Abstract
Salinity is one of the most important abiotic factors directly affecting the reproduction, molting, growth, immune, physiological and metabolic activities of Chinese mitten crab (Eriocheir sinensis). This species has strong osmoregulatory capacity and can maintain stringent internal homeostasis. However, the mechanisms conferring tolerance to salinity fluctuations are not well understood. To reveal the genes and pathways involved in osmoregulation, adult male crabs (body weight = 110 ± 5 g) were acclimated for 144 h in freshwater (FW, 0 ppt) or seawater (SW, 25 ppt). Changes in the transcriptome of crab gills were then analysed by RNA-Seq, and 174,903 unigenes were obtained. Comparison of genes between FW- SW-acclimated groups identified 932 genes that were significantly differentially expressed in the gill, comprising 433 and 499 up- and downregulated transcripts. Gene Ontology functional enrichment analysis revealed that important biological processes related to salt stress were significantly enriched, including energy metabolism, ion transport, signal transduction and antioxidant activity. Kyoto Encyclopaedia of Genes and Genomes enrichment analysis mapped the differentially expressed genes to 241 specific metabolic pathways, and pathways related to energy metabolism, oxidative phosphorylation and the tricarboxylic acid (TCA)/citrate cycle were significantly enriched. Salinity stress altered the expression of many enzymes involved in energy metabolism, ion transport, signal transduction and antioxidant pathways, including citrate synthase (CS), Na+/K+-ATPase (NKA), Na+-K+-2Cl cotransporter-1 (NKCC1), dopamine receptor D1 (DRD1), synaptic binding protein 1 (STXBP1), Cu2+/Zn2+ superoxide dismutase (SOD1) and glutathione S-transferase (GST). Additionally, the obtained transcriptomic sequencing data provided a useful resource for identification of novel genes, and further physiological analysis of Chinese mitten crab.
Introduction
Chinese mitten crab (Eriocheir sinensis) is one of the most economically important species for freshwater aquaculture in China, from the Eastern Pacific coast to the Korean Peninsula [1]. This freshwater crab species is native to China, but can also be found as an invasive species in Europe and America [2]. After puberty molting, sexually mature crabs migrate downstream to brackish water for reproduction, and juvenile crabs migrate from the sea to freshwater to grow into adult crabs [3, 4]. E. sinensis is a euryhaline species and a strong osmoregulator [5, 6]. During reproductive and growth migrations, E. sinensis can adjust its hemolymph osmotic concentration to adapt the changes in environmental salinity [7].
A number of studies have shown that salinity affects energy metabolism, ion transport, signal transduction and oxidative stress in juvenile and adult crabs [8, 9, 10, 11, 12]. A lot of energy is consumed to regulate osmotic pressure and ionic balance in response to changes in environmental salinity. At the isosmotic point, the pressure of infiltration is minimal and crustaceans can regulate homeostatic osmotic pressure using minimal energy consumption, whereas more energy is needed for osmotic adjustment [13, 14]. However, the underlying mechanisms of energy metabolism related to osmoregulation in E. sinensis are poorly understood, especially at the molecular level, and the genes and pathways involved remain largely unknown.
Inorganic ions and several specific free amino acids such as glycine, proline and alanine are the main contributors to hemolymph osmolality in crustaceans [6, 11, 15]. Ion transport is mainly achieved by the action of various ion transport enzymes such as Na+/K+-ATPase (NKA), V-type H+-ATPase (VHA), carbonic anhydrase and HCO3—ATPase [16, 17, 18]. Ion exchange mainly occurs in the posterior gill [19, 20]. However, studies on ion transporters in E. sinensis have mainly focused on changes in ion transport enzyme activity, and few studies have been carried out at the molecular level in crustaceans.
The osmotic adjustment process in aquatic animals is complicated [21]. Previous studies have demonstrated that in order to cope with salinity stress, aquatic animals can activate signal transduction pathways to maintain osmotic homeostasis [22, 23, 24]. Studies have also shown that NKA can be activated by the cAMP-dependent pathway to adjust osmotic pressure under salinity stress [9]. However, information about the signal transduction events controlling osmoregulation in aquatic animals is limited, and further work is needed to remedy this [23]. Many studies have shown that salinity stress can increase reactive oxygen species (ROS) in aquatic animal tissues, resulting in oxidative damage [25, 26]. In response, antioxidant systems are stimulated to scavenge ROS [10]. However, the underlying mechanisms of ROS generation induced by salinity stress are unclear.
Transcriptomics approaches have been widely applied to study osmotic adjustment in aquatic animals [12, 27, 28, 29]. Advances in transcriptome technology have greatly improved our ability to analyse non-model species. Therefore, in order to further investigate the potential mechanisms of osmotic regulation in E. sinensis at the transcriptional level, we performed transcriptome sequencing analysis of osmoregulation and physiological responses in E. sinensis exposed to seawater conditions, with an emphasis on energy metabolism, transporters, signal transduction and antioxidants.
Materials and methods
Animals and experimental conditions
Chinese mitten crabs (body weight = 110 ± 5 g) were obtained from the Chongming Research Base of Shanghai Ocean University and kept in a freshwater tank for 1 week for acclimation. After acclimation, healthy crabs deprived of food for 24 h were randomly divided into a freshwater group FW (0 ppt) and a seawater group SW (25 ppt) in six tanks (80 cm × 40 cm × 40 cm), with 10 crabs in each tank, and all treatments were performed in triplicate. The details of the experiment were deposited in protocols.io. http://dx.doi.org/10.17504/protocols.io.v4ve8w6 [PROTOCOL DOI]
There was no feeding during the experiment, which had a 12 h light / 12 h dark photoperiod. Water quality parameters were monitored twice daily to maintain conditions of 24.5–30.0°C, pH 8.0 ± 0.4, dissolved oxygen >5 mg/L, and total ammonia nitrogen <0.01 mg/L. After salinity stress for 144 h, six crabs per tank were treated with tricaine methane sulfonate (MS 222, 200 mg/L) and the three posterior gill tissues were harvested immediately and stored at -80°C for RNA extraction.
Hemolymph osmolarity measurement
After acclimation, healthy crabs were selected and transferred to tanks (80cm×40cm×40cm) for experiment. A salinity group SW (ppt 25) and a control group FW (ppt 0) were set up, 10 crabs were placed in each tank and all treatments were performed in triplicate. Samples were taken at 0, 3, 6, 12, 24, 48, 72, 96 and 144 hours of the experiment. At each sampling, three crabs were taken from each experimental group. Crabs were placed on ice for 15 minutes for anesthesia. Hemolymph samples were extracted from the base joints of the third foot with a 2 ml syringe. Hemolymph osmolarity was measured by OSMOMAT 3000 (Gonotec). The experimental data were analyzed by SPASS 18.0 software, and p-value <0.05 was significant difference.
RNA extraction, transcriptome library preparation, and RNA-Seq
Total RNA was extracted from gill tissue using TRIzol (Invitrogen) according to the manufacturer’s instructions. The concentration and quality of total RNA were examined using Agilent 2100 and NanoDrop 2000 instruments prior to subsequent experiments. Only high-quality RNA samples with optical density (OD) 260/280 values >1.8 and <2.2, a 28S:18S ratio >1.0, and RNA yield >5 μg were used for subsequent transcriptome analysis.
