Abstract
The western corn rootworm (WCR, Diabrotica virgifera virgifera) gene, dvssj1, is a putative homolog of the Drosophila melanogaster gene, snakeskin (ssk). This gene encodes a membrane protein associated with the smooth septate junction (SSJ) which is required for the proper barrier function of the epithelial lining of insect intestines. Disruption of DVSSJ integrity by RNAi technique has been shown previously to be an effective approach for corn rootworm control, by apparent suppression of production of DVSSJ1 protein leading to growth inhibition and mortality. To understand the mechanism that leads to the death of WCR larvae by dvssj1 double-stranded RNA, we examined the molecular characteristics associated with SSJ functions during larval development. Dvssj1 dsRNA diet feeding results in dose-dependent suppression of mRNA and protein; this impairs SSJ formation and barrier function of the midgut and results in larval mortality. These findings suggest that the malfunctioning of the SSJ complex in midgut triggered by dvssj1 silencing is the principal cause of WCR death. This study also illustrates that dvssj1 is a midgut-specific gene in WCR and its functions are consistent with biological functions described for ssk.
Introduction
RNA interference (RNAi) pathways are common among many eukaryotes including insects [1], and transgenic crops utilizing RNAi represent a promising new tool for insect pest management [2]. The western corn rootworm (WCR), Diabrotica virgifera virgifera (Coleoptera: Chrysomelidae), is one of the most economically important pests of maize in the United States, and Europe where it is an invasive species [3, 4]. Currently, WCR damage is managed by combinations of crop rotation, broad-spectrum soil insecticides [5], and transgenic crops expressing crystalline (Cry) proteins from Bacillus thuringiensis (Bt) [4]. Insect resistance to existing transgenic traits threatens the durability of Bt crops[6], which highlights the importance of developing new technologies for protection from WCR [7, 8].
The general mechanism through which RNAi works has been well described [9], as has its potential for a new transgenic approach to rootworm control [2, 10]. Many genes have been reported to be effective insecticidal targets in WCR following the provision of double-stranded RNA (dsRNA) in diet bioassay [11–13]. Effective control of WCR in transgenic maize plants has been achieved with RNAi targeting the α-tubulin gene [11], the V-ATPase subunit A [11] and C [14] genes, an intracellular protein trafficking pathway gene, snf7 [11, 15, 16], and a midgut expressed gene, dvssj1[17]. Once dsRNA is ingested by a susceptible insect and taken up by the midgut epithelial cells, dicer RNase III type enzymes bind and digest cytoplasmic dsRNA into small RNAs (siRNAs), that then associate with an RNA-induced silencing complex (RISC) and lead to specific suppression of the target mRNA [18]. While it has been reported that WCR larvae mortality can be caused by Dvsnf7 [15] and Dv v-ATPase C [14] dsRNAs, this RNAi response was observed only with dsRNAs greater than 60 bp in length. A lack of uptake of short Dvsnf7 dsRNA (<60 bp) or 21 bp siRNA was observed in larval midgut cells [15]. Previously, we reported that both dietary delivery of dsRNA and transgenic plants expressing dsRNA targeting dvssj1 result in mRNA suppression leading to growth inhibition and mortality of WCR by apparent suppression of smooth septate junction (SSJ) protein located within the intestinal lining [17].
The epithelia of most invertebrate species possess specialized cell-cell junctions, known as septate junctions (SJ) [19, 20], that typically form circumferential belts around the apicolateral regions of epithelial cells and form the paracellular barrier between adjacent cells [20]. Arthropods have two types of SJs: pleated SJs (PSJs) and smooth SJs (SSJs), found in ectodermally and endodermally derived epithelia, respectively [21]. SSJs appear to regulate the paracellular pathway of the intestine and renal system in arthropods [22]. The molecular composition of SSJs is different from that of PSJs[21]. More than 20 PSJ-related proteins have been identified and characterized in Drosophila melanogaster [21]. In contrast, only three SSJ-specific proteins encoded by D. melanogaster genes snakeskin (ssk), mesh and tetraspanins (tsp2A) have been reported [23–25]. SSK, MESH, and TSP2A form a complex that are mutually interdependent for their correct SSJ localization [24, 25]. Several PSJ components, including Discs large (Dlg), Lethal giant larvae (Lgl), Coracle (Cora) and Fasciclin III (FasIII), have been confirmed to localize to SSJs. In ssk-and mesh-deficient midguts, Lgl, Cora, and FasIII are mislocalized but Dlg is not [24]. The functions of these PSJ proteins in SSJs remain uncertain since Dlg, Lgl, Cora, and FasIII are not required for the SSJ localization of TSP2A, MESH, and SSK, and are dispensable for SSJ formation [21]. Genetic studies in D. melanogaster have shown that normally impermeant fluorescent-labeled dextrans (10 kDa) are able to access the paracellular route in mutant flies that are defective for smooth septate formation[21]. The ssk-RNAi and ssk-deletion mutants are lethal at the late stage 17 of D. melanogaster embryo. Ssk, mesh, and tsp2A are essential to D. melanogaster development, SSJ formation, and midgut paracellular barrier function [23–25].
The goal of the current study is to understand the mechanism of dsRNA insecticidal activity when targeting the SSJ of WCR by analyzing: (1) silencing effects on molecular expression as it relates to larval growth and survival; and (2) dvssj1 function by replacing ssk with dvssj1 in D. melanogaster utilizing CRISPR/Cas9 gene modification. Our results show that compromised dvssj1/ssk expression caused by either RNAi- or CRISPR/Cas9-mediated systems is associated with defects in SSJ structure, changes in the localization of certain SSJ proteins, and impaired barrier function of the midgut in both WCR and D. melanogaster strains which culminate in impaired larval development and survival.
Method and materials
Double-stranded RNA production by in vitro transcription
The gene-specific primers (S1 Table) contained T7 RNA polymerase sites at the 5’ end of each primer were used to generate PCR product that served as the template for dsRNA synthesis by in vitro transcription (IVT) using a MEGAscript kit (Life Technologies, Carlsbad, CA). For Cy3-labelling of dsRNA, 25% of CTP and UTP were replaced with Cy3-CTP and Cy3-UTP (GE Healthcare, UK). DsRNAs were purified by Megaclear kit (Life Technologies, Carlsbad, CA) and examined by 48-well E-gel electrophoresis (Life Technologies) to ensure dsRNA integrity and quantified using Phoretix 1D (Cleave Scientific) or a NanoDrop 8000 Spectrophotometer (ThermoFisher Scientific,).
Target expression during WCR different life stage
To assess levels of dvssj1 mRNA and protein expression at different life stages, WCR rearing, total RNA extraction, and complementary DNA (cDNA) synthesis were prepared as in a previous study [26]. WCR were reared on artificial diet, collected at different life stages and/or kept in 10% neutral buffered formalin (4% formaldehyde) for 48 to 72 hours and processed for paraffin embedding.
