Abstract
Enterohemorrhagic Escherichia coli (EHEC) colonize intestinal epithelium by generating characteristic attaching and effacing (AE) lesions. They are lysogenized by prophage that encode Shiga toxin 2 (Stx2), which is responsible for severe clinical manifestations. As a lysogen, prophage genes leading to lytic growth and stx2 expression are repressed, whereas induction of the bacterial SOS response in response to DNA damage leads to lytic phage growth and Stx2 production both in vitro and in germ-free or streptomycin-treated mice. Some commensal bacteria diminish prophage induction and concomitant Stx2 production in vitro, whereas it has been proposed that phage-susceptible commensals may amplify Stx2 production by facilitating successive cycles of infection in vivo. We tested the role of phage induction in both Stx production and lethal disease in microbiome-replete mice, using our mouse model encompassing the murine pathogen Citrobacter rodentium lysogenized with the Stx2-encoding phage Φstx2dact. This strain generates EHEC-like AE lesions on the murine intestine and causes lethal Stx-mediated disease. We found that lethal mouse infection did not require that Φstx2dact infect or lysogenize commensal bacteria. In addition, we detected circularized phage genomes, potentially in the early stage of replication, in feces of infected mice, confirming that prophage induction occurs during infection of microbiota-replete mice. Further, C. rodentium (Φstx2dact) mutants that do not respond to DNA damage or express stx produced neither high levels of Stx2 in vitro or lethal infection in vivo, confirming that SOS induction and concomitant expression of phage-encoded stx genes are required for disease. In contrast, C. rodentium (Φstx2dact) mutants incapable of prophage genome excision or of packaging phage genomes retained the ability to produce Stx in vitro, as well as to cause lethal disease in mice. Thus, in a microbiome-replete EHEC infection model, lytic induction of Stx-encoding prophage is essential for lethal disease, but actual phage production is not.
Author summary
Enterohemorrhagic Escherichia coli (EHEC), a food-borne pathogen that produces Shiga toxin, is associated with serious disease outbreaks worldwide, including over 390 food poisoning outbreaks in the U.S. in the last two decades. Humans acquire EHEC by ingesting contaminated food or water, or through contact with animals or their environment. Infection and toxin production may result in localized hemorrhagic colitis, but may progress to life-threatening systemic hemolytic uremic syndrome (HUS), the leading cause of kidney failure in children. Treatment for EHEC or HUS remains elusive, as antibiotics have been shown to exacerbate disease. Shiga toxin genes reside on a dormant bacterial virus present in the EHEC genome, but are expressed when the virus is induced to leave its dormant state and begin to replicate. Extensive virus replication has been thought necessary to produce sufficient toxin to cause disease. Using viral and bacterial mutants in our EHEC disease mouse model, we showed that whereas an inducing signal needed to begin viral replication was essential for lethal disease, virus production was not: sufficient Shiga toxin was produced to cause lethal mouse disease, even without viral replication. Future analyses of EHEC-infected human samples will determine whether this same phenomenon applies, potentially directing intervention strategies.
Introduction
Shiga toxin-producing Escherichia coli (STEC) is a food-borne zoonotic agent associated with worldwide disease outbreaks that pose a serious public health concern. Enterohemorrhagic Escherichia coli (EHEC), a subset of STEC harboring specific virulence factors that promote a specific mode of colonization of the intestinal epithelium (see below), is acquired by humans by ingestion of contaminated food or water, or through contact with animals or their environment. EHEC serotype O157:H7 is a major source of E. coli food poisoning in the United States, accounting for more than 390 outbreaks in the last two decades [1–5]. EHEC infection usually presents as localized hemorrhagic colitis, and may progress to the life-threatening systemic hemolytic uremic syndrome (HUS), characterized by the triad of hemolytic anemia, thrombocytopenia, and renal failure [5, 6]. HUS is the leading cause of renal failure in children [7].
EHEC, along with enteropathogenic E. coli and Citrobacter rodentium belong to the family of bacteria known as attaching and effacing (AE) pathogens that are capable of forming pedestal-like structures beneath bound bacteria by triggering localized actin assembly [8–10]. While this ability of EHEC leads to colonization of the large intestine, production of prophage-encoded Shiga toxin (Stx) promotes intestinal damage resulting in hemorrhagic colitis [11–17]. Shiga toxin may further translocate across the colonic epithelium into the bloodstream, leading to systemic disease. Distal tissue sites, including the kidney, express high levels of the Shiga toxin-binding globotriosylceramide (Gb3) receptor, potentially leading to HUS [14, 15, 18–21].
Genes encoding EHEC Shiga toxin are typically encoded in the late gene transcription region of integrated lambdoid prophages [22, 23] and their expression is thus predicted to be temporally controlled by phage regulons [24–27]. Early studies showed that high levels of Stx production and release from the bacterium in vitro required prophage induction, i.e., the mechanism by which quiescent prophages of lysogenic bacteria are induced to replicate intracellularly and released as phage particles by host cell lysis [27, 28]. Lambdoid phage inducers are most commonly agents that damage DNA or interfere with DNA synthesis, such as ultraviolet light or mitomycin C. These inducing stimuli trigger activation of the bacterial RecA protein, ultimately leading to the cleavage of the prophage major repressor protein, CI, allowing expression of phage early and middle genes. Late gene transcription, which requires the Q antiterminator, results in the expression of many virion structural genes and of endolytic functions S and R, which lyse the bacterium and release progeny phage [29]. Other signaling pathways involving quorum sensing or stress responses have also been implicated in lysogenic induction [30, 31].
Unfortunately, antibiotics commonly used to treat diarrheal diseases in children and adults are known to induce the SOS response. Trimethoprim-sulfamethoxazole and ciprofloxacin have been shown to enhance Stx production in vitro [32–34], and antibiotic treatment of EHEC-infected individuals is associated with an increased risk of HUS [35]. Hence, antibiotics are contraindicated for EHEC infection and current treatment is limited to supportive measures [36].
A more detailed understanding of the role of prophage induction and Stx production and disease has been pursued in animal models of EHEC infection. Although some strains of conventional mice can be transiently colonized by EHEC, colonization is not robust and typically diminishes over the course of a week [13, 37], necessitating use of streptomycin-treated [16] or germ-free mice [38, 39] to investigate disease manifestations that require efficient, longer-term intestinal colonization. In streptomycin-treated mice colonized with EHEC, administration of ciprofloxacin, a known SOS inducer, induces the Stx prophage lytic cycle, leading to increased Stx production in mouse intestines and to Stx-mediated lethality [40]. Conversely, an EHEC strain encoding a mutant CI repressor incapable of inactivation by the SOS response was also incapable of causing disease in germ-free mice [41].
A potential limitation of the antibiotic-treated or germ-free mouse infection models is the disruption or absence, respectively, of microbiota, with concomitant alterations in immune and physiological function [42]. For example, a laboratory-adapted E. coli strain that lacks the colonization factors of commensal or pathogenic E. coli is capable of stably colonizing streptomycin-treated mice [43], and, when overproducing Stx2, is capable of causing lethal infection in antibiotic-treated mice [17]. Further, as up to 10% of human gut commensal E. coli were found to be susceptible to lysogenic infection by Stx phages in vitro [44], it has been postulated that commensals may play an amplifying role in EHEC disease by fostering successive rounds of lytic phage growth [44–47]. Finally, gut microbiota may also directly influence expression of stx genes. For example, whereas a genetic sensor of phage induction suggests that the luminal environment of the germ-free mouse intestine harbors a prophage-inducing stimulus [41], several commensal bacteria have been shown to inhibit prophage induction and/or Stx production in vitro [48–50]. Alternatively, colicinogenic bacteria produce DNAse colicins that may trigger the SOS response, increasing Stx production [51].
Our laboratory previously developed a murine model for EHEC using the murine AE pathogen C. rodentium [52, 53], which efficiently colonizes conventionally raised mice and allows the study of infection in mice with intact microbiota. The infecting C. rodentium is lysogenized with E. coli Stx2-producing phage Φ1720a-02 [52, 54] encoding Stx variant Stx2dact (Stx2d activatable), which is particularly potent in mice [55, 56]. Infection of C57BL/6 mice with C. rodentium(Φ1720a-02), (herein referred to as C. rodentium(Φstx2dact)), produces many of the features of human EHEC infection, including colitis, renal damage, weight loss, and potential lethality, in an Stx2dact-dependent manner [52].
In the current study, we address phage, bacterial, and host factors that lead to lethal EHEC infection. We found that C. rodentium(Φstx2dact) strains lacking RecA, which is required for induction of an SOS response, or phage Q protein, which is required for efficient transcription of the late phage genes, did not produce high levels of Stx in vitro or cause lethal disease in mice. In contrast, mutants defective in prophage excision, phage assembly, or phage-induced bacterial lysis retained the ability to both produce Stx2dact upon prophage induction in vitro and to cause lethal disease. Excised phage genomes, potentially undergoing DNA replication leading to phage production or representing packaged phage, were detected, albeit at low levels, in fecal samples of mice infected with wild type C. rodentium(Φstx2dact), but not in mice infected with excision-defective C. rodentium(Φstx2dact). Thus, in a microbiome-replete EHEC infection model, lytic induction of Stx-encoding prophage, but not actual production of viable phage particles, is essential for Stx production and lethal disease.
Results
Gene map and features of Φstx2dact prophage
Lambdoid phage Φ1720a-02 was originally isolated from EC1720a-02, a STEC strain found in packaged ground beef [54]. Our novel C. rodentium-mediated mouse model of EHEC infection encompasses C. rodentium DBS100 (also known as C. rodentium strain ICC 168 (GenBank accession number NC_013716.1)), lysogenized with phage Φ1720a-02 marked with a chloramphenicol (cam)-resistance cassette inserted into the phage Rz gene, creating strain DBS770 [52, 53]. A second lysogen, DBS771, was lysogenized with the same phage but with an additional kanamycin (kan)-resistance cassette inserted into and inactivating the prophage stx2A gene. For simplicity, strains DBS770 and DBS771 will herein be referred to as C. rodentium (Φstx2dact), and C. rodentium(ΦΔstx2dact::kanR), respectively (Table 1).
Table 1. Bacterial strains and plasmids.
| Strain | Description | Reference |
|---|---|---|
| C. rodentium wild type | Strain DBS100 (also known as ICC 168). |
[57, 58] |
| C. rodentium (Φstx2dact) | DBS770, i.e., DBS100 (Φ1720a-02 ΔRz::cat), chloramphenicolR |
[59] and GenBank accession number KF030445 |
| C. rodentium (Φstx2dact::kanR) | DBS771, i.e., DBS770 with a kanamycin resistance cassette inserted into the stx2A gene, chloramphenicol and kanamycin resistant |
[59] |
| C. rodentium (Φstx2dact Δint) | DBS770 deleted for prophage int gene |
This study |
| C. rodentium (Φstx2dact ΔSR) | DBS770 deletion for prophage SR genes |
This study |
| C. rodentium (Φstx2dact ΔB) | DBS770 deleted for prophage B gene |
This study |
| C. rodentium (Φstx2dactΔQ) | DBS770 with deleted for prophage Q gene |
This study |
| C. rodentium ΔrecA (Φstx2dact) | DBS770 deleted for host recA gene |
This study |
| C. rodentium ΔrpoS (Φstx2dact) | DBS770 deleted for host rpoS gene |
This study |
| C. rodentium ΔqseC (Φstx2dact) | DBS770 deleted for host qseC gene |
This study |
| C. rodentium ΔqseF (Φstx2dact) | DBS770 deleted for host qseF gene |
This study |
| E. coli K12 DH5α |
fhuA2 lac(del)U169 phoA glnV44 Φ80' lacZ(del)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17 |
[60] |
| Plasmids | Description | Reference |
| pKD46 | Phage Lambda-red recombinase, bla |
[61] |
| pTOPO-Q | pCR4 TOPO vector encoding Q gene and 100 bp region upstream |
This study |
To identify phage genes critical for lethal mouse infection, we sought to inactivate specific prophage genes and then assess their resulting phenotypes in the C. rodentium mouse model. As a first step, we sequenced the parental strain DBS100 and the genomes of C. rodentium (Φstx2dact) and C. rodentium (ΦΔstx2dact::kanR), revealing that the three genomes were identical except for prophage sequences present in C. rodentium (Φstx2dact) and C. rodentium (ΦΔstx2dact::kanR) (see Materials and Methods).
