Abstract
Objective
Liver metastasis, which contributes substantially to high mortality, is the most common recurrent mode of colon carcinoma. Thus, it is necessary to identify genes implicated in metastatic colonization of the liver in colon carcinoma.
Methods
We compared mRNA profiling in 18 normal colon mucosa (N), 20 primary tumors (T) and 19 liver metastases (M) samples from the dataset GSE49355 and GSE62321 of Gene Expression Omnibus (GEO) database. Gene ontology (GO) and pathways of the identified genes were analyzed. Co-expression network and protein-protein interaction (PPI) network were employed to identify the interaction relationship. Survival analyses based on The Cancer Genome Atlas (TCGA) database were used to further screening. Then, the candidate genes were validated by our data.
Results
We identified 22 specific genes related to liver metastasis and they were strongly associated with cell migration, adhesion, proliferation and immune response. Simultaneously, the results showed that C-X-C motif chemokine ligand 14 (CXCL14) might be a favorable prediction factor for survival of patients with colon carcinoma. Importantly, our validated data further suggested that lower CXCL14 represented poorer outcome and contributed to metastasis. Gene set enrichment analysis (GSEA) showed that CXCL14 was negatively related to the regulation of stem cell proliferation and epithelial to mesenchymal transition (EMT).
Conclusions
CXCL14 was identified as a crucial anti-metastasis regulator of colon carcinoma for the first time, and might provide novel therapeutic strategies for colon carcinoma patients to improve prognosis and prevent metastasis.
Keywords: Colon carcinoma, liver metastasis, mRNA profiling, functions annotation
Introduction
Colon carcinoma is one of the most common malignant diseases with 945,000 new cases every year and is the fourth cause of cancer-related deaths worldwide (1). Unfortunately, about 70% of colon carcinoma patients develop liver metastases. Curative-intent resections can be performed in only 10%−15% of liver metastases (2). In the majority of metastatic patients, the standard treatment remains palliative chemotherapy. However, most colon cancer patients with active metastasis appear to be resistant, or even non-responsive, to current treatments. A major clinical challenge is to explore possible therapeutic targets that are specifically expressed in liver metastatic settings.
There have been many attempts to determine predictive factors or explain the underlying mechanisms for distant metastasis. MicroRNA 34a, microRNA-34a-5p, microRNA-340 are associated with colon carcinoma cell proliferation and metastasis (3,4). In addition, the CpG island methylator phenotype (CIMP) is concordant between primary colon carcinoma and distant metastases (5). Mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K) signaling pathways inhibit metastasis to the liver (6). Alterations in gene expression, protein expression, posttranslational modification, microRNA and linc-RNA have been reported to act a part of role in tumor progression. However, these have not revealed effective predicted factor which is specific to liver metastasis. Transcriptomic changes inherit from genomic information and take place before protein level. Therefore, we attempt to investigate the malignant features of hepatic metastasis microenvironment by RNA-sequencing.
Gene expression profiling has become a strategy to identify genes involved in the progression and the prognosis of different cancers. Few attentions were focused on the gene signatures associated with metastatic disease (7). Two studies presented gene signatures associated with metastatic disease containing more than 400 genes. Such long lists of genes are difficult to be used for the development of new therapies (8,9). Pairs of primary and metastatic tumors were analyzed and the samples clustered by patients but not the tissue origin (10,11). The identified genes are specific to colon carcinoma and hepatic metastases, but the precise target is still unknown (12). Comparative profiling of primary colon carcinomas and liver metastases identifies lymphoid enhancer factor-1 (LEF1) as a prognostic biomarker (13). However, this research only focused on the development of diagnostic and prognostic markers without trying to identify gene signatures able to distinguish metastatic from primary cancer tissues (13). Therefore, it is most important for us to investigate effective targets for the treatment of liver metastasis.
To identify genes implicated in metastatic colonization of the liver in colon carcinoma, we compared mRNA expression between groups of normal colon mucosa (N), primary tumors (T) and liver metastases (M) samples which from Gene Expression Omnibus (GEO) database. The expression of the differential genes was processed by gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) orthology and Signal network, which are all effective bioinformatics analytical methods. We then verified the clinical significance of identified genes using clinical samples. Our data provide novel information and help further understanding of the liver metastasis cascade of colon carcinoma.
Materials and methods
Microarray data
The transcriptional expression data (GSE49355 and GSE62321) of human colon tumor were downloaded from the GEO database. They were from the same set of patients. It contained 18 normal colon mucosa (N), 20 primary tumors (T) and 19 liver metastases (M) samples. Platforms information were GPL96 [HG-U133A] and GPL97 [HG-U133B] Affymetrix Human Genome U133A/B Array and the datasets were already normalized.
Investigating of differential expression genes (DEGs)
Genes were standardized and interpreted functionally before comparison. Using random variance model (RVM) t-test (14) and the normal colon mucosa group as the control group, the P value and the false discovery rate (FDR) were calculated for each DEG. FDR was calculated to correct the P-value, which controls type I errors. With a threshold of P<0.05, FDR<0.05 and fold change (FC) >2, DEGs were picked out.
Hierarchical cluster analysis
Hierarchical cluster analysis was performed to ensure good characterizations of screened DEGs between different groups (15). In hierarchical cluster analysis, Pearson correlation was used to calculate the correlation between the genes and samples.
Venn analysis
To identify specific genes of liver metastasis, genes expression in each tissue were input to the web tool Venn Diagrams (http://bioinformatics.psb.ugent.be/webtools/Venn).
GO annotation analysis
Functional analysis of differentially expressed genes was carried out by the GO project (http://www.geneontology.org) on the basis of biological process (16).
Pathway annotation analysis
Pathway analysis was used to identify significant pathways involving DEGs, according to KEGG, BioCarta, and Reactome.
Co-expression network analysis
For each pair of genes, the Pearson correlation coefficient was calculated, and 0.8 was defined as the threshold to construct the network. Within the network analysis, degree of the association is an important factor to determine the relative importance of a gene. We have employed different colors and sizes of node to discriminate the degree of the associations for one gene with the surrounding nodes. The co-expression networks were constructed by Cytoscape (17).
