Abstract
The huge increase in the availability of bacterial genomes led us to a point in which we can investigate and query pan-genomes, for example, the full set of genes of a given bacterial species or clade. Here, we used a data set of 1,311 high-quality genomes from the human pathogen Pseudomonas aeruginosa, 619 of which were newly sequenced, to show that a pan-genomic approach can greatly refine the population structure of bacterial species, provide new insights to define species boundaries, and generate hypotheses on the evolution of pathogenicity. The 665-gene P. aeruginosa core genome presented here, which constitutes only 1% of the entire pan-genome, is the first to be in the same order of magnitude as the minimal bacterial genome and represents a conservative estimate of the actual core genome. Moreover, the phylogeny based on this core genome provides strong evidence for a five-group population structure that includes two previously undescribed groups of isolates. Comparative genomics focusing on antimicrobial resistance and virulence genes showed that variation among isolates was partly linked to this population structure. Finally, we hypothesized that horizontal gene transfer had an important role in this respect, and found a total of 3,010 putative complete and fragmented plasmids, 5% and 12% of which contained resistance or virulence genes, respectively. This work provides data and strategies to study the evolutionary trajectories of resistance and virulence in P. aeruginosa.
Keywords: genome sequencing, comparative genomics, cystic fibrosis, antibiotic resistance, virulence factors, core genome
Introduction
One of the outcomes of the post-genomic era of bacterial genomics is the increased interest in the study and the comparison of pan-genomes (Vernikos et al. 2015). A pan-genome is the full set of genes of a given bacterial species or clade (Tettelin et al. 2005). During the last decade, several pan-genomes have been reported. These studies allowed the characterization of the population structure (Lefebure and Stanhope 2007), biogeography (Bosi 2016; Vincent and Charette 2017), evolutionary trajectories (Lefebure and Stanhope 2007), and pathogenicity (Jacobsen et al. 2011; Bosi 2016) of different bacterial species of ecological or medical interest. Whole-genome sequencing (WGS) yields high-quality data for hundreds of bacterial isolates of a given bacterial species and permits detailed, in-depth genome comparisons.
Here we sought to perform such comparisons using as a model system the opportunistic human pathogen Pseudomonasaeruginosa. This ubiquitous microorganism can be found in water, soil, plants, animals, contaminated, and anthropic environments (Hardalo and Edberg 1997). Pseudomonasaeruginosa is the third most frequent opportunistic pathogen found in hospitals, is resistant to most classes of antibiotics, and causes major infections in immunocompromised patients (cancer, HIV, organ transplants), patients with burns and individuals with cystic fibrosis (Bodey et al. 1983; Hauser and Rello 2003). This capacity of P. aeruginosa to thrive in a repertoire of hosts and environments has often been associated with its relatively large genome (∼6–7 Mb). However, the molecular details of which genes are involved in the adaptation to different niches, what is the detailed population structure of this species and how it could be implicated in encoding traits of clinical interest have not yet been fully investigated at the pan-genome level. Only 2 studies using limited sets of 17 and 181 strains partly defined the pan-genome and molecular evolution of P. aeruginosa (Valot et al. 2015; Mosquera-Rendon et al. 2016). Here, we took advantage of a new data set of 1,311 strains, 619 of which are reported here as part of the International Pseudomonas Consortium (IPC) (Freschi et al. 2015), to: 1) Update the pan-genome of P. aeruginosa; 2) redefine its population structure; 3) attempt to define the links between population structure and pathogenicity; and 4) study the contribution of horizontal gene transfer (HGT) in P. aeruginosa genome evolution.
Materials and Methods
DNA Preparation, Sequencing, and Assembly (IPC Isolates)
Bacterial colonies were isolated on Difco Pseudomonas Isolation Agar (BD, Sparks, MD). Genomic DNA was extracted from overnight cultures using the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany). Genomic DNA (500 ng) was mechanically fragmented for 40 s using a Covaris M220 (Covaris, Woburn, MA) with default settings. Fragmented DNA was transferred to a polymerase chain reaction tube and library synthesis was performed with the Kapa Hyperprep kit (Kapa Biosystems, Wilmington, MA) according to the manufacturer’s instructions. TruSeq HT adapters (Illumina, SanDiego, CA) were used to barcode the libraries, which were each sequenced in 1/48 of an Illumina MiSeq 300-bp paired-end run at the Plateforme d’Analyses Génomiques of the Institut de Biologie Intégrative et des Systèmes (Université Laval, Québec, Canada). Each data set was assembled de novo with the A5 pipeline version A5-miseq 20140521. Raw reads, assembly data and metadata were uploaded on IPCD (https://ipcd.ibis.ulaval.ca).
Genome Data Set
We downloaded all P. aeruginosa genome assemblies present on NCBI on January 17, 2016 (https://www.ncbi.nlm.nih.gov/genome/genomes/187) as well as those present on IPCD on February 26, 2016. For the IPCD assemblies, if one isolate was sequenced two times, we chose the assembly with the least number of scaffolds. We then selected from both these sets only the assemblies that had less than 100 scaffolds to define our final data set of 1,311 P. aeruginosa genomes. The complete list of genome assemblies, their IDs and sequencing statistics are available in supplementary file S1 (IPCD) and file S2 (NCBI), Supplementary Material Online. Metadata for the NCBI isolates were retrieved from NCBI by downloading the attributes of each BioSample. Metadata for the IPCD isolates can be found on the IPCD web site (IPCD, https://ipcd.ibis.ulaval.ca; last accessed December 2018). In the analyses where we studied the P. aeruginosa species boundaries or the evolutionary history of specific genes, we also used the following genomes: P. composti (gis: 1098292672 and 1000298361), P. fluorescens (NC_016830.1), P. knackmussii (NZ_HG322950.1), P. nitroreducens (gis: 738540191, 573593795, 422814577), P. pseudoalcaligenes (NZ_HG916826.1), P. putida (NC_002947.4), P. resinovorans (gis: 523399098; NC_021499.1), P. stutzeri (NZ_CP007441.1), P. syringae (NC_007005.1), and P. thermotolerans (gis: 478735095, 570972061).