Quantitative reverse transcription PCR (qRT-PCR)
Gene expression was analyzed using two-step real-time quantitative RT-PCR. The assay was run, with 3 replicates per sample, using a singleplex set up with Bioline Sensifast Probe Lo Rox kit (Taunton, MA) and analyzed using the 2-ΔΔCt method based on relative expression of the dvssj1 gene and a reference gene dvrps10 (S1 Table). Data from qRT-PCR assays were analyzed using JMP (Version 12. SAS Institute Inc., Cary, NC) and statistical differences were detected using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison tests; P<0.05 was considered statistically significant.
In situ hybridization and immunohistochemistry analyses
Paraffin sections were cut 5 μm thick, collected on Superfrost Plus Gold slides (Fisher Scientific), air-dried overnight, and baked for 1 hour at 60°C. Sections were processed for RNA in situ hybridization (ISH) with the RNAScope Detection Kit (Chromogenic) according to the manufacturer’s standard protocol (Advanced Cell Diagnostics, Hayward, CA) or as in a previous study [26]. Slide images were acquired using a Leica Aperio CS2 digital scanner and captured at 40x magnification with a resolution of 0.25 μm per pixel. For immunohistochemistry (IHC), sections were incubated with primary antibodies (1:500 dilution) at 4°C overnight as described in S1 Method. Following five washes, they were incubated with secondary antibodies (goat anti-mouse antibody Alexa Fluor 488; 1:500 dilution) for overnight. After five additional washes, they were coverslipped and imaged with a confocal laser scanning microscope (TCP SP2; Leica) or EVOS FL Auto Imaging System (ThermoFisher) at room temperature.
Correlation between molecular expression and larval dose-response bioassay
Larval bioassays
Larval bioassays were conducted in three independent replicates, with each replicate consisting of nine treatments: two negative controls (water & Escherichia coli GUS dsRNA) and seven dvssj1 dsRNA doses. Overall diet preparation method followed a modified version of the manufacturer’s recommendations for Diabrotica artificial diet (Frontier Scientific, Inc., Newark, DE) [27]. Two 96-well diet plates per treatment were prepared as described previously [17], with seven dosing solutions containing purified dvssj1 dsRNA at concentrations of 1.7 to 1.7×10−6 ng/μl. Control diet containing GUS dsRNA was prepared at the highest dose of 1.7 ng/μl. Newly hatched WCR were acclimated to neutral diet for 24 hours prior to transfer to treatment plates at infest rates of either 1 per well or 1–3 per well for all treatments. Infested plates were sealed in bags containing moist paper towels and placed in an incubator (Percival Scientific, Inc., Perry, IA) set at 27.5°C and 0:24 light to dark hours. Plates infested with 1–3 insects per well were collected after 48 hours, assessed for development based on head capsule size [28] and mortality, and live insects were used for analysis of dvssj1 transcript and protein levels. Plates infested with 1 insect per well were assessed at the end of seven days for growth inhibition as previously described [17], along with development and mortality.
RNA isolation and qRT-PCR
Insects were collected from larval bioassays for dvssj1 expression analysis at 48 hours post-dsRNA-exposure. Three samples per treatment per bioassay replicate were collected at a rate of 15 live whole insects per sample, homogenized in Buffer RLT from the RNeasy Mini Kit (Qiagen N.V., Hilden, Germany), and stored frozen at -80°C. Total RNA isolation was guided by the kit-provided protocol with an on-column DNase treatment (RNase-free DNase kit, Qiagen N.V.). Total RNA was quantified and cDNA was prepared on the day of RNA isolation using the SensiFAST cDNA Synthesis Kit (Bioline, London, England). Reaction conditions were as described in kit instructions, and reverse transcription (RT) was conducted in a C1000 Touch instrument (Bio-Rad Laboratories, Inc., Hercules, CA). Resulting cDNA was analyzed by a MIQE-compliant triplex assay between dvssj1 and two genes (α-tubulin and elongation factor 1a, EF1α) previously validated as suitable RT-qPCR references in WCR [29–31]. The SensiFAST Probe Lo-ROX Kit (Bioline) was used to amplify cDNA on 384-well PCR microplates (Corning, Inc., Corning, NY). In addition to insect samples, each plate contained three standard curves of synthetic DNA fragments (one per assay target) and no amplification controls. All standards, controls, and samples were loaded in triplicate. Quantitative PCR was performed using the Life Technologies Viia 7 Real-Time PCR System (ThermoFisher Scientific, Inc.).
Gene expression analysis
Quantitative PCR data were generated using the QuantStudio Real-Time PCR Software (v. 1.1) (ThermoFisher Scientific, Inc.) with a standard curve experimental design. Expression of dvssj1 in each treatment group was determined by first interpolating concentration values from the appropriate standard curve, then calculating a ratio between the value obtained for the target gene relative to each reference gene within each sample. The geometric mean of the ratios within a sample was determined and the natural log of each resulting sample value was calculated. All log-transformed values within a treatment group were averaged and the final value per treatment was determined by back-transforming the difference between the treatment and water control group and multiplying by 100 to express final values as a percentage of the water control group (S5 and S7 Figs). These calculations were performed within each of the three bioassay replicates and final values from all three replicates were plotted using Prism 7 (ver. 7.02, GraphPad Software, Inc., La Jolla, CA).
Western blot analyses
Extracted guts from 15 larvae were combined and homogenized in 15μl of ice-cold buffer (50 mM Na2HPO4-NaH2PO4, 50 mM NaCl, 5 mM EGTA, 5 mM EDTA, pH 7.5) that contained Complete protease inhibitors at 2.2X standard concentration (Roche) and 1mM PMSF using a hand-held motorized homogenization Pellet Pestle (Thermo Cat # 12-141-361 and Kimble 749520–0000). Homogenate (10 μl) was mixed with sample buffer 26 μl for the carcass of 2X NuPAGE LDS Sample Buffer and Reducing Agent (Life Technologies, Carlsbad, CA) and heated to 100°C for 10 min. Prepared samples were separated on 4–12% Bis-Tris polyacrylamide gels (NuPAGE, Life Technologies) using MES running buffer (NuPage, Life Technologies). Proteins were then transferred to nitrocellulose membrane using an iBlot2 system (Life Technologies, Carlsbad, CA). After transfer, membranes were blocked overnight in 5% non-fat milk in PBS, 0.1% Tween-20, and 5% glycerol. Membranes were then incubated with anti-DVSSJ1 mouse antibody (Genescript, USA) at 1: 2,500 in 1% non-fat milk in PBS with 0.1% Tween-20 for at least 90 minutes, followed by 1-hour incubation in HRP-conjugated goat anti-mouse (GAM) secondary Ab at 1: 12,500 (Bio-Rad, Hercules, CA). Western blots were developed for 5 minutes using SuperSignal West Dura Extended Duration Substrate (Life Technologies, Carlsbad, CA). Band densities from chemiluminescence were measured after digital imaging with a LAS-4000 system (GE Healthcare, Pittsburg, PA) using the TotalLab software suite (Nonlinear Dynamics, Durham, NC). Protein concentrations for midgut homogenates were determined using Pierce Coomassie Plus Assay with BSA standards (ThermoFisher Scientific, Waltham, MA). Western blot band volumes for each treatment were adjusted for total protein loaded per sample and then normalized to the values determined for water treated control samples. The data were plotted and analyzed using Prism 7 (ver. 7.02, GraphPad Software, Inc., La Jolla, CA). The concentration-response data for transcript and protein expression were fit using a standard logistic equation (y = A1 + (A2-A1)/(1 + 10^((LOGx0-x)*p).