PPI network construction
In order to reveal functional associations between proteins in a genome-wide scale, STRING online tool (18,19) was used to construct a PPI network. In the PPI network, each node represents a protein, and each edge represents an interaction of pairwise proteins. The nodes with a relatively large number of edges were defined as hub proteins.
Gene set enrichment analysis (GSEA)
GSEA was performed by the GSEA software and gene sets used in this work were downloaded from the Molecular Signatures Database. The MSigDB collects various types of gene set and the online pathway database included 1,320 Canonical pathways derived from the pathway databases of BioCarta, KEGG, PID, Reactome and others databases. The data for GSEA analysis is from The Cancer Genome Atlas (TCGA).
TCGA database analysis
TGCA database was derived from UCSC Cancer Browser (https://genome-cancer.ucsc.edu). Overall survival (OS) analysis of colon cancer patients with high and low levels of different genes was shown by using a Kaplan-Meier survival plot. The cut-off values for the genes were the median respectively. We used Kaplan-Meier curves to present the prognosis of the high and low groups. The Wilcoxon log-rank test was then conducted on the Kaplan-Meier curves to detect the survival difference between these two groups. All survival analysis was conducted using the R software.
Clinical specimens
Specimens were from colon carcinoma patients who were diagnosed and received operation in the Department of Anus and Intestine Surgery of the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China) from 2011 to 2013. Pre- and post-operative clinical data and other survival-related data were perfected by reviewing the medical records and following-up the patients by telephone. All postoperative specimens were examined by one pathologist and reviewed by another pathologist. Of them, all patients were used as the basis of the present study. The clinical data of the patients are shown in Table 1. Collection of samples in this study was approved by Institutional Ethics Committee of the First Affiliated Hospital of Zhengzhou University (Ethics approval number: Science-2010-LW-1213), and informed consent was obtained from each patient with available follow-up information.
1.
Characteristics of patients with colon carcinoma
| Characteristics | No. of cases | % |
| Gender | ||
| Male | 66 | 64.1 |
| Female | 37 | 35.9 |
| Age (year) | ||
| <60 | 56 | 54.4 |
| ≥60 | 47 | 45.6 |
| Tumor size (cm) | ||
| <4 | 64 | 62.1 |
| ≥4 | 39 | 37.9 |
| Pathological type | ||
| Adenocarcinoma | 88 | 85.4 |
| Others | 15 | 14.6 |
| Lymph node metastasis | ||
| Yes | 67 | 65.0 |
| No | 36 | 35.0 |
| Liver metastasis | ||
| Yes | 86 | 83.5 |
| No | 17 | 16.5 |
| TNM stage | ||
| I | 26 | 25.2 |
| II | 39 | 37.9 |
| III | 22 | 21.4 |
| IV | 16 | 15.5 |
| Histological differentiation | ||
| Low | 16 | 15.5 |
| Low-moderate | 14 | 13.6 |
| Moderate | 73 | 70.9 |
Quantitative real-time polymerase chain reaction (qRT-PCR)
Tumor or marginal tissues were cut into 20 mm of pieces and mechanically grinded. Then, total RNA was extracted using Trizol solution (Invitrogen, Waltham, MA, USA). qRT-PCR was performed using specific primers and SYBR Green qPCR Master Mix (Takara, Japan). Listed primers were used: 5’-GGAGCCAAAAGGGTCATCATCTC-3’ sense primer and 5’-GAGGGGCCATCCACAGTCTTCT-3’ antisense primer for GAPDH, 5’- CGCTACAGCGACGTGAAGAA-3’ sense primer and 5’-GTTCCAGGCGTTGTACCAC-3’ antisense primer for CXC chemokine ligand 14 (CXCL14). GAPDH was used as an internal control. With the 2-ΔΔCt method, we compared the expression level of clinical samples (20). For each sample, the expression of CXCL14 as well as GAPDH was examined, the relative expression of CXCL14 was calculated by using the 2-ΔCt value of CXCL14 dividing the 2-ΔCt value of GAPDH (20).
Immunohistochemistry
Paraffin-embedded tissues of 45 colon cancer samples were examined for the expression of CXCl14 protein (Abcam, Cambridge, UK; 1:200). Sections were treated with 3% H2O2 and 5% bull serum albumin (BSA) and incubated with primary antibodies overnight at 4 °C. After incubation with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at 37 °C, sections were washed and counterstained with hematoxylin, and visualized under a microscope (Olympus, Shinjuku, Japan) (21).
Statistical analysis
Clinicopathologic factors were compared by using the χ2 test and continuous variables were compared by using the Student t test or one-way analysis of variance (ANOVA) analysis. Kaplan-Meier analysis and the log-rank test were used for survival analysis. Univariate and multivariate logistic regression models identified the association between CXCL14 expression and clinical characteristics. P<0.05 was considered statistically difference. All statistics associated with clinical samples were performed using Prism 7 (GraphPad Software Inc., La Jolla, USA). Statistical analysis of significance was calculated by ANOVA followed by Tukey’spost hoc test with SPSS 16.0 for Windows (SPSS Inc., Chicago, IL, USA). The bioinformatics analysis was used by using R software (Version 3.4; R Foundation for Statistical Computing, Vienna, Austria).