An Illumina TruseqTM RNA sample prep kit was used to construct an RNA-Seq transcriptome library. PolyA mRNA was separated from total RNA using magnetic beads with Oligo (dT). Upon addition of fragmentation buffer, enriched mRNA can be randomly broken into small fragments of ~200 bp. First-stand complementary DNA (cDNA) was synthesised using random hexamer primers with mRNA as template. Subsequently, second-strand cDNA was synthesised and a stable double-stranded structure was formed. End Repair Mix (Illumina) was used to repair cohesive ends of second-strand cDNA, and a single A base was added at the 3’ end for adapter ligation. Fifteen cycles of PCR amplification were carried out to enrich the cDNA library. Target bands were recovered from 2% agarose gel electrophoresis experiments (Certified Low Range Ultra Agarose). TBS-380 was used to quantify cDNA, and cDNA fragments were amplified to clusters by bridge PCR. Finally, a HiSeq4000 platform (Illumina) was used for sequencing.
De novo assembly of sequencing reads
Raw data arising from Illumina sequencing were firstly quality-trimmed and adapter-clipped using SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle). In this process, reads containing adapter and poly-N sequences were removed, and reads with a quality percentage (Q value) <5 bases higher than 50% were also removed. The resulting clean data were used for RNA de novo assembly with Trinity (http://trinityrnaseq.sourceforge.net/, Version: trinityrnaseq-r20140413). All sequence reading segments were assembled from scratch to generate overlapping groups and single sequences. This analysis was the basis for follow-up and biological functional analyses.
Functional annotation of transcripts
Transcripts obtained from gills subjected to different salinity treatments were scanned against NCBI non-redundant (Nr) protein, NCBI nucleotide (Nt), Swiss-Prot, and Kyoto Encyclopaedia of Genes and Genomes (KEGG) databases using BLASTx (version 2.2.25) with an E-value cut-off of <1.0×10−5. Blast2GO (http://www.Blast2go.com/b2ghome, version2.5.0) was used to obtain Gene Ontology (GO) annotation information, and the annotation results were classified into biological processes, molecular functions, and cellular components. Pathways in which transcripts were involved were analysed by KEGG (http://www.genome.jp/kegg/), a database that integrates genomic, chemical, and functional information.
Differential gene expression and enrichment analysis
Differentially expressed genes (DEGs) were identified based on read count data obtained from the analysis of gene expression levels. Gene expression levels were measured according to the Fragments Per Kilobase of transcript per Million fragments mapped (FPKM) method. Trimmed means of M-values (TMM) was used to standardise read count data, and differences were analysed using the DEGseq package for R. Filtering thresholds were q-value <0.005 and |log2FoldChange| >1. Cluster analysis was performed based on DEG levels. GO enrichment analysis of DEGs was performed using GOseq based on the Wallenius non-central hypergeometric distribution. This approach allows the probability of GO terms enriched for different genes to be more accurately calculated.
Quantitative real-time PCR (qRT-PCR)
Ten DEGs were randomly selected for validation of the Illumina sequencing data by qRT-PCR using primers listed in Table 1. RNA from gill tissue was extracted from FW and SW groups according to the manufacturer’s instructions. After reverse-transcription, primers were used to carry out qRT-PCR. The 18S rRNA gene was used as an internal reference. First-stand cDNA was synthesised from 1 μg of RNA using a PrimeScrip RT reagent kit with gDNA Eraser (Cat. No. RR036A; TaKaRa). The qRT-PCR reaction system contained 10 μL 2×SYBR Premix Ex Taq (Cat. No. RR420A; TaKaRa), 0.2 μmol/L primers. and 2 μL cDNA template. Reactions were performed on an ABI 7500 Real-Time PCR System (Life Tech, Applied Biosystems). There were six biological repeats at each data point and each sample was repeated in triplicate in order to reduce PCR system error. The 2 –ΔΔCt method was employed to determinate relative expression levels for each sample.
Table 1. Primers used for qRT-PCR.
| Genes | Primers (5’–3’) |
|---|---|
| NDUFV1 |
F: TTGGACAAGGGTGTCGACTG R: CAGTGGGAAATCCAGCACCT |
| GAPDH |
F: GACATTGTGCTCTCCAACGC R: CGACGAGGGGATGATGTTGT |
| ACSBG |
F: CAGGGAGGGAGATTCTTGCG R: ATACGATTTGGTCCTCGGGC |
| ABCC3 |
F: AAACCCGACGACTCAACTGT R: GCCAATGTCTGCAAAATCTACT |
| DRD1 |
F: GAGAGACGCTTCCAACCTCC R: CCAGCGACTCCAGAGTTACC |
| ARF1 |
F: CTGCATCAGTATCGACCGCT R: GTAGGCCATGTTCACCACGA |
| SOD1 |
F: TTGGAGATGGTCCCCTCGAT R: AACTACCCCTCGCTGCTCTA |
| MIF1 |
F: GAATGCGACCGCAGAATGAG R: CCCACCATATGAGGCATCGG |
| NKCC1 |
F: GGTTAAGCACGTGGTGAGGA R: TCCTTGGCTTCAATGCCGAT |
| ATP1B |
F: CACGTGACTGAGTGTTCCGA R: ATGCATGGGGATTCACGGTT |
| NKA |
F: TGAATGACTCCCCAGCTCTCAAG R: CAGAATCATGTCAGCAGCCTGCT |
| 18S |
F: GGGTCCGAAGCGTTTACT R: TCACCTCTAGCGGCACAA |
NDUFV1, NADH dehydrogenase (ubiquinone) flavoprotein 1; GAPDH, Glyceraldehyde 3-phosphate dehydrogenase; ACSBG, Long-chain-fatty-acid-CoA ligase; ABCC3, Canalicular multispecific organic anion transporter 2; DRD1, Dopamine receptor D1; ARF1, ADP-ribosylation factor 1; SOD1, Cu2+/Zn2+ Superoxide dismutase-1; MIF1, Macrophage migration inhibitory factor; NKCC1, Na+-K+-2Cl cotransporter-1; ATP1B, Sodium/potassium-transporting ATPase subunit beta; NKA, Na+/K+ATPase.
Results
Hemolymph osmolarity of E.sinensis
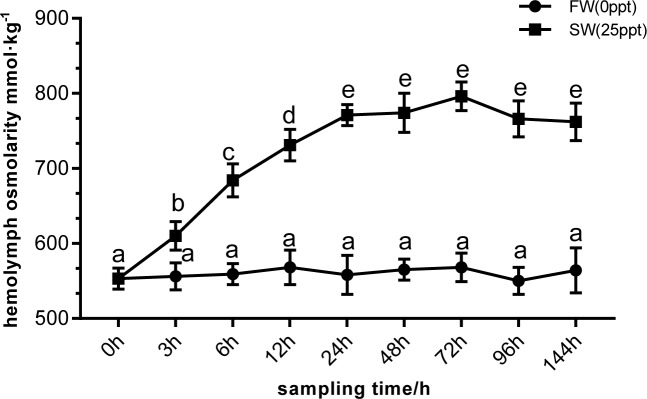
The hemolymph osmolarity of crabs in SW group was higher than that of control group FW at corresponding sampling time. Hemolymph osmolarity in SW group increased rapidly from 0 hour to 24 hour, with the effect of salinity, hemolymph osmolarity first increased and then decreased. Hemolymph osmolarity decreases after reaching the highest point at 72 hour (Fig 1).
