.78 (s, 1H, H6), 7.94 (d, J = 8.5 Hz, 2H, H18, H22), 8.66 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 21.5 (Me), 56.5 (2 × OMe), 60.5 (CH2), 61.2 (OMe), 105.4 (C5), 107.3 (C12, C16), 116.3 (C4), 124.2 (C6), 127.0 (C18, C22), 127.1 (C7), 128.1 (C3), 129.9 (C19, C21), 132.2 (C11), 132.3 (C17), 134.3 (C8), 140.9 (C20), 142.1 (C14), 152.2 (C2), 153.1 (C13, C15), 163.8 (COO), 183.7 (C10). Anal. calcd. for C27H26N2O6: C, 68.34; H, 5.52; N, 5.90%. Found: C, 68.35; H, 5.47; N, 5.94%.
Ethyl 2-(p-tolyl)-7–(3,5-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8r. Beige solid, Yield: 40%; m.p. 142–144 ˚C; IR (KBr, cm−1): 2999, 2918, 1686, 1649, 1593, 1452, 1302, 1236, 1159, 1053, 804, 754; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 2.41 (s, 3H, Me), 3.84 (s, 6H, 2 × OMe), 4.40 (q, J = 7.0 Hz, 2H, CH2), 6.97 (bs, 1H, H14), 7.05 (bs, 2H, H12, H16), 7.29 (d, J = 8.0 Hz, 2H, H19, H21), 7.60 (d, J = 9.5 Hz, H4), 7.79 (s, 1H, H6), 7.95 (d, J = 8.5 Hz, 2H, H18, H22), 8.65 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 21.5 (Me), 55.8 (2 × OMe), 60.5 (CH2), 104.7 (C14), 105.4 (C5), 107.4 (C12, C16), 116.4 (C4), 124.9 (C6), 127.0 (C7), 127.1 (C18, C22), 128.0 (C3), 129.9 (C19, C21), 132.3 (C8), 132.5 (C17), 140.8 (C20), 141.3 (C11), 152.3 (C2), 160.8 (C13, C15), 163.8 (COO), 184.2 (C10). Anal. calcd. for C26H24N2O5: C, 70.26; H, 5.44; N, 6.30%. Found: C, 70.29; H, 5.39; N, 6.33%.
Ethyl 2-(p-tolyl)-7–(3,4-dimethoxybenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8s. Beige solid, Yield: 40%; m.p. 150–151 ˚C; IR (KBr, cm−1): 3088, 2974, 2929, 1721, 1681, 1614, 1514, 1457, 1272, 1148, 1088; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 3.96 (s, 3H, OMe), 3.98 (s, 3H, OMe), 4.41 (q, J = 7.0 Hz, 2H, CH2), 6.95 (d, J = 8.0 Hz, 1H, H15), 7.28 (d, J = 8.0 Hz, 2H, H19, H21), 7.59 (m, 3H, H12, H16, H4), 7.74 (s, 1H, H6), 7.94 (d, J = 8.0 Hz, 2H, H18, H22), 8.65 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 56.2 (OMe), 56.3 (OMe), 60.5 (CH2), 105.0 (C5), 110.0 (C15), 111.8 (C12), 116.1 (C4), 123.9 (C6), 124.7 (C16), 127.1 (C18, C22), 127.2 (C7), 128.0 (C3), 129.9 (C19, C21), 131.9 (C11), 132.1 (C17), 132.4 (C8), 140.7 (C20), 149.2 (C13), 152.1 (C2), 153.1 (C14), 164.0 (COO), 183.5 (C10). Anal. calcd. for C26H24N2O5: C, 70.26; H, 5.44; N, 6.30%. Found: C, 70.30; H, 5.40; N, 6.35%.
Ethyl 2-(p-tolyl)-7–(4-bromobenzoyl)pyrrolo[1,2-b]pyridazine-5-carboxylate8t. Yellow solid, Yield: 40%; m.p. 160–162 ˚C; IR (KBr, cm−1): 3072, 2974, 1697, 1620, 1503, 1452, 1219, 1211, 1082, 816; 1H NMR (500 MHz CDCl3) δ 1.42 (t, J = 7.0 Hz, 3H, CH3), 2.42 (s, 3H, Me), 4.41 (q, J = 7.0 Hz, 2H, CH2), 7.29 (d, J = 8.5 Hz, 2H, H19, H21), 7.62 (d, J = 9.5 Hz, H4), 7.67 (d, J = 7.5 Hz, 2H, H12, H16), 7.75 (s, 1H, H6), 7.76 (d, J = 8.0 Hz, 2H, H13, H15), 7.89 (d, J = 8.0 Hz, 2H, H18, H22), 8.67 (d, J = 9.5 Hz, 1H, H3). 13 C NMR (125 MHz CDCl3) δ 14.7 (CH3), 21.6 (Me), 60.6 (CH2), 105.7 (C5), 116.5 (C4), 124.6 (C6), 127.0 (C14), 127.1 (C18, C22), 127.2 (C7), 128.1 (C3), 130.0 (C19, C21), 132.2 (C17), 132.5 (C8), 131.1 (C13, C15), 131.9 (C12, C16), 138.0 (C11), 141.0 (C20), 152.4 (C2), 163.7 (COO), 183.6 (C10). Anal. calcd. for C24H19BrN2O3: C, 62.22; H, 4.13; N, 6.05%. Found: C, 62.25; H, 4.05; N, 6.08%.
2–(2-Oxo-2–(3,4,5-trimethoxyphenyl)ethyl)phthalazin-2-ium bromide10a. Brown solid, Yield: 77%; m.p. 150–152 ˚C; IR (KBr, cm−1): 2976, 1670, 1584, 1339, 1125, 764. 1H NMR (500 MHz DMSO-d6) δ 3.80 (s, 3H, OMe), 3.91 (s, 6H, 2 × OMe), 6.87 (s, 2H, H11), 7.44 (s, 2H, H14, H18), 8.50 (t, J = 7.5 Hz, 1H, H6), 8.62 (t, J = 8.0 Hz, H5), 8.69 (d, J = 8.0 Hz, 1H, H4), 8.76 (d, J = 8.0 Hz, 1H, H7), 10.24 (s, 1H, H3), 10.77 (s, 1H, H8). 13 C NMR (125 MHz DMSO-d6) δ 56.4 (2 × OMe), 60.4 (OMe), 69.0 (C11), 106.3 (C14, C18), 127.3 (C10), 127.5 (C9), 128.6 (C13), 128.7 (C4), 130.8 (C7), 136.6 (C6), 139.9 (C5), 143.3 (C16), 153.1 (C15, C17), 153.5 (C8), 154.9 (C3), 189.6 (C12). Anal. calcd. for C19H19BrN2O4: C, 54.43; H, 4.57; N, 6.68%. Found: C, 54.47; H, 4.54; N, 6.71%.
