Abstract
Trichoderma asperellum (NAIMCC-F-03167) and Hypocrea nigricans (NAIMCC-F-03168) were isolated from the acidic soil of the vicinity of Litchi orchard, Ranchi, Jharkhand and were characterized on the basis of morphological, molecular and biochemical features. Both strains are fast growing, light to dark green, highly sporulative and have ability to cover 90 mm Petri dish within 96 h of inoculation. Biochemcial estimation of both isolates indicated significant cellulase and phosphate solubilisation activity. Highest cellulase activity was observed in T. asperellum (5.63 cm) followed by H. nigricans (5.10 cm) and phosphate solubilisation index was observed maximum in T. asperellum (1.93) followed by H. nigricans (1.39). Moreover, these isolates were molecularly identified on the basis of ribosomal DNA based sequences database and phylogenetic analysis in NCBI GenBank as T. asperellum (NCBI-KM 438015) and H. nigricans (NCBI-KJ910335). Negetive effect on sporulation of Lead (Pb) and Cadmium (Cd) was observed while in heavy metal scavenging potential, T. asperellum (88.9% Cd) showed highest scavenging potential followed by H. nigricans (87.2% Cd) while in Pb scavenging potential, H. nigricans (88% Pb) followed highest scavenging potential followed by T. asperellum (81.30% Pb) after 21 days of inoculation from 30 µg/ml heavy metals concentrated broth medium. If both potential bioagents can apply in Cd and Pb affected soil/water will be helpful in scavenging of heavy metals as well as management of phosphorus deficiency and soilborne fungal diseases.
Keywords: T. asperellum, H. nigricans, Cellulase activity, Phosphate solubilisation, Heavy metals scavenging potential
Introduction
The rhizosphere is a dynamic interface between plant and soil harbouring numerous microbial populations valued for releasing chelating agents, acidification, phosphate solubilisation and rhizoremediation through multifaceted mode of actions including mycoparasitism, plant growth promotion, organic substrate decomposition, nutrient solubilisation, mobilization of nutrient and heavy metal scavenging. Heavy metals are well known hazardous environmental toxicants and a potential threat to the very life of ecosphere ranging from microbes to human as causing DNA breakage, failure of vital organs like kidney and damage of brain tissues in the human being [1–3]. Higher concentrations of heavy metal in soil are potential threat to human health as they get entered into food chain through plant uptake [4] and persistent long term exposure of heavy metals like cadmium may reduce human life span up to 10–30 years [5]. Besides, they influence germination, growth and development of the annual as well as perennial crop plants [6]. Zaltauskaite and Sodiene [7] further elaborated the negative impact of Cadmium (Cd) and Lead (Pb) on the growth, reproduction and survival of the earthworm (Eisenia foetida). A report indicated that maximum lead toxicity is common in soils receiving sewage water for irrigation, industrial effluents waterway and high traffic areas [8]. Krishnaveni et al. [9] extended the negative impact of Lead toxicity on biochemical and amino acid contents in Vigna unguiculata. Srinivasan et al. [10], reported the negative consequence of lead toxicity that include leaves chlorosis, rapid inhibition of root growth, stunted plant shoot, inactivation of oxidative enzymes etc. Moreover, several reports extended the negative impact of lead toxicity to reduced seed germination, root/shoot growth, tolerance index and dry mass of root and shoots [11]. Verma and Dubey [12] precisely quantified that 1 mM of lead may cause 14–30% reduction in seed germination and 13–45% retardation in crop seedling growth. It also inhibits chlorophyll synthesis by impairing plant uptake of essential elements like Mg and Fe [13]. Ling et al. [14] highlighted affects of heavy metal toxicity particularly Hg on seed germination, coleoptiles growth and elongation in vegetable crops.
All these literatures comprehensively outlined emerging contours of threat domain emanating from heavy metal toxicity in our surroundings and warrants immediate scientific intervention to reduce their burden below threshold limits. Although over decades, number of approaches including chemical precipitation, filtration, electrochemical treatment, evaporation, ion exchanges and reverse osmosis process were reported by several workers to rectify the problem [15, 16]. However, scavenge heavy metals from contaminated soil by the above mentioned processes found practically difficult and economically unsustainable. The limitation inspired to explore using bacteria, yeast, fungi, algae and higher plants to scavenge heavy metals from the environments and many such microbes found eco-friendly, cost effective and economically sustainable [17]. Phytoremediation/bioremediation of heavy metals in contaminated soil and water by plants is a potential alternative cleanup strategy promising for moderate and low contamination level of heavy metals. There efficiency could be further enhanced by inoculation of selected rhizospheric microbes in the plant rhizosphere [18, 19]. Petrovic et al. [20] isolated a Cu tolerance Trichoderma species capable of scavenging toxic Cu from the soil. Preliminary success inspired several researches to search novel yet potential microbial inoculants which have multifaceted mode of action including scavenging of heavy metals toxicity from the contaminated soil and water. Among the microbial bio-inoculants, Trichoderma species (Teliomorph: Hypocrea) widely used as bio-inoculants in agriculture for the management of soil borne phytopathogens, cellulase activities and plant growth promotion. However, there potential to scavenge heavy metals and solubilising been explored fully. In this backdrop present study conducted with three objectives namely: (1) to establish molecular identification of novel bioagents isolated from acidic soil ecosystem, (2) to verify the heavy metal scavenging potential and cellulase activity of the selected potential bioagents and (3) to assess the ability of selected biogents to check soil borne fungal phytopathogens as part of their multifaceted/broad-spectrum actions.
Materials and Methods
Isolation of Bio-control Agent from the Soil
Rhizospheric soil (1 g) at tertiary root region of litchi trees was collected and then serial dilutions (10−3) were made for isolation of fungal mycoflora. During isolation 6 fungi were predominantly observed in the rhizopsheric soil viz., Trichoderma sp., Aspergillus sp., Penicillium sp. Mucor sp., Rhizopus sp. and Fusarium sp. Among the fungal mycoflora, Trichoderma and Hypocrea species were isolated, purified and selected for further experimentation. Identification of the Trichoderma and Hypocrea isolates was done on the basis of colony morphology, microscopic features and on molecular basis.
Molecular Analysis of T. asperellum and H. nigricans
Pure cultures of the selected fungal isolates were grown on liquid Potato Dextrose Broth medium for the isolation of genomic DNA from fungal mycelium. The total genomic DNA was extracted from both the fungal isolates namely T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 based on cetrimide tetradecyl trimethyl ammonium bromide (CTAB) mini extraction method of Cramer et al. [21] with minor modification. Three microlitre of the DNA sample was then analyzed by gel electrophoresis using 1.0% agarose with ethidium bromide at 110 V for 45 min and the DNA bands were visualized under UV trans-illuminator.