Fig 1. Hemolymph osmolarity of E.sinensis at different time points under high salinity stress.
The different letters mean significant differences.
Sequencing and de novo assembly
A total of 47,446,444 raw reads were generated from E. sinensis gill tissue. After removing low-quality sequences and adapter clipping, 45,517,342 clean reads remained, including 24.1 million clean reads from FW samples and 21.4 million clean reads from SW samples (Table 2). Using Trinity for de novo assembly, 210,835 transcripts were obtained. The longest transcript of each gene was taken as a unigene for subsequent analysis, and 174,903 unigenes were obtained. This process yielded 164,908 transcripts (78.22%) and 144,562 unigenes (82.65%) ranging from 200 bp to 500 bp in length, and the average length of transcripts and unigenes was 494 bp and 433 bp, respectively (Table 3; S1 Fig).
Table 2. Statistics for Illumina short reads from the E. sinensis gill transcriptome.
| Sample name | Raw reads | Clean reads | Q20 (%)a | Q30 (%)b | GC Content (%)c |
|---|---|---|---|---|---|
| FW1 | 25,161,634 | 24,136,230 | 96.39 | 92.28 | 50.77 |
| SW1 | 22,284,810 | 21,381,112 | 96.81 | 92.86 | 53.10 |
| Total | 47,446,444 | 45,517,342 |
a Q20%, percentage of bases with Phred value >20.
b Q30%, percent of bases with Phred value >30.
c GC%, percentage of G and C bases among total bases.
Table 3. Summary of RNA-Seq de novo assembly results.
| Min length | Mean length | Max length | N50a | N90b | Total nucleotides | |
|---|---|---|---|---|---|---|
| Transcripts | 201 | 494 | 15,986 | 615 | 230 | 104,198,495 |
| Unigenes | 201 | 433 | 15,986 | 456 | 224 | 75,751,538 |
aN50, Transcripts sorted by length from long to short, for which the sum of the length of transcripts was not less than 50% of the total length of mosaic transcripts.
bN90, Transcripts sorted by length from long to short, for which the sum of the length of transcripts was not less than 90% of the total length of mosaic transcripts.
Annotation and functional analysis of gill transcripts
Seven databases, namely Nr, Nt, Protein family (PFAM), Clusters of Orthologous Groups (COG), SwissProt, KEGG, and GO, were used to obtain comprehensive gene functional information. A total of 28,296 (16.17%) unigenes were matched using the NR database, 12,448 (7.11%) unigenes were matched using the NT database, 21,985 (12.56%) unigenes were matched using the SwissProt database, and 44,672 (25.54%) unigenes were annotated in at least one database (Table 4).
Table 4. Summary of E. sinensis gill transcriptome annotation information.
| Category | Number of unigenes | Percentage (%) |
|---|---|---|
| Annotated in NR | 28,296 | 16.17 |
| Annotated in NT | 12,448 | 7.11 |
| Annotated in KO | 13,347 | 7.63 |
| Annotated in SwissProt | 21,985 | 12.56 |
| Annotated in PFAM | 30,611 | 17.5 |
| Annotated in GO | 30,578 | 17.48 |
| Annotated in COG | 15,241 | 8.71 |
| Annotated in all databases | 4,238 | 2.42 |
| Annotated in at least one database | 44,672 | 25.54 |
| Total unigenes | 174,903 | 100 |
GO database analysis facilitated division of DEGs into Biological Process (BP), Cellular Component (CC) and Molecular Function (MF) categories, and annotated unigenes were further assigned to 49 subcategories (S2 Fig). Analysis of level 2 GO terms showed that in the BP category, the most common annotation terms were cellular process (GO:0009987), metabolic process (GO:0008152), and single-organism process (GO:0044699); in the CC category, the most common annotation terms were cell (GO:0005623), cell part (GO:0044464), macromolecular complex (GO:0032991), organelle (GO:0043226), membrane (GO:0016020), and membrane part (GO:0044425); in the MF category, the most common annotation terms were binding (GO:0005488), catalytic activity (GO:0003824), and transporter activity (GO:0005215).
The 174,903 assembled unigenes were aligned to the COG database to further investigate and classify possible gene functions. In total, 15,241 unigenes were classified into 26 COG categories based on functions according to COG number (S3 Fig). Among the matched sequences, general function prediction only (2,868) represented the largest group of unigenes, followed by signal transduction mechanisms (2,097), posttranslational modification, protein turnover, chaperones (1,833), and translation, ribosomal structure and biogenesis (1,384).
In order to better understand the relationships between DEGs under high salinity conditions, we assigned unigenes based on KEGG pathways (S4 Fig). A total 174,903 unigenes were assigned to Cellular Processes (A), Environmental Information Processing (B), Genetic Information Processing (C), Metabolism (D), and Organismal Systems (E) categories. These five main categories included 32 Hierarchy 2 and 277 KEGG pathways. Among Hierarchy 2 pathways, many unigenes were strongly associated with signal transduction (1,550), translation (1,411), transport and catabolism (902), and carbohydrate metabolism (892).
