Abstract
The enormous health benefits associated with probiotics has resulted in an increased consumption of probiotic supplements. Several factors like regular sub-culturing, storage, unfavourable conditions etc. might compromise the efficacy and/or safety of lactic acid bacteria which are the major components of many probiotic preparations available in the market. The present study evaluated the probiotic characteristics and safety of probiotic bacteria isolated from two preparations available commercially in India. The products did not specify the genera, species or strains of the bacteria used. These were cultured using standard microbiological methods for cultivation of lactic acid bacteria. Bacteria were identified by PCR amplification and sequence analysis of 16S rRNA gene. Microbiological and molecular analyses revealed that both preparations contained homogenous population of Enterococcus faecium and Pediococcus acidilactici respectively. Assessment for several essential and desirable probiotic properties revealed that both the probiotic strains were safe and resistant to salt, lysozyme, bile salt and common antibiotics. The probiotic preparation containing P. acidilactici was better than that containing E. faecium as it survived in low pH and showed bile salt hydrolase activity. The probiotic preparation containing P. acidilactici also exhibited cholesterol-lowering activity.
Electronic supplementary material
The online version of this article (10.1007/s12088-018-0762-9) contains supplementary material, which is available to authorized users.
Keywords: Cholesterol-lowering, Commercial products, Enterococcus faecium, Pediococcus acidilactici, Probiotics
The World Health Organization defines probiotics as viable microorganisms which when consumed in adequate amounts confer health benefits on the consumer. Bacteria which are used as probiotics include lactic acid bacteria (LAB), Streptococcus, Weissella, Lactococcus, Enterococcus, Pediococcus and yeast like Saccharomyces boulardii [1]. As in other developing economies, the probiotic industry in India has experienced a great expansion with newer probiotic supplements being added every year in the market. Various global studies have reported a mismatch between the information on the label and laboratory safety assessment of many commercial probiotic products [2–4].Very few studies have correctly evaluated the quality and safety standards of probiotic products available in India [5]. For any probiotic to deliver beneficial effects to the consumer it should adhere to the gut epithelial cells, show resistance to common antibiotics, tolerance to lysozyme, low pH, gastric juice, bile and should be safe for human use. Also, certain characters like cholesterol assimilation and production of bacteriocins confer selective advantage to certain probiotics over the others. Thus, the present study was conducted to identify, characterize and assess probiotic characteristics of strains present in the commercial probiotic products marketed in India.
Two commercial probiotic preparations which claimed to contain LAB were collected from the local retailers and analysed separately using standard microbiological and molecular criteria, prior to their expiry date. Since, the labels on both the commercial probiotics did not specify actual identities of the bacterial species; hence in our laboratory these were assigned names as Probiotic LAB 1(PLAB1) and Probiotic LAB 2 (PLAB2). These were propagated and maintained in MRS broth (HiMedia, India) at 37 °C. The isolates were biochemically characterized and identified by PCR-amplification and sequencing of 16S rRNA gene using universal eubacterial primers as described earlier [6].The isolates were assessed for several desirable and essential probiotic characteristics. Survival at different pH and temperatures was studied by incubating the isolates in MRS broths of pH 1–10 and MRS broth at 16 °C, 28 °C, 37 °C, 45 °C and 60 °C, respectively. Survival in salt was assessed by incubating the isolates in MRS broth containing increasing concentrations of NaCl (0.5%, 2%, 3%, 4%, 5%, 7% and 10%) at 37 °C. Resistance to lysozyme was determined following a published protocol [5]. Phenol tolerance was determined by incubating the LAB in MRS broth containing 0.4% phenol for 24 h. Acid tolerance was determined by incubating LAB in three separate flasks of MRS broth at pH 1.5, 2 and 3 at 37 °C. Bile tolerance was determined by incubating isolates in MRS broth containing increasing amounts of bile salts—0.06%, 0.125%. 0. 25%, 0. 5% and 1% (w/v), at 37 °C. Resistance to gastric juice was determined by incubating the isolates in simulated gastric juice (pH 3.0) prepared by dissolving pepsin (0.3%, w/v) in saline (0.5%, v/v) and determining the viable cell counts at 0, 2 and 4 h at 37 °C. For determining bile salt hydrolase (BSH) activity LAB were grown in MRS plates supplemented with 0.5% bile salts at 37 °C for 24–48 h and visually checked for white zones of precipitation. The cell surface hydrophobicity and auto-aggregation capabilities were assessed following the published protocols [7, 8]. The co-aggregation capability of PLAB1 and PLAB2 was determined by examining their co-aggregation with E.coli NG9 following the method described by Golowczyc et al. [9]. Antibiotic susceptibilities were assessed by antibiotic disc diffusion method. Results were interpreted as per the guidelines of Clinical Laboratory Standards Institute [10]. Antagonistic/antimicrobial activity of PLAB1 and PLAB2 against other bacteria viz. E.coli NG9, Bacillus subtilis, B. amyloliquefaciens and B. pseudomycoides was assessed by agar well diffusion method [11]. The isolates were investigated for their ability to assimilate cholesterol in MRS broth with and without bile salts using the protocol of Rudel and Morris [12]. Haemolytic activity and DNase production was determined using published protocols [13, 14].