PCR Amplification of ITS Region of T. asperellum and H. nigricans
The internal transcribed spacer (ITS) regions of the rDNA repeat from the 3′end of the 28S and the 5′end of the 28S gene were amplified using the two primers, ITS 1 primer: 5′-CCTCCGCTTATTGATATGC3′ and ITS4 primer: 5′GGAAGTAAAAGTCGTAACAAGG3 which were synthesized on the basis of conserved regions of the eukaryotic rRNA gene [22]. The PCR-amplification reactions were performed in a 50 μl mixture containing 50 mM KCl, 20 mM Tris HCl (pH 8.4), 2.0 mM MgCl2, 200 μM of each of the four deoxynucleotide triphosphates (dNTPs), 0.2 mM of each primer, 40 ηg/μl of template and 2.5 U of Taq polymerase. The cycle parameters included an initial denaturation at 94 °C for 5 min, followed by 40 cycles of denaturation at 94 °C for 1 min, primer annealing at 55 °C for 2 min and primer extension at 72 °C for 3 min and a final extension for 10 min at 72 °C. Amplified products were separated on 1.2% agarose gel in TAE buffer, pre-stained with ethidium bromide (1 μg/ml) and electrophoresis was carried out at 110 volts for 45 min in TAE buffer. One Kb ladder (HiMedia, India) was used as a marker. The gel was observed in a UV transilluminator. Amplified PCR products using ITS specific primers were freeze dried (CHRIST ALPHA I-2LD) and custom sequenced (ABI PRISM 310TM Genetic Analyzer, Applied Biosystems, USA) using same upstream and downstream primers (Xcelris Labs Limited, India).
Nucleotide Sequences Analysis of T. asperellum and H. nigricans
The 18S rDNA sequences of fungal isolates were identified using online Blast program from NCBI (http://www.ncbi.nih.gov/blast). Twenty-eight 18S rDNA sequences of T. asperellum and H. nigricans with high sequence similarity were selected for sequence comparison from the Gene Bank Nucleotide Database, NCBI, USA. The submitted sequences along with selected sequences were aligned by the ClustalW program using website http://www.ebi.ac.uk/clustalw/. The evolutionary history was inferred using the Neighbor-Joining method [23]. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history of the taxa analyzed [24]. All positions containing gaps and missing data were eliminated. There were a total of 533 positions in the final dataset. Phylogenetic analyses were conducted using MEGA version 6 [25]. The best fitting model was chosen based on lowest BIC score values (3452.255) using MEGA version 6 to find best DNA/protein models.
Antagonistic Efficacy Against Different Phytopathogens
Four different plant pathogens, S. rolfsii, S. sclerotiorum, F. oxysporum f. sp. pisi and R. solani were isolated, from their respective host collected from the experimental field of the Research Centre, Ranchi. The isolates of T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 were evaluated for their bio-control potentials through dual culture technique. The mycelial bits of 5 mm diameter of T. asperellum and H. nigricans were placed opposite to each pathogen on Petri plates containing sterilized PDA and were incubated at 27 ± 2 °C. Data on radial mycelial growth of the targeted fungal pathogen was recorded after 24 h interval. The inhibition (%) of mycelial growth over control was calculated using following equation [26].
where, I = Inhibition of targeted fungal pathogen (%), C = Growth (mm) of targeted fungal pathogen in control, T = Growth (mm) of targeted fungal pathogens in treatment.
Phosphate Solubilizing Activity of T. asperellum and H. nigricans
Phosphorus is a plant nutrient which is rapidly made immobile and less available for plant even addition to the soluble phophoric fertilizers. As we know, phosphate solubilising microorganisms may be able to improve the P nutrition of plants as well as it stimulates plant growth promotion. An In vitro study was conducted to test the inorganic phosphate solubilisation potential of the selected biocontrol agents, T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168. A disc of 5 mm of the fungal mycelium of T. asperellum and H. nigricans was placed on PVK agar medium (Glucose-10gm, Ca (PO)4-5gm, (NH)SO4-0.5gm, NaCl-0.2gm, MgSO4.7H2O-0.1gm, KCl-0.2gm, Yeast extract-0.5gm, 4Mn SO4.7H O-0.002gm, FeSO4.7H2O-0.002gm, Agar-15gm, Rose Bengal-0.0018gm, Distilled water-1L) in Petri dishes and incubated for 3 days at 25 ± 2 °C and data on colony diameter and halo zone formation was recorded. Solubilizing Index was calculated using the formula:
Cellulase Activity of T. asperellum and H. nigricans
Qualitative evaluation of cellulase activity of the selected T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 were also studied on Carboxymethyl cellulose (CMC) agar media (Carboxymethyl cellulose-0.5gm, NaNO3-0.1 gm, K2HPO4-0.1g, KCl-0.1gm, MgSO4-0.05gm, Yeast extract-0.05gm, Glucose-0.1gm, and Agar-1.7% w/v) were evaluated using agar plate based clearing assays [27]. Two wells of 5 mm diameter were prepared on solidified CMC agar media; wells were further loaded with culture filtrate of T. asperellum and H. nigricans (100 µl) separately and other with sterile water (control). The loaded culture filtrates Petri plates were incubated for up to a week at 25–30 °C and then after Petri dishes were flooded with Congo red solution (0.1%) for 15 min, and then de-stained with the NaCl solution for 10–15 min. Unstained areas indicated where the CMC has been broken down to b 1–4 glucans that contain seven or fewer glucose residues. The diameter of the clear zone (mm) was measured to provide a quantitative comparison of cellulolytic activity.
Heavy Metal Scavenging Potential of T. asperellum and H. nigricans
Screening of heavy metal scavenging potential was assessed by Atomic absorption spectroscopy. Both fungal bioagents, T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 were grown in 250 ml Erlenmeyer flasks containing 100 ml of PD broth amended with a non-inhibitory concentration of two heavy metals in triplicates i.e. Cadmium and Lead separately (@ 10, 20 and 30 µg) and were incubated at 25 ± 2 °C. Mycelial mat were harvested at 7, 14 and 21 days intervals in order to determine their dry weights and culture filtrates were retained in order to measure the metal concentration in the filtrates. The standard concentrations (5, 10 and 15 µg) of Cadmium (Cd) and Lead (Pb) were prepared from Cadmium nitrate and Lead nitrate, respectively. The available heavy metals like Cd and Pb in culture filtrate was estimated with the help of Atomic Absorption Spectrophotometer (ECIL; Model—AAS4141).
