Abstract
In this study, the circulating miRNome from diagnostic neuroblastoma serum was assessed for identification of noninvasive biomarkers with potential in monitoring metastatic disease. After determining the circulating neuroblastoma miRNome, 743 miRNAs were screened in 2 independent cohorts of 131 and 54 patients. Evaluation of serum miRNA variance in a model testing for tumor stage, MYCN status, age at diagnosis, and overall survival revealed tumor stage as the most significant factor impacting miRNA abundance in neuroblastoma serum. Differential abundance analysis between patients with metastatic and localized disease revealed 9 miRNAs strongly associated with metastatic stage 4 disease in both patient cohorts. Increasing levels of these miRNAs were also observed in serum from xenografted mice bearing human neuroblastoma tumors. Moreover, murine serum miRNA levels were strongly associated with tumor volume. These findings were validated in longitudinal serum samples from metastatic neuroblastoma patients, where the 9 miRNAs were associated with disease burden and treatment response.
Keywords: Genetics, Oncology
Keywords: Noncoding RNAs
Nine miRNAs, identified in serum from neuroblastoma patients and strongly associated with metastatic disease burden, have application potential in monitoring response to treatment.
Introduction
Neuroblastoma (NB) is a childhood cancer arising from the developing sympathetic nervous system (1). The disease is responsible for 11% of pediatric cancer deaths and is ranked third among cancers with the highest mortality rate (2). Approximately 60% of the NB patients are diagnosed with metastatic disease, mostly represented in the high-risk patient subgroup. High-risk patients have low survival rates of about 50%, and 2 thirds of these children will ultimately relapse, eventually entering a second line of intensive treatment, enrolling in an early-stage clinical trial or receiving palliative therapy (1). Monitoring of treatment response is essential for assessment of its efficacy and for guiding the procedure of follow-up treatment after refractory or relapsed NB.
Current methods for monitoring disease and evaluation of treatment response are mostly based on detection of minimal residual disease (MRD), which is determined by estimating the number of residual tumor cells in peripheral blood or BM (3). While several cellular MRD markers have been proposed, biomarkers for noninvasive cell-free determination of disease burden are currently lacking.
MicroRNAs — a class of small noncoding RNAs involved in various aspects of health and disease, including NB pathophysiology — seem to be particularly stable and abundant in serum and plasma (4, 5). Several studies have reported a relationship between serum miRNAs and disease characteristics such as MYCN amplification or increased risk for adverse outcome (6, 7). The interest in pursuing the investigation of the miRNome from liquid biopsy samples is also fortified by the fact that collection, processing, and storage of these samples is relatively straightforward and less invasive. For these reasons, the circulating miRNome may be a valuable source for development of more advanced or refined tests for longitudinal treatment follow-up of NB patients.
In the present study, we quantified the circulating miRNome from 185 diagnostic serum samples and assessed the association of 4 disease characteristics with serum miRNA abundance: the international NB staging system (INSS), gene copy number status of the MYCN oncogene, age at diagnosis, and overall survival. We found a strong positive correlation between serum miRNA abundance and tumor stage and selected 9 miRNAs with a proportional increase from stage 1 to stage 4 NB patients. As tumor stage reflects cancer spread, we propose this set of miRNAs as serum markers for assessment of disease burden in human NB.
Results
Defining the human NB circulating miRNome.
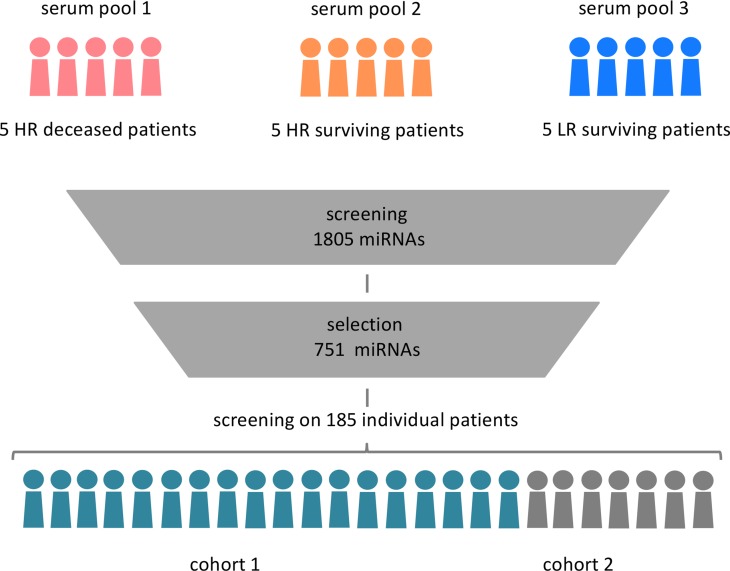
As only a subset of miRNAs end up in circulation, we decided to first define the circulating miRNome (i.e., the ensemble of miRNAs that are detectable in serum from NB patients). To this end, we created 3 serum pools, each composed of 5 patient samples (Figure 1). Pooled samples were either high-risk patients who died of the disease (pool 1), high-risk patients who survived (pool 2), or low-risk patients who survived the disease (pool 3). Since the fraction of low-risk NB patients who die of disease is very low (approximately 2%), no such pool was included in the study (1).
Figure 1. Defining the human neuroblastoma circulating miRNome.
Three serum pools were prepared, each containing 5 serum samples from 3 neuroblastoma subgroups: 5 high-risk (HR) deceased patients, 5 high-risk surviving patients, and 5 low-risk (LR) surviving patients for serum pool 1, 2, and 3, respectively. Expression of 1,805 miRNAs was measured by qPCR; 751 well-expressed miRNAs were selected and profiled on 2 independently collected and processed patient cohorts of 131 patients and 54 patients, respectively.
The pooled samples were screened for 1,805 miRNAs in order to select detectable miRNAs in at least 1 of the pools. A set of 751 miRNAs was ultimately selected, hereby defining the circulating NB miRNome. We argue that these serum pools are representative for the majority of the NB patient population, resulting in a comprehensive selection of circulating NB miRNAs. Expression of the NB miRNome was subsequently analyzed by quantitative PCR (RT-qPCR) in 2 NB patient cohorts of 131 and 54 serum samples, respectively.
Before normalization, 8 miRNAs with missing values in more than 75% of the samples were excluded from the dataset. All analyses were performed on the first cohort, initially. The second cohort was used to validate the findings from the first cohort.
Metastatic disease status has the largest impact on circulating miRNA levels in serum.
Generalized additive modeling (GAM) was used on normalized miRNA expression values in order to quantify the association with tumor stage, MYCN status, age at diagnosis, and overall survival. A straightforward model was chosen, whereby interaction terms were excluded, since our aim was to determine which of these features has the largest impact on serum miRNA expression and, hence, is responsible for the largest variance in the data set.
