SUMMARY
Maternal mRNAs synthesized during oogenesis initiate the development of future generations. Some maternal mRNAs are either somatic or germline determinants and must be translationally repressed until embryogenesis. However, the translational repressors themselves are temporally regulated. We used polar granule component (pgc), a Drosophila maternal mRNA, to ask how maternal transcripts are repressed while the regulatory landscape is shifting. pgc, a germline determinant, is translationally regulated throughout oogenesis. We find that different conserved RNA-binding proteins bind a 10-nt sequence in the 3′ UTR of pgc mRNA to continuously repress translation at different stages of oogenesis. Pumilio binds to this sequence in undifferentiated and early-differentiating oocytes to block Pgc translation. After differentiation, Bruno levels increase, allowing Bruno to bind the same sequence and take over translational repression of pgc mRNA. We have identified a class of maternal mRNAs that are regulated similarly, including zelda, the activator of the zygotic genome.
In Brief
Flora et al. show that pgc, a germline determinant, is translationally regulated throughout oogenesis. Different conserved RBPs bind a 10-nt sequence in the 3′ UTR to continuously repress translation throughout oogenesis. This mode of regulation applies to a class of maternal mRNAs, including zelda, the activator of the zygotic genome.
Graphical Abstract

INTRODUCTION
The germline gives rise to eggs and sperm that launch the next generation. Upon fertilization, the egg differentiates into every cell lineage of the adult organism, including a new germline, and is therefore totipotent (Seydoux and Braun, 2006; Cinalli et al., 2008). Pivotal to the task of kick-starting the next generation is a maternally synthesized trust fund of mRNAs deposited into the egg during oogenesis (Lasko 2012). After fertilization, and prior to zygotic genome activation, translation of these maternally supplied mRNAs helps power early development (Zhang and Smith, 2015; Tadros and Lipshitz, 2009; Lee et al., 2014). Some of the maternally supplied mRNAs code for key determinants of both somatic and germline cell fate and thus need to be exquisitely regulated during oogenesis and early embryogenesis.
RNA-binding proteins (RBPs) regulate the maternal mRNAs through interactions with sequences within the 3′ UTRs of their target mRNAs (Rosario et al., 2017; Slaidina and Lehmann, 2014; Johnstone and Lasko, 2001). Loss of RBPs during oogenesis results in death, sterility, or germline to soma trans-differentiation (Ciosk et al., 2006; Forbes and Lehmann, 1998). This suggests that RBPs are critical for silencing key somatic and germline determinants during oogenesis. Consistent with this observation, it has been shown that gene regulation during oogenesis and early embryogenesis relies primarily on the 3′ UTRs of mRNAs rather than on their promoters (Merritt et al., 2008; Rangan et al., 2009). Additionally, loss of specific sequences in the 3′ UTR of maternal mRNAs results in their dysregulation (Kim-Ha et al., 1995; Wharton and Struhl, 1991). However, several RBPs that are regulators of translation also fluctuate in levels of expression, with these fluctuations promoting critical developmental transitions. For example, during C. elegans oogenesis, GLD-1 and MEX-3, two RBPs whose loss results in germline to soma trans-differentiation, have a reciprocal expression pattern (Mootz et al., 2004; Ciosk et al., 2006; Draper et al., 1996). In human fetal ovary, RBPs such as deleted in azoospermia-like (DAZL) play an important role in regulating RNA targets, such as TEX11, a gene required for recombination and DNA repair, via its 3′ UTR (Rosario et al., 2017). During human oogenesis, DAZL has a dynamic expression pattern; it is robustly expressed in the pre-meiotic and post-meiotic germ cells but absent during meiotic stages (Anderson et al., 2007; He et al., 2013). The conundrum remains as to how mRNAs can be continually silenced during oogenesis when the RBPs that regulate them fluctuate.
Drosophila oogenesis is an excellent model to investigate how maternal mRNAs are continuously regulated. Oogenesis in Drosophila begins when germline stem cells (GSCs) divide to both self-renew and give rise to a stem cell daughter called a cystoblast (CB) (Figures 1A and 1B) (Chen and McKearin, 2003). The CB differentiates by undergoing four incomplete mitotic divisions to give rise to 2-, 4-, 8-, and 16-cell cysts (Figure 1B) (McKearin and Ohlstein, 1995; McKearin and Spradling, 1990). Of these 16 cells, one is designated as the oocyte and the others become nurse cells (Figure 1A) (Spradling et al., 1997); the maternal mRNAs and proteins synthesized by the nurse cells are deposited into the oocyte (Spradling 1993). The oocyte and surrounding nurse cells are encapsulated by somatic cells to form an egg chamber, which progresses through successive developmental stages (Margolis and Spradling, 1995; Gilboa and Lehmann, 2004). These maternal mRNAs that are deposited into the oocyte need to be post-transcriptionally regulated to promote proper oogenesis and embryogenesis (Richter and Lasko, 2011; Lasko 2012; Laver et al., 2015).
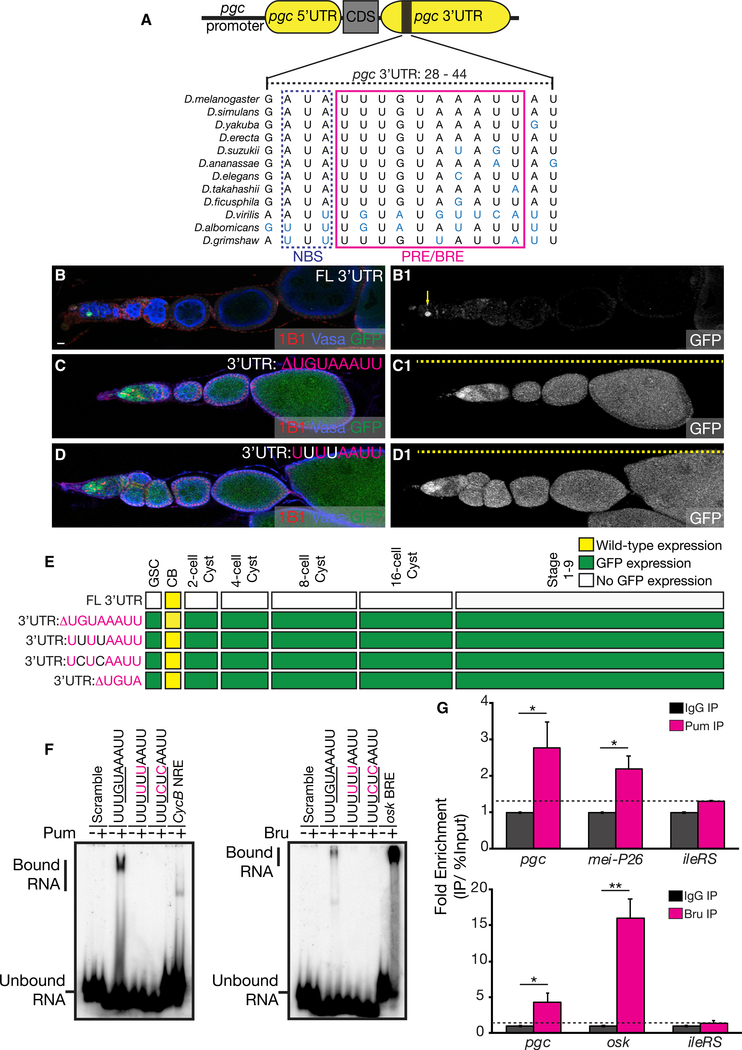
Figure 1. Pgc Is Translationally Regulated via Its UTRs.
(A) Schematic representation of a Drosophila ovariole.
(B) Schematic representation of a germarium housing the germline stem cells (GSCs) (blue), pre-cystoblast (pre-CB) (green), and differentiating cysts. The singlecells of the germarium can be identified by spectrosomes and the differentiating cysts can be identified by fusomes.
(C) The ovariole of a pgcGFP ovary stained with 1B1 (red), which marks the spectrosomes and fusomes; Vasa (blue), which marks the germline; and GFP (green), which marks Pgc-expressing cells. Expression of GFP is restricted to the pre-CB (arrow).
(D) The ovariole of a wild-type fly probed for pgc RNA (magenta) using FISH shows that pgc RNA is present throughout oogenesis.
(E) RNA-seq track of pgc in nosGAL4 > UAS-tkv ovaries.
(F) The ovariole of a transgenic fly (pgc promoter-nos 5′ UTR-GFP-K10 3′ UTR) stained with 1B1 (red), Vasa (blue), and GFP (green). GFP expression shows that pgc promoter is active throughout oogenesis (dashed line).
(G) The ovariole of a transgenic fly (pgc promoter-nos 5′ UTR-GFP- pgc 3′ UTR) stained with 1B1 (red), Vasa (blue) and GFP (green) shows GFP expression only in the earliest stages of oogenesis (dashed line).
(H) The ovariole of a transgenic fly (pgc promoter-pgc 5′ UTR-GFP-K10 3′ UTR) stained with 1B1 (red), Vasa (blue), and GFP (green) shows GFP expression throughout oogenesis (dashed line).
Scale bars, 10 μm. See also Figure S1.
Polar granule component (pgc) is a superb candidate to address how maternal mRNAs are regulated during oogenesis developmental transitions. During oogenesis, pgc is synthesized and provided to the oocyte, where it localizes to the germ plasm (Nakamura et al., 1996). While pgc mRNA is continuously present, Pgc is only translated in two short pulses: in the CB during oogenesis and in the germ cells during embryogenesis (Hanyu-Nakamura et al., 2008; Flora et al., 2018). Pgc expression in the CB is required to promote timely differentiation (Flora et al., 2018), while expression of Pgc in the germ cells is required to repress the expression of somatic genes that could interfere with germline specification (Hanyu-Nakamura et al., 2008). Pgc performs these tasks by causing global transcriptional silencing through targeting the basal transcriptional elongation machinery of RNA polymerase II (Martinho et al., 2004; Hanyu-Nakamura et al., 2008; Flora et al., 2018). pgc can even suppress transcription in other cell types upon ectopic expression (Timinszky et al., 2008). The strong effects of Pgc on transcription lead to a requirement for strict regulation of pgc translation in cells where it is normally found. It is known that the 3′ UTR of pgc mRNA is sufficient to mediate translational control after GSC differentiation into an oocyte (Rangan et al., 2008); however, it is not known whether pgc is regulated transcriptionally or translationally prior to differentiation nor what trans-acting factors regulate pgc translation after differentiation.
Temporally restricted RBPs that bind to 3′ UTRs regulate developmental transitions during Drosophila oogenesis by controlling translation of their targets. Pumilio (Pum), an RBP that belongs to the conserved Pum- and Fem-3-binding factor (PUF) family of proteins, is present at high levels in the undifferentiated cells in the ovary, including GSCs, CBs, and early-differentiating cysts (Lin and Spradling 1993; Forbes and Lehmann, 1998). Pum represses the translation of differentiation-promoting mRNAs in GSCs, thereby preventing stem cell loss (Forbes and Lehmann, 1998; Joly et al., 2013). Pum expression is attenuated in the differentiated stages, allowing for the expression of the differentiation-promoting mRNAs (Forbes and Lehmann, 1998; CarreiraRosario et al., 2016). Drosophila Bruno 1 (Bru), a CUGBP and ETR-3-like factor (CELF) superfamily protein, is expressed at increasing levels during differentiation and is then maintained for the rest of oogenesis (Xin et al., 2013; Sugimura and Lilly, 2006; Webster et al., 1997). Bru regulates several maternal mRNAs post-differentiation during oogenesis (Schüpbach and Wieschaus, 1991; Webster et al., 1997; Snee et al., 2014). Thus, Pum and Bru have reciprocal temporal regimes and could act jointly to repress targets throughout oogenesis. However, it is not known whether further repression is required of Pum targets after differentiation or Bru targets prior to differentiation.
Pum and Bru can use various cofactors to mediate translational repression using distinct mechanisms. Pum partners with Nanos (Nos) to recruit translation modulators such as Twin, a deadenylase causing a shortening of the poly(A)-tail (Joly et al., 2013). Pum can also recruit brain tumor (Brat), which is known to modulate translation by interacting with Drosophila eukaryotic translation initiation factor 4E homologous protein (d4EHP), a cap-binding protein (Cho et al., 2006; Harris et al., 2011). Bru can form oligomers to form silencing particles or can partner with Cup, which associates with the 5′ cap-binding initiation factor eIF4E, to regulate mRNAs (Nakamura et al., 2004; Kim-Ha et al., 1995; Chekulaeva et al., 2006). Why certain mechanisms are preferred over others is not known.
Here, we elucidate a control mechanism that ensures handoff of translational repression of a germline determinant, pgc, from one set of regulators to another. This governs the critical expression of Pgc just in CBs, ensuring proper maintenance of GSCs and their conversion into differentiated progeny. We demonstrate that this control depends on a 10-nt sequence in the 3′ UTR of pgc mRNA. In the undifferentiated stages, we find that Pum binds the 10-nt sequence and partners with Nos and the CCR4-Not complex to regulate pgc mRNA in a poly(A)-dependent manner. When Nos levels drop in CBs, pgc is expressed. After CB differentiation, Pum switches partners to use Brat to suppress pgc in the early-differentiating cysts in a cap-dependent manner. However, when Pum levels diminish, pgc mRNA is bound by Bru via the same 10-nt sequence to translationally regulate it. Bru recruits Cup to silence pgc translation also in a cap-dependent manner. We find that a class of maternal mRNAs, including zelda, which play pivotal roles during development, are also regulated by both Pum and Bru and contain this 10-nt sequence. This suggests that the sequential handoff of mRNAs between Pum and Bru is broadly utilized to control translation of maternal RNAs. We propose that this handoff from one set of trans-acting factors utilizing a poly(A)-shortening mechanism to another set of trans-acting factors that utilizes a cap-dependent mechanism is required to protect mRNAs post-differentiation and prime them for translation during embryogenesis.