siRNA profile analysis in WCR larvae exposed to dvssj1 dsRNA in diet
WCR neonates were transferred to artificial WCR diet containing 210-bp dvssj1 dsRNA at 180 ng/μl in the diet for 48 hours and collected for total RNA isolation. Total RNA was prepared using the mirVana RNA isolation kit (Life Technologies), and 1μg of RNA used to generate small RNA libraries using the TruSeq small RNA kit (Illumina). RNA 3′ and 5′ adapters were ligated in consecutive reactions with T4 RNA ligase. Ligated RNA fragments were primed with an adapter-specific RT primer and reverse transcribed with Superscript II reverse transcriptase (Life Technologies) followed by eleven cycles of amplification with adapter specific primers. Resulting cDNA libraries were separated on a 6% TBE gel and library fragments with inserts of 15–50 bp excised. Recovered cDNA libraries were validated by QC on an Agilent Bioanalyzer HiSens DNA chip (Agilent Technologies Inc.) and were sequenced for 50 cycles on the Illumina GAIIx according to Illumina protocols with one sample per lane. Trimmed reads 18 to 41-nt in length were aligned to 210-bp dvssj1 sequence. Only perfectly matched reads were included for analysis of sequence length distribution of siRNA. The 18- and 41-nt dvssj1 reads were visualized using Integrative Genomics Viewer software (Broad Institute, Cambridge, MA, USA) or aligned to 210 bp dvssj1 using Sequencher (v 4.8, Gene Codes Corporation) to identify siRNA (>20 counts) regions (S5 Fig).
RNA binding to midgut cells
Fifty diet-raised 2nd instar WCR were treated and surface-sterilized as previously described [15], with modifications (S1 Method). Twenty midguts were dissected and rinsed in sterile 1x PBS to eliminate gut contents. Pretreated midguts were incubated with Cy3-labeled dvssj1 dsRNA and siRNAs at 10 ng/μl in insect medium (EX-CELL 420 Serum-Free Medium; Sigma) for 15 hours at 25°C protected from light. Unconjugated Cy3 dye was used as a control at the same concentration. Midguts were washed twice with 1x PBS and fixed in 4% paraformaldehyde for 1 hour at room temp. Then midguts were washed three times with 1x PBST (PBS +0.1% Tween) for 5 minutes each. Samples were counterstained with DAPI (Sigma, 10 μg/μl of stock diluted to 1:1000) for 5 min and washed once in 1x PBST. Individual guts were placed on glass slides with 2x SlowFade concentrated Slow Fade antifade reagent in PBS, then coverslipped and imaged on a Leica TCS SPE. The 405 nm laser line was used for excitation of DAPI staining and the 532 nm laser line was used for excitation of Cy3 staining. Image analyses were carried out as described in S1 Method.
Double-stranded RNA treatment for target suppression analyses
To evaluate SSJ protein and mRNA expression after dvssj1 dsRNA treatments, 4-day old larvae were transferred to 96-well plate containing 167 ng/μl of dvssj1 dsRNA (dsssj1) or gfp dsRNA (dsgfp) in 30 μl of an artificial diet [17] for 48 hours. Treated larvae were then transferred to the new plates containing 167 ng/μl of dsgfp for an additional five days. The larvae were then harvested for IHC and ISH microscopic studies as described in S1 Method and S12 and S13 Figs.
Generation of D. melanogaster ssk knockout and dvssj1 replacement lines via CRISPER/Cas9
CRISPR/Cas9-mediated genome editing by homology-dependent repair (HDR) using two guide RNAs and a dsDNA plasmid donor was used for replacing ssk with homolog dvssj1 or making an ssk deletion mutant at the same breakpoint. The ScarlessDsRed system [32, 33] was employed to facilitate genetic screening. Edited ssk deletion and dvssj1-replacing lines were generated and confirmed by WellGenetics Inc (Taiwan) as described in S1 Method and S9, S10, S11 Figs and S2 Table. Since ssk is a lethal gene, edited lines were maintained as heterozygous in TM6B (containing visual marker). Crossing experiments before and after excision of DsRed were conducted for dvssj1 or ssk viability evaluation.
Results and discussion
DVSSJ1 interacts with other SSJ proteins for essential midgut functions
The insect midgut is the primary organ for digestion and nutrient absorption [34]. The midgut epithelium mediates nutrient uptake and serves as the barrier separating the extracellular fluid of the body from the gut lumen and thereby protects the body from toxic substances and infectious organisms present in the environment. We previously reported the discovery of the dvssj1 gene through dsRNA diet-based screening for targets that have the potential for WCR control [17]. However, the gene function was unknown at the time of our discovery and remained so until the report that its D. melanogaster ortholog, ssk, is required for intestinal barrier function by contributing to SSJ formation [21, 22]. The SSJ-specific membrane proteins, Ssk and Mesh, were identified by screening monoclonal antibodies raised against SSJ-containing membrane fractions[23, 24]. Recently, a genetic screen based on microscopic observation of SSJ formation in D. melanogaster identified Tsp2A (a homolog of dvssj3; S1 Fig) as a novel SSJ-specific membrane protein [25]. All three D. melanogaster SSJs form a protein complex that contributes to the specialization between epithelial cell apical and basolateral membranes and are required for D. melanogaster SSJ formation and intestinal barrier function [21].
Midgut-specific expression of dvssj1 during different life stages
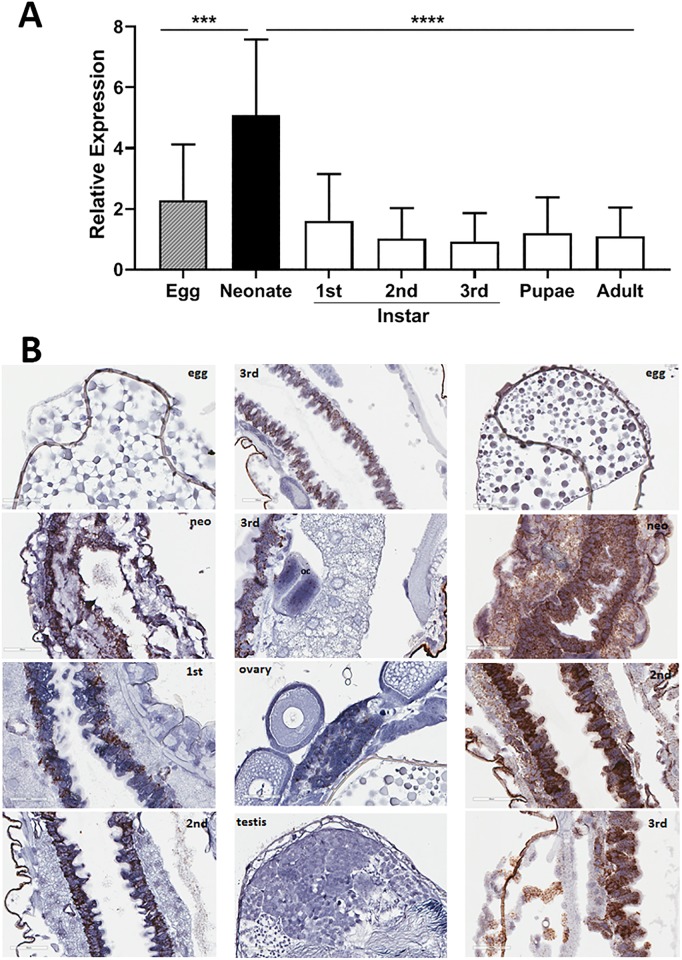
Expression of dvssj1 mRNA was analyzed by real-time quantitative reverse transcription PCR (qRT-PCR) using individual whole insects representing different life stages (Fig 1A), and by in situ hybridization (ISH) analysis of specific life stages, and dissected reproductive tissues from the WCR adults (Fig 1B, S2 Fig). The mRNA expression of dvssj1 showed clear differences depending on the life stage. For example, dvssj1 mRNA expression was about 4- or 5-fold higher in newly hatched neonates than in larvae, pupae, or adults. Localization of dvssj1 mRNA occurred predominantly in the cells of the midgut epithelium (Fig 1B) during different stages of larval development which was in contrast to ribosomal protein s10 (dvrps10)[17], which was highly expressed in every visible cell type (Fig 1B). Expression of dvssj1 mRNA in WCR has also been detected in oenocyte cells [35] and cells within Malpighian tubules (MTs) (Fig 1B) which are thin fingerlike extensions attached to the exterior intestinal tract between the midgut and the posterior gut or hindgut [36, 37].