Metal uptake was estimated as the amount of metal (mg) per unit of mycelium dry weight (g) [28] as:
where, Q = Metal uptake (mg metal/mg biomass), Ci = Initial metal concentration (mg/L), Cf = Final metal concentration (mg/l), m = quantity of dry biomass (mg), V = suspension volume (ml).
Results and Discussion
Isolation of T. asperellum and H. nigricans
T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 were isolated from the acidic soil (pH 4.4–5.5) collected from the vicinity of Litchi (Litchi chinensis) plants, ICAR-Research complex for Eastern Region, Research Centre, Ranchi, Jharkhand (23o16′48″ N Latitude and 85o 24′ 41″ E longitude, 650 M altitude). The soil characteristically contains low organic carbon ranging from 0.9 to 1.2%. Isolated bioagents were comprehensively characterized on the basis of their morphological, molecular and biochemical features. Isolates of both T. asperellum and H. nigricans were deposited at National Agriculturally Important Microbial Culture Collection at NBAIM Mau, Uttar Pradesh, India and allotted national identity number namely, T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168, respectively.
Antagonistic, Competitive Saprophytic Ability, Cellulase and Phosphate Solubilisation Efficacy of T. asperellum and H. nigricans
Both isolates namely T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 found fast growing microbe covers 90 mm Petri dishes on PDA within four days of inoculation (Table 1),
Table 1.
Periodical growth of T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 on PDA medium at 27 ± 2 °C
| Isolates | Periodical growth (mm) | |||
|---|---|---|---|---|
| 24 h | 48 h | 72 h | 96 h | |
| T. asperellum NAIMCC-F-03167 | 23.33 ± 2.08 | 68.0 ± 2.64 | 82.0 ± 3.60 | 90.0 ± 0.0 |
| H. nigricans NAIMCC-F-03168 | 21.0 ± 1.00 | 66.67 ± 2.51 | 79.0 ± 4.04 | 90.0 ± 0.0 |
± Standard error of mean
Both of them recorded strong antagonistic efficacy against four dreaded soil borne phytopathogens viz., F. oxysporum f. sp. pisi, S. rolfsii, S. sclerotiorum and R. solani under in vitro as well as in vivo (applied in the field after 21 days of inoculation) (Fig. 1). Highest cellulase activity was observed in NAIMCC-F-03167 (5.63 cm) followed by NAIMCC-F-03168 (5.10 cm) by forming a clear zone in corboxy methyl cellulose medium (Table 2, Fig. 2). For phosphate solubilisation index 1.93 value scored by T. asperellum NAIMCC-F-03167 while 1.39 was recorded by H. nigricans NAIMCC-F-03168 (Table 2, Fig. 3).
Fig. 1.
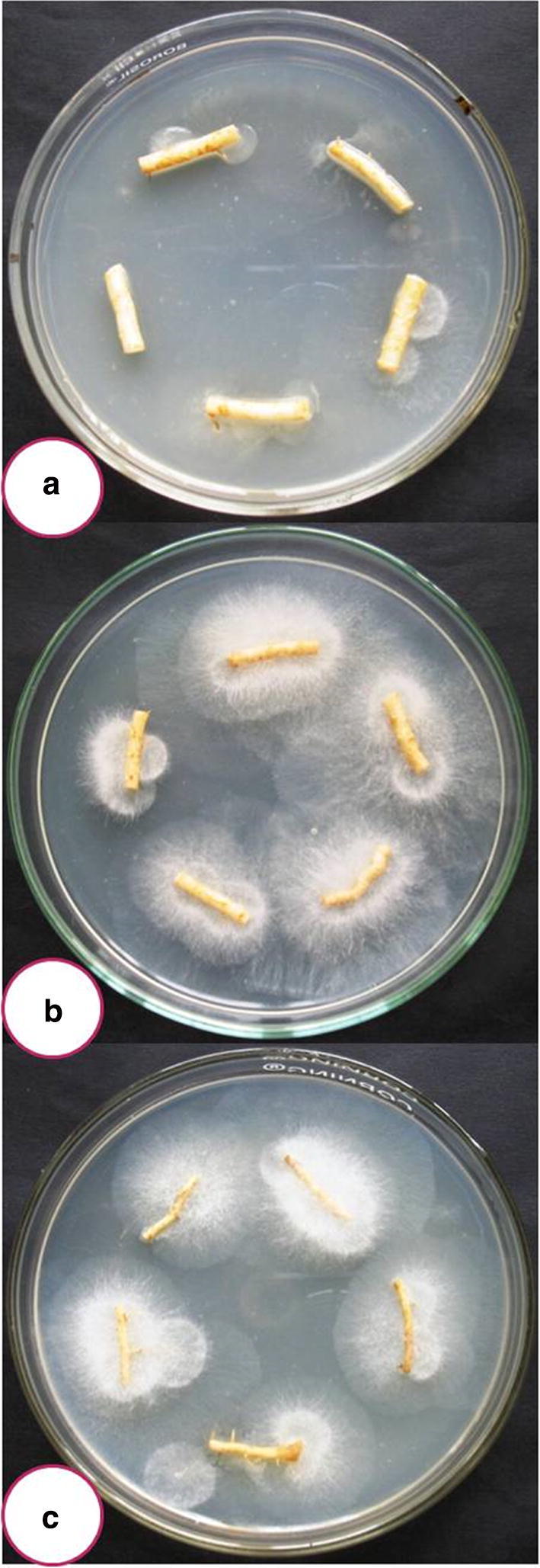
Competitive saprophytic ability of both bioagents with the roots of bottle gourd after 21 days of inoculation. a non-inoculated, b Inoculated with T. asperellum NAIMCC-F-03167, c Inoculated with H. nigricans NAIMCC-F-03168
Table 2.
Qualitative analysis of cellulose activity and phosphate solubilisation by T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168
| Isolates | Cellulase activity | Phosphate Solubilisation | Phosphate solubilisation index | |
|---|---|---|---|---|
| Diameter of clear zone (mm) | Diameter of colony growth (mm) | Diameter of halo zone (mm) | ||
| T. asperellum NAIMCC-F-03167 | 56.33 ± 2.88 | 9.00 ± 3.46 | 17.33 ± 6.02 | 1.93 |
| H. nigricans NAIMCC-F-03168 | 51.0 ± 2.64 | 8.33 ± 1.15 | 11.67 ± 2.88 | 1.39 |
± Standard error of mean
Fig. 2.