RESULTS
Pgc Is Translationally Regulated via Its UTRs
During oogenesis, Pgc is expressed in CBs, where it promotes timely differentiation (Figure 1C) (Flora et al., 2018). To assess if this temporal specificity of Pgc protein production is due to transcriptional or translational regulation, we carried out fluorescent in situ hybridization (FISH) for pgc in wild-type ovaries and for GFP in ovaries of flies carrying a reporter for Pgc (Flora et al., 2018). pgc transcription in the GSCs was difficult to discern because of the low resolution of FISH in the germarium; however, we did detect pgc mRNA in all later-differentiated stages (Figures 1D and S1A–S1C). To assess pgc mRNA expression in the GSCs through an alternate method, we overexpressed the self-renewal signaling receptor, thick veins receptor (TKV), to enrich for GSCs and then sequenced their transcriptome (Xie and Spradling, 1998). We detected 88 transcripts per million (TPM) of pgc, indicating that the mRNA is transcribed in the GSCs (Figures 1E and S1D). To further substantiate that the pgc promoter is active in the GSCs, we created a reporter construct in which the pgc promoter drives the expression of GFP flanked by the nos 5′ UTR and K10 3′ UTR, which are not translationally silenced during oogenesis (Figure 1F) (Serano et al., 1994; Gavis and Lehmann, 1992, 1994). We observed GFP expression throughout oogenesis, including in the GSCs. This suggests that the maternal pgc mRNA is transcribed from the GSCs onward throughout oogenesis and is under strict translational regulation pre- and post-differentiation (Rangan et al., 2008).
The 5′ UTR and 3′ UTR of an mRNA are commonly recognized by sequence-specific RBPs to regulate translation (Wilkie et al., 2003). We wanted to test the potential role of both the 5′ UTR and 3′ UTR of pgc in repressing translation in the GSCs. pgc mRNA has two annotated 5′ UTRs; to determine which one was expressed in the GSCs, we designed primers that distinguish these two forms. We carried out PCR on RNA enriched from GSCs by overexpressing TKV, and for CBs, by using a mutation for differentiation factor, bag-of-marbles (bam) (Xie and Spradling, 1998; McKearin and Ohlstein, 1995). We found that only the short form was expressed in the GSCs and CBs (Figure S1E). To determine if this short pgc 5′ UTR is required for translational regulation of pgc, we swapped it with the nos 5′ UTR in a GFP reporter construct that still retained the pgc 3′ UTR and the pgc promoter. We found that the absence of the pgc 5′ UTR results in upregulation of GFP protein expression in the GSCs, but not in later stages (Figure 1G). Our results indicate that in GSCs, the pgc 5′ UTR is required for translational regulation, while the 3′ UTR is not sufficient (Figure 1G). In differentiated stages, the 3′ UTR alone is sufficient to mediate translational regulation (Figure 1G). To test if the 5′ UTR is sufficient for translational regulation in GSCs, we created a construct with the pgc 5′ UTR and non-repressed K10 and tubulin (tub) 3′ UTRs flanking GFP under the control of the pgc promoter (Figures 1H and S1F). GFP was expressed in the GSCs as well as in later-differentiating stages and egg chambers, demonstrating that the 5′ UTR alone is not sufficient for translational regulation (Figures 1H and S1F). Taken together, we conclude that both the pgc 5′ UTR and 3′ UTR are required for translational control pre-differentiation in the GSCs and that the 3′ UTR alone is sufficient post-differentiation in the cysts and egg chambers.
A cis-Element in the pgc 3′ UTR that Binds Both Pum and Bru Is Required for Translational Control throughout Oogenesis
We predicted that cis-acting sequences in either the 5′ or 3′ UTRs of pgc could regulate translation during oogenesis by recruiting trans-acting factors. To identify these sequences, we carried out a phylogenetic analysis of the pgc 5′ and 3′ UTR in Drosophilids separated by 40 million years of evolution and discovered several regions of conservation in the 3′ UTR (Figure S1G). We could not identify unique conserved regions in the pgc 5′ UTR, as the sequence overlaps with the coding region of type III alcohol dehydrogenase (T3dh). We also used algorithms that search for RBP-binding sequences and did not find any in the short form 5′ UTR of pgc (Bailey et al., 2009). In the 3′ UTR, a conserved 10-nt sequence, UUUGUAAAUU, stood out (Figures 2A and S1G). This sequence closely matches the sequences that have been previously described as the Pumilio response element (PRE), which is part of the Nanos response element (NRE) in hunchback and Cyclin B (CycB), respectively (Weidmann et al., 2016; Murata and Wharton, 1995; Kadyrova et al., 2007). PREs are known to bind Pum, which then recruits Nos, to bind to the Nanos-binding sequence (NBS), resulting in translational regulation of RNAs (Figure 2A) (Murata and Wharton, 1995; Kadyrova et al., 2007). This sequence in the pgc 3′ UTR can also bind another conserved RBP, Bru. Pum binds to the UGUA motif, while Bru binds to a uUG/AUG/AUG/AUu motif, which is described as the Bruno response element (BRE) (Kim-Ha et al., 1995; Wharton and Struhl, 1991).
Figure 2. A cis-Element in the pgc 3′ UTR that Binds Pum and Bru Is Required for Translational Control throughout Oogenesis.
(A) The NBS and PRE and/or BRE sequence identified in the pgc 3′ UTR is conserved in 12 species of Drosophilids.
(B) An ovariole of a pgcGFP fly stained with 1B1 (red), Vasa (blue), and GFP (green) showing that GFP expression is restricted to the pre-CB (arrow in B1).
(C) An ovariole of a pgcGFP reporter that lacks the PRE and/or BRE sequence in the 3′ UTR stained with 1B1 (red), Vasa (blue), and GFP (green). GFP regulation was lost throughout oogenesis (dashed line in C1).
(D) An ovariole of a pgcGFP reporter in which the PRE and/or BRE core UGUA motif was mutated stained with 1B1 (red), Vasa (blue), and GFP (green). GFP regulation was lost throughout oogenesis (dashed line in D1).
(E) A developmental profile of GFP expression in different stages of oogenesis of transgenes in which the PRE and/or BRE sequence was either deleted ormutated.
(F) EMSAs show that purified Pum and Bru proteins bind to the PRE and/or BRE of the pgc 3′ UTR, the NRE of the CycB 3′ UTR, and the BRE of the osk 3′ UTR, respectively.
(G) qPCR of pgc, mei-P26 (positive control), and ileRS (negative control) carried out on RNA samples extracted after an IP with Pum antibody (top). qPCR of pgc, osk (positive control), and ileRS (negative control) carried out on RNA samples extracted after an IP with Bru antibody (bottom). RIP-qPCR graphs represent an average generated from three independent biological samples. The error bars represent SE. A Student’s t test analysis was performed. * and ** indicate a p value < 0.05 and < 0.005, respectively.
Scale bar, 10 μm. See also Figures S1 and S2.
We asked if this conserved 10-nt sequence that is predicted to bind two RBPs can regulate pgc translation. To test this, we generated a reporter construct that deleted 8 nt of the conserved sequence including the UGUA motif that is known to bind Pum and the uUG/AUG/A motif that binds Bru. This resulted in an upregulation of translation throughout oogenesis (Figures 2B, 2C, 2E, and S1J). We also generated three transgenes in which we mutated the core UGUA motif to UUUU or UCUC and also deleted the core UGUA motif. We found that all these changes resulted in loss of translational control (Figures 2D, 2E, and S1H–S1J). To test if this 10-nt PRE and/or BRE was sufficient for translation regulation, we generated a reporter construct where we inserted the conserved sequence into the tub 3′ UTR (tub 3′UTR: NBS + PRE and/or BRE), fused it to GFP and pgc 5′ UTR, and drove it under the control of pgc promoter. We found that the inclusion of this sequence in the 3′ UTR of tub is sufficient to repress GFP translation throughout oogenesis, but it is not sufficient for GFP expression in the pre-CB (Figures S2A–S2C). Thus, we conclude that the conserved 10-nt sequence in the pgc 3′ UTR that is predicted to bind Pum and Bru is required and sufficient for translation repression of pgc during oogenesis.
To determine if the conserved sequence binds Pum and Bru as predicted, we purified the recombinant RNA-binding domain of Pum and full-length Bru and carried out electrophoresis mobility shift assay (EMSA) experiments (Figure S2D) (Chekulaeva et al., 2006; Weidmann et al., 2016). As positive controls, we utilized the NRE in CycB and the BRE in Oskar’s (osk) 3′ UTR and demonstrated that our recombinant Pum and Bru bound the NRE and BRE, respectively (Figure 2F) (Kim-Ha et al., 1995; Kadyrova et al., 2007). Both Pum and Bru also bound the PRE and/or BRE in the 3′ UTR of pgc. This binding was lost when the core UGUA sequence was mutated to UCUC or UUUU (Figure 2F). To test if Pum and Bru also bind to pgc mRNA in vivo, we performed an RNA immunoprecipitation (RIP)-qPCR experiment with anti-immunoglobulin G (anti-IgG), anti-Pum, and anti-Bru antibody in lysates from wild-type ovaries. We observed that along with known RNA targets, mei-P26 for Pum and osk for Bru, pgc RNA was significantly enriched in both Pum and Bru pull-downs relative to non-specific IgG pull downs. There was no significant enrichment of a non-target RNA, isoleucyl-tRNA synthetase (ileRS), in either of these pull-downs (Figures 2G and S2E). Thus, we conclude that Pum and Bru bind to the 10-nt PRE and/or BRE of pgc 3′ UTR in vitro and to pgc mRNA in vivo.
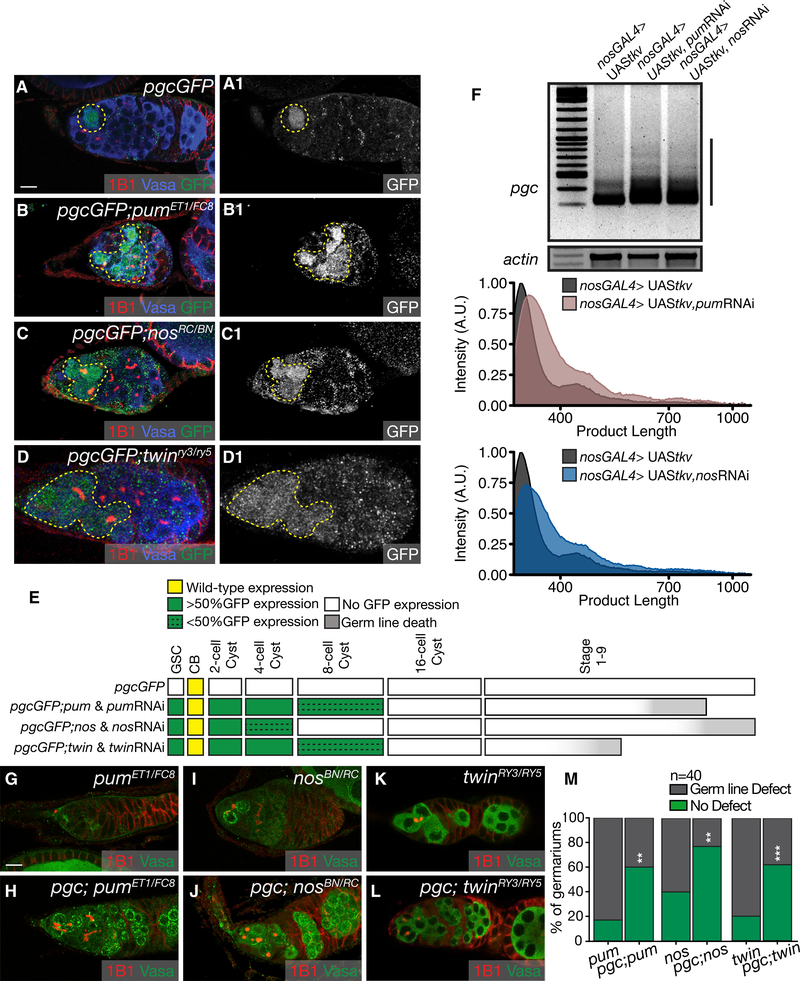
Pum and Its Cofactor, Nos, Regulate Pgc Translation in the GSCs and Early-Differentiating Cysts
We asked if pgc was translationally regulated by Pum and Bru during oogenesis, and in particular, given their inverse expression patterns, if they might each govern distinct phases. Pum is expressed from GSCs to the 8-cell cyst stage and is attenuated from the 16-cell cyst onward (Figures S2F–S2F2′) (Forbes and Lehmann, 1998; Carreira-Rosario et al., 2016). Bru levels are low from GSCs to the 8-cell cyst stage but are high in the 16-cell cyst stage and throughout later oogenesis (Figures S2F–S2F2′) (Webster et al., 1997; Sugimura and Lilly, 2006; Xin et al., 2013). Thus, we hypothesized that Pum may regulate pgc translation until the 8-cell cyst and Bru thereafter. We first focused on Pum and its potential role in regulating pgc translation during early oogenesis. Pum requires co-factors to regulate translation and can use distinct partners and multiple mechanisms. Pum is known to recruit Nos and Twin, a deadenylase, to NRE-containing 3′ UTRs to induce poly(A)-tail shortening in Drosophila embryonic germ cells (Sonoda and Wharton, 1999; Kadyrova et al., 2007). During oogenesis, Twin is ubiquitously expressed (Temme et al., 2010; Joly et al., 2013) and Nos protein is present in all stages, except for in the pre-CB where Pgc is expressed (Figures S3A–S3B1) (Forbes and Lehmann, 1998; Li et al., 2009). We therefore hypothesized that Pum might be regulating Pgc expression with Nos and Twin only until the cyst stages, during which time a drop in Nos expression in the pre-CBs would allow for Pgc expression.