2–(2-Oxo-2–(3,5-dimethoxyphenyl)ethyl)phthalazin-2-ium bromide10b. Brown solid, Yield: 76%; m.p. 178–180 ˚C; IR (KBr, cm−1): 2976, 1703, 1589, 1319, 1011. 1H NMR (500 MHz DMSO-d6) δ 3.86 (s, 6H, 2 × Me), 6.79 (s, 2H, H11), 6.93 (t, J = 2.0 Hz, 1H, H16), 7.26 (d, J = 2.0 Hz, 2H, H14, H18), 8.51 (t, J = 8.0 Hz, 1H, H6), 8.61 (t, J = 8.0 Hz, H5), 8.67 (d, J = 8.0 Hz, 1H, H4), 8.75 (d, J = 8.0 Hz, 1H, H7), 10.21 (s, 1H, H3), 10.69 (s, 1H, H8). 13 C NMR (125 MHz DMSO-d6) δ 55.8 (2 × OMe), 69.1 (C11), 106.3 (C14, C18), 106.6 (C16), 127.3 (C10), 127.5 (C9), 128.6 (C4), 130.8 (C7), 135.3 (C13), 136.6 (C6), 139.9 (C5), 153.6 (C8), 154.9 (C3), 160.9 (C15, C17), 190.5 (C12). Anal. calcd. for C18H17BrN2O3: C, 55.54; H, 4.40; N, 7.20%. Found: C, 55.55; H, 4.37; N, 7.22%.
2–(2-Oxo-2–(3,4-dimethoxyphenyl)ethyl)phthalazin-2-ium bromide10c. Brown solid, Yield: 74%; m.p. 224–226 ˚C; IR (KBr, cm−1): 3017, 2974, 1701, 1589, 1313, 1204, 1011. 1H NMR (500 MHz DMSO-d6) δ 3.86 (s, 3H, OMe), 3.92 (s, 3H, OMe), 6.74 (s, 2H, H11), 6.98 (d, J = 8.0 Hz, 1H, H17), 7.47 (overlapped signals, 2H, H14, H18), 8.47 (t, J = 8.0 Hz, 1H, H6), 8.60 (t, J = 8.0 Hz, H5), 8.68 (d, J = 8.0 Hz, 1H, H4), 8.76 (d, J = 8.0 Hz, 1H, H7), 10.25 (s, 1H, H3), 10.72 (s, 1H, H8). 13 C NMR (125 MHz DMSO-d6) δ 55.8 (OMe), 56.1 (OMe), 68.9 (C11), 110.5 (C14), 111.4 (C18), 123.8 (C17), 126.2 (C13), 127.5 (C9), 128.7 (C4), 130.9 (C7), 136.6 (C6), 139.9 (C5), 148.9 (C15), 153.5 (C8), 154.9 (C3), 154.5 (C16), 188.9 (C12). Anal. calcd. for C18H17BrN2O3: C, 55.54; H, 4.40; N, 7.20%. Found: C, 55.55; H, 4.37; N, 7.22%.
2–(2-Oxo-2–(4-bromophenyl)ethyl)phthalazin-2-ium bromide10d. White solid, Yield: 76%; m.p. 222–224 ˚C; IR (KBr, cm−1): 3013, 1684, 1613, 1580, 1352. 1H NMR (500 MHz DMSO-d6) δ 6.83 (s, 2H, H11) 7.89 (d, J = 8.5 Hz, 2H, H15, H17), 8.08 (d, J = 8.5 Hz, 2H, H14, H18), 8.48 (t, J = 8.0 Hz, 1H, H6), 8.61 (t, J = 8.0 Hz, H5), 8.68 (d, J = 8.0 Hz, 1H, H4), 8.75 (d, J = 8.0 Hz, 1H, H7), 10.23 (s, 1H, H3), 10.76 (s, 1H, H8). 13 C NMR (125 MHz DMSO-d6) δ 68.8 (C11), 127.3 (C10), 127.5 (C9), 128.6 (C4), 129.2 (C16), 130.5 (C14, C18), 130.8 (C7), 132.3 (C15, C17), 132.5 (C13), 136.6 (C6), 139.9 (C5), 153.5 (C8), 154.9 (C3), 190.1 (C12). Anal. calcd. for C16H12Br2N2O: C, 47.09; H, 2.96; N, 6.86%. Found: C, 48.11; H, 2.94; N, 6.88%.
Ethyl 3–(3,4,5-trimethoxybenzoyl)pyrrolo[2,1-a]phthalazine-1-carboxylate11a. White solid, Yield: 40%; m.p. 235–237 ˚C; IR (KBr, cm−1): 3041, 2933, 2835, 1711, 1650, 1583, 1261, 1172, 1130, 1042, 785; 1H NMR (500 MHz CDCl3) δ 1.41 (t, J = 7.0 Hz, 3H, CH3), 3.90 (s, 6H, 2 × OMe), 3.90 (s, 3H, OMe), 4.41 (q, J = 7.0 Hz, 2H, CH2), 7.24 (s, 2H, H16, H20), 7.73 (s, 1H, H2), 7.76 (d, J = 7.5 Hz, 1H, H7), 7.90 (overlapped signals, 2H, H8, H6), 8.75 (s, 1H, H5), 9.84 (d, J = 8.0 Hz, 1H, H9). 13 C NMR (125 MHz CDCl3) δ 14.6 (CH3), 56.5 (2 × OMe), 60.9 (CH2), 61.2 (OMe), 107.6 (C16, C20), 108.3 (C1), 122.3 (C11), 124.2 (C2), 127.0; 127.1 (C12, C3), 127.6 (C9), 127.7 (C6), 129.8 (C7), 130.2 (C13), 133.1 (C8), 134.1 (C15), 142.3 (C18), 146.5 (C5), 153.1 (C17, C19), 164.4 (COO), 183.8 (C14). Anal. calcd. for C24H22N2O6: C, 66.35; H, 5.10; N, 6.45%. Found: C, 66.58; H, 5.05; N, 6.48%.
Ethyl 3–(3,5-dimethoxybenzoyl)pyrrolo[2,1-a]phthalazine-1-carboxylate11b. White solid, Yield: 41%; m.p. 220–222 ˚C; IR (KBr, cm−1): 3040, 2969, 2835, 1707, 1649, 1595, 1458, 1383, 1173, 1096, 756; 1H NMR (500 MHz CDCl3) δ 1.41 (t, J = 7.0 Hz, 3H, CH3), 3.85 (s, 6H, 2 × OMe), 4.41 (q, J = 7.0 Hz, 2H, CH2), 6.70 (bs, 1H, H18), 7.08 (d, J = 2.0 Hz, 2H, H16, H20), 7.74 (overlapped signals, 2H, H2, H7), 7.90 (overlapped signals, 2H, H8, H6), 8.76 (s, 1H, H5), 9.83 (d, J = 8.5 Hz, 1H, H9). 13 C NMR (125 MHz CDCl3) δ 14.6 (CH3), 55.8 (2 × OMe), 60.9 (CH2), 105.0 (C18), 107.8 (C16, C20), 108.4 (C1), 122.3 (C11), 124.8 (C2), 127.0; 127.1 (C12, C3), 127.6 (C9), 127.7 (C6), 129.8 (C7), 130.4 (C13), 133.0 (C8), 141.1 (C15), 146.6 (C5), 160.7 (C17, C19), 164.4 (COO), 184.3 (C14). Anal. calcd. for C23H20N2O5: C, 68.31; H, 4.98; N, 6.93%. Found: C, 68.34; H, 4.95; N, 6.96%.