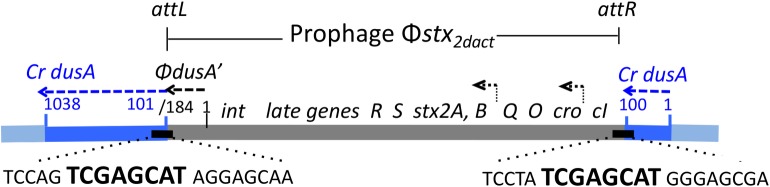
We then annotated the entire Φstx2dact prophage (GenBank accession number KF030445.1; Figs 1 and S1). As is typical of Stx phages, the sequence revealed a lambdoid phage with a mosaic gene organization that does not precisely match that of phage λ, but is nevertheless somewhat syntenic with other lambdoid phages [62], (S2 Fig). Further, although lysogenized independently, C. rodentium(Φstx2dact) and C. rodentium(ΦΔstx2dact::kanR) prophages were integrated at the same location, i.e. 100 bp into the coding sequence of dusA (encoding tRNA-dihydroxyuridine synthase A). A recent study revealed that known integrase genes, at least half of which belong to prophages, were found adjacent to the host dusA gene in over 200 bacterial species [63]. Furthermore, a 21 base pair motif found at the prophage-host DNA junctions in many bacteria was present at the prophage junctions, attL and attR, of C. rodentium(Φstx2dact) and C. rodentium(ΦΔstx2dact::kanR), as well as at the presumed attB phage insertion site in the parental C. rodentium dusA gene (Fig 1). A seven-base segment within this 21-base sequence is completely conserved between attL, attR, and attB and likely represents the ‘core’ recombination site for integration or excision (Fig 1, bolded sequence; [64]). Note that, although the Φstx2dact and ΦΔstx2dact::kanR prophages interrupt the dusA gene, they encode a 184 bp ORF (designated “ΦdusA’” in Fig 1) that is in frame with the 3’ 937 nucleotides (positions 101 to 1038) of dusA
Fig 1. Prophage Φstx2dact in C. rodentium (Φstx2dact).
The Φstx2dact prophage (gray), flanked by attL and attR upon insertion into C. rodentium dusA sequence (blue, “Cr dusA”), was determined by whole genome sequencing of C. rodentium(ΦΔstx2dact::kanR). The 3’ end of the prophage (nucleotides 1–184) encodes the N-terminal 61 residues of “ΦdusA,’” in the same reading frame as the 3’ end (nucleotides 101–1038) of the C. rodentium dusA gene (“Cr dusA”). Bent arrows indicate direction of transcription of Q, stx, and phage late genes. Depicted are attL and attR sequence motifs, characteristic of other prophages inserted within the host dusA gene ([63]). Within this sequence, a seven-base “core” sequence (bolded), perfectly conserved in attL and attR, as well as in the Φstx2dact attP sequence shown here and in the parental attB sequence in C. rodentium dusA (TCCAGTCGAGCATGGGAGC), is the cross-over site for phage integration and excision.
A prior analysis of the host C. rodentium DBS100 genome sequence revealed the presence of 10 additional partial and intact prophages distributed around the genome [65], although it is not known if any of these prophages can give rise to intact phage. Sequence analysis showed only 2 regions of homology between Φstx2dact and these prophages (S3 Fig): one resident prophage encoded 70% homology to a region encoding Φstx2dact Cro, CI repressor, and a hypothetical protein, and a second resident prophage showed 79% homology to another Φstx2dact gene encoding a hypothetical protein.
Survey of prophage integration (att) sites during murine infection reveals Φstx2dact prophage excision from C. rodentium(Φstx2dact), but no secondary lysogeny of commensal bacteria
Although Φstx2dact harbors a cat insertion in the Rz gene, a gene that contributes to phage λ lysis under some conditions [2], prophage induction with mitomycin C resulted in lysis of C. rodentium(Φstx2dact) (S4 Fig), suggestive of lytic phage induction. Nevertheless, pilot experiments revealed that Φstx2dact plaques were not detectable on any of numerous E. coli K12 and other indicator strains (Materials and Methods). This finding is not unusual for Stx-producing phages [66–68]. To more rigorously test whether this phage can infect E. coli K12, we selected for E. coli K12 lysogens by infecting E. coli K12 strain DH5α with supernatants of mitomycin C -induced cultures of C. rodentium (Φstx2dact), then selecting for kanamycin-resistant clones. These clones were verified as lysogens by PCR-detection of phage genes (S5A Fig). DH5α lacks RecA and thus cannot undergo an SOS response to trigger prophage induction. However, when the RecA-producing plasmid pER271 was introduced to the DH5α lysogens, they were more sensitive to UV light than non-lysogens containing the same plasmid (S5B Fig), consistent with lysogenic induction. Hence, Φstx2dact is a functional phage that is capable of infecting bacteria, including E. coli K12.
In the course of EHEC infection of streptomycin-treated mice, Stx phage can be induced by antibiotic treatment to lysogenize other E coli strains in the intraluminal environment [40] [69]. It has been postulated that successive cycles of infection of non-pathogenic commensal E. coli could amplify Stx production and exacerbate disease [38, 44, 45, 47]. We first addressed this question by testing whether lysogeny of commensal bacteria by phage Φstx2dact was detectable following oral C. rodentium(Φstx2dact) infection of mice. Mice orally gavaged with C. rodentium(Φstx2dact) normally exhibit weight loss and lethal disease [52], typically succumbing to disease after day 7 post-infection. DNA was extracted from fecal samples of a group of five mice at days 1 and 6 post-infection. The DNA samples were used as a template to generate a library of sequences encompassing the sequence downstream of attL (specifically, spanning the region from the phage int gene, through Φ dusA and into the adjacent host sequence; see Fig 1). This strategy is a modification of that used for Tn-seq library analysis ([70], Materials and Methods).
Although we were unable to obtain detectable amplified DNA from fecal samples produced on day 1 post-infection, consistent with the low titer of C. rodentium(Φstx2dact) in the stool at this early time point, the day 6 post-infection sample yielded a DNA library, which was subjected to massively parallel sequencing to identify the origin of the host DNA into which the prophage was integrated. Of 17,142,098 readable sequences generated, 99.56% showed homology to C. rodentium (Φstx2dact), i.e. included C. rodentium (Φstx2dact) attL and the adjacent C. rodentium dusA gene sequence, indicating prophage integration into the original C. rodentium strain (Table 2; see Materials and Methods). For the remaining 0.44% of sequences, the C. rodentium dusA sequences adjacent to the attL core sequence were replaced by phage-specific attR sequences, thus regenerating attP. These latter sequences likely reflect excised circular phage genomes generated following induction of the C. rodentium(Φstx2dact) lysogen. Thus, C. rodentium(Φstx2dact) undergoes lytic induction in the murine host, consistent with previous findings of EHEC infection in streptomycin-treated mice. Furthermore, no integration of the Φstx2dact prophage into either a different site in C. rodentium, or into a different bacterial host was observed, leading to the conclusion that lysogeny of intestinal bacteria by Φstx2dact is not a common event in this model.
Table 2. Comprehensive survey of prophage attachment (integration) sites reveals prophage excision but not secondary lysogeny of commensal bacteria during murine infection by C. rodentium (Φstx2dact).
| Sequence identity | Number | Percent of total |
|---|---|---|
| C. rodentium(Φstx2dact) attL | 17,066,136 | 99.56 |
| Φstx2dact attP (replicative form) | 75,962 | 0.44 |
| Total sequences1 | 17,142,098 | 100.00 |
1Of a total of 17,868,095 sequences, 725,997 were of poor quality, resulting in a total of 17,142,098 readable sequences.
C. rodentium RecA and Φstx2dact proteins integrase, Q, endolysins, and portal protein are required for efficient phage production and release in vitro
Prophage induction of lambdoid phages is often initiated by DNA damage, in which SOS pathway activation leads to RecA-promoted autocleavage of CI repressor, followed by transcription of early genes from the from PL and PR promoters. Subsequent temporally programmed transcription of the prophage genome results in the production of delayed early (middle) proteins such as Int (integrase), essential for prophage integration and excision, and antiterminator protein Q. Production of Q in turn mediates the transcription of late genes, including portal protein gene B, responsible for translocation of phage DNA into the virion protein capsid, and lysis genes S and R, encoding endolysins that disrupt the bacterial plasma membrane causing release of intact phage progeny (for a review, see Gottesman and Weisberg [71]). Late genes in EHEC phages also encompass stx.
To uncover the roles of specific phage and bacterial functions in EHEC disease, we used lambda red recombination (Materials and Methods) to construct C. rodentium(Φstx2dact) strains defective for prophage genes SR, int, B, or Q, or the host gene recA, which is well documented to be central to the SOS response and lytic induction. In addition, we inactivated three other genes that have been implicated as having more subtle roles in the lytic induction of Shiga toxin-encoding phage [30, 31]: rpoS, which controls the bacterial stress response, and qseC and qseF, which control quorum sensing pathways (Materials and Methods, Table 1). The production of Shiga toxin phage has been shown to be influenced by growth medium [66], but none of the mutants displayed a growth defect upon in vitro culture in LB or DMEM medium (S6 Fig).
We then tested C. rodentium(Φstx2dact) and several of the mutant derivatives predicted to have dramatic effects on phage production for the ability to generate Φstx2dact following SOS induction. Given that Φstx2dact was found to not form plaques on indicator strains tested, we instead utilized qPCR to quantify phage [72–74]. Specifically, we employed primers flanking the phage attP site to distinguish integrated and excised phage DNA, as only the latter will have reconstituted the attP site [71]. This technique detects both unpackaged phage genomes and those packaged in phage capsids, as in our initial experiments lysates were not treated with DNase prior to qPCR enumeration. Note that protease digestion of the capsid prior to qPCR quantitation was also eliminated, as capsid undergoes melting during the high heating steps of the PCR procedure [75]
Supernatants of mid-logarithmic phase (t = 0h) LB cultures contained 1.3×109–3.8×109 attP copies (phage genomes) per ml (Table 3 legend), compared to approximately 108 viable bacteria per ml, indicating significant spontaneous prophage induction during the period leading to mid-log growth. After four additional hours (t = 4h), supernatant phage concentration increased 3.2-fold relative to t = 0h, consistent with continued spontaneous prophage induction (Table 3, “Relative attP production”, “- Mito C”). Prophage induction of the wild type lysogen with the SOS inducer mitomycin C led to a 234-fold increase in relative attP production (Table 3, “+ Mito C”), a 73-fold increase above baseline levels. As predicted [76, 77], the generation of circular phage genomes required Int recombinase, as at all time points tested, attP copies were below the level of detection of 1× 104/ml in uninduced or mitomycin C-induced cultures of the C. rodentium(Φstx2dactΔint) mutant (Table 3).
Table 3. C. rodentium RecA and Φstx2dact proteins integrase, Q, endolysins, and portal protein are required for efficient phage production and release in vitro.
| Strain | Function deleted | Relative attP (phage) production | ||
|---|---|---|---|---|
| - Mito Ca | + Mito Cb | +Mito C + DNAsec | ||
| WT | None (WT) | 3.2 (±0.01) | 234.6 (±24.4) | 162.5 (±1.1) |
| Δint | Phage integrase | Not detected | Not detected | Not determined |
| ΔrecA | Host RecA | 0.5 (±0.6)* | 29.4 (±11.4)* | Not determined |
| ΔQ | Phage late gene transcription anti-terminator | 0.5 (±0.3)* | 6.0 (±0.3)* | Not determined |
| ΔSR | Phage endolysin | 0.6 (±0.2)* | 6.3 (±2.0)* | Not determined |
| ΔB | Phage portal protein | 4.1 (±0.08) | 208.7 (±17.2) | 9.2 (±1.3)* |
aSupernatants from mid-log (t = 0h) cultures or parallel cultures grown for an additional 4 hours (t = 4h) were analyzed for attP copies by qPCR. Shown are average values of t = 4h/t = 0h (+/- SEM) for each lysogen, derived from the values of three different dilutions of each supernatant (see Materials and Methods). For all lysogens except C. rodentium(Φstx2dactΔint), absolute numbers of attP molecules at t = 0h ranged from 1.3×109 to 3.8×109/ml. For C. rodentium(Φstx2dactΔint), attP copies were below the limit of detection, i.e., <1× 104/ml.
bSupernatants from mid-log (t = 0h) cultures, and parallel cultures subsequently exposed to 0.25 μg/ml mitomycin C for 4 hours (t = 4h) were analyzed for attP copies by qPCR and the ratios of the two values determined as above. For C. rodentium(Φstx2dact int), attP copies were below the limit of detection, i.e., <1× 104/ml.
cSupernatants from mid-log (t = 0h) cultures or parallel cultures subsequently exposed to 0.25 μg/ml mitomycin C for 4 hours (t = 4h) were analyzed for attP copies by qPCR after treatment with DNase (1 hr, according to manufacturer’s instructions), to remove unpackaged DNA. The ratios of the two values were determined as above. Note that DNase treatment longer than 1 hr did not significantly alter the results.
*indicates statistical significance (p<0.05) compared to identically treated WT, calculated by one-way Anova.