To test this hypothesis, we separately assayed for PgcGFP expression in pum, nos, and twin mutants. We observed that in the absence of each of these genes, the reporter was ectopically expressed in the GSCs, as marked by pMAD, and in 2- and 4-cell cysts (Figures 3A–3D1 and S3C–S3F). Ectopic expression in the GSCs was also observed upon germline depletion of pum, nos, and twin via RNAi (Figures S3G–S3I and S3N). We confirmed that Pum RNAi depleted Pum in the germline (Figures S3J–S3K1). Twin is a deadenylase and is part of the CCR4-Not complex (Morris et al., 2005; Temme et al., 2010; Fu et al., 2015). To determine if other members of this complex were involved in regulating pgc translation, we depleted Pop2 and Not1 in the germline using RNAi and assayed for GFP expression. Compared to pgcGFP, depletion of Pop2 and Not1 resulted in ectopic expression of the reporter from the GSCs to the 4-cell cysts, consistent with what we observed in the nos, pum, and twin mutants (Figures S3L–S3N). We also observed that loss of pum and twin results in an elevated GFP expression in the 8-cell cyst. Differences of ectopic pgcGFP reporter expression is not due to nosGAL4 activity in the germline (Figures S3O–S3P1). We generated a developmental profile to show the temporal loss of translational regulation of GFP at each stage of development in pum, nos, and twin when compared to control pgcGFP ovarioles (Figure 3E). Taken together, we can conclude that pgc is regulated by Nos, Pum, and Twin from GSCs to the 4-cell cyst stage via the CCR4-Not complex. In the pre-CB, when Nos is absent, Pgc is expressed even though Pum and Twin proteins are still present. This suggests that Pum and Twin alone are not sufficient for regulating pgc in the pre-CB and require the presence of their co-regulator Nos.
Figure 3. Pum and Its Cofactor, Nos, Regulate Pgc Translation in the GSCs and Early-Differentiating Cysts.
(A) A germarium of a pgcGFP ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows expression of GFP only in the pre-CB (dashed circle).
(B) A germarium of a pgcGFP; pumET1/FC8 ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant GFP expression from GSCs to the 8-cell cyst (100% from GSC to 4-cell cyst, 32% in 8-cell cyst, n = 25) (dashed outline).
(C) A germarium of a pgcGFP; nosRC/BN ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant GFP expression from GSCs to the 4-cell cyst (100% from GSCs to 2-cell cyst, 13% in 4-cell cyst, n = 25) (dashed outline).
(D) A germarium of a pgcGFP; twinRY3/RY5 ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant GFP expression from GSCs to the 8-cell cyst (100% from GSC to 4-cell cyst, 40% in 8-cell cyst, n = 25) (dashed outline). The GFP channel is shown in A1–D1.
(E) A developmental profile of GFP expression when Pum, Nos, and Twin are depleted in the germline.
(F) PAT assay of pgc poly(A)-tail length in GSC tumors and in GSC tumors lacking Pum and Nos.
(G, I, and K) Germaria of pumET1/FC8 (G), nos RC/BN (I), and twin RY3/RY5 (K) mutants stained with 1B1 (red) and, Vasa (green).
(H, J, and L) Germaria of pgc; pumET1/FC8 (H) pgc; nosRC/BN (J) and pgc; twinRY3/RY5 (L) double mutants stained with 1B1 (red) and Vasa (green).
(M) A graphical representation of the rescue experiment (n = 40). A population proportion z-test was performed. ** and *** indicate a p value < 0.005 and < 0.0005, respectively.
Scale bars, 10 μm. See also Figures S3 and S4.
To test if Pum and Nos control translation of pgc mRNA by shortening poly(A)-tail length, we utilized the poly(A)-tail-length (PAT) assay (Sallé s and Strickland, 1999). We performed this assay on RNA extracted from GSC-enriched tumors and GSC tumors depleted of Nos and Pum to eliminate the stage of oogenesis in which pgc is translationally repressed (Figures S4A–S4C1). In the absence of these RBPs, we detected an increase in the length of the poly(A)-tail compared to the control (Figure 3F). Together, these observations suggest that Pum, Nos, and Twin are recruited to pgc’s 3′ UTR to suppress its translation in the GSCs by a mechanism that involves shortening its poly(A)-tail.
We next asked if this regulation of pgc by Pum, Nos, and Twin is biologically meaningful. Loss of pum and nos results in failure to maintain GSCs, and this defect is thought to be the result of dysregulation of differentiation-promoting mRNAs in the GSCs (Forbes and Lehmann, 1998; Wang and Lin, 2005). We have previously shown that pgc promotes timely differentiation in the pre-CBs (Flora et al., 2018). Thus, we hypothesized that in nos, pum, and twin mutants, Pgc is upregulated in the GSCs, forcing premature differentiation. To test this hypothesis, we made double mutants of pgc with nos, pum, and twin, respectively. Lowering pgc levels in all three mutants rescued germline defects (Figures 3G–3M). Together, our results suggest that Pgc is translationally repressed by Pum, Nos, and Twin in the GSCs to ensure appropriate GSC self-renewal and maintenance.
Me31B Cooperates with the Decapping Protein dGe-1 and the pgc 5′ UTR to Mediate Repression in GSCs and Early-Differentiating Cysts
Our results suggest that Pum, Nos, and Twin regulate pgc translation via a conserved sequence in the pgc 3′ UTR. However, we also found a requirement for the pgc 5′ UTR in the regulation of pgc in undifferentiated cells (Figure 1G). Does the 5′ UTR and 3′ UTR of pgc cooperate to mediate repression? It has been shown that recruitment of the CCR4-Not complex also facilitates the recruitment of the decapping complex to the 5′ UTR of mRNAs (Meyer et al., 2004; Garneau et al., 2007; Behm-Ansmant et al., 2006) and that these two complexes at the 5′ UTR and 3′ UTR can be bridged by an RNA helicase, DDX6, or maternal expression at 31B (Me31B) (Ozgur et al., 2015; Nakamura et al., 2001). This allows “masking” of the mRNAs, making them inaccessible to the ribosome. We therefore hypothesized that Pum, Nos, and Twin at the pgc 3′ UTR could recruit decapping complex members, such as EDC4 or Drosophila Ge-1 (dGe-1), to the cap at the 5′ UTR to promote translational repression by masking through the bridging action of Me31B (Fan et al., 2011; Eulalio et al., 2007).
To test this model, we first asked if Me31B associates with pgc mRNA. We used wild-type ovaries from a Me31B protein-GFP trap construct and carried out a RIP-qPCR experiment with both anti-GFP and anti-IgG antibodies. We found that there was a significant enrichment of pgc mRNA bound to Me31B-GFP protein comparable to those of the positive control, osk mRNA (Figure 4A) (Nakamura et al., 2001) and no significant enrichment of a non-target RNA, ileRS. Next, we assayed for pgcGFP expression upon germline depletion of me31B and found a loss of GFP repression from the GSC to the 4-cell cyst (Figures 4B–4C1 and 4E). me31B RNAi results in depletion of Me31B (Figures S4D–S4E1). We also observed ectopic pgcGFP reporter expression from the GSC to the 8-cell cyst stage in the presence of the dGe-1 RNAi (Figures 4D, 4E, and S4F). Our results suggest that pgc 5′ and 3′ UTRs together with Me31B and proteins of the decapping complex such as dGe-1 regulate its translation.
Figure 4. Me31B Cooperates with the Decapping Protein dGe-1 and the pgc 5′ UTR to Mediate Repression in GSCs and Early-Differentiating Cysts.
(A) Western blot shows pull-down of GFP from me31BGFP-trap fly ovary lysates (top). qPCR of pgc, mei-P26 (positive control), and ileRS (negative control) carried out on RNA samples extracted after the IP (bottom). The graph represents an average generated from three independent biological samples. The error bars represent SE. A Student’s t test analysis was performed. * and ** indicate a p value < 0.05 and < 0.005, respectively..
(B) A germarium of a pgcGFP ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows expression of GFP only in the pre-CB (dashed circle)..
(C) A germarium of a pgcGFP; nosGAL4>me31BRNAi ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant GFP expression in GSCs to the 4-cell cyst (100%, n = 20) (dashed outline)..
(D) A germarium of pgcGFP; nosGAL4>dGe-1RNAi stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant GFP expression in GSCs to the 8-cell cyst stages (100%, n = 20) (dashed outline). The GFP channel is shown in B1–D1..
(E) A developmental profile of GFP expression when Me31B and dGe-1 are depleted in the germline..
Scale bar, 10 μm. See also Figure S4.
Pum and Its Cofactor, Brat, Regulate Pgc Translation in the 4- to 16-Cell Cysts
Pum can also mediate translational repression via an alternate mechanism by recruiting Brat (Sonoda and Wharton, 2001; Muraro et al., 2008; Olesnicky et al., 2012; Harris et al., 2011). Brat engages the cap-binding protein d4EHP, which competes with the cap-binding protein eIF4E, to prevent translational initiation (Cho et al., 2005). Pum is present from the GSCs until the 8-cell cyst and is attenuated from the 16-cell cyst onward, while Brat is expressed only after the CB differentiates and persists throughout all later cyst stages (Carreira-Rosario et al., 2016; Harris et al., 2011). To test if Pum regulates pgc via Brat, we assayed for pgcGFP expression in the pum680 mutant, a separation-of-function mutant that disrupts the interaction between Pum and Brat without affecting the interaction between Pum and Nos (Wharton et al., 1998; Sonoda and Wharton 1999). We found that in pum680 mutants, there was ectopic pgcGFP reporter expression from the 4- to 16-cell cyst, but not in the earlier stages (Figures 5A–5B1 and S5A). This observation suggested that Pum may be interacting with Brat and its partner, d4EHP, to repress pgc translation in the differentiating cysts. To test this, we depleted brat and d4EHP in the germline using RNAi. We observed that loss of Brat and d4EHP also results in ectopic expression of GFP from 4- to 16-cell cyst, but not in the earlier stages (Figures 5C–5D1 and S5A). Although we do not see an upregulation of reporter expression in the 16-cell cyst in a pum mutant and a pumRNAi ovary (Figures 3B and S3G), we do see ectopic expression of GFP in the 16-cell cyst when Brat and d4EHP are depleted in the germline. Brat can act independent of d4EHP during oogenesis and independent of Pum during embryogenesis (Harris et al., 2011; Laver et al., 2015). We do not think Brat acts independent of either Pum or d4EHP to regulate pgc during oogenesis, as we see ectopic reporter expression from the 4- to 16-cell cyst when the Pum-Brat interaction is specifically perturbed in a pum680 mutant and upon loss of d4EHP. We think that the reason why pum mutant alleles and RNAi lines repress pgc in the 16-cell cysts could be due to their hypomorphic nature. A developmental profile of GFP expression in pgcGFP, pgcGFP; pum680, pgcGFP; nosGAL4 > bratRNAi and pgcGFP; nosGAL4 > d4EHPRNAi shows that compared to the control, loss of Brat and d4EHP results in the loss of pgcGFP regulation restricted to the 4- and 16-cell cysts (Figure 5E). We conclude that Pum, Brat, and d4EHP regulate Pgc translation in the 4- to 16-cell cysts. To determine whether Pum-Brat complex affects the poly(A)-tail length of pgc, we performed a PAT assay on pgc RNA in pum680 mutants and germline depletions of brat and d4EHP. We observed no significant change in these mutants (Figure S5B). These results suggest that Pum switches not only binding partners but also the mode of regulation from a poly(A)-tail-dependent mechanism to a cap-dependent mechanism to regulate pgc translation pre- and post-differentiation, respectively.
Figure 5. Pum and Its Cofactor, Brat, Regulate Pgc Translation in 4- to 16-Cell Cysts.
(A) A germarium of a pgcGFP ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows expression of GFP in the pre-CB (dashed circle).
(B) A germarium of a pgcGFP; pum680 ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant expression of GFP in the differentiating cysts (25% in the 4-cell cyst, 75% in the 8-cells cyst, and 10% in the 16-cell cyst, n = 20) (dashed outline).
(C) A germarium of a pgcGFP; nosGAL4>bratRNAi ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant expression of GFP in the differentiating cysts (38% in the 4-cell cyst, 54% in the 8-cells cyst, and 18% in the 16-cell cysts, n = 30) (dashed outline).
(D) A germarium of a pgcGFP; nosGAL4>d4EHPRNAi ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant expression of GFP in the differentiating cysts (34% in the 4-cell cyst, 62% in the 8-cells cyst, and 15% in the 16-cell cyst, n = 32) (dashed outline). The GFP channel is shown in A1–D1.
(E) A developmental profile of GFP expression when the Pum-Brat interaction is ablated and Brat and d4EHP are depleted in the germline.
Scale bar, 10 μm. See also Figure S5.