Fig 1. Expression and localization of dvssj1 mRNA at different life stages of western corn rootworm (WCR).
(A) Relative expression of the whole insect of WCR was measured at different life stages by qRT-PCR. Relative expression analysis (mean ± SE) was based on dvssj1 expression in individual insects (n = 12) at each life stage, after being normalized to the expression of the reference gene, dvrps10. Expression data were subjected to one-way analysis of variance using JMP (v12. SAS Institute Inc, Cary, NC) followed by Tukey’s multiple comparison tests. Neonate stage showed statistically significant high expression (P = 0.004[***] or < 0.0001[****]). (B) Visualization of dvssj1 mRNA expression during different life stages by in situ hybridization (ISH). Representative WCR sections were collected from the egg, neonate (neo; first 24-hrs after hatch), 1st, 2nd and 3rd instar of larvae, and dissected testis and ovary from adults. All samples were hybridized with the dvssj1 probe and a control probe (dvrps10) were included for egg, neonate, 2nd and 3rd instars to compare (S2 Fig). Images were captured at 40x magnification with 60 μm scale bars.
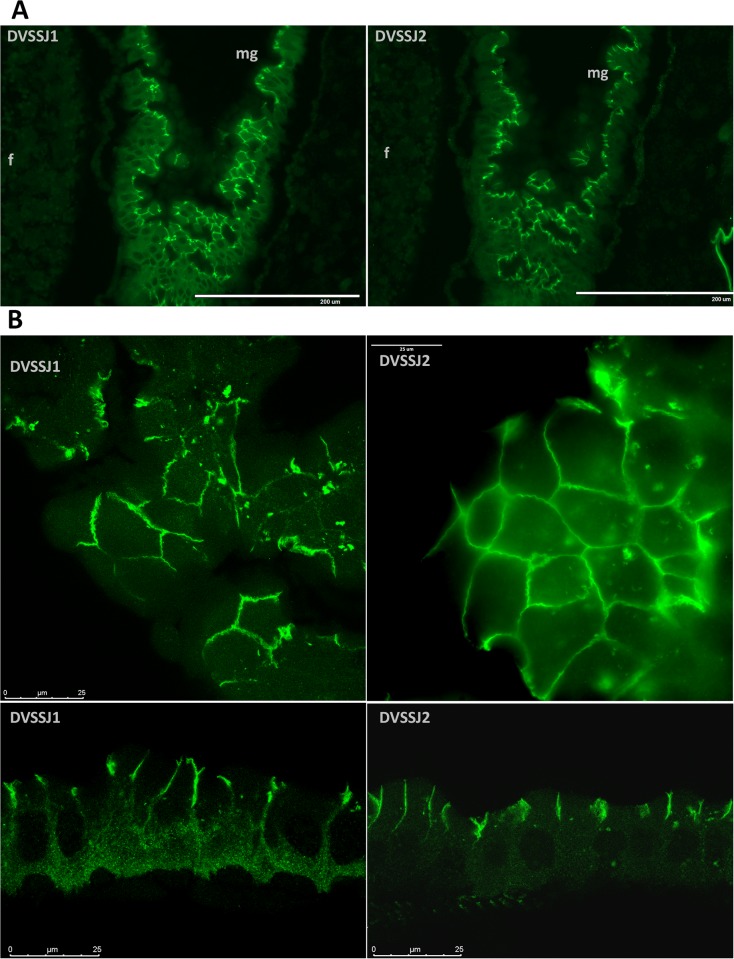
WCR SSJ proteins were visualized by immunohistochemistry (IHC) using both peptide (DVSSJ2)[17] and total protein (DVSSJ1) antibodies as described in the S1 Method. Expression of DVSSJ1 and DVSSJ2 in the midgut (Fig 2) was observed in the longitudinal sections of whole larvae showing patterns that were reminiscent of the D. melanogaster orthologs snakeskin [23] and mesh [24]. The strongest immunostainings of SSJ1 or SSJ2 in midgut were observed at the apico-lateral part of epithelial column cells (Fig 2B), while weak or no signal was found in the foregut or hindgut (S3 and S4 Figs). Neither SSJ protein was observed in fat body cells, but consistent with their mRNA expression patterns they were detected in SSJs in oenocytes [35] and MTs (S4 Fig), which are responsible for lipid processing and detoxification [38], or excretory and osmoregulatory functions [39, 40], respectively. In D. melanogaster, SSK expression appears at stage 12 embryos in midgut rudiments and its expression is sustained until the adult stage throughout the midgut and MTs [23].
Fig 2. Immunofluorescence microscopic analysis of DVSSJ1 and DVSSJ2 in WCR larvae.
(A) two adjacent sections were selected to compare DVSSJ 1 (left) and DVSSJ2 (right) expression. Images are representative sections (S3 Fig) of midguts (mg) collected from the 3rd instar of WCR larvae and hybridized with antibodies as described in Method. (B) confocal images of selected midgut cells showing DVSSJ1 and DVSSJ2 localization. Scale bar = 200 um (A) and bar = 25 um (B).