Host and phage functions contributed to the amount of phage production. In the absence of inducer (Table 3, “- MitoC”), the concentration of attP copies in culture supernatants of C. rodentiumΔrecA(Φstx2dact), predicted to be defective for SOS induction, did not increase between t = 0h and t = 4h, with an average relative attP production of 0.5. Lysogens deficient in the antiterminator Q, required for late gene transcription, or deficient in the S and R endolysins, which promote the efficient release of phage from infected bacteria, were also deficient in relative attP production in the absence of inducer (Table 3). Finally, C. rodentium(Φstx2dactΔB), predicted to replicate but not package phage genomes, showed no defect in the production of attP copies in the culture supernatant in the absence of inducer, with relative phage production ratio of 4.1. However, as described below, DNAse sensitivity assays suggested that these attP sequences are likely not packaged into phage particles.
The mutants defective in baseline phage production were similarly defective in the titer of attP copies after induction with mitomycin C (Table 3, “+ Mito C”). Induction of C. rodentiumΔrecA (Φstx2dact) resulted in an increase in attP production, consistent with low levels of phage production by RecA-deficient λ lysogens of E. coli following induction [78], but the relative attP value of 29 was eight-fold lower than wild type. C. rodentium(Φstx2dactΔQ) and C. rodentium(Φstx2dactΔSR) each also demonstrated dramatically diminished attP copies in mitomycin C-induced culture supernatants, with relative attP production of approximately 6. The small increase in attP levels for each of these mutants upon induction is consistent with readthrough of early transcription of Q-deficient λ mutants [79] and low level bacterial lysis in the absence of phage-encoded endolysins, respectively.
Finally, C. rodentium (Φstx2dactΔB), generated wild type levels of phage genome copies, with a 209-fold increase in relative attP production. However, DNAse treatment of supernatants diminished this value more than 23-fold, whereas parallel treatment diminished the relative attP production by wild type C. rodentium(Φstx2dact) less than 1.5-fold (Table 3, “+Mito C + DNAse”), consistent with a defect in packaging of Φstx2dact genomes in the absence of the B portal protein.
Proteins required for the SOS response and/or late gene transcription are essential for Stx2dact production
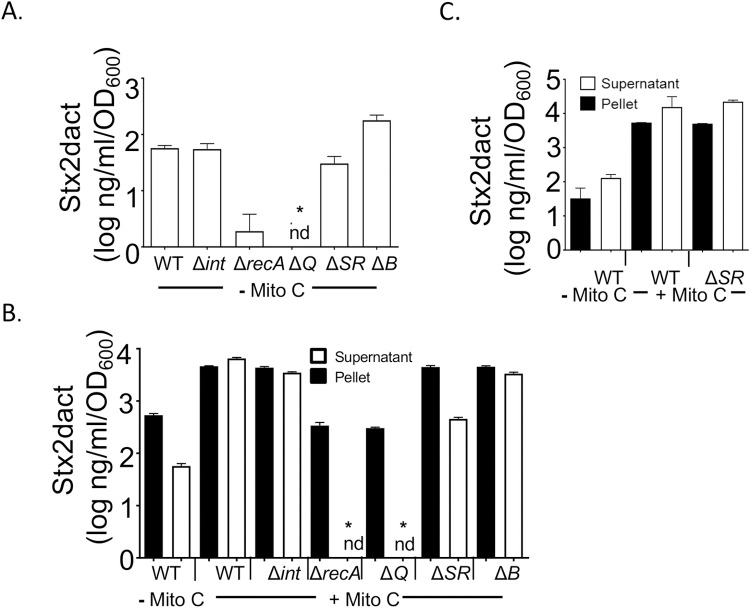
To determine which host or phage functions are required for production of Stx2dact in vitro, we measured Stx2dact in culture supernatants by ELISA [53]. To quantitate non-induced levels of toxin, and to provide ample time for toxin to accumulate, we grew triplicate cultures of the C. rodentium(Φstx2dact) or the mutant derivatives described above for four hours (t = 4h) beyond mid-log phase (defined as t = 0h). Stx2dact was present in the culture supernatants of wild type C. rodentium(Φstx2dact) at approximately 50 ng/ml/OD600 unit, consistent with previous measurements [53] (Fig 2A, “WT”). Prophage excision and phage production were not required for this basal level of Stx2dact: culture supernatants of C. rodentium(Φstx2dactΔint), which did not harbor detectable phage (Table 3), contained equivalent amounts of toxin (Fig 2A, “Δint”). Uninduced culture supertants of C. rodentium(Φstx2dactΔSR) contained levels of Stx2dact two-fold lower than (and statistically indistinguishable from) wild type, consistent with the moderately (5-fold) lower levels of phage found in cultures of wild type C. rodentium (Φstx2dact) (Table 3, “- MitoC”). Supernatants of C. rodentium(Φstx2dactΔB), which contained attP DNA but relatively few packaged phage (Table 3), also produced levels of Stx2dact statistically indistinguishable from wild type. Finally, in contrast, C. rodentiumΔrecA (Φstx2dact), which is unable to mount an SOS response, and C. rodentium(Φstx2dactΔQ), which cannot transcribe phage late genes, including stx2dactA and stx2dactB, were defective for basal levels of Stx2dact production (Fig 2A, “ΔrecA”, “ΔQ”).
Fig 2. SOS responsiveness and lytic induction-dependent transcription of stx genes are required for wild type basal and induced levels of Stx2 production in vitro.
A. The indicated lysogens were grown in the absence of mitomycin C until t = 4h, i.e., four hours after attaining approximately mid-log phase (which was designated as t = 0h; “- Mito C”), and culture supernatants were subjected to capture ELISA to determine the basal level of Stx2 production (see Materials and Methods). Quantities are expressed relative to the specific OD600 at t = 0h. nd: not detected. B. The indicated lysogens were grown to mid-log phase (t = 0h) and cultured for four more hours (t = 4h) either in the absence (“- Mito C”) or presence of 0.25 μg/ml mitomycin C (“+ Mito C”). Pellets (filled bars) or supernatants (open bars) were subjected to capture ELISA to determine the level of Stx2 production. Quantities are expressed relative to the specific OD600 at t = 0h. nd: not detected. C. Wild type C. rodentium(Φstx2dact) and C. rodentium(Φstx2dact ΔRS)) were grown to mid-log phase (designated as t = 0h) and cultured for 16 more hours (t = 16h) either in the absence (“- Mito C”) or presence of 0.25 μg/ml mitomycin C (“+ Mito C”). Pellets (filled bars) or supernatants (open bars) were subjected to capture ELISA to determine the level of Stx2dact production. Quantities are expressed relative to the specific OD600 at t = 0h. For all panels, results are averages ± SEM of triplicate samples, and are a representative of at least two experiments involving independently derived mutants. Asterisks (*) indicate Stx level significantly (p <0.05) different from wild type C. rodentium (Φstx2dact) calculated using Kruskal–Wallis one-way analysis of variance followed by Dunn's nonparametric comparison.
To test whether the defect in Stx2dact production was due to the lesion in the Q gene, we complemented C. rodentium(Φstx2dactΔQ) with plasmid pTOPO-Q, the wild type Q gene (Table 1). The complemented strain indeed increased Stx2dact production 286-fold (S7 Fig, “ΔQ + pTOPO-Q”). Nevertheless, this level of Stx2dact was 12-fold lower than that produced by the wild type C. rodentium (Φstx2dact) strain, a defect that is likely due to unregulated Q production in trans [80] because we found that pTOPO-Q similarly diminished Stx production by the WT strain (S7 Fig, “WT + pTOPO-Q”). The exquisite developmental control of gene expression during the lysogenic and lytic cycle is a hallmark of lambdoid phages [23], making complementation of many of phage mutants technically challenging [80].Hence, to minimize the risk that phenotypes observed were due to off-target lesions, we isolated two independent clones of each mutant and tested both clones for each of the phenotypes observed throughout this study.
We also assessed toxin production by wild type C. rodentium(Φstx2dact) and mutant derivatives after 4h of mitomycin C induction. Given that mitomycin C-induced Φstx2dact functions may be involved in the release of toxin from the bacterial host [27], we assessed toxin in cell pellets and in culture supernatants separately. As previously observed [52], mitomycin C induction resulted in a more than 100-fold increase of Stx2dact in culture supernatants (Fig 2B, “WT"). A nearly equivalent amount of toxin remained associated with the bacterial cell pellet, suggesting that under these conditions, a significant fraction of bacteria remained unlysed. Culture supernatants or cell pellets of the C. rodentiumΔrpoS (Φstx2dact) mutant predicted to be defective in the bacterial stress response, or the C. rodentiumΔqseC(Φstx2dact) and C. rodentiumΔqseF(Φstx2dact) mutants defective for quorum sensing, showed wild type levels of Stx2dact (S8 Fig), indicating that neither the bacterial stress response nor the QseC- or QseF-mediated quorum responses were required for toxin production. Culture supernatants of C. rodentium(Φstx2dactΔint) and C. rodentium(Φstx2dactΔB), which showed no defect in basal levels of toxin production (Fig 2A), also contained amounts of cell-associated toxin and supernatant-associated Stx2dact indistinguishable from wild type (Fig 2B,”Δint” and” ΔB”), despite the lack of prophage excision and/or phage production in these mutant strains. The ΔSR lysogen, defective for phage endolytic functions, produced wild type levels of cell-associated Stx2dact at 4 h post-induction, but supernatant-associated toxin was approximately ten-fold lower than wild type levels (Fig 2B, “ΔSR”). This difference is consistent with a defect in bacterial lysis and Stx2dact release, but did not reach statistical significance. In addition, by 16 h post-induction of the ΔSR lysogen, Stx2dact was detected in supernatants at levels similar to that of the WT strain (Fig 2C), suggesting that any defect in R and S proteins results in a delay rather than an absolute block in toxin release. Finally, however, deficiency in the RecA or Q proteins was associated with a near-complete absence of Stx2dact in cell supernatants (Fig 2B,”ΔrecA” and” ΔQ”), reinforcing the notion that these proteins, which are required for the SOS response and/or transcription of the stx2dact genes ([81] [71]), are essential for large amounts of Stx2dact production.
C. rodentium(Φstx2dact) undergoes lytic induction during murine infection
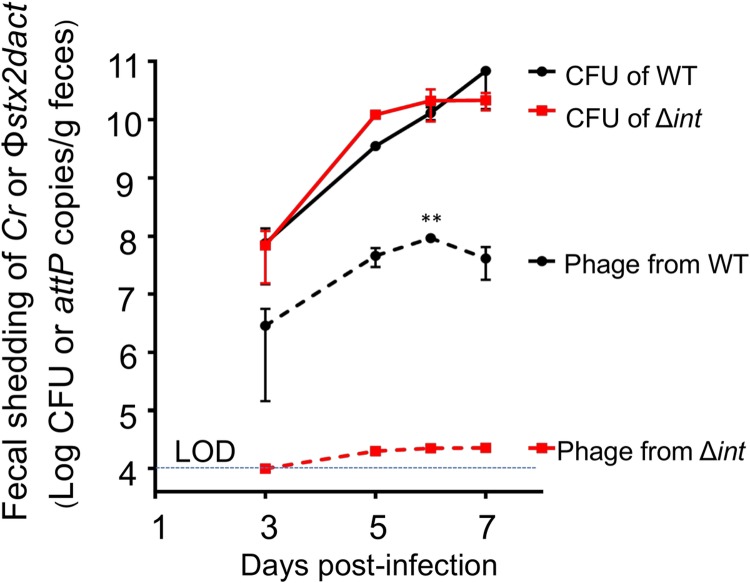
Stx-encoding prophages undergo lytic induction during EHEC infection of germ-free or antibiotic-treated mice [40, 41, 69], and our comprehensive survey of prophage integration sites in fecal microbiota (Table 2) indicated that C. rodentium(Φstx2dact) undergoes some degree of lytic induction during infection of conventional mice. To assess this induction further, we infected conventionally raised C57BL/6 mice with C. rodentium(Φstx2dact) by oral gavage and measured fecal shedding of both the infecting strain, by plating for CFU, and Φstx2dact, by quantitating attP (non-integrated phage) copies by qPCR. As previously observed, by day 3 post-infection, C. rodentium(Φstx2dact) was detected in the stool at 8 x 107 per gram, and reached 9 x 1010 per gram by day 6 post-infection ([82]; Fig 3, “CFU of WT"). Further, murine infection by this strain was indeed associated with lytic induction, as excised phage genomes were detected in stool at all time points (Fig 3, “Phage from WT”).
Fig 3. C. rodentium(Φstx2dact) undergoes lytic induction during murine infection.
Eight-week old female C57BL/6 mice were infected by oral gavage with C. rodentium(Φstx2dact) or C. rodentium(Φstx2dact Δint). At the indicated time points, attP copies, reflecting excised prophages, and viable bacteria were determined by qPCR or plating for CFU, respectively (see Materials and Methods). Shown are averages ± SEM of 5 mice per group of a representative of two experiments. Level of detection of attP was 1 x 104 copies/g feces. Asterisks (**) indicate significance differences (p <0.01) between the WT and C. rodentium (Φstx2dact Δint) calculated using 2-way ANOVA followed by Bonferroni post tests.