Ethyl 3–(3,4-dimethoxybenzoyl)pyrrolo[2,1-a]phthalazine-1-carboxylate11c. White solid, Yield: 41%; m.p. 226–228 ˚C; IR (KBr, cm−1): 3101, 2959, 2932, 1713, 1645, 1466, 1412, 1263, 1177, 1024, 764; 1H NMR (500 MHz CDCl3) δ 1.41 (t, J = 7.0 Hz, 3H, CH3), 3.96 (s, 3H, OMe), 3.97 (s, 3H, OMe), 4.40 (q, J = 7.0 Hz, 2H, CH2), 6.94 (d, J = 8.0 Hz, 1H, H19), 7.58 (overlapped signals, 2H, H16, H20), 7.68 (s, 1H, H2), 7.73 (t, J = 7.5 Hz, 1H, H7), 7.88 (overlapped signals, 2H, H8, H6), 8.71 (s, 1H, H5), 9.82 (d, J = 8.0 Hz, 1H, H9). 13 C NMR (125 MHz CDCl3) δ 14.6 (CH3), 56.22 (OMe), 56.23 (OMe), 60.8 (CH2), 108.1 (C1), 110.1 (C16), 112.1 (C19), 122.2 (C11), 123.5 (C2), 125.1 (C20), 127.1 (C3), 127.3 (C12), 127.5 (C9), 127.7 (C6), 129.7 (C7), 129.9 (C13), 131.7 (C15), 133.0 (C8), 146.4 (C5), 149.2 (C17), 153.3 (C18), 164.5 (COO), 183.8 (C14). Anal. calcd. for C23H20N2O5: C, 68.31; H, 4.98; N, 6.93%. Found: C, 68.32; H, 4.97; N, 6.94%.
Ethyl 3–(4-bromobenzoyl)pyrrolo[2,1-a]phthalazine-1-carboxylate11d35. White solid, Yield: 51%; m.p. 180–182 ˚C; IR (KBr, cm−1): 2980, 1722, 1653, 1587, 1464, 1379, 1242; 1H NMR (500 MHz CDCl3) δ 1.38 (t, J = 7.0 Hz, 3H, CH3), 4.37 (q, J = 7.0 Hz, 2H, CH2), 7.62 (d, J = 8.5 Hz, 2H, H17, H19), 7.65 (s, 1H, H2), 7.74 (t, J = 8.0 Hz, 1H, H7), 7.78 (d, J = 8.5 Hz, 2H, H16, H20), 7.87 (overlapped signals, 2H, H8, H6), 8.73 (s, 1H, H5), 9.80 (d, J = 8.5 Hz, 1H, H9). 13 C NMR (125 MHz CDCl3) δ 14.5 (CH3), 60.9 (CH2), 108.5 (C1), 122.3 (C11), 124.7 (C2), 126.7 (C12); 126.8 (C18), 127.5 (C3), 127.6 (C9), 127.7 (C6), 129.9 (C7), 130.5 (C13), 131.3 (C16, C20), 131.7 (C17, C19), 133.1 (C8), 137.9 (C15), 146.6 (C5), 164.2 (COO), 183.5 (C14). Anal. calcd. for C21H15BrN2O3: C, 59.59; H, 3.57; N, 6.62%. Found: C, 59.62; H, 3.55; N, 6.64%.
Molecular modelling
Flexible docking experiments were carried out in Autodock Vina40, using a 18x22x22 Å3 grid box centered on the colchicine binding site of the α,β-tubulin heterodimer crystal structure (PDB: 1SA0)41. The 3 D structures of the compounds were constructed in Avogadro v1.2.042 and were subjected to 10,000 steepest descent steps of energy minimisation in the MMFF94 force field. One hundred poses were generated for each ligand, and the best-ranked models were chosen for further visual inspection in order to assess the consistency of the generated docking solutions relative to the docking poses of known inhibitor colchicine. Molecular graphics and visual analyses were performed in The PyMOL Molecular Graphics System, Version 1.8.2. (Schrödinger, LLC). Logp values were calculated using the ChemAxon/Chemicalize server (www.chemicalize.com).
Cell proliferation assay
The compounds were tested against a panel of 60 human cancer cell lines at the National Cancer Institute, Rockville, MD. The cytotoxicity experiments were realised using a 48 h exposure protocol using sulphorhodamine B assay43–45.
Results and discussion
Chemistry
The chosen method for the assembly of pyrrolo[1,2-b]pyridazine moieties relied on 1,3-dipolar cycloaddition of different pyridazinium ylides to ethyl propiolate.
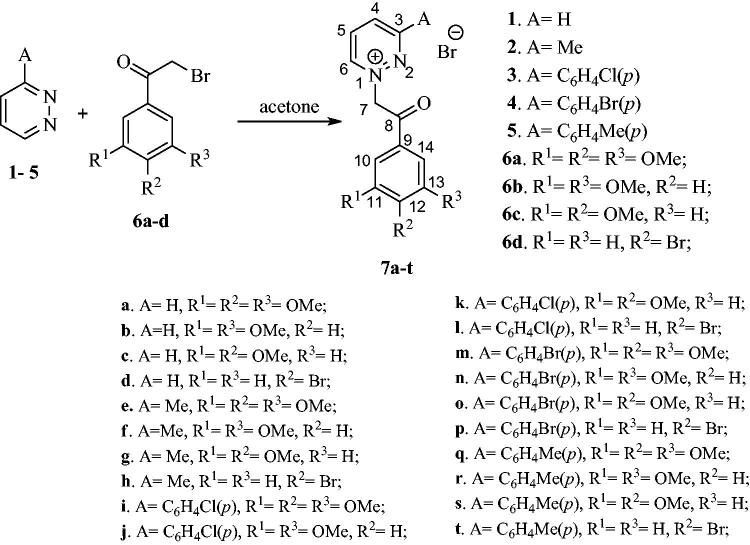
First, pyridazines 1–5 (Scheme 1) were used for the synthesis of their monoquaternary salts with 2-bromoacetophenones 6a–d. While compound 6d is commercially available, compounds 6a–c were synthesised using reported procedures46. The quaternisation reactions were carried out at room temperature (r.t.) in a minimal amount of acetone, leading to the formation of salts 7a–t (Scheme 1).
Scheme 1.
Synthesis of pyridazin-1-ium quaternary salts 7a–t.
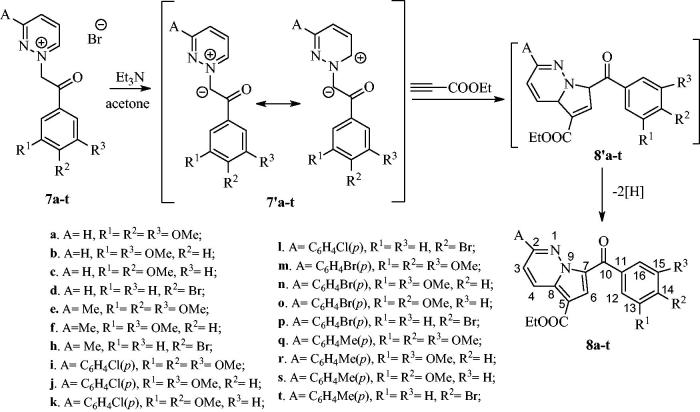
As shown in Scheme 2, ethyl propiolate was reacted with the corresponding pyridazinium ylides 7′a–t (in situ generated in basic medium from salts 7a–t) to give the intermediate dihydropyrrolo[1,2-b]pyridazines 8′a–t, which in turn underwent oxidative dehydrogenation under atmospheric conditions, yielding the final compounds 8a–t in moderate yields (40–52%) (Scheme 2).
Scheme 2.
Synthesis of pyrrolo[1,2-b]pyridazines 8a–t.