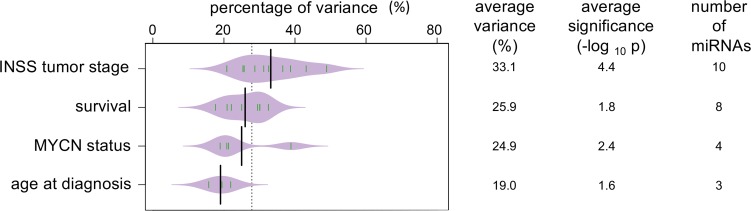
The analysis returned the level of significance (P value) and the percentage of variance in the dataset explained by each of the tested features. The level of significance was used to measure the prediction ability of disease features for the 743 tested miRNAs. The percentage of variance was used to define the importance of each feature in influencing miRNA expression in serum. The average statistical significance, the number of significant miRNAs, and the average variance explained by each disease feature is provided in Figure 2.
Figure 2. Tumor stage has the largest impact on circulating miRNA levels in serum.
Bean plot of the average variance per disease feature (%), average significance (–log10 of the P value), and number of miRNAs significantly influenced by disease features. Green lines in the violin plot represent percentage of variance for individual miRNAs. The evaluated disease features were tumor stage (1,2, 3, 4, and 4S), overall survival (survival), MYCN status, and age at diagnosis (18 moths cut-off). P values were corrected for multiple-testing error by the Benjamini-Hochberg method prior to averaging.
INSS tumor stage explains, on average, 33.1% of the variance in this serum miRNA dataset. Ten miRNAs were identified as being strongly significantly affected by tumor stage. The average level of significance was P < 0.0001 (range: 3.65 × 10–2 to 3.19 × 10–11). Overall survival status (alive or dead of disease) explained an average 25.9% of the variance. Eight miRNAs were identified with an average P < 0.05 (range: 3.58 × 10–2 to 5.88 × 10–3). MYCN copy number status and age at diagnosis were responsible for 24.9% and 19.0% of the expression variance, respectively, with an average P < 0.01 and < 0.05, respectively. The number of miRNAs to be significantly influenced by MYCN status or age at diagnosis were 4 and 3, respectively.
None of the identified miRNAs were significantly influenced by 2 or more disease characteristics. Based on the overall percentage of variance explained and the average level of significance for the identified sets of miRNAs for each disease feature, we concluded that INSS tumor stage had the largest impact on circulating serum miRNA levels.
Identification of serum miRNAs as potential markers for disease burden.
Mann-Whitney U test was used to select differentially expressed miRNAs between serum from patients with metastatic disease (stage 4 tumors) and serum from patients with localized disease (stage 1 and stage 2 tumors).
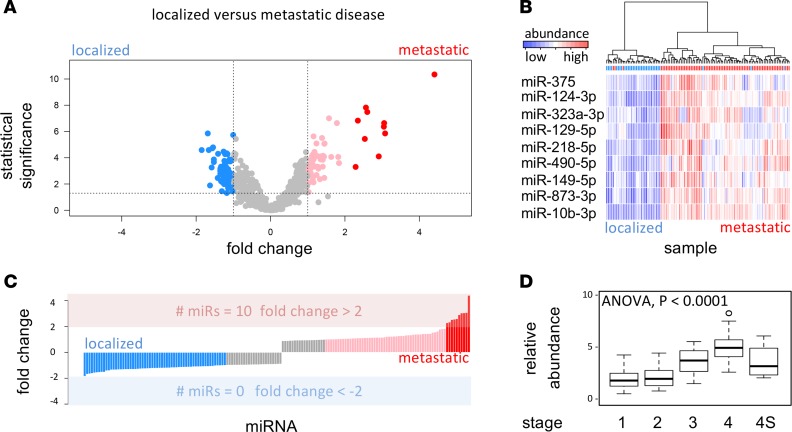
The analysis revealed 327 differentially expressed miRNAs (after correction for multiple-testing). One hundred eighty-four and 143 miRNAs were more abundant in serum from metastatic and localized disease, respectively (Figure 3A). The magnitude of the differences was much larger for miRNAs circulating in metastatic serum samples (Figure 3C). In fact, metastatic serum revealed 10 miRNAs with minimal fold-changes higher than 2 ΔCq units (~4-fold difference), while the maximum fold-change in serum from localized disease was lower than 2 ΔCq units.
Figure 3. Identification of serum miRNAs as potential markers for disease burden in patient cohort 1.
(A) Fold change (log2 scale) of miRNA abundance in serum (x axis) against statistical significance (y axis, –log10 of the P value) between patients with metastatic (stage 4) neuroblastoma (n = 76) and patients with localized (stage 1 and stage 2) neuroblastoma (n = 33). Higher-fold change denotes higher abundance in metastatic serum. Pink dots represent miRNAs with at least 2-fold (1 log2 unit) higher abundance, and red dots represent miRNAs with at least a 4-fold higher abundance in serum from metastatic patients; blue dots represent miRNAs with at least 2-fold higher abundance in serum from patients with localized disease. For the selection of differentially abundant miRNAs, a Mann-Whitney U test was used at 0.05 significance level (corrected for multiple-testing by the Benjamini-Hochberg method). (B) Heatmap depicting hierarchical clustering of serum samples (cohort 1) based on 9 miRNAs with at least 4-fold higher abundance in serum from metastatic patients. (C) Fold change distribution of miRNA abundance in localized versus metastatic serum. The bar colors correspond to dot colors in plot A. (D) Boxplot of average miRNA expression for the selected set of 9 miRNAs in serum from stage 1, 2, 3, 4, and 4S patients.
Interestingly, 9 of the top 10 differential miRNAs were also put forward by the GAM analysis. These overlapping miRNAs were miR–873-3p, miR–149-5p, miR–124-3p, miR–218-5p, miR–490-5p, miR–323a-3p, miR–10b-3p, miR-375, and miR–129-5p (Figure 3B).
We further evaluated the serum levels of the 9 overlapping miRNAs in patients stratified according to INSS tumor stage. Average miRNA abundance was calculated per sample; samples were grouped per stage and depicted by a boxplot in Figure 3D.
A proportional increase of miRNA levels in serum from stage 1 to stage 4 patients was observed. One-way ANOVA was used to quantify the significance of the difference between stage groups. This analysis revealed a significant P value of 2.0 × 10–16. All differences were significant, except for the difference between stage 1 and stage 2 tumors and the difference between stage 4S and all other tumor stages.
Even though we would label the selected miRNAs as being indicators of metastatic tumor load (stage 4 disease), there is a discrepancy with respect to how the miRNAs behave in serum from stage 4S patients. Stage 4S patients have a particular pattern of metastatic disease with good outcome due to spontaneous regression; however, the expression of the selected set of 9 miRNAs is not significantly different from the localized tumor stages. This may be due to the fact that this cohort only contained a limited number of samples. Alternatively, it may reflect the difference in biology between stage 4 and stage 4S tumors, which may result in a different miRNA signature in patient serum samples. Based on these results, we assume that the identified set of miRNAs qualify as candidate markers of metastatic, nonstage 4S disease (hereafter referred to as markers for metastatic disease).
Validation of serum miRNA markers for metastatic disease in independent patient cohort.
In order to evaluate the robustness of the identified markers for metastatic disease, we measured all 743 microRNAs in an independent cohort consisting of sera from 54 NB patients.
There is a significantly positive correlation of localized versus metastatic (stage 4) fold change abundance between samples from the first and second cohort (r = 0.7, P < 0.001). If only differential miRNAs between localized and metastatic disease are taken into account, the correlation between the 2 independent sample cohorts is even higher (r = 0.91, P < 0.001).