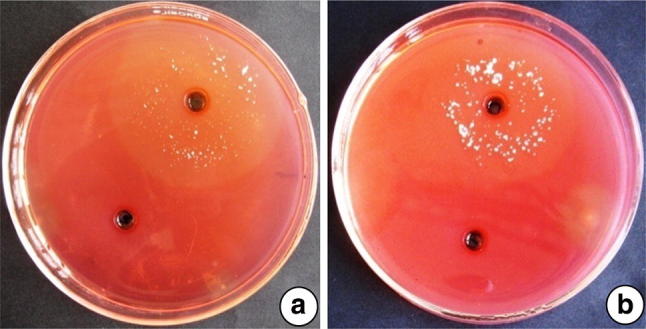
Cellulase activities of tested isolates in CMC amended medium indicating positive reaction by hallo zone formation aT. asperellum NAIMCC-F-03167, bH. nigricans NAIMCC-F-03168
Fig. 3.
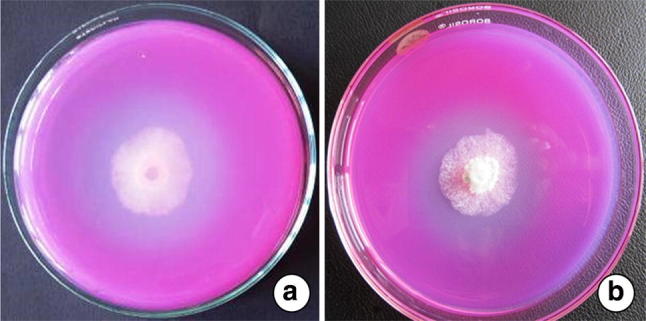
Phosphate solubilisation activities of the tested isolates in the PVK medium forming halo zone indicating positive reaction aT. asperellum NAIMCC-F-03167, bH. nigricans NAIMCC-F-03168
Trichoderma species comprise a well known cosmopolitan soilborne fungal bioagent naturalised across ecosystems through acquiring broad spectrum environmental adoptability. They are fast growing and colonize in their habitats by utilization of the available substrates, secreting antimicrobial metabolites besides extracellular enzymes as reported in other strains of Trichoderma species [29, 30]. These advantages associated with isolates of Trichoderma encouraged researchers to utilise the microbe in agriculture for the management of soilborne diseases, plant growth promotion and bioremediation of waste management. Several species of Trichoderma likewise, T. viride, T. harzianum, T. atroviride, T. virens etc. showed strong mycoparasitic and antagonistic ability against several soilborne phytopathogenic fungi, viz., Fusarium, Rhizoctonia, Sclerotium, Pythium, Phytophthora and Botrytis [31–33]. A report indicated that T. harzianum was found an effective bioagent which has ability to recycling and bioconservation of solid waste [34].
Morphological Characteristics of T. asperellum and H. nigricans
Both isolates are very much frequent and readily isolated from the rhizosphere of the organic carbon rich acid soils of Jharkhand. As per morphological characteristics, both isolates of T. asperellum and H. nigricans are compact, light–dark green, fast growing, and covering 90 mm within 72–96 h. Both strains are initially whitish green which later turn dark green in colour with high sporulation patterns were observed in both isolates of T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168.
Molecular Identification of T. asperellum and H. nigricans
18S rRNA gene sequencing of both T. asperellum NAIMCC-F-03167 (KM 438015) and H. nigricans NAIMCC-F-03168 (KJ910335) internal transcribed spacer 1, partial sequence; 5.8S ribosomal RNA gene and internal transcribed spacer 2, complete sequence; and 28S ribosomal RNA gene, partial sequence. Homology analysis of both fungal bioagents which have heavy metal scavenging activity with the blastn program, GenBank, NCBI revealed that T. asperellum KM 438015 showed maximum homology with maximum identity 99%) that’s why its identified as T. asperellum while H. nigricans KJ910335 showed maximum homology with Maximum identity 100%) that’s why its identified as H. nigricans KJ910335. 28 S rRNA gene nucleotide sequences of these bioagents were submitted to Gene Bank database under accession number KM 438015 (T. asperellum) and KJ910335 (H. nigricans). The bold letter indicates the phylogenic position of T. asperellum (KM 438015) and H. nigricans (KJ910335). They phylogenetically belong to ancestral of the mycoparasitic genus Trichoderma (Teleomorph Hypocrea, Ascomycota, Dikarya) and species asperellum (T. asperellum, KM 438015) while H. nigricans KJ910335 phylogenetically belongs to ancestral of the genus Hypocrea and species nigricans (H. nigricans KJ910335). Neighbor-joining phylogenetic tree of the identified fungal isolates and their gene accession number T. asperellum KM 438015) and H. nigricans (KJ910335) based on 18S rDNA sequences (Fig. 4).
Fig. 4.
Neighbor-joining phylogenetic tree of the identified fungal isolates and their gene accession number with T. asperellum (KM438015) and H. nigricans (KJ910335) based on 18S rDNA sequences. The bold letters indicates the phylogenic position of T. asperellum (KM 438015) and H. nigricans (KJ910335)
The ribosomal DNA genes (rDNA) possess characterstics that are suitable for identification of fungal isolates at species level. rDNA are highly stable and exhibit a mosaic of conserved and diverse regions within the genome [35]. They also occure in multiple copies up to 200 copies per haploid genome, arranged in tandem repeats with each repeat consisting of 18S small subunit (SSU), and 5.8S and 28S large subunit (LSU) genes [36]. Internal transcribed spacer (ITS) regions have been successfully used to generate specific primers capable of differentiating closely related fungal species [37]. In the present study, the test fungal strains isolated from the soil samples were subjected to molecular analysis for identification. ITS region of rDNA were amplified using genus specific ITS 1 and ITS 4 primers. When the 18S rDNA sequences (838 and 565 bp) of these isolates were compared using the ‘Blast’ analysis tool on NCBI, the samples were found to be T. asperellum KM438015 and H. nigricans KJ910335 with 99% and 100% sequences similarity respectively (Table 3). The ITS sequence was submitted to the NCBI GenBank, USA. From the sequence aligment obserevd between isolates and homologus sequences, phylogenetic tree was constructed based on maximum likelihood algorithm with Kimura 2-parameter method of analysis see the Fig. 4 [38]. In conclusion, above results indicate both isolates NAIMCC-F-03167 and NAIMCC-F-03168 were identified as T. asperellum and H. nigricans respectively on the basis of similar 18S rDNA based sequences database of NCBI GenBank and phylogenetic analysis through BLAST and registered in NCBI as NCBI-KM438015 and NCBI-KJ910335.
Table 3.