Analysis of DEGs
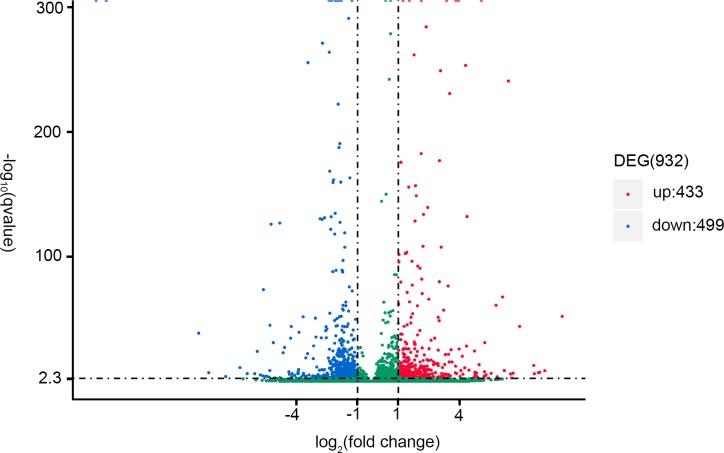
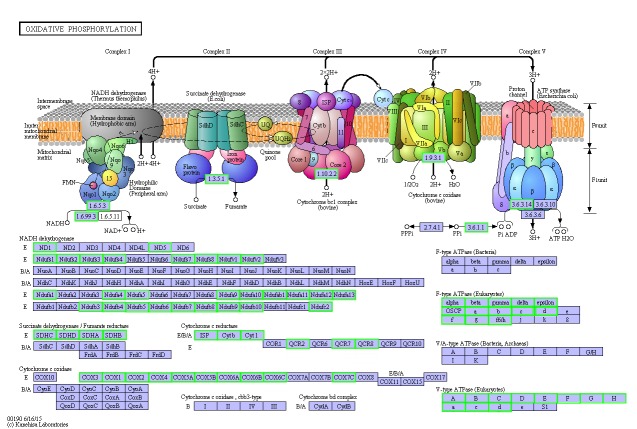
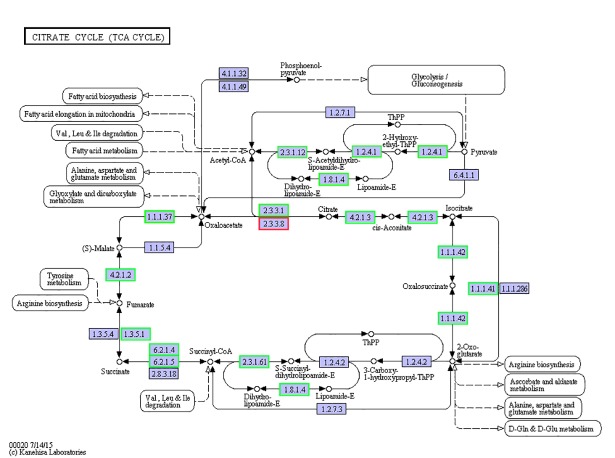
A total of 932 DEGs were identified from SW and FW gill tissue, of which 433 and 499 were up- and downregulated, respectively (Fig 2). Using the GO database, DEGs were classified according to their biological processes, cellular components and molecular functions (S5 Fig). Based on DEGs of FW vs SW, GO annotations were categorised into 2,021 subcategories within the three major categories. Among biological processes, many DEGs were associated with organophosphate metabolic process (GO:0019637), nucleoside phosphate metabolic process (GO:0006753), nucleoside metabolic process (GO:0009116), ribose phosphate metabolic process (GO:0019693), and nucleoside metabolic process (GO:0009116). Among cellular components, many DEGs were associated with membrane part (GO:0044425), mitochondrium (GO:0005739), organelle envelope (GO:0031967), envelope (GO:0031975), and mitochondrial part (GO:0044429). Among molecular functions, most DEGs were enriched in catalytic activity (GO:0003824) and oxidoreductase activity (GO:0016491). In order to further investigate the relationships between DEGs identified in FW and SW groups, KEGG pathway analysis was performed, and DEGs were categorised into 134 specific KEGG pathways (S6 Fig). Significantly enriched pathways included oxidative phosphorylation (Fig 3), the tricarboxylic acid (TCA) cycle (Fig 4), and collecting duct acid secretion (S7 Fig). The top 13 most enriched KEGG pathways are listed in Table 5. Candidate DEGs potentially associated with salinity adaptation and osmoregulation were categorised into four energy metabolism, transporters, signal transduction and antioxidant pathways based on a combination of enrichment analysis, annotation, and manual literature searches (Table 6).
Fig 2. Volcano plot comparing saltwater (SW) and freshwater (FW) treatment groups.
Fig 3. DEGs related to oxidative phosphorylation.
Fig 4. DEGs related to the TCA/citrate cycle.
Table 5. Differentiation of pathway analysis of E. sinensis gill.
| Pathway terms | KEGG ID | Number of DEGs Number | Background numbera | p-value |
|---|---|---|---|---|
| Oxidative phosphorylation | ko00190 | 75 | 322 | 3.52E-27 |
| Cardiac muscle contraction | ko04260 | 19 | 72 | 1.15E-07 |
| Valine, leucine and isoleucine degradation | ko00280 | 20 | 110 | 8.45E-06 |
| TCA/citrate cycle | ko00020 | 23 | 155 | 2.39E-05 |
| Carbon metabolism | ko01200 | 42 | 456 | 0.000145 |
| Rheumatoid arthritis | ko05323 | 13 | 79 | 0.00166 |
| Collecting duct acid secretion | ko04966 | 10 | 51 | 0.002887 |
| Synaptic vesicle cycle | ko04721 | 14 | 105 | 0.005537 |
| Propanoate metabolism | ko00640 | 10 | 66 | 0.014234 |
| Benzoate degradation | ko00362 | 6 | 26 | 0.022755 |
| Fatty acid degradation | ko00071 | 12 | 107 | 0.042323 |
| Caprolactam degradation | ko00930 | 4 | 13 | 0.041195 |
a Background number: the number of total genes assigned to the pathway
Table 6. DEGs potentially associated with salinity adaptation and osmoregulation.
| Functional group | Gene name | Gene ID | Fold change (SW/FW) | Gene function | Cellular component |
|---|---|---|---|---|---|
| Energy metabolism | Cytochrome c oxidase subunit 2 | c119126_g1 | -2.4084 | Transcription coactivator activity | Mitochondrial inner membrane |
| NADH dehydrogenase (ubiquinone) flavoprotein 1 | c173489_g1 | -2.1231 | Oxidoreductase activity, acting on NAD(P)H | Mitochondrial inner membrane | |
| Glyceraldehyde-3-phosphate dehydrogenase | c169636_g1 | -5.0203 | Oxidoreductase activity | ||
| Branched-chain aminotransferase | c157822_g2 | -1.8577 | Catalytic branched-chain amino acid metabolism | ||
| Citrate synthase | c134754_g1 | -2.6232 | Transferring acyl groups | Mitochondrial matrix | |
| Isocitrate dehydrogenase | c179743_g1 | -2.183 | Mitochondrial matrix | ||
| Succinate dehydrogenase | c168548_g1 | -1.7838 | Electron transport | Mitochondrial matrix | |
| Long-chain-fatty-acid-CoA ligase | c172027_g1 | -2.8424 | Fatty acid degradation | Mitochondrial matrix | |
| Transporters | Na+/K+ATPase | c173400_g1 | -2.0157 | Ion transport | Potassium-exchanging ATPase complex |
| V-type H+-transporting ATPase subunit G | c163080_g1 | -2.3686 | Hydrogen ion transmembrane transporter activity | Membrane | |
| V-type H+-transporting ATPase subunit F | c170449_g1 | -2.2102 | Hydrogen ion transmembrane transporter activity | Membrane | |
| Na+-K+-2Cl-contransporter | c165384_g3 | 2.4757 | Sodium ion transport | Membrane | |
| Chloride channel 2 | c182730_g1 | -1.6306 | Chloride transport | Membrane | |
| ATP-binding cassette, subfamily C | c179421_g1 | 4.3496 | Transmembrane transport | Integral component of membrane | |
| Signal transduction | Dopamine receptor D1 | c167704_g1 | 4.0794 | G-protein coupled receptor activity | Integral component of membrane |
| Synaptotagmin-1 | c181649_g2 | 2.2691 | Regulation of neurotransmitter release | Membrane | |
| Syntaxin-binding protein 1 | c179461_g2 | 1.3491 | Regulation of neurotransmitter release | Membrane | |
| ADP-ribosylation factor 1 | c170851_g2 | 6.1241 | Cell vesicle transport process | Membrane | |
| Phospholipase D1/2 | c161757_g1 | 1.8775 | Phospholipid signal transduction | ||
| Antioxidant activity | Cu2+/Zn2+ Superoxide dismutase | c153237_g1 | 6.5184 | Antioxidant activity | |
| Glutathione S-transferase | c163902_g1 | 1.4387 | |||
| Glucose-6-phosphate 1-dehydrogenase | c178975_g1 | 1.5714 | Oxidation-reduction process | ||
| Glutathione peroxidase | c153117_g1 | -1.2138 | Response to oxidative stress | ||
| Macrophage migration inhibitory factor | c142195_g1 | -4.2166 | Immunoregulatory |
qRT-PCR validation of RNA-Seq data
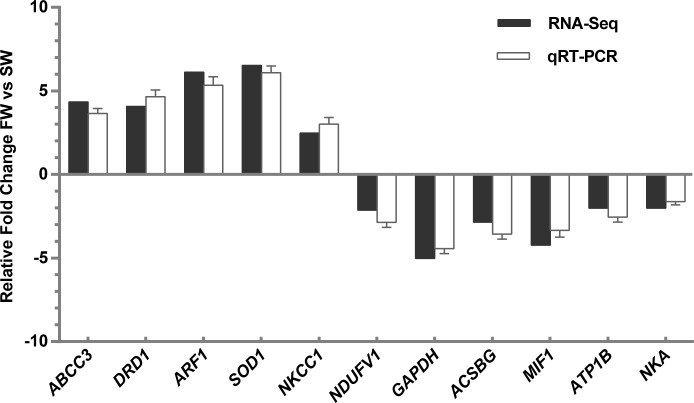
To validate our Illumina sequencing results, five upregulated DEGs (ABCC3, DRD1, ARF1, SOD1 and NKCC1) and six downregulated genes (NDUFV1, GAPDH, ACSBG, MIF1, ATP1B and NKA) were selected or verified by qRT-PCR. The results showed that the expression trends of all ten DEGs were consistent with the high-throughput RNA-Seq results, indicating that the sequencing results were reliable (Fig 5).
