Abstract
The T cell compartment is phenotypically and functionally heterogeneous; subsets of naive and memory cells have different functional properties, and also differ with respect to homeostatic potential and the ability to persist in vivo. Human stem cell memory T (TSCM) cells, which possess superior immune reconstitution and antitumor response capabilities, can be identified by polychromatic flow cytometry on the basis of the simultaneous expression of several naive markers together with the memory marker CD95. We describe here a protocol based on the minimum set of markers required for optimal identification of human and nonhuman primate (NHP) TSCM cells with commonly available flow cytometers. By using flow sorters, TSCM cells can thereby be isolated efficiently at high yield and purity. With the use of the 5.5-h isolation procedure, depending on the number of cells needed, the sorting procedure can last for 2–15 h. We also indicate multiple strategies for their efficient expansion in vitro at consistent numbers for functional characterization or adoptive transfer experiments.
INTRODUCTION
The T cell compartment is highly heterogeneous, and dozens of phenotypically and functionally distinct subsets can be identified in the peripheral blood by polychromatic flow cytometry1,2. Conventionally, memory cells have been divided into central memory (TCM) and effector memory (TEM) subsets according to the expression of C-C chemokine receptor 7 (CCR7) and CD62L (L-selectin); these cells home to secondary lymphoid or peripheral tissues, respectively3. More recently, a subset of memory T cells with stem cell–like properties (TSCM) has been identified4. These cells are the least differentiated of all distinct memory populations, expressing multiple naive markers and the memory antigen CD95 (Fig. 1). Functionally, TSCM cells can generate multiple memory T cell populations, and they possess an enhanced self-renewal capacity4. In addition, they are endowed with superior immune reconstitution potential in immunodeficient hosts and mediate superior antitumor immunity in a humanized mouse model4. Thus, the translational applications of TSCM cells for the development of new vaccines or adoptive T cell therapies are considerable. In mice, TSCM cells specific to the Y antigen have been identified in the setting of allogeneic bone marrow transplantation5. Furthermore, cells with TSCM properties could be induced in vitro by the activation of the Wnt/β-catenin pathway in naive T cells stimulated through the T cell receptor (TCR)6. However, mouse TSCM cells specific for viral or tumor antigens have not been described so far, and the identification of a mouse TSCM population with relevance in physiology and pathology remains elusive. The NHP is probably the best animal model for human immunity; as far as T cells are concerned, the immune systems are phenotypically similar, and with a few exceptions the same human antibody reagents can be used to delineate T cell subsets in rhesus macaques7. Indeed, a population of naive-like cells expressing CD95 can be identified in rhesus macaques (Fig. 1), thus allowing the study of TSCM cells in vivo.
Figure 1|.
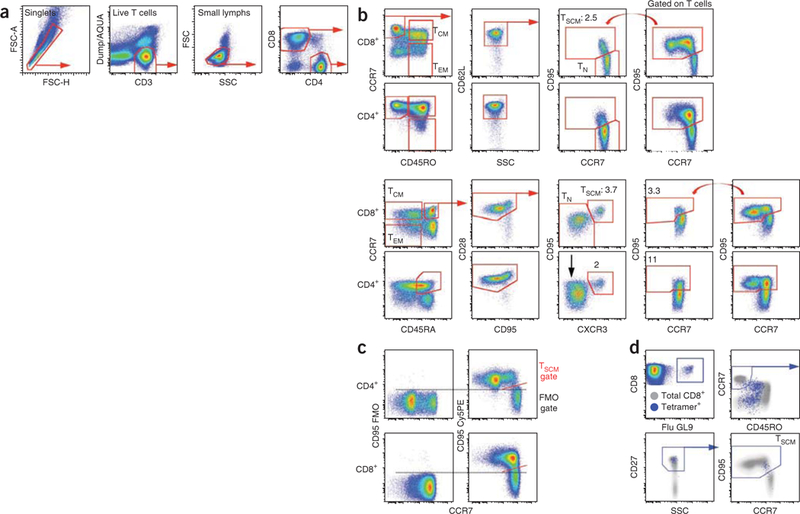
Gating strategy for the identification of human and rhesus TSCM cells. (a,b) Human and NHP PBMCs were stained as indicated in Table 1. In this example, both panels included anti-CD4 conjugated to Qdot 585 in addition to anti-CD8 conjugated to Pacific Blue in order to allow the simultaneous identification of CD4+ and CD8+ T cells. These T cells were identified by first gating on singlets (FSC-H versus FSC-A), live CD3+ T cells (CD3 versus Dump/AQUA ) and lymphocytes (SSC versus FSC). Naive-like T cells were defined as CD45RO–CCR7+ CD62L+ in humans and as CD45RA+ CCR7+ CD28+ in rhesus macaques. In these gated populations, TSCM cells express CD95, whereas TN cells are CD95–. In NHP CD8+ T cells, CXCR3 is coexpressed with CD95 and thus helps identify CD8+ TSCM cells but not CD4 + TSCM cells, as not all CD95+ TSCM in naive-like CD4+ cells express CXCR3+ (black arrow). Red straight arrows indicate the sequential gating strategy. The curved red arrows indicate the gate to be copied on gated ‘naive-like’ cells. (c) CD95 FMO control in human T cells. Dashed bars indicate the threshold for positivity for CD95 expression, whereas the diagonal red bar indicates the TSCM gate. (d) Identification of human antigen–specific TSCM cells. Human PBMCs were stained as indicated in Table 1. CD8+ T cells were identified as in a. T cells specific for the influenza matrix protein58–66 (Flu GL9) were identified using an MHC class I tetramer (blue dots overlaid on total CD8+ T cells in gray). In this case, frozen PBMCs were used, and CD27 replaced CD62L for the identification of TSCM cells. Blue straight arrows indicate the sequential gating strategy. FSC, forward scatter; SSC, side scatter.
Development of the protocol
Human and NHP TSCM cells are relatively rare, comprising about 2–4% of the total CD4+ or CD8+ T cells in the blood. There is no unique marker to phenotypically identify these cells; the expression of any given marker is shared with naive or memory cells. This presents a challenge for the isolation of these cells at high yield and high purity for further functional analyses. By polychromatic flow cytometry, we originally characterized human TSCM cells as simultaneously expressing multiple naive markers including CD45RA, CCR7, CD62L, CD27, CD28, CD127 (IL-7Rα) and CD11adim and lacking CD45RO; unlike naive T cells (TN), they also express the memory antigen CD95 (ref. 4). However, the simultaneous analysis of all of these nine markers is not critical for the identification of human TSCM cells (Fig. 2); seven- or eight-color panels (Table 1) accurately identify and allow for sorting of human and NHP TSCM cells using commonly available flow cytometers. All antibody and fluorochrome combinations described are commercially available.