Dose-dependent effects of dsRNA exposure are correlated to target suppression, larval development and survival
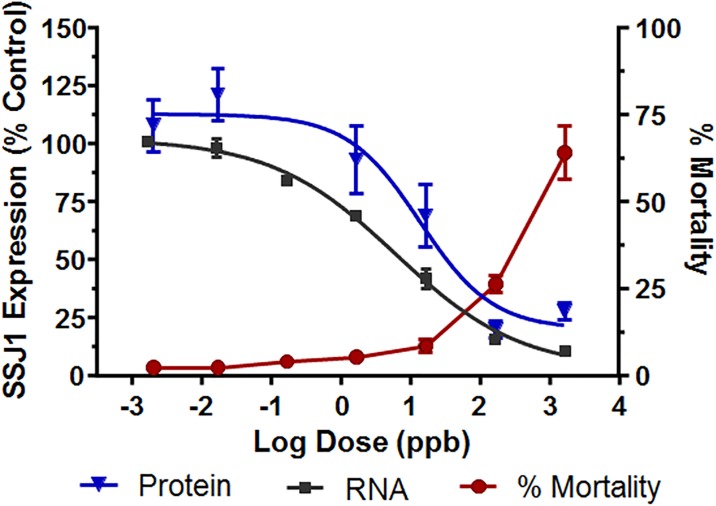
To examine the relationship between exposure to dvssj1 dsRNA and suppression of dvssj1 transcript and protein levels, WCR neonates were provided diet containing a range of dvssj1 dsRNA concentrations. Larvae were assessed for survival and extent of growth and developmental inhibition after two and seven days of exposure. Larval development and mortality were consistent across all treatments and not different from controls after two days, whereas nearly all larvae showed growth inhibition or mortality after seven days exposure to the highest dsRNA dose (S6 Fig). As expected, the mortality and growth inhibition observed at seven days increased with increasing dsRNA dose (S5B Fig). To minimize the impact of natural variability in development and mortality between treatment and control groups, dvssj1 transcript and protein levels were measured after two days of exposure. These results revealed a direct correlation between the dose of dvssj1 dsRNA and suppression of mRNA, protein, and the mortality observed after 7-day treatments (S7 Fig). Fitting of the data revealed 50% suppression of transcript and protein at approximately 6.1×10−2 ng/μl and 1.4×10−2 ng/μl, respectively, with less than 50% mortality at these concentrations observed at the end of seven days exposure (Fig 3 and S7 Fig).
Fig 3. Dvssj1 transcript and protein suppression correlate to dsRNA dose, as measured two days post-exposure.
Larval mortality correlates with the extent of suppression, as measured seven days post-exposure. Larvae were infested on artificial diet alone, or with GUS dsRNA, or with increasing concentrations of dvssj1 dsRNA as described in methods. After 48 hours feeding, larvae were collected and processed as described in order to assess the expression of the dvssj1 transcript (black squares) or protein (blue triangles). Curves fit to the data were generated using a square logistic equation as described in methods. Duplicate sister plates were infested in order to follow larval growth inhibition (see S6 Fig) and mortality (red) for five additional days allowing the impact caused by the dose-dependent suppression of dvssj1 transcript and protein expression to be quantified. The data reflect 3 independent determinations. Errors bars for each curve are explained in methods.
Dvssj1 mRNA expression patterns (Fig 1B) and dvssj1 mRNA knockdown (S12C Fig) were also demonstrated in WCR larvae using RNAscope ISH. Both mRNA and protein of dvssj1 were observed predominantly in midgut epithelial cells and expression patterns varied slightly between different regions of the midgut (S2, S3 and S4 Figs). Similar expression patterns were also observed in guts dissected from WCR larvae (S5 Fig). The gene expression of dvssj1 mRNA in midgut epithelial cells of WCR is consistent with a functional role of dvssj1 analogous to D. melanogaster ssk [23]. Microscopic observations of nearly whole insects in section (S12C Fig) showed a significant difference in the overall size of dvssj1-treated and control individuals. Previous ultrastructural examination of midgut epithelial cells revealed apparent occlusion of the gut lumen, and numerous examples of enterocytes blebbing into the gut lumen, after dvssj1 dsRNA consumption [17]. These observations are consistent with the notion that suppression of dvssj1 mRNA and its protein accumulation are the cause of WCR growth inhibition and mortality. Furthermore, the toxic effect to WCR resulting from oral exposure to dvssj1 dsRNA in diet or expressed in planta [17] can be attributed to suppression of dvssj1 mRNA leading to a reduction in DVSSJ1 expression/accumulation, loss of the midgut epithelium diffusional barrier, and cellular deformities due to improper intercellular contacts [22].
Double-strand RNA processing and uptake in WCR midgut
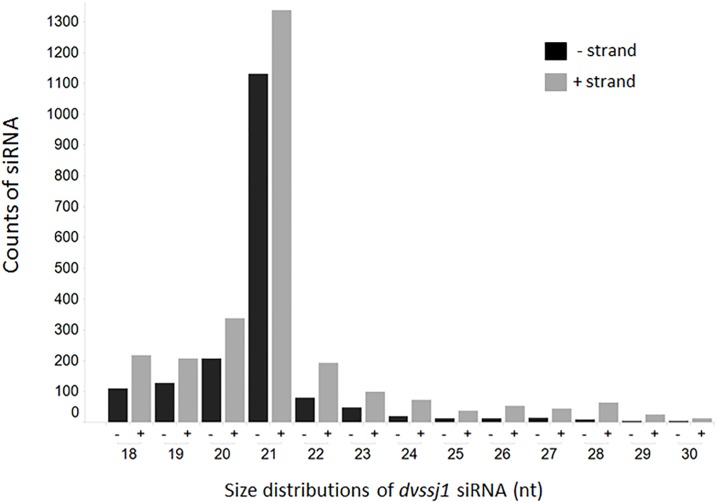
To understand dsRNA processing in WCR midgut, we performed an RNAseq study to characterize small RNA generated from 3rd instar larvae exposure to 210-bp dsRNA of dvssj1. The siRNA sequences mapped to the sense and antisense strands of the 210-bp dvssj1 sequence (Fig 4 and S8 Fig). These small RNAs are distributed across the 210 bp-length of the dvssj1 sequence, with three regions that have dominant siRNA accumulation over other regions (S8 Fig). The top siRNA represented 14.1% of all dvssj1 siRNA (Table 1).
Fig 4. Size distribution of dvssj1 small RNAs between 18 and 30 nucleotides.
Small RNA sequencing of WCR larvae fed with 210-bp dvssj1 dsRNA was conducted to identify siRNAs mapped to 210-bp region of dvssj1. Sense (grey) and antisense (black) siRNA distribution was compared between 18 and 30 nucleotides.
Table 1. Summary of small RNAs (18 to 41 nt) mapped to 210 bp of dvssj1.
| siRNA summary | Count |
|---|---|
| total count of all dvssj1 siRNA | 4543 |
| total number of siRNA type | 818 |
| number of minus strand | 1790 |
| number of plus strand | 2751 |
| siRNA with above 20 counts* | |
| region-1 (32-76nt) | 488 |
| region-2 (93-132nt) | 1538 |
| region-3 (169-191nt) | 170 |
| Top siRNA in region-2 | |
| TCCTTGATATCCGGTTCGGTA | 641 |
*Three regions contain small RNAs with more than 20 counts were grouped based on their mapped locations to 210 bp of dvssj1 (S8 Fig).