Results
Gene expression analysis
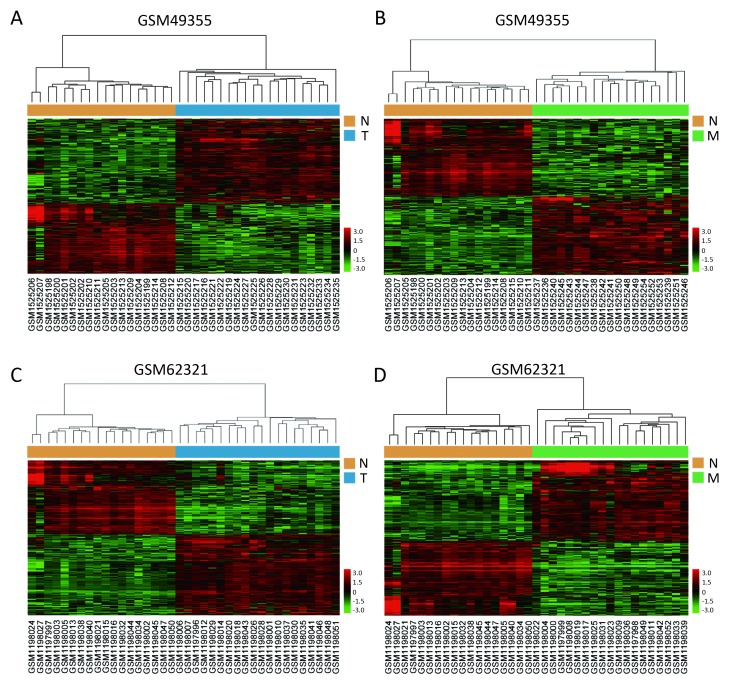
We used the public transcriptome sequencing dataset (GSE49355 and GSE62321) from GEO database, including 18 normal colon mucosa (N), 20 primary tumors (T) and 19 liver metastases (M) samples. Detailed sample information could be found in Supplementary Table S1. Expression profiling of the 57 samples was conducted on Affymetrix human U133A/B chips. Expression profiling of the 57 samples was conducted on Affymetrix human U133A chips containing 22,200 probes corresponding to about 12,700 genes. These gene expression data have been performed normalization and log2 transformation. Hierarchical cluster analysis showed that normal samples clustered together and were relatively well separated from T and M samples in GSE49355 and GSE62321 (Figures 1A−D).
S1.
| Samples | Normal_colon | Primary_tumor | Liver_metastasis |
| 016_MV | Yes | Yes | Yes |
| 003_JCP | No | No | Yes |
| 005_JME | No | No | Yes |
| 022_JB | Yes | Yes | No |
| 026_SA | Yes | No | No |
| 044_MB | Yes | Yes | Yes |
| 045_JC | No | Yes | No |
| 050_NC1B | No | No | Yes |
| 056_MC | No | Yes | Yes |
| 059_MT | Yes | Yes | Yes |
| 061_CM | Yes | Yes | No |
| 073_PD | Yes | Yes | Yes |
| 094_AM | No | Yes | Yes |
| 089_NC | Yes | No | Yes |
| 109_JC | No | No | Yes |
| 115_CB | Yes | Yes | Yes |
| 119_PM | Yes | Yes | No |
| 130_YL | No | Yes | No |
| 149_JGI | Yes | Yes | Yes |
| 179_AB | No | No | Yes |
| 189_JR | Yes | Yes | No |
| 196_TD | Yes | Yes | Yes |
| 213_RG | Yes | Yes | Yes |
| 222_PEC | Yes | Yes | Yes |
| 227_SS | Yes | Yes | No |
| 234_YC | Yes | No | No |
| 223_GB | Yes | Yes | Yes |
| 244_FP | No | Yes | Yes |
1.
Expression differences of genes in normal colon mucosa (N), primary tumors (T) and liver metastases (M) samples. Hierarchical clustering analysis of differentially expressed genes in 20 T samples vs. 18 N samples and 19 M samples vs. 18 N samples from GSE49355 (A, B) and GSE62321 (C, D).
Identification of specific gene signatures
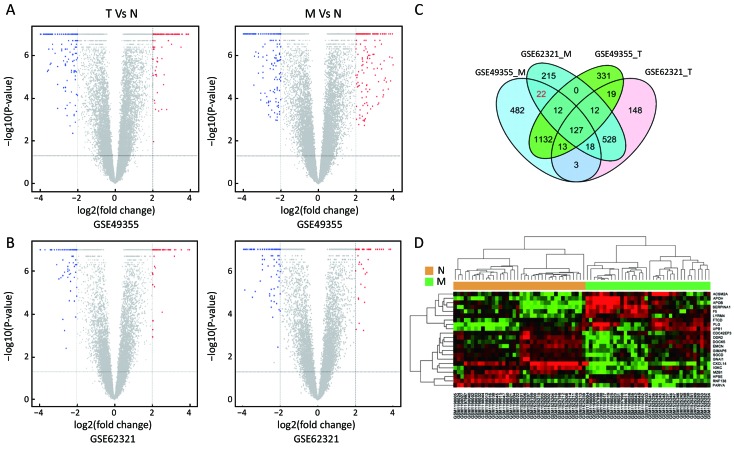
To identify molecular signatures that regulate distant metastasis in colon carcinoma, we compared mRNA expression levels in T vs. N and M vs. N. After analyzing the transcriptomic changes of T vs. N, a total of 1,646 DEGs including 861 up-regulated and 785 down-regulated transcription factors were screened out from GSE49355, and a total of 868 DEGs including 477 up-regulated and 391 down-regulated transcription factors were also identified in GSE62321. Of 1,809 DEGs, 869 were down-regulated and 940 overexpressed in M vs. N in GSE49355. The volcano plot of DEGs distribution was also presented 934 DEGs including 468 up-regulated and 466 down-regulated when comparing M with N in GSE62321 (Figure 2A, B) (P<0.05, FDR<0.05, FC>2, respectively). Based on the fact that the GSE49355 and GSE62321 were from the same panel of patients but different platform, union analysis was first performed and 719 specific genes related to liver metastasis of colon carcinoma were identified (Supplementary Table S2). However, taken into account that some of the 719 genes might be due to a single platform error, we took the intersection analysis here for obtaining higher accurate genes. The results showed that 179 genes might play an important role in the metastasis of cancer and were altered in M vs. N. Excluding 157 genes associated with tumor development, 22 genes were specific for liver metastasis (Figure 2C). Subsequently, unsupervised hierarchical cluster analysis was performed on selected 22 genes expression data using Pearson correlation-based distance and average clustering. Considerable patients’ non-pairing of N and M samples was observed in the dendrogram. Most of the specific genes showed a significantly differential expression between N and M samples (Figure 2D). Details were shown in Supplementary Table S3.