Figure 2|.
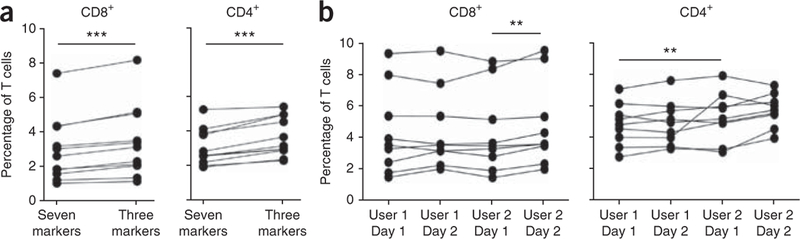
Reproducibility and robustness of three versus seven marker-based TN cell definitions for TSCM identification. (a) Naive-like T cells were defined as CD45RO–CCR7+ CD45RA+ CD62L+ CD27+ CD11adim CD127 (seven markers) or as CD45RO–CCR7+ CD62L+ (three markers). TSCM cells were subsequently identified as CD95+. The proportion among CD8+ and CD4+ is shown (n = 11). (b) Interuser and interexperiment variability of the strategy for the identification of human TSCM cells. Data were analyzed by the same user to minimize subjectivity in the gating procedure, **P < 0.01; ***P < 0.001.
TABLE 1|.
Panels and reagents for TSCM cell identification.
| Panel no. | 1 | 2 | 3 |
|---|---|---|---|
| Purpose | Human TSCM (bulk or antigen-specific) | Human TSCM (bulk or antigen-specific) | NHP TSCM |
| Source of cells | Fresh | Cryopreserved | Fresh or cryopreserved |
| AmCyan | AQUA LIVE/DEAD | AQUA LIVE/DEAD | AQUA LIVE/DEAD |
| APC-H7 or APC-Cy7 | CD3 | CD3 | CD3 |
| Pacific Blue | CD4 or CD8 | CD4 or CD8 | CD4 or CD8 |
| APC | CD45RO | CD45RO | CD95 |
| FITC | CCR7 | CCR7 | CCR7 |
| PE-Cy7 | CD62L | CD27 | CD45RA |
| PE-Cy5 | CD95 | CD95 | |
| ECD | CD28 | ||
| PE | MHC class I tetramer CD58 CD122 |
MHC class I tetramer CD58 CD122 |
CXCR3 |
For human cells (Table 1), the panels include the following:(i)a ‘dump’ channel to exclude dead cells with a viability dye; (ii)antibodies to CD3, CD8 and CD4 to define the lineage of interest; (iii)antibodies to CD45RO, CCR7, and either CD62L or a different marker expressed by naive cells (e.g., CD27, CD28 or CD45RA) to identify naive-like cells and subsets of memory cells8; (iv) and anti-CD95 to discriminate CD95– TN from CD95+ TSCM cells. These panels leave the phycoerythrin (PE) channel open to accommodate an additional antibody of interest, a major histocompatibility complex (MHC) class I tetramer for the identification of antigen-specific CD8+ T cells (Fig. 1d), or anti-CD58 or anti-CD122 (Fig. 3). CD58, the lymphocyte function–associated antigen 3, belongs to the immunoglobulin superfamily and mediates the interaction between lymphocytes and CD2, expressed on a variety of cell types including the endothelium. CD122 is the β-chain of the IL-2/IL-15 receptor complex, which forms a low-affinity receptor together with the γ-chain. Both CD58 and CD122 are found at higher levels on conventional memory cells and TSCM cells than on TN cells. This differential expression can be utilized to better identify TSCM cells (Fig. 1a). In our hands, CD95 is preferred to CD58 and CD122, as CD95-specific antibodies are available through multiple vendors and as conjugates to different fluorochromes, thus allowing the design of complex and interchangeable multicolor panels. Most notably, staining for CD95 provides a better separation of TSCM cells from TN cells by flow cytometry compared with CD58 and CD122. However, the vast majority of CD95+ TSCM cells also coexpress these markers. As an obvious consequence, TSCM cells identified on the basis of the increased expression of CD58 or CD122 also have increased expression of the TSCM core phenotypic marker CD95 (Fig. 3b), thus indicating that CD58 and CD122 are valid markers for the identification of true TSCM cells. A similar combination of antibodies can be used to identify NHP TSCM cells, e.g., CD45RA, CCR7, CD28, CD95 and CXCR3 (Table 1).
Figure 3|.
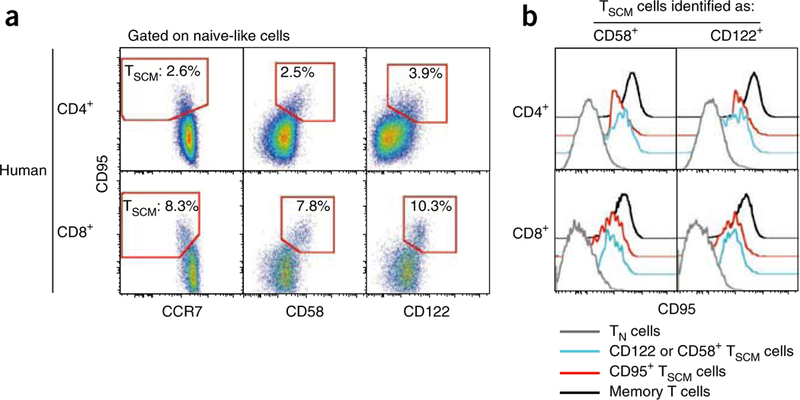
Co-staining strategies to improve the identification and isolation of human TSCM cells. (a) Human CD4+ and CD8+ naive-like cells were identified as described previously4. The expression of CCR7 (left), CD58 (center) or CD122 (right) versus CD95 in human CD4+ and CD8+ T cells is shown. The gate identifies TSCM cells as depicted in Figure 1b. Numbers indicate the percentage of cells identified by the gates. (b) Expression of CD95 by CD4+ and CD8+ TSCM cells identified by the increased expression of CD58 and CD122. T cell subsets are defined as in Figure 1. Memory T cells are defined as positive for CD45RO.