Interestingly, given the relatively high phage production by induced C. rodentium(Φstx2dact) in vitro, the amount of phage detected in stool was quite low. At day 3 post-infection, 5 x 106 attP copies were detected per gram of stool, a value 16-fold lower than the concentration of viable C. rodentium(Φstx2dact) in stool at that time point. By day 6 post-infection, attP copies had increased to 5 x 107 per gram of feces, but were approximately 600-fold lower than the fecal bacterial counts. These results indicate that C. rodentium(Φstx2dact) thus undergoes lytic induction and growth in this murine model, although not to the degree seen upon induction in vitro.
Lethal disease in mice correlates with the ability to produce Stx2dact but not with the ability to produce phage
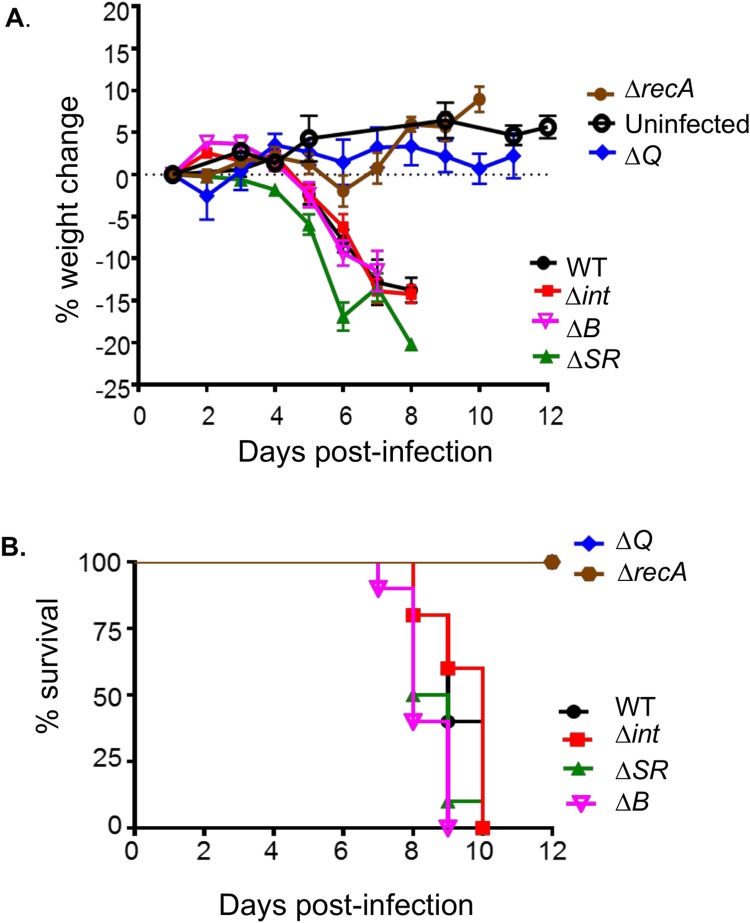
To test the importance of SOS induction and phage functions on disease in our microbiota-replete model of infection, we infected C57BL/6 mice with C. rodentium(Φstx2dact) and mutant derivatives by oral gavage. The wild type and the mutant lysogens colonized mice similarly, although the ΔB and Δint mutant lysogens appeared to colonize at somewhat higher levels (S9 Fig). C. rodentium ΔrecA(Φstx2dact) and C. rodentium(Φstx2dactΔQ), the two mutant lysogens that displayed dramatic defects in basal and mitomycin C-induced levels of Stx2dact in vitro, were the only ones incapable of causing sickness or death (Fig 4,”ΔrecA” and”ΔQ”), supporting the hypothesis that induction of an SOS response and the subsequent expression of phage late genes, including stx genes, are required for Shiga toxin production during infection of a microbiota-replete host.
Fig 4. Lethal disease in mice correlates with the ability to produce Stx2dact but not with the ability to produce phage.
Eight-week old female C57BL/6 mice were infected by oral gavage with the indicated lysogens. A. Percentage weight change was determined at indicated post-infection time. Data shown are averages ± SEM of 10 mice per group. Asterisks (*, **) indicate significance (p <0.05, <0.01) determined by 2-way ANOVA followed by Bonferroni post tests. B. Percent survival at the indicated post-infection time was monitored in 10 mice per group. Data represent cumulative results of 3 separate experiments.
The RpoS-deficient and QseC-deficient C. rodentium(Φstx2dact) mutants that are compromised in bacterial stress and quorum-sensing responses, respectively, retained the ability to cause weight loss and lethality with kinetics that were indistinguishable from that of WT C. rodentium(Φstx2dact) (S10 Fig). Thus, although previous results indicated that some quorum sensing mutants display diminished virulence during infection by non-Stx-producing C. rodentium [83], our results are consistent with the the ability of these strains to produce wild type levels of Stx2dact after SOS induction (S8 Fig). In addition, the lack of endolysins that appeared to somewhat delay release of Stx2dact into supernatants by C. rodentium (Φstx2dactΔSR) was not reflected by any delay in the kinetics of weight loss or lethality in infected mice (Fig 3,”ΔSR”), consistent with the ability of this strain to produce wild type levels of Stx2dact upon extended culture in vitro. Thus, C. rodentium (Φstx2dactΔSR) is capable of triggering Stx2dact–mediated disease in the absence of phage-induced lysis.
Finally, the production of intact phage is not essential to disease in this model. C. rodentium (Φstx2dact-ΔB), which is unable to generate intact phage in vitro, and C. rodentium(Φstx2dactΔint), which can neither generate excised phage genomes in vitro or in vivo, both retained full virulence in this model. We conclude that in this microbiota-replete model of EHEC infection, disease progression correlates exclusively with the ability to produce Stx2dact, regardless of the lysogen’s ability to amplify the stx2 genes by phage excision and genome amplification, or by the production of phage that are capable of secondary infection of commensal bacteria.
Discussion
Commensal organisms have the potential to suppress or enhance phage induction and Stx production. Although a role for induction of stx-encoding prophages in the production of Stx and serious disease during animal infection has been well documented in antibiotic-treated and germ-free mice [40, 41, 69], we used a murine model of EHEC infection that features an intact microbiome.
To investigate phage functions required for C. rodentium(Φstx2dact) to produce Stx and cause disease in conventional mice, we first characterized prophage genetic structure. Φstx2dact prophage was integrated into the C. rodentium dusA gene, an integration site utilized by prophages in over 200 bacterial species [63]. Although the orientation of the regulatory and late genes within the Φstx2dact prophage is noncanonical with respect to attL and attR (with int adjacent to attL; Fig 1), this orientation has been previously observed in at least one other lambdoid phage. In addition, Φstx2dact genes encoding several key phage proteins were identified by homology, and their inactivation had the predicted effects on phage development and production (Table 3; [77]). For example, antiterminator Q and integrase were required for phage production, as measured by detection of attP, and portal protein B was required for packaging of phage DNA into DNAse-resistant virions.
Stx production in vitro by the prophage mutants, as well as by a host recA mutant, confirmed that prophage induction, i.e., the SOS-dependent process required to initiate a temporal program of phage gene expression that normally leads to phage lytic growth, is essential for high-level Stx2 production in vitro. Mitomycin C treatment of C. rodentium(Φstx2dact) resulted in a greater than 100-fold increase in Stx2dact in culture supernatants, similar to the mitomycin C-mediated increase in Shiga toxin production by EHEC ([41]; Fig 2). Three signaling pathways, mediated by RpoS, QseC, and QseF, previously demonstrated to influence SOS induction of EHEC in vitro, had no effect on Stx2dact production by C. rodentium (Φstx2dact). In contrast, and as expected, RecA, required for mounting an SOS response, was necessary for this enhanced production of Stx2dact (Fig 2). It was previously shown that inactivation of the EHEC prophage repressor CI, a key step in the SOS response, is required for the increase in EHEC Stx production upon mitomycin C induction in vitro [41].
Despite the previous observation that the increase in phage genome copy number plays the most quantitatively important role in mitomycin C-enhanced Stx1 production by Stx phage H-19B [27], we found that integrase-deficient C. rodentium(Φstx2dact), which is deficient in phage excision and replication (Table 3; [76]), produced levels of Stx2dact indistinguishable from wild type (Fig 2). Apparently, enhanced expression of late genes stx2dactA and stx2dactB still occurs in the absence of integrase and is sufficient for wild type levels of Stx2dact production. As expected, antiterminator protein Q, required for the transcription of late genes including stx, was essential for Stx2dact production by C. rodentium (Φstx2dact), consistent with previous findings for the Stx2 phage Φ361 [26]. Finally, the S endolysin of Stx phage H-19B was previously shown to promote the timely release of toxin after mitomycin C induction [27]; we found that deficiency of the RS endolysins encoded by Φstx2dact appeared to diminish the release of Stx2dact into culture supernatants at 4 hours post-induction (Fig 2B). However, the decrease was not statistically significant, and RS-deficiency had no discernible effect on toxin release by 16 hours (Fig 2C). Dead and dying E. coli cells are known to release their contents into the surroundings at the end of stationary phase [84]; additionally, E. coli O157:H7 has been shown to release Shiga toxin via outer membrane vesicles [85].
Whereas previous work in streptomycin-treated or gnotobiotic murine models has demonstrated that induction of the lytic developmental program of Stx phage occurs during infection and is required for disease [40, 41, 69], we document here that prophage induction occurs during infection of mice with intact microbiota. attP sequences (indicative of excised, uningegrated phage genomes) were detected in the feces of infected mice, as revealed by deep sequencing (Table 3), or by qPCR (Fig 3).
Deep sequencing of phage genomes in the stool of mice revealed no evidence of Φstx2dact lysogeny of commensal bacteria during C. rodentium(Φstx2dact) murine infection, suggesting that secondary infection of commensals by this phage is rare. Furthermore, C. rodentium(Φstx2dact) mutants deficient in phage integrase or portal protein B, which retained the ability to produce Stx2dact, but were incapable of generating phage or infecting commensal bacteria, caused weight loss and lethality of mice with kinetics indistinguishable from wild type C. rodentium(Φstx2dact) (Fig 4). Indeed, the only C. rodentium(Φstx2dact) derivatives incapable causing disease in animals were those with a demonstrated defect in the production of Stx2dact in vitro (Table 3 and Fig 4). For example, RecA, essential for the initiation of the SOS response that leads to prophage induction, was required for lethality after oral inoculation of C. rodentium (Φstx2dact), consistent with the previous finding that RecA was required for lethality following intravenous EHEC infection of conventional mice [74]. We conclude that amplification of Stx2dact production by successive rounds of lytic infection of commensal bacteria, as has been postulated [38, 44, 45, 47], is not required for toxin-mediated disease in this microbiota-replete model.
We detected more than 1 x 109 phage/ml in uninduced mid-log cultures, suggesting that there is a high level of spontaneous induction under in vitro culture conditions. In contrast, despite severe Stx2dact-mediated disease manifestations during productive infection by C. rodentium(Φstx2dact), the number of attP sequences detected in feces was extremely low, suggesting that the level of prophage induction during infection may also be low. On day 6 post-infection, only 0.44% of all phage genomes detected were excised, compared to 99.66% that were integrated, reflecting intact prophage (Table 3). Depending on the day post-infection, excised phage detected by qPCR numbered 20- to 1000-fold fewer than viable C. rodentium(Φstx2dact) cells (Fig 3). Notably, previous work using a genetic reporter to indicate activation of lytic promoters of EHEC Stx phage 933W showed that the intestinal environment of a gnotobiotic mouse was strongly inducing [41]. While we cannot rule out the possibility that the low number of Φstx2dact attP sequences detected in feces reflects an instability of phage particles or some other factor in the intestinal milieu, our findings are consistent with the possibility that a low rate of Φstx2dact induction may be sufficient to promote disease in this model. Given that the methods to measure phage particles utilized in this study can be applied to patient samples, future studies will focus on the extent of lytic induction of Stx phage during human infection, and how it may correlate with disease outcome.
Materials and methods
Ethics statement
Mice were purchased from Jackson Laboratories and maintained in the Tufts University animal facility. All procedures were performed in compliance with Tufts University IACUC protocol B2014-87. If examination revealed signs of suffering, manifested by greatly diminished activity, poor grooming/appearance, biting, greatly increased respiratory rate or diminished appetite, or weight loss greater than 15% of body weight, then the animal was euthanized. Primary euthanasia method: CO2 asphyxiation or CO2 followed by cardiac stick. Secondary euthanasia method. Cervical dislocation, decapitation, thoracotomy or major organ removal is performed following the primary method."
Bacterial strains and plasmids
Strains and plasmids used in this study are listed in Table 1.