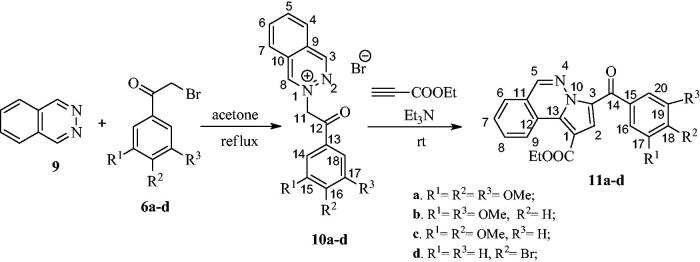
For the synthesis of compounds 11, in a similar manner, phthalazine was first reacted with 2-bromoacetophenones 6a–d, to give monoquaternary salts 10a–d (Scheme 3). Phthalazinium salts 10a–d furnished pyrrolo[2,1-a]phthalazines 11a–d when treated with triethylamine and ethyl propiolate in acetone at room temperature (Scheme 3).
Scheme 3.
Synthesis of pyrrolo[2,1-a]phthalazines 11a–d from phthalazine via quaternary phthalazinum salts 10a–d.
Biological activity
Fourteen of the synthesised compounds (8a, b, d, e, f, h, i, j, k, n, q, and 11a–c) were selected by the National Cancer Institute (NCI) for screening against a panel of 60 human tumour cell lines at a single dose of 10 μM43, the representative results for the active compounds being summarised in Table 1.
Table 1.
Results of the in vitro growth inhibition (GI %) caused by compounds 8a, b, d, e, f, h and 11a-c against human cancer cell lines in the single-dose assaya.
| Cell type | Compound |
8a |
8b |
8d |
8e |
8f |
8h |
11a |
11b |
11c |
|---|---|---|---|---|---|---|---|---|---|---|
| Cell line | GI (%) (10−5M)a | |||||||||
| Leukemia | CCRF-CEM | 89 | 76 | 23 | 83 | 86 | 4 | 30 | 77 | 11 |
| K-562 | 89 | 78 | 79 | 90 | 90 | 6 | 77 | 94 | 11 | |
| SR | 82 | 68 | 72 | 77 | 82 | 18 | 77 | 95 | 27 | |
| HL-60(TB) | 100b (33) | 100b (22) | 47 | 100b (24) | 100b (17) | 11 | 71 | 100b (8) | 18 | |
| MOLT-4 | 81 | 67 | 31 | 82 | 70 | 23 | 38 | 71 | 31 | |
| RPMI-8226 | 79 | 71 | 51 | 82 | 88 | 22 | 18 | 67 | 21 | |
| Non-small Cell lung cancer |
A549/ATCC | 76 | 65 | 27 | 73 | 73 | 23 | 45 | 82 | 20 |
| HOP-62 | 68 | 69 | 23 | 49 | 52 | 21 | 10 | 57 | 10 | |
| NCI-H460 | 90 | 85 | 0 | 90 | 89 | 0 | 49 | 93 | 0 | |
| NCI-H522 | 65 | 75 | 84 | 96 | 100b (11) | 34 | 60 | 97 | 16 | |
| Colon cancer | COLO205 | 100b (25) | 100b (13) | 29 | 100b (39) | 100b (11) | 1 | 40 | 81 | 0 |
| HCT-116 | 88 | 86 | 54 | 85 | 75 | 22 | 49 | 96 | 19 | |
| HCT-15 | 76 | 78 | 30 | 75 | 68 | 8 | 41 | 70 | 23 | |
| HT-29 | 96 | 93 | 40 | 92 | 96 | 27 | 70 | 98 | 0 | |
| SW-620 | 66 | 81 | 60 | 73 | 82 | 6 | 64 | 92 | 1 | |
| KM12 | 79 | 72 | 48 | 72 | 70 | 8 | 60 | 84 | 4 | |
| CNS cancer | SF-295 | 86 | 72 | 26 | 59 | 76 | 1 | 27 | 91 | 13 |
| SF-539 | 100b (12) | 80 | 22 | 85 | 96 | 6 | 13 | 89 | 4 | |
| SNB-75 | 66 | 76 | 31 | 52 | 82 | 21 | 21 | 100b (10) | 14 | |
| U251 | 85 | 75 | 35 | 81 | 77 | 28 | 19 | 100b (13) | 14 | |
| Melanoma | LOX IMVI | 57 | 60 | 9 | 69 | 51 | 11 | 34 | 90 | 4 |
| M14 | 96 | 100b (16) | 45 | 79 | 89 | 0 | 50 | 84 | 6 | |
| MDA-MB-435 | 100b (57) | 99 | 88 | 97 | 100b (16) | 0 | 93 | 96 | 1 | |
| UACC-62 | 70 | 74 | 39 | 41 | 53 | 7 | 30 | 58 | 1 | |
| SK-MEL-2 | 42 | 79 | 55 | 72 | 84 | 19 | 22 | 85 | 3 | |
| SK-MEL-5 | 74 | 74 | 37 | 97 | 69 | 10 | 66 | 81 | 34 | |
| Ovarian cancer | OVCAR-3 | 100b (15) | 99 | 22 | 87 | 99 | 0 | 53 | 100b (13) | 2 |
| NCI/ADR-RES | 97 | 93 | 50 | 79 | 81 | 7 | 36 | 81 | 12 | |
| SK-OV-3 | 73 | 80 | 27 | 76 | 94 | 25 | 17 | 62 | 7 | |
| OVCAR-8 | 70 | 62 | 24 | 75 | 70 | 10 | 22 | 66 | 12 | |
| OVCAR-4 | 38 | 38 | 48 | 43 | 19 | 16 | 100b (18) | 0 | ||
| Renal cancer | A498 | 100b (5) | 100b (3) | 15 | 77 | 100b (2) | 7 | 26 | 84 | 18 |
| ACHN | 48 | 47 | 51 | 40 | 1 | 21 | 98 | 9 | ||
| RXF393 | 100b (4) | 64 | 25 | 66 | 86 | 21 | 12 | 71 | 10 | |
| TK-10 | 46 | 36 | 22 | 35 | 20 | 3 | 100b (9) | 15 | ||
| Breast cancer | MCF7 | 80 | 75 | 51 | 78 | 79 | 12 | 70 | 84 | 14 |
| MDA-MB-468 | 100b (12) | 73 | 22 | 63 | 70 | 8 | 1 | 57 | 5 | |
| Prostate cancer | PC-3 | 78 | 64 | 33 | 69 | 75 | 24 | 45 | 68 | 24 |
| DU-145 | 76 | 63 | 6 | 79 | 78 | 16 | 6 | 54 | 2 | |
The most active compounds are highlighted in bold.
Data obtained from NCI’s in vitro 60 cell one dose screening at 10−5M concentration.
Cytotoxic effect; lethality percent is represented in brackets.
Pyrrolo[2,1-b]pyridazines 8a, 8b, 8e, 8f, and pyrrolo[2,1-a]phthalazine 11b showed a very good growth inhibition effect on almost all 60 cell lines, the best results being registered on leukemia HL-60 (TB) cell, colon cancer COLO205 cell, melanoma MDA-MB-435 cell, ovarian cancer cell OVCAR-3, and renal cancer A498 cell. Compound 8a also showed a moderate cytotoxic effect, notably on melanoma MDA-MB-435 cells (57% cytotoxic). Mild to moderate cytotoxic effects were also observed for compounds 8b, 8e, 8f, and 11b against several cell types. Interestingly, the substitution of pyrrolo[2,1-b]pyridazine heterocycle at position 2 with a 4-substituted phenyl group resulted in the loss of the activity, compounds 8i–j, 8k, 8n, and 8q showing almost no inhibition effect against the 60 cell tested lines (data not shown). In contrast, 2-methylpyrrolo[2,1-b]pyridazines 8e–f showed similar activity to unsubstituted compounds 8a–b. Another interesting aspect is that substitution of the 3,4,5-trimethoxyphenyl ring of with a 3,5-dimethoxyphenyl one, did not diminish the inhibitory activity. In fact, pyrrolo[2,1-a]phthalazine compound 11b showed better growth inhibitory properties when compared with 11a. Substitution of the 3,4,5-trimethoxyphenyl ring with 3,4-dimethoxyphenyl or 4-bromophenyl also caused a reduction in biological activity, with the exception of compound 8d, which maintained moderate GI% values on most tested cell lines, although lower than 3,4,5-trimethoxyphenyl-substituted analogues 8a–b.