Fig 5. qPCR validation of RNA-Seq data.
Discussion
In recent years, transcriptome analysis technology has developed rapidly, and has been applied to the study of aquatic animals. E. sinensis is a representative species for analysis of hyperosmotic regulation [5, 30]. In this species, osmoregulatory mechanisms are activated in response to changes in environmental salinity during reproductive migration [31]. To further investigate the potential mechanisms underpinning osmotic regulation and identify osmotic-regulated genes, we compared the transcriptomes of E. sinensis gill tissue in freshwater (FW; 0 ppt) and seawater (SW; 25 ppt) treatment groups. A total of 932 DEGs were identified, comprising 433 and 499 up- and downregulated genes. We analysed these DEGs with an emphasis on genes related to energy metabolism, ion channel regulation, signal transduction, and oxidative stress.
Energy metabolism
Osmotic pressure regulation is achieved by several ion transporters and ion transport channels, which require a large amount of energy [32]. However, in our present study, most DEGs related to two important energy metabolism pathways, oxidative phosphorylation and the TCA cycle, were downregulated. Genes related to nicotinamide adenine dinucleotide (NADH) dehydrogenase, ubiquinone cytochrome C reductase, cytochrome C oxidase, and ATP synthase were downregulated. These enzymes are located on the mitochondrial membrane and constitute electron transfer chains that synthesise ATP through biological oxidation [33]. The same trend was found for genes related to the TCA cycle; Citrate synthase (CS), isocitrate dehydrogenase and succinate dehydrogenase were all downregulated. As the first rate-limiting enzyme of the TCA cycle, CS catalyses the condensation of oxaloacetic acid and acetyl coenzyme A to produce citric acid and coenzyme A for ATP production [34]. Isocitrate dehydrogenase catalyses the oxidative decarboxylation of isocitrate to produce α-ketoglutaric acid, CO2 and NADH, providing energy for organisms and biosynthetic precursors. NADH is an important coenzyme in cells that is involved in most oxidation-reduction reactions of sugars, fats and proteins [35]. Succinic dehydrogenase converts succinic acid into fumaric acid, accompanied by the formation of ATP. Unlike other enzymes in the TCA cycle, succinic dehydrogenase is the only enzyme embedded in the mitochondrial inner membrane, and is an important part of the mitochondrial inner membrane [36]. Our current findings appear to contradict those of a previous study in which osmoregulatory processes are accompanied by an increase in energy demand [11], but these results were consistent with those of Zhang et al. (2018) [12]. The process of osmotic pressure regulation in E. sinensis can also be divided into two stages; in the initial stage (1–3 days), the energy requirement of osmotic pressure regulation increases, hence energy metabolism increases; in the subsequent regulatory stage (at 3 days after transfer), the energy requirement decreases, and energy metabolism returns to normal levels. We speculate that after 144 h of high salinity stress, osmotic pressure regulation in E. sinensis enters a relatively stable stage, during which the energy required for osmotic regulation is decreased (Fig 1) [37]. Additionally, a salinity of 25 ppt is close to the isosmotic state for E. sinensis (33 ppt). At the isosmotic point, the energy used by organisms to maintain osmotic pressure is minimal [38]. This results in the downregulation of genes encoding enzymes are related to energy metabolism. Further studies should be performed, including time courses, to probe the link between energy metabolism and osmotic adjustment in E. sinensis.
Transporters
The ion transport epithelium in the gill is the main location for osmotic adjustment and ion transport in crustaceans [39]. Ion transport is accomplished mainly by NKA and other ion transport enzymes [40]. Gill epithelium NKA is essential for hyperosmotic crustaceans. It is mainly involved in osmotic regulation and ion regulation, can switch between Na+ and K+ transport, and can also affect the activity of other enzyme systems related to ionic regulation on the gill epithelium [41]. In our current transcriptome analysis, expression of NKA in the gill filament of the SW group was significantly downregulated compared with the control group. E. sinensis is a hyperosmoregulator, which means it needs to take up salt from the medium when salinity fluctuates between freshwater and seawater. We conclude that the downregulation of NKA is most likely due to E. sinensis is close to isosmotic to the seawater (25ppt), which would reduce NKA activity [42, 43].
Another classic ion transporter, NKCC1, was also included among the identified DEGs. NKCC1 is particularly important for regulating ion transport, and belongs to the SLC12A family that includes two subtypes, NKCC1 and NKCC2 [44]. NKCC1 is closely related to the secretion of Cl− in epithelial cells, and is considered the secretion subtype [45]. In the present study, expression of NKCC1 was upregulated in the gill of E. sinensis, which reflects the activation of chloride secretion. Crabs may accelerate the secretion of Cl- from the body by increasing NKCC1 levels in a high salinity environment, thereby achieving salt and water retention.
Furthermore, the gene encoding V-type H+-ATPase (VHA) was downregulated following salinity stress. During osmoregulation in teleosts, VHA is a major ion-regulating enzyme, second only to NKA, which powers the uptake of sodium and chloride ions by excreting protons [46]. Our results imply that VHA may be downregulated in response to high salinity in the surrounding water body. This reflects a reduction in sodium chloride intake by the body.