Experimental design
As described, human TSCM cells can be identified as expressing multiple markers of TN cells and also CD95, which is preferentially found on the surface of memory cells. Three markers, i.e., CD45RO, CCR7 and CD62L, are sufficient for the identification of TN-like cells (defined as CD45RO–CCR7+ CD62L+ ) and for the exclusion of memory T cell contaminants of unknown function, as can occur when only two markers are used to identify TN cells8. Even though statistically significant differences are found, the proportion of CD8+ and CD4+ TSCM cells changes only minimally when TN-like cells are defined on the basis of seven (mean ± s.e.m, CD8+ : 2.95 ± 0.56; CD4+ : 2.81 ± versus three markers (CD8+ : 3.40±0.62; CD4+ : 3.59 ± 0.45; Fig. 2a). These differences can be ascribed, at least in part, to the fact that transition from CD45RA to CD45RO expression is not as complete as for other memory-defining antigens, and thus some cells with intermediate expression of CD45RA but negative for CD45RO might be included in the final population. If a flow cytometer with a limited number of detectors is used (e.g., eight detectors), we suggest using anti-CD45RO instead of anti-CD45RA, as it allows the exclusion of CD45RO+ /CD45RA+ activated cells. However, if more detectors are available, the additional inclusion of anti-CD45RA helps to better delineates the human TSCM cells. When cryopreserved cells are used, CD62L staining is not reliable because expression is lost with the freeze-thaw procedure1. In this case, we recommend including a different marker for the identification of naive-like cells, such as CD27, CD28 or CD127 (IL-7Rα), as depicted in Figure 1d and Table 1.
Unfortunately, the gating procedure in flow cytometry is highly user dependent. Moreover, given the dim expression of CD95 in TSCM cells compared with conventional memory cells (Fig. 3b), the separation of TSCM cells from CD95 − TN cells can be difficult. To minimize this problem and to identify TSCM cells consistently, a standardized gating strategy has been developed (Fig. 1a,b). We noted that TSCM cells (especially in humans) have slightly lower levels of CCR7 compared with TN cells. This property allows better delineation of TN and TSCM cells when CCR7 is plotted against CD95 expression, and it can be exploited by positioning the sorting gate on a diagonal alongside the TSCM population (Fig. 1a). Indeed, clear-cut separation of positive and negative expression of CD95 can be visualized when using this approach. The gate identifying CD95+ cells can then be copied in the same bivariate plot after gating for multiple naive markers, i.e., CD45RO, CCR7 and CD62L/CD27 for human cells and CD45RA, CCR7 and CD28 for rhesus macaques (Fig. 1a,b). Negligible differences can be observed between experiments performed by different users and on different days by using the strategy we describe (Fig. 2b).
Alternatively, the inclusion of more markers in the panel can improve separation of the TSCM cell population; for example, higher levels of CD58 and CD122 are found on TSCM cells compared with TN cells (Fig. 3a). CD58- or CD122-specific antibodies are available conjugated to PE, and can be included in both panels 1 and 2 (Table 1). For general advice on the development of more complex panels, refer to published guidelines9.
Anti-CD95 clone DX2 antibody (like other CD95-specific antibodies) is capable of inducing apoptosis in target cells. Despite quiescent lymphocytes being generally resistant to CD95-induced apoptosis10,11, we suggest including sodium azide (NaN3) in the staining buffer and keeping the sample cold during long FACS sorting procedures to minimize cellular metabolism.
When quantifying TSCM cells in human patient samples, peripheral blood lymphocytes from a healthy donor should ideally be included as a control to help set gates; we have found that T cells from patients with different pathologies or receiving different therapies may exhibit substantially altered representation of the subsets, making it difficult to judge delineation gates ‘by eye’ (E.L. and M.R., unpublished observation).
We use a similar combination of antibodies to track TSCM cells in rhesus macaques, based on the expression of CD45RA, CCR7, CD28 and CD95 (note that no anti-rhesus CD45RO reagents are currently available). In rhesus macaques, CD95 expression in the TSCM versus TN populations is not as distinct as in humans (Fig. 1b), making isolation of TSCM cells for sorting more difficult. The addition of anti-CXCR3 to the panel can improve the identification of TSCM cells in CD8+ but not the CD4+ T cell lineage, as all NHP CD8+ but not CD4+ TSCM cells express CXCR3 (Fig. 1b). Indeed, CXCR3 can replace CD95 for the identification and isolation of CD8+ TSCM cells in NHP (Fig. 1b).
If flow cytometry sorting is planned, cell staining can be preceded by negative magnetic isolation of the target lineage (CD4+ or CD8+ ) to shorten the sorting time. Sorted cells can be subsequently expanded by stimulating them with a combination of IL-7, which preferentially expands TN, TSCM and TCM cells4, IL-15, which selectively expands memory cells12, or CD3/CD2/CD28-specific antibody-coated beads. In contrast to the latter, IL-7- and IL-15–mediated expansions partly maintain the initial phenotype of the population4,13,14 rather than inducing excessive proliferation, acquisition of effector function and in vitro–induced senescence15.
To identify CD95+ cells, fluorescence minus one (FMO) controls (i.e., samples stained with all fluorochromes except the one of interest1) are not fully informative in this particular example, as some TN cells can express low levels of CD95 (Fig. 1c). These cells are not included in the TSCM cell gate because of the impossibilityto clearly separate CD95 dull and CD95– TN cells. Clear-cut separation of CD95 expression can be easily visualized by plotting CD95 versus CCR7, as described above. However, FMO controls can still be important to guide the gating procedure and to reveal compensation artifacts.
In this manuscript, flow cytometer setup is not discussed, as it has been thoroughly described in detail elsewhere16. In particular, the procedure described by Perfetto et al.16 defines methods for setting detector photomultiplier (PMT) voltages in advance using quality control reagents, such as prestained beads, thereby ensuring the greatest signal-to-background separation. By using such procedures, no changes in PMT settings are needed before the initiation of the experiment, and thus the time spent at the machine is limited to the acquisition of the sample. Importantly, quality control of laser alignment, laser delays and PMT transmission should be checked before every experiment by running Rainbow beads, as described16. To minimize the time spent at the flow cytometer on the day of the experiment, we recommend that the experiment template be set up in advance. A rigorous quality control (QC) program for instrument alignment and settings is critical for reproducible evaluation of TSCM cells using polychromatic flow cytometry.
The MHC class I tetramer used here was synthesized and conjugated in our laboratory. The antibodies are commercially available from several companies, as listed in the Reagents section. All antibodies and tetramers should be carefully titrated before use, whether they are obtained commercially or synthesized in the laboratory. The titer giving the best separation over the background should be chosen. However, in some cases, a lower concentration of the antibody can be used to minimize ‘spreading error’ to other fluorochromes (i.e., after compensation)17. Detailed theoretical considerations and practical procedures regarding antibody-antigen binding for flow cytometric analyses have been discussed elsewhere18.
Applications of the protocol
The protocol described here has been optimized for the identification, isolation and in vitro expansion of human TSCM cells. NHP TSCM cells can be isolated with a similar panel. However, the same procedure, but with different monoclonal antibodies, can be used to identify and sort any T cell population from the human body for further studies and applications.