Pangenome
The pangenome analysis was performed using SaturnV (v1.1.0; https://github.com/ejfresch/saturnV; last accessed May 18, 2017), a software we developed to determine and study bacterial pan-genomes. SaturnV was developed using a modular concept, meaning that several independent modules are available to analyze and study pan-genomes. The long-term objective of this software project is to build a platform where it is easy to integrate new modules and algorithms that allow quick, accurate, and standardized analyses of pan-genomes. To determine the P. aeruginosa pan-genome, we used SaturnV’s “core” module, which takes genome assemblies as input and provides a matrix of gene presence/absence as output as well and their orthology/paralogy relationships. Several algorithms can be used to generate the matrix. For this work, we used the “lazy” algorithm. Briefly, we annotated the assemblies using Prodigal (Hyatt et al. 2010) (Prodigal provides the gene and protein sequences present in each genome assembly as well as the coordinates of their genomic locations); we took the protein sequences of the first isolate and we performed usearch (Edgar 2010) searches against each set of proteins of all other isolates, separately. We filtered the results: Two proteins needed to produce an alignment that covered 85% or more of the length of the two sequences (query and subject) and have a percentage of sequence identity of 50% or more to be considered as hits. For paralogs, the two proteins needed to produce an alignment that covered 85% or more of the length of the two sequences (query and subject) and had a percentage of sequence identity of 90% or more to be considered as hits. We built a graph where nodes were the ids of the protein sequences of all genomes. Edges were assigned according to the filtered usearch results. We identified all the nodes of the graph that did not have any connection with other nodes and we proceeded with a second iteration of usearch searches against the protein sequences of all isolates using these sequences as queries (the set of proteins of each isolate was queried separately). Again, we filtered the results and added the edges to our graph. Finally, we determined the connected components of the graph and wrote them into a text file: A matrix of gene presence/absence and homology (orthologous and paralogous) relationships. All usearch searches were performed using the “usearch_local” option, using the following command:
usearch8 -usearch_local <query_genes> -threads 1 -db <current_genome>.udb -id <perc_identity_ali> -userout <current_genome>_lazy_i<iter>.txt -userfields query+target+id+alnlen+mism+opens+qlo+qhi+tlo+thi+evalue+bits+ql+tl
Functional Enrichment
Functional enrichment was determined using the COG database (Tatusov et al. 2000). We used the matrix generated by SaturnV to generate a fasta file containing all protein sequences corresponding to the P. aeruginosa pan-genome. For unique genes, there was no ambiguity on which sequence to take. For flexible and core genes, we took the first sequence present on the table (going from left to right) as representative of all other orthologous sequences. We then queried the COG database (downloaded on July 2016) to determine the functions of these sequences using LAST (http://last.cbrc.jp; last accessed November 22, 2016) (Kielbasa et al. 2011). We filtered the LAST results by selecting only the hits that had 50% sequence identity or more. Using the associations present in the COG database between sequences and functions and our filtered LAST results, one or more functions were assigned to gene sequences. Those for which we could not assign a function were flagged as “unknown.” To study the functional enrichment of core and dispensable genes, we sampled our sequence-functions associations 1,000 times (respecting the proportion of core/dispensable genes), we calculated the null distributions for each biological function and we compared them with the values we observed in the core/dispensable genes. If the number of observed genes associated with a particular biological function was greater than the mean of the null distribution the P-value was calculated by dividing the number of values in the null distribution that were greater the number of observed genes divided by 1,000 (number of samplings). If the number of observed genes associated with a particular biological function was less than the mean of the null distribution the P-value was calculated by dividing the number of values in the null distribution that were less the number of observed genes divided by 1,000 (number of samplings).
Phylogeny (Core SNPs and Flexible Genes) and Genetic Distances between Isolates
In order to determine the core genome phylogeny, we took all 1:1 core genes, that is, the core genes for which we found one and only one ortholog in each of the other genomes. We aligned them using Prank (v.150803) (Loytynoja 2014) and removed uninformative positions with BMGE (v.1.12) (Criscuolo and Gribaldo 2010). Finally, we generated the phylogenetic tree using FastTree (Price et al. 2010) (v. 2.1.8; boostraps: 1000; model: GTR). To draw the tree based on flexible gene presence/absence, we used the matrix generated by SaturnV. The rows of flexible genes were converted to a binary format (presence = 1; absence = 0). We used iqtree (Nguyen et al. 2015) (v. 1.4.4; bootstraps: 1000; model: GTR2) to calculate the final tree. Average Nucleotide Identities were calculated using pyani (https://github.com/widdowquinn/pyani; last accessed November 18, 2018), whereas MuMi distances were calculated using harvest (Treangen et al. 2014). Heatmaps were generated using R scripts (R Core Team 2017), taking advantage of the gplots library (Warnes et al. 2016). The outgroup species used to study the boundaries of the P. aeruginosa clade (listed above) were chosen according to the phylogeny proposed by Gomila et al. (2015).
Antibiotic Resistance
Prediction of antibiotic resistance genes was performed using RGI (v.3.1.1) (Jia et al. 2017). We used the best hit ARO field to determine the resistance profiles. Principal component analysis (PCA) and discriminant analyses of principal components (DAPC) analyses were performed with R, using the ade4 (Dray and Dufour 2007) and adegenet (Jombart 2008) packages.
In Silico Serotyping
In silico serotyping was performed using PAst (Thrane et al. 2016) (v.1.0), in order to compare our results with those of Thrane et al. (2015).
Virulence Factors
Prediction of virulence genes was performed through usearch searches querying each proteome against the Virulence Factor core DataBase (Chen et al. 2016) that includes genes associated with experimentally verified virulence factors (downloaded on November 2016). The threshold was 50% sequence identity at the protein level. PCA and DAPC analyses were performed with R, using the ade4 and adegenet packages.
Gene Trees of the Exo Genes
We downloaded the sequences of the exo genes of UCBPP-PA14 or PAO1 (locus tags: PA3841, PA14_00560, PA14_51530, PA14_36345) from the Pseudomonas Genome DB (Winsor et al. 2016) database. We performed LAST searches to find the orthologous sequences in our isolates and set the threshold for a match at 90% sequence identity and 90% of query coverage. We performed sequence alignments with Prank and generated the trees with FastTree.
Mobilome
In order to detect potential phage genes present in our genomes we queried all protein sequences present in each of our isolates against the PHAST database (http://phast.wishartlab.com/Download.html, last accessed February 8, 2017., prophage and virus database, downloaded on Jun 2018) (Zhou et al. 2011). To generate the matrix of presence/absence of phage proteins we performed LAST searches and set the threshold for a match at 90% sequence identity. The hierarchical clustering of the matrix of presence/absence of phage proteins was performed using the gplots R library.