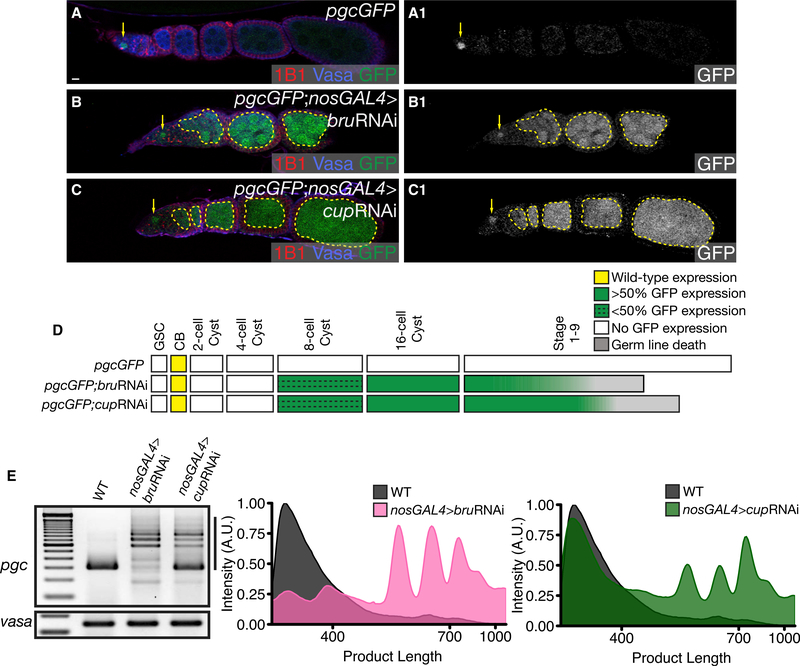
Bru and Cup Regulate Pgc Translation in the Later Stages of Oogenesis
After differentiation, levels of Pum diminish and levels of Bru increase (Figures S2F–S2F2′). We have shown that Bru binds to the 10-nt conserved sequence in the 3′ UTR that is required for pgc translational control throughout oogenesis (Figures 2C and 2F). Therefore, we asked if Bru and its binding partner, Cup, can repress Pgc translation post-differentiation (Nakamura et al., 2004; Chekulaeva et al., 2006; Kim et al., 2015b). Assaying for the pgc reporter in both bru mutants and germline depletion of Bru via RNAi, we found that translation was de-repressed primarily from the 16-cell cyst stage onward (Figures 6A–6B1, S6A, and S6B). We confirmed that bruRNAi depleted Bru in the germline (Figures S6C–S6D1). To determine if Bru recruits Cup to mediate this regulation, we depleted cup in the germline via RNAi and observed similar ectopic expression of GFP from the 16-cell cyst stage (Figure 6C). A developmental profile of GFP expression in pgcGFP; nosGAL4, pgcGFP; nosGAL4 > bruRNAi and pgcGFP; nosGAL4 > cupRNAi shows that compared to the control, loss of bru and cup results in loss of pgcGFP regulation primarily from the 16-cell cyst stage onward (Figure 6D). To test if Bru and Cup’s mode of regulation affected the poly(A)-tail length of pgc, we performed a PAT assay on pgc RNA in germline depletion of Bru and Cup. We observed that Bru and Cup depletion results in an increase of pgc poly(A)-tail length with depletion of bru showing a more dramatic change (Figure 6E). As Bru can act independent of Cup to form RNA oligomers that “mask” transcripts from the translation initiation machinery (Chekulaeva et al., 2006), we think that in the absence of Cup, Bru can independently regulate a subset of pgc mRNAs. As loss of components of the CCR4Not complex does not show loss of translational control in later stages and poly(A)-tail length increase has been shown as directly correlated to increased translational efficiency (Eichhorn et al., 2016; Sachs and Wahle, 1993), we favor the model that pgc is regulated in the differentiated stages by Bru and its binding partner, Cup, via a cap-dependent mechanism that restricts access to both cap and poly-adenylation machinery.
Figure 6. Bru and Cup Regulate Pgc Translation in the Later Stages of Oogenesis.
(A) An ovariole of a pgcGFP ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows expression of GFP in the pre-CB (arrow).
(B) An ovariole of a pgcGFP; nosGAL4>bruRNAi ovary stained with 1B1 (red), Vasa (blue), and GFP (green) aberrant expression of GFP beyond the 16-cell cyst (12% from 8-cell cyst onward, 100% from 16-cell cyst onward, n = 25) (dashed outline).
(C) An ovariole of a pgcGFP; nosGAL4>cupRNAi ovary stained with 1B1 (red), Vasa (blue), and GFP (green) shows aberrant expression of GFP from the later cyst stages (20% from 8-cell cyst onward, 100% from 16-cell cyst onward, n = 30) (dashed outline). The GFP channel is shown in A1–C1.
(D) A developmental profile of GFP expression when Bru and Cup are depleted in the germline.
(E) PAT assay analysis of pgc poly(A)-tail length of pgc RNA when Bru and Cup are depleted in the germline.
Scale bars, 10 μm. See also Figure S6.
A Class of Germline RNAs Are Similarly Regulated by Both Pum and Bru
Our results show that the conserved RBPs Pum and Bru can recognize and bind the same cis-element in the pgc 3′ UTR to mediate repression throughout oogenesis. We wondered if this mechanism could be applicable for regulation of other maternally deposited mRNAs. To address this, we carried out a polysomesequencing (Poly-seq) experiment to calculate the translational efficiency (TE) of transcripts (Kronja et al., 2014). We utilized this method to identify transcripts that are actively translated in the ovaries of nosGAL4>pumRNAi and nosGAL4>bruRNAi flies when compared to young nosGAL4 flies. We used young nosGAL4 ovaries as controls because they do not have mature later stages (stage 10 and onward) comparable to germline depletion of both Pum and Bru. We conducted RNA sequencing (RNA-seq) of transcripts extracted from the polysome fractions (Figure S7A). We found that when Pum and Bru are depleted in the germline, 1,081 and 908 transcripts have higher TE, respectively, than in the control (Figures 7A–7C; Tables S1 and S2). 436 of these transcripts display an increase in TE when either pum or bru is depleted, suggesting that these targets may be co-regulated (Figure 7C; Table S3). 212 of the 436 shared transcripts contained a sequence similar to the 10-nt PRE and/or BRE sequence identified in the pgc 3′ UTR (Figure S7B; Table S4). 368 of the 436 transcripts and 179 of the 212 transcripts are maternally provided mRNAs that are also present in mature eggs (Kronja et al., 2014). Gene Ontology analysis of the 212 shared targets show these genes are required for gastrulation and cell motility; processes mediated by maternally deposited RNAs and occurring prior to the maternal-to-zygotic transition of Drosophila embryogenesis (Figure 7D). One such gene identified to be co-regulated by Pum and Bru throughout oogenesis was zelda, a maternally provided mRNA that plays the role of master regulator during early Drosophila embryogenesis (Figures 7A and 7B) (Harrison et al., 2011; Nien et al., 2011; Liang et al., 2008). It is a transcription factor that is required to activate early-developmental somatic genes essential for cellularization, sex determination, and body patterning. We do not know if these maternal mRNAs are expressed in the CBs, like pgc, or if additional translational regulatory mechanisms silence these mRNAs there. Taken together, our results demonstrate that key determinants for somatic and germline fate, such as zelda and pgc, respectively, are translationally suppressed by Pum and Bru to ensure their repression during oogenesis.
Figure 7. A Class of Germline RNAs Are Similarly Regulated by Both Pum and Bru.
(A and B) A bi-plot representing the translational efficiencies (TEs) of expressed mRNAs in nosGAL4>pumRNAi (A) and nosGAL4>bruRNAi (B) versus young wild-type ovaries. The lines represent cutoffs, which are 1 SD above and below the median ratio. Pink points represent shared targets of Pum and Bru containing a PRE and/or BRE sequence.
(C) A Venn diagram showing the shared targetsthat have a higher TE upon the germline depletion of pum and bru. The targets in the pink set contain a PRE and/or BRE similar to that of pgc’s in their 3′ UTR.
(D) Gene Ontology analysis of the 212 sharedtargets.
(E) A model accounting for the sequential regulationof pgc RNA by different RBPs throughout oogenesis.
See also Figure S7 and Tables S1–S4.
DISCUSSION
Here, we report that a maternal mRNA, pgc, is translationally repressed via different temporally restricted RBPs that use the same cis-acting sequence during oogenesis. We find that prior to differentiation, pgc 5′ and 3′ UTRs cooperate to regulate translation. In contrast, after differentiation, the 3′ UTR of pgc is necessary and sufficient for translational control. We find that a 10-nt conserved sequence in this 3′ UTR is essential for pgc regulation during the entirety of oogenesis. Surprisingly, two distinct RBPs whose expression is temporally restricted, Pum and Bru, both recognize and bind this conserved sequence to regulate translation. We find that regulation by these RBPs during oogenesis is not unique to pgc but that a large class of maternal mRNAs also lose translational control in the absence of both Pum and Bru. Our results indicate that 212 members of this class of mRNAs also share in their 3′ UTR a version of the 10-nt conserved sequence necessary for Pum and Bru regulation of pgc. These findings suggest that we have identified a broadly utilized mechanism that prevents the translation of specific mRNAs during oogenesis. The fact that some of these mRNAs affect gastrulation and developmental patterning argues that this mechanism evolved to prevent the translation of mRNAs that govern the key early steps of embryogenesis but could be deleterious if translated during oogenesis.
We find that a dynamic and diverse landscape of translational regulators has evolved to allow fine-scale control of maternal mRNAs. mRNAs can be regulated either by the CCR4-Not complex shortening the poly(A) tail or by the decapping machinery or other proteins that bind the cap interfering with cap recognition (Meyer et al., 2004; Garneau et al., 2007; Temme et al., 2014). CCR4-Not complex members as well as decapping machinery proteins are expressed continuously during Drosophila germline development and thus cannot mediate dynamic translational control on their own (Temme et al., 2010; Joly et al., 2013; Fan et al., 2011). However, carefully choreographed expression of specific RBPs that recognize and bind sequences in the UTRs recruit these regulatory proteins to target transcripts at different stages. Our studies show that Pum, whose expression is restricted to the earliest stages of oogenesis, associates with Nos to recruit the CCR4-Not complex to regulate pgc mRNA poly(A)-tails in the GSCs. After differentiation, Pum switches binding partners and complexes with Brat, a protein only expressed in the differentiating stages, and d4EHP, an adaptor protein that binds to the mRNA cap to mask pgc transcript from the translation initiation factors. As Pum levels diminish, this mode of regulation is handed over to Bru, which is robustly expressed from the 16-cell cyst onward, and its partner, Cup, which binds to eIF4E at the mRNA cap to mask pgc transcript from the translation initiation factors. Thus, we posit that by utilizing temporally restricted RBPs that bind the 3′ UTR at a single conserved sequence in a combinatorial fashion, the germline can sculpt differential expression of maternal mRNAs.
Why does pgc use the same sequence to bind the two trans-acting factors, Pum and Bru, as opposed to utilizing two distinct sequences? We observed that Pum recruits Brat, which complexes with d4EHP, to bind the cap and prevent the initiation machinery from accessing the mRNA. Bru accomplishes this by recruiting Cup, which binds eIF4E at the cap. If Pum and Bru are present at the same time, as in the 8- to 16-cell cyst stage, and are bound to different sequences, then they will recruit two proteins that compete to bind to the mRNA cap. In the presence of Pum, its partner d4EHP can outcompete the cap partner eIF4E (Cho et al., 2005), which would make the handoff from Pum to Bru difficult. How then is repression of pgc mRNA seamlessly transitioned from one RBP to another? We also observed a temporal overlap in repression in the 4- and 8-cell cysts mediated by Pum with its two distinct partner complexes (Figure S7C). From the GSCs through 8-cell cyst stage, Pum partners with Nos, Twin, Me31B, and dGe-1 to repress pgc, while it partners with Brat and d4EHP to regulate pgc from the 4- through 16-cell cyst stages (Figures 7E and S7C). The overlap between Pum- and Bru-mediated repression occurs between the 8- and 16-cell cyst stages (Figures 7E and S7C). We hypothesize that to maintain seamless translational regulation during the 4- to 16-cell cyst stages, instead of competing for the cap, the RBPs compete to bind the same cis-element of their target mRNAs. When levels of one RBP diminish and those of another increase, the RBP present at a lower concentration could be displaced from its binding site on the mRNA, allowing for a smooth transition. Thus, we favor the idea that seamless transitions are mediated by overlapping trans-acting factor regimes and competition for the binding site.
pgc is transcribed continuously from the GSC stage onward and accumulates in the oocyte post differentiation. We find that there is a switch in mode of pgc regulation from a Twin (CCR4)-dependent mechanism mediated by Pum, which can destabilize mRNAs in the GSCs, to a Twin (CCR4)-independent mode mediated by Bru in the later-differentiated stages. Loss of Bru during oogenesis results in a dramatic increase in poly-adenylation of the pgc mRNA as well as translation of Pgc. This suggests that Bru-mediated regulation not only translationally represses pgc mRNA during oogenesis but also could maintain it in a state poised for poly-adenylation and translation. We also show that this mode of regulation is not unique to pgc and that there is a large set of maternally deposited germline mRNAs, including zelda, that seem to be regulated similarly. zelda, a transcription factor that activates the zygotic genome, is expressed at low levels in early embryos and increases as development proceeds concurrent with attenuation of Bru levels (Harrison et al., 2011; Nien et al., 2011; Webster et al., 1997). We hypothesize that post-differentiation, it is advantageous to switch the mode of translational regulation to a cap-dependent mechanism mediated by proteins such as Bru to prime these mRNAs to be translated during early embryonic development.
During mammalian development, maternally synthesized mRNAs are deposited into the egg to support embryonic development and need to be translationally regulated. Pum and CELF and/or Bruno-like proteins are both expressed in the mammalian germline and required for fertility (Kress et al., 2007; Mak et al., 2016). The mammalian homologs of Pum, PUMILIO 1 and 2 also bind to a sequence similar to the Drosophila NRE, and CELF1 and/or Bruno-like proteins bind to an “EDEN” sequence similar to Drosophila BREs (Wang et al., 2001; Vlasova et al., 2008; Jenkins et al., 2009). Pum and CELF and/or Bruno-like proteins are required not only in the germline but also for the development of other organs, including the CNS in mice (Spassov and Jurecic, 2003; Barreau et al., 2006; Wagnon et al., 2011; Zhang et al., 2017). Whether Pum and Bru function together on similar targets in the mammalian germline and nervous system as they do in the Drosophila ovary is not known. Our data suggest that such a handoff mechanism could be acting in these vertebrate systems as well.