MATERIALS
REAGENTS
Human or NHP lymphocytes isolated from the peripheral blood or from a different site of acquisition by Ficoll gradient centrifugation according to standard techniques (refs. 12,19), which takes 2 h. If you are working with frozen cells, thaw the cells as described in ref. 12.
Human peripheral blood or NHP peripheral blood, lymph node or spleen ! CAUTION Universal precautions must be taken and experiments must be carried out in (at least) category 2 biological safety cabinets and using appropriate personal protection equipment. ! CAUTION Experiments must conform to all relevant institutional and governmental ethics regulations, and appropriate informed consent must be obtained for the proposed use of human blood.
Ficoll-Paque PLUS (GE Healthcare, cat. no. 17–1440-02)
Dulbecco’s PBS (DPBS; Life Technologies, cat. no. 14120)
Heat-inactivated FBS (Life Technologies, cat. no. 10438–026)
Penicillin-streptomycin/l-glutamine (Life Technologies, cat. no. 10378–016)
RPMI 1640–phenol red (Life Technologies, cat. no. 21870–076)
RPMI 1640–no phenol red (Life Technologies, cat. no. 00–0327DK)
HEPES (Life Technologies, cat. no. 15630–080)
Sodium azide (NaN3; Sigma-Aldrich, cat. no. S2002) ! CAUTION Avoid contact and inhalation. Wear gloves, safety glasses and a lab coat.
CD4+ T cell isolation kit II (Miltenyi Biotec, cat. no. 130–091-055)
CD8+ T cell isolation kit (Miltenyi Biotec, cat. no. 130–094-156)
CD4+ T cell isolation kit, NHP (Miltenyi Biotec, cat. no. 130–092-144)
CD8+ T cell isolation kit, NHP (Miltenyi Biotec, cat. no. 130–092-143)
Fluorescently conjugated anti-human monoclonal antibodies (all listed antibodies are whole immunoglobulin; refer to Table 1 for specific antibodies required for particular staining conditions): anti-human CD3 APC-H7 (clone SK7, IgG1 κ; BD Pharmingen, cat. no. 560176; used to stain human T cells), anti-human CD4 Brilliant Violet 421 (clone OKT4, IgG2b K; Biolegend, cat. no. 317433; used to stain human and NHP T cells), anti-human CD8 Pacific Blue (clone RPA-T8, IgG1 K; BD Pharmingen, cat. no. 558207; used to stain human and NHP T cells), anti-human CD45RO APC (clone UCHL1, IgG2a K; BD Pharmingen, cat. no. 559865; used to stain human T cells), anti-human CCR7 FITC (clone 150503, IgG2a K; BD Pharmingen, cat. no. 561271; used to stain human and NHP T cells), anti-human CD62L PE-Cy7 (clone DREG-56, IgG1 K; Biolegend, cat. no. 304822; used to stain human T cells), anti-human CD27 PE-Cy7 (clone 1A4CD27, IgG1 K; Beckman Coulter, cat. no. A54823; used to stain human T cells), anti-human CD95 PE-Cy5 (clone DX2, IgG1 K; Biolegend, cat. no. 305610; used to stain human T cells), anti-human CD3 APC-Cy7 (clone SP34–2, IgG1 λ; BD Pharmingen, cat. no. 557757; used to stain NHP T cells) anti-human CD45RA PE-Cy7 (clone L48, IgG1 K; BD Biosciences, cat. no. 337167; used to stain NHP T cells), anti-human CD28 ECD (clone CD28.2, IgG1; Beckman Coulter, cat. no. 6607111; used to stain NHP T cells), anti-human CD95 APC (clone DX2, IgG1 K; BD Pharmingen, cat. no. 558814; used to stain NHP T cells), anti-human CXCR3 PE (clone 1C6/CXCR3, IgG1 K; BD Pharmingen, cat. no. 550633; used to stain NHP T cells), anti-human CD58 PE (clone L306.4, IgG2a K; BD Biosciences, cat. no. 340295; used to stain human T cells), anti-human CD122 PE (clone Mik-B3, IgG1 K; BD Pharmingen, cat. no. 554525; used to stain human T cells) ▲CRITICAL Each lot of antibody must be titrated before use.
MHC class I tetramers from the NIH tetramer core facility (http://tetramer.yerkes.emory.edu/) or produced as described in ref. 20. They are available from multiple vendors; however, we have only used those from the NIH core facility
Mouse anti-monkey CD3 antibody (Life Technologies, clone FN18, IgG1, cat. no. APS0301), used to expand NHP T cells
Mouse anti-human CD28 antibody (BD Biosciences, clone CD28.2, IgG1 K, cat. no. 555725), used to expand NHP T cells
LIVE/DEAD AQUA fluorescent-reactive dye (Life Technologies, cat. no. L34957) ! CAUTION AQUA may be an irritant for the eyes. Wear gloves and safety glasses. ▲ CRITICAL Each lot of dye must be titrated before use.
BD CompBeads anti-mouse IgK (BD Biosciences, cat. no. 552843)
SPHERO COMPtrol goat anti-mouse Ig (heavy + light) particles (Spherotech, cat. no. CMIgP-50–5H)
R-NH2 Beads (SMPLX Amine active beads; Bangs Laboratories; for proprietary reasons, this reagent can only be ordered by phone, and is not available in the Bangs Laboratories catalog)
Formaldehyde, 20% (vol/vol) aqueous (Tousimis, cat. no. 1008B)
-
Carboxy-fluorescein diacetate succinimidyl ester (CFSE; Life Technologies, cat. no. C1165)! ! CAUTION CFSE may be an irritant for the eyes.
Wear gloves and safety glasses.
Recombinant human IL-7 (Peptrotech, cat. no. 200–07)
Recombinant human IL-15 (Peptrotech, cat. no. 200–15)
Human T cell activation and expansion kit (Miltenyi Biotec, cat. no. 130–091-441)
Ethidium bromide (EB; Life Technologies, cat. no. E-1305)
! CAUTION Avoid contact and ingestion. Wear gloves, safety glasses and a lab coat.
Acridine Orange (AO; Life Technologies, cat. no. A-1301) ! CAUTION Avoid contact and ingestion. Wear gloves, safety glasses and a lab coat.