In our analysis about the plasmidome all protein sequences of the isolates present in our data set were queried against the NCBI database of plasmid proteins (ftp://ftp.ncbi.nih.gov/refseq/release/plasmid/; downloaded on November 2016). We performed the searches with usearch and set the threshold at 50% sequence identity. We used the Prodigal annotation (gffs) to find modules of “plasmid” genes, that is, adjacent genes with the same orientation (positive or negative strand). To detect bacterial genera other than Pseudomonas among this potentially transferred material, we selected all modules that contained five or more plasmid genes and used the NCBI database of plasmid proteins to associate a module to one or more bacterial genera. We then built a network in which the central hub is Pseudomonas and the other nodes correspond to the other genera. To visualize the graph we used Cytoscape (Shannon et al. 2003), using a force-directed layout so that the closer a node is to the central one, the more that genus is likely to have an important role in HGT with P. aeruginosa.
Results
Genome Sequencing and Pan-Genome Analysis
To maximize and improve the overall sampling of P. aeruginosa genetic diversity, we sequenced 619 P. aeruginosa isolates (supplementary file S1, Supplementary Material online) from 19 clinical (including patients with cystic fibrosis, burns, wounds, pneumonia, chronic obstructive pulmonary disease, and cancer) and 7 environmental sources (including plants, rivers, soil, animals, and oil sands). The isolates are available upon request for further experimental studies (http://ipcd.ibis.ulaval.ca/index/about/). Assembled genomes are available as part of BioProject PRJNA325248 (International Pseudomonas aeruginosa Consortium [IPC] genome sequencing project). To further enhance this data set, 692 high-quality genomes (<100 scaffolds; supplementary file S2, Supplementary Material online) from NCBI were used to build a final data set of 1,311 assemblies. Using the associated metadata, it was possible to draw a map describing the current geographical status of the P. aeruginosa data set used (supplementary fig. S1, Supplementary Material online). The present study contributes sequencing data for 13 countries that were not previously represented among high-quality genome assemblies available on NCBI.
Two studies completed during the course of this work attempted to define the core and pan-genome size of P. aeruginosa (Valot et al. 2015; Mosquera-Rendon et al. 2016). However, the number of isolates considered was limited and an order of magnitude lower than our data set. To calculate the pan-genome of P. aeruginosa, we developed a new bioinformatics tool, called SaturnV (available at https://github.com/ejfresch/saturnV; last accessed May 18, 2017). SaturnV can determine the pan-genome of thousands of isolates very quickly, at the expense of relatively high memory requirements, to overcome the calculation challenges due to the large number of isolates considered.
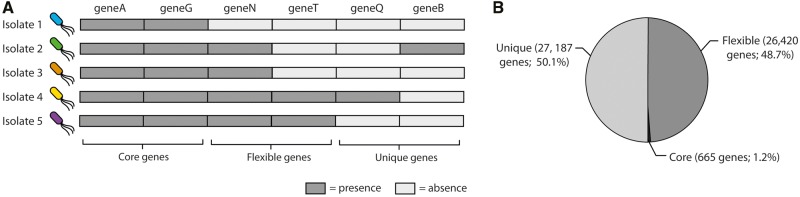
As described by Tettelin et al. (2005), a pan-genome is constituted by core genes, that is, genes found in all strains of one species; flexible genes, found in more than one strain but not all of them, and unique genes, found in one strain only (fig. 1A). The P. aeruginosa pan-genome of our data set consists of 54,272 genes: 665 of them are core genes, 26,420 are flexible genes, and 27,187 are unique genes (fig. 1B). Core genes represent only 1% of the P. aeruginosa pan-genome and their number (665) is in the same order of magnitude as the set of essential genes in laboratory media determined by Turner et al. (2015) (n = 336).
Fig. 1.
—The P. aeruginosa pan-genome (A) a pan-genome is constituted by three types of genes: core, flexible, and unique. Core genes are present in all isolates of a given bacterial species, flexible genes are present in more than one isolates but not all of them, unique genes are present in one single isolate. (B) Pie chart showing the proportions of core, flexible and unique genes determined by SaturnV (https://github.com/ejfresch/saturnV; last accessed May 18, 2017). Unique genes constitute 51% of the P. aeruginosa pan-genome, whereas flexible genes constitute 48% of it. Core genes constitute only 1% of the pan-genome.
We characterized the functions of core and dispensable (flexible + unique) genes, and found that core genes were enriched in RNA processing, chromatin structure, cell division and partitioning, metabolism and transport of amino acids, nucleotides, coenzymes and lipids, transcription and translation (supplementary fig. S2A, Supplementary Material online). Core genes were expected to be enriched in housekeeping functions. However, core genes were also enriched in genes with unknown function, which points out that our knowledge on the functions of P. aeruginosa genes is still far from being complete even for genes with housekeeping roles and found in thousands of isolates. Dispensable genes functions were enriched for secondary metabolites biosynthesis and metabolism, intracellular trafficking and secretion, and mobile elements. They were also enriched in unknown genes, that is, in genes that do not match any gene present in the COG database (see Materials and Methods for further details). In fact, we found that 33.1% of P. aeruginosa pan-genome genes do not match any sequence present in the COG database (supplementary fig. S2B–D, Supplementary Material online).