Signal transduction
Some studies have shown that neuroendocrine systems can regulate the activity of NKA, VHA, and cystic fibrosis transmembrane conductance regulator in epithelial cells via cAMP-dependent pathways [47, 48, 49]. In the present study, DRD1 was significantly upregulated after salinity stress. Dopamine binds to dopamine receptors and plays an important role in maintaining the balance of ions in kidney [50]. DRD1 enhances the role of cAMP in cells by activating adenylate cyclase [51]. NKA activity can be inhibited by phosphorylation of NKA subunits mediated by protein kinase A [52]. Thus, we infer that under high salinity stress, E. sinensis might reduce the activity of NKA by upregulating the expression of DRD1, thereby achieving ionic regulation.
In addition, we also found that STXBP1 was upregulated significantly. STXBP1 is a very important protein in synaptic vesicles, which is fixed to the membrane of secretory vesicles via a single transmembrane domain at its N-terminus. It can sense the flow of Ca2+ and cause the fusion of vesicles and the release of neurotransmitters [53]. This further indicates that during osmotic adjustment, the neuroendocrine system may regulate ion transport in the gill epithelium through neurotransmitter signalling.
Antioxidant pathways
Changes in salinity can cause an increase in ROS in fish that may result in oxidative damage if not cleared quickly [54]. Antioxidant mechanisms are the main barrier against oxidative damage in fish. Fish can scavenge ROS and enhance resistance through antioxidant mechanisms [55]. In the present study, various antioxidant genes were included among the identified DEGs, including GST (upregulated), glucose-6-phosphate dehydrogenase (G6PDH) (upregulated), and SOD1 (upregulated). GST is an important detoxifying enzyme in organisms that attaches glutathione to various electrophilic compounds and increases their solubility, thereby facilitating their excretion from cells. GST also possesses glutathione-dependent peroxidase activity and protects the body from endogenous peroxide damage [56]. G6PDH provides the reductive NADH coenzyme for biosynthesis, and produces glutathione, which regulates the redox state of the organism [57]. We speculate that high salinity stress may cause oxidative stress in E. sinensis, and GST and G6PDH likely play important roles in preventing oxidative damage in crabs.
Conclusions
The present study demonstrates that brackish water conditions decrease energy metabolism and enhance the antioxidant capacity of Chinese mitten crab. E. sinensis can mobilise the neural transmission system to alter the activity of ion transport enzymes and osmotic regulation. However, the details of osmotic regulation in E. sinensis remain unclear, and protein expression should also be analysed to elucidate the mechanisms responsible.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Acknowledgments
We would like to thank the native English speaking scientists of Elixigen Company (Huntington Beach, California) for editing our manuscript.
Data Availability
Our next-generation sequencing data has been uploaded to NCBI Sequence Read Archive (SRA), and the BioProject ID is PRJNA508867. There is the link for reviewing our SRA submission data: https://submit.ncbi.nlm.nih.gov/subs/sra/SUB4871716/overview (Account: junyz, password: 992325zjy).
Funding Statement
National Natural Science Foundation of China [grant number 31472287] and the China Agriculture Research System-48 are all the funding of support received during this study; there is no additional external funding received for this study.
References
- 1.Sui L, Zhang F, Wang X, Bossier P, Sorgeloos P, Hänfling B. Genetic diversity and population structure of the Chinese mitten crab (Eriocheir sinensis) in its native range. Marine Biology. 2009; 156(8): 1573–1583. 10.1007/s00227-009-1193-2 [DOI] [Google Scholar]
- 2.Anger K. Effects of temperature and salinity on the larval development of the Chinese mitten crab Eriocheir sinensis (Decapoda: Grapsidae). Marine Ecology Progress. 1991; 72(1–2): 103–110. 10.3354/meps072103 [DOI] [Google Scholar]
- 3.Cheng Y, Wu X, Yang X, Hines AH. Current trends in hatchery techniques and stock enhancement for Chinese mitten crab, Eriocheir japonica sinensis. Reviews in Fisheries Science. 2008; 16(1–3): 377–84. 10.1080/10641260701681698 [DOI] [Google Scholar]
- 4.Zhang T, Li Z, Cui Y. Survival, growth, sex ratio, and maturity of the Chinese mitten crab (Eriocheir sinensis) reared in a chinese pond. Journal of Freshwater Ecology. 2001; 16(4): 633–640. 10.1080/02705060.2001.9663855 [DOI] [Google Scholar]
- 5.Rathmayer M, Siebers D. Ionic balance in the freshwater-adapted Chinese crab, Eriocheir sinensis. Journal of Comparative Physiology B Biochemical Systemic & Environmental Physiology. 2001; 171(4): 271–281. 10.1007/s003600100 [DOI] [PubMed] [Google Scholar]
- 6.Wang R, Zhuang P, Feng G, Zhang L, Huang X, Jia X. Osmotic and ionic regulation and Na+/K+-ATPase, carbonic anhydrase activities in mature Chinese mitten crab, Eriocheir sinensis H. Milne Edwards, 1853 (Decapoda, Brachyura) exposed to different salinities. Crustaceana. 2012; 85(12–13): 1431–1447. 10.1163/15685403-00003125 [DOI] [Google Scholar]
- 7.Dittel AI, Epifanio CE. Invasion biology of the Chinese mitten crab Eriochier sinensis: A brief review. Journal of Experimental Marine Biology & Ecology. 2009; 374(2): 79–92. 10.1016/j.jembe.2009.04.012 [DOI] [Google Scholar]
- 8.Torres G, Charmantier-Daures M, Chifflet S, Anger K. Effects of long-term exposure to different salinities on the location and activity of Na+/K+-ATPase in the gills of juvenile mitten crab, Eriocheir sinensis. Comparative Biochemistry & Physiology Part A Molecular & Integrative Physiology. 2007; 147(2): 460–465. 10.1016/j.cbpa.2007.01.020 [DOI] [PubMed] [Google Scholar]
- 9.Mo JL, Devos P, Trausch G. Active absorption of Cl−and Na+ in posterior gills of Chinese crab Eriocheir sinensis: modulation by dopamine and cAMP. Journal of Crustacean Biology. 2003; 23(August 2003): 505–512. 10.1651/C-2295 [DOI] [Google Scholar]