The most active compounds 8a, 8b, 8e, 8f, and 11b were selected for the second stage five dose-response studies42–44 selected results being presented in Table 2.
Table 2.
Results of the 5-dose in vitro human cancer cell growth inhibitiona for compounds 8a–b, e–f and 11b and compared with standard drug Doxorubicin.
| Cell type | Compound → |
8a |
8b |
8e |
8f |
11b |
Doxorubicinc |
|---|---|---|---|---|---|---|---|
| Cell line ↓ | GI50 (nM)b | ||||||
| Leukemia | CCRF-CEM | 261 | 2510 | 212 | 348 | n.d. | 79 |
| HL-60(TB) | 228 | 1380 | 160 | 248 | 820 | 126 | |
| K-562 | 90.6 | 538 | n.d. | n.d. | n.d. | 200 | |
| MOLT-4 | 443 | 2630 | 396 | 527 | n.d. | 32 | |
| RPMI-8226 | 246 | 1820 | n.d. | n.d. | n.d. | 79 | |
| SR | 46.1 | 573 | 48.1 | 75.9 | 442 | 25 | |
| Non-small Cell Lung cancer |
A549/ATCC | 487 | 11100 | 223 | 767 | n.d. | 63 |
| HOP-62 | 363 | 10200 | 398 | 691 | n.d. | 63 | |
| NCI-H460 | 312 | 2580 | 133 | 365 | 494 | 16 | |
| NCI-H522 | 343 | 346 | 171 | 303 | 236 | 32 | |
| Colon cancer | COLO205 | 193 | 797 | n.d. | n.d. | n.d. | 200 |
| HCT-116 | 276 | n.d. | 164 | 331 | 455 | 79 | |
| HCT-15 | 171 | 587 | 84.4 | 280 | 484 | 6310 | |
| HT-29 | 208 | 403 | 133 | 401 | 384 | 126 | |
| KM12 | 216 | n.d. | 57.7 | 254 | 351 | 251 | |
| SW-620 | 155 | 518 | 68.7 | 280 | 483 | 100 | |
| CNS cancer | SF-268 | 733 | 26500 | 550 | 676 | 1590 | 100 |
| SF-295 | 180 | 2100 | 65.7 | 311 | 483 | 100 | |
| SF-539 | 276 | 1850 | 130 | 349 | 1060 | 126 | |
| SNB-19 | 769 | 4260 | 420 | 752 | 1570 | 40 | |
| SNB-75 | 211 | 399 | n.d. | 384 | 471 | 63 | |
| U251 | 402 | 2000 | 331 | 549 | 730 | 40 | |
| Melanoma | MALME-3M | 247 | >100000 | n.d. | n.d. | 1070 | 126 |
| M14 | 176 | 394 | 136 | 251 | 485 | 159 | |
| MDA-MB-435 | 31.4 | 221 | 25.6 | 45.6 | 188 | 251 | |
| SK-MEL-2 | 385 | 738 | 495 | 494 | n.d. | 159 | |
| SK-MEL-5 | 269 | 508 | 58.5 | 276 | 623 | 79 | |
| UACC-62 | 176 | 523 | 61.5 | 477 | 692 | 159 | |
| Ovarian cancer | OVCAR-3 | 145 | 402 | 64.9 | 289 | 341 | 398 |
| NCI/ADR-RES | 200 | 463 | 123 | 308 | 476 | 7943 | |
| SK-OV-3 | 426 | 4900 | 546 | 878 | n.d. | 200 | |
| Renal cancer | 786-0 | 395 | 11400 | 335 | 523 | n.d. | 126 |
| A498 | 46.6 | n.d. | 76.2 | 388 | n.d. | 100 | |
| CAKI-1 | 301 | 2976 | n.d. | n.d. | n.d. | 1000 | |
| RXF 393 | 185 | 1640 | 116 | 239 | 1070 | 100 | |
| Prostate cancer | PC-3 | 166 | 7450 | 93.9 | 317 | 839 | 316 |
| DU-145 | 333 | 3960 | 391 | 906 | n.d. | 100 | |
| Breast cancer | MCF7 | 94.6 | 1310 | 48.1 | 313 | 410 | 40 |
| HS 578T | 236 | 1990 | 190 | 284 | 1840 | 316 | |
| BT-549 | 437 | 1220 | 878 | 1990 | 1180 | 251 | |
| T-47D | n.d.c | 17200 | >100000 | 501 | n.d. | 63 | |
| MDA-MB-468 | 281 | 1110 | 66.8 | 297 | 403 | 50 | |
The most active compounds are highlighted in bold.
Data obtained from NCI’s in vitro 60 cell 5-dose screening43–45.
GI50 – the molar concentration of tested compound causing 50% growth inhibition of tumor cells. Determined at five concentration levels (100, 10, 1.0, 0.1 and 0.01 μM).
GI50 data for Doxorubicin tested at a highest concentration of 100 μM were obtained from NCI database: https://dtp.cancer.gov/dtpstandard/dwindex/index.jsp.
n.d.: Not determined.
All five tested compounds confirmed the preliminary results by displaying good antiproliferative properties. The best candidate, 2-methyl-pyrrolo[2,1-b]pyridazine 8e exhibited GI50 values <100 nM in thirteen cell lines, notably on melanoma MDA-MB-435 cell (GI50 = 25.6 nM), leukemia SR cell (GI50 = 48.1 nM) and breast cancer MCF7 cell (GI50 = 48.1 nM). Compound 8e showed better GI50 values against melanoma MDA-MB-435, SK-MEL-5, and UACC-62 and colon cancer HCT-15, KM12 and SW-620 cell than Doxorubicin (NSC: 123127 code), the NCI standard drug for this type of analysis.
Interestingly, even if it exhibits an overall inhibitory activity lower than the 2-methyl substituted compound 8e, compound 8a shows an excellent inhibitory activity on melanoma MDA-MB-435 cell (GI50 = 31.4 nM), leukemia SR cell (GI50 = 46.1 nM) and renal cancer A498 (GI50 = 46.6 nM). Also, compound 8f displayed very good activity against melanoma MDA-MB-435 cell (GI50 = 45.6 nM).
Notably, compound 8e showed a very good cytostatic activity on melanoma MDA-MB-435 cell with a total growth inhibition level of effect (TGI) of 76.3 nM and leukemia HL-60(TB) (TGI = 588 nM), whereas compounds 8a and 8f showed the best cytostatic activity on melanoma MDA-MB-435 cell with a TGI of 117 nM and 420 nM, respectively. Significant cytotoxic activity was exhibited only by compound 8a on MDA-MB-435 melanoma cell with a lethal concentration (LC50) value of 438 nM.