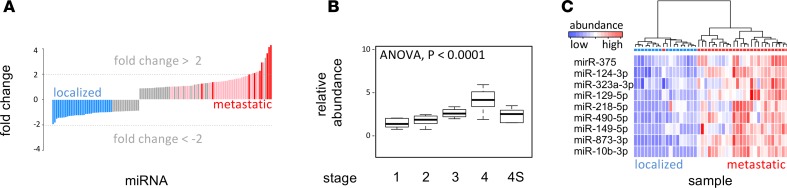
In Figure 4A, a distribution of abundance fold changes is shown for localized versus metastatic disease. Seven of the 9 miRNAs identified on the first cohort have a 4-fold abundance difference (2 log2 units) in the second sample cohort, as well. The remaining 2 miRNAs show fold differences larger than 2.5 (1.4 log2 units).
Figure 4. Validation of serum miRNA markers for metastatic disease in patient cohort 2.
(A) Fold change distribution of miRNA abundance between sera from metastatic (stage 4) neuroblastoma (n = 27) and localized (stage 1 and 2) neuroblastoma (n = 17) patients (cohort 2, n = 54). Red bars indicate the position of the 9 miRNAs identified in the first cohort. (B) Boxplot of miRNA abundance for the selected 9 miRNAs in sera from stage 1, 2, 3, 4, and 4S disease, in the cohort 2. (C) Heatmap depicting hierarchical clustering of serum samples based on the selected set of 9 miRNAs.
A proportional increase in the average expression from the 9 miRNAs from stage 1 to stage 4 was confirmed in the second cohort (Figure 4B). An ANOVA test revealed a significant P value of 5.8 × 10–14. All differences between stage 4 samples and samples from other tumor stages were significant (P < 0.01). In Figure 4C, the ability of the 9-miRNA set to classify the independent cohort into localized and metastatic disease is shown.
Serum markers for metastatic disease are high but nondifferential in primary tumors.
Two public datasets of miRNA expression in fresh-frozen diagnostic NB tumors were used to evaluate expression of the serum markers for metastatic disease in the tumor. The tumor expression datasets were generated using different miRNA expression profiling technologies than used in the present study: (a) stem-loop RT-qPCR miRNA expression method (Invitrogen), as described in ref. 8, and (b) small RNA sequencing, as described in ref. 9.
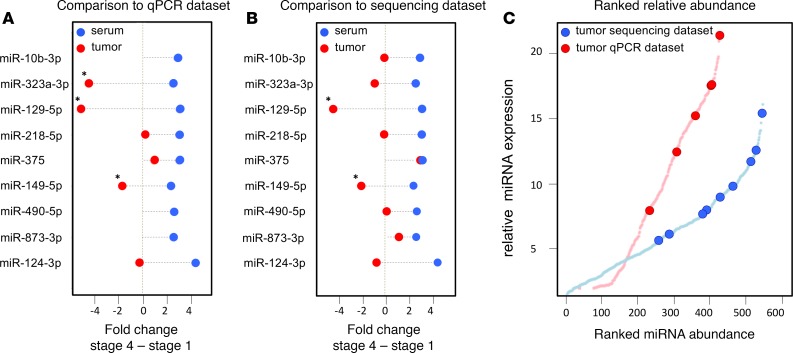
There were 201 microRNAs measured in common between our serum dataset and the stem-loop RT-qPCR tumor dataset, including 6 of the 9 serum markers for metastatic disease. Fold changes were determined between 25 stage 4 tumors from patients with fatal outcome and 25 stage 1 tumors from patients with favorable outcome (Figure 5A). No significant correlation was found between serum and tumor microRNA fold changes (r = –0.08, P = 0.22). Only 3 of the 6 miRNAs (miR–129-5p, miR–149-5p, and miR–323a-3p) were significantly differentially expressed between metastatic and localized disease in the tumor dataset. However, the fold changes are in opposite direction as to what is observed in serum (i.e., while these miRNAs were more abundant in serum from metastatic disease, they were found to be higher expressed in localized disease in tumor). The 6 miRNAs were among the top 55% miRNAs with highest expression in the tumor RT-qPCR data (Figure 5C).
Figure 5. Serum markers for metastatic disease are highly but not differentially expressed in primary tumors.
(A) miRNA abundance differences in serum and in tumor tissue measured by stem-loop qPCR. (B) miRNA abundance differences in serum and in tumor tissue measured by small RNA sequencing. The serum markers for metastatic disease that are differentially expressed between stage 4 and stage 1 tumor are marked by asterisk (Mann–Whitney U test P < 0.05, Benjamini-Hochberg correction for multiple-testing); red dots represent fold changes in tumor tissue, and blue dots represent fold changes in serum. (C) Ranked miRNA abundance in the qPCR data (red) and in the small RNA sequencing data (blue).
In the small RNA sequencing dataset, there were 5 patients with metastatic (stage 4) disease and fatal outcome and 5 patients with localized disease (stage 1) and favorable outcome. All 9 serum markers for metastatic disease were present in the tumor dataset. No significant correlation was found between serum and tumor microRNA fold changes (r = –0.13, P = 0.01). Only 2 of the 9 miRNAs (miR–129-5p and miR–149-5p) showed a significantly differential expression between metastatic and localized tumors (Figure 5B). Again, while these miRNAs were more abundant in metastatic disease in serum, they were higher expressed in localized disease in tumor. The 9 miRNAs were among the top 54% miRNAs with highest expression in the tumor sequencing data (Figure 5C).
Serum microRNA markers for metastatic disease are specific for NB.
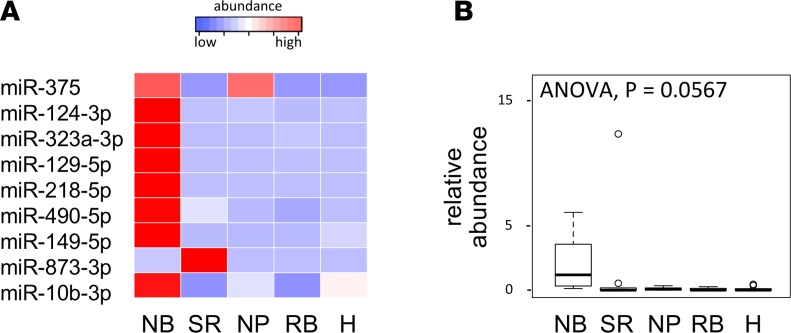
We further evaluated the specificity of the selected set of 9 miRNAs in serum pools from healthy children and other pediatric cancer entities (sarcoma, nephroblastoma, and rhabdomyosarcoma), as depicted in Figure 6. Low to absent levels were observed in healthy serum. When compared with serum from other pediatric cancer entities, expression levels for 8 of 9 miRNAs were higher in NB serum than in the pools from other pediatric cancer entities.
Figure 6. Serum microRNA markers for metastatic disease are specific for neuroblastoma.