STAR⋆METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to Lead Contact, Dr. Prashanth Rangan (prangan@albany.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Fly strains
Drosophila was grown on corn flour and agar media with brewer’s yeast. All strains were grown at 25˚C, except RNAi crosses, which were grown at 29˚C. pgcGFP and pgc⊿ used in this study have been previously reported (Martinho et al., 2004; Flora et al., 2018). liprin-γH1 flies were a gift from the Triesman Lab (Astigarraga et al., 2010). nos mutants were generated by crossing the nosRC and nosBN alleles (Arrizabalaga and Lehmann 1999). pum mutants were created by crossing the pumFC8 and pumET1 alleles (Forbes and Lehmann 1998). twin mutants were created by crossing the twinry3 and twinry5 (Morris et al., 2005). The pum680 allele is described in Wharton et.al.,1998. aret mutants were created by crossing the aretPA and aretQB (Schüpbach and Wieschaus 1991). nosGAL4::VP16 and nosGAL4.NGT was gifted by the Lehmann lab. w1118, nosRNAi, pumRNAi, twinRNAi, bratRNAi, d4EHPRNAi, not1RNAi, pop2RNAi, Me31BRNAi, dGe-1RNAi, bruRNAi and cupRNAi lines were acquired from the Bloomington Drosophila Stock Center, Bloomington, IN. The transgenic flies in this paper were generated in the Rangan Lab. They are as follows: P-P-P/pgcGFP (pgc promoter-pgc 5′UTR-eGFP-pgc3′UTR) (Flora et al., 2018), P-P-T (pgc promoter-pgc 5′UTR-eGFP-α-tubulin84B 3′UTR), P-P-K (pgc promoter-pgc 5′UTR-eGFP-K10 3′UTR), P-N-K (pgc promoter-nos 5′UTR-eGFP-K10 3′UTR), generate P-N-P (pgc promoter-nos 5′UTR-eGFP-pgc3′UTR), P-P-T:NBS+PRE/BRE (pgc promoter-pgc 5′UTR-eGFP-α-tubulin84B 3′UTR: NBS+PRE/BRE), P-P-P: ΔUGUAAAUU (pgc promoter-pgc 5′UTR-eGFP-pgc 3′UTR: ΔUGUAAAUU), P-P-P: ΔUGUA (pgc promoter-pgc 5′UTR-eGFP-pgc 3′UTR: ΔUGUA), P-P-P: UUUUAAUU (pgc promoter-pgc 5′UTR-eGFP-pgc 3′UTR: UUUUAAUU), P-P-P: UCUCAAUU (pgc promoter-pgc 5′UTR-eGFP-pgc 3′UTR: UCUCAAUU).
METHOD DETAILS
Generation of transgenic fly strains
The P-P-P/pgcGFP construct was generated by cloning eGFP coding sequence into a plasmid with the pgc 5′UTR and pgc 3′UTR as previously described (Flora et al., 2018).The P-P-T and P-P-K constructs were assembled by PCR amplifying a XhoI-KpnI fragment containing the α-tubulin84B (tub) 3′UTR or K10 3′UTR was then cloned into the XhoI-KpnI site of the P-P-P plasmid, respectively. In order to allow for interchanging of the 700 bp pgc promoter and pgc 5′UTR region of P-P-K, AgeI site was created between of those regions of P-P-K via GenScript by Fisher Scientific. The P-N-K construct was then generated by inserting the nos 5′UTR with Agel and Spel overhangs into the AgeI-SpeI site of the P-P-K plasmid. The pgc 3′UTR fragment was cloned downstream of eGFP at the XhoI-KpnI site of P-N-K to generate P-N-P. The P-P-P: ΔUGUAAAUU, P-P-P: ΔUGUA, P-P-P: UUUUAAUU and P-P-P: UCUCAAUU transgenes in (Figures 2 and S1) was created by site-directed mutagenesis using Phusion High-Fidelity DNA Polymerase. The primers used are listed separately. For the sufficiency experiment the P-P-T: NBS+PRE/BRE construct was generated by inserting the PRE/BRE sequence was added at the same location (after nucleotide 28 of tub 3′UTR) of that of pgc 3′UTR into α -tubulin 84B 3′UTR. These gene fragments were created from gBlock gene fragment service by Integrated DNA technology with XhoI and KpnI sites. The plasmids for injections were then constructed by cloning those gBlock fragments via restriction digest.
Immuno-fluorescence Staining
Female Drosophila ovaries were dissected in cold 1X PBS and fixed in 4% paraformaldehyde for 20 minutes at room temperature (RT). The tissue was permeabilized in 1mL of PBST (1X PBS, 0.2% Tween and 1% Triton-X) for 1 hour at RT. After permeabilization the tissues were blocked in 1mL of BBT (0.5% BSA in PBST) for 2 hours at RT. Then 0.5mL of primary antibody was added and tissues were placed on a nutator at 4˚C overnight. The following steps were then carried out at RT. After incubation, ovaries were washed three times in 1mL of BBT for 10, 15, 30 minutes. An additional wash for 30 minutes was carried on by adding 2% Donkey serum to 1mL of BBT. After the last wash secondary antibody in 0.5ml of BBT with 4% Donkey serum was added and incubated for 2 hours protected from light. After the incubation, ovaries were washed in 1mL of PBST for five times. After the washed onedrop of Vectashield was added and then the tissue was mounted on a glass slide and a coverslip was placed on the slide (Flora et al., 2018). The antibodies used and dilution are listed as follows: Rabbit anti-Vasa (1:4000 dilution), chicken anti-Vasa (1:500 dilution), mouse anti-1B1 (1:20), rabbit anti-GFP (1:2000), rabbit anti-pSmad3 (1:150), rabbit anti-Nanos (1:500), rabbit anti-Bruno (Lehmann Lab) (1:500), rabbit anti-Pumilio (1:150), Alexa 488, Cy3 and Cy5 conjugated secondary antibodies were used at a concentration of 1:500.
Fluorescent in situ hybridization (FISH)
FISH of the ovaries was carried out probes against pgc and GFP, which were a gift from the Lehmann lab (Trcek et al., 2017). The ovaries were dissected in 1XPBS, fixed in 3% methanol-free paraformaldehyde in PBS for 20 minutes and washed 3 times with PBST. Next, they were treated with 3 ug/ml Proteinase K in PBS and placed on a nutator for 13 minutes at RT, and then placed on ice for 30 minutes. The tissue was then blocked in 2 mg/ml glycine in PBST twice for 10 minutes each and rinsed twice with PBST for 2 minutes. The ovaries were post-fixed for 20 minutes in 3%. The tissue was then washed with PBST 5 times for 2 minutes and washed with pre-warmed fresh pre-hybridization mix (10% deionized formamide in 2X SSC) for 10 minutes. 60 μL per sample of hybridization mix (10% deionized formamide, 0.5 μL of yeast t-RNA, 0.5 μL of salmon sperm DNA, 1 μM of probe, 10% Dextran sulfate, 2 mg/ml BSA, 2X SSC and 1 μL of RNase Out) was added and the sample was incubated overnight at 37˚C for at least 12 hours and no more than 16 hours. After incubation, 1 mL of pre-warmed pre-hybridization solution was added to the tissues. After 10 minutes, the pre-hybridization solution was removed, and the ovaries were washed 5 times with 1XPBS for 15 minutes each. After the last wash, PBS was aspirated out and a drop of Vectashield (Vector Labs, Inc.) was added to the tissue before preparing the slide.
Imaging
All images were taken on a Carl Zeiss 710 Meta confocal microscope using 20X or 40X oil immersion objectives. Scale bars were added using Zen Blue image processing software.
Western Blot
Twenty wild-type ovaries or 40 mutant ovaries were dissected in 1XPBS. Tissue was homogenized in 30 μL of RIPA buffer and centrifuged at 13,000 rpm for 15 minutes at 4˚C. 1 mL of the protein extract was used to carry out a Bradford assay. 25 μg of protein sample was denatured with 4X Laemmli Sample Buffer and β-marcepthanol at 95˚C for 5 minutes. The samples were loaded in a Mini-PROTEAN TGX 4%–20% gradient SDS-PAGE gels and run at 110V for 1 hour. The proteins were then transferred to a 0.20 μm nitrocellulose membrane at 100V for 1 hour at 4˚C. After transfer, the membrane was blocked in 5% milk in PBST for 2 hours at RT and 1˚ antibody prepared in 5% milk in PBST was added to the membrane and incubated at 4˚C O/N. The membrane was rinsed in 0.5% milk in PBST 4–5 times before adding 2˚ antibody prepared in 5% milk in PBST. After 2 hours the membrane was rinsed in PBST 4–5 times. Chemiluminescence ECL kit was used to develop the membrane. The membrane was stripped prior to re-probing for loading control. Antibodies used for Western Blots are listed below:
Primary antibody rat anti-HA was used at 1:3000 dilution. Anti-rat HRP (1:10,000) was used at 1:10,000 dilution. Rabbit anti-Vasa (1:6000) was used as a loading control. Anti-rabbit HRP was used at 1:10,000 dilution.
For Western Blot analysis pgcHA levels were normalized to Vasa levels of each genotype. Then the fold change was calculated for each genotype by subtracting fold change of wild-type control from all experimental samples.
For RIP western blots, rabbit anti-Pum, rabbit anti-Bru and rabbit anti-GFP was used at a 1:4000, 1:6000 and 1:5000 dilution respectively. Anti-rabbit HRP was used at 1:10,000 dilution.
RNA Extraction
Wild-type ovaries were dissected in 1XPBS. After dissection, 100 μL of Trizol reagent was added to the tissue and homogenized. Additional, 900 μL of Trizol was added, mixed and incubated at RT for 3 minutes. After incubation, 200 μL of Chloroform was added to each sample and mixed vigorously and incubated at RT for 5 minutes before centrifugation at 13,000 rpm for 20 minutes at 4˚C. 2 volumes of 100% ethanol, 10% volume 3 M sodium acetate and 0.5 ul of glycol blue was added to aqueous layer and incubated at 20˚C for 1 hour. The samples were centrifuged at 13,000 rpm for 20 minutes at 4˚C. The pellet was washed with 75% ethanol, airdried and re-suspended in RNase free H2O. 10 μg of nucleic acid was then taken and subjected to a DNase treatment using the TURBO DNA-free Kit.
Real Time-PCR (RT-PCR) and quantitative Real Time-PCR (qRT-PCR)
500ng of DNase treated RNA was reverse transcribed using Super Script III. For RT-PCR experiments, 1.5 μL of cDNA was amplified using 0.5 μL of 10 μM of each reverse and forward primers, 0.5 μL of 10 μM (d)NTP and 0.125 μL Taq Polymerase and 2.5 μL 10XTaq Polymerase Buffer. The thermal cycling conditions for PCR was 95˚C for 30 s, 32 cycles of 95˚C for 30 s, 3˚below the Tm of the lowest Tm primer for 30 s, 68˚C for 1 minute, and 1 cycle of 68˚C for 4 minutes. After PCR, 2.8 μL of Orange-G dye was added to each sample and 10 μL of PCR product was ran on a 1% agarose gel stained with ethidium bromide to visualize bands.
For qRT-PCR experiments, 0.5 μL of cDNA was amplified using 5 μL of SYBR green Master Mix, 0.3 μL of 10 μM of each reverse and forward primers. The thermal cycling conditions were as follows: 50˚C for 2 min, 95˚C for 10 min, 40 cycles at 95˚C for 15 s, and 60˚C for 60 s. The experiments were carried out in technical triplicate and three biological replicates for each data point.
Pumilio Protein Purification
Pumilio expression plasmid pFN18K Pum RNA-binding domain (aa 1091–1426) was gifted to us by the Goldstrohm lab. Pumilio was purified following the protocol adapted from Weidmann et.al, 2016. The vector was transformed into KRX cells. A single colony from the plate was picked and inoculated in 100 mL of LB containing 25 μg/mL of kanamycin and incubated in a shaker at 37˚C overnight. 20 mL of this starter culture was inoculated in 1L of 2xYT (16 g Bacto Tryptone, 10 g Bacto Yeast Extract, 5g NaCl, pH 7.0 adhusted with 5N NaOH) media containing 2mM MgSO4 and 25 μg/mL of kanamycin and incubated in a shaker at 37˚C till OD600 was between 0.7 and 0.9. Protein was induced for 3 hours in a shaker at 37˚C by adding 5 mL of 20% w/v L-rhamnose (0.1% final). The cells were split into 500 mL aliquots and pelleted. Pumilio was purified from one pellet of 500 mL culture. Pellet was resuspended in 30 mL of filtered Bugwash (50mM Tris-HCl, pH 8.0, 10% w/v Sucrose) and centrifuged again. Supernatant was discarded. The pellet was resuspended in 25 mL of filtered Binding buffer (50mM Tris pH 8.0, 2mM MgCl2, 150 mM NaCl) that contained freshly added 1mM DTT, 0.05% v/v NP-40 and 1x Protease inhibitor Cocktail (50X: 50 mM PMSF, 500 μg/ml aprotinin, 500 μg/ml pepstatin, 500 μg/ml leupeptin, dissolve in 10% v/v ethanol). After pellet was resuspended 1.25 mL of 10mg/ml lysozyme was added, mixed by inversion and incubated at 4˚C for 30 minutes. Then 140 μL of 1M MgCl2 and 26 μL of DNase I was added and incubated at 4˚C for 20 minutes. The lysate was then centrifuged at 50,000Xg for 30 minutes at 4˚C. Supernatant was transferred to a new tube and 50 μL of equilibrated HaloLink Resin beads were added and incubated for 4–6 hours at 4˚C. After incubation, lysate was centrifuged, and resin was transferred to a new tube. Resin was washed in filtered Wash buffer (50 mM Tris pH 8.0, 1M NaCl, 2 mM MgCl2) four times and eluted in 250 μL of Binding Buffer. For cleavage of AcTEV tag, 3 μL (30 units) of AcTEV protease was added to the eluted beads and incubated on a nutator at 4˚C overnight. The next day tube was centrifuged and the supernatant containing purified Pumilio was transferred to new tube and 100% glycerol was added to the eluted protein for a final glycerol concentration of 20%. Protein was aliquoted, and flash frozen in liquid nitrogen and stored at 80˚C.