Although pyrrolo[2,1-a]phthalazine 11b displayed the best mean growth inhibitory effect in preliminary single dose evaluation (Table 1), it did not exhibit GI50 values under the 100 nm threshold, as was the case for the more simple pyrrolo[1,2-b]pyridazines. Therefore, the introduction of a bulkier heterocycle, such as pyrrolo[2,1-a]phthalazine, in place of the 3′-hydroxy-4′-methoxyphenyl ring of phenstatin is less favourable in terms of antiproliferative activity than pyrrolo[1,2-b]pyridazine.
Molecular modelling
Because both computational and biological models of 3,4,5-trimethoxyphenyl-containing phenstatin analogues supported the hypothesis that the antiproliferative effects of these compounds are induced by inhibiting tubulin polymerisation5,7,33,47,48, docking experiments were performed on the colchicine binding site of the α,β-tubulin heterodimer (PDB:1SA0), in order to evaluate the shape and electrostatic complementarity between ligands and the α,β-tubulin heterodimer interface, which could account for the observed antiproliferative effects.
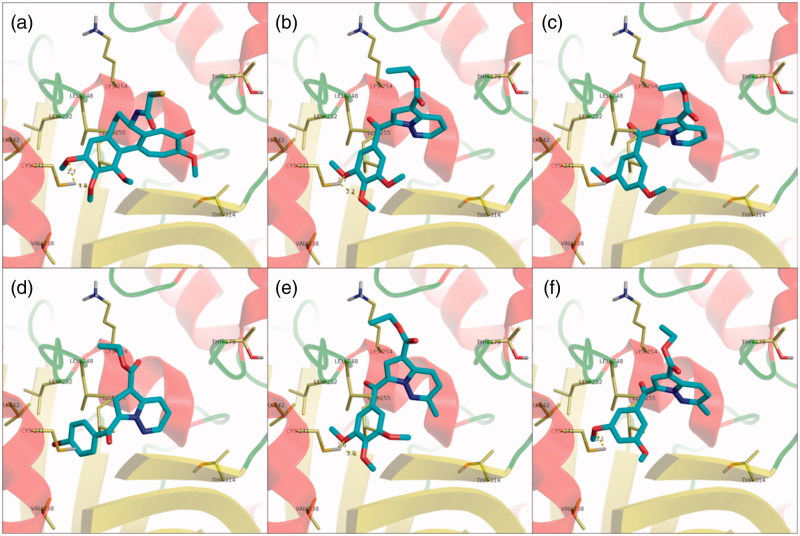
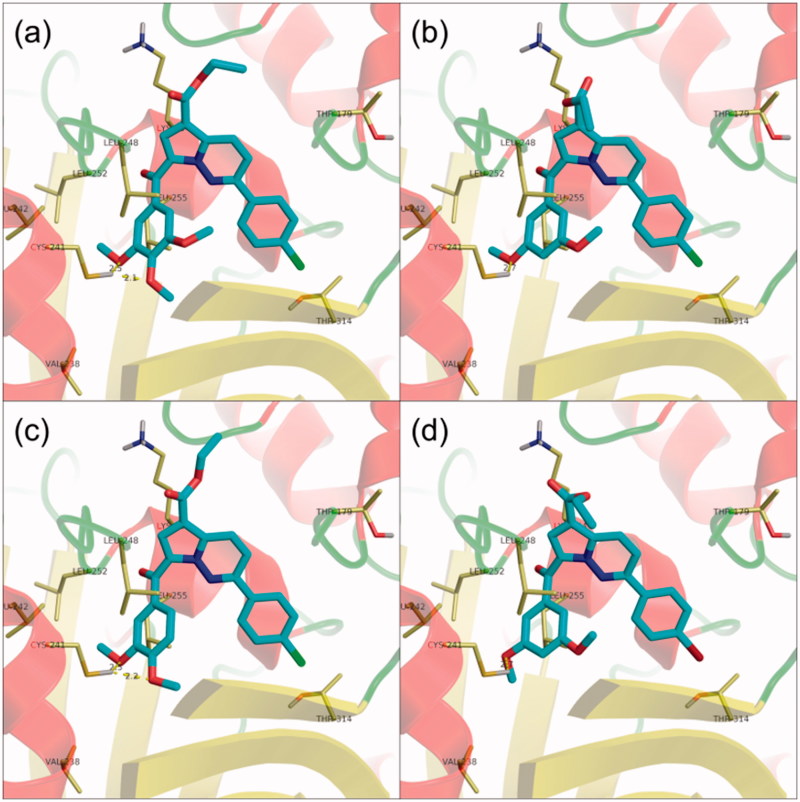
Compounds 8a and 8b displayed similar docking conformations grouped into two distinct clusters, both having the trimethoxyphenyl subunit overlapping with the one in the co-crystallised DAMA-colchicine ligand (Figure 2(a)), and interacting with the protein through hydrogen bonding with βCys241. The ligands are further stabilised in the binding pocket through hydrophobic interactions with βLeu242, βLeu248, βAla250, βLeu252, βLeu255, and βVal238. The diazine moiety either extended on top of the binding pocket, with the ester functional group orienting towards the dimer interface (Figure 2(b,c)), or was flipped at about 180°, to have the ester group roughly overlapping with the third colchicine ring in the crystal structure. Interestingly, the 4-bromo-substituted compound 8d, which displayed a less pronounced biological activity than 8a and 8b, adopted a conformation in which the p-bromo substituted phenyl was accommodated more deeply in the colchicine binding pocket, resulting in a shift in the position of the central heterocyclic moiety towards the center of the colchicine binding site, which led to the disruption of the hydrogen bond with βCys241 (Figure 2(d)).
Figure 2.
Structure and docking of diazines in the tubulin binding site: (a) DAMA-colchicine, (b) 8a, (c) 8b, (d) 8d, (e) 8e, (f) 8f; the α,β-tubulin heterodimer is represented as ribbons; amino acids in the binding site are represented as sticks.
Overall, the docking experiments suggest that the removal of the 4-methoxy group does not influence the accommodation of the ligand in the binding pocket, in agreement with the biological data, while the introduction of a bromine atom as substituent can induce a different binding conformation which leads to the disruption of the hydrogen bond between the ligand and βCys241, which could account for the reduced antiproliferative activity of compound 8d.
The 2-methyl-substituted analogues of 8a and 8b (8e and 8f) were accommodated in a similar fashion to that of parent compounds (Figure 3(e,f)), suggesting that the introduction of a methyl substituent does not influence the binding preferences of the compounds in the colchicine binding site, in agreement with the biological data in terms of antiproliferative activity. Compound 8h, which displayed a marked reduction in biological activity when compared to parent compound 8d, did not form the two expected well-defined clusters of conformations, but rather had a broad range of unrelated docking poses, the most energetically favourable being similar to the second cluster of compound 8d.
Figure 3.
Structure and docking of diazines in the tubulin binding site: (a) 8i, (b) 8j, (c) 8k, (d) 8n; the α,β-tubulin heterodimer is represented as ribbons; amino acids in the binding site are represented as sticks.
Interestingly, 2-(p-halogeno-phenyl)-substituted compounds 8i, 8j, 8k, and 8n, which showed a marked decrease in growth inhibition activity when compared to unsubstituted analogues, were compatible with the colchicine binding site, and were accommodated in a similar fashion to their unsubstituted or 2-methyl-substituted analogues with biological activity to the α,β-tubulin heterodimer (Figures 2 and 3). A closer inspection of the basic physicochemical properties of these four compounds reveals, however, a violation of Lipinski’s rule of five in terms of logp values49 (Table 3), which could account for the loss in antiproliferative efficacy in spite of apparent activity at the colchicine binding site50.