AutoMACS running buffer (Miltenyi Biotec, cat. no. 130–091-221)
Ethanol (Sigma-Aldrich, cat. no. 676829)
Dimethyl sulfoxide (DMSO; Sigma-Aldrich, cat. no. D8418)
EQUIPMENT
Polystyrene round-bottom tubes, 5 ml (BD Falcon, cat. no. 352052)
Conical tubes, 15 ml (BD Falcon, cat. no. 352097)
Conical tubes, 50 ml (BD Falcon, cat. no. 352098)
Microcentrifuge tubes, 1.5 ml (Eppendorf, cat. no. 022363204)
QuadroMACS starting kit (Miltenyi Biotec, cat. no. 130–091-051)
Miltenyi LS columns (Miltenyi Biotec, cat. no. 130–042-401)
Flow cytometer or cell sorter equipped with a violet, a blue and a red laser, capable of collecting eight different fluorescences
Tissue culture six-well plates (Corning, cat. no. 3516)
Tissue culture 24-well plates (Corning, cat. no. 3524)
Tissue culture 96-well plates (Corning, cat. no. 3799)
Benchtop ultrasonic cleaner (Branson, cat. model 1510)
Cellometer automated cell counter (Nexcelom Bioscience, Vision)
Cellometer disposable counting chambers (Nexcelom Bioscience, cat. no. CHT4-PD100)
REAGENT SETUP
Complete culture medium (R10)
Prepare R10 by making up 10% (vol/vol) FBS, 1% (vol/vol) penicillin-streptomycin/l-glutamine in RPMI 1640 medium with phenol red. Prepare the medium in advance and store it at 4 °C for up to 2 weeks.
Staining buffer I
Mix 4% (vol/vol) FBS in RPMI 1640 medium with no phenol red.
Staining buffer II
Mix 4% (vol/vol) FBS and 0.02% (vol/vol) NaN3 in RPMI 1640 medium with no phenol red.
Sorting buffer
Mix 4% (vol/vol) FBS and 25 mM HEPES in RPMI 1640 medium with no phenol red. ▲ CRITICAL staining buffer I, staining buffer II and sorting buffer can be stored at 4 °C for up to 2 weeks.
MACS buffer
Mix 5 ml of 0.5 M EDTA stock (5 mM) and 2.5 g of BSA, add PBS to adjust the volume to 500 ml, and then filter-sterilize and degas the solution. Prepare the buffer in advance and store it at 4 °C for up to 6 months.
EB stock solution
Mix 3 mg ml−1 of EB in ethanol and store it in a dark bottle for 6 months at 4 °C.
AO stock solution
Mix 5 mg ml−1 of AO in ethanol and store it in a dark bottle for 6 months at 4 °C.
EB/AO working solution
Add 10 Ml of EB stock to 10 µl of AO stock and dilute it to 1 ml with PBS (final concentration of EB = 30 µg ml−1; final concentration of AO = 50 µg ml−1). Store the solution at 4 °C for up to 6 months.
AQUA viability dye
Thaw AQUA powder at 37 °C for 30 s, add 50 µl of DMSO, pipette thoroughly and store the mixture at − 20 °C for up to 3 months.
Bead medium
Mix 2% (vol/vol) FBS and 0.02% (wt/vol) NaN3 in PBS. Store the medium at 4 °C for up to 1 month.
R-NH2 AQUA CompBeads
Make a 1:5 dilution of the bead stock with bead medium (~46.2 × 106 beads per ml); take 350 µl of this mixture (16 × 106), wash it in PBS and resuspend the beads in 300 Ml of PBS. Add 100 µl of AQUA dye and incubate for 1.5 h. Wash the beads twice with bead medium and resuspend them in a 2-ml volume. Spike in an equal concentration (350 µl) of unstained amine beads. Add bead medium to obtain a final volume of 4 ml. Store the beads in a glass vial for up to 6 months at 4 °C.
CFSE stock
Thaw the powder and resuspend it in DMSO at a final concentration of 5 mM. Store the stock in aliquots at − 20 °C for up to 6 months.
Anti-monkey CD3 antibody solution (necessary to stimulate NHP T cells)
Dilute the antibody to a final concentration of 10 µg ml−1 in PBS immediately before use. Coat the plate overnight at 4 °C. Do not store the diluted antibody solution.
Formaldehyde working solution
Mix 1% (vol/vol) formaldehyde in PBS. Store it at 4 °C for up to 1 month.
PROCEDURE
Cell isolation and staining ● TIMING 2 h for magnetic separation and 1.5 h for fluorescent staining
-
1|
Determine the cell number and viability with Cellometer Vision. Add 20 Ml of EB/AO working solution to 20 µl of cell suspension and count the number of cells. Use at least 0.5 × 106 cells for simple phenotyping and 4 × 106 cells for the analysis of antigen-specific TSCM cells. If sorting is planned, start with enough cells to obtain the desired number of TSCM cells. On the basis of our experience, the yield after sorting is one TSCM cell per 250 peripheral blood mononuclear cells (PBMCs) for CD4+ T cells and one TSCM cell per 500–1,000 PBMCs for CD8 + T cells, depending on the donor. Similar numbers can be obtained for NHP TSCM cells.
▲ CRITICAL STEP If you are performing simple phenotyping or sorting small numbers of TSCM cells, we recommend that thawing and staining be performed on the same day. If fixed, samples can be run the following day. If considerable numbers (millions) of TSCM cells are needed, such as for adoptive transfer experiments, flow cytometric sorting will take many hours. Enriched cells can be left at 37 °C overnight, and surface staining can be performed the following day, before sorting. If you are analyzing mRNA expression by gene array, cells should be recovered without interruption and kept at 4 °C to avoid changes in gene expression.
-
2|
If flow cytometry sorting is planned, enrich CD4+ or CD8+ T-cell populations by negative selection according to the kit manufacturer’s instructions.
-
3|
Add PBS to the cells to remove any residual proteins.
-
4|
Centrifuge the cell suspension for 5 min at 400g at room temperature (RT; 24 °C).
-
5|
Prepare AQUA working solution in excess (15% more than the volume needed for the experiment) by diluting the stock solution in water. Vortex the solution and add PBS to obtain the desired concentration as determined by titration. Vortex the solution again.
▲ CRITICAL STEP The amounts of AQUA and of antibody needed for the experiment are determined by titration experiments performed in advance. Use 100 µl of AQUA staining solution if up to 10 × 106 cells are stained. If more cells are used, consider that a 100 × 106 cell pellet corresponds to a volume of ~100 µl. If 100 µl of staining solution is used to stain such a number of cells, the final concentration of the dye (or of the antibody) will be diluted. Therefore, on the basis of our experience, we suggest preparing a staining solution containing 3× or 4× the concentration of the reagent to obtain a final volume of ~200 µl. The staining volume should be scaled up according to the number of cells. In any case, the optimal titer of antibodies to be used in sorting experiments can be determined by a titration experiment, where for instance 1×, 2×, 4× or 8× the amount of the antibody optimal for staining 106 cells is used. Detailed theoretical considerations and practical procedures regarding antibody binding to antigen for flow cytometric analyses can be found elsewhere18.
-
6|
Remove the supernatant from pelleted cells.