Phylogeny
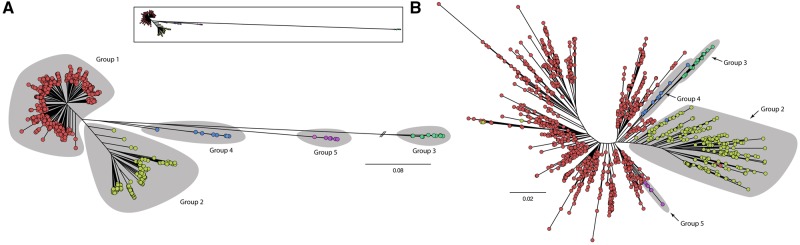
The list of core genes was used to analyze phylogenetic relationships among the 1,311 isolates based on core SNPs. Previous studies (Stewart et al. 2014; Freschi et al. 2015; Vincent et al. 2017) defined three major groups of P. aeruginosa isolates. A recent study (Thrane et al. 2015) suggested that a fourth group might exist, but this putative group consisted of only two isolates. Here, we clearly define five groups among P. aeruginosa isolates (fig. 2A). Two main groups (groups 1 and 2) include most of the isolates (n = 986 and 297, respectively; 98% of all isolates combined). A third group (group 3, n = 14) is very distant with respect to groups 1 and 2. Finally, the remaining two groups (group 4, n = 11; group 5, n = 7) have an intermediate position between groups 1–2 and group 3. To support this, we performed an average nucleotide identity (ANI) analysis to calculate genetic identity between isolates. If our phylogeny correctly identified groups of isolates, we expected to observe blocks with high ANI scores corresponding to those five groups. The ANI scores confirmed that groups 3, 4, and 5 are separate groups (supplementary fig. S3, Supplementary Material Online). Furthermore, group 3 isolates are rather distantly related to the other isolates, with a range of ANI values (93–94%) that is expected at the boundary of a bacterial species (Konstantinidis and Tiedje 2005). To confirm this, we performed another ANI analysis with six closely related Pseudomonas species as outgroups. We found that there is a clear gap between all isolates that were classified as P. aeruginosa, including group 3 isolates, and those belonging to sister species of P. aeruginosa (supplementary fig. S4A, Supplementary Material Online). An independent analysis based on MUMi genetic distances also confirmed that group 3 isolates belong to P. aeruginosa (supplementary fig. S4B, Supplementary Material Online). To determine the robustness of the population structure observed using the core genome, we examined genome architecture using a matrix of flexible gene presence/absence to generate a tree (fig. 2B). We used only flexible genes because core and unique genes are uninformative. This confirmed the five groups from the core genome phylogeny. Only six isolates were an exception to this rule. Their evolutionary trajectories are of interest because according to core SNPs they belong to a given group, but according to gene presence/absence they belong to another. Two scenarios could explain these shifts: Either the isolate was at the border between two groups or sharing a mobile element with isolates from a different group. We found examples of both scenarios. IPCD isolate 436, that was assigned to group 4 in the core SNP tree and was found in group 2 in the flexible gene presence/absence tree, was indeed at the border between groups 4 and 2 in the core SNP tree. IPCD isolates 1,125 and 1,133, that belonged to group 2 according to the core SNP tree, but belonged to group 1 according to the flexible gene presence/absence tree, shared a long stretch of genes that matches plasmid pBM413. This plasmid is present in some group 1 isolates, thus explaining the shift. Results on the presence/absence of flexible genes also show that group 3 is closer to the other groups than in the core genome phylogeny, providing further support that group 3 isolates are part of the P. aeruginosa species.
Fig. 2.
—Five groups of P. aeruginosa (A) Phylogenetic tree of all P. aeruginosa isolates calculated with all SNPs (n = 55,664) present in the 448 core genes that do not have paralog ambiguities (1:1 core genes). The different groups of isolates are highlighted by gray areas. Isolates belonging to each group are identified by a specific color (group 1: red; group 2: green; group 3: aquamarine; group 4: blue; group 5: violet). The small tree on the top of the panel shows the actual genetic distance between group 3 isolates and the other groups of isolates. (B) Tree representing the distances between isolates based on their genome architecture (calculated using flexible gene presence/absence, n = 26,420). Isolates belonging to each group are identified by a specific color (group 1: red; group 2: green; group 3: aquamarine; group 4: blue; group 5: violet).
Antimicrobial Resistance and Virulence Genes
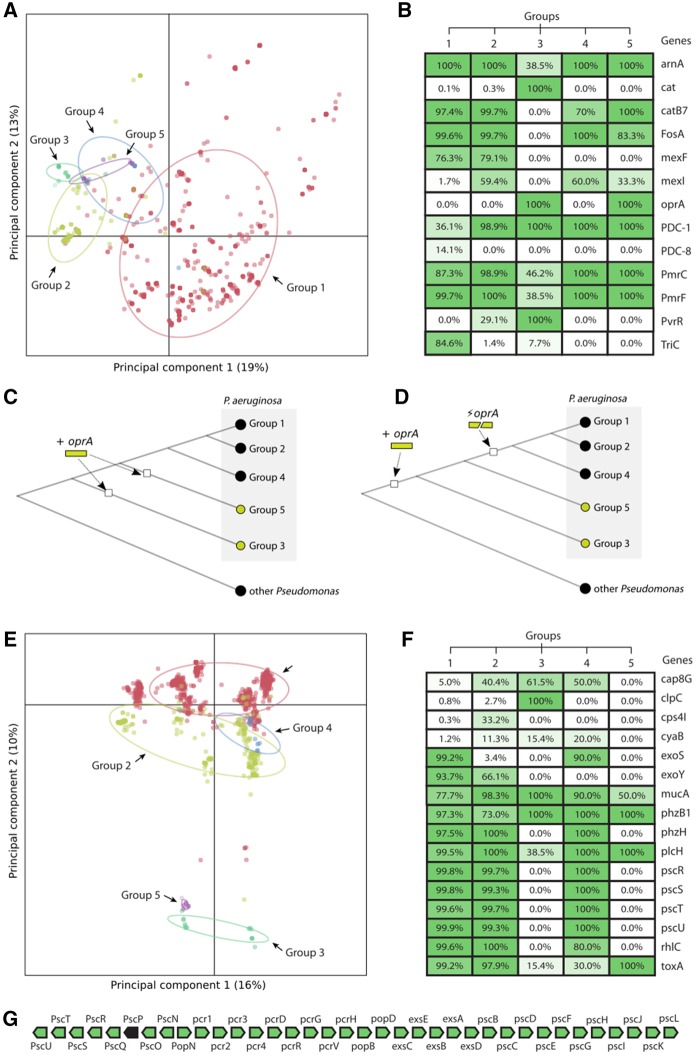
Given that P. aeruginosa is an opportunistic pathogen involved in clinical infections, it is of prime importance to understand the full extent of the variation in antimicrobial resistance (AMR) gene content. We previously predicted AMR gene profiles for 389 P. aeruginosa strains using the resistance gene identifier (RGI) (Freschi et al. 2015), and found 334 unique profiles. Analysis of 1,311 strains (raw results of the matrices of predicted resistance genes are available in supplementary files S3 and S4, Supplementary Material Online) confirmed there is a huge variation in P. aeruginosa AMR gene profiles, with the most frequent profile found in only 6% of the strains; supplementary fig. S5, Supplementary Material Online). As for SNPs conferring antibiotic resistance, we observed a widespread presence of SNPs involved in rifampicin resistance on gene rpoB (supplementary fig. S6, Supplementary Material Online). Several combinations of SNPs involved in fluoroquinolone resistance were identified in genes gyrA, gyrB, parE, parC as well as in genes encoding efflux pump subunits. To confirm whether AMR gene profiles are linked to the population structure of P. aeruginosa, we performed a PCA using the matrix of AMR gene presence/absence. We then mapped the groups found in the phylogeny depicted in figure 2 onto the PCA data (fig. 3A). By using the first two principal components, which summarized 32% of the overall variance, we could partly separate the five groups. Subsequent DAPC (supplementary fig. S7, Supplementary Material Online) identified candidate AMR genes (fig. 3B) present at different frequencies in these five groups. Although these results are solely based on the calls performed by RGI, we were able to validate two candidate lineage-specific genes. For instance, our analyses showed that the chloramphenicol acetyltransferase gene (cat) was present in group 3 isolates only. We confirmed this result by performing LAST searches (Kielbasa et al. 2011) using the cat gene from PA7 (PSPA7_4187) as query. PSPA7_4187 was identified and characterized for the first time by Roy et al. (2010) in the PA7 strain. These authors noted that other P. aeruginosa reference strains including PAO1 did not possess this gene, although they did have other genes involved in chloramphenicol resistance. Our analyses show that this gene is present without exception in all group 3 isolates (n = 14) of our data set, whereas it is not present in any other strain belonging to other groups or in the reference sequences of other Pseudomonas species, closely related to P. aeruginosa (P. composti, P. knackmussii, P. nitroreducens, P. thermotolerans, P. resinovorans, P. pseudoalcaligenes, P. stutzeri, P. putida, P. syringae, and P. fluorescens). From an evolutionary perspective, this means that this gene was likely acquired from the ancestor of group 3 isolates. Another piece of evidence that points toward this scenario is that other P. aeruginosa strains belonging to groups 1 and 2 (including PA14 and LESB58) have orthologs of the genes flanking the cat gene with putative phage sequences inserted between them, suggesting that insertions can occur at that specific genomic location. The other candidate lineage-specific