(A) Standardized relative abundance for individual circulating miRNAs in serum pools from neuroblastoma (NB), sarcoma (SR), nephroblastoma (NP), and rhabdomyosarcoma (RB) patients and healthy children (H). (B) Relative abundance of circulating miRNAs in serum pools from NB, SR, NP, and RB patients and H.
These results suggest that the increased levels of the identified set of miRNAs in serum from children with NB is likely caused by the presence of NB tumor cells in the patient’s body.
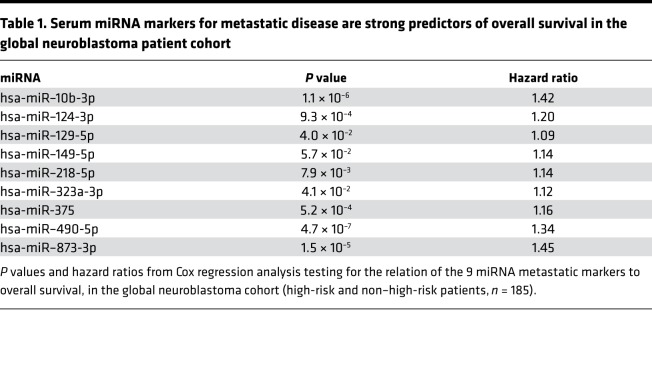
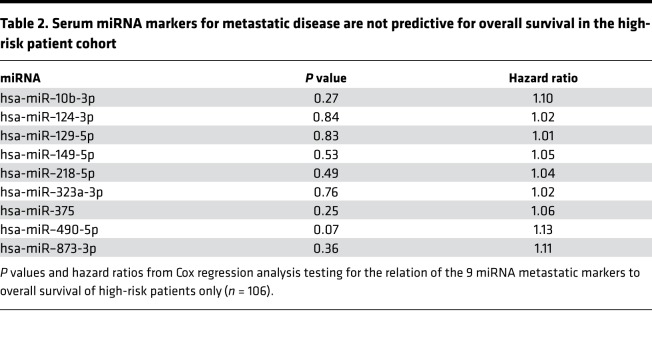
Serum miRNA markers for metastatic disease are strong predictors of overall survival in a global NB patient cohort but not in the high-risk subgroup.
Given the importance of biomarkers for more accurate outcome prediction, the abundance of the 9 miRNAs in serum was assessed for potential association with overall survival. To this purpose, Kaplan-Meier and Cox regression analyses were performed. The results showed a significant prediction of overall survival in a patient cohort including NB patients from the low-, intermediate-, and high-risk group (Table 1 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.97021DS1). However, when tested in the high-risk patient cohort only, none of the miRNAs was significantly associated with overall survival (Table 2 and Supplemental Figure 2).
Table 1. Serum miRNA markers for metastatic disease are strong predictors of overall survival in the global neuroblastoma patient cohort.
Table 2. Serum miRNA markers for metastatic disease are not predictive for overall survival in the high-risk patient cohort.
Proportional increase of metastatic disease miRNA markers in serum from mice engrafted with human NB cells.
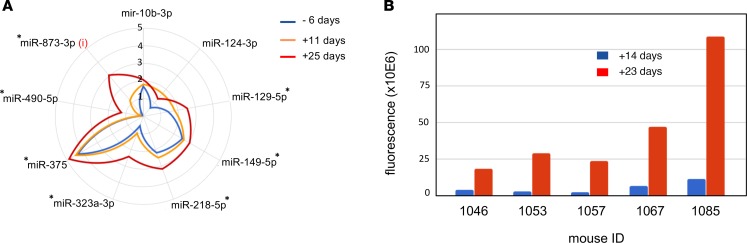
To evaluate to what extent tumor volume may impact the levels of miRNA markers for metastatic disease, we engrafted 4- to 6-week-old immunodeficient mice with human luciferase-positive SH-SY5Y NB cells. Small RNA sequencing was performed on serum samples collected 6 days before engraftment, as well as 11 and 25 days after engraftment. Abundance levels of the selected set of 9 miRNAs were evaluated, in order to determine how the tumor graft impacted levels of these miRNAs in murine serum. Significantly increased miRNA levels over time were observed for the selected miRNAs (Figure 7A and Supplemental Figure 3). Of note, miR–873-3p — the only miRNA that is not conserved between human and mouse — was not detectable prior to engraftment and showed increasing levels 11 and 25 days after engraftment.
Figure 7. Proportional increase of metastatic disease miRNA markers in serum from mice engrafted with human neuroblastoma cells.
(A) Average miRNA abundance (log10 normalized values) in murine serum from 8 engrafted mice 6 days before engraftment and 11 days and 25 days after engraftment. i, miR-873-3p is a human specific miRNA assay; asterisks represent significant increase of miRNA abundance over time based on linear mixed-effects model analysis (P < 0.05, 1-way ANOVA).(B) Luciferase fluorescence assessment of tumor volume 14 days and 23 days after engraftment for 5 of 8 mice in A.
The higher detection levels of the selected miRNAs between 11 and 25 days after engraftment were associated with substantial tumor growth, as evidenced by increased luciferase bioluminescence (Figure 7B).
These findings indicate that an increase in tumor volume could result in increased abundance of the 9 miRNAs in murine serum; at least 1 of the miRNAs (human-specific miR–873-3p) must directly originate from the growing human tumor in the immunodeficient mice.
Serum abundance of disease burden miRNA markers changes during treatment of metastatic NB patients.
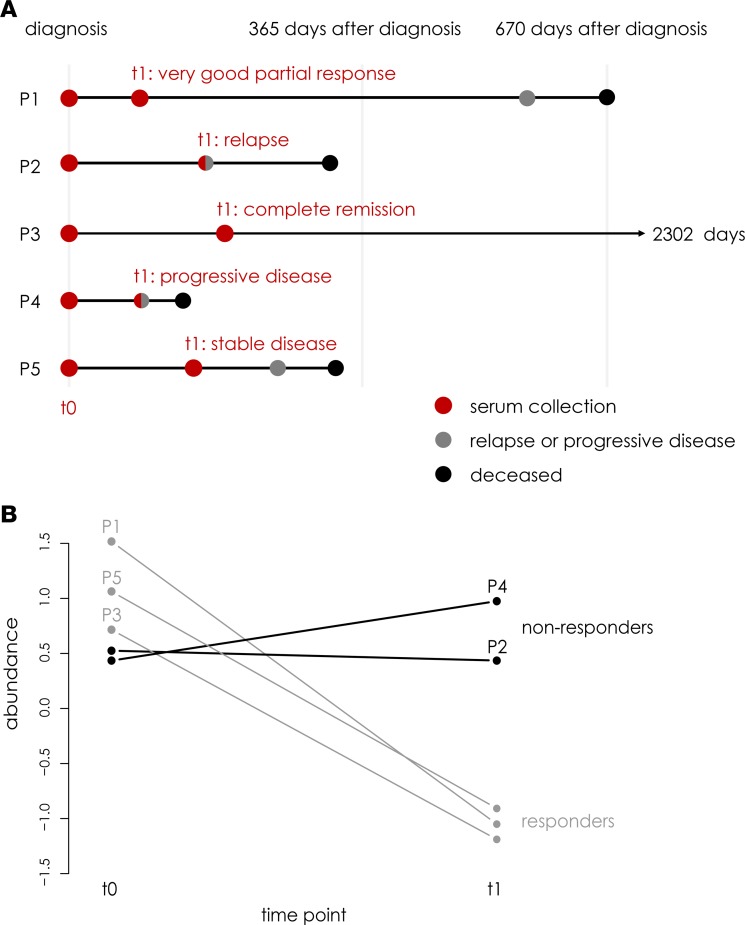
Subsequently, we set out to explore the use of the 9-miRNA signature to monitor disease burden during treatment of NB patients diagnosed with metastatic disease. To this purpose, the abundance levels of the 9 miRNAs were measured using RT-qPCR in 2 serial serum samples from 5 stage 4 NB patients, 1 collected at diagnosis (t0) and 1 during treatment (t1; Figure 8A and Supplemental Table 1). Patients who demonstrated treatment response and presented with reduced tumor volume at t1 show a clear decrease in the average abundance of the 9 disease burden miRNA markers (Figure 8B; Supplemental Figure 4). In contrast, abundance levels of these 9 miRNAs remain high or increase in nonresponders (i.e., patients who show relapse or progressive disease at t1). These proof-of-concept data demonstrate that the identified 9-miRNA signature can potentially be used for disease monitoring in NB.