Phage Φstx2dact whole genome sequencing, assembly, and integration site determination
Genomic DNA was isolated from 5 ml of strain C. rodentium(Φstx2dact::kanR) (Table 1) grown overnight at 37°C in LB broth containing chloramphenicol (12.5 μg/ml) and kanamycin (25 μg/ml). DNA was extracted using a DNeasy kit (Qiagen), according to the manufacturer’s protocol for Gram negative bacteria. A library of this DNA was then constructed for Illumina sequencing using Illumina TruSeq DNA Sample Preparation Kit per the manufacturer’s instructions. Following sequencing, the bacterial genome was assembled de novo into 1500 contigs using assemblers ABySS [86], and Edena [87]. The Bowtie2 program [88] was then used to map the stx2 gene against this assembled genome and the contig containing this gene was identified. When aligned to the C. rodentium genome, a 69594-bp contig revealed a 47,343 bp prophage containing the stx2 gene and other phage lambda-like gene sequences inserted into the host dusA gene. (Although the C. rodentium dusA gene is interrupted by the prophage genome, a potentially functional dusA gene is reconstituted at the attL bacterial/phage DNA junction by fusion with a prophage-derived open reading frame that we term “ΦdusA’” in Fig 1.) The prophage sequence was deposited in GenBank as Φ1720a-02, accession number KF030445.1.
Integration of the prophage in both C. rodentium(Φstx2dact) and C. rodentium (ΦΔstx2dact::kanR) into the host dusA gene was verified by PCR amplification of the attL and attR phage-host junctions using primers DusF/PhageR and DusR/PhageF, respectively (Table 4), then DNA sequencing of the amplified junctions. Subsequent whole genome sequencing of C. rodentium(Φstx2dact) and C. rodentium(ΦΔstx2dact::kanR) showed that, except for the Φstx2dact prophage sequences, they are identical to C. rodentium ICC 168, also known as strain DBS100 (GenBank accession number NC_013716.1), and to each other. The encoded Φstx2dact prophage sequences were identical except for the presence of the kanR gene in stxA of strain C. rodentium (ΦΔstx2dact::kanR) (S1 Fig) flanked by the sequence TCCCCGGGTCATTATTCCCT CCAGGTA upstream of the kanR gene and the sequence CTTATTCCTCCTAGTTAGTCACCCGGGA downstream of the kanR gene.
Table 4. Primers used in this study.
| Primer | --------> |
|---|---|
| Primers for Mutant Construction and Validation | |
| Cr (ΦΔ SR) F | ATCGGTGTGTGCCGGTGGTCTTTATATTGTTGTGAGCTTCC GGATTGCGGGAGACGGGGTGGTCATGATCAGCACGTGTT GACAATTAATCATCGG |
| Cr (ΦΔ SR) R | CAGCCCATAACAGACAGACGATGATGCAGATAACCAGAG CGTAAATAATCGCGGTTACTCTTCTCAGTCCTGCTCCTCG GCCACGAAGTGCACGCAG |
| Cr (ΦΔ SR) validation F | CAACGAGAAAATCCCATGTCAGAAATTACATCCCTGGTC |
| Cr (ΦΔ SR) validation R | CTCATCAGCTTACTCTCCCCGCGCCGC |
| Cr (ΦΔ int) F | CGTTAGGTTCCCGCACAGGTTCCCACGTTTTATGGGAACC CGAAATAACGAGGTCGTGTAGGTCATGATCAGCACGTGTT GACAATTAATCATCGG |
| Cr (ΦΔ int) R | ATACTGTGTTTGTATACAGTATCATTTTTAACTGTATGGATA AACAGTGTCAGTCCTGCTCCTCGGCCACGAAGTGCACGC AG |
| Cr (ΦΔ int) validation F | GGGAACCCGAAATAACGAGGTCGTGTA |
| Cr (ΦΔ int) validation R | CATTTTTAACTGTATGGATAAACAGTG |
| Cr (ΦΔ Q) F | AGTAACCACTCTTAACATACTGACATACTTTTTGCGGACC GCGCTAATCATTTTGGTCATGATCAGCACGTGTTGACAATT AATCATCGG |
| Cr (ΦΔ Q) R | CGTTTTATCGATCGCGCGCTGGCGATTGGTGTGCTGTCCT GATTTTGTGGAGAAAGTTGTCAGTCCTGCTCCTCGGCCAC GAAGTGCACGCAG |
| Cr (ΦΔ Q)500bpextn F | ACCAGCCGCCCATTTACCAC |
| Cr (ΦΔ Q)500bpextn R | CCGGAAAGTGCAGCCCGTAAG |
| Cr (ΦΔ Q) validation F | TGCGGACCGCGCTAATCATTTT |
| Cr (ΦΔ Q) validation R | CCTGATTTTGTGGAGAAAGTTG |
| Q100 R | CGGATACCGTGGCATTTGA |
| Cr (ΦΔ B) F | GCCGCGATGGTGAGCCGCAGGCGGGGAAAACCGGGATT TAAACTGGCGAGGTTTTAGGTCATGATCAGCACGTGTTGA CAATTAATCATCGG |
| Cr (ΦΔ B) R | TCGTCATAAATATAAATATCCGCGTCACCCGGCCCCCCA GCCTGCATCCTGAACCAGGATTCAGTCCTGCTCCTCGGC CACGAAGTGCACGCAG |
| Cr (ΦΔ B) validation F | ACCGGGATTTAAACTGGCGAGGTTTTA |
| Cr (ΦΔ B) validation R | CCCCCCAGCCTGCATCCTGAACCAGGAT |
| Cr (Φ)Δ recA F | AATTGCTTCAACAGTACAGAATTCACTATCCGGATAAGCG CAAGCGGAACCCGGCATGACAGGAGTAGTTAGGTCATGA TCAGCACGTGTTGACAATTAATCATCGG |
| Cr (Φ)Δ recA R | ACCCTGAGTTGTAACTTACCTTCTTGCCGGACGGCAGCTT TGCGCCATCCGGCTTGCGGTTACCTGAAAATCAGTCCTG CTCCTCGGCCACGAAGTGCACGCAG |
| Cr (Φ)Δ recA validation F | ACTGTATGAGCATACAGTAT |
| Cr (Φ)Δ recA validation R | GCAAAAGGGCCGCATAAGCG |
| Cr(Φ)Δ qseC F | CTGGGCAGCGATTTTATTCGTACCGTTCACGGCATCGGCT ATACCCTTAGCGAGGCATAAAAGGTCATGATCAGCACGTG TTGACAATTAATCATCGG |
| Cr(Φ)Δ qseC validation F | ACGCCGTTGAGGTTCACGTCC |
| Cr(Φ)Δ qseC validation R | GCAAAATGCGTTTGAGGCT |
| Δ ROD24971 F | GTGTTCCTGTTTTAGTCGCGTAACCGGTTGCTAACCGTATC ATATCTTGCGGTATGTTGCGGAGGGTCATGATCAGCACGT GTTGACAATTAATCATCGG |
| Δ ROD24971 R | ACACGCCTGACGCGATACACGGTGATGACCACCCCGCCG CGCCGGTATCGCCTGACGAAGAGGTATCTCAGTCCTGCTC CTCGGCCACGAAGTGCACGCAG |
| Δ ROD24971 validation F | GGTTTAATAATCGCATCAATC |
| Δ ROD24971 validation R | CGTAAGCCAGGCGGGAGCTAC |
| Cr (Φ)Δ rpoS F | CGCAGCGATAAATCGACGGAGCAGGCTGACACGGGCTTG TTTTGTCAAGGGATCACGGGTAGGAGCCACCTTGGTCATG ATCAGCACGTGTTGACAATTAATCATCGG |
| Cr (Φ)Δ rpoS R | AGCGGGCAATAATGCAGCCAAAGAAAAAGACCAGCCTCAC AGAGACTGGTCTTTTCTGATGGAACGGTGCTCAGTCCTGC TCCTCGGCCACGAAGTGCACGCAG |
| Cr (Φ)Δ rpoS validation F | ATAGCGACTATGGGTAGCAC |
| Cr (Φ)Δ rpoS validation R | CCCGCCAGATCTGATAAGCG |
| PCR and qPCR Primers | |
| AttP F | CTTTGGATAGGTTCCCAATAGGC |
| AttP R | GGGTTCCCATAAAACGTGGG |
| RecA F | CGCTGACGTTACAGGTGATCGC |
| RecA R | CCATAGAGGATCTGGAACTCGG |
| Dus F | CCTTCGGGCTAAGCCCGG |
| Dus R | GCGCCGTCCACGCGAGG |
| Phage F | GTGACCAAGGCGTACCTGGC |
| Phage R | CCATCACTTTCTGTGTGCCCC |
| Primers for Construction of Sequencing Library | |
| PCR Primer 1 | TTGCTTTCCCTGTAAGTGATAACACC |
| PCR Primer 2 | GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGGGGG GGGGGGGGGGG |
| PCR Primer 3 | AATGATACGGCGACCACCGAGATCTACACTCTTTTTTACTG GAATTCTCGGTTTAGCATTGCTCCT |
| PCR Primer 4 | CAAGCAGAAGACGGCATACGAGATTAAGGCGAGTGACTGG AGTTCAGACGTGTGCTCTTCCGATCT |
| Seq-P | ATCTACACTCTTTTTTACTGGAATTCTCGGTTTAGCATTGCT Cct |
Cr = Citrobacter rodentium
Phage Φstx2dact genome annotation
The Φstx2dact genome sequence was first annotated using the program RAST (http://rast.nmpdr.org/ [89]). The annotation was further refined by analyzing each open reading frame using the NCBI program MEGABLAST against the GenBank nucleotide database. Note that although the insertion of the marker into the Rz, gene affects lysis by phage λ lysogens in the presence of high magnesium, this gene has been altered in other studies of Stx phage [66] and in this study, lysis of C. rodentium (Φstx2dact) occurred upon in vitro induction (S4 Fig).
Characterization of phage and prophage sequences in murine stool by massively parallel sequencing and analysis
DNA was extracted from fecal samples of 5 infected sick mice at 6 days post-infection, according to the method of Yang et al. [90]. Twenty mg stool samples were suspended in 5 ml PBS, pH7.2, and centrifuged at 100 × g for 15 min at 4°C. The supernatant was centrifuged at 13,000 × g for 10 min at 4°C, and the resulting pellet was washed 3 times in 1.5 ml acetone, centrifuging at 13,000 × g for 10 min at 4°C after each wash step. Two hundred μl of 5% Chelex-100 (Bio-Rad) and 0.2 mg proteinase K were added to the pellet and the sample was incubated for 30 min at 56°C. After vortexing briefly, the sample was centrifuged at 10,000 × g for 5 min and the supernatant containing the DNA was harvested and stored.
To characterize bacteria that harbor the Φstx2dact prophage, we sequenced the bacterial bacterial-host attL prophage junction and adjacent bacterial DNA by following, with slight modifications, the methodology of Klein et al. [70] for constructing high-throughput sequencing libraries that contain a repetitive element (in this case, the phage int (integrase) gene). Briefly, genomic DNA was sheared by sonication to a size of 100–600 bp, followed by addition of ~20 deoxycytidine nucleotides to the 3’ ends of all molecules using Terminal deoxynucleotidyl Transferase. Two rounds of PCR using a poly-C-specific and phage int gene-specific primer pair (PCR primers 1 and 2, Table 4) were used to amplify attL and to add on sequences necessary for high-throughput sequencing (PCR primers 3 and 4, Table 4).
Amplicons were sequenced using the MiSeq desktop sequencer (Ilumina) and primer Seq-P (Table 4), providing reads of up to 300 bp. As amplicons spanned the region from the phage int gene, through attL, and into the adjacent host genome (see Fig 1), reads of this length were required. 17,868,095 sequences encompassing 5 Gb were downloaded to the Galaxy server (https://usegalaxy.org/) and analyzed (Table 3). We first excluded sequences that clearly reflected attL (i.e., contained the 184 bp of ΦdusA’ followed by C. rodentium dusA), indicating the prophage inserted into the C. rodentium genome. Of the remaining 801,959 sequences, 75,962 (0.44% of the total) encoded the intact attP site, implying that they were circular. These latter sequences presumably reflected excised circular phage genomes, possibly undergoing early theta DNA replication, ultimately leading to phage production. The remaining 725,997 sequences encoded only strings of A’s and/or C’s, and were eliminated from consideration.
Generation of C. rodentium(Φstx2dact) deletion constructs
Deletion mutants of C. rodentium(Φstx2dact) in the prophage or the host genome were generated using a modified version of a one-step PCR-based gene inactivation protocol [61, 82]. Briefly, a PCR product of the zeocin-resistance gene and its promoter region flanked by 70–500 bp homology of the region upstream and downstream of the targeted gene was generated using the primers listed in Table 4. The chromosomal DNA served as template when the flanking regions were 500 bp in length on either side of the zeocin cassette. The PCR product was electroporated into competent C. rodentium(Φstx2dact) cells containing the lambda red plasmid pKD46 and recombinants were selected on plates containing chloramphenicol and zeocin (75 μg/ml). Replacement of the gene of interest with the zeocin resistance cassette was confirmed using specific primers (Table 4). At least two independent clones, validated using PCR, were obtained and subsequently analyzed.