Listed NCBI GenBank accession numbers of T. asperellum and H. nigricans strains were used for homology and phylogenetic analysis of the isolated bioagents
| GenBank accession no. | Identified as | Country of origin | Identity (%) |
|---|---|---|---|
| KF804099 | Trichoderma asperellum strain SBTT-076 | India | 99 |
| JN128894 | Trichoderma asperellum strain T42 | India | 99 |
| KJ432865 | Trichoderma asperellum isolate SI14 | Pakistan | 99 |
| KF766525 | Trichoderma asperellum strain SBTT-028 | India | 99 |
| KC898194 | Trichoderma asperellum strain ASP5 | Egypt | 99 |
| KC561056 | Trichoderma asperellum strain CEN162 | Brazil | 99 |
| KC561057 | Trichoderma asperellum strain CEN201 | Brazil | 99 |
| EU077227 | Trichoderma asperellum strain T34 | Spain | 99 |
| AY857217 | Trichoderma asperellum strain TUB F-755 | Austria | 99 |
| AF278788 | Trichoderma asperellum strain 433.97 | Austria | 99 |
| GU198318 | Trichoderma asperellum strain GJS 05-328 | USA | 99 |
| GU198303 | Trichoderma asperellum strain GJS 05-199 | USA | 99 |
| GU198304 | Trichoderma asperellum strain GJS 06-292 | USA | 99 |
| EU264001 | Trichoderma asperellum strain GJS 05-328 | USA | 99 |
| JN941680 | Hypocrea nigricans strain NBRC 31285 | Japan | 100 |
| JN941679 | Hypocrea nigricans strain NBRC 31286 | Japan | 100 |
| JN941678 | Hypocrea nigricans strain NBRC 31289 | Japan | 100 |
| JN941681 | Hypocrea nigricans strain NBRC 30611 | Japan | 100 |
| JX242484 | Hypocrea atroviridis strain SMF-H08 | China | 100 |
| JQ806366 | Hypocrea lixii | Sweden | 100 |
| JN941682 | Hypocrea muroiana strain NBRC 31293 | Japan | 100 |
| EU722404 | Hypocrea koningii strain JH | Korea | 100 |
| AF548103 | Hypocrea jecorina strain RutC-30 | Sweden | 100 |
| AF548102 | Hypocrea jecorina strain QM 9414 | Sweden | 100 |
| AB027338 | Hypocrea lutea | Japan | 100 |
| FJ716243 | Hypocrea spp. | Ireland | 100 |
Effect of Heavy Metal and Scavenging Potential of T. asperellum and H. nigricans
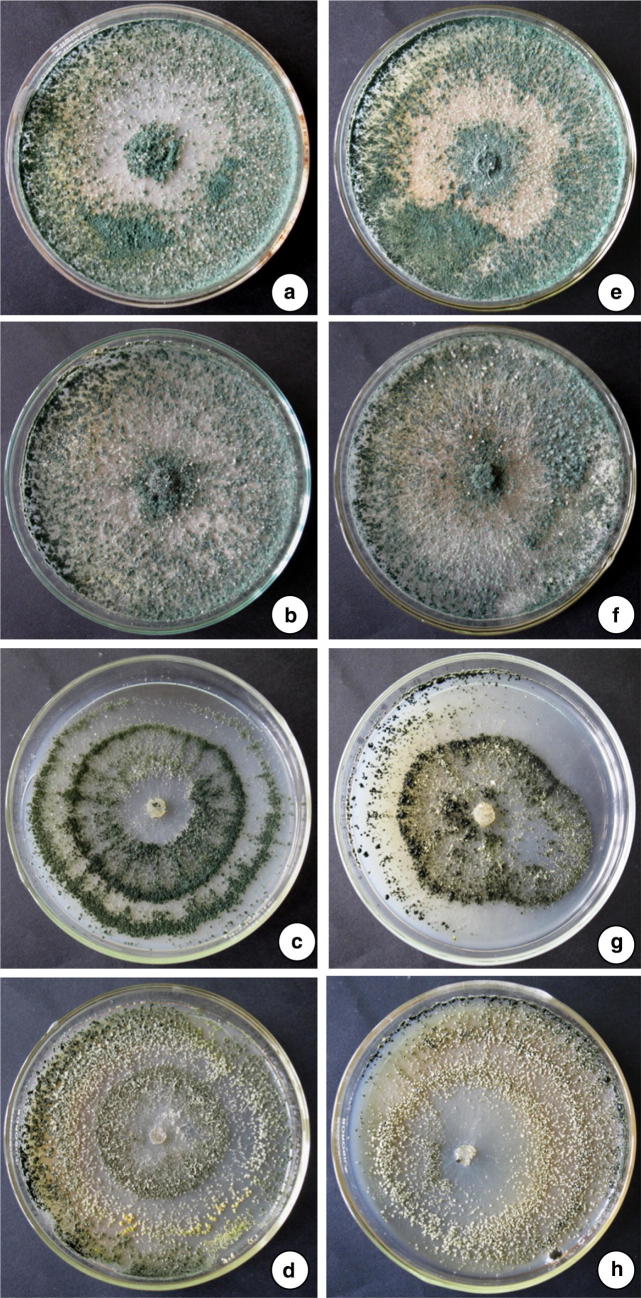
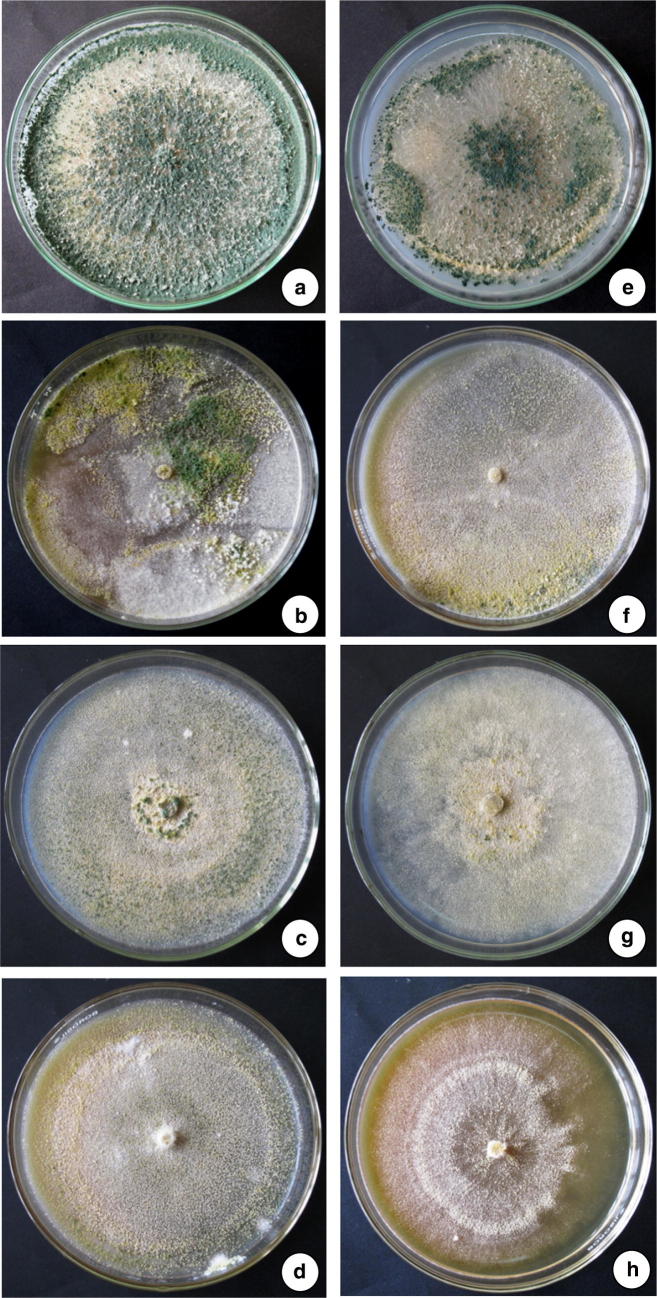
Toxicological effect of heavy metals (Lead and Cadmium) on the biomass accumulation and sporulation of T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 were also recorded during the experimentation. Cd and Pb caused negative effect on biomass accumulation and sporulation at higher concentration. Cd and Pb (20 and 30 µg/ml) was showed negative effect on sporulation (Fig. 5, 6) while biomass accumulation was least affected (Table 4). In the experiment conducted for studying the heavy metals scavenging potential of T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 in heavy metal amended broth medium, it was observed that both the isolates have efficacy in scavenging with varied potential. T. asperellum showed highest scavenging efficacy which scavenged 88.9% Cd while H. nigricans scavenged 87.2% Cd from 30 µg/ml concentrated medium after 21 days of inoculation. In 20 µg/ml concentrated medium, T. asperellum scavenged 83.3% and H. nigricans scavenged 82.3 Cd. However, maximum scavenging potential of lead was observed in T. asperellum (88%) followed by H. nigricans (81.30%) from 30 µg/ml concentrated medium after 21 days of inoculation. After 14 days of inoculation, 81.0% Pb was scavenged by T. asperellum and 80.3% Pb by H. nigricans (Table 5).
Fig. 5.
Effect of heavy metals on sporulation of T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 in Pb amended medium, Column 1. T. asperellum, a Control, b 10 µg/ml, c 20 µg/ml, d 30 µg/ml, Column 2. H. nigricans, e Control, f 10 µg/ml, g 20 µg/ml, h 30 µg/ml
Fig. 6.
Effect of heavy metals on sporulation of T. asperellum and H. nigricans in Cd amended medium, Column 1. T. asperellum NAIMCC-F-03167, a Control, b 10 µg/ml, c 20 µg/ml, d 30 µg/ml, Column 2. H. nigricans NAIMCC-F-03168, e Control, f 10 µg/ml, g 20 µg/ml, h 30 µg/ml
Table 4.
Biomass accumulation of T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 under heavy metal stress condition in broth medium
| Isolates | Treatment | Dry weight of fungal mycelium (gm) | |||||
|---|---|---|---|---|---|---|---|
| Cadmium | Lead | ||||||
| 7 days | 14 days | 21 days | 7 days | 14 days | 21 days | ||
| T. asperellum NAIMCC-F-03167 | CONTROL | 1.793 | 1.86 | 1.948 | 1.722 | 1.889 | 1.914 |