Figure 8. Serum abundance of disease burden miRNA markers changes during treatment of stage 4 neuroblastoma patients.
(A) The disease course for each of the patients (P1–P5) is depicted, including the time points of serum collection and occurrence of disease events (relapse, progressive disease, and death). (B) For each of the individual patients, the change in average abundance level of the 9 disease burden miRNA markers in serum is shown. Note that at t1, P1, P3, and P5 are considered treatment responders, while P2 and P4 are considered nonresponders.
Discussion
In this study, the abundance levels of 743 human miRNAs were evaluated in 185 diagnostic serum samples from 2 NB patient cohorts. The association between circulating miRNA abundance variance and patient characteristics in the first cohort revealed that tumor stage had the strongest impact on miRNA levels. By taking the overlap from the differential abundance analysis when comparing metastatic stage 4 and localized stages 1 and 2 disease and the outcome of the variance modeling, a core set of 9 miRNAs was defined. These miRNAs were highly abundant in serum from patients with metastatic disease in both tested patient cohorts. Evaluation of the selected set of 9 miRNAs in serum from stage 1,2, 3, 4, and 4S disease showed that the more the disease is spread throughout the body, the higher the levels of the selected miRNAs in serum are. This significant association to tumor stage was confirmed in both patient cohorts, and we therefore decided to label this set of miRNAs as potential circulating biomarkers for metastatic disease in NB.
To evaluate to what extent tumor load could influence circulating miRNA levels, we measured their abundance in serum from mice orthotopically engrafted with human NB cells. A clear increase of the core set of 9 serum miRNA levels was observed 11 days and 25 days after orthotopic injection. Interestingly, there was 1 nonconserved (human-specific) miRNA, which was only detected in murine serum after human NB engraftment (first time point, 11 days after engraftment), with even more pronounced abundance 2 weeks later (25 days after engraftment). Furthermore, the increased levels of the core set of 9 miRNAs in murine serum were accompanied by substantial growth of the human tumor in the murine host. Not only do these findings show that the increased levels of the core set of miRNAs in serum is triggered by the growing NB tumor, but they also provide clear evidence that at least 1 of these miRNAs is derived from the tumor tissue itself.
Comparison of NB serum with serum from healthy children or from children with other cancer types showed that the core set of miRNAs is primarily detected at high levels in NB serum, indicating that these miRNAs may be specific to NB disease. The average expression of the 9 miRNAs in the 3 NB pools was higher than in the 4 other non-NB pools, each of them composed of 5 individual samples. While the number of representative samples from the other pediatric entities and the healthy children is limited, the low levels (or even absence) of these miRNAs in 3 different cancer entities and healthy children is an indication of these miRNAs potentially being specific to NB disease. Nevertheless, a larger study with more samples is required to confirm the presumed specificity of these miRNAs for NB patient sera.
In a previous study, 5 miRNAs were shown to be specifically abundant in serum from MYCN-amplified high-risk NB compared with serum from non–MYCN-amplified low-risk NB and 9 other pediatric cancer entities (6). Three of these miRNAs (miR–124-3p, miR–218-5p, and miR–490-5p) were also part of the core set of miRNAs identified in the present study. These confirmatory results show that the findings in our study are robust and highlight the potential of using serum miRNAs in future clinical practice, since their abundance may be reliably measured independently from sample processing methodologies and miRNA profiling technologies.
In order to assess whether the circulating biomarker miRNAs are overexpressed in metastatic tumors compared with localized NB, we evaluated their expression in 2 public datasets from diagnostic primary NB samples — one established using stem-loom RT-qPCR and the other by deep sequencing of small RNAs. The findings in tumor tissue are not in line with our observations in serum: (a) expression of the majority of the identified set of 9 miRNAs is not significantly different in tumor tissue between stage 4 and stage 1 NB, and (b) for the 2 miRNAs that were differentially expressed in tumor (miR–129-5p and miR–149-5p), increased levels were found in stage 1 tumors, while the opposite was true in serum.
This finding strongly suggests that the miRNA levels in serum are not upregulated due to differential expression in the tumor but are likely proportional with tumor volume and, as such, reflect disease burden and metastatic status. The fact that these miRNAs were among the most abundant miRNAs in the primary tumor further supports this.
There are a few studies that report the use of circulating tumor DNA in NB, 2 of which report the utility of circulating cell-free DNA for noninvasive detection copy number alterations (10, 11). These studies mainly assessed the concordance between array comparative genomic hybridization (CGH) and circulating cell-free DNA profiling, as circulating DNA may provide a more cost effective and less invasive alternative for copy number evaluation of NB tumors. In a third study, performed by Combaret and colleagues, levels of MYCN DNA were measured in peripheral blood and proposed as prognostic biomarker at the time of diagnosis or for follow-up of patients after chemotherapy (12). From a technology point of view, both circulating miRNAs and circulating tumor DNA (ctDNA) are stable molecules and are relatively easy to quantify by various techniques, such as RT-qPCR, digital PCR, or massive parallel sequencing. In this respect, no superiority is claimed of circulating miRNA over circulating ctDNA (or vice versa).
Given the importance of prognostic markers for accurate outcome prediction of high-risk NB patients, the abundance of the 9 miRNAs in serum was evaluated to assess their relation to overall survival. When tested in the high-risk patient cohort only, none of the miRNAs were significantly associated with overall survival. However, we observed a strong and significant association with overall survival in a cohort including NB patients from all risk groups. This is an interesting finding, as it supports the association of these circulating miRNAs with more advanced NB stage and disease. Nevertheless, as currently used prognostic factors (such as age at diagnosis, tumor stage, and MYCN amplification status) are effective in distinguishing between non–high-risk and high-risk patients, the relevance of such miRNAs in clinical practice is rather low.