To complement C. rodentium(Φstx2dactΔQ), the only phage mutant with a defect in Stx2dact production, the region encoding the anti-terminator Q was amplified from WT genomic DNA using primers Cr (ΦΔQ) validation F and Q100 R (Table 4), cloned into the pCR4-TOPO vector and transformed into Top 10 cells using the TOPO TA cloning kit (Life Technologies). Kanamycin-resistant colonies were screened for the presence of vector carrying the Q gene (pTOPO-Q). pTOPO-Q was then transformed into electrocompetent wild type C. rodentium(Φstx2dact) or C. rodentium(Φstx2dact ΔQ), using standard cloning techniques.
Quantification of Stx2dact produced in vitro
Overnight 37°C cultures of C. rodentium(Φstx2dact) or deletion derivatives were diluted 1:25 into 10 ml of fresh medium with appropriate antibiotics. Two independently derived clones for each mutant were tested, with indistinguishable results. The cultures were grown at 37°C with aeration to an OD600 of 0.4, and one ml of each culture was set aside (Table 3, “t = 0h”). The remaining culture was split into 2 cultures. These cultures were grown for a further 4 hours (Table 3, “t = 4h”) in the absence or presence of 0.25 μg/ml mitomycin C. (We first measured phage and Stx2 production at various times post-induction and found the 4-hour time point to be optimal for obtaining maximal phage and Stx2 following mitomycin C induction). Supernatants depleted of intact bacteria were then harvested by centrifugation at 17,800 × g for 5 minutes at room temperature. For C. rodentium(Φstx2dact) and C. rodentium (Φstx2dactΔSR), a portion of each culture was also collected after ~16 h of incubation (“t = 16h”). Supernatants and pellets were quantitated for Stx2dact by ELISA, as described previously [52].
Quantification of phage genomes by qPCR
Attempts to quantitate phage using plaque titers were unsuccessful. Attempts included the use of various host strains, including C. rodentium non-lysogens, E. coli K12 strains Epi300, LE392, or DH5α, E. coli OP50, and Shigella. Plate modifications included the addition of subinhibitory concentrations of antibiotics, addition of 10 mM CaCl2 or MgCl2 or both, addition of 5% glycerol to bottom agar, addition of tetrazolium to bottom agar, or Sybr staining and fluorescence microscopy of phage. Instead, excised phage genomes in cell supernatants were quantitated by qPCR. Protease digestion of capsids prior to qPCR quantitation was not required, as capsid undergoes melting during the high heating steps of the PCR procedure [75]. Supernatants were serially diluted 1:10, 1:100 and 1:1000 in distilled water. Separate reactions using two μl of the various dilutions as a template were carried out in duplicate. qPCR master-mix (Bio-Rad) was prepared according to the manufacturer’s instructions, using the attP primer set (Table 4) to detect copies of excised phage DNA. Results were compared to a standard curve, derived from a known concentration of a template fragments generated from amplifying C. rodentium(Φstx2dact) DNA using attP primers. The template was serially diluted, in duplicate, to detect copy numbers ranging from 1010 to 102. qPCR reactions were carried out as follows: 95°C for 3 min, followed by 35 cycles of 95°C for 1 min, 58°C for 30 sec, and 72°C for 1 min. Phage genomes in the supernatant of C. rodentium (Φstx2dactΔB), which lacks the portal protein required for genome packaging, was diminished 18-fold by DNAse treatment, supporting our method of qPCR quantitation of phage (see Table 3).
Mouse infection studies
Mice were purchased from Jackson Laboratories and maintained in the Tufts University animal facility. Seven to eight-week-old female C57BL/6J mice were gavaged with PBS or ∼5×108 CFU of overnight culture of C. rodentium(Φstx2dact) or deletion derivatives in 100 μl PBS. Inoculum concentrations were confirmed by serial dilution plating. Fecal shedding was determined by plating dilutions of fecal slurry on either chloramphenicol, to detect wild type C. rodentium(Φstx2dact), or chloramphenicol-zeocin plates, to detect deletion derivatives marked with a zeomycin resistance gene [52]. Body weights were monitored daily, and mice were euthanized upon losing >15% of their body weight.
DNA from infected mouse fecal pellets was isolated using the QIAGEN DNeasy Blood and Tissue kit with modifications. Fecal pellets were incubated with buffer ATL and proteinase K overnight at 55°C. Buffer AL was added, and after mixing, pellets were further incubated at 56°C for 1 h. Pellet mixtures were then centrifuged at 8000 rpm for 1 min and the pellets were discarded. Ethanol was added to the supernatants, which were processed according to the manufacturer’s protocol. DNA concentrations were determined using a NanoDrop spectrophotometer. qPCR was performed as described above.
Statistical tests
Data were analyzed using GraphPad Prism software. Comparison of multiple groups were performed using the Kruskal-Wallis test with Dunn's multiple comparison post-test, or 2-way ANOVA with Bonferroni’s post-tests. In all tests, P values below 0.05 were considered statistically significant. Data represent the mean ± SEM in all graphs.
Supporting information
The 47,239 bp prophage DNA sequence (gray), flanked by attL and attR upon insertion into C. rodentium dusA sequence (blue, “Cr dusA”), was determined by whole genome shotgun sequencing of C. rodentium (Φstx2dact::kanR) and annotated, as described in Materials and Methods. Names of encoded proteins are shown. Unannotated ORFs indicate hypothetical proteins. At the far left end is a phage sequence that encodes the N-terminal 112 amino acids of an open reading frame (“ΦdusA’”) in the same reading frame as the 3’ end of the C. rodentium dusA gene. Strain C. rodentium(Φstx2dact) encodes a chloramphenicol acetyl transferase protein (“cat”) inserted into the prophage Rz gene. The sequence of C. rodentium(Φstx2dact::kanR) is identical to C. rodentium (Φstx2dact) except that the gene encoding the A subunit of Stx2dact (“Stx2A”) contains an 894 bp insertion encoding kanamycin resistance (“kan”), plus an additional 27 bp upstream and 28 bp downstream. Prophage genes studied in this work are shown in bold. Cr: C. rodentium.
(TIFF)
The location of the host integration site and genome size for each prophage is indicated in parentheses after the phage name. Open reading frames of prophages λ, 933W, and Sp5 [28, 91, 92] are shown in comparison to those of prophage 1720a-02, and are depicted as arrows pointing in the direction of transcription. The site of insertion of the chloramphenicol cassette (cat) in phage 1720a-02 is indicated with an open triangle. The dotted blue line indicates the presence of additional prophage genes. Genes and genomes are not drawn to scale.
(TIF)
The sequence of phage Φstx2dact was used as a query to interrogate the C. rodentium DBS100 genome (i.e., the parent of the Φstx2dact lysogen) for regions of homology, using the program Megablast (NCBI). Two regions of homology, each to a different endogenous C. rodentium prophage, were identified. A. Region of homology between Φstx2dact and a hypothetical protein. B. Region of homology encompassing a gene encoding a hypothetical protein (upstream of cro), the cro gene (demarcated by red arrows), and a large portion of the cI gene, (demarcated by a blue arrow). Black arrows indicate the direction of transcription.
(TIFF)
Cultures were grown in LB medium to OD600 = 0.4 (T = 0), then each was divided into two cultures. One culture was induced with mitomycin C (0.25 μg/ml) and the other was left uninduced. OD600 culture readings were followed for 4 hours. Two independent isolates of strain C. rodentium (Φstx2dactΔQ) (“ΔQ”), both unable to produce large bursts of phage on induction, were used as the control. Black arrow indicates time at which mitomycin C was added.
(TIFF)
Lysogens of strain DH5α were obtained by infecting a log phase culture with Φstx2dact at high multiplicity of infection, according to the method of Ray and Sakalka [93], and lysogens were isolated by selecting for kanamycin-resistant survivors. A. Agarose gel of PCR analysis showing that a putative DH5α(Φstx2dact) lysogen and C. rodentium control lysogens encode Φstx2dact genes SR, whereas a DH5α non-lysogen did not. B. Strains DH5α containing the recA-bearing plasmid pER271, and the same strain harboring the Φstx2dact prophage, were streaked on LB plates. One half of the plate was shielded with aluminum foil (-UV), while the unprotected half (+UV) was illuminated for 15 seconds using a UVP model UVGL-25 Mineralight UV lamp at 254 nm wavelength from a distance of 8 inches, then incubated overnight at 37°C in the dark.
(TIF)
The indicated wild type or mutant C. rodentium (Φstx2dact) strains were grown in LB broth or DMEM (Gibco, GlutaMAX) without antibiotics. Growth was measured over time by optical density (OD600), and growth curves are the average of duplicate samples. Doubling times were calculated based on the exponential growth regions of each curve. Representative results from one of two experiments are shown.
(TIF)
Q-deficient C. rodentium(Φstx2dactΔQ) (“ΔQ”), C. rodentium(Φstx2dactΔQ)/pTOPO-Q (“ΔQ+pQ”), wild type C. rodentium(Φstx2dact)/pTOPO-Q (““WT+pQ”) and wild type C. rodentium(Φstx2dact) (“WT") were grown to mid-log phase and cultured for four more hours either in the absence (“-“) or presence (“+”) of 0.25 μg/ml mitomycin C. Pellets (filled bars) or supernatants (open bars) were subjected to capture ELISA to determine the level of Stx2dact production. Quantities are expressed relative to the specific OD600 at t = 0h. Results are averages ± SEM of triplicate samples, and are a representative of one of two experiments. nd, not detected. Asterisks indicate Stx levels significantly (p<0.05) different from C. rodentium(Φstx2dactΔQ) calculated using Kruskal–Wallis one-way analysis of variance followed by Dunn's nonparametric comparison.
(TIFF)
The indicated lysogens were grown to mid-log phase (designated as t = 0h) and cultured for four more hours (t = 4h) either in the absence (“-“) or presence (“+”) of 0.25 μg/ml mitomycin C. Pellets (filled bars) or supernatants (open bars) were subjected to capture ELISA to determine the level of Stx2dact production. Quantities are expressed relative to the specific OD600 at t = 0h. Results are averages ± SEM of triplicate samples, and are a representative of at least two experiments. Stx levels of the C. rodentium qseC, qseF, or rpoS mutant strains were not significantly different from wild type C. rodentium(Φstx2dact), calculated using Kruskal–Wallis one-way analysis of variance followed by Dunn's multiple comparisons test.
(TIF)
Eight-week old female C57BL/6 mice were infected by oral gavage with the indicated lysogens. Fecal shedding of the lysogens was determined by plating for viable counts (see Materials and Methods). No significant differences were observed, as determined by 2-way ANOVA. A. Colonization of mice by wild type or recA- or prophage mutant lysogens. B. Colonization of mice by wild type or quorum-sensing mutant lysogens.
(TIF)
Eight-week old female C57BL/6 mice were infected by oral gavage with the indicated lysogens. A. Percentage weight change was determined at indicated post-infection time. Data shown are averages ± SEM of 10 mice per group. No significant differences were observed, as determined by 2-way ANOVA. B. Percent survival at the indicated post-infection time was monitored in 10 mice per group. Data represent cumulative results of 3 separate experiments.
(TIF)
Acknowledgments
We are grateful to Sara Roggensack for suggesting the PhageSeq methodology, David Lazinski for help with preparation of PhageSeq libraries, Martin Marinus for critical review of the manuscript, and Michael Pereira, Martin Marinus, and members of the Leong Lab for many useful suggestions.
Data Availability
All relevant data are within the paper and its Supporting Information files, except that prophage sequence data are available from GenBank (https://www.ncbi.nlm.nih.gov/genbank/), accession number KF030445.