The results of morphological, biochemical and 16S rRNA gene sequencing of PLAB1 and PLAB2 are summarized (Sup Table 1). 16S rRNA gene sequencing and BLAST analysis confirmed that PLAB1 was Enterococcus faecium and PLAB2 was Pediococcus acidilactici. Both, E. faecium and P. acidilactici could grow well in the temperature range of 28–45 °C, however, optimum growth was observed at 37 °C (Fig. 1). Both isolates thrived in increasing salt concentrations, but growth decreased as the concentration of salt increased from 0.5 to 10% (Fig. 2). Both isolates showed high tolerance for lysozyme. Survival percentage of E. faecium and P. acidilactici were ~ 80% and ~ 93% respectively, even after 120 min of incubation. Both isolates could not survive at pH 1.5 and 2.0. E. faecium survived at pH 3.0 for only 1 h but, P. acidilactici survived at pH 3.0 even after 3 h. The number of colonies of both E. faecium and P. acidilactici decreased as the concentration of bile salts increased from 0.06 to 1%, although both isolates survived in 0.5% bile salts even after 2–4 h (Sup Table 2). P. acidilactici produced BSH while E. faecium did not, as evident from the white zone of precipitation around the wells in MRS supplemented with bile. Both P. acidilactici and E. faecium displayed a higher adherence for chloroform (35.53% and 25.87%, respectively) than iso-octane (16.47% and 15.04%, respectively). P. acidilactici showed lower auto- and co-aggregation capabilities, about 9 percent each. However, E. faecium showed higher auto-aggregation (29.93%) and co-aggregation (10.30%) than P. acidilactici.
Fig. 1.
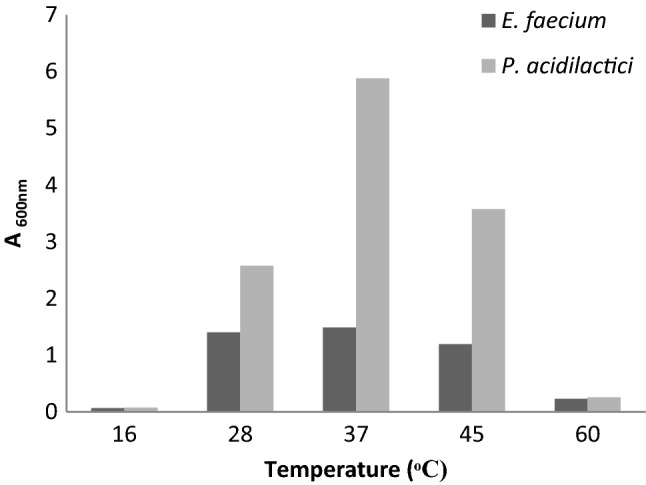
Survival of E. faecium and P. acidilactici at different temperatures
Fig. 2.