2.
Identification of specific genes associated with liver metastasis in colon carcinoma. (A, B) With a threshold of P<0.05, false discovery rate (FDR)<0.05 and fold change >2, differential expression genes (DEGs) were picked out by volcano plot when comparing 20 primary tumors (T) samples with 18 normal colon mucosa (N) samples and 19 liver metastases (M) samples with 18 N samples from GSE49355 and GSE62321; (C) Venn diagram of commonly DEGs in comparison groups; (D) Hierarchical clustering analysis of specific genes associated with liver metastasis in the two datasets.
S2.
| Gene symbol | Gene ID | Description | Style |
| A1BG | 1 | alpha-1-B glycoprotein | up |
| AADAC | 13 | arylacetamide deacetylase | up |
| ABCC2 | 1244 | ATP binding cassette subfamily C member 2 | up |
| ABCG5 | 64240 | ATP binding cassette subfamily G member 5 | up |
| ABHD5 | 51099 | abhydrolase domain containing 5 | down |
| ACE | 1636 | angiotensin I converting enzyme | down |
| ACER3 | 55331 | alkaline ceramidase 3 | down |
| ACSM2A | 123876 | acyl-CoA synthetase medium-chain family member 2A | up |
| ACSM5 | 54988 | acyl-CoA synthetase medium-chain family member 5 | up |
| ACTL10 | 170487 | actin like 10 | up |
| ADAM8 | 101 | ADAM metallopeptidase domain 8 | up |
| ADAMTS8 | 11095 | ADAM metallopeptidase with thrombospondin type 1 motif 8 | down |
| ADGRL3 | 23284 | adhesion G protein-coupled receptor L3 | down |
| ADH4 | 127 | alcohol dehydrogenase 4 (class II), pi polypeptide | up |
| ADRA2A | 150 | adrenoceptor alpha 2A | down |
| AGXT | 189 | alanine-glyoxylate aminotransferase | up |
| AHSG | 197 | alpha-2-HS-glycoprotein | up |
| AIFM3 | 150209 | apoptosis inducing factor, mitochondria associated 3 | down |
| AKR1C4 | 1109 | aldo-keto reductase family 1, member C4 | up |
| AKR1D1 | 6718 | aldo-keto reductase family 1, member D1 | up |
| ALB | 213 | albumin | up |
| ALCAM | 214 | activated leukocyte cell adhesion molecule | up |
| ALDH8A1 | 64577 | aldehyde dehydrogenase 8 family member A1 | up |
| ALDOB | 229 | aldolase, fructose-bisphosphate B | up |
| AMBP | 259 | alpha-1-microglobulin/bikunin precursor | up |
| AMDHD1 | 144193 | amidohydrolase domain containing 1 | up |
| AMIGO2 | 347902 | adhesion molecule with Ig-like domain 2 | up |
| AMPD2 | 271 | adenosine monophosphate deaminase 2 | up |
| ANAPC11 | 51529 | anaphase promoting complex subunit 11 | up |
| ANGPTL3 | 27329 | angiopoietin like 3 | up |
| ANKRD37 | 353322 | ankyrin repeat domain 37 | up |
| ANKZF1 | 55139 | ankyrin repeat and zinc finger domain containing 1 | up |
| AOX1 | 316 | aldehyde oxidase 1 | up |
| AP5M1 | 55745 | adaptor related protein complex 5 mu 1 subunit | down |
| APCS | 325 | amyloid P component, serum | up |
| APOA1 | 335 | apolipoprotein A1 | up |
| APOA2 | 336 | apolipoprotein A2 | up |
| APOA5 | 116519 | apolipoprotein A5 | up |
| APOB | 338 | apolipoprotein B | up |
| APOC3 | 345 | apolipoprotein C3 | up |
| APOE | 348 | apolipoprotein E | up |
| APOH | 350 | apolipoprotein H | up |
| APOL1 | 8542 | apolipoprotein L1 | up |
| APOM | 55937 | apolipoprotein M | up |
| AQP3 | 360 | aquaporin 3 (Gill blood group) | up |
| AQP7 | 364 | aquaporin 7 | down |
| AQP9 | 366 | aquaporin 9 | up |
| ARG1 | 383 | arginase 1 | up |
| ARHGAP6 | 395 | Rho GTPase activating protein 6 | down |
| ARMCX1 | 51309 | armadillo repeat containing, X-linked 1 | down |
| ARRB2 | 409 | arrestin beta 2 | up |
| ASB9 | 140462 | ankyrin repeat and SOCS box containing 9 | up |
| ASGR2 | 433 | asialoglycoprotein receptor 2 | up |
| ATOX1 | 475 | antioxidant 1 copper chaperone | up |
| ATP2B4 | 493 | ATPase plasma membrane Ca2+ transporting 4 | down |
| ATP6V0D1 | 9114 | ATPase H+ transporting V0 subunit d1 | down |
| B3GNT8 | 374907 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 | down |
| BAD | 572 | BCL2 associated agonist of cell death | down |
| BAIAP2L1 | 55971 | BAI1 associated protein 2 like 1 | up |
| BCAP29 | 55973 | B-cell receptor-associated protein 29 | down |
| BCL7A | 605 | B-cell CLL/lymphoma 7A | up |
| BDKRB2 | 624 | bradykinin receptor B2 | down |
| BEX4 | 56271 | brain expressed X-linked 4 | down |
| BHLHB9 | 80823 | basic helix-loop-helix domain containing, class B, 9 | up |
| BMP3 | 651 | bone morphogenetic protein 3 | down |
| BMP5 | 653 | bone morphogenetic protein 5 | down |
| BNC2 | 54796 | basonuclin 2 | down |
| BNIP3 | 664 | BCL2/adenovirus E1B 19kDa interacting protein 3 | up |
| BOC | 91653 | BOC cell adhesion associated, oncogene regulated | down |
| BOD1L1 | 259282 | biorientation of chromosomes in cell division 1 like 1 | up |