Finally, in order to obtain first insights into whether this 9-miRNA signature can be used for disease monitoring in NB patients, abundance levels of the 9 miRNAs were measured in serially collected serum samples from metastatic stage 4 NB patients. As such, we could demonstrate that the serum abundance of the 9-miRNA signature changes during the treatment of NB patients and that these changes are related to the disease status of the patient and tumor volume at the time of serum collection. Although these findings and their clinical use need to be validated in larger patient cohorts, they clearly indicate that serum abundance of the 9-miRNA signature can potentially be used for disease monitoring in NB patients diagnosed with metastatic disease.
With the present study, we provide unique first insights into the poorly understood mechanisms behind serum miRNA levels and its relation to tumor miRNA expression. In summary, we have identified 9 miRNAs and convincingly demonstrated that their elevated levels in serum are correlated with tumor volume in serum samples from murine xenografts and disease burden in human NB patients. Further studies are required in order to evaluate the utility of these miRNAs as potential biomarkers for disease burden in a longitudinal disease monitoring or treatment efficacy study design.
Methods
Patient demographics.
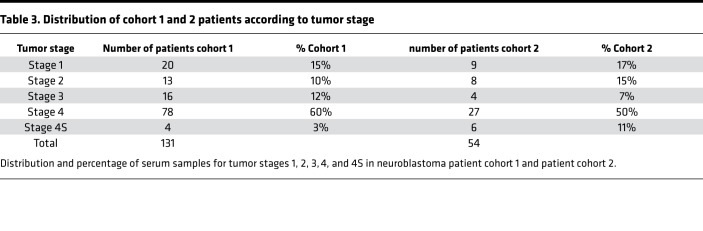
Serum was collected from 185 NB patients at diagnosis from 6 different clinical centers: Ghent, Essen and Cologne, Lyon, Tel Aviv, and Brno. Clinico-pathological properties are listed in Supplemental Table 2. The serum samples were collected and processed in 2 independent cohorts. Cohort 1 contained 131 samples from all 6 centers; cohort 2 contained 54 samples from Cologne. The number of samples over different tumor stages for each sample cohort is shown in the Table 3. Serial serum samples, collected at diagnosis (t0) and during treatment (t1), from 5 patients diagnosed with metastatic disease (stage 4) NB were collected at the Centre Léon Bérard (Lyon, France). Serum from healthy children (5 samples) and diagnostic serum for 3 additional cancer entities (sarcoma, nephroblastoma, and rhabdomyosarcoma; 5 samples each) were obtained from Ghent University Hospital.
Table 3. Distribution of cohort 1 and 2 patients according to tumor stage.
Serum sample pooling.
Three NB serum pools were prepared from human diagnostic samples (Supplemental Table 3), each containing 200 μl from 5 serum samples (40 μl each): pool 1 contained 5 high-risk patients who died of the disease within 36 months after diagnosis; pool 2 contained 5 high-risk surviving patients with at least 5 years follow-up time; and pool 3 contained 5 low-risk surviving patients with at least 5 years follow-up time.
The same approach was used for preparation of 3 additional pools from diagnostic serum from the other pediatric cancer entities and healthy controls.
RNA isolation, reverse transcription, and RT-qPCR reaction.
The serum pools were screened for 1,805 microRNAs using RT-qPCR (miRBase release version 20), as described in ref. 13. The individual samples were screened for 754 microRNAs, including 751 human miRNAs and 3 spike-in control miRNAs: cel-miR–39-3p, miRTC (miRNA reverse transcription control, included in the reverse transcription (RT) kit), and PPC (positive PCR control, synthetic spike-in control included in the SYBR Green PCR mix). Briefly, the commercial miRNeasy serum/plasma kit (Qiagen) was used to extract RNA from 200 μl serum. The serum samples were lysed with 1,000 μl Qiazol and spiked with 3.5 μl synthetic cel-miR–39-3p (1.6 × 108 copies/μl). Chloroform (200 μl; MilliporeSigma) was used for phase separation at 4°C and 12,000 g. The aqueous phase (600 μl) was transferred into an RNeasy MiniElute spin column and subsequently washed with 100% ethanol, buffer RWT, buffer RPE, and 80% ethanol (VWR). The total RNA fraction was eluted with 14 μl RNase-free water (MilliporeSigma).
For reverse transcription (miScript II RT kit, Qiagen), 1.5 μl RNA was mixed with 1 μl reverse transcriptase, 2 μl RT buffer, 1 μl nucleic acid mix, and 4.5 μl RNase-free water. Samples were incubated for 60 minutes at 37°C and 5 minutes at 95°C.
The RT product (2 μl) was diluted 22-fold with 42 μl nuclease-free water and used for quantification of synthetic spike-in RNA molecules: cel-miR–39-3p and miRNA reverse transcription control (miRTC) in 10 μl RT-qPCR reactions. To this purpose, 2 μl of the diluted RT product was mixed with 5 μl SYBR Green PCR mix, 1 μl universal primer, 1 μl miRNA specific primer, and 1 μl RNase-free water. Samples were incubated at 95°C for 15 minutes, followed by 40 cycles of denaturation (15 seconds at 94°C), annealing (30 seconds at 55°C), and extension (30 seconds at 70°C).
For the preamplification reaction (miScript PreAMP PCR Kit, Qiagen), 5 μl of 5-fold diluted RT product was mixed with 5 μl preamp buffer, 2 μl DNA polymerase, 5 μl target specific assay pool, 1 μl universal primer mix, and 7 μl RNase-free water to obtain a 25 μl preamplification reaction. Samples were incubated at 95°C for 15 minutes, followed by 2 cycles of denaturation (30 seconds at 94°C), annealing (60 seconds at 55°C), and extension (60 seconds at 70°C) and 10 additional cycles of denaturation (30 seconds at 94°C) and 1-step annealing/extension (3 minutes at 60°C). Nuclease-free water (100 μl) was added to the 25 μl preamplification product in order to obtain a 5-fold dilution.
For miRNA quantification on the preamplified product, 5 μl RT-qPCR reactions were prepared for each miRNA and each sample. Plates (384-well) prespotted with 1× miRNA RT-qPCR primer assays were used. RT-qPCR mix was prepared for each multiwell plate by mixing 1,025 μl SYBR Green PCR mix, 205 μl universal primer mix, 50 μl preamplified template cDNA, and 770 μl nuclease-free water and dispensed by a Tecan Evo75 liquid handler (5 μl per well). The cycling conditions for quantitative PCR consisted of an initial activation step (15 minutes at 95°C) and 40 cycles of denaturation (15 seconds at 94°C), annealing (30 seconds at 55°C), and extension (30 seconds at 70°C). The synthetic spike-in control PPC was included in the SYBR Green PCR mix PPC.
All RT-qPCR reactions were performed on a CFX384 Real Time PCR Detection System (Bio-Rad). Cq values were determined by the CFX software manager version 3.1 with the regression Cq value determination mode.
Quality control of the RT-qPCR data.
Quantification of cel-miR–39-3p, added to the serum samples during lysis, was used as processing control of the serum RNA isolation. Two samples with no detection of cel-miR–39-3p were excluded from the study. miRTC level differences between serum samples and negative control samples (water mixed with MS2 phage RNA) were used to evaluate RT efficiency and RT-qPCR inhibition. Twenty-two serum samples were excluded due to a miRTC Cq value of 1 cycle higher than the negative control samples. PPC levels across all 384-well plates were used to assess deviating global Cq shifts as result of interrun variation, batch-effect, or technical failure.