| 10 µg | 1.654 | 2.325 | 1.806 | 2.104 | 1.919 | 1.825 | |
| 20 µg | 1.548 | 2.012 | 1.662 | 1.908 | 1.83 | 1.842 | |
| 30 µg | 1.555 | 1.983 | 1.509 | 1.544 | 1.795 | 1.676 | |
| H. nigricans NAIMCC-F-03168 | CONTROL | 1.905 | 2.175 | 1.883 | 1.745 | 2.278 | 1.942 |
| 10 µg | 1.492 | 1.87 | 1.661 | 1.601 | 2.683 | 1.882 | |
| 20 µg | 1.559 | 2.422 | 1.59 | 1.841 | 2.127 | 1.865 | |
| 30 µg | 1.494 | 2.016 | 1.509 | 1.537 | 2.113 | 1.772 | |
Table 5.
Heavy metal scavenging potential of T. asperellum NAIMCC-F-03167 and H. nigricans NAIMCC-F-03168 from the heavy metal amended broth medium
| Isolates | Weekly scavenging potential of heavy metals from the broth medium | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Initial concentration (µg) | Cadmium (µg) | Lead (µg) | |||||||
| 7 days | 14 days | 21 days | % Cd scavenging after 21 days | 7 days | 14 days | 21 days | % Pb scavenging after 21 days | ||
| T. asperellum NAIMCC-F-03167 | 10 | 5.32 | 6.37 | 7.01 | 70.1 | 2.8 | 6.33 | 7.02 | 70.2 |
| 20 | 14.27 | 15.63 | 16.66 | 83.3 | 7.85 | 15.94 | 16.06 | 80.3 | |
| 30 | 23.95 | 25.54 | 26.66 | 88.9 | 2.84 | 23.38 | 24.39 | 81.3 | |
| H. nigricans NAIMCC-F-03168 | 10 | 4.91 | 6.24 | 6.97 | 69.7 | 1.36 | 4.98 | 5.52 | 55.2 |
| 20 | 14.22 | 15.54 | 16.46 | 82.3 | 2.29 | 13.38 | 16.19 | 81.0 | |
| 30 | 23.95 | 25.29 | 26.16 | 87.2 | 7.83 | 13.51 | 26.44 | 88.1 | |
| SD | 8.43 | 8.55 | 8.69 | 2.90 | 6.72 | 8.60 | |||
| SEM | 3.44 | 3.49 | 3.55 | 1.18 | 2.74 | 3.51 | |||
Report indicated that use of soil microbes in scavenging heavy metals from the contaminated soil as well as they also reported that the Pseudomonads have proven as potential bio-adsorption which has potential to adsorb and mobilize heavy metals from the polluted effluents. They also used A. niger (M3) for bioabsorption of Nickel from the nickel polluted effluents [17]. Paranthaman and Karthikeyen [39] successfully used Pseudomonads for the bioremediation of heavy metals from the effluents of paper mill. Besides bacterial microflora, several fungal species also reported to play important role in scavenging heavy metals [40, 41]. Moreover, Mcaskie et al. [42] reported that Citrobactor spp. secretes inorganic metabolic products such as sulphide, carbonate or phosphate ions in their respiratory metabolism which precipitate toxic metals (Cd) in a form of non-enzymatic detoxification. Fouvert and Roux [43] also studied the mycelial mat of Mucor niceri, A. niger and P. chrysogenum and reported their role in removal of nickel, zinc, cadmium and lead by bio-adsorption. Anand et al. [44] also reported that T. viride scavenge Cu from the medium by binding with cell wall. Ng et al. [45, 46] reported that the T. reesei has synergistic efficacy on the cellulases activity and adsorption of heavy metals. They also concluded that it has also capacity to remove toxic Cr(VI) from the aqueous solutions and extend its role in utilization of tea waste. Several reports also stressed the role of research in finding ecofriendly technologies to remove heavy metals from the aquatic system [47, 48]. Mohan et al. [49] reported that the bio-absorbents (microbial inoculants) have been considered as the cheapest, most abundant and eco-friendly options for removal of heavy metals from the aquatic system. Besides beneficial effects in bio-adsorption of heavy metals, it also caused several negative impacts on microbial growth and development. Pb and Cd showed negatively effect on mycelial growth and sporulation which inhibited more than > 30% on their growth and sporulation. As per results and literature reviewed and surveyed biocide market of Eastern region of India indicated that there are no native formulations available in the market which have multifaceted mode of action viz., biocontrol potential, scavenging potential of heavy metals, phosphate solubilisation and plant growth promotion activity. So, both these novel fungal organisms viz., T. asperellum and H. nigricans can be exploited for the management of soilborne phytopathogens, plant growth promotion, sustainable management of phosphorus deficiency and management of heavy metal toxicity (especially Cadmium and Lead affected soils) on wider areas to nullified the toxic effect of these metals on plants, animals and human beings.