| BSG | 682 | basigin (Ok blood group) | down |
| BTC | 685 | betacellulin | down |
| C10orf10 | 11067 | chromosome 10 open reading frame 10 | up |
| C11orf31 | 280636 | chromosome 11 open reading frame 31 | up |
| C14orf132 | 56967 | chromosome 14 open reading frame 132 | down |
| C15orf65 | 145788 | chromosome 15 open reading frame 65 | down |
| C16orf62 | 57020 | chromosome 16 open reading frame 62 | down |
| C17orf75 | 64149 | chromosome 17 open reading frame 75 | up |
| C1QTNF2 | 114898 | C1q and tumor necrosis factor related protein 2 | down |
| C1orf109 | 54955 | chromosome 1 open reading frame 109 | up |
| C2 | 717 | complement component 2 | up |
| C20orf194 | 25943 | chromosome 20 open reading frame 194 | down |
| C3 | 718 | complement component 3 | up |
| C4A | 720 | complement component 4A (Rodgers blood group) | up |
| C4BPA | 722 | complement component 4 binding protein alpha | up |
| C4orf3 | 401152 | chromosome 4 open reading frame 3 | up |
| C5 | 727 | complement component 5 | up |
| C5AR1 | 728 | complement component 5a receptor 1 | up |
| C6 | 729 | complement component 6 | up |
| C6orf1 | 221491 | chromosome 6 open reading frame 1 | up |
| C7orf13 | 129790 | chromosome 7 open reading frame 13 | up |
| C8A | 731 | complement component 8 alpha subunit | up |
| C8B | 732 | complement component 8, beta polypeptide | up |
| C8orf4 | 56892 | chromosome 8 open reading frame 4 | down |
| C9 | 735 | complement component 9 | up |
| C9orf142 | 286257 | chromosome 9 open reading frame 142 | up |
| CA9 | 768 | carbonic anhydrase 9 | up |
| CACNA2D1 | 781 | calcium voltage-gated channel auxiliary subunit alpha2delta 1 | down |
| CALD1 | 800 | caldesmon 1 | down |
| CAMK2B | 816 | calcium/calmodulin dependent protein kinase II beta | down |
| CAST | 831 | calpastatin | down |
| CBLN2 | 147381 | cerebellin 2 precursor | down |
| CBX8 | 57332 | chromobox 8 | up |
| CCL11 | 6356 | C-C motif chemokine ligand 11 | down |
| CCL18 | 6362 | C-C motif chemokine ligand 18 | up |
| CD163 | 9332 | CD163 molecule | up |
| CD19 | 930 | CD19 molecule | down |
| CD27 | 939 | CD27 molecule | down |
| CD69 | 969 | CD69 molecule | down |
| CD79B | 974 | CD79b molecule | down |
| CDC42EP3 | 10602 | CDC42 effector protein 3 | down |
| CDH2 | 1000 | cadherin 2 | up |
| CDKL1 | 8814 | cyclin dependent kinase like 1 | down |
| CDS1 | 1040 | CDP-diacylglycerol synthase 1 | down |
| CEP76 | 79959 | centrosomal protein 76 | up |
| CFHR2 | 3080 | complement factor H related 2 | up |
| CFHR4 | 10877 | complement factor H related 4 | up |
| CHDH | 55349 | choline dehydrogenase | up |
| CHMP1B | 57132 | charged multivesicular body protein 1B | down |
| CIDEC | 63924 | cell death inducing DFFA like effector c | down |
| CLIC4 | 25932 | chloride intracellular channel 4 | down |
| CLIP3 | 25999 | CAP-Gly domain containing linker protein 3 | down |
| CLMP | 79827 | CXADR-like membrane protein | down |
| CLRN3 | 119467 | clarin 3 | down |
| CLTB | 1212 | clathrin light chain B | down |
| CNDP1 | 84735 | carnosine dipeptidase 1 (metallopeptidase M20 family) | up |
| CNPY2 | 10330 | canopy FGF signaling regulator 2 | up |
| CNRIP1 | 25927 | cannabinoid receptor interacting protein 1 | down |
| COL14A1 | 7373 | collagen type XIV alpha 1 | down |
| COL6A1 | 1291 | collagen type VI alpha 1 | down |
| COL6A2 | 1292 | collagen type VI alpha 2 | down |
| COLEC11 | 78989 | collectin subfamily member 11 | up |
| COLEC12 | 81035 | collectin subfamily member 12 | down |
| COX20 | 116228 | COX20 cytochrome c oxidase assembly factor | up |
| COX7A1 | 1346 | cytochrome c oxidase subunit 7A1 | down |
| CPA3 | 1359 | carboxypeptidase A3 | down |
| CPB2 | 1361 | carboxypeptidase B2 | up |
| CPNE2 | 221184 | copine 2 | down |
| CPOX | 1371 | coproporphyrinogen oxidase | up |
| CPS1 | 1373 | carbamoyl-phosphate synthase 1 | up |
| CPXM2 | 119587 | carboxypeptidase X (M14 family), member 2 | down |
| CRACR2A | 84766 | calcium release activated channel regulator 2A | down |
| CRAT | 1384 | carnitine O-acetyltransferase | down |
| CREB3L1 | 90993 | cAMP responsive element binding protein 3-like 1 | down |
| CRK | 1398 | v-crk avian sarcoma virus CT10 oncogene homolog | down |
| CRP | 1401 | C-reactive protein, pentraxin-related | up |
| CTNND1 | 1500 | catenin delta 1 | down |
| CTSB | 1508 | cathepsin B | up |
| CUL7 | 9820 | cullin 7 | up |
| CWF19L1 | 55280 | CWF19-like 1, cell cycle control (S. pombe) | up |
| CXCL14 | 9547 | C-X-C motif chemokine ligand 14 | down |
| CXCL16 | 58191 | C-X-C motif chemokine ligand 16 | up |