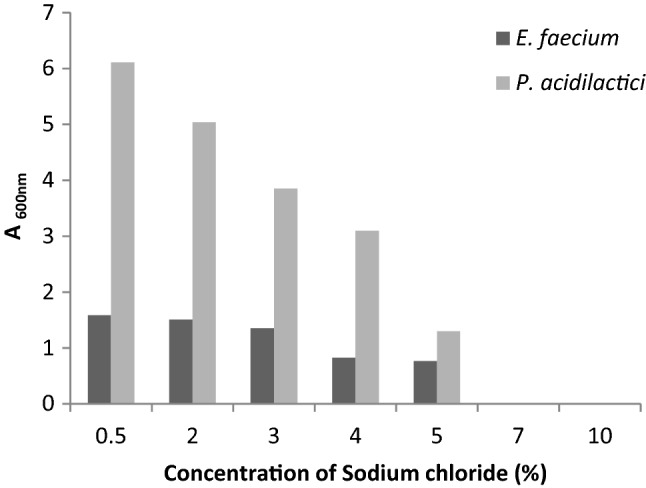
Tolerance of E. faecium and P. acidilactici to different concentrations of salt
The viable cell count (log10 cfu/ml) for E. faecium reduced from 5.01 ± 0.038 (mean ± SEM) to 3.04 ± 0.049 (mean ± SEM), while for P. acidilactici it increased from 5.65 ± 0.06 (mean ± SEM) to 6.98 ± 0.11 (mean ± SEM) indicating P. acidilactici was resistant to phenol even after 24 h. Both E. faecium and P. acidilactici were resistant to most of the common antibiotics, whereas sensitive to gentamicin, sparfloxacin, norfloxacin and ampicillin/sulbactam (Sup Table 3). E. faecium showed a zone of inhibition only against E.coli NG9 but not against B. subtilis, B. amyloliquefaciens, and B. pseudomycoides. However, P. acidalacti displayed zones of inhibition against all the tested bacteria, except B. pseudomycoides (Sup Table 4). P. acidilactici showed more cholesterol removal in both bile and non-bile medium i.e. 93.63% and 66.01% respectively. However, E. faecium assimilated only 56.82% and 30.64% of the cholesterol from bile and non-bile containing media, respectively. Both E. faecium and P. acidilactici did not show hemolytic and DNAse activities.
The study confirmed that the two probiotic preparations PLAB1 and PLAB2 available in the market contained homogenous populations of E. faecium and P. acidilactici respectively. Assessment of the probiotic isolates for several essential and desirable probiotic properties revealed some interesting observations. It was observed that both E. faecium and P. acidilactici were resistant to lysozyme, however, P. acidilactici survived well (> 90%) even after 2 h of contact with lysozyme. Another attribute important for probiotics to sustain inside the human gut, is survival in the low pH of the stomach (pH 1.5–2.0) and gastric acid (pH 3.0) for at least 3–4 h. Though, E. faecium could not survive at pH 3.0 > 3 h, but P. acidilactici maintained its viability even after 3 h, at pH 3. It also showed greater survival in simulated gastric juice experiment even after 24 h. Following their passage through the stomach, in the intestine, probiotic bacteria have to counteract the effect of bile juices. Both E. faecium and P. acidilactici showed good survival in 0.5% bile salts even after 2–4 h of exposure. P. acidilactici tested positive for BSH activity implying that it might survive better in the host GI tract, because bacterial BSH mediates deconjugation of bile salts which improves the intestinal viability of probiotics [15].Thus, the probiotic preparation containing P. acidilactici seemed to possess an edge over that containing E. faecium as it survived better in the harsh conditions of the human GI tract and possessed an added advantage as a cholesterol-lowering bio-therapeutic.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Mr. Rajesh Kumar Mahto for technical help. NS is supported by Start up Research Grant for Young Scientists (SB/YS/LS-156/2014), Science and Engineering Research Board of Department of Science and Technology. NSS is supported by ICMR-SRF.