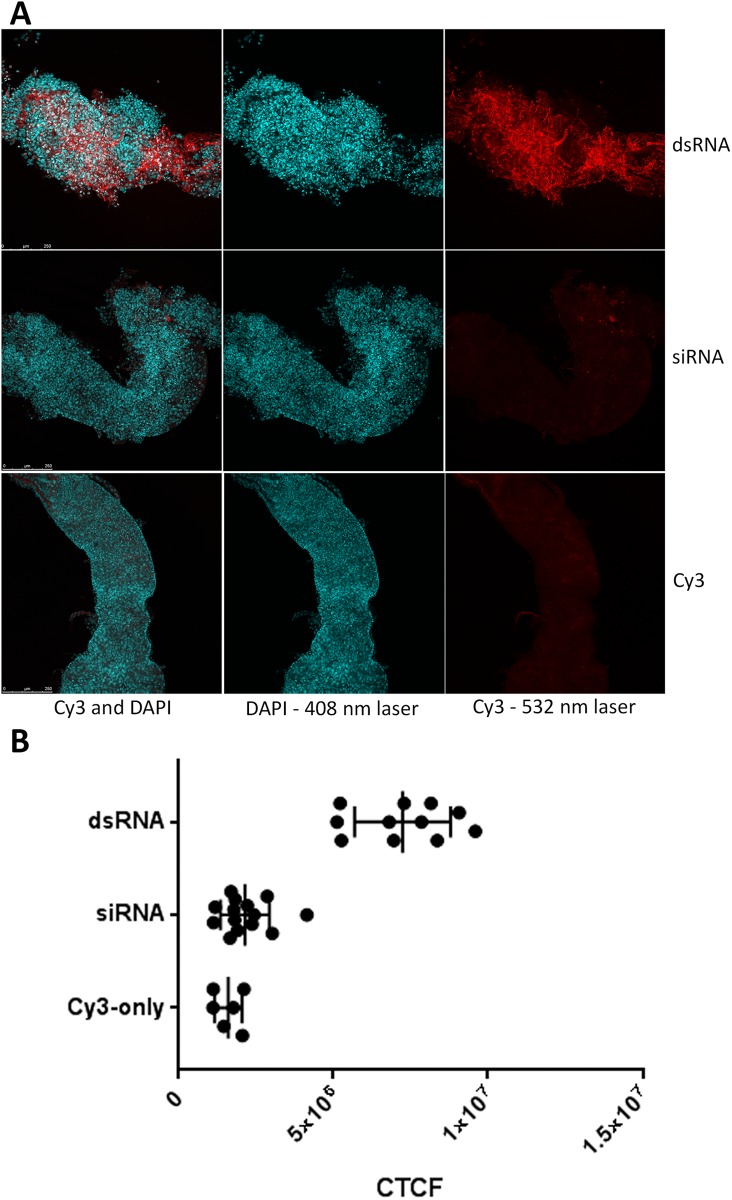
To evaluate dsRNA uptake in WCR midgut, Cy3-labelled 210 bp dvssj1 dsRNA and Cy3-labelled siRNA (21-mer) representing one of the top siRNAs were incubated with dissected WCR midgut tissues. The 210 bp Cy3-dsRNA was able to clearly bind to midgut cells, while in contrast, Cy3-siRNA did not bind to the midgut as determined by the absence of a detectable Cy3 label (Fig 5A). Controls with insect medium containing unincorporated Cy3 dye showed no fluorescence binding to the midgut (Fig 5A). Two independent experiments were conducted and a summary of results shown in Fig 5B. These results under test conditions are consistent with previous reports of dsRNA uptake and ineffectiveness of exogenous siRNA (21-mer) binding to WCR midgut [15].
Fig 5. RNA binding to midgut cells of western corn rootworm (WCR).
(A) Cy3-labeled 210 dvssj1 dsRNA is binding to WCR midgut cells (Top panel), but 21-bp siRNA of dvssj1 is not observed (middle panel). Commercial Cy3 dye was used as a control, which showed no binding also. (B) Summary of the fluorescent intensity of midgut cells. Corrected Total Cell Fluorescence (CTCF) was calculated in ImageJ. DsRNA treatment (n = 11) is significantly different (P<0.0001) from control and siRNA treatment (n = 14).
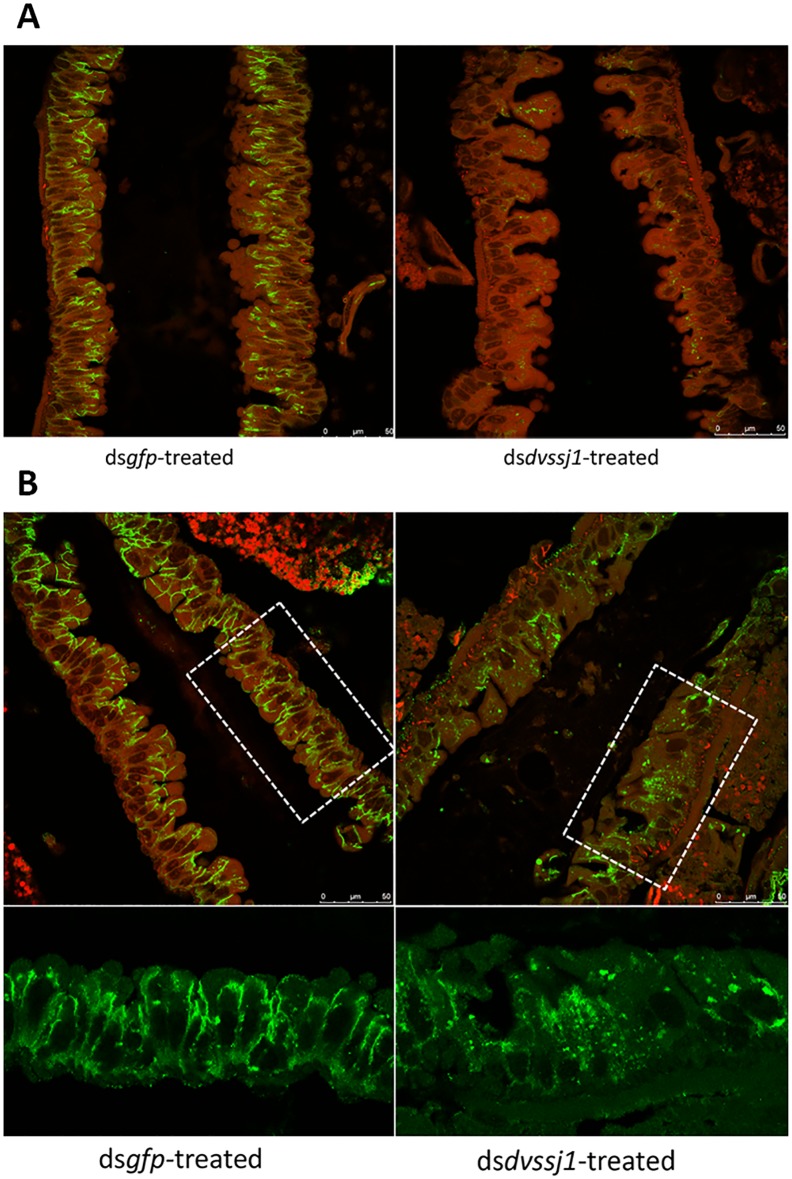
Since molecular characterizations of SSJ has been reported in D. melanogaster [23] and technical challenges have hindered conducting similar midgut barrier functional evaluations in WCR, we generated D. melanogaster ssk knock-out and dvssj1 knock-in substitution (S9, S10 and S11 Figs) lines using CRISPR-Cas9-mediated genome editing. As expected, homozygous ssk knockout resulted in a lethal phenotype at the larval stage (Table 2). Molecular substitution of ssk by dvssj1 resulted in only survival of heterozygous dvssj1 knock-ins suggesting that there is insufficient homology (54.9%) of the WCR protein [17] to allow interaction with other fly SSJ components and substitute in SSJ formation (Table 2 and S11 Fig). Furthermore, it has been reported that D. melanogaster SSK interacts with MESH for midgut barrier functions and these proteins are mutually interdependent for their localization [24]. Examination of SSJ protein expression in WCR midgut cells after dvssj1 dsRNA-treatment showed clear DVSSJ1 protein knockdown and the consequent mislocalization of DVSSJ2 protein (Fig 6). We previously reported that suppression of dvssj2 [17] and dvssj3 [41] expression can cause WCR mortality in artificial diet delivery bioassays. These results indicate that WCR SSJ proteins have a similar function like D. melanogaster SSJs, contributing to a protein complex that is required for SSJ formation and normal intestinal barrier function [22].