| CYP1B1 | 1545 | cytochrome P450 family 1 subfamily B member 1 | up |
| CYP2C8 | 1558 | cytochrome P450 family 2 subfamily C member 8 | up |
| CYP2E1 | 1571 | cytochrome P450 family 2 subfamily E member 1 | up |
| CYP4A11 | 1579 | cytochrome P450 family 4 subfamily A member 11 | up |
| CYP8B1 | 1582 | cytochrome P450 family 8 subfamily B member 1 | up |
| CYSLTR1 | 10800 | cysteinyl leukotriene receptor 1 | down |
| CYSTM1 | 84418 | cysteine rich transmembrane module containing 1 | down |
| CYYR1 | 116159 | cysteine and tyrosine rich 1 | down |
| DAAM2 | 23500 | dishevelled associated activator of morphogenesis 2 | down |
| DACT3 | 147906 | dishevelled binding antagonist of beta catenin 3 | down |
| DDR2 | 4921 | discoidin domain receptor tyrosine kinase 2 | down |
| DDX55 | 57696 | DEAD-box helicase 55 | up |
| DHPS | 1725 | deoxyhypusine synthase | up |
| DHRS2 | 10202 | dehydrogenase/reductase (SDR family) member 2 | up |
| DHX8 | 1659 | DEAH-box helicase 8 | up |
| DIRC2 | 84925 | disrupted in renal carcinoma 2 | down |
| DIXDC1 | 85458 | DIX domain containing 1 | down |
| DLL1 | 28514 | delta like canonical Notch ligand 1 | down |
| DNAJB9 | 4189 | DnaJ heat shock protein family (Hsp40) member B9 | up |
| DOCK5 | 80005 | dedicator of cytokinesis 5 | down |
| DOK4 | 55715 | docking protein 4 | down |
| DPP7 | 29952 | dipeptidyl peptidase 7 | up |
| DPYS | 1807 | dihydropyrimidinase | up |
| DQX1 | 165545 | DEAQ-box RNA dependent ATPase 1 | down |
| DSG3 | 1830 | desmoglein 3 | up |
| DUS4L | 11062 | dihydrouridine synthase 4 like | up |
| E2F3 | 1871 | E2F transcription factor 3 | up |
| EBF1 | 1879 | early B-cell factor 1 | down |
| ECH1 | 1891 | enoyl-CoA hydratase 1, peroxisomal | down |
| EDNRB | 1910 | endothelin receptor type B | down |
| EFNA2 | 1943 | ephrin A2 | down |
| EFTUD2 | 9343 | elongation factor Tu GTP binding domain containing 2 | up |
| EGFR | 1956 | epidermal growth factor receptor | down |
| EHD1 | 10938 | EH domain containing 1 | down |
| EHD2 | 30846 | EH domain containing 2 | down |
| EIF4G3 | 8672 | eukaryotic translation initiation factor 4 gamma 3 | down |
| EMCN | 51705 | endomucin | down |
| EMID1 | 129080 | EMI domain containing 1 | down |
| ENGASE | 64772 | endo-beta-N-acetylglucosaminidase | up |
| ENO2 | 2026 | enolase 2 | up |
| ENO3 | 2027 | enolase 3 | up |
| ENPEP | 2028 | glutamyl aminopeptidase | up |
| ENPP3 | 5169 | ectonucleotide pyrophosphatase/phosphodiesterase 3 | down |
| ENY2 | 56943 | enhancer of yellow 2 homolog (Drosophila) | up |
| EPAS1 | 2034 | endothelial PAS domain protein 1 | down |
| EPB41L4A-AS1 | 114915 | EPB41L4A antisense RNA 1 | up |
| EPHA4 | 2043 | EPH receptor A4 | down |
| EPHB4 | 2050 | EPH receptor B4 | up |
| EPSTI1 | 94240 | epithelial stromal interaction 1 (breast) | up |
| ERBIN | 55914 | erbb2 interacting protein | down |
| ERCC2 | 2068 | excision repair cross-complementation group 2 | up |
| ETV5 | 2119 | ETS variant 5 | up |
| EVA1A | 84141 | eva-1 homolog A, regulator of programmed cell death | up |
| F10 | 2159 | coagulation factor X | up |
| F13B | 2165 | coagulation factor XIII B chain | up |
| F2 | 2147 | coagulation factor II, thrombin | up |
| F3 | 2152 | coagulation factor III, tissue factor | down |
| F5 | 2153 | coagulation factor V | up |
| F9 | 2158 | coagulation factor IX | up |
| FAM114A1 | 92689 | family with sequence similarity 114 member A1 | down |
| FAM131B | 9715 | family with sequence similarity 131 member B | up |
| FAM149B1 | 317662 | family with sequence similarity 149 member B1 | up |
| FAM214A | 56204 | family with sequence similarity 214 member A | down |
| FAM21C | 253725 | family with sequence similarity 21 member C | down |
| FAM43A | 131583 | family with sequence similarity 43 member A | down |
| FAM63A | 55793 | family with sequence similarity 63 member A | down |
| FANCE | 2178 | Fanconi anemia complementation group E | up |
| FBLIM1 | 54751 | filamin binding LIM protein 1 | down |
| FBLN2 | 2199 | fibulin 2 | down |
| FBXL2 | 25827 | F-box and leucine-rich repeat protein 2 | up |
| FCGR1A | 2209 | Fc fragment of IgG receptor Ia | up |
| FCGR1B | 2210 | Fc fragment of IgG receptor Ib | up |
| FCGR3A | 2214 | Fc fragment of IgG receptor IIIa | up |
| FCN3 | 8547 | ficolin 3 | up |
| FCRLB | 127943 | Fc receptor like B | up |
| FERMT2 | 10979 | fermitin family member 2 | down |
| FGA | 2243 | fibrinogen alpha chain | up |
| FGB | 2244 | fibrinogen beta chain | up |
| FGF7 | 2252 | fibroblast growth factor 7 | down |
| FGFBP1 | 9982 | fibroblast growth factor binding protein 1 | down |