Table 3.
Theoretical logp values of biologically tested compounds.
| Compound | logp | Compound | logp | Compound | logp |
|---|---|---|---|---|---|
| 8a | 2.77 | 8h | 4.14 | 8q | 5.32 |
| 8b | 2.93 | 8i | 5.41 | 11a | 3.76 |
| 8d | 4.01 | 8j | 5.56 | 11b | 3.92 |
| 8e | 2.90 | 8k | 5.56 | 11c | 3.92 |
| 8f | 3.06 | 8n | 5.73 |
Values were calculated using the ChemAxon/Chemicalize server (www.chemicalize.com).
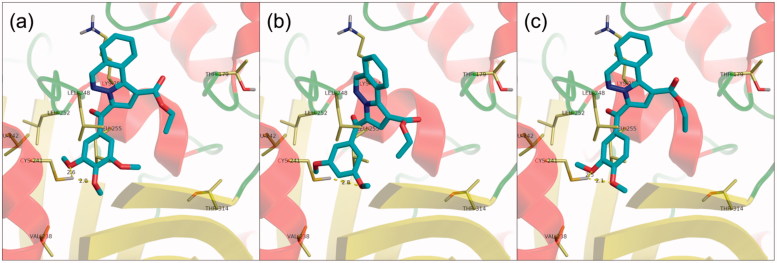
Docking of pyrrolo[2,1-a]phthalazines 11a–c revealed a single cluster of conformations for each compound, similar to the second cluster obtained for pyrrolo[1,2-b]pyridazines 8a, 8b, 8e, and 8f, in which the heterocyclic subunit is oriented as to have the ester group roughly overlapping with the third colchicine ring in the crystal structure (Figure 4). The methoxyphenyl subunit is stabilised by a hydrogen bond interaction with βCys241, similar to the case of pyrrolo[1,2-b]pyridazine analogues. Notably, compound 11b adopts a conformation slightly deeper in the hydrophobic pocket, which induces a rotation of the heterocyclic core and facilitates a hydrophobic interaction with βLeu248, which is unique among the three docked pyrrolo[2,1-a]phthalazines. A tighter hydrophobic interaction between 11b and the protein could account for the pronounced antiproliferative activity exerted by 11b among the three tested pyrrolo[2,1-a]phthalazines.
Figure 4.
Structure and docking of diazines in the tubulin binding site: (a) 11a, (b) 11b, (c) 11c; the α,β-tubulin heterodimer is represented as ribbons; amino acids in the binding site are represented as sticks.
However, for all compounds, complementary tubulin polymerisation assays are needed in order to confirm the proposed molecular mechanism.
Conclusion
In summary, five of the newly synthesised pyrrolo[1,2-b]pyridazine and pyrrolo[2,1-a]phthalazine phenstatin analogues showed in vitro antiproliferative activity, the most potent being compounds 8f with GI50 values <100 nM on thirteen cell lines including colon, ovarian, renal, prostate, brain and breast cancer, melanoma and leukemia. Notably, compound 8a showed a very good antiproliferative effect on melanoma MDA-MB-435 cell, renal cancer A498, and leukemia SR cell. The substitution of position 2 of pyrrolo[1,2-b]pyridazine with a methyl group generally appears to increase the antiproliferative potency of the compounds, while the introduction of a more bulkier substituent is completely detrimental for the growth inhibitory properties, despite the fact that docking studies showed a good compatibility with the colchicine binding site of tubulin. The lack of proliferative activity in the case of the bulkier 2–(4-X-phenyl)-pyrrolo[1,2-b]pyridazines could be explained by the suboptimal lipophilicity and solubility of these compounds. However, further assaying in terms of tubulin polymerisation is needed in order to confirm the proposed antiproliferative mechanism of action of the newly synthetised compounds. Compound 8f could serve as a useful lead compound for further structural optimisation in the development of new anticancer agents.
Funding Statement
The authors are thankful for financial support to Romanian Ministry of Research and Innovation, Program 1- Development of the national R & D system, Subprogram 1.2 - Institutional performance - RDI excellence financing projects, Grant no. 34PFE
Acknowledgements
The authors acknowledge to National Cancer Institute (NCI) for biological evaluation of compounds on their 60- cell panel: the testing was performed by the Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis (the URL to the Program's website: http://dtp.cancer.gov/). We also thank CERNESIM Research Centre from Alexandru Ioan Cuza University of Iasi, for the NMR experiments.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013;13:717–26. [DOI] [PubMed] [Google Scholar]
- 2.Hurwitz M. Chemotherapy in prostate cancer. Curr Oncol Rep 2015;17:44. [DOI] [PubMed] [Google Scholar]
- 3.Prasad V, De Jesús K, Mailankody S. The high price of anticancer drugs: origins, implications, barriers, solutions. Nat Rev Clin Oncol 2017;14:381–90. [DOI] [PubMed] [Google Scholar]
- 4.Li WL, Sun HH, Xu ST, et al. Tubulin inhibitors targeting the colchicine binding site: a perspectiveof privileged structures. Future Med Chem 2017;9:1765–94. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Chen J, Xiao M, et al. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm Res 2012;29:2943–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez EA. Microtubule inhibitors: differentiating tubulin-inhibiting agents based on mechanisms of action, clinical activity, and resistance. Mol Cancer Therap 2009;8:2086–95. [DOI] [PubMed] [Google Scholar]
- 7.Kaur R, Kaur G, Gill RK, et al. Recent developments in tubulin polymerization inhibitors: an overview. Eur J Med Chem 2014;87:89–124. [DOI] [PubMed] [Google Scholar]
- 8.Bhattacharyya B, Panda D, Gupta S, Banerjee M. Anti-mitotic activity of colchicine and the structural basis for its interaction with tubulin. Med Res Rev 2008;28:155–83. [DOI] [PubMed] [Google Scholar]
- 9.Lin CM, Ho HH, Pettit GR, Hamel E. Antimitotic natural products combretastatin A-4 and combretastatin A-2: studies on the mechanism of their inhibition of the binding of colchicine to tubulin. Biochemistry 1989;28:6984–91. [DOI] [PubMed] [Google Scholar]
- 10.Gigant B, Wang C, Ravelli RBG, et al. Structural basis for the regulation of tubulin by vinblastine. Nature 2005;435:519–22. [DOI] [PubMed] [Google Scholar]
- 11.Sherbet GV. Suppression of angiogenesis and tumour progression by combretastatin and derivatives. Cancer Lett 2017;403:289–95. [DOI] [PubMed] [Google Scholar]
- 12.Pettit GR, Toki B, Herald DL, et al. Antineoplastic agents. 379. Synthesis of phenstatin phosphate. J Med Chem 1998;41:1688–95. [DOI] [PubMed] [Google Scholar]
- 13.Pettit GR, Grealish MP, Herald DL, et al. Antineoplastic agents. 443. Synthesis of the cancer cell growth inhibitor hydroxyphenstatin and its sodium diphosphate prodrug. J Med Chem 2000;43:2731–7. [DOI] [PubMed] [Google Scholar]
- 14.Dumontet C, Jordan MA. Microtubule-binding agents: a dynamic field of cancer therapeutics. Nat Rev Drug Disc 2010;9:790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer 2004;4:253–65. [DOI] [PubMed] [Google Scholar]