gene involved in antibiotic resistance we validated is oprA. This gene is part of the MexXY-OprA/OprM multidrug efflux system and was considered to be a signature of strains belonging to serotype O12 (Morita et al. 2012a), PA7 being again the prototype strain where this gene was discovered (Roy et al. 2010). We performed LAST searches and found that oprA (PSPA7_3271) was present in all group 3 and 5 isolates, but not in isolates from groups 1, 2, and 4. Furthermore, this gene was not found in other Pseudomonas species, closely related to P. aeruginosa. Two possible scenarios could explain this finding: 1) oprA was acquired independently by the ancestors of group 3 and 5 isolates or 2) it was acquired by the ancestor of all modern P. aeruginosa isolates and subsequently lost by the ancestor of group 1, 2, and 4 isolates (fig. 3C and D). In order to test which one of these scenarios was the most likely to explain the evolutionary history of oprA, we performed LAST searches on all isolates present in our data set using the five genes flanking oprA (PSPA7_3271) on each side as queries. We found that orthologs of these genes were present in almost all strains of our data set, but not on the P. aeruginosa sister species. Also, the order of these genes was always respected, suggesting that the scenario that involves the acquisition of oprA by the ancestor of all modern P. aeruginosa and its subsequent loss of in the ancestor of groups 1, 2, and 4 is the most likely one to explain the evolutionary trajectory of this gene (fig. 3D). This scenario is also in agreement with the observation made by Morita et al. (2012b) that PAO1 and other strains actually do seem to have a small portion of oprA downstream of mexY. By looking at our genome annotation data and by performing LAST searches we found that this sequence was present in the vast majority of isolates belonging to groups 1, 2, and 4 (99.5% of group 1 isolates, 96% of group 2 isolates and 100% of group 4 isolates). With respect to the association between oprA and the serotype O12, Thrane et al. (2015) showed that some isolates belonging to group 1 had serotype O12 (serotype switching), suggesting that the presence of oprA is not necessarily linked to the serotype O12 as it was previously thought. To confirm this finding, we performed in silico serotyping for group 3 isolates, taking advantage of our newly sequenced strains. Our analyses showed that even within group 3 isolates we can find isolates belonging to different serotypes. This finding supports the hypothesis that the presence of oprA is not linked to the O12 serotype and shows as well that we still know little about the biological variation present outside of groups 1 and 2. The in silico serotyping results for all strains of our data set are available in supplementary file S5, Supplementary Material Online.
Fig. 3.
—Links between population structure and the occurrence of resistance as well as virulence genes (A) PCA analysis of the profiles of predicted antibiotic resistance genes. Isolates belonging to each group are identified by a specific color (group 1: red; group 2: green; group 3: aquamarine; group 4: blue; group 5: violet). (B) Candidate genes that explain the differences between the five groups of isolates. The genes were hits in DAPC analyses. Percentage values represent isolates of a given group in which one particular antibiotic resistance gene was found, according to RGI (best hit ARO field). (C) First scenario to explain the evolutionary history of oprA: it has been acquired independently by the ancestors of group 3 and 5 isolates. (D) Second scenario to explain the evolutionary history of oprA: it has been acquired by the ancestor of all P. aeruginosa modern isolates and subsequently lost in the ancestor of group 1, 2, and 4 isolates. (E) PCA analysis of the profiles of predicted virulence factors. Isolates belonging to each group are identified by a specific color (group 1: red; group 2: green; group 3: aquamarine; group 4: blue; group 5: violet). (F) Candidate genes that explain the differences between the five groups of isolates. The genes were best hits of DAPC analyses. Percentage values represent isolates of a given group in which one particular virulence factor was found, according to usearch searches. (G) Stretch of 36 genes that codes for a type-three secretion system, which is missing from group 3 and 5 isolates (green: found in our DAPC analysis; black: not found in our DAPC analysis).
We also predicted virulence genes for all isolates by querying the virulence factor database (VFDB) using protein sequences. The results are available in supplementary file S6, Supplementary Material Online. Like for AMR gene profiles, we hypothesized that there could be a link between virulence factor profiles and population structure in P. aeruginosa. We first performed a PCA analysis showing that 26% of the variance in virulence factor profiles can be explained by the first two principal components (fig. 3E). Subsequent DAPC analysis (supplementary fig. S8, Supplementary Material Online) revealed a list of genes encoding virulence factors (fig. 3F) that differentiate the five groups of P. aeruginosa isolates. We found that several psc genes (pscR, pscS, pscT, and pscU), all sharing the same pattern of presence/absence in the different groups of isolates, were part of a 36-gene fragment encoding a type-three secretion system, which is missing from group 3 and 5 isolates (fig. 3G). Although this difference was described for PA7, that belongs to group 3 (Filloux 2011), the pan-genome analysis shown here demonstrates that this is a characteristic of two entire groups of isolates, including the newly described group 5. We also looked for these genes in other Pseudomonas species closely related to P. aeruginosa and we were not able to find any ortholog. This finding suggests that these genes were likely acquired by the ancestor of groups 1, 2, and 4 isolates. Two other genes present in our list were exoS and exoY that code for two of the four effector proteins (ExoS, ExoT, ExoU, and ExoY) secreted by the type-three secretion system (Hauser 2009). ExoS was found only in group 1 and 4 isolates (with the exception of 10 isolates belonging to group 2), whereas exoY was present only in group 1 and 2 isolates. The exo genes are known to be characterized by a huge genetic diversity between isolates. We sought to take advantage of out data set of isolates to evaluate the extent of sequence diversity of these genes and detect potential links between sequence diversity and the position of the isolates in the phylogeny. To this aim we generated a gene tree for each one of the exo genes (we performed the analysis also for exoT and exoU). We first found that exoT was present in all isolates from groups 1, 2, and 4 (with the exception of 2 isolates, both belonging to group 2), whereas exoU was present in 94% (277/293 isolates) of the isolates of group 2 and in one single isolate of group 4 (1/11). All gene trees show that indeed there is a lot of genetic diversity between isolates, even between isolates belonging to the same group (supplementary figs. S9–S12, Supplementary Material online). By comparing the different gene trees, we observed that 1) for exoU the number of sequence variants is significantly reduced compared with the other exo genes; and 2) the version of the exoS gene present in group 4 isolates is different from the exoS genes of the isolates belonging to group 1 and 2. The biological and evolutionary significance of these findings and the impact on the virulence of P. aeruginosa should now be investigated.