| FGG | 2266 | fibrinogen gamma chain | up |
| FGL1 | 2267 | fibrinogen like 1 | up |
| FGR | 2268 | FGR proto-oncogene, Src family tyrosine kinase | up |
| FLRT2 | 23768 | fibronectin leucine rich transmembrane protein 2 | down |
| FLVCR1 | 28982 | feline leukemia virus subgroup C cellular receptor 1 | up |
| FMO3 | 2328 | flavin containing monooxygenase 3 | up |
| FOXF1 | 2294 | forkhead box F1 | down |
| FRG1HP | 100132352 | FSHD region gene 1 family member H, pseudogene | up |
| FSTL1 | 11167 | follistatin like 1 | down |
| FTCD | 10841 | formimidoyltransferase cyclodeaminase | up |
| FUT3 | 2525 | fucosyltransferase 3 (Lewis blood group) | down |
| FUT6 | 2528 | fucosyltransferase 6 | down |
| G6PC | 2538 | glucose-6-phosphatase catalytic subunit | up |
| GABRE | 2564 | gamma-aminobutyric acid type A receptor epsilon subunit | up |
| GALNT7 | 51809 | polypeptide N-acetylgalactosaminyltransferase 7 | down |
| GAMT | 2593 | guanidinoacetate N-methyltransferase | up |
| GATM | 2628 | glycine amidinotransferase | up |
| GC | 2638 | GC, vitamin D binding protein | up |
| GCFC2 | 6936 | GC-rich sequence DNA-binding factor 2 | up |
| GFRA3 | 2676 | GDNF family receptor alpha 3 | down |
| GIMAP6 | 474344 | GTPase, IMAP family member 6 | down |
| GLA | 2717 | galactosidase alpha | up |
| GLYATL1 | 92292 | glycine-N-acyltransferase like 1 | up |
| GM2A | 2760 | GM2 ganglioside activator | up |
| GNAI1 | 2770 | G protein subunit alpha i1 | down |
| GNAL | 2774 | G protein subunit alpha L | down |
| GNAO1 | 2775 | G protein subunit alpha o1 | down |
| GNG2 | 54331 | G protein subunit gamma 2 | down |
| GNG4 | 2786 | G protein subunit gamma 4 | up |
| GOLGA8A | 23015 | golgin A8 family member A | up |
| GPATCH2 | 55105 | G-patch domain containing 2 | up |
| GPR137B | 7107 | G protein-coupled receptor 137B | up |
| GPR89B | 51463 | G protein-coupled receptor 89B | up |
| GRB7 | 2886 | growth factor receptor bound protein 7 | up |
| GREM1 | 26585 | gremlin 1, DAN family BMP antagonist | down |
| GSKIP | 51527 | GSK3B interacting protein | down |
| GTF2IP20 | 441124 | general transcription factor IIi pseudogene 20 | up |
| GULP1 | 51454 | GULP, engulfment adaptor PTB domain containing 1 | down |
| GUSBP11 | 91316 | glucuronidase, beta pseudogene 11 | down |
| HADH | 3033 | hydroxyacyl-CoA dehydrogenase | down |
| HAMP | 57817 | hepcidin antimicrobial peptide | up |
| HAND1 | 9421 | heart and neural crest derivatives expressed 1 | down |
| HBA1 | 3039 | hemoglobin subunit alpha 1 | down |
| HBG1 | 3047 | hemoglobin subunit gamma 1 | down |
| HEXIM1 | 10614 | hexamethylene bis-acetamide inducible 1 | down |
| HFE2 | 148738 | hemochromatosis type 2 (juvenile) | up |
| HHIP | 64399 | hedgehog interacting protein | down |
| HOTAIRM1 | 100506311 | HOXA transcript antisense RNA, myeloid-specific 1 | down |
| HOXA13 | 3209 | homeobox A13 | down |
| HOXD1 | 3231 | homeobox D1 | down |
| HP | 3240 | haptoglobin | up |
| HPD | 3242 | 4-hydroxyphenylpyruvate dioxygenase | up |
| HPN | 3249 | hepsin | up |
| HPR | 3250 | haptoglobin-related protein | up |
| HPS4 | 89781 | HPS4, biogenesis of lysosomal organelles complex 3 subunit 2 | up |
| HPSE | 10855 | heparanase | down |
| HPX | 3263 | hemopexin | up |
| HRG | 3273 | histidine rich glycoprotein | up |
| HSD11B1 | 3290 | hydroxysteroid (11-beta) dehydrogenase 1 | up |
| HSPA6 | 3310 | heat shock protein family A (Hsp70) member 6 | up |
| HSPB7 | 27129 | heat shock protein family B (small) member 7 | down |
| HYAL1 | 3373 | hyaluronoglucosaminidase 1 | up |
| ID1 | 3397 | inhibitor of DNA binding 1, HLH protein | down |
| ID3 | 3399 | inhibitor of DNA binding 3, HLH protein | down |
| IDH3A | 3419 | isocitrate dehydrogenase 3 (NAD(+)) alpha | down |
| IFI44L | 10964 | interferon induced protein 44 like | up |
| IGF2BP3 | 10643 | insulin like growth factor 2 mRNA binding protein 3 | up |
| IGFBP1 | 3484 | insulin like growth factor binding protein 1 | up |
| IGFBP2 | 3485 | insulin like growth factor binding protein 2 | up |
| IGHA1 | 3493 | immunoglobulin heavy constant alpha 1 | down |
| IGHD | 3495 | immunoglobulin heavy constant delta | down |
| IGHG1 | 3500 | immunoglobulin heavy constant gamma 1 (G1m marker) | down |
| IGK | 50802 | immunoglobulin kappa locus | down |
| IGKC | 3514 | immunoglobulin kappa constant | down |
| IGLL3P | 91353 | immunoglobulin lambda like polypeptide 3, pseudogene | down |
| IGLL5 | 100423062 | immunoglobulin lambda like polypeptide 5 | down |
| IGLV1-44 | 28823 | immunoglobulin lambda variable 1-44 | down |