Acknowledgements
The authors are grateful to Dr. B. P. Bhatt, Director, ICAR-Research Complex for Eastern Region, Patna, India, for providing facility for doing this research is duly acknowledged. Authors are also thankful to Dr. S. B. Choudhary, NBPGR, Ranchi, Jharkhand for critical review of the manuscript.
Funding
Funding was provided by Indian Council of Agricultural Research, New Delhi, India.
References
- 1.Kristanti RA, Hadibarata T, Toyama T, Tanaka Y, Kazuhiro M. Bioremediation of crude oil by white rot fungi Polyporus sp. S133. J Microbiol Biotechnol. 2011;21:995–1000. doi: 10.4014/jmb.1105.05047. [DOI] [PubMed] [Google Scholar]
- 2.Sarma H. Metal hyper accumulation in plants: a review focusing on phytoremediation technology. J Environ Sci Technol. 2011;4:118–138. doi: 10.3923/jest.2011.118.138. [DOI] [Google Scholar]
- 3.Singh R, Gautam N, Mishra A, Gupta R. Heavy metals and living system: an overview. Indian J Pharmacol. 2011;43(3):246–253. doi: 10.4103/0253-7613.81505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alvarez-Ayuso E. Cadmium in soil-plant systems: an overview. Int J Environ Pollut. 2008;33:275–291. doi: 10.1504/IJEP.2008.019399. [DOI] [Google Scholar]
- 5.Jain S, Bhatt A. Molecular and In situ characterization of cadmium-resistant diversified extremophilic strains of Pseudomonas for their bioremediation potential. Biotech. 2014;4:297–304. doi: 10.1007/s13205-013-0155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sethy SK, Ghosh S. Effect of heavy metals on germination of seeds. J Nat Sci Biol Med. 2013;4:272–275. doi: 10.4103/0976-9668.116964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaltauskaite J, Sodiene I. Effects of total cadmium and lead concentrations in soil on the growth, reproduction and survival of earthworm Eisenia fetida. Ekologija. 2010;56:10–16. doi: 10.2478/v10055-010-0002-z. [DOI] [Google Scholar]
- 8.Ogundele DT, Adio AA, Oludele OE. Heavy metal concentrations in plants and soil along heavy traffic roads in North Central Nigeria. J Environ Anal Toxicol. 2015;5:334. [Google Scholar]
- 9.Krishnaveni M, Kumar JS, Shravanan PS. Influence of Lead on biochemicals and proline contents of Vigna unguiculata (L.) Walp. Int J Plant Sci. 2015;10:142–151. doi: 10.15740/HAS/IJPS/10.2/142-151. [DOI] [Google Scholar]
- 10.Srinivasan M, Sahi SV, Paulo JCF, Venkatachalam P. Lead heavy metal toxicity induced changes on growth and antioxidative enzymes level in water hyacinths [Eichhornia crassipes (Mart.)] Bot Stud. 2016;55:54. doi: 10.1186/s40529-014-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma P, Dubey RS. Lead toxicity in plants. Braz J Plant Physiol. 2005;17:35–52. doi: 10.1590/S1677-04202005000100004. [DOI] [Google Scholar]
- 12.Verma S, Dubey RS. Lead toxicity induces lipid peroxidation and alters the activities of antioxidants enzymes in growing rice plants. Plant Sci. 2003;164:645–655. doi: 10.1016/S0168-9452(03)00022-0. [DOI] [Google Scholar]
- 13.Aliu S, Gashi B, Rusinovci I, Fetahu S, Vataj R. Effects of some heavy metals in some morpho-physiological parameters in maize seedlings. Am J Biochem Biotech. 2013;9:27–33. doi: 10.3844/ajbbsp.2013.27.33. [DOI] [Google Scholar]
- 14.Ling T, Fangke Y, Jun R. Effect of Mercury to seed germination, coleoptile growth and root elongation of four vegetables. Res J Phytochem. 2010;4:225–233. doi: 10.3923/rjphyto.2010.225.233. [DOI] [Google Scholar]
- 15.Rajasulochana P, Preethy V. Comparison on efficiency of various techniques in treatment of waste and sewage water—a comprehensive review. Resour Eff Technol. 2016;2:175–184. [Google Scholar]
- 16.Mishra A, Malik A. Recent advances in microbial metal bioaccumulation. J Crit Rev Env Sci Tech. 2004;43:1162–1222. doi: 10.1080/10934529.2011.627044. [DOI] [Google Scholar]
- 17.Gupta A, Joia J, Sood A, Sood R, Sidhu C, Kaur G. Microbes as potential tool for remediation of heavy metals: a review. J Microb Biochem Technol. 2016;8:364–372. [Google Scholar]
- 18.Pilon-Smits E. Phytoremediation. Annu Rev Plant Biol. 2005;56:15–39. doi: 10.1146/annurev.arplant.56.032604.144214. [DOI] [PubMed] [Google Scholar]
- 19.Aken BV, Correa PA, Schnoor JL. Phytoremediation of polychlorinated biphenyls: new trends and promises. Environ Sci Technol. 2010;44:2767–2776. doi: 10.1021/es902514d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petrovic JJ, Danilovic G, Curcic N, Milinkovic M, Stosic N, Pankovic D, Raicevic Vera. Copper tolerance of Trichoderma species. Arch Biol Sci Belgrade. 2014;66:137–142. doi: 10.2298/ABS1401137J. [DOI] [Google Scholar]
- 21.Cramer RA, Byrne PF, Brick MA, Panella L, Wickliffe E, Schwartz HF Characterization of Fusarium oxysporum isolates from common bean and sugar beet using pathogenicity assays and random-amplified polymorphic DNA markers. J Phytopathol. 2003;151:352–360. doi: 10.1046/j.1439-0434.2003.00731.x. [DOI] [Google Scholar]
- 22.Borneman J, Hartin RJ. PCR primers that amplify fungal rRNA genes from environmental samples. Appl Environ Microbiol. 2000;66:4356–4360. doi: 10.1128/AEM.66.10.4356-4360.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biology Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent JM. Distortion of fungal hyphae in presence of certain inhibitors. Nature. 1947;159:850. doi: 10.1038/159850b0. [DOI] [PubMed] [Google Scholar]