-
7|
Add AQUA working solution to the cell pellet, resuspend by pipetting and incubate the mixture for 15 min at RT in the dark.
-
8|
Wash the cells by adding staining buffer I (use a volume that dilutes the staining solution by 20- to 30-fold).
-
9|
Spin the mixture for 5 min at 400g at RT. In the meantime, prepare CCR7 staining solution in excess (15% more than the volume needed for the experiment as indicated in Step 5) in staining buffer 1. If NHP cells are to be stained, prepare CXCR3 staining solution at this step in the same buffer.
▲ CRITICAL STEP CCR7 and other chemokine receptors recycle through the plasma membrane. Do not include NaN3 in the staining buffer, as it prevents the internalization of surface antigens and can produce a loss of fluorescence intensity. Refer to Step 5 for the amount of antibody needed to stain a large number of cells.
-
10|
Spin antibody staining solution in a microcentrifuge at 15,000g for 5 min at RT to remove antibody aggregates. Use the supernatant only to stain cells.
-
11|
After the cells have been pelleted (Step 10), discard the supernatant and add CCR7 staining solution, resuspend the cell pellet by pipetting and then incubate it for 20 min at 37 °C in the dark. If NHP cells are being used, perform CXCR3 staining at this stage.
▲ CRITICAL STEP Incubation at 37 °C allows CCR7 and CXCR3 to recycle through the plasma membrane and improves their detection by producing a gain of fluorescence. However, for rhesus macaques, we do not see any difference by staining for CCR7 at 37 °C versus RT.
-
12|
Add staining buffer II to wash the cells (use a volume that dilutes the staining solution by 20–30-fold) and spin the mixture for 5 min at 400g at RT. In the meantime, prepare surface staining antibody mix in excess (15% more than the volume needed for the experiment, as indicated in Step 5). Centrifuge the antibody mix as indicated in Step 10.
▲ CRITICAL STEP Prepare this mix using staining buffer II containing NaN3 to minimize cellular metabolism. Refer to Step 5 for the amount of antibody needed to stain a large number of cells.
-
13|
Remove the supernatant from pelleted cells.
-
14|
Add surface staining antibody mix to the cell pellet, resuspend it by pipetting and incubate the mixture for 20 min at RT in the dark. In the meantime, prepare compensation controls. Vortex CompBeads and aliquot 30 µl to each tube. Prepare a tube for each fluorochrome plus a tube with beads only (unstained negative control). For each type of CompBead used in the experiment, include the relative negative control.
▲ CRITICAL STEP CompBeads tend to form aggregates over time. Before use, sonicate CompBeads for 2 min.
-
15|
Add the fluorescently conjugated antibody to the tubes containing CompBeads at the same titer that is used for the staining; vortex the tubes and incubate them for 15 min at RT.
-
16|
Wash the sample and compensation controls by adding 2 ml of staining buffer II to each tube. Spin the sample for 5 min at 400g at RT.
-
17|
If you are phenotyping, resuspend the cells in formaldehyde working solution. If you are sorting, resuspend the cells in sorting buffer. Keep the cells on ice and in the dark. Resuspend the compensation tube contents in the same buffer.
▲ CRITICAL STEP If you are performing a long sort, it is preferable to resuspend the cells in RPMI 1640 medium supplemented with HEPES. Indeed, CO2-based buffers will lose pH under high sort pressures, thus reducing cell survival after sorting.
Acquisition and cell sorting ● TIMING ~2–15 h, depending on the number of samples and cells required for the experiment
-
18|
Run the compensation controls. Create the compensation matrix by following the cell sorter–specific instructions as defined in the operator’s manual, and apply to tubes if sorting is to be performed.
? TROUBLESHOOTING
-
19|
Run the sample(s).
-
20|
Use the gating strategy depicted in Figure 1.
? TROUBLESHOOTING
-
21|
By using a 70-µm nozzle, proceed with sorting the population(s) of interest. In our example, these are human TN cells (CD45RO–CCR7+ CD62L+ CD95-); human TSCM cells (CD45RO–CCR7+ CD62L+ CD95+); human TCM cells (CD45RO+ CCR7+ ); human TEM cells (CD45RO+ CCR7− ); NHP TN cells (CD45RA+ CCR7+ CD28+ CD95–); NHP TSCM cells (CD45RA+ CCR7+ CD28+ CD95+ ); NHP TCM cells (CD45RA–CCR7+ ); and NHP TEM cells (CD45RA–CCR7–). Sort the cells into a 5-ml conical tube or a 1.5-ml microcen-trifuge tube containing R10 complete medium, if cell culture is planned afterward. Other buffers can be used for different applications. In our experience, 250,000 TSCM cells per hour are obtained by flow cytometry sorting.
▲ CRITICAL STEP Keep the sample and sorted cells chilled in order to minimize cellular metabolism. However, to avoid cell nonresponsiveness, do not chill the sample if a short stimulation is planned.
-
22|
Check the purity of sorted subsets (in general it should be > 95%).
? TROUBLESHOOTING
(Optional) T cell expansion in vitro ● TIMING 7–14 d
-
23|
If you are stimulating NHP cells, coat the plate with anti-monkey CD3 antibody solution overnight at 4 °C (see Reagent Setup). Remove the antibody solution and wash it three times with cold PBS before adding NHP T cell suspension.
-
24|
Wash the cells in R10 if you are proceeding directly to cell culture and stimulation. If you are performing CFSE staining to track cell proliferation, wash with PBS to remove any traces of proteins.
-
25|
Pellet the cells by centrifuging for 5 min at 400g at RT. If you are not performing CFSE staining, proceed directly to Step 33.
-
26|
If you are performing CFSE staining, proceed with the remainder of the procedure. First prepare the CFSE working solution by adding 2 µl of the stock to 1 ml of PBS (final concentration = 10 µM). Prewarm the solution to 37 °C before adding it to the cell pellet.
-
27|
Discard the supernatant from the cell pellet.
-
28|
Add the appropriate volume of CFSE to achieve ~107 cells per ml. Vortex the mixture.
-
29|
Incubate the mixture for 7 min in a 37 °C water bath.
-
30|
Add 1–2 ml of cold FBS to stop the reaction. Vortex and top up with R10.
-
31|
Centrifuge the tubes for 5 min at 400g at RT.
-
32|
Discard the supernatant and resuspend it in R10 at a density of 2.5 × 105 cells per ml.
-
33|
Culture the cells in the presence of the appropriate stimuli. Leave some extra wells with unstimulated CFSE-stained cells to be used as a compensation control at the time of analysis. Unstained PBMCs will provide the appropriate negative control. If desired, collect the cells and stain for surface antigens as indicated in Steps 3–17.