The Cq detection cut-off was determined based on evaluation of single positive signals obtained from replicate miRNA expression data, as described in ref. 14. In the current study, 2 NB sample pools were used instead of technical replicate samples. The fraction of single positive results was reduced by 95% at Cq value 29, which was chosen as Cq detection cut-off.
The RT-qPCR reproducibility was assessed for 4 representative sequences in all tested samples: hsa-miR–1224-5p, endogenous human microRNA; cel-miR–39-3p, synthetic C. elegans microRNA spiked into each serum sample to assess RNA extraction efficiency; miRTC, short synthetic RNA sequence that is spiked by the manufacturer into the reverse transcription master mix; and PPC, short synthetic DNA sequence that is spiked by the manufacturer into the RT-qPCR master mix. For 97% of the duplicate reactions, ΔCq values lower than 0.5 were observed, indicating a high RT-qPCR repeatability.
Collection and processing of murine serum samples.
miRNA expression profiles from murine serum were generated through small RNA sequencing of serum samples from 8 female mice bearing orthotopic NB xenografts and 8 control (nonengrafted) female mice. Human luciferase-SH-SY5Y NB cells (1 × 106) were surgically implanted in athymic immunodeficient NCr nude mice under the subrenal capsule (15). Tumor size was assessed 14 days and 23 days after injection of NB cells, by measurement of luciferase intensity. Blood (100 μl) was collected 6 days before, 11 days after, and 25 days after tumor cell injection in serum BD Vacutainer tubes, allowed to clot at room temperature for 45 minutes, and then centrifuged at 2,000 g for 15 minutes at 4°C using a fixed angle rotor. The resulting serum (50 μl) was used for RNA isolation with the miRNeasy serum/plasma kit (Qiagen). Subsequently, transfer RNA (tRNA) fragment depletion was performed as described in ref. 16. The tRNA depleted RNA samples were then suspended in 7.5 μl RNase-free water and used for small RNA library preparation by TruSeq small RNA library preparation kit v2 (Illumina). Library size selection was performed on a Pippin Prep (Sage Science) device with a specified collection size range of 125–153 bp. Libraries were further purified and concentrated by precipitation and quantified using RT-qPCR. Subsequently, equimolar library pools were prepared and further diluted to 4 nM. The pooled library was sequenced at a final concentration of 1.2 pM on a NextSeq 500 using high output v2 kits (single-end, 75 cycles, Illumina). Quantification of small RNAs was done using Cobra, Biogazelle.
Processing of the serial NB serum samples.
RNA from the serially collected serum samples (200 μl) was extracted using the miRNeasy serum/plasma kit (Qiagen). Additionally, 10 RNA samples from cohort 1 that have the highest and lowest average abundance level of the 9 disease-burden miRNA markers were reprofiled to serve as control samples. miRNA abundance levels of these samples were profiled by RT-qPCR, using miScript Primer Assays (Qiagen) for the 9 disease-burden miRNA markers (miR-375 [MS00031829], miR–873-3p [MS00037835], miR–149-5p [MS00003570], miR–124-3p [MS00006622], miR–218-5p [MS00006769], miR–490-5p [MS00009786], miR–323a-3p [MS00037219], miR–10b-3p [MS00008421], and miR–129-5p [MS00006643]) and 8 reference miRNAs (miR-3909 [MS00023513], miR-1203 [MS00014154], miR–4800-5p [MS00038962], miR–4755-5p [MS00039417], miR–877-3p [MS00010654], miR-1976 [MS00016751], miR–766-3p [MS00005439], and miR–532-3p [MS00010052]).
RT and preamplification was performed as described in ref. 13. For preparation of the miScript PreAmp Primer Mix, 70 μl of RNase-free water was mixed with 10 μl miRTC miScript Primer Assay and 10 μl of each of the 10× target miScript Primer Assays (diluted in 550 μl TE, pH 8.0).
miRNA quantification was performed in 10 μl RT-qPCR reactions, consisting of 5 μl SYBR Green PCR mix, 1 μl 10× miScript Universal Primer, 1 μl 10× target miScript Primer Assay, 1 μl nuclease-free water, and 2 μl 5-fold diluted preamplification product. RT-qPCR reactions were performed on a LightCycler 480 (LC480) system (Roche Diagnostics). Cycling conditions were as described above. Cq values were determined by the LC480 software, using the second derivative maximum method.
Statistics.
In order to define the NB serum miRNome, pairwise comparisons of miRNA expression of the 3 serum pools was performed to select miRNAs detected in at least 1 of the serum pools based on 3 selection criteria. First, miRNAs with ΔCq larger than 1.5 between 2 pools were selected (303 miRNAs). Second, miRNAs showing Cq values below 27 in 1 sample and an absent signal (Cq > 29) in the other sample were selected (90 miRNAs). The final selection criterion was based on detection (Cq < 28) in the high-risk serum pools (358 miRNAs). This resulted in a total of 751 unique miRNAs that were subsequently profiled in a first patient cohort of 131 individual serum samples. A second patient cohort containing 54 NB serum samples was independently collected at a later stage of the study and processed by a different batch of miRNA expression profiling reagents.
A threshold Cq value of 29 was determined based on the evaluation of single positive signals obtained from replicate miRNA expression data, as described in ref. 14.
After removal of Cq values above 29 cycles, miRNAs with missing values in more than 75% (an empirically chosen threshold) of the samples were excluded from the dataset. This led to exclusion of 8 miRNAs.
Raw Cq data were normalized as described in ref. 17. Briefly, the arithmetic mean Cq for each miRNA across all samples was determined and subtracted from individual Cq values (i.e., mean centering of the miRs). Then, the arithmetic mean Cq value per sample was calculated across all miRNAs and subtracted from each individual Cq value to obtain normalized miRNA expression values per sample.
GAM, Mann-Whitney U test and 1-way ANOVA was performed using R statistical programing tool version 3.4.3.
For the evaluation of the variance of miRNA abundance in serum by GAM analysis (18), the following mathematic model was used:
miRNA expressioni = (β1 × INSS tumor stage) + (β2 × MYCN status) + (β3 × survival) + (β4 × age at diagnosis),
Where i represents miRNA 1 to miRNA 743, β(1–4) represents the coefficient of the prediction model for INSS tumor stage (stage 1, 2, 3, 4, 4S coded from 1–5); MYCN status (normal=0, amplified=1), survival (alive=0, dead=1), and age at diagnosis (younger than 18 months=0, older than 18 months=1).
GAM analysis was performed by R statistical programming, version 3.4.3, shown in Supplemental Data 1.
For evaluation of the significance of miRNA abundance increase in murine serum in function of time, linear mixed-effects model analysis was used, assuming random effects (Supplemental Data 1).
In order to evaluate the increase in hazard ratio in function of miRNAs levels in serum, Cox proportional hazards regression model was applied (Supplemental Data 1). Comparison of median survival times between patients with low and high abundance of the metastatic disease markers in serum and for construction of Kaplan-Meier plots is shown in Supplemental Data 1.