Horizontal Gene Transfer
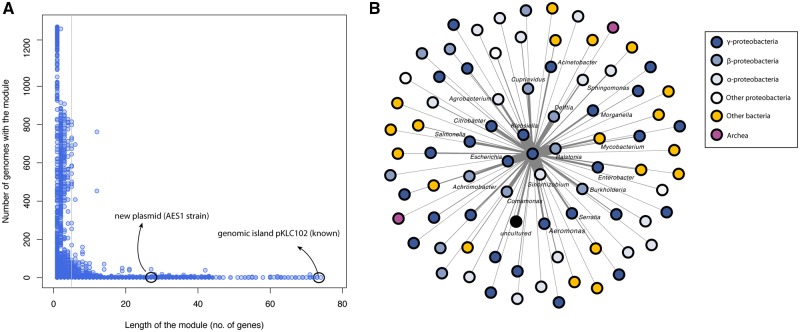
To study the role of HGT in the evolution of the P. aeruginosa genome we first looked at the flexible genes and determined the proportion of flexible genes that were found in a single group of isolates as well as the proportion of flexible genes shared between multiple groups. We found that 10,515 (40%) of the 26,420 flexible genes were present in 1 group only. Eighty-five genes (all found in group 3 isolates) were present in more than 90% of the isolates of a single group and were not found in the other groups. The majority of flexible genes (15,905, 60%) were therefore present in isolates belonging to multiple groups, a finding that provides an upper estimate of how many genes could be potentially related to HGT events. We then sought to estimate the prevalence of phages in our data set of isolates, because phages are often related to HGT events. To this aim, we identified all genes that matched sequences present the PHAST database of phage genes (see Materials and Methods). We found that 4,209 (7.8%) out of 54,272 genes in the pan-genome matched sequences from the PHAST database. Phage genes were found in the pan-genome with the following proportions: 15 among core genes (2.3% of all core genes), 2,017 among flexible genes (7.6% of all flexible genes) and 2,177 among unique genes (8% of all unique genes). A clustering analysis using the profiles of presence/absence of phage genes present in our isolates showed that we can divide them into two main clusters (supplementary fig. S13, Supplementary Material online). Both clusters contain isolates from all five groups, suggesting that the presence these two distinct pools of phage genes is not related to the phylogeny. The genomic regions that contain phage genes present in the different isolates should now be screened to find potential cases of HGT. Finally, we sought to detect the presence of potential plasmids in our isolates. To this aim we identified all genes that matched sequences present in the NCBI plasmid database (hereafter referred to as plasmid genes; see Materials and Methods for further details). We found that 9,923 (18%) out of 54,272 genes in the pan-genome matched sequences from the plasmid database. Plasmid genes were found in the pan-genome with the following proportions: 102 among core genes (15% of all core genes), 4,410 among flexible genes (17% of all flexible genes), and 5,411 among unique genes (20% of all unique genes). To identify segments of the genome that were likely to have been acquired through HGT, we identified all the modules of plasmid genes present in all genomes, that is, two or more plasmid genes adjacent to one another and located on the same strand. We found 11,334 genetic elements that followed these rules. Module frequency distribution as a function of module length is shown in figure 4 A. The large variance in module length and in the number of strains in which they are present suggests that HGT has a non-negligible role in the evolution of the P. aeruginosa genome. We considered only modules that contained 5 or more plasmid genes (3,010 modules) for further analyses. We found examples of known and new genetic elements linked to HGT (fig. 4A; supplementary file S7, Supplementary Material Online). Associations between plasmid genes and bacterial species available in the NCBI plasmid database were used to predict the bacterial genera that are likely to be involved in HGT with P. aeruginosa. We then built a network where the central node is P. aeruginosa to show that Pseudomonas is expected to have undergone the most HGT events with Sinorhizobium, Ralstonia, Klebsiella and Escherichia. All, like Pseudomonas, belong to phylum Proteobacteria. Although Ralstonia and Sinorhizobium are mostly known as environmental bacteria, Escherichia and Klebsiella are well-known pathogens. We also searched for antibiotic resistance genes in plasmid-gene modules using the RGI. We found that 164 of 3,010 modules (5%) contained one or more predicted AMR genes, indicating that HGT contributes to AMR in P. aeruginosa. One example is a previously unreported plasmid of 39,473 bp in strain AES-1 of the Pseudomonas international reference panel, which carries a gene predicted to be implicated in sulfonamide resistance. We also investigated the link between the modules of plasmid genes and virulence factors. We found that 363 of 3,010 modules (12%) contained sequences matching known virulence genes.
Fig. 4.
—Pan-genomic analysis of plasmid-mediated HGT (A) Frequency distribution of plasmid-gene modules, that is, adjacent genes with the same orientation (positive or negative strand) that match one or more sequences present on the NCBI database of plasmid proteins (sequence comparisons were performed at protein level). The gray line defines the threshold used to perform analyses (modules had to include 5 or more genes). Black circles define two examples (a known genetic island and an unknown plasmid) of regions we identified using this module approach that are related to HGT. (B) Network of bacterial genera that are likely to have exchanged plasmid genes with P. aeruginosa. The network was generated by getting the species information of the best match in the NCBI plasmid protein database for each of the genes present in the modules (sequence comparisons were performed at protein level). A force directed layout was applied to the graph so that the closer the nodes are to the center node, the more genes they exchanged with P. aeruginosa. Node colors reflects taxonomy. For clarity, only the names of the top candidate species are shown.
These results show that HGT events are widespread in P. aeruginosa, and are potentially implicated in the emergence of antibiotic resistance and virulence.