▲ CRITICAL STEP Human T cell subsets can be efficiently expanded with CD3/CD2/CD28-specific beads, IL-7 or IL-15. NHP T cell subsets can be expanded by stimulating with plate-bound anti-CD3 and soluble anti-CD28 (final concentration = 1 µg ml−1). Moreover, NHP CD8+ T cell subsets, with the exception of TN cells, can be expanded in the presence of human IL-15. CD3/CD2/CD28-specific beads should be used at a 1:2 bead-to-cell ratio to ensure optimal stimulation. IL-7 and IL-15 can both be used at a concentration of 25 ng ml−1. However, we recommend that antibody and cytokine concentrations be optimized according to the experimental need.
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
TABLE 2|.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| Reagent Setup | Lymphocytes clump after thawing | Excessive cell death | Include DNase in thawing medium. If aggregates are still seen, filter sample over a 70-µm strainer |
| 18 | CompBeads aggregates | Sonication was not effective | Increase sonication time to 10 min |
| 20 | Poor CCR7 staining | Presence of NaN3 in the buffer | Use buffer without NaN3 |
| Staining performed at RT | Stain for CCR7 at 37 °C for 20 min in a separate step | ||
| Antibody aggregates are seen after staining | The 2-min antibody mix centrifugation was not effective | Spin the antibody mix for 5–10 min before use | |
| 22 | Low purity of sorted subsets | Poor separation of antigen expression because of very high initial cell number | Increase the antibody concentration in the mix |
| Poor separation of antigens because of titration issues, fluorochrome spreading, and so on | Carefully test the panel before use. Include FMO controls to check if compensation is correct and reveal spreading errors | ||
| 33 | CFSE dilution is not observed despite an increase in cell numbers in culture | CFSE aliquot has expired or has been frozen/thawed multiple times | Use a new batch of CFSE and test it on fresh PBMCs stimulated with anti-CD3/CD2/CD28 beads for 4 d before use |
| Sorted cells do not expand after stimulation | Expired anti-CD3/CD2/CD28 beads or cytokines | Use a new batch of anti-CD3/CD2/CD28 beads or cytokines | |
| Cytokine concentration is too low | Titrate cytokines to optimize concentration |
● TIMING
Steps 1–17, cell isolation and staining: 3.5 h; Ficoll separation (detailed in Reagent Setup) takes an additional 2 h
Steps 18–22, acquisition and cell sorting: ~2–15 h
Steps 23–33, (optional) T cell expansion in vitro: 7–14 d
ANTICIPATED RESULTS
The panels indicated here allow the correct identification of human and NHP CD4+ and CD8+ TSCM cells (Fig. 1). Naive-like cells, which include both true naive cells and TSCM cells, must be identified using at least three markers and are defined here as CD45RO–CCR7+ CD62L+ in humans and as CD45RA+ CCR7+ CD28+ in rhesus macaques (Table 1). In humans, if cryopreserved cells are being used, CD62L is replaced by a different marker (e.g., CD27, Fig. 1d and Table 1). Within naive-like cells, a subset expressing CD95, the TSCM population, can be identified (Fig. 1a,b). Adding an MHC class I tetramer allows the identification of antigen-specific TSCM cells by using the same gating strategy (Fig. 1d).
The expected frequency of TSCM cells should be ~2–4% of the total CD4+ and CD8+ T-cell populations and does not change appreciably with the age of the donor (E.L., L.G., N.P.R. and M.R., unpublished observations). In our experience, the expected yield after sorting is one CD4+ TSCM cell per 250 PBMCs and one CD8+ TSCM cell per 500–1,000 PBMCs.
For human PBMCs, improved identification of the TSCM population can be achieved by including CD58 or CD122 in the staining panel, as these markers are differentially expressed in TSCM versus TN cells (Fig. 3a). In rhesus macaques, adding CXCR3 ensures better separation of the CD8+ TSCM subset, as virtually all CD8+ TSCM cells are also CXCR3+ (Fig. 1b).
Bulk TSCM cells, as well as other subsets, can be sorted by flow cytometry at high purity (Fig. 4) for subsequent genetic analysis, in vitro expansion and genetic manipulation. Indeed, stimulation with beads coated with CD3/CD2/CD28-specific antibodies or homeostatic cytokines induce cell cycle entry, thus allowing the transduction with retroviral vectors21. Genetically modified cells can then be used for adoptive transfer experiments.
Figure 4|.
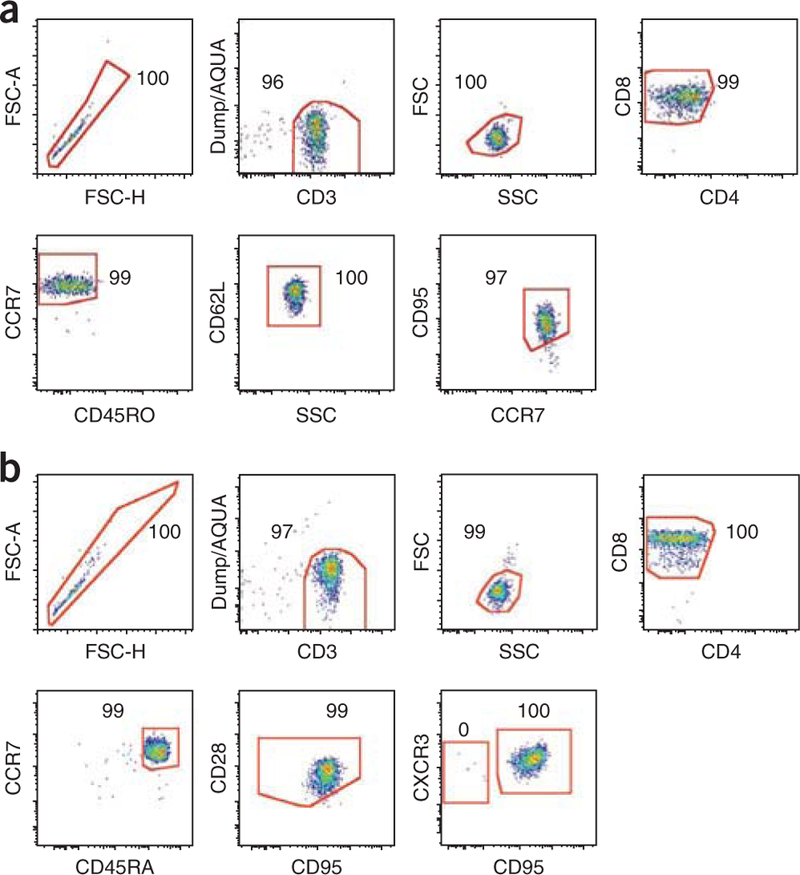
Flow cytometric sorting of TSCM cells. (a,b) Representative post-sort purity analyses of human (a) and NHP (b) CD8+ TSCM cells. TSCM cells are identified as described in Figure 1. Numbers indicate the percentage of cells in each gate.