For data analysis of the serial NB serum samples and control samples that were reprofiled, raw Cq data were analyzed using qbase+ version 3.1 (Biogazelle) (19). Reference target stability was evaluated using geNorm analysis in qbase+; only 4 stably expressed reference miRNAs (miR–4755-5p, miR–4800-5p, miR-3909, and miR–766-3p) were retained for normalization. Log10 transformed normalized data were exported from qbase+ and used for further analysis in R.
Author contributions
FZ, KDP, JV, TVM, AVG, PM, and JV conceptualized and designed the study. Data was acquired and analyzed by FZ, AD, KV, FD, AVG, HHL, PM, and JV. Biomaterials were provided by TL, RN, APB, VC, GS, GL, AS, JHS, SR, JB, PM, MJ, SA, MDH, MM, EB, JS, FB, and MF. All authors contributed to interpretation of the results. The manuscript was drafted by FZ, AD, PM, and JV and was approved and revised by all authors.
Supplementary Material
Acknowledgments
The authors express their special gratitude to the Fournier-Majoie foundation for the generous support and guidance throughout the course of the study. The authors also thank Kom op tegen Kanker (Stand up to Cancer, the Flemish cancer society), Foundation against Cancer (STK), Belgian National Lottery, the National Health Institute Carlos III, and the European Regional Development Fund (ISCIII&FEDER).
Version 1. 12/06/2018
Electronic publication
Funding Statement
https://www.fournier-majoie.org/nl/portfolio/academische-portfolio
Vlaamse Liga Tegen Kanker (VLK)
Stichting Tegen Kanker (STK)
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: JCI Insight. 2018;3(23):e97021. https://doi.org/10.1172/jci.insight.97021.
Contributor Information
Fjoralba Zeka, Email: Fjoralba.zeka@hotmail.be.
Anneleen Decock, Email: a.decock@ugent.be.
Alan Van Goethem, Email: alanvangoethem@hotmail.com.
Katrien Vanderheyden, Email: Katrien.vanderheyden@ugent.be.
Fleur Demuynck, Email: fleur.demuynck@ugent.be.
Tim Lammens, Email: tim.lammens@ugent.be.
Hetty H. Helsmoortel, Email: hetty.helsmoortel@ugent.be.
Joëlle Vermeulen, Email: joellevermeulen@hotmail.com.
Rosa Noguera, Email: rosa.noguera@uv.es.
Ana P. Berbegall, Email: ana.berbegall@uv.es.
Valérie Combaret, Email: valerie.combaret@lyon.unicancer.fr.
Gudrun Schleiermacher, Email: gudrun.schleiermacher@curie.net.
Geneviève Laureys, Email: genevieve.laureys@uzgent.be.
Alexander Schramm, Email: alexander.schramm@uni-due.de.
Sven Rahmann, Email: Sven.Rahmann@uni-due.de.
Julie Bienertová-Vašků, Email: vasku.julie@seznam.cz.
Pavel Mazánek, Email: Mazanek.Pavel@seznam.cz.
Marta Jeison, Email: MARTAJ@clalit.org.il.
Shifra Ash, Email: shifra.ash@gmail.com.
Michael D. Hogarty, Email: hogartym@email.chop.edu.
Eveline Barbieri, Email: exbarbie@txch.org.
Frank Berthold, Email: frank.berthold@uk-koeln.de.
Tom Van Maerken, Email: Tom.VanMaerken@UGent.be.
Frank Speleman, Email: franki.speleman@ugent.be.
Matthias Fischer, Email: matthias.fischer@uk-koeln.de.
Katleen De Preter, Email: katleen.depreter@ugent.be.
Pieter Mestdagh, Email: Pieter.Mestdzagh@UGent.be.
Jo Vandesompele, Email: jo.vandesompele@ugent.be.
References
- 1.Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13(6):397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith MA, Altekruse SF, Adamson PC, Reaman GH, Seibel NL. Declining childhood and adolescent cancer mortality. Cancer. 2014;120(16):2497–2506. doi: 10.1002/cncr.28748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartomo TB, et al. Minimal residual disease monitoring in neuroblastoma patients based on the expression of a set of real-time RT-PCR markers in tumor-initiating cells. Oncol Rep. 2013;29(4):1629–1636. doi: 10.3892/or.2013.2286. [DOI] [PubMed] [Google Scholar]
- 4.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blondal T, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59(1):S1–S6. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 6.Murray MJ, et al. Solid tumors of childhood display specific serum microRNA profiles. Cancer Epidemiol Biomarkers Prev. 2015;24(2):350–360. doi: 10.1158/1055-9965.EPI-14-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramraj SK, Aravindan S, Somasundaram DB, Herman TS, Natarajan M, Aravindan N. Serum-circulating miRNAs predict neuroblastoma progression in mouse model of high-risk metastatic disease. Oncotarget. 2016;7(14):18605–18619. doi: 10.18632/oncotarget.7615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mestdagh P, et al. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36(21):e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schulte JH, et al. Deep sequencing reveals differential expression of microRNAs in favorable versus unfavorable neuroblastoma. Nucleic Acids Res. 2010;38(17):5919–5928. doi: 10.1093/nar/gkq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chicard M, et al. Genomic Copy Number Profiling Using Circulating Free Tumor DNA Highlights Heterogeneity in Neuroblastoma. Clin Cancer Res. 2016;22(22):5564–5573. doi: 10.1158/1078-0432.CCR-16-0500. [DOI] [PubMed] [Google Scholar]
- 11.Van Roy N, et al. Shallow Whole Genome Sequencing on Circulating Cell-Free DNA Allows Reliable Noninvasive Copy-Number Profiling in Neuroblastoma Patients. Clin Cancer Res. 2017;23(20):6305–6314. doi: 10.1158/1078-0432.CCR-17-0675. [DOI] [PubMed] [Google Scholar]
- 12.Combaret V, et al. Circulating MYCN DNA as a tumor-specific marker in neuroblastoma patients. Cancer Res. 2002;62(13):3646–3648. [PubMed] [Google Scholar]
- 13.Zeka F, Mestdagh P, Vandesompele J. RT-qPCR-based quantification of small non-coding RNAs. Methods Mol Biol. 2015;1296:85–102. doi: 10.1007/978-1-4939-2547-6_9. [DOI] [PubMed] [Google Scholar]
- 14.Mestdagh P, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
- 15.Agarwal S, et al. Transmembrane adaptor protein PAG1 is a novel tumor suppressor in neuroblastoma. Oncotarget. 2016;7(17):24018–24026. doi: 10.18632/oncotarget.8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Goethem A, et al. Depletion of tRNA-halves enables effective small RNA sequencing of low-input murine serum samples. Sci Rep. 2016;6:37876. doi: 10.1038/srep37876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.D’haene B, Mestdagh P, Hellemans J, Vandesompele J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol Biol. 2012;822:261–272. doi: 10.1007/978-1-61779-427-8_18. [DOI] [PubMed] [Google Scholar]
- 18. Wood. Generalized Additive Models: An Introduction with R. Second Edition. Boca Ratan, FL:Chapman and Hall/CRC;2017. [Google Scholar]
- 19.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.