Bruno Protein Purification
Bruno expression plasmid pETM-82 was acquired from EMBL (Chekulaeva et al., 2006). 5 mL of Bruno in pETM-82 in BL21(DE3) was grown overnight at 37˚C. This culture was added to 1000 mL of LB-Kanamycin media. Cells were shaken at 220 rpm at 37˚C for 2–3 hr or until OD600~0.8. The culture was then cooled down to 25˚C.0.5 mM IPTG was added to induce the cells and shaken at 220 rpm at 25˚C for 3 hours. The cells were then centrifuged at 4000xg for 20 minutes at 4˚C in 50 mL aliquots. The pellet was re-suspended in 3 mL of re-suspension buffer (20 mM Na phosphate, 50 mM NaCl, 20 mM imidazole, 10 ul of 500 mg/ml pH 7.4) and sonicated at 20% intensity for 20 s for 3 times and pulsed for 20 s for 3 times using 1/8 probe, making sure the cell suspension is on ice throughout sonication. The suspension was then centrifuged at 10,000xg for 10 minutes for 4˚C. Meanwhile, the His GraviTrap column was equilibrated with 10 mL binding buffer (20 mM Na phosphate, 50 mM NaCl, 20 mM imidazole, 10 ul of 500 mg/ml pH 7.4). The supernatant was added to the column and washed with increments of 1 ml, 4 mL and 5 mL of binding buffer. The protein was then eluted using the following washes; twice with 1 mL of elution buffer (1), twice with 1 mL of elution buffer (2) and three times with 1 mL of elution buffer (3).
Elution Buffer (1): 20 mM NaPO4, 50 mM NaCl, 150 mM imidazole, pH 7.4
Elution Buffer (2): 20 mM NaPO4, 50 mM NaCl, 300 mM imidazole, pH 7.4
Elution Buffer (3): 20 mM NaPO4, 50 mM NaCl, 500 mM imidazole, pH 7.4
The last two fractions contained purified Bruno protein. 100% glycerol was added to the eluted protein for a final glycerol concentration of 20%. The eluted protein sample was de-salted using the PD-10 column. Protein was aliquoted, and flash frozen in liquid nitrogen and stored at 80˚C.
Electrophoretic mobility shift assays (EMSA)
RNA oligonucleotides were end-labeled using T4 Kinase with ATP [γ−32P]. Excess ATP was eliminated by using G-25 Sephadix Columns. All RNA-binding reaction was performed in 1X Binding Buffer (50mM Tris pH 7.5, 150mM NaCl, 2mM DTT, 0.1mg/μl BSA, 0.001% Igepal CA-630, 0.5 μL of dIdC and 0.5 μL of yeast t-RNA). RNA and purified protein were incubated for 20 minutes at RT and then ran on a 6% native polyacrylamide TBE gel at 150V for 4 hours at 4˚C. The gel was then dried onto Whatmann filter paper and exposed to a phosphor screen overnight. A Typhoon Trio imager was used to image the EMSAs.
Poly(A) tail length (PAT) Assay
500ng of DNase treated RNA was reverse transcribed using Super Script III but instead of using oligo (dT), 5 μL of anchored Oligo (dT) primer was used for each sample (Rangan et al., 2008). 2 μL of cDNA was then amplified using 0.5 μL of gene specific forward primer, 0.5 μL of anchored Oligo(d)T, 0.5 μL of 10 μM dNTP and 0.125 μL Taq Polymerase and 2.5 μL 10XTaq Polymerase Buffer. The thermal cycling conditions for PCR was 95˚C for 30 s, 30 cycles of 95˚C for 30 s, 2˚ below Tm of primer for 30 s, 65˚C for 1.5 minutes, and 1 cycle of 65˚C for 4 minutes. After PCR, 2.8 μL of Orange-G dye was added to each sample and 10 μL of PCR product was ran on a 2.5% agarose gel. The gel was post-stained with ethidium bromide for 20 minutes, and then washed three times with H2O prior to imaging.
RNA-Immuno-precipitation (RIP)-qPCR
Each IP experiment was carried out in 100 pairs of wild-type ovaries. Ovaries were dissected in RNase free 1XPBS. After dissection, PBS was aspirated and 100 μL of RIPA lysis buffer was added to the tissues and homogenized. Another 200 μL of RIPA lysis buffer was added to the lysate and mixed well. The lysate was then centrifuged at 13,000 rpm for 20 minutes at 4˚C. 5% of cleared lysate was set aside for Western Blot analysis. 10% of the lysate was set aside and frozen in Trizol as RNA Input for each IP experiment. Remaining lysate was divided equally; one was for IgG control and the other for antibody of interest (AI). 100 μL of Dynabeads Protein A was rinsed 3 times with 400 μL of 1:10 dilution of NP-40 buffer. 25 μL of resuspended beads were added to each AI and IgG containing lysate samples and incubated overnight at 4˚C. After incubation, the beads were washed 4 times with 1:10 dilution of NP-40 buffer for 1 minute. An additional two washes for 5 minutes were carried out before re-suspending the beads in 25 μL of NP-40 buffer. 10 μL of beads from each of the samples were used to perform a Western Blot analysis to confirm pull-down. The other 15 μL was used to extract RNA to perform qRT-PCR experiments to show association of RNA with pulled-down protein. Buffers and antibodies used are described below:
RIPA lysis buffer: 10mM Tris-Cl Buffer (pH 8.0), 1mM EDTA, 1% Triton X-100,0.1% Sodium deoxycholate, 0.1% SDS, 140mM NaCl, 1mM PMSF, 1 cOmplete, EDTA-free Protease Inhibitor Cocktail Pill RNase free H2O.
NP-40 buffer: 50mM Tris-Cl Buffer (pH 8.0), 150mM NaCl, 10% NP-40, 1 cOmplete, EDTA-free Protease Inhibitor Cocktail Pill, RNase free H2O.
The following antibodies were added to the lysate and incubated at 4˚C for 3 hours; 2.5 μL of rabbit anti-GFP, 1.25 μL of Rabbit IgG, 1 μL of rabbit anti-Bru (Dr. Lilly) or 2 μL rabbit anti-Pum (Lehmann lab).
RNA sequencing and sample library preparation
Total RNA was extracted with Trizol, treated with Turbo DNase and poly(A)+ RNA was isolated by double selection with poly-dT beads, using ~6μg total RNA, which is then followed by first- and second-strand synthesis. Sequencing libraries were prepared using NEXTflex Rapid Illumina DNA-Seq Library Prep Kit. 75 base-pair single-end mRNA sequencing was performed an Illumina NextSeq 500 by the Center for Functional Genomics.
Polysome profiling and Polysome-seq
~80 ovaries were dissected in PBS supplemented with cycloheximide and frozen immediately with liquid nitrogen. Tissue was homogenized in 200 μL of cold lysis buffer consisting of 1x Polysome buffer supplemented with 1% Triton-X and 1 protease inhibitor pill per 10 mL of buffer. The lysate was centrifuged at 15,000 x g at 4˚C for 10 minutes. 20% of lysate was kept aside for “Input RNA” libraries. 750 μL of cleared lysate was loaded onto 10%–50% sucrose gradients (500 mM KCl; 15 mM Tris-HCl, pH 7.5; 15 mM MgCl2; and 100 μg/ml cycloheximide) in Beckman Coulter 9/16×3.5 PA tubes (Cat. #331372). Gradients were centrifuged at 35,000xg using a SW41 rotor for 3 hours at 4˚C. Gradients were fractionated on a Brandel flow cell (Model #621140007) at 0.75 mls/min and 750 μL was collected for each fraction with the sensitivity settings at 0.5 Abs. RNA was extracted from the fractions using standard acid phenol: chloroform extraction. The RNA pellet was washed with 80% ethanol and air-dried. After air-drying the pellet was dissolved in 10 μL of nuclease-free water. Turbo DNase treatment and library preparation was carried out as described above.
QUANTIFICATION AND STATISTICAL ANALYSIS
Western Blot Analysis
To calculate relative change in HA protein expression of the various transgenes reported in this study, first, ImageJ was used to calculate the arbitrary units (A.U) of PgcHA bands and loading control Vasa bands. Then the HA A.U was divided by the Vasa A.U to calculate relative fold change. Wild-type control A.U was subtracted from each ratio to eliminate background. Western blots were repeated three times with independent biological samples.
Quantitative Real Time-PCR (qRT-PCR) analysis
To calculate fold change in GFP mRNA levels to RP49 mRNA levels, first, the Ct values of technical replicates of each trial was averaged. ΔCt was calculated by subtracting RP49 Ct average from the Ct average of GFP. Then the of the 2^-ΔCt was calculated for each trial. To diminish background, 2^-ΔCt valued form wild-type control was subtracted from GFP and RP49 2^-ΔCt values.
To calculate relative protein levels to mRNA levels, the fold protein change was divided by fold RNA change from qRT-PCR experiment for each biological trial. The average, standard deviation and standard error was then calculated for the three trials.
RNA-Immuno-precipitation (RIP) qPCR analysis
The following calculation was adapted from the Sigma-aldrich Imprint RIP Kit protocol.
- Each RIP RNA fractions’ Ct value was normalized to each of the Input RNA fraction Ct value for the same qPCR Assay (ΔCt) to account for RNA sample preparation differences.
- The % Input for each RIP fraction (linear conversion of the normalized RIP ΔCt) was calculated.
- The normalized RIP fraction Ct value for the normalized background [IgG Ab] fraction Ct value (first ΔΔCt) was adjusted.
- IP Fold Enrichment above the sample specific background (linear conversion of the first ΔΔCt) was calculated.
Statistical Analysis
A student’s two-tailed t test or population proportion z-test were carried out to calculate significance of results. Standard error was calculated from three independent biological samples for each experiment and is represented by the error bar. *, ** and *** denotes p values less than 0.05, 0.005 and 0.005 respectively. All analysis was carried out using Microsoft Excel.
RNA-seq data analysis
After quality of reads was assessed the RNA-seq reads were aligned via HISAT2 (version 2.1.0) (Kim et al., 2015a) set to be splice aware to UCSC dm6 release 6.01. Count tables were generated using featureCounts (version 3.16.5) (Liao et al., 2014).
Translation Efficiency (TE) Analysis
To determine translation efficiencies (TE), CPMs (counts per million) values were calculated for all polysome-seq libraries. Any transcript having zero reads in any library was discarded from analysis. The log2 ratio of CPMs between the polysome fraction and total mRNA was calculated and averaged between replicates. This ratio represents TE. After TE of each sample was calculated and replicates were averaged, TE of pumRNAi and bruRNAi were compared to that of Control. This ratio represents ΔTE. Targets were defined as transcripts falling greater or less than one standard deviation from the median of ΔTE (Kronja et al., 2014). To discover sequences similar to the pgc BRE in the 3′UTR of targets, all annotated 3′UTRs were downloaded from Flybase for all analyzed targets. A list of BREs and PREs that contain the core sequence UGUA was compiled manually through a literature search. Using the R package Biostrings this list was used to generate and apply a position weight matrix (pwm). This pwm was used to score all 10-mers in all of the previously mentioned 3′UTRs. A minimum score of 90% was set as cutoff. Additionally, we manually ensured that the core sequence UGUA was present in all targets above the cutoff. Targets identified from polysome-seq were subsetted from the list of RNAs containing a pgc-like BRE in their 3′UTR using a custom R script.
Gene Ontology (GO) Enrichment Analysis
Significant over-represented functional categories of the 212 PRE/BRE containing shared targets of Pum and Bru was carried out using the PANTHER Gene List Analysis tool. Selected GO terms with p value < 0.05 have been shown in Figure 7D.