Effective TSCM expansion in vitro is achieved by stimulating with beads coated with CD3/CD2/CD28-specific antibodies (human), plate-bound anti-CD3 and soluble anti-CD28 (NHP) or the homeostatic cytokines IL-7 and IL-15. In vitro, CD4+ T cells are preferentially expanded by IL-7, whereas CD8+ T cells respond to both IL-7 and IL-15 (ref. 22). A differential response of human TN and memory cells is seen with these stimuli, as depicted in Figure 5. Thus, we recommend that expansion conditions be optimized before proceeding with the experiment. A combination of both IL-7 and IL-15 can be used to maximize T cell stimulation21. Stimulation through CD3, CD2 and CD28 expands cells much more efficiently than the use of homeostatic cytokines, but it also causes a drastic change in the cell phenotype, including downregulation of CD45RA, CCR7 and CD62L with progressive proliferation as measured by CFSE dilution4. Thus, the user should choose the appropriate stimuli depending on the application.
Figure 5|.
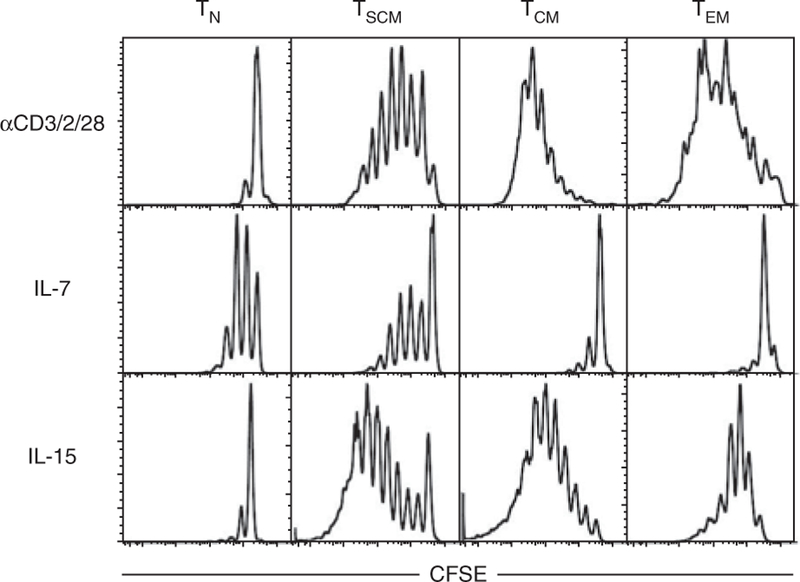
Differential response of T cell subsets to different stimuli in vitro. Human TN, TSCM, TCM and TEM cells were sorted as in Figure 1, stained with CFSE and stimulated with beads coated with CD3/CD2/CD28-specific antibodies for 6 d, 25 ng ml−1 IL-7 for 14 d or 25 ng ml−1 IL-15 for 10 d.
ACKNOWLEDGMENTS
We thank J. Yu and M. Beddall (ImmunoTechnology Section, VRC) for antibody conjugation; S.P. Perfetto, D. Ambrozak and R. Nguyen (Flow Cytometry Core, VRC) for assistance with cell sorting; M. F. Quigley (Immunology Laboratory, VRC) for providing the HLA-A*0201+ donor; and the other members of the ImmunoTechnology section for continuous discussion. This work was supported by the NIH Intramural Research Program and by the Associazione Italiana per la Ricerca sul Cancro to E.L.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Perfetto SP, Chattopadhyay PK & Roederer M Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol 4, 648–655 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Lugli E, Troiano L & Cossarizza A Investigating T cells by polychromatic flow cytometry. Methods Mol. Biol 514, 47–63 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Sallusto F, Lenig D, Forster R, Lipp M & Lanzavecchia A Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401, 708–712 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Gattinoni L et al. A human memory T cell subset with stem cell-like properties. Nat. Med 17, 1290–1297 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Joe G, Hexner E, Zhu J & Emerson SG Host-reactive CD8+ memory stem cells in graft-versus-host disease. Nat. Med 11, 1299–1305 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Gattinoni L et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med 15, 808–813 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitcher CJ et al. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol 168, 29–43 (2002). [DOI] [PubMed] [Google Scholar]
- 8.De Rosa SC, Herzenberg LA & Roederer M 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat. Med 7, 245–248 (2001). [DOI] [PubMed] [Google Scholar]
- 9.Mahnke YD & Roederer M Optimizing a multicolor immunophenotyping assay. Clin. Lab. Med 27, 469–485 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitz I et al. An IL-2-dependent switch between CD95 signaling pathways sensitizes primary human T cells toward CD95-mediated activation-induced cell death. J. Immunol 171, 2930–2936 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Lugli E et al. Quercetin inhibits lymphocyte activation and proliferation without inducing apoptosis in peripheral mononuclear cells. Leuk. Res 33, 140–150 (2009). [DOI] [PubMed] [Google Scholar]
- 12.Lugli E et al. Transient and persistent effects of IL15 on lymphocyte homeostasis in nonhuman primates. Blood 116, 3238–3248 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geginat J, Sallusto F & Lanzavecchia A Cytokine-driven proliferation and differentiation of human naive, central memory, and effector memory CD4+ T cells. J. Exp. Med 194, 1711–1719 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geginat J, Lanzavecchia A & Sallusto F Proliferation and differentiation potential of human CD8+ memory T cell subsets in response to antigen or homeostatic cytokines. Blood 101, 4260–4266 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Gattinoni L et al. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J. Clin. Invest 115, 1616–1626 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay P & Roederer M Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat. Protoc 1, 1522–1530 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Roederer M Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry 45, 194–205 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Kantor AB & Roederer M FACS analysis of lymphocytes. in Handbook of Experimental Immunology, Vol. 49 (eds. Herzenberg LA, Weir DM, Herzenberg LA & Blackwell C) 1–13 (Blackwell Science, 1997). [Google Scholar]
- 19.English D & Andersen BR Single-step separation of red blood cells. Granulocytes and mononuclear leukocytes on discontinuous density gradients of Ficoll-Hypaque. J. Immunol. Methods 5, 249–252 (1974). [DOI] [PubMed] [Google Scholar]
- 20.Altman JD et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996). [PubMed] [Google Scholar]
- 21.Cavalieri S et al. Human T lymphocytes transduced by lentiviral vectors in the absence of TCR activation maintain an intact immune competence. Blood 102, 497–505 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Surh CD & Sprent J Homeostasis of naive and memory T cells. Immunity 29, 848–862 (2008). [DOI] [PubMed] [Google Scholar]