- 10.Long X, Wu X, Zhao L, Ye H, Cheng Y, Zeng C. Effects of salinity on gonadal development, osmoregulation and metabolism of adult male Chinese mitten crab, Eriocheir sinensis. Plos One. 2017; 12(6): e0179036 10.1371/journal.pone.0179036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li E, Wang S, Li C, Wang X, Chen K, Chen L. Transcriptome sequencing revealed the genes and pathways involved in salinity stress of Chinese mitten crab, Eriocheir sinensis. Physiological Genomics. 2014; 46(5): 177–190. 10.1152/physiolgenomics.00191.2013 [DOI] [PubMed] [Google Scholar]
- 12.Zhang D, Liu J, Qi T, Ge B, Liu Q, Jiang S, et al. Comparative transcriptome analysis of Eriocheir japonica sinensis response to environmental salinity. Plos One. 2018; 13(9): e0203280 10.1371/journal.pone.0203280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Via GJD. Effects of salinity and temperature on oxygen consumption in a freshwater population of Palaemonetes antennarius (Crustacea, Decapoda). Comparative Biochemistry & Physiology Part A Physiology. 1987; 88(2): 299–305. 10.1016/0300-9629(87)90487-7 [DOI] [Google Scholar]
- 14.Kutty MN, Murugapoopathy G, Krishnan TS. Influence of salinity and temperature on the oxygen consumption in young juveniles of the Indian prawn Penaeus indicus. Marine Biology. 1971; 11(2): 125–131. 10.1007/BF00348761 [DOI] [Google Scholar]
- 15.Chen JC, Chia PG. Osmotic and ionic concentrations of Scylla serrata (Forskål) subjected to different salinity levels. Comparative Biochemistry & Physiology Part A Physiology. 1997; 117(2): 239–244. 10.1016/S0300-9629(96)00237-X [DOI] [Google Scholar]
- 16.Tsai JR, Lin HC. V-type H+-ATPase and Na+/K+-ATPase in the gills of 13 euryhaline crabs during salinity acclimation. Journal of Experimental Biology. 2007; 210(Pt 4): 620–627. 10.1242/jeb.02684 [DOI] [PubMed] [Google Scholar]
- 17.Riestenpatt S, Petrausch G, Siebers D. Cl− influx across posterior gills of the Chinese crab (Eriocheir sinensis): potential energization by a V-type H+ -ATPase. Comparative Biochemistry & Physiology Part A Physiology. 1995; 110(3): 235–241. 10.1016/0300-9629(94)00169-T [DOI] [Google Scholar]
- 18.Pequeux A, Marchal A, Wanson S, Gilles R. Kinetic characteristics and specific activity of gill Na+/K+-ATPase in the euryhaline Chinese crab, Eriocheir sinensis during salinity acclimation. Marine Biology Letters. 1984; 5(1): 35–43. [Google Scholar]
- 19.Barra JA, Pequeux A, Humbert W. A morphological study on gills of a crab acclimated to fresh water. Tissue & Cell. 1983; 15(4): 583–596. 10.1016/0040-8166(83)90009-5 [DOI] [PubMed] [Google Scholar]
- 20.Gilles R, Pequeux A, Bianchini A. Physiological-aspects of NaCl movements in the gills of the euryhaline crab, Eriocheir sinensis, acclimated to fresh-water. Comparative Biochemistry and Physiology Part A. 1988; 90(1): 201–207. 10.1016/0300-9629(88)91028-6 [DOI] [Google Scholar]
- 21.Kultz D, Fiol D, Valkova N, Gomez-Jimenez S, Chan SY, Lee J. Functional genomics and proteomics of the cellular osmotic stress response in 'non-model' organisms. Journal of Experimental Biology. 2007; 210(9): 1593–1601. 10.1242/jeb.000141 [DOI] [PubMed] [Google Scholar]
- 22.Evans, David H. Cell signaling and ion transport across the fish gill epithelium. Journal of Experimental Zoology Part A Ecological Genetics & Physiology. 2010; 293(2002): 336–347. 10.1002/jez.1857 [DOI] [PubMed] [Google Scholar]
- 23.Evans TG, Somero GN. A microarray-based transcriptomic time-course of hyper- and hypo-osmotic stress signaling events in the euryhaline fish Gillichthys mirabilis: osmosensors to effectors. Journal of Experimental Biology. 2009; 211(2): 3636–3649. 10.1242/jeb.022160 [DOI] [PubMed] [Google Scholar]
- 24.Fiol DF, Kültz D. Osmotic stress sensing and signaling in fishes. Febs Journal. 2010; 274(22): 5790–5798. 10.1111/j.1742-4658.2007.06099.x [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Wang WN, Wang AL, Wang JM, Sun RY. Effects of dietary vitamin E supplementation on antioxidant enzyme activities in Litopenaeus vannamei (Boone, 1931) exposed to acute salinity changes. Aquaculture. 2007; 265(1): 351–358. 10.1016/j.aquaculture.2007.02.010 [DOI] [Google Scholar]
- 26.Sies H. 1-Oxidative Stress: Introductory Remarks. Oxidative Stress. 1985: 1–8. [DOI] [Google Scholar]
- 27.Zhang X, Wen H, Wang H, Ren Y, Zhao J, Li Y. RNA-Seq analysis of salinity stress-responsive transcriptome in the liver of spotted sea bass (Lateolabrax maculatus). Plos One. 2017; 12(3): e0173238 10.1371/journal.pone.0173238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S, Chen G, Xu H, Zou W, Yan W, Wang Q, et al. Transcriptome analysis of mud crab (Scylla paramamosain) gills in response to Mud crab reovirus (MCRV). Fish & Shellfish Immunology. 2016; 60(2): 545–553. 10.1016/j.fsi.2016.07.033 [DOI] [PubMed] [Google Scholar]
- 29.Leguen I, Cam AL, Montfort J, Peron S, Fautrel A. Transcriptomic analysis of trout gill ionocytes in fresh water and sea water using laser capture microdissection combined with microarray analysis. Plos One. 2015; 10(10): e0139938 10.1371/journal.pone.0139938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Péqueux A. Osmotic regulation in crustaceans. Journal of Crustacean Biology. 1995; 15(1): 1–60. 10.2307/1549010 [DOI] [Google Scholar]
- 31.Jia XY, Zhuang P, Feng GP, Wang RF, Jun LU, Huang XR. The relationship between hemolymph biochemical parameters and salinity in female parent Chinese mitten crab (Eriocheir sinensis). Journal of Fisheries of China. 2012; 36(1): 91–97. (in Chinese with English abstract) [Google Scholar]
- 32.Tseng Y, Hwang P. Some insights into energy metabolism for osmoregulation in fish. Comparative Biochemistry & Physiology Toxicology & Pharmacology Cbp. 2008; 148(4): 419–429. 10.1088/0268-1242/18/6/311 [DOI] [PubMed] [Google Scholar]
- 33.Dahout-Gonzalez C, Nury H, Trézéguet V, Lauquin GJ, Pebay-Peyroula E, Brandolin G. Molecular, functional, and pathological aspects of the mitochondrial ADP/ATP carrier. Physiology. 2006; 21(21): 242–249. 10.1152/physiol.00005.2006 [DOI] [PubMed] [Google Scholar]
- 34.Schnarrenberger C, Martin W. Evolution of the enzymes of the citric acid cycle and the glyoxylate cycle of higher plants. Febs Journal. 2010; 269(3): 868–883. 10.1046/j.0014-2956.2001.02722.x [DOI] [PubMed] [Google Scholar]
- 35.Ying W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: regulation and biological consequences. Antioxidants & Redox Signaling. 2008; 10(2): 179–206. 10.1089/ars.2007.1672 [DOI] [PubMed] [Google Scholar]
- 36.King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006; 25(34): 4675–4682. 10.1038/sj.onc.1209594 [DOI] [PubMed] [Google Scholar]