Funding Statement
This work was supported by National Institute of Health (https://www.nih.gov/) grants R21AI107587 and 2R01AI046454 to JML. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Karmali M.A., Gannon V., and Sargeant J.M., Verocytotoxin-producing Escherichia coli (VTEC). Vet Microbiol, 2010. 140(3–4): p. 360–70. 10.1016/j.vetmic.2009.04.011 [DOI] [PubMed] [Google Scholar]
- 2.Kaper J.B., Nataro J.P., and Mobley H.L., Pathogenic Escherichia coli. Nat Rev Microbiol, 2004. 2(2): p. 123–40. 10.1038/nrmicro818 [DOI] [PubMed] [Google Scholar]
- 3.Pennington H., Escherichia coli O157. Lancet, 2010. 376(9750): p. 1428–35. 10.1016/S0140-6736(10)60963-4 [DOI] [PubMed] [Google Scholar]
- 4.Sperandio V. and Hovde C.J., eds. Enterohemorrhagic Escherichia coli and Other Shiga Toxin-Producing E. coli. 2015, ASM Press: Washington, D.C. [Google Scholar]
- 5.Karmali M.A., Host and pathogen determinants of verocytotoxin-producing Escherichia coli-associated hemolytic uremic syndrome. Kidney Int Suppl, 2009(112): p. S4–7. 10.1038/ki.2008.608 [DOI] [PubMed] [Google Scholar]
- 6.Tarr P.I., Gordon C.A., and Chandler W.L., Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet, 2005. 365(9464): p. 1073–86. 10.1016/S0140-6736(05)71144-2 [DOI] [PubMed] [Google Scholar]
- 7.Scheiring J., et al. , Outcome in patients with recurrent hemolytic uremic syndrome. Pediatric Transplantation, 2005. 9: p. 48–48. 10.1111/j.1399-3046.2005.00442.x [DOI] [PubMed] [Google Scholar]
- 8.Brady M.J., et al. , Enterohaemorrhagic and enteropathogenic Escherichia coli Tir proteins trigger a common Nck-independent actin assembly pathway. Cell Microbiol, 2007. 9(9): p. 2242–53. 10.1111/j.1462-5822.2007.00954.x [DOI] [PubMed] [Google Scholar]
- 9.Vingadassalom D., et al. , Insulin receptor tyrosine kinase substrate links the E. coli O157:H7 actin assembly effectors Tir and EspF(U) during pedestal formation. Proc Natl Acad Sci U S A, 2009. 106(16): p. 6754–9. 10.1073/pnas.0809131106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai Y., et al. , Intimate host attachment:e nteropathogenic and enterohaemorrhagic Escherichia coli. Cell Microbiol, 2013. 15(11): p. 1796–808. 10.1111/cmi.12179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Proulx F., Seidman E.G., and Karpman D., Pathogenesis of Shiga toxin-associated hemolytic uremic syndrome. Pediatr Res, 2001. 50(2): p. 163–71. 10.1203/00006450-200108000-00002 [DOI] [PubMed] [Google Scholar]
- 12.Thorpe C.M. and Acheson D.W., Testing of urinary Escherichia coli isolates for Shiga toxin production. Clin Infect Dis, 2001. 32(10): p. 1517–8. 10.1086/320173 [DOI] [PubMed] [Google Scholar]
- 13.Robinson C.M., et al. , Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc Natl Acad Sci U S A, 2006. 103(25): p. 9667–72. 10.1073/pnas.0602359103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obrig T.G., Escherichia coli Shiga Toxin Mechanisms of Action in Renal Disease. Toxins (Basel), 2010. 2(12): p. 2769–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melton-Celsa A., et al. , Pathogenesis of Shiga-toxin producing escherichia coli. Curr Top Microbiol Immunol, 2012. 357: p. 67–103. 10.1007/82_2011_176 [DOI] [PubMed] [Google Scholar]
- 16.Wadolkowski E.A., Burris J.A., and O'Brien A.D., Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect Immun, 1990. 58(8): p. 2438–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wadolkowski E.A., et al. , Acute renal tubular necrosis and death of mice orally infected with Escherichia coli strains that produce Shiga-like toxin type II. Infect Immun, 1990. 58(12): p. 3959–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keepers T.R., et al. , A murine model of HUS: Shiga toxin with lipopolysaccharide mimics the renal damage and physiologic response of human disease. J Am Soc Nephrol, 2006. 17(12): p. 3404–14. 10.1681/ASN.2006050419 [DOI] [PubMed] [Google Scholar]
- 19.Davis T.K., Van De Kar N.C., and Tarr P.I., Shiga Toxin/Verocytotoxin-Producing Escherichia coli Infections: Practical Clinical Perspectives. Microbiol Spectr, 2014. 2(4): p. EHEC-0025-2014. [DOI] [PubMed] [Google Scholar]
- 20.Melton-Celsa A.R. and O'Brien A.D., New Therapeutic Developments against Shiga Toxin-Producing Escherichia coli. Microbiol Spectr, 2014. 2(5). [DOI] [PubMed] [Google Scholar]
- 21.Freedman S.B., et al. , Shiga Toxin-Producing Escherichia coli Infection, Antibiotics, and Risk of Developing Hemolytic Uremic Syndrome: A Meta-analysis. Clin Infect Dis, 2016. 62(10): p. 1251–1258. 10.1093/cid/ciw099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizutani S., Nakazono N., and Sugino Y., The so-called chromosomal verotoxin genes are actually carried by defective prophages. DNA Res, 1999. 6(2): p. 141–3. [DOI] [PubMed] [Google Scholar]
- 23.Tyler J.S., Livny J., and Friedman D.I., Lambdoid Phages and Shiga Toxin, in Phages; Their role in Pathogenesis and Biotechnology., Waldor M.K., Friedman D.I., and Adhya S., Editors. 2005, ASM Press: Washington, D.C: p. 131–164. [Google Scholar]
- 24.Neely M.N. and Friedman D.I., Arrangement and functional identification of genes in the regulatory region of lambdoid phage H-19B, a carrier of a Shiga-like toxin. Gene, 1998. 223(1–2): p. 105–13. [DOI] [PubMed] [Google Scholar]
- 25.Neely M.N. and Friedman D.I., Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol, 1998. 28(6): p. 1255–67. [DOI] [PubMed] [Google Scholar]
- 26.Wagner P.L., et al. , Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J Bacteriol, 2001. 183(6): p. 2081–5. 10.1128/JB.183.6.2081-2085.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner P.L., et al. , Bacteriophage control of Shiga toxin 1 production and release by Escherichia coli. Mol Microbiol, 2002. 44(4): p. 957–70. [DOI] [PubMed] [Google Scholar]
- 28.Tyler J.S., Mills M.J., and Friedman D.I., The operator and early promoter region of the Shiga toxin type 2-encoding bacteriophage 933W and control of toxin expression. J Bacteriol, 2004. 186(22): p. 7670–9. 10.1128/JB.186.22.7670-7679.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casjens S.R. and Hendrix R.W., Bacteriophage lambda: Early pioneer and still relevant. Virology, 2015. 479–480: p. 310–30. 10.1016/j.virol.2015.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hughes D.T., et al. , The QseC adrenergic signaling cascade in Enterohemorrhagic E. coli (EHEC). PLoS Pathog, 2009. 5(8): p. e1000553 10.1371/journal.ppat.1000553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamovic L., et al. , Heterogeneity in phage induction enables the survival of the lysogenic population. Environ Microbiol, 2016. 18(3): p. 957–69. 10.1111/1462-2920.13151 [DOI] [PubMed] [Google Scholar]
- 32.Karch H., Strockbine N.A., and O'Brien A.D., Growth of Escherichia coli in the presence of trimethoprim-sulfamethoxazole facilitates detection of Shiga-like toxin producing strains by colony blot assay FEMS Microbiol Lett, 1986. 35(2–3): p. 141–145. [Google Scholar]
- 33.Walterspiel J.N., et al. , Effect of subinhibitory concentrations of antibiotics on extracellular Shiga-like toxin I. Infection, 1992. 20(1): p. 25–9. [DOI] [PubMed] [Google Scholar]
- 34.Matsushiro A., et al. , Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J Bacteriol, 1999. 181(7): p. 2257–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong C.S., et al. , The Risk of the Hemolytic–Uremic Syndrome after Antibiotic Treatment of Escherichia coli O157:H7 Infections. The New England Journal of Medicine, 2000. 342: p. 1930–1936. 10.1056/NEJM200006293422601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dundas S., et al. , Using antibiotics in suspected haemolytic-uraemic syndrome: antibiotics should not be used in Escherichia coli O157:H7 infection. BMJ, 2005. 330(7501): p. 1209; author reply 1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohawk K.L., et al. , Pathogenesis of Escherichia coli O157:H7 strain 86–24 following oral infection of BALB/c mice with an intact commensal flora. Microb Pathog, 2010. 48(3–4): p. 131–42. 10.1016/j.micpath.2010.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goswami K., et al. , Coculture of Escherichia coli O157:H7 with a Nonpathogenic E. coli Strain Increases Toxin Production and Virulence in a Germfree Mouse Model. Infect Immun, 2015. 83(11): p. 4185–93. 10.1128/IAI.00663-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taguchi H., et al. , Experimental infection of germ-free mice with hyper-toxigenic enterohaemorrhagic Escherichia coli O157:H7, strain 6. J Med Microbiol, 2002. 51(4): p. 336–43. 10.1099/0022-1317-51-4-336 [DOI] [PubMed] [Google Scholar]
- 40.Zhang X., et al. , Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis, 2000. 181(2): p. 664–70. 10.1086/315239 [DOI] [PubMed] [Google Scholar]
- 41.Tyler J.S., et al. , Prophage induction is enhanced and required for renal disease and lethality in an EHEC mouse model. PLoS Pathog, 2013. 9(3): p. e1003236 10.1371/journal.ppat.1003236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fiebiger U., Bereswill S., and Heimesaat M.M., Dissecting the Interplay Between Intestinal Microbiota and Host Immunity in Health and Disease: Lessons Learned from Germfree and Gnotobiotic Animal Models. Eur J Microbiol Immunol (Bp), 2016. 6(4): p. 253–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Myhal M.L., Laux D.C., and Cohen P.S., Relative colonizing abilities of human fecal and K 12 strains of Escherichia coli in the large intestines of streptomycin-treated mice. Eur J Clin Microbiol, 1982. 1(3): p. 186–92. [DOI] [PubMed] [Google Scholar]
- 44.Gamage S.D., et al. , Commensal bacteria influence Escherichia coli O157:H7 persistence and Shiga toxin production in the mouse intestine. Infect Immun, 2006. 74(3): p. 1977–83. 10.1128/IAI.74.3.1977-1983.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gamage S.D., et al. , Nonpathogenic Escherichia coli can contribute to the production of Shiga toxin. Infect Immun, 2003. 71(6): p. 3107–15. 10.1128/IAI.71.6.3107-3115.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamage S.D., et al. , Diversity and host range of Shiga toxin-encoding phage. Infect Immun, 2004. 72(12): p. 7131–9. 10.1128/IAI.72.12.7131-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iversen H., et al. , Commensal E. coli Stx2 lysogens produce high levels of phages after spontaneous prophage induction. Front Cell Infect Microbiol, 2015. 5: p. 5 10.3389/fcimb.2015.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Sablet T., et al. , Human microbiota-secreted factors inhibit shiga toxin synthesis by enterohemorrhagic Escherichia coli O157:H7. Infect Immun, 2009. 77(2): p. 783–90. 10.1128/IAI.01048-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asahara T., et al. , Probiotic bifidobacteria protect mice from lethal infection with Shiga toxin-producing Escherichia coli O157:H7. Infect Immun, 2004. 72(4): p. 2240–7. 10.1128/IAI.72.4.2240-2247.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carey C.M., et al. , The effect of probiotics and organic acids on Shiga-toxin 2 gene expression in enterohemorrhagic Escherichia coli O157:H7. J Microbiol Methods, 2008. 73(2): p. 125–32. 10.1016/j.mimet.2008.01.014 [DOI] [PubMed] [Google Scholar]
- 51.Toshima H., et al. , Enhancement of Shiga toxin production in enterohemorrhagic Escherichia coli serotype O157:H7 by DNase colicins. Appl Environ Microbiol, 2007. 73(23): p. 7582–8. 10.1128/AEM.01326-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallick E.M., et al. , A novel murine infection model for Shiga toxin-producing Escherichia coli. J Clin Invest, 2012. 122(11): p. 4012–24. 10.1172/JCI62746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mallick E.M., et al. , The ability of an attaching and effacing pathogen to trigger localized actin assembly contributes to virulence by promoting mucosal attachment. Cell Microbiol, 2014. 16(9): p. 1405–24. 10.1111/cmi.12302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gobius K.S., Higgs G.M., and Desmarchelier P.M., Presence of activatable Shiga toxin genotype (stx(2d)) in Shiga toxigenic Escherichia coli from livestock sources. J Clin Microbiol, 2003. 41(8): p. 3777–83. 10.1128/JCM.41.8.3777-3783.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melton-Celsa A.R., Darnell S.C., and O'Brien A.D., Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect Immun, 1996. 64(5): p. 1569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teel L.D., et al. , One of two copies of the gene for the activatable shiga toxin type 2d in Escherichia coli O91:H21 strain B2F1 is associated with an inducible bacteriophage. Infect Immun, 2002. 70(8): p. 4282–91. 10.1128/IAI.70.8.4282-4291.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barthold S.W., et al. , The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci., 1976. 26: p. 889–893. [PubMed] [Google Scholar]