DATA AND SOFTWARE AVAILABILITY
The accession number for the data reported in this paper is GEO: GSE119458.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-GFP | abCam | Cat# ab6556 |
| Chicken polyclonal anti-GFP | abCam | Cat# ab13970 |
| Rabbit polyclonal anti-pMad | abCam | Cat# ab52903 |
| Mouse monoclonal anti-1B1 | Developmental studies Hybridoma Bank | Antibody Registry ID:528070 |
| Rat monoclonal anti-HA high affinity | Roche Diagnostics | REF:11867423001 |
| Rabbit polyclonal anti-Vasa | Rangan Lab | N/A |
| Chicken polyclonal anti-Vasa | Rangan Lab | N/A |
| Rabbit polyclonal anti-Pumilio | Gift from Lehmann Lab | N/A |
| Rabbit polyclonal anti-Bruno | Gift from Lehmann Lab | N/A |
| Rabbit polyclonal anti-Nanos | Gift from Buszczak Lab | N/A |
| Rabbit polyclonal anti-Bruno | Gift from Lily Lab (Sugimura and Lilly, 2006) | N/A |
| Anti-rabbit Alexa 488 | Jackson ImmunoResearch Labs | Code:711–546-152 |
| Anti-chicken Alexa 488 | Jackson ImmunoResearch Labs | Code:703–546-155 |
| Anti-rabbit Alexa Cy3 | Jackson ImmunoResearch Labs | Code:711–166-152 |
| Anti-mouse Alexa Cy3 | Jackson ImmunoResearch Labs | Code:715–546-150 |
| Anti-chicken Alexa 647 | Jackson ImmunoResearch Labs | Code:703–606-155 |
| Anti-mouse Alexa 647 | Jackson ImmunoResearch Labs | Code:715–606-150 |
| Anti-Rat HRP | Jackson ImmunoResearch Labs | Code:112–035-003 |
| Anti-Rabbit HRP | Jackson ImmunoResearch Labs | Code:111–035-144 |
| ChromePure Rabbit IgG | Jackson ImmunoResearch Labs | Code: 011–000-003 |
| Bacterial and Virus Strains | ||
| BL21(DE3) competent E.coli | New England Biolabs Inc. | Cat# C25271 |
| KRX E.coli competent cells | Promega | Cat# L3002 |
| DH5α competent cells | Invitrogen | Cat# 18265017 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde (Methanol Free),10% Ultrapure | Polysciences Inc. | Cat# 04018–1 |
| Donkey Serum | Sigma-Aldrich | SKU: D9663 |
| Vectashield Antifade Mounting Medium with DAPI | Vector Laboratories | Cat# H-1200 |
| T4 Polynucleotide Kinase | New England Biolabs Inc. | Cat# M0201S |
| Restriction Endonuclease Xhol | New England Biolabs Inc. | Cat# R0146S |
| Restriction Endonuclease Kpnl | New England Biolabs Inc. | Cat# R0142S |
| Restriction Endonuclease Agel | New England Biolabs Inc. | Cat# R0552S |
| Restriction Endonuclease Spel | New England Biolabs Inc. | Cat# R0133S |
| Restriction Endonuclease Notl | New England Biolabs Inc. | Cat# R0189S |
| Phusion High-Fidelity DNA Polymerase | New England Biolabs Inc. | Cat# M0530S |
| HaloLink Resin | Promega | Cat# G1912 |
| L-Rhamnose monohydrate | Sigma-Aldrich | SKU: R3875 |
| IPTG | Invitrogen | Cat# 15529019 |
| AcTEV Protease | Invitrogen | Cat# 12575015 |
| LightShift Poly (dI-dC) | ThermoFisher Scientific | Cat# 20148E |
| Yeast tRNA | ThermoFisher Scientific | Cat# AM7119 |
| Salmon Sperm DNA | ThermoFisher Scientific | Cat# 15632011 |
| Nonidet P-40 (NP-40) substitute | IBI Scientific | Cas# 9016–45-9 |
| Tween-20 detergent | VWR | Cat# 97062–332 |
| Triton X-100 detergent | VWR | Cat# 97062–208 |
| Igepal CA-630 detergent | Sigma-Aldrich | SKU: I8896 |
| DNase I | Roche | Cat# 04 716 728 001 |
| Aprotinin | Sigma-Aldrich | SKU: 10236624001 |
| PMSF | Sigma-Aldrich | SKU: 10837091001 |
| Leupeptin protease inhibitor | ThermoFisher Scientific | Cat# 78435 |
| Pepstatin A protease inhibitor | ThermoFisher Scientific | Cat# 78436 |
| TRIzol | Invitrogen | Cat# 15596026 |
| Dynabeads Protein A | Invitrogen | Cat# 10002D |
| cOmplete, EDTA-free Protease Inhibitor Cocktail Pill | Sigma-Aldrich | SKU: 11873580001 |
| Bradford reagent | Bio-Rad | Cat. #500–0205 |
| 4X Laemmli Sample Buffer | Bio-Rad | Cat. #161–0747 |
| Ultrapure Sucrose | Amresco | Code: 0335–1KG |
| Bruno expression plasmid pETM-82 | EMBL (Chekulaeva et al., 2006) | N/A |
| Pumilio expression plasmid pFN18K | Goldstrohm Lab (Weidmann et al., 2016) | N/A |
| Critical Commercial Assays | ||
| G-25 Sephadix Columns | Roche | Cat# 11273990001 |
| PD-10 column | GE Health care Life Sciences | Cat# 17–0851-01 |
| His GraviTrap | GE Health care Life Sciences | Cat# 11–0033-99 |
| TURBO DNA-free Kit | Life Technologies | Cat# AM1907 |
| Super Script III | Life Technologies | Cat# 1808051 |
| SYBR Green Master Mix | Applied Biosystems | Cat# 4367659 |
| NEXTflex Rapid Illumina DNA-Seq Library Prep Kit | Bioo Scientific | Cat# NOVA-5138–11 |
| Mini-PROTEAN TGX 4–20% gradient SDS- PAGE gels | Bio-Rad | Cat# 456–1094 |
| Western ECL Substrate | Bio-Rad | Cat# 1705060 |
| Deposited Data | ||
| RNA-seq Data | This paper | GEO: GSE119458 |
| Polysome-seq Data | This paper | GEO: GSE119458 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: w*; pgc⊿ | (Martinho et al., 2004; Flora et al., 2018) | N/A |
| D. melanogaster: w*; Df(2R)Liprin-γH1, P{neoFRT}42D Liprin-γH1/CyO | Bloomington Drosophila Stock Center (Astigarraga et al., 2010) | BDSC:63813; FlyBase: FBst0063813 |
| D. melanogaster: w*; P{UAS-tkv.CA}3 | Bloomington Drosophila Stock Center | BDSC:36537; FlyBase: FBst0036537 |
| D. melanogaster: pumFC8 mutant | (Forbes and Lehmann, 1998) | N/A |
| D. melanogaster: pumET1 mutant | (Forbes and Lehmann, 1998) | N/A |
| D. melanogaster: RNAi for pum: y1 v1; P{TRiP.JF02267}attP2 | Bloomington Drosophila Stock Center | BDSC:26725; FlyBase: FBst0026725 |
| D. melanogaster: RNAi for pum: y1 sc* v1; P{TRiP.HMS01685}attP40 | Bloomington Drosophila Stock Center | BDSC:38241; FlyBase: FBst0038241 |
| D. melanogaster: st1 pum680/TM3, Sb1 Ser1 | Bloomington Drosophila Stock Center (Wharton et al., 1998) | BDSC:3260; FlyBase: FBst0003260 |
| D. melanogaster: nosRC mutant | (Arrizabalaga and Lehmann, 1999) | N/A |
| D. melanogaster: nosBN mutant | (Arrizabalaga and Lehmann, 1999) | N/A |
| D. melanogaster: RNAi for nos: y1 sc* v1; P{TRiP.HMS00930}attP2 | Bloomington Drosophila Stock Center | BDSC:33973; FlyBase: FBst0033973 |
| D. melanogaster: RNAi for nos: y1 sc* v1; P{TRiP.GLC01867}attP40 | Bloomington Drosophila Stock Center | BDSC:57700 FlyBase: FBst0057700 |
| D. melanogaster: twinRY3 mutant | (Morris et al., 2005) | N/A |
| D. melanogaster: twinRY5 mutant | (Morris et al., 2005) | N/A |
| D. melanogaster: RNAi for twin: y1 sc* v1; P{TRiP.HMS00493}attP2 | Bloomington Drosophila Stock Center | BDSC:32490; FlyBase: FBst0032490 |
| D. melanogaster: RNAi for not: y1 sc* v1; P{TRiP.HMS00526}attP2 | Bloomington Drosophila Stock Center | BDSC:32836; FlyBase: FBst0032836 |
| D. melanogaster: RNAi for pop2: y1 sc* v1; P{TRiP.HM05235}attP2 | Bloomington Drosophila Stock Center | BDSC:30492; FlyBase: FBst0030492 |
| D. melanogaster: RNAi for me31B: y1 v1; P{TRiP.HM05052}attP2 | Bloomington Drosophila Stock Center | BDSC:28566; FlyBase: FBst0028566 |
| D. melanogaster: RNAi for dGe1: y1 sc* v1; P{TRiP.HMS00340}attP2 | Bloomington Drosophila Stock Center | BDSC:32349; FlyBase: FBst0032349 |
| D. melanogaster: RNAi for Brat: y1 sc* v1; P{TRiP.HMS01121}attP2 | Bloomington Drosophila Stock Center | BDSC:34646; FlyBase: FBst0034646 |
| D. melanogaster: RNAi for d4eHP: y1 sc* v1; P{TRiP.GL01035}attP2 | Bloomington Drosophila Stock Center | BDSC:36876; FlyBase: FBst0036876 |
| D. melanogaster: aretQB (bruno) mutant | Schupbach Lab (Schüpbach and Wieschaus, 1991) | N/A |
| D. melanogaster: aretPA (bruno) mutant | Schupbach Lab (Schüpbach and Wieschaus, 1991) | N/A |
| D. melanogaster: RNAi for bruRNAi: y1 v1; P{TRiP.HMS01899}attP40 | Bloomington Drosophila Stock Center | BDSC:38983; FlyBase: FBst0038983 |
| D. melanogaster: RNAi for cupRNAi: y1 sc* v1; P{TRiP.GL00327}attP2/TM3, Sb1 | Bloomington Drosophila Stock Center | BDSC:35406; FlyBase: FBst0035406 |
| D. melanogaster: nosGAL4::VP16 | Lehmann Lab (NYUMC) | N/A |
| D. melanogaster: nosGAL4.NGT | Lehmann Lab (NYUMC) | N/A |
| D. melanogaster: pumGFP transgene | Gift from Salz Lab (Case Western) | N/A |
| D. melanogaster: me31BGFP-TRAP transgene | Gift from Nakamura Lab (RIKEN) | N/A |
| D. melanogaster: pgcGFP transgene (P-P-P) | Rangan Lab (Flora et al., 2018) | N/A |
| D. melanogaster: pgc promoter-pgc 5′UTR-eGFP-tubulin 3′UTR transgene (P-P-T) | This paper | N/A |
| D. melanogaster: pgc promoter-pgc 5′UTR-eGFP-K10 3′UTR (P-P-K) | This paper | N/A |
| D. melanogaster: pgc promoter-nos 5′UTR-eGFP-K10 3′UTR (P-N-K) | This paper | N/A |
| D. melanogaster: pgc promoter-nos 5′UTR-eGFP-pgc3′UTR (P-N-P) | This paper | N/A |
| D. melanogaster: pgc promoter-pgc 5′UTR-eGFP-tubulin 3′UTR+(NBS+PRE/BRE) transgene (P-P-T: NBS+PRE/BRE) | This paper | N/A |
| D. melanogaster: pgcGFP transgene (P-P-P: ΔUGUAAAUU) | This paper | N/A |
| D. melanogaster: pgcGFP transgene (P-P-P: UUUUAAUU) | This paper | N/A |
| D. melanogaster: pgcGFP transgene (P-P-P: UCUCAAUU) | This paper | N/A |
| D. melanogaster: pgcGFP transgene (P-P-P: ΔUGUA) | This paper | N/A |
| Oligonucleotides | ||
| Primers used for generating transgenes see Table S5 | This paper | N/A |
| Primers for site-directed mutagenesis see Table S5 | This paper | N/A |
| Oligonucleotides for EMSA see Table S5 | This paper | N/A |
| Primers for RT-PCR see Table S5 | This paper | N/A |
| Primers for qRT-PCR see Table S5 | This paper | N/A |
| Primers for PAT assay see Table S5 | This paper | N/A |
| GFP RNA FISH probe labeled with CALFluor590 | (Trcek et al., 2017) | N/A |
| pgc RNA FISH probe labeled with CALFluor590 | (Trcek et al., 2017) | N/A |
| Recombinant DNA | ||
| Plasmid: pCaSpeR2 P element transformation vector | Drosophila Genomics Resource Center | Stock Number: 1066 |
| Software and Algorithms | ||
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| MEME Suite | (Bailey et al., 2009) | http://meme-suite.org/doc/overview.html |
| HISAT2 | (Kim et al., 2015a) | https://ccb.jhu.edu/software/hisat2/index.shtml |
| featureCounts | (Liao et al., 2014) | http://bioinf.wehi.edu.au/featureCounts/ |
| R package Biostrings | Bioconductor | https://bioconductor.org/packages/release/bioc/html/Biostrings.html |
| PANTHER Gene Analysis | Gene Ontology Reference Genome Project | http://www.pantherdb.org/ |
Highlights.
pgc, a germline RNA, is translationally regulated throughout Drosophila oogenesis
A conserved 10-nt sequence in the pgc 3′ UTR is required for its regulation
Pum and Bru, conserved RBPs, sequentially repress pgc translation via this sequence
A class of maternal RNAs are also regulated by Pum and Bru during oogenesis
ACKNOWLEDGMENTS
We would like to thank Dr. Buszczak (UT Southwestern) for anti-Nanos, Dr. Lilly (NIH), anti-Bruno (IP), and Dr. Lehmann (NYUMC) for anti-Pumilio and anti-Bruno (IF) antibodies; Dr. Salz (Case Western) and Dr. Nakamura (RIKEN) for the pumGFP and me31BGFP transgenes, respectively; Dr. Goldstrohm (University of Minnesota) for Pum protein expression vector; and Dr. Lehmann for GFP in situ probes. P.R. is funded by the NIH/NIGMS (grant RO1 1119484–1-68857) and the Pew Biomedical Scholars Program.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and five tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.12.007.