- 27.Johnsen HR, Krause K. Cellulase activity screening using pure carboxymethyl cellulose: application to soluble cellulolytic samples and to plant tissue prints. Int J Mol Sci. 2014;15:830–838. doi: 10.3390/ijms15010830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gadd GM. Biosorption: critical review of scientific rationale, environmental importance and significance for pollution treatment. J Chem Technol Biotechnol. 2009;84:13–28. doi: 10.1002/jctb.1999. [DOI] [Google Scholar]
- 29.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 30.Harman GE. Overview of mechanisms and uses of Trichoderma sp. Phytopathol. 2006;96:190–194. doi: 10.1094/PHYTO-96-0190. [DOI] [PubMed] [Google Scholar]
- 31.Schuster A, Schmoll M. Biology and biotechnology of Trichoderma. Appl Microbiol Biotechnol. 2010;87:787–799. doi: 10.1007/s00253-010-2632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khaledi N, Taheri P. Biocontrol mechanisms of Trichoderma harzianum against soybean charcoal rot caused by Macrophomina phaseolina. J Plant Prot Res. 2016;56:21–31. doi: 10.1515/jppr-2016-0004. [DOI] [Google Scholar]
- 33.Guilger M, Pasquoto-Stigliani T, Bilesky-Jose N, Grillo R, Abhilash PC, Fernandes FL, de Lima R. Biogenic silver nanoparticles based on Trichoderma harzianum: synthesis, characterization, toxicity evaluation and biological activity. Sci Rep. 2017;7:44421. doi: 10.1038/srep44421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman A, Begum D, Rahman M, Bari MA, Ilias GNM, Alam MF. Isolation and identification of Trichoderma species and their use of bioconversion of solid waste. Turk J Biol. 2011;35:183–194. [Google Scholar]
- 35.Raja HA, Miller AN, Pearce CJ, Oberlies NH. Fungal identification using molecular tools: a primer for the natural products research community. J Nat Prod. 2017;80:756–770. doi: 10.1021/acs.jnatprod.6b01085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bruns TD, White TJ, Taylor JW. Fungal molecular systematic. Annu Rev Ecol Syst. 1991;22:525–564. doi: 10.1146/annurev.es.22.110191.002521. [DOI] [Google Scholar]
- 37.Freeman J, Ward E. Gaeumannomyces graminis, the take-all fungus and its relatives. Mol Plant Pathol. 2004;5:235–252. doi: 10.1111/j.1364-3703.2004.00226.x. [DOI] [PubMed] [Google Scholar]
- 38.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 39.Paranthaman SR, Karthikeyen B. Bioremediation of heavy metal in paper mill effluents using Pseudomonas spp. Int J Microbiol. 2015;1:1–5. [Google Scholar]
- 40.Chaturvedi AD, Pal D, Penta S, Kumar A. Ecotoxic heavy metals transformation by bacteria and fungi in aquatic ecosystem. World J Microbiol Biotechnol. 2015;31:1595–1603. doi: 10.1007/s11274-015-1911-5. [DOI] [PubMed] [Google Scholar]
- 41.Ayangbenro AS, Babalola OO. A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int J Environ Res Public Health. 2017;14:94. doi: 10.3390/ijerph14010094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naz N, Young HK, Ahmed N, Gadd GM. Cadmium accumulation and DNA homology with metal resistance genes in sulfate-reducing bacteria. Appl Environ Microbiol. 2005;71:4610–4618. doi: 10.1128/AEM.71.8.4610-4618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai C-X, Xu J, Deng N-F, Dong X-W, Tang H, Liang Y, Fan X-W, Li Y-Z. A novel approach of utilization of the fungal conidia biomass to remove heavy metals from the aqueous solution through immobilization. Sci Rep. 2016;6:36546. doi: 10.1038/srep36546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anand P, Isar J, Saran S, Saxena RK. Bioaccumulation of copper by Trichoderma viride. Bioresour Technol. 2006;97:1018–1025. doi: 10.1016/j.biortech.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 45.Ng IS, Tsai SW, Ju YM, Yu SM, Ho TH. Dynamic synergistic effect on Trichoderma reesei cellulases by novel beta-glucosidases from Taiwanese fungi. Bioresour Technol. 2011;102:6073–6081. doi: 10.1016/j.biortech.2010.12.110. [DOI] [PubMed] [Google Scholar]
- 46.Ng IS, Wu X, Yang X, Xie Y, Lu Y, Chen C. Synergistic effect of Trichoderma reesei cellulases on agricultural tea waste for adsorption of heavy metals Cr(VI) Bioresour Technol. 2013;145:297–301. doi: 10.1016/j.biortech.2013.01.105. [DOI] [PubMed] [Google Scholar]
- 47.Malaviya P, Singh A. Bioremediation of chromium solutions and chromium containing wastewaters. J Critical Rev Microbiol. 2014;42:607–633. doi: 10.3109/1040841X.2014.974501. [DOI] [PubMed] [Google Scholar]
- 48.Li H, Li Z, Liu T, Xiao X, Peng Z, Deng L. A novel technology for biosorption and recovery hexavalent chromium in wastewater by biofunctional magnetic beads. Bioresour Technol. 2008;99:6271–6279. doi: 10.1016/j.biortech.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Mohan D, Rajput S, Singh VK, Steele PH, Pittman CU., Jr Modelling and evaluation of chromium remediation from water using low cost bio-char, a green adsorbent. J Hazard Mater. 2010;188:319–333. doi: 10.1016/j.jhazmat.2011.01.127. [DOI] [PubMed] [Google Scholar]