- 58.Schauer D.B. and Falkow S., The eae gene of Citrobacter freundii biotype 4280 is necessary for colonization in transmissible murine colonic hyperplasia. Infect. Immun, 1993. 61: p. 4654–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mallick E.M., et al. , A novel murine infection model for Shiga toxin-producing Escherichia coli. The Journal of clinical investigation, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor R.G., Walker D.C., and McInnes R.R., E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res, 1993. 21(7): p. 1677–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Datsenko K.A. and Wanner B.L., One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A, 2000. 97(12): p. 6640–5. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith D.L., et al. , Comparative genomics of Shiga toxin encoding bacteriophages. BMC Genomics, 2012. 13: p. 311 10.1186/1471-2164-13-311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farrugia D.N., et al. , A novel family of integrases associated with prophages and genomic islands integrated within the tRNA-dihydrouridine synthase A (dusA) gene. Nucleic Acids Res, 2015. 43(9): p. 4547–57. 10.1093/nar/gkv337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nash H.A., Integration and excision of bacteriophage lambda: the mechanism of conservation site specific recombination. Annu Rev Genet, 1981. 15: p. 143–67. 10.1146/annurev.ge.15.120181.001043 [DOI] [PubMed] [Google Scholar]
- 65.Petty N.K., et al. , The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J Bacteriol, 2010. 192(2): p. 525–38. 10.1128/JB.01144-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hernandez-Doria J.D. and Sperandio V., Bacteriophage Transcription Factor Cro Regulates Virulence Gene Expression in Enterohemorrhagic Escherichia coli. Cell Host Microbe, 2018. 23(5): p. 607–617 e6. 10.1016/j.chom.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Islam M.R., et al. , A sensitive and simple plaque formation method for the Stx2 phage of Escherichia coli O157:H7, which does not form plaques in the standard plating procedure. Plasmid, 2012. 67(3): p. 227–35. 10.1016/j.plasmid.2011.12.001 [DOI] [PubMed] [Google Scholar]
- 68.McDonald J.E., et al. , High-throughput method for rapid induction of prophages from lysogens and its application in the study of Shiga Toxin-encoding Escherichia coli strains. Appl Environ Microbiol, 2010. 76(7): p. 2360–5. 10.1128/AEM.02923-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Acheson D.W., et al. , In vivo transduction with shiga toxin 1-encoding phage. Infect Immun, 1998. 66(9): p. 4496–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klein B.A., et al. , Identification of essential genes of the periodontal pathogen Porphyromonas gingivalis. BMC Genomics, 2012. 13: p. 578 10.1186/1471-2164-13-578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gottesman M.E. and Weisberg R.A., Little lambda, who made thee? Microbiol Mol Biol Rev, 2004. 68(4): p. 796–813. 10.1128/MMBR.68.4.796-813.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anderson B., et al. , Enumeration of bacteriophage particles: Comparative analysis of the traditional plaque assay and real-time QPCR- and nanosight-based assays. Bacteriophage, 2011. 1(2): p. 86–93. 10.4161/bact.1.2.15456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edelman D.C. and Barletta J., Real-time PCR provides improved detection and titer determination of bacteriophage. Biotechniques, 2003. 35(2): p. 368–+. 10.2144/03352rr02 [DOI] [PubMed] [Google Scholar]
- 74.Fuchs S., et al. , Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb Pathog, 1999. 27(1): p. 13–23. 10.1006/mpat.1999.0279 [DOI] [PubMed] [Google Scholar]
- 75.Qiu X., Heat induced capsid disassembly and DNA release of bacteriophage lambda. PLoS One, 2012. 7(7): p. e39793 10.1371/journal.pone.0039793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stevens W.F., Adhya S., and Szybalski W., Origin and bidirectional orientation of DNA Replication in coliphage lambda, in The Bacteriophage Lambda., Hershey A.D., Editor. 1971, Cold Spring Harbor Laboratory: Cold Spring Harbor, NY: p. 515–533. [Google Scholar]
- 77.Gottesman M.E. and Yarmolinsky M.B., Integration-Negative Mutants of Bacteriophage Lambda. Journal of Molecular Biology, 1968. 31(3): p. 487–+. [DOI] [PubMed] [Google Scholar]
- 78.Brooks K. and Clark A.J., Behavior of lambda bacteriophage in a recombination deficienct strain of Escherichia coli. J Virol, 1967. 1(2): p. 283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Herskowitz I., Control of gene expression in bacteriophage lambda. Annu Rev Genet, 1973. 7: p. 289–324. 10.1146/annurev.ge.07.120173.001445 [DOI] [PubMed] [Google Scholar]
- 80.Burt D.W. and Brammar W.J., The cis-specificity of the Q-gene product of bacteriophage lambda. Mol Gen Genet, 1982. 185(3): p. 468–72. [DOI] [PubMed] [Google Scholar]
- 81.Dove W.F., Action of the lambda chromosome. I. Control of functions late in bacteriophage development. J Mol Biol, 1966. 19(1): p. 187–201. [DOI] [PubMed] [Google Scholar]
- 82.Mallick E.M., et al. , Allele- and tir-independent functions of intimin in diverse animal infection models. Front Microbiol, 2012. 3: p. 11 10.3389/fmicb.2012.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moreira C.G., et al. , Bacterial Adrenergic Sensors Regulate Virulence of Enteric Pathogens in the Gut. MBio, 2016. 7(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Navarro Llorens J.M., Tormo A., and Martinez-Garcia E., Stationary phase in gram-negative bacteria. FEMS Microbiol Rev, 2010. 34(4): p. 476–95. 10.1111/j.1574-6976.2010.00213.x [DOI] [PubMed] [Google Scholar]
- 85.Bielaszewska M., et al. , Host cell interactions of outer membrane vesicle-associated virulence factors of enterohemorrhagic Escherichia coli O157: Intracellular delivery, trafficking and mechanisms of cell injury. PLoS Pathog, 2017. 13(2): p. e1006159 10.1371/journal.ppat.1006159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Simpson J.T., et al. , ABySS: A parallel assembler for short read sequence data. Genome Research, 2009. 19(6): p. 1117–1123. 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hernandez D., et al. , De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Research, 2008. 18(5): p. 802–809. 10.1101/gr.072033.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Langmead B. and Salzberg S.L., Fast gapped-read alignment with Bowtie 2. Nat Methods, 2012. 9(4): p. 357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aziz R.K., et al. , The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 2008. 9: p. 75 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang J.-L., et al. , A simple and rapid method for extracting bacterial DNA from intestinal microflora for ERIC-PCR detection. World J. Gastroenterol., 2008. 14: p. 2872–2876. 10.3748/wjg.14.2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mondal S.I., et al. , Genes essential for the morphogenesis of the Shiga toxin 2-transducing phage from Escherichia coli O157:H7. Sci Rep, 2016. 6: p. 39036 10.1038/srep39036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ogura Y., et al. , The Shiga toxin 2 production level in enterohemorrhagic Escherichia coli O157:H7 is correlated with the subtypes of toxin-encoding phage. Sci Rep, 2015. 5: p. 16663 10.1038/srep16663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ray U. and Sakalka A., Lysogenization of Escherichia coli by bacteriophage Lambda: complementary activity of the host's DNA polymerase I and ligase and bacteriophage replication proteins Q and P. J Virol, 1976. 18(2): p. 511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The 47,239 bp prophage DNA sequence (gray), flanked by attL and attR upon insertion into C. rodentium dusA sequence (blue, “Cr dusA”), was determined by whole genome shotgun sequencing of C. rodentium (Φstx2dact::kanR) and annotated, as described in Materials and Methods. Names of encoded proteins are shown. Unannotated ORFs indicate hypothetical proteins. At the far left end is a phage sequence that encodes the N-terminal 112 amino acids of an open reading frame (“ΦdusA’”) in the same reading frame as the 3’ end of the C. rodentium dusA gene. Strain C. rodentium(Φstx2dact) encodes a chloramphenicol acetyl transferase protein (“cat”) inserted into the prophage Rz gene. The sequence of C. rodentium(Φstx2dact::kanR) is identical to C. rodentium (Φstx2dact) except that the gene encoding the A subunit of Stx2dact (“Stx2A”) contains an 894 bp insertion encoding kanamycin resistance (“kan”), plus an additional 27 bp upstream and 28 bp downstream. Prophage genes studied in this work are shown in bold. Cr: C. rodentium.
(TIFF)
The location of the host integration site and genome size for each prophage is indicated in parentheses after the phage name. Open reading frames of prophages λ, 933W, and Sp5 [28, 91, 92] are shown in comparison to those of prophage 1720a-02, and are depicted as arrows pointing in the direction of transcription. The site of insertion of the chloramphenicol cassette (cat) in phage 1720a-02 is indicated with an open triangle. The dotted blue line indicates the presence of additional prophage genes. Genes and genomes are not drawn to scale.
(TIF)
The sequence of phage Φstx2dact was used as a query to interrogate the C. rodentium DBS100 genome (i.e., the parent of the Φstx2dact lysogen) for regions of homology, using the program Megablast (NCBI). Two regions of homology, each to a different endogenous C. rodentium prophage, were identified. A. Region of homology between Φstx2dact and a hypothetical protein. B. Region of homology encompassing a gene encoding a hypothetical protein (upstream of cro), the cro gene (demarcated by red arrows), and a large portion of the cI gene, (demarcated by a blue arrow). Black arrows indicate the direction of transcription.
(TIFF)
Cultures were grown in LB medium to OD600 = 0.4 (T = 0), then each was divided into two cultures. One culture was induced with mitomycin C (0.25 μg/ml) and the other was left uninduced. OD600 culture readings were followed for 4 hours. Two independent isolates of strain C. rodentium (Φstx2dactΔQ) (“ΔQ”), both unable to produce large bursts of phage on induction, were used as the control. Black arrow indicates time at which mitomycin C was added.
(TIFF)
Lysogens of strain DH5α were obtained by infecting a log phase culture with Φstx2dact at high multiplicity of infection, according to the method of Ray and Sakalka [93], and lysogens were isolated by selecting for kanamycin-resistant survivors. A. Agarose gel of PCR analysis showing that a putative DH5α(Φstx2dact) lysogen and C. rodentium control lysogens encode Φstx2dact genes SR, whereas a DH5α non-lysogen did not. B. Strains DH5α containing the recA-bearing plasmid pER271, and the same strain harboring the Φstx2dact prophage, were streaked on LB plates. One half of the plate was shielded with aluminum foil (-UV), while the unprotected half (+UV) was illuminated for 15 seconds using a UVP model UVGL-25 Mineralight UV lamp at 254 nm wavelength from a distance of 8 inches, then incubated overnight at 37°C in the dark.
(TIF)
The indicated wild type or mutant C. rodentium (Φstx2dact) strains were grown in LB broth or DMEM (Gibco, GlutaMAX) without antibiotics. Growth was measured over time by optical density (OD600), and growth curves are the average of duplicate samples. Doubling times were calculated based on the exponential growth regions of each curve. Representative results from one of two experiments are shown.
(TIF)
Q-deficient C. rodentium(Φstx2dactΔQ) (“ΔQ”), C. rodentium(Φstx2dactΔQ)/pTOPO-Q (“ΔQ+pQ”), wild type C. rodentium(Φstx2dact)/pTOPO-Q (““WT+pQ”) and wild type C. rodentium(Φstx2dact) (“WT") were grown to mid-log phase and cultured for four more hours either in the absence (“-“) or presence (“+”) of 0.25 μg/ml mitomycin C. Pellets (filled bars) or supernatants (open bars) were subjected to capture ELISA to determine the level of Stx2dact production. Quantities are expressed relative to the specific OD600 at t = 0h. Results are averages ± SEM of triplicate samples, and are a representative of one of two experiments. nd, not detected. Asterisks indicate Stx levels significantly (p<0.05) different from C. rodentium(Φstx2dactΔQ) calculated using Kruskal–Wallis one-way analysis of variance followed by Dunn's nonparametric comparison.
(TIFF)
The indicated lysogens were grown to mid-log phase (designated as t = 0h) and cultured for four more hours (t = 4h) either in the absence (“-“) or presence (“+”) of 0.25 μg/ml mitomycin C. Pellets (filled bars) or supernatants (open bars) were subjected to capture ELISA to determine the level of Stx2dact production. Quantities are expressed relative to the specific OD600 at t = 0h. Results are averages ± SEM of triplicate samples, and are a representative of at least two experiments. Stx levels of the C. rodentium qseC, qseF, or rpoS mutant strains were not significantly different from wild type C. rodentium(Φstx2dact), calculated using Kruskal–Wallis one-way analysis of variance followed by Dunn's multiple comparisons test.
(TIF)
Eight-week old female C57BL/6 mice were infected by oral gavage with the indicated lysogens. Fecal shedding of the lysogens was determined by plating for viable counts (see Materials and Methods). No significant differences were observed, as determined by 2-way ANOVA. A. Colonization of mice by wild type or recA- or prophage mutant lysogens. B. Colonization of mice by wild type or quorum-sensing mutant lysogens.
(TIF)
Eight-week old female C57BL/6 mice were infected by oral gavage with the indicated lysogens. A. Percentage weight change was determined at indicated post-infection time. Data shown are averages ± SEM of 10 mice per group. No significant differences were observed, as determined by 2-way ANOVA. B. Percent survival at the indicated post-infection time was monitored in 10 mice per group. Data represent cumulative results of 3 separate experiments.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files, except that prophage sequence data are available from GenBank (https://www.ncbi.nlm.nih.gov/genbank/), accession number KF030445.