Discussion
Redefining the Population Structure of P. aeruginosa
The newly sequenced, high-quality genomes presented here constitute a resource for the scientific community focusing on bacterial genomics, as all isolates have been sequenced using the same protocol and instrumentation, and genome assemblies have been generated using the same bioinformatics tools. Moreover, these isolates have been selected to maximize diversity. Of course, this was done within the limits of the collection available, which is biased towards developed countries. This is mainly due to significant economic resources invested in attempts to control opportunistic infections caused by antibiotic-resistant P. aeruginosa, coupled with the higher incidence of cystic fibrosis among Caucasians (O’Sullivan and Freedman 2009). Future sequencing initiatives should focus on improving the sampling of many regions of Africa and Asia for which we currently do not have high-quality data (supplementary fig. S1, Supplementary Material Online). Our collection is also biased towards clinical strains, which explains at least in part why phylogenetic groups 1 and 2 are the most frequently sampled.
The P. aeruginosa pan-genome was determined from over a thousand isolates, that is, an order of magnitude more than previously published for P. aeruginosa and many other bacterial species. Our results show that core genes (665) constitute only 1% of the entire pan-genome. It is important to notice that this is a conservative estimate, because only the genes that we were able to find in all isolates were included in the core genome. This set of genes can now be compared with the sets of candidate essential genes in Pseudomonas (Turner et al. 2015) as well as Mycoplasma genitalium (Glass et al. 2006), considered as representative of the minimal set of genes required for a living bacterium. These analyses may lead to better understanding of the minimal set of genes for P. aeruginosa. Flexible and unique genes also warrant further investigation, as they are largely responsible for the ability of P. aeruginosa to adapt to different ecological niches. Functional analysis of the pan-genome shows that, although P. aeruginosa is a well-studied organism, one third of its genes are still poorly characterized.
To our knowledge, this study is the first to present a five-group population structure of P. aeruginosa, that is, with strong evidence for two previously undescribed groups of isolates (groups 4 and 5). These new groups are intermediate between groups 1–2 and group 3, which is genetically very distant from all other groups. Ongoing sequencing efforts will confirm whether other groups of isolates exist and fill this gap between group 3 and the remainder of the phylogeny. The long branches observed in the tree built from core SNPs were not observed in the tree build from the presence/absence of flexible genes. This result is in agreement with the hypothesis that the differences between the five groups could be mostly due to changes at sequence level rather changes in the overall genome architecture. Considering the current genus level evolutionary tree (Gomila et al. 2015), where group 3 is the most basal P. aeruginosa group, these changes could be key to explain the apparent success of group 1 and 2 isolates, which are the most frequently sampled in the clinic.
Finally, results presented here contribute to validate the genomic species boundaries for P. aeruginosa. The approach used can be applied to study species boundaries in other bacteria, identify incorrect taxonomic assignment, and even assign taxonomic categories to unclassified genomes, as shown by Jeukens et al. (2017).
Linking Population Structure and the Potential for Pathogenicity
WGS and comparative genomics focusing on AMR and virulence genes showed variations in resistance and virulence profiles among P. aeruginosa isolates, some of which are linked to population structure. For instance, our analyses showed that the genes cat and oprA are found in group 3 and group 3 and 5 isolates only, respectively. Our analysis also allowed us to come out with hypotheses on the evolutionary trajectories of these genes: cat was likely acquired by the ancestor of group 3 isolates, whereas the oprA gene was likely acquired by the ancestor of all P. aeruginosa modern isolates and subsequently lost in the ancestor of group 1, 2, and 4 isolates. By performing the same kind of analysis on virulence genes, we found that a stretch of 36 genes encoding for a type-three secretion system was missing from isolates belonging to groups 3 and 5. Furthermore, we found evidence that this stretch of genes was likely acquired by the ancestor of group 1, 2, and 4 isolates. The mechanisms by which this stretch of genes was acquired as well as its impact for the pathogenicity of group 1, 2, and 4 isolates should now be investigated. Finally, by comparing the orthologous sequences of the exo genes we were able to provide a global picture of their sequence diversity in P. aeruginosa and we found that the version of the exoS gene present in lineage four isolates significantly differs from the one present in group 1 and 2 isolates. The examples presented above show how the present study constitutes a resource to design experiments or bioinformatics analyses that aim to understand the evolution of antibiotic resistance and virulence in P. aeruginosa. From another point of view, these data can contribute to fuel the development of personalized treatments of infections, because by combining an accurate phylogeny with AMR predictions and phenotypic data about antibiotic resistance it will be possible to get a more detailed picture of the specific strain responsible for a given patient’s infection, which is not currently available. In this respect, future studies should focus in particular on group 1 and 2 strains that are more often found in human infections.
In our study, we also hypothesized that HGT had an important role in the emergence of antibiotic resistance. We first established an upper bound for the number of genes that could be potentially involved in HGT, by calculating the number of genes in the accessory genome shared between all groups. We found that 10,515 flexible genes matched this criterion. Furthermore, we looked at the presence of phage proteins in the genomes of our isolates and we observed that the isolates can be divided in two clusters. Both clusters contain isolates from multiple groups. Finally, we looked at the presence of plasmid proteins in our isolates and we found 3,010 modules of consecutive genes that matched known plasmid genes. A fraction of them did contain AMR and/or virulence genes (5% and 12%, respectively). This paves the way for further studies that should investigate which genes have been exchanged between Pseudomonas and other microbes, and how these events could have affected Pseudomonas pathogenicity.
The results presented here show that determining new bacterial pan-genomes, or updating existing ones with larger and more representative data sets, gives a better understanding of the population structure, biogeography, and evolution of microbes. Moreover, by linking virulence and antibiotic resistance profiles to pan-genomic data, we can generate hypotheses on the evolutionary trajectories of resistance and virulence genes. Finally, combining pan-genomic data (e.g., the phylogeny) with antibiotic resistance data and real-time sequencing data could have important implications for the development of diagnostic tools as well as to improve current therapy regimens to treat bacterial infections. Public databases should find new ways to incorporate, update, link, and visualize pan-genomic data to accelerate this process.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by Cystic Fibrosis Canada (CF Canada grant ID number 2610 to R.C.L. and a postdoctoral fellowship to J.J.), an Alexander Graham Bell Canada Graduate Scholarships from the Natural Sciences and Engineering Research Council of Canada (NSERC) for A.T.V. and the Natural Sciences and Engineering Research Council of Canada (NSERC, grant to S.J.C.). S.J.C. is a research scholar of the Fonds de la Recherche du Québec-Santé (FRQS). This study includes cystic fibrosis clinical isolates from Dr Jane Burns (CF Isolate Core at Seattle Children’s Research Institute [Grant NIH P30 DK089507]). We also acknowledge Stephan Heeb, Dao Nguyen, Craig Winstanley, François Malouin, André Cantin, Timothy Kidd, Scott Bell, Jim Manos, Eric Déziel, Tyrone Pitt, Jean-Paul Pirnay, Gabriel Perron, Iain Lamont, Jane Turton, Dervla Kenna, Joe Harrison, Simon Rousseau, Pedro Santos, Robert Hancock, Julie Milot, Burkhard Tümmler, Jean Barbeau, and Karen Liljebjelke for providing bacterial strains used in this study.