Author Contributions
Conceived and designed the experiments: NS, NSS, JSV. Performed the experiments: NS, SM, PS, NSS. Wrote the paper: NS, JSV.
Compliance with Ethical Standards
Conflict of interest
The authors declare that no competing interest exists.
References
- 1.Kurzak P, Ehrman MA, Bauer J, Vogel RF. Characterization of Lactobacilli towards their use as probiotic adjuncts in poultry. J Appl Microbiol. 2002;92:966–975. doi: 10.1046/j.1365-2672.2002.01608.x. [DOI] [PubMed] [Google Scholar]
- 2.Fasoli S, Marzotto M, Rizzotti L, Rossi F, Dellaglio F, Torriani S. Bacterial composition of commercial probiotic products as evaluated by PCR-DGGE analysis. Int J Food Microbiol. 2003;82:59–70. doi: 10.1016/S0168-1605(02)00259-3. [DOI] [PubMed] [Google Scholar]
- 3.Elliot E, Teversham K. An evaluation of nine probiotics available in South Africa, August 2003. S Afr Med J. 2004;94:121–124. [PubMed] [Google Scholar]
- 4.Theunissen J, Britz TJ, Torriani S, Witthuhn RC. Identification of probiotic microorganisms in South African products using PCR-based DGGE analysis. Int J Food Microbiol. 2005;98:11–21. doi: 10.1016/j.ijfoodmicro.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 5.Zago M, Fornasari ME, Carminati D, Burns P, Suàrez V, Vinderola G, Reinheimer J, Giraffa G. Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol. 2011;28:1033–1040. doi: 10.1016/j.fm.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee J, Yun HS, Cho KW, Oh S, Kim SH, Chun T, Kim B, Whang KY. Evaluation of probiotic characteristics of newly isolated Lactobacillus spp.: immune modulation and longevity. Int J Food Microbiol. 2011;148:80–86. doi: 10.1016/j.ijfoodmicro.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Kos B, Susković J, Vuković S, Simpraga M, Frece J, Matosić S. Adhesion and aggregation ability of probiotic strain Lactobacillus acidophilus M92. J Appl Microbiol. 2003;94:981–987. doi: 10.1046/j.1365-2672.2003.01915.x. [DOI] [PubMed] [Google Scholar]
- 9.Golowczyc MA, Mobili P, Garrote GL, Abraham AG, De Antoni GL. Protective action of Lactobacillus kefir carrying S-layer protein against Salmonella enterica serovar enteritidis. Int J Food Microbiol. 2007;118:264–273. doi: 10.1016/j.ijfoodmicro.2007.07.042. [DOI] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (2010) Performance standards for antimicrobial susceptibility testing. 20th informational supplement (M100–S20) Clinical and Laboratory Standards Institute, Wayne, PA
- 11.Ridwan BU, Koning CJ, Besselink MG, Timmerman HM, Brouwer EC, Verhoef J, Gooszen HG, Akkermans LM. Antimicrobial activity of a multispecies probiotic (Ecologic 641) against pathogens isolated from infected pancreatic necrosis. Lett Appl Microbiol. 2008;46:61–67. doi: 10.1111/j.1472-765X.2007.02260.x. [DOI] [PubMed] [Google Scholar]
- 12.Rudel LL, Morris MD. Determination of cholesterol using o-phthalaldehyde. J Lipid Res. 1973;14:364–366. [PubMed] [Google Scholar]
- 13.Pieniza S, Andreazzab R, Anghinonic T, Camargoc F, Brandelli A. Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control. 2014;37:251–256. doi: 10.1016/j.foodcont.2013.09.055. [DOI] [Google Scholar]
- 14.Gupta H, Malik RK. Incidence of virulence in bacteriocin-producing enterococcal isolates. Le Lait. 2007;87:587–601. doi: 10.1051/lait:2007031. [DOI] [Google Scholar]
- 15.Pisano M, Casula M, Corda A, Fadda M, Deplano M, Cosentino S. In vitro probiotic characteristics of lactobacillus strains isolated from fiore sardo cheese. Ital J Food Sci. 2008;20:505–516. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