- 37.Sangiao-Alvarellos S, Arjona FJ, Mp MDR, Míguez JM, Mancera JM, Soengas JL. Time course of osmoregulatory and metabolic changes during osmotic acclimation in Sparus auratus. Journal of Experimental Biology. 2005; 208(Pt 22): 4291–4304. 10.1242/jeb.01900 [DOI] [PubMed] [Google Scholar]
- 38.Roast SD, Rainbow PS, Smith BD, Nimmo M, Jones MB. Trace metal uptake by the Chinese mitten crab Eriocheir sinensis: the role of osmoregulation. Marine Environmental Research. 2002; 53(5): 453–464. 10.1016/S0141-1136(02)00090-9 [DOI] [PubMed] [Google Scholar]
- 39.Onken H. Active NaCl absorption across split lamellae of posterior gills of Chinese crabs (Eriocheir sinensis) adapted to different salinities. Comparative Biochemistry & Physiology Part A Molecular & Integrative Physiology. 1999; 123(4): 377–384. [Google Scholar]
- 40.Charmantier G, Charmantier-Daures M, Bouaricha N, Thuet P, Aiken DE. Ontogeny of osmoregulation and salinity tolerance in two decapod crustaceans: Homarus americanus and Penaeus japonicus. Biological Bulletin. 1988; 175(1): 102–110. 10.2307/1541897 [DOI] [Google Scholar]
- 41.Castilho PC, Martins IA, Bianchini A. Gill Na+/K+-ATPase and osmoregulation in the estuarine crab, Chasmagnathus granulata Dana, 1851 (Decapoda, Grapsidae). J Exp Mar Bio Ecol. 2001; 256(2): 215–227. 10.1016/S0022-0981(00)00315-4 [DOI] [PubMed] [Google Scholar]
- 42.Mendonça NN, Masui DC, Mcnamara JC, Leone FA, Furriel RPM. Long-term exposure of the freshwater shrimp Macrobrachium olfersii to elevated salinity: Effects on gill Na+/K+-ATPase α-subunit expression and K + -phosphatase activity. Comparative Biochemistry & Physiology Part A Molecular & Integrative Physiology. 2007; 146(4): 534–543. 10.1016/j.cbpa.2006.01.019 [DOI] [PubMed] [Google Scholar]
- 43.Belli NM, Faleiros RO, Firmino KC, Masui DC, Leone FA, Mcnamara JC, et al. Na+/K+-ATPase activity and epithelial interfaces in gills of the freshwater shrimp Macrobrachium amazonicum (Decapoda, Palaemonidae). Comparative Biochemistry & Physiology Part A Molecular & Integrative Physiology. 2009; 152(3): 431–439. 10.1016/j.cbpa.2008.11.017 [DOI] [PubMed] [Google Scholar]
- 44.Cutler CP, Cramb G. Two isoforms of the Na+/K+/2Cl− cotransporter are expressed in the European eel (Anguilla anguilla). BBA—Biomembranes. 2002; 1566(1): 92–103. [DOI] [PubMed] [Google Scholar]
- 45.Lytle C, Xu JC, Biemesderfer D, Rd FB. Distribution and diversity of Na-K-Cl cotransport proteins: a study with monoclonal antibodies. American Journal of Physiology. 1995; 269(1): 1496–1505. 10.1152/ajpcell.1995.269.6.C1496 [DOI] [PubMed] [Google Scholar]
- 46.Beyenbach KW, Wieczorek H. The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. Journal of Experimental Biology. 2006; 209(Pt 4): 577–589. 10.1242/jeb.02014 [DOI] [PubMed] [Google Scholar]
- 47.Holt MEV, King SA, Cala PM, Pedersen SF. Regulation of the Pleuronectes americanus Na+/H+ exchanger by osmotic shrinkage, β-adrenergic stimuli, and inhibition of Ser/Thr protein phosphatases. Cell Biochemistry & Biophysics. 2006; 45(1): 1–18. 10.1385/CBB:45:1:1 [DOI] [PubMed] [Google Scholar]
- 48.Ferraris JD, Persaud P, Williams CK, Chen Y, Burg MB. cAMP-independent role of PKA in tonicity-induced transactivation of tonicity-responsive enhancer/osmotic response element-binding protein. Proceedings of the National Academy of Sciences of the United States of America. 2002; 99(26): 16800–16805. 10. 10.1073/pnas.222659799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yip YY, Yeap YY, Bogoyevitch MA, Ng DC. cAMP-dependent protein kinase and c-Jun N-terminal kinase mediate stathmin phosphorylation for the maintenance of interphase microtubules during osmotic stress. Journal of Biological Chemistry. 2014; 289(4): 2157–2169. 10.1074/jbc.M113.470682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strange PG. D1/D2 dopamine receptor interaction at the biochemical level. Trends in Pharmacological Sciences. 1991; 12(2): 48–49. 10.1016/0165-6147(91)90494-D [DOI] [PubMed] [Google Scholar]
- 51.Bertorello AM, Hopfield JF, Aperia A, Greengard P. Inhibition by dopamine of Na+/K+-ATPase activity in neostriatal neurons through D1 and D2 dopamine receptor synergism. Nature. 1990; 347(6291): 386–388. 10.1038/347386a0 [DOI] [PubMed] [Google Scholar]
- 52.LA H, RB F, DM C, M S, DR S. Reciprocal modulation of function between the D1 and D2 dopamine receptors and the Na+/K+-ATPase. Journal of Biological Chemistry. 2008; 283(52): 36441–36453. 10.1074/jbc.M805520200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen GH, Wang YJ, Qin S, Yang QG, Zhou JN, Liu RY. Age-related spatial cognitive impairment is correlated with increase of synaptotagmin 1 in dorsal hippocampus in SAMP8 mice. Neurobiology of Aging. 2007; 28(4): 611–618. 10.1016/j.neurobiolaging.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 54.Martínez-Álvarez RM, Morales AE, Sanz A. Antioxidant defenses in fish: biotic and abiotic factors. Reviews in Fish Biology & Fisheries. 2005; 15(1–2): 75–88. 10.1007/s11160-005-7846-4 [DOI] [Google Scholar]
- 55.Rudneva II. Blood antioxidant system of black sea elasmobranch and teleost. 1997; 118(2): 255–260. 10.1016/S0742-8413(97)00111-4 [DOI] [Google Scholar]
- 56.Yang Y, Cheng JZ, Singhal SS, Saini M, Pandya U, Awasthi S, et al. Role of glutathione S-transferases in protection against lipid peroxidation. Overexpression of hGSTA2-2 in K562 cells protects against hydrogen peroxide-induced apoptosis and inhibits JNK and caspase 3 activation. Journal of Biological Chemistry. 2001; 276(22): 19220–19230. 10.1074/jbc.M100551200 [DOI] [PubMed] [Google Scholar]
- 57.Mejía SÁ, Baiza Gutman LA, Camarillo CO, Navarro RM, Sánchez Becerra MC, Santana LD, et al. Nicotinamide prevents sweet beverage-induced hepatic steatosis in rats by regulating the G6PD, NADPH/NADP+ and GSH/GSSG ratios and reducing oxidative and inflammatory stress. European Journal of Pharmacology. 2017; 818(5): 499–507. 10.1016/j.ejphar.2017.10.048 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
Data Availability Statement
Our next-generation sequencing data has been uploaded to NCBI Sequence Read Archive (SRA), and the BioProject ID is PRJNA508867. There is the link for reviewing our SRA submission data: https://submit.ncbi.nlm.nih.gov/subs/sra/SUB4871716/overview (Account: junyz, password: 992325zjy).