Table 2. Summary of viability test for edited ssk-D. melanogaster lines.
| Edited line | dvssj1 knockin | ssk deletion |
|---|---|---|
| Before DsRed excision | ||
| Heterozygous | 207 | 216 |
| Homozygous | 0 | 0 |
| After DsRed excision | ||
| Heterozygous | 289 | 241 |
| Homozygous | 0 | 0 |
Two separate viability tests were conducted before and after DsRed maker excision. Both Drosophila ssk (deletion) knock-out and dvssj1 knock-in substitution lines were crossed and maintained in the balance line (S1 Method and S10 Fig). A number of survival insects were counted based on Tubby phenotype (TM6B). No homozygous edited fly was recovered.
Fig 6. Immunohistochemistry detection of SSJ proteins in dsRNA-treated midgut cells.
WCR larvae (4-day old) were treated with 167 ng/μl dsRNA in the diet for 48 hours (gfp left panel and dvssj1 right panel), and collected 7 days after treatment, and hybridized with the DVSSJ1 (A) and DVSSJ2 (B) antibodies. Confocal images were captured with combined GFP and DAPI channels. Boxed areas were enlarged to show DVSSJ2 expression pattern changes (GFP channel only) after dvssj1 dsRNA treatment. Scale bar = 25 μm.
Conclusion
Our findings show that disruption of SSJ function by down-regulation of dvssj1 expression can be correlated directly with insect mortality making it a well-suited gene target for RNAi silencing and providing an alternative molecular target for control of corn rootworm. This study also illustrates that dvssj1 is a midgut-specific gene in WCR and its functions are consistent with biological functions described for ssk in Drosophila.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Adane Kassa, Jonathan Robeson and Lisa A. Procyk for bioassay support; Kimberly Lang, James Peitzman, Angel Ortiz and Erick Hernandez for preparing midguts, collecting and/or staging insects; Katherine Thilges and Bliss M. Kernodle for microscopic analysis; Caitlin Farris and Layton Weishaar for RNA isolation and molecular supports; John Ding for DVSSJ1 protein expression; DuPont Pioneer Genomics group for RNA sequencing; Ian Lamb for thoughtful comments and suggestions.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
Funding for this work was provided by DuPont Pioneer. The funder provided support in the form of salaries for XH, JPS, DMKK, NMR, CDV, JPM, MEN, ALL, and GW, as well as research materials, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘Author Contributions’ section.
References
- 1.Katoch R, Sethi A, Thakur N, Murdock L. RNAi for Insect Control: Current perspective and future challenges. Appl Biochem Biotech. 2013:1–27. [DOI] [PubMed] [Google Scholar]
- 2.Baum JA, Roberts JK. Progress towards RNAi-mediated insect pest management. Insect Midgut and Insecticidal Proteins. 2014;47:249–95. 10.1016/B978-0-12-800197-4.00005-1 [DOI] [Google Scholar]
- 3.Gray ME, Sappington TW, Miller NJ, Moeser J, Bohn MO. Adaptation and invasiveness of western corn rootworm: intensifying research on a worsening pest. Annu Rev Entomol. 2009;54:303–21. Epub 2008/12/11. 10.1146/annurev.ento.54.110807.090434 . [DOI] [PubMed] [Google Scholar]
- 4.Narva KE, Siegfried BD, Storer NP. Transgenic approaches to western corn rootworm control. Adv Biochem Eng Biotechnol. 2013;136:135–62. 10.1007/10_2013_195 . [DOI] [PubMed] [Google Scholar]
- 5.Levine E, Oloumi-Sadeghi H. Management of Diabroticite rootworms in corn. Annu Rev Entomol. 1991;36(1):229–55. 10.1146/annurev.en.36.010191.001305 [DOI] [Google Scholar]
- 6.Tabashnik BE, Brevault T, Carriere Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotech. 2013;31(6):510–21. 10.1038/nbt.2597 [DOI] [PubMed] [Google Scholar]
- 7.Gassmann AJ, Petzold-Maxwell JL, Keweshan RS, Dunbar MW. Field-evolved resistance to Bt maize by western corn rootworm. PLoS One. 2011;6(7):e22629 10.1371/journal.pone.0022629 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andow DA, Pueppke SG, Schaafsma AW, Gassmann AJ, Sappington TW, Meinke LJ, et al. Early detection and mitigation of resistance to Bt maize by western corn rootworm (Coleoptera: Chrysomelidae). J Econ Entomol. 2016;109(1):1–12. 10.1093/jee/tov238 . [DOI] [PubMed] [Google Scholar]
- 9.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–58. 10.1038/nrm3089 [DOI] [PubMed] [Google Scholar]
- 10.Scott JG, Michel K, Bartholomay LC, Siegfried BD, Hunter WB, Smagghe G, et al. Towards the elements of successful insect RNAi. J Insect Physiol. 2013;59(12):1212–21. 10.1016/j.jinsphys.2013.08.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, et al. Control of coleopteran insect pests through RNA interference. Nat Biotech. 2007;25(11):1322–6. [DOI] [PubMed] [Google Scholar]
- 12.Rangasamy M, Siegfried BD. Validation of RNA interference in western corn rootworm Diabrotica virgifera virgifera LeConte (Coleoptera: Chrysomelidae) adults. Pest Manag Sci. 2012;68(4):587–91. Epub 2012/04/14. . [DOI] [PubMed] [Google Scholar]
- 13.Alves AP, Lorenzen MD, Beeman RW, Foster JE, Siegfried BD. RNA interference as a method for target-site screening in the western corn rootworm, Diabrotica virgifera virgifera. J Insect Sci. 2010;10:162 Epub 2010/11/12. 10.1673/031.010.14122 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Khajuria C, Rangasamy M, Gandra P, Fitter M, Geng C, et al. Long dsRNA but not siRNA initiates RNAi in western corn rootworm larvae and adults. J Appl Entomol. 2015;139(6):432–45. [Google Scholar]