- 16.Nepali K, Ojha R, Sharma S, et al. Tubulin inhibitors: a patent survey. Recent Pat Anticancer Drug Discov 2014;9:176–220. [DOI] [PubMed] [Google Scholar]
- 17.Marx MA. Small-molecule, tubulin-binding compounds as vascular targeting agents. Expert Opin Ther Pat 2002;12:769–76. [Google Scholar]
- 18.Ojha R, Sharma S, Nepali K. Anticancer agents targeting tubulin In: Zaman K Atta-Ur-Rahman, eds. Topics in anticancer research. Oak Park, Illinois: Bentham Science; 2015:156–270. [Google Scholar]
- 19.Gholap SS. Pyrrole: an emerging scaffold for construction of valuable therapeutic agents. Eur J Med Chem 2016;110:13–31. [DOI] [PubMed] [Google Scholar]
- 20.Zbancioc GN, Mangalagiu II. Pyrrolopyridazine derivatives as blue organic luminophores: synthesis and properties. Part 2. Tetrahedron 2010;66:278–82. [Google Scholar]
- 21.Mitsumori T, Bendikov M, Sedo J, Wudl F. Synthesis and properties of novel highly fluorescent pyrrolopyridazine derivatives. Chem Mater 2003;15:3759–68. [Google Scholar]
- 22.El Guesmi N, Ahmed SA, Althagafi II, Khairou KS. Photochromism of dihydroindolizines. Part XXI: multiaddressable photochromic performances based on pyrrolo[1,2-b]pyridazine photochromes: kinetics, substituent effect and solvatochromism. J Photochem Photobiol A 2017;346:287–95. [Google Scholar]
- 23.Mangalagiu II. Recent achievements in the chemistry of 1,2-diazine. Curr Org Chem 2011;15:730–52. [Google Scholar]
- 24.Wermuth CG. Are pyridazines privileged structures?. Med Chem Comm 2011;2:935–41. [Google Scholar]
- 25.Butnariu RM, Mangalagiu II. New pyridazine derivatives: synthesis, chemistry and biological activity. Bioorg Med Chem 2009;17:2823–9. [DOI] [PubMed] [Google Scholar]
- 26.Caprosu MD, Butnariu RM, Mangalagiu II. Synthesis and antimicrobial activity of some new pyridazine derivatives. Heterocycles 2005;65:1971–879. [Google Scholar]
- 27.Antoci V, Mantu D, Cozma DG, et al. Hybrid anticancer 1,2-diazine derivatives with multiple mechanism of action. Part 3. Med Hypotheses 2014;82:11–5. [DOI] [PubMed] [Google Scholar]
- 28.Mantu D, Maftei D, Iurea D, et al. Synthesis, structure and in vitro anticancer activity of new polycyclic 1,2-diazine. Med Chem Res 2014;3:2909–15. [Google Scholar]
- 29.Duan JJ, Lu Z, Jiang B, et al. Discovery of pyrrolo[1,2-b]pyridazine-3-carboxamides as Janus kinase (JAK) inhibitors. Bioorg Med Chem Lett 2014;24:5721–6. [DOI] [PubMed] [Google Scholar]
- 30.Tang PC, Feng J, Huang J, et al. Discovery of pyrrolopyridazines as novel DGAT1 inhibitors. Bioorg Med Chem Lett 2009;20:6030–3. [DOI] [PubMed] [Google Scholar]
- 31.Lim J, Altman MD, Baker J, et al. Identification of N-(1H-pyrazol-4-yl)carboxamide inhibitors of interleukin-1 receptor associated kinase 4: bicyclic core modifications. Bioorg Med Chem Lett 2015;25:5384–8. [DOI] [PubMed] [Google Scholar]
- 32.Chen Z, Kim SH, Barbosa SA, et al. Pyrrolopyridazine MEK inhibitors. Bioorg Med Chem Lett 2006;16:628–32. [DOI] [PubMed] [Google Scholar]
- 33.Ghinet A, Abuhaie CM, Gautret P, et al. Studies on indolizines. Evaluation of their biological properties as microtubule-interacting agents and as melanoma targeting compounds. Eur J Med Chem 2015;89:115–27. [DOI] [PubMed] [Google Scholar]
- 34.Frolova LV, Magedov IV, Romero AE, et al. Exploring natural product chemistry and biology with multicomponent reactions. Discovery of a novel tubulin-targeting scaffold derived from the rigidin family of marine alkaloids. J Med Chem 2013;56:6886–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zbancioc GN, Caprosu MC, Moldoveanu CC, et al. Microwave assisted 1,3-dipolar cycloaddition reactions of 2-(4-halobenzoyl) phthalazinium methylides. Rev Roum Chim 2005;50:353–8. [Google Scholar]
- 36.Zbancioc G, Mangalagiu II. Microwave-assisted synthesis of highly fluorescent pyrrolopyridazine derivatives. Synlett 2006;5:804–6. [Google Scholar]
- 37.Caprosu MC, Zbancioc GN, Moldoveanu CC, Mangalagiu II. 3-dipolar cycloaddition reactions of p-halogenophenyl-phthalazinium ylides to activated alkenes and alkynes. Collect Czech Chem C 2004;69:426–34. [Google Scholar]
- 38.Roman M, Mangalagiu II, Caprosu M, Petrovanu M. Studies on 3-methylpyridazinium ylides. Analele Stiintifice Ale Universitatii “Al. I. Cuza” Din Iasi, Chimie 1999;7:117–22. [Chem. Abstr. 132 (2000) 207810]. [Google Scholar]
- 39.Mangalagiu II, Druta I, Constantinescu M, et al. Pyridazinium ylides. Regiochem Tetrahedron 1996;52:8853–62. [Google Scholar]
- 40.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravelli RBG, Gigant B, Curmi PA, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 2004;428:198–202. [DOI] [PubMed] [Google Scholar]
- 42.Hanwell MD, Curtis DE, Lonie DC, et al. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J Cheminform 2012;4:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6:813–23. [DOI] [PubMed] [Google Scholar]
- 44.Skehan P, Storeng R, Scudiero D, et al. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst 1990;82:1107–12. [DOI] [PubMed] [Google Scholar]
- 45.Boyd RB. The NCI in vitro anticancer drug discovery screen: concept, implementation, and operation In: Teicher B, ed. Anticancer drug development guide: preclinical screening, clinical trials, and approval. Totowa, NJ: Humana Press Inc; 1997:23–42. [Google Scholar]
- 46.Zbancioc G, Zbancioc AM, Mangalagiu II. Ultrasound and microwave assisted synthesis of dihydroxyacetophenone derivatives with or without 1,2-diazine skeleton. Ultrason Sonochem 2014;21:802–11. [DOI] [PubMed] [Google Scholar]
- 47.Kamal A, Kumar GB, Vishnuvardhan MV, et al. Synthesis of phenstatin/isocombretastatin–chalcone conjugates as potent tubulin polymerization inhibitors and mitochondrial apoptotic inducers. Org Biomol Chem 2015;13:3963–81. [DOI] [PubMed] [Google Scholar]
- 48.Badhani B, Kakkar R. In silico studies on potential MCF-7 inhibitors: a combination of pharmacophore and 3D-QSAR modeling, virtual screening, molecular docking, and pharmacokinetic analysis. J Biomol Struct Dyn 2017;35:1950–67. [DOI] [PubMed] [Google Scholar]
- 49.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3–26. [DOI] [PubMed] [Google Scholar]
- 50.Bennion BJ, Be NA, McNerney MW, et al. Predicting a drug’s membrane permeability: a computational model validated with in vitro permeability assay data. J Phys Chem B 2017;121:5228–37. [DOI] [PubMed] [Google Scholar]