Authors’ Contributions
L.F., A.T.V., J.J., J.G.E.R., I.K.I, B.B., S.J.C., and R.C.L. designed the study; J.J., J.G.E.R., and I.K.I. performed the experiments; L.F., A.T.V., J.J., J.G.E.R., and M.J.D. analyzed the data; L.F., A.T.V., J.J., J.G.E.R., I.K.I, B.B., S.J.C., and R.C.L. drafted and reviewed the manuscript.
Literature Cited
- Bodey GP, Bolivar R, Fainstein V, Jadeja L. 1983. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 5(2):279–313. [DOI] [PubMed] [Google Scholar]
- Bosi E. 2016. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc Natl Acad Sci U S A. 113(26):E3801–E3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 44(D1):D694–D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo A, Gribaldo S. 2010. BMGE (block mapping and gathering with entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 10(1):210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray S, Dufour A. 2007. The ade4 package: implementing the duality diagram for ecologists. J Stat Softw. 22:1–20. [Google Scholar]
- Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461. [DOI] [PubMed] [Google Scholar]
- Filloux A. 2011. Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front Microbiol. 2:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freschi L, et al. 2015. Clinical utilization of genomics data produced by the international Pseudomonas aeruginosa consortium. Front Microbiol. 6:1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, et al. 2006. Essential genes of a minimal bacterium. Proc Natl Acad Sci U S A. 103(2):425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomila M, Peña A, Mulet M, Lalucat J, García-Valdés E. 2015. Phylogenomics and systematics in Pseudomonas. Front Microbiol. 6:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardalo C, Edberg SC. 1997. Pseudomonas aeruginosa: assessment of risk from drinking water. Crit Rev Microbiol. 23(1):47–75. [DOI] [PubMed] [Google Scholar]
- Hauser AR, Rello J. 2003. Severe infections caused by Pseudomonas aeruginosa. Boston: Kluwer Academic Publishers. [Google Scholar]
- Hauser AR. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol. 7(9):654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, et al. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11(1):119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen A, Hendriksen RS, Aaresturp FM, Ussery DW, Friis C. 2011. The Salmonella enterica pan-genome. Microb Ecol. 62(3):487–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeukens J, et al. 2017. A pan-genomic approach to understand the basis of host adaptation in Achromobacter. Genome Biol Evol.9(4):1030–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia B, et al. 2017. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45(D1):D566–D573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T. 2008. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24(11):1403–1405. [DOI] [PubMed] [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. 2011. Adaptive seeds tame genomic sequence comparison. Genome Res. 21(3):487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. 2005. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A. 102(7):2567–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebure T, Stanhope MJ. 2007. Evolution of the core and pan-genome of Streptococcus: positive selection, recombination, and genome composition. Genome Biol. 8:R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A. 2014. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 1079:155–170. [DOI] [PubMed] [Google Scholar]
- Morita Y, Tomida J, Kawamura Y. 2012a. MexXY multidrug efflux system of Pseudomonas aeruginosa. Front Microbiol. 3:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita Y, Tomida J, Kawamura Y. 2012. Primary mechanisms mediating aminoglycoside resistance in the multidrug-resistant Pseudomonas aeruginosa clinical isolate PA7. Microbiology 158(Pt 4):1071–1083. [DOI] [PubMed] [Google Scholar]
- Mosquera-Rendon J, et al. 2016. Pangenome-wide and molecular evolution analyses of the Pseudomonas aeruginosa species. BMC Genomics 17:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan BP, Freedman SD. 2009. Cystic fibrosis. Lancet 373:1891–1904. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5(3):e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2017. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; Available from: http://www.r-project.org/, last accessed December 20, 2018. [Google Scholar]
- Roy PH, et al. 2010. Complete genome sequence of the multiresistant taxonomic outlier Pseudomonas aeruginosa PA7. PLoS One 5(1):e8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart L, et al. 2014. Draft genomes of 12 host-adapted and environmental isolates of Pseudomonas aeruginosa and their positions in the core genome phylogeny. Pathog Dis. 71(1):20–25. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28(1):33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci U S A. 102(39):13950–13955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrane SW, et al. 2015. The widespread multidrug-resistant serotype O12 Pseudomonas aeruginosa clone emerged through concomitant horizontal transfer of serotype antigen and antibiotic resistance gene clusters. MBio 6(5):e01396–e01315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrane SW, Taylor VL, Lund O, Lam JS, Jelsbak L. 2016. Application of whole-genome sequencing data for O-specific antigen analysis and in silico serotyping of Pseudomonas aeruginosa isolates. J Clin Microbiol. 54(7):1782–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KH, Wessel AK, Palmer GC, Murray JL, Whiteley M. 2015. Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc Natl Acad Sci U S A. 112(13):4110–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valot B, et al. 2015. What it takes to be a Pseudomonas aeruginosa? The core genome of the opportunistic pathogen updated. PLoS One 10(5):e0126468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G, Medini D, Riley DR, Tettelin H. 2015. Ten years of pan-genome analyses. Curr Opin Microbiol. 23:148–154. [DOI] [PubMed] [Google Scholar]
- Vincent AT, Charette SJ. 2017. Phylogenetic analysis of the fish pathogen Aeromonas salmonicida underlines the dichotomy between European and Canadian strains for the salmonicida subspecies. J Fish Dis. 40(9):1241–1247. [DOI] [PubMed] [Google Scholar]
- Vincent AT, et al. 2017. Genomic characterisation of environmental Pseudomonas aeruginosa isolated from dental unit waterlines revealed the insertion sequence ISPa11 as a chaotropic element. FEMS Microbiol Ecol. 93:fix106. [DOI] [PubMed] [Google Scholar]
- Warnes GR, et al. 2016. gplots: various r programming tools for plotting data. R package version 3.0.1. Available from: https://cran.r-project.org/package=gplots, last accessed March 30, 2016. [Google Scholar]
- Winsor GL, et al. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44(D1):D646–D653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res. 39(Suppl):W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.