REFERENCES
- Anderson RA, Fulton N, Cowan G, Coutts S, and Saunders PT (2007). Conserved and divergent patterns of expression of DAZL, VASA and OCT4 in the germ cells of the human fetal ovary and testis. BMC Dev. Biol 7, 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrizabalaga G, and Lehmann R (1999). A selective screen reveals discrete functional domains in Drosophila Nanos. Genetics 153, 1825–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astigarraga S, Hofmeyer K, Farajian R, and Treisman JE (2010). Three Drosophila liprins interact to control synapse formation. J. Neurosci 30, 15358–15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, and Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Méreau A, and Osborne HB (2006). Mammalian CELF/Bruno-like RNA-binding proteins: molecular characteristics and biological functions. Biochimie 88, 515–525. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, and Izaurralde E (2006). mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 20, 1885–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Rosario A, Bhargava V, Hillebrand J, Kollipara RK, Ramaswami M, and Buszczak M (2016). Repression of Pumilio protein expression by Rbfox1 promotes germ cell differentiation. Dev. Cell 36, 562–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, and Ephrussi A (2006). Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124, 521–533. [DOI] [PubMed] [Google Scholar]
- Chen D, and McKearin DM (2003). A discrete transcriptional silencer in the bam gene determines asymmetric division of the Drosophila germline stem cell. Development 130, 1159–1170. [DOI] [PubMed] [Google Scholar]
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, and Sonenberg N (2005). A new paradigm for translational control: inhibition via 5′−3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 121, 411–423. [DOI] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, and Sonenberg N (2006). Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr. Biol 16, 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinalli RM, Rangan P, and Lehmann R (2008). Germ cells are forever. Cell 132, 559–562. [DOI] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, and Priess JR (2006). Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science 311, 851–853. [DOI] [PubMed] [Google Scholar]
- Draper BW, Mello CC, Bowerman B, Hardin J, and Priess JR (1996). MEX-3 is a KH domain protein that regulates blastomere identity in early C. elegans embryos. Cell 87, 205–216. [DOI] [PubMed] [Google Scholar]
- Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, and Bartel DP (2016). mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife 5, 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang S-F, Doerks T, Dorner S, Bork P, Boutros M, and Izaurralde E (2007). Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 21, 2558–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan S-J, Marchand V, and Ephrussi A (2011). Drosophila Ge-1 promotes P body formation and oskar mRNA localization. PLoS ONE 6, e20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora P, Schowalter S, Wong-Deyrup S, DeGennaro M, Nasrallah MA, and Rangan P (2018). Transient transcriptional silencing alters the cell cycle to promote germline stem cell differentiation in Drosophila. Dev. Biol. 434, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes A, and Lehmann R (1998). Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125, 679–690. [DOI] [PubMed] [Google Scholar]
- Fu Z, Geng C, Wang H, Yang Z, Weng C, Li H, Deng L, Liu L, Liu N, Ni J, and Xie T (2015). Twin promotes the maintenance and differentiation of germline stem cell lineage through modulation of multiple pathways. Cell Rep. 13, 1366–1379. [DOI] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, and Wilusz CJ (2007). The highways and byways of mRNA decay. Nat. Rev. Mol. Cell Biol 8, 113–126. [DOI] [PubMed] [Google Scholar]
- Gavis ER, and Lehmann R (1992). Localization of nanos RNA controls embryonic polarity. Cell 71, 301–313. [DOI] [PubMed] [Google Scholar]
- Gavis ER, and Lehmann R (1994). Translational regulation of nanos by RNA localization. Nature 369, 315–318. [DOI] [PubMed] [Google Scholar]
- Gilboa L, and Lehmann R (2004). How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development 131, 4895–4905. [DOI] [PubMed] [Google Scholar]
- Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, and Nakamura A (2008). Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature 451, 730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RE, Pargett M, Sutcliffe C, Umulis D, and Ashe HL (2011). Brat promotes stem cell differentiation via control of a bistable switch that restricts BMP signaling. Dev. Cell 20, 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MM, Li X-Y, Kaplan T, Botchan MR, and Eisen MB (2011). Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 7, e1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Stewart K, Kinnell HL, Anderson RA, and Childs AJ (2013). A developmental stage-specific switch from DAZL to BOLL occurs during fetal oogenesis in humans, but not mice. PLoS ONE 8, e73996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins HT, Baker-Wilding R, and Edwards TA (2009). Structure and RNA binding of the mouse Pumilio-2 Puf domain. J. Struct. Biol 167, 271–276. [DOI] [PubMed] [Google Scholar]
- Johnstone O, and Lasko P (2001). Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu. Rev. Genet 35, 365–406. [DOI] [PubMed] [Google Scholar]
- Joly W, Chartier A, Rojas-Rios P, Busseau I, and Simonelig M (2013). The CCR4 deadenylase acts with Nanos and Pumilio in the fine-tuning of Mei-P26 expression to promote germline stem cell self-renewal. Stem Cell Reports 1, 411–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova LY, Habara Y, Lee TH, and Wharton RP (2007). Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development 134, 1519–1527. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, and Salzberg SL (2015a). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, Pai CI, Sato K, Person MD, Nakamura A, and Macdonald PM (2015b). Region-specific activation of oskar mRNA translation by inhibition of Bruno-mediated repression. PLoS Genet 11, e1004992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, and Macdonald PM (1995). Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81, 403–412. [DOI] [PubMed] [Google Scholar]
- Kress C, Gautier-Courteille C, Osborne HB, Babinet C, and Paillard L (2007). Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol. Cell. Biol 27, 1146–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, and Orr-Weaver TL (2014). Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 7, 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko P (2012). mRNA localization and translational control in Drosophila oogenesis. Cold Spring Harb. Perspect. Biol. 4, a012294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laver JD, Marsolais AJ, Smibert CA, and Lipshitz HD (2015). Regulation and function of maternal gene products during the maternal-to-zygotic transition in Drosophila. Curr. Top. Dev. Biol 113, 43–84. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, and Giraldez AJ (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annu. Rev. Cell Dev. Biol 30, 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Minor NT, Park JK, McKearin DM, and Maines JZ (2009). Bam and Bgcn antagonize Nanos-dependent germ-line stem cell maintenance. Proc. Natl. Acad. Sci. USA 106, 9304–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H-L, Nien C-Y, Liu H-Y, Metzstein MM, Kirov N, and Rushlow C (2008). The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Lin H, and Spradling AC (1993). Germline stem cell division and egg chamber development in transplanted Drosophila germaria. Dev. Biol 159, 140–152. [DOI] [PubMed] [Google Scholar]
- Mak W, Fang C, Holden T, Dratver MB, and Lin H (2016). An important role of Pumilio 1 in regulating the development of the mammalian female germline. Biol. Reprod 94, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis J, and Spradling A (1995). Identification and behavior of epithelial stem cells in the Drosophila ovary. Development 121, 3797–3807. [DOI] [PubMed] [Google Scholar]
- Martinho RG, Kunwar PS, Casanova J, and Lehmann R (2004). A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr. Biol 14, 159–165. [DOI] [PubMed] [Google Scholar]
- McKearin D, and Ohlstein B (1995). A role for the Drosophila bag-of-marbles protein in the differentiation of cystoblasts from germline stem cells. Development 121, 2937–2947. [DOI] [PubMed] [Google Scholar]
- McKearin DM, and Spradling AC (1990). bag-of-marbles: a Drosophila gene required to initiate both male and female gametogenesis. Genes Dev. 4 (12B), 2242–2251. [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, and Seydoux G (2008). 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr. Biol 18, 1476–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, and Wahle E (2004). Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol 39, 197–216. [DOI] [PubMed] [Google Scholar]
- Mootz D, Ho DM, and Hunter CP (2004). The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development 131, 3263–3272. [DOI] [PubMed] [Google Scholar]
- Morris JZ, Hong A, Lilly MA, and Lehmann R (2005). twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development 132, 1165–1174. [DOI] [PubMed] [Google Scholar]
- Muraro NI, Weston AJ, Gerber AP, Luschnig S, Moffat KG, and Baines RA (2008). Pumilio binds para mRNA and requires Nanos and Brat to regulate sodium current in Drosophila motoneurons. J. Neurosci 28, 2099–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, and Wharton RP (1995). Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80, 747–756. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Mukai M, Kobayashi S, and Lasko PF (1996). Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science 274, 2075–2079. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, and Kobayashi S (2001). Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128, 3233–3242. [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, and Hanyu-Nakamura K (2004). Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev. Cell 6, 69–78. [DOI] [PubMed] [Google Scholar]
- Nien C-Y, Liang H-L, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, and Rushlow C (2011). Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 7, e1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesnicky EC, Bhogal B, and Gavis ER (2012). Combinatorial use of translational co-factors for cell type-specific regulation during neuronal morphogenesis in Drosophila. Dev. Biol 365, 208–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozgur S, Basquin J, Kamenska A, Filipowicz W, Standart N, and Conti E (2015). Structure of a human 4E-T/DDX6/CNOT1 complex reveals the different interplay of DDX6-binding proteins with the CCR4-NOT complex. Cell Rep. 13, 703–711. [DOI] [PubMed] [Google Scholar]
- Rangan P, DeGennaro M, and Lehmann R (2008). Regulating gene expression in the Drosophila germ line. Cold Spring Harb. Symp. Quant. Biol 73, 1–8. [DOI] [PubMed] [Google Scholar]
- Rangan P, DeGennaro M, Jaime-Bustamante K, Coux R-X, Martinho RG, and Lehmann R (2009). Temporal and spatial control of germ-plasm RNAs. Curr. Biol 19, 72–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, and Lasko P (2011). Translational control in oocyte development. Cold Spring Harb. Perspect. Biol 3, a002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario R, Childs AJ, and Anderson RA (2017). RNA-binding proteins in human oogenesis: Balancing differentiation and self-renewal in the female fetal germline. Stem Cell Res. (Amst.) 21, 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs A, and Wahle E (1993). Poly(A) tail metabolism and function in eucaryotes. J. Biol. Chem. 268, 22955–22958. [PubMed] [Google Scholar]
- Sallés FJ, and Strickland S (1999). Analysis of poly(A) tail lengths by PCR: the PAT assay. Methods Mol. Biol 118, 441–448. [DOI] [PubMed] [Google Scholar]
- Schüpbach T, and Wieschaus E (1991). Female sterile mutations on the second chromosome of Drosophila melanogaster. II. Mutations blocking oogenesis or altering egg morphology. Genetics 129, 1119–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serano TL, Cheung H-K, Frank LH, and Cohen RS (1994). P element transformation vectors for studying Drosophila melanogaster oogenesis and early embryogenesis. Gene 138, 181–186. [DOI] [PubMed] [Google Scholar]
- Seydoux G, and Braun RE (2006). Pathway to totipotency: lessons from germ cells. Cell 127, 891–904. [DOI] [PubMed] [Google Scholar]
- Slaidina M, and Lehmann R (2014). Translational control in germline stem cell development. J. Cell Biol 207, 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snee M, Benz D, Jen J, and Macdonald PM (2014). Two distinct domains of Bruno bind specifically to the oskarmRNA. RNA Biol. 5, 49–57. [PubMed] [Google Scholar]
- Sonoda J, and Wharton RP (1999). Recruitment of Nanos to hunchback mRNA by Pumilio. Genes Dev. 13, 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda J, and Wharton RP (2001). Drosophila Brain Tumor is a translational repressor. Genes Dev. 15, 762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov DS, and Jurecic R (2003). Mouse Pum1 and Pum2 genes, members of the Pumilio family of RNA-binding proteins, show differential expression in fetal and adult hematopoietic stem cells and progenitors. Blood Cells Mol. Dis 30, 55–69. [DOI] [PubMed] [Google Scholar]
- Spradling AC (1993). Developmental Genetics of Oogenesis, Volume 1 (Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Spradling AC, de Cuevas M, Drummond-Barbosa D, Keyes L, Lilly M, Pepling M, and Xie T (1997). The Drosophila germarium: stem cells, germ line cysts, and oocytes. Cold Spring Harb. Symp. Quant. Biol 62, 25–34. [PubMed] [Google Scholar]
- Sugimura I, and Lilly MA (2006). Bruno inhibits the expression of mitotic cyclins during the prophase I meiotic arrest of Drosophila oocytes. Dev. Cell 10, 127–135. [DOI] [PubMed] [Google Scholar]
- Tadros W, and Lipshitz HD (2009). The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. [DOI] [PubMed] [Google Scholar]
- Temme C, Zhang L, Kremmer E, Ihling C, Chartier A, Sinz A, Simonelig M, and Wahle E (2010). Subunits of the Drosophila CCR4-NOT complex and their roles in mRNA deadenylation. RNA 16, 1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme C, Simonelig M, and Wahle E (2014). Deadenylation of mRNA by the CCR4-NOT complex in Drosophila: molecular and developmental aspects. Front. Genet 5, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timinszky G, Bortfeld M, and Ladurner AG (2008). Repression of RNA polymerase II transcription by a Drosophila oligopeptide. PLoS ONE 3, e2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Lionnet T, Shroff H, and Lehmann R (2017). mRNA quantification using single-molecule FISH in Drosophila embryos. Nat. Protoc 12, 1326–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova IA, Tahoe NM, Fan D, Larsson O, Rattenbacher B, Sternjohn JR, Vasdewani J, Karypis G, Reilly CS, Bitterman PB, and Bohjanen PR (2008). Conserved GU-rich elements mediate mRNA decay by binding to CUG-binding protein 1. Mol. Cell 29, 263–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagnon JL, Mahaffey CL, Sun W, Yang Y, Chao HT, and Frankel WN (2011). Etiology of a genetically complex seizure disorder in Celf4 mutant mice. Genes Brain Behav. 10, 765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, and Lin H (2005). The division of Drosophila germline stem cells and their precursors requires a specific cyclin. Curr. Biol 15, 328–333. [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, and Hall TM (2001). Crystal structure of a Pumilio homology domain. Mol. Cell 7, 855–865. [DOI] [PubMed] [Google Scholar]
- Webster PJ, Liang L, Berg CA, Lasko P, and Macdonald PM (1997). Translational repressor bruno plays multiple roles in development and is widely conserved. Genes Dev. 11, 2510–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidmann CA, Qiu C, Arvola RM, Lou T-F, Killingsworth J, Campbell ZT, Tanaka Hall TM, and Goldstrohm AC (2016). Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio. eLife 5, 1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton RP, and Struhl G (1991). RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell 67, 955–967. [DOI] [PubMed] [Google Scholar]
- Wharton RP, Sonoda J, Lee T, Patterson M, and Murata Y (1998). The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1, 863–872. [DOI] [PubMed] [Google Scholar]
- Wilkie GS, Dickson KS, and Gray NK (2003). Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci 28, 182–188. [DOI] [PubMed] [Google Scholar]
- Xie T, and Spradling AC (1998). decapentaplegic is essential for the maintenance and division of germline stem cells in the Drosophila ovary. Cell 94, 251–260. [DOI] [PubMed] [Google Scholar]
- Xin T, Xuan T, Tan J, Li M, Zhao G, and Li M (2013). The Drosophila putative histone acetyltransferase Enok maintains female germline stem cells through regulating Bruno and the niche. Dev. Biol 384, 1–12. [DOI] [PubMed] [Google Scholar]
- Zhang K, and Smith GW (2015). Maternal control of early embryogenesis in mammals. Reprod. Fertil. Dev 27, 880–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Chen D, Xia J, Han W, Cui X, Neuenkirchen N, Hermes G, Sestan N, and Lin H (2017). Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev. 31, 1354–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.