- 15.Bolognesi R, Ramaseshadri P, Anderson J, Bachman P, Clinton W, Flannagan R, et al. Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (Diabrotica virgifera virgifera LeConte). PLoS One. 2012;7(10):e47534 10.1371/journal.pone.0047534 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramaseshadri P, Segers G, Flannagan R, Wiggins E, Clinton W, Ilagan O, et al. Physiological and cellular responses caused by RNAi- mediated suppression of Snf7 orthologue in western corn rootworm (Diabrotica virgifera virgifera) larvae. PLoS ONE. 2013;8(1):e54270 10.1371/journal.pone.0054270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu X, Richtman NM, Zhao JZ, Duncan KE, Niu X, Procyk LA, et al. Discovery of midgut genes for the RNA interference control of corn rootworm. Sci Rep. 2016;6:30542 Epub 2016/07/29. 10.1038/srep30542 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huvenne H, Smagghe G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. Journal of Insect Physiology. 2010;56(3):227–35. 10.1016/j.jinsphys.2009.10.004 [DOI] [PubMed] [Google Scholar]
- 19.Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends Cell Biol. 2006;16(4):181–8. Epub 2006/03/16. 10.1016/j.tcb.2006.02.006 . [DOI] [PubMed] [Google Scholar]
- 20.Behr M, Riedel D, Schuh R. The Claudin-like Megatrachea is essential in septate junctions for the epithelial barrier function in Drosophila. Developmental Cell. 2003;5(4):611–20. 10.1016/S1534-5807(03)00275-2. [DOI] [PubMed] [Google Scholar]
- 21.Izumi Y, Furuse M. Molecular organization and function of invertebrate occluding junctions. Semin Cell Dev Biol. 2014;36:186–93. Epub 2014/09/23. 10.1016/j.semcdb.2014.09.009 . [DOI] [PubMed] [Google Scholar]
- 22.Furuse M, Izumi Y. Molecular dissection of smooth septate junctions: understanding their roles in arthropod physiology. Ann N Y Acad Sci. 2017;1397(1):17–24. Epub 2017/06/22. 10.1111/nyas.13366 . [DOI] [PubMed] [Google Scholar]
- 23.Yanagihashi Y, Usui T, Izumi Y, Yonemura S, Sumida M, Tsukita S, et al. Snakeskin, a membrane protein associated with smooth septate junctions, is required for intestinal barrier function in Drosophila. J Cell Sci. 2012;125(Pt 8):1980–90. 10.1242/jcs.096800 . [DOI] [PubMed] [Google Scholar]
- 24.Izumi Y, Yanagihashi Y, Furuse M. A novel protein complex, Mesh-Ssk, is required for septate junction formation in the Drosophila midgut. J Cell Sci. 2012;125(Pt 20):4923–33. Epub 2012/08/03. 10.1242/jcs.112243 . [DOI] [PubMed] [Google Scholar]
- 25.Izumi Y, Motoishi M, Furuse K, Furuse M. A tetraspanin regulates septate junction formation in Drosophila midgut. J Cell Sci. 2016;129(6):1155–64. Epub 2016/02/06. 10.1242/jcs.180448 . [DOI] [PubMed] [Google Scholar]
- 26.Niu X, Kassa A, Hu X, Robeson J, McMahon M, Richtman NM, et al. Control of western corn rootworm (Diabrotica virgifera virgifera) reproduction through plant-mediated RNA interference. Sci Rep. 2017;7(1):12591 Epub 2017/10/05. 10.1038/s41598-017-12638-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao JZ, Oneal MA, Richtman NM, Thompson SD, Cowart MC, Nelson ME, et al. mCry3A-selected western corn rootworm (Coleoptera: Chrysomelidae) colony exhibits high resistance and has reduced binding of mCry3A to midgut tissue. J Econ Entomol. 2016. Epub 2016/03/27. 10.1093/jee/tow049 . [DOI] [PubMed] [Google Scholar]
- 28.Hammack L, Ellsbury MM, Roehrdanz RL, Pikul JL Jr. Larval sampling and instar determination in field populations of northern and western corn rootworm (Coleoptera: Chrysomelidae). J Econ Entomol. 2003;96(4):1153–9. Epub 2003/09/25. . [DOI] [PubMed] [Google Scholar]
- 29.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–22. Epub 2009/02/28. 10.1373/clinchem.2008.112797 . [DOI] [PubMed] [Google Scholar]
- 30.Miyata K, Ramaseshadri P, Zhang Y, Segers G, Bolognesi R, Tomoyasu Y. Establishing an in vivo assay system to identify components involved in environmental RNA interference in the western corn rootworm. PLoS ONE. 2014;9:1–15. 10.1371/journal.pone.0101661 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigues TB, Khajuria C, Wang H, Matz N, Cardoso DC, Valicente FH, et al. Validation of Reference Housekeeping Genes for Gene Expression Studies in Western Corn Rootworm (Diabrotica virgifera virgifera). PLoS ONE. 2014;9 10.1371/journal.pone.0109825 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bier E, Harrison MM, O’Connor-Giles KM, Wildonger J. Advances in Engineering the Fly Genome with the CRISPR-Cas System. Genetics. 2018;208(1):1–18. 10.1534/genetics.117.1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, et al. Highly Specific and Efficient CRISPR/Cas9-Catalyzed Homology-Directed Repair in Drosophila. Genetics. 2014;196(4):961–71. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dow JA. Insect midgut function. Adv Insect Physiol. 1986;19:187–328. [Google Scholar]
- 35.Chien CH, Chen WW, Wu JT, Chang TC. Investigation of lipid homeostasis in living Drosophila by coherent anti-stokes raman scattering microscopy. J Biomed Opt. 2012;17(12):126001 Epub 2012/12/05. 10.1117/1.JBO.17.12.126001 . [DOI] [PubMed] [Google Scholar]
- 36.Beyenbach KW, Skaer H, Dow JA. The developmental, molecular, and transport biology of malpighian tubules. Annu Rev Entomol. 2010;55:351–74. Epub 2009/12/08. 10.1146/annurev-ento-112408-085512 . [DOI] [PubMed] [Google Scholar]
- 37.King B, Denholm B. Malpighian tubule development in the red flour beetle (Tribolium castaneum). Arthropod Struct Dev. 2014;43(6):605–13. Epub 2014/09/23. 10.1016/j.asd.2014.08.002 . [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445(7125):275–80. Epub 2006/12/01. 10.1038/nature05382 . [DOI] [PubMed] [Google Scholar]
- 39.Pannabecker T. Physiology of the malpighian tubule. Annual Review of Entomology. 1995;40(1):493–510. 10.1146/annurev.en.40.010195.002425 [DOI] [Google Scholar]
- 40.Green LF, Bergquist PR, Bullivant S. The structure and function of the smooth septate junction in a transporting epithelium: the malpighian tubules of the New Zealand glow-worm Arachnocampa luminosa. Tissue Cell. 1980;12(2):365–81. Epub 1980/01/01. . [DOI] [PubMed] [Google Scholar]
- 41.HU X, NIU X, Richtman NM. Compositions and methods to control insect pests. International Patent Application No WO 2017/218207. 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.