| IKZF3 | 22806 | IKAROS family zinc finger 3 | down |
| IL18 | 3606 | interleukin 18 | down |
| IL1RAP | 3556 | interleukin 1 receptor accessory protein | up |
| IL7R | 3575 | interleukin 7 receptor | down |
| INAFM2 | 100505573 | InaF motif containing 2 | down |
| INHBE | 83729 | inhibin beta E | up |
| INO80B | 83444 | INO80 complex subunit B | up |
| ITFG2 | 55846 | integrin alpha FG-GAP repeat containing 2 | up |
| ITGA8 | 8516 | integrin subunit alpha 8 | down |
| ITIH1 | 3697 | inter-alpha-trypsin inhibitor heavy chain 1 | up |
| ITIH2 | 3698 | inter-alpha-trypsin inhibitor heavy chain 2 | up |
| ITIH3 | 3699 | inter-alpha-trypsin inhibitor heavy chain 3 | up |
| ITIH4 | 3700 | inter-alpha-trypsin inhibitor heavy chain family member 4 | up |
| ITSN1 | 6453 | intersectin 1 | down |
| JADE3 | 9767 | jade family PHD finger 3 | up |
| JAM3 | 83700 | junctional adhesion molecule 3 | down |
| JAML | 120425 | junction adhesion molecule like | down |
| JMJD7-PLA2G4B | 8681 | JMJD7-PLA2G4B readthrough | up |
| JPH2 | 57158 | junctophilin 2 | down |
| JUND | 3727 | JunD proto-oncogene, AP-1 transcription factor subunit | down |
| KANK2 | 25959 | KN motif and ankyrin repeat domains 2 | down |
| KBTBD12 | 166348 | kelch repeat and BTB domain containing 12 | down |
| KCNAB2 | 8514 | potassium voltage-gated channel subfamily A regulatory beta subunit 2 | up |
| KCNH2 | 3757 | potassium voltage-gated channel subfamily H member 2 | up |
| KDM3A | 55818 | lysine demethylase 3A | up |
| KIF12 | 113220 | kinesin family member 12 | up |
| KIF20B | 9585 | kinesin family member 20B | up |
| KLHL13 | 90293 | kelch like family member 13 | down |
| KLHL23 | 151230 | kelch like family member 23 | up |
| KLHL7 | 55975 | kelch like family member 7 | up |
| KLRD1 | 3824 | killer cell lectin like receptor D1 | down |
| KNG1 | 3827 | kininogen 1 | up |
| KRT6B | 3854 | keratin 6B | up |
| L1CAM | 3897 | L1 cell adhesion molecule | down |
| L3MBTL1 | 26013 | l(3)mbt-like 1 (Drosophila) | up |
| LAMC2 | 3918 | laminin subunit gamma 2 | up |
| LBP | 3929 | lipopolysaccharide binding protein | up |
| LECT2 | 3950 | leukocyte cell derived chemotaxin 2 | up |
| LETM1 | 3954 | leucine zipper and EF-hand containing transmembrane protein 1 | down |
| LINC00094 | 266655 | long intergenic non-protein coding RNA 94 | up |
| LINC00911 | 100996280 | long intergenic non-protein coding RNA 911 | up |
| LINC00959 | 387723 | long intergenic non-protein coding RNA 959 | down |
| LINC00982 | 440556 | long intergenic non-protein coding RNA 982 | up |
| LIPA | 3988 | lipase A, lysosomal acid type | up |
| LOC100288911 | 100288911 | uncharacterized LOC100288911 | down |
| LOC100505501 | 100505501 | uncharacterized LOC100505501 | down |
| LOC101927263 | 101927263 | uncharacterized LOC101927263 | down |
| LOC101928881 | 101928881 | uncharacterized LOC101928881 | up |
| LOC284112 | 284112 | uncharacterized LOC284112 | down |
| LOC389834 | 389834 | ankyrin repeat domain 57 pseudogene | down |
| LOC389906 | 389906 | zinc finger protein 839 pseudogene | up |
| LOC81691 | 81691 | exonuclease NEF-sp | up |
| LOXL1 | 4016 | lysyl oxidase like 1 | up |
| LPA | 4018 | lipoprotein(a) | up |
| LPGAT1 | 9926 | lysophosphatidylglycerol acyltransferase 1 | up |
| LRCH2 | 57631 | leucine-rich repeats and calponin homology (CH) domain containing 2 | down |
| LRG1 | 116844 | leucine-rich alpha-2-glycoprotein 1 | up |
| LRP3 | 4037 | LDL receptor related protein 3 | up |
| LRP4 | 4038 | LDL receptor related protein 4 | up |
| LRRC32 | 2615 | leucine rich repeat containing 32 | up |
| LRRTM2 | 26045 | leucine rich repeat transmembrane neuronal 2 | down |
| LRSAM1 | 90678 | leucine rich repeat and sterile alpha motif containing 1 | up |
| LSAMP | 4045 | limbic system-associated membrane protein | down |
| LUZP1 | 7798 | leucine zipper protein 1 | down |
| LXN | 56925 | latexin | down |
| LYRM4 | 57128 | LYR motif containing 4 | up |
| MAFB | 9935 | MAF bZIP transcription factor B | up |
| MAGI1 | 9223 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | down |
| MAGI3 | 260425 | membrane associated guanylate kinase, WW and PDZ domain containing 3 | down |
| MAGOHB | 55110 | mago homolog B, exon junction complex core component | up |
| MAP7D2 | 256714 | MAP7 domain containing 2 | up |
| MAPK6 | 5597 | mitogen-activated protein kinase 6 | down |
| MARCO | 8685 | macrophage receptor with collagenous structure | up |
| MAT1A | 4143 | methionine adenosyltransferase 1A | up |
| MAX | 4149 | MYC associated factor X | down |
| MB21D2 | 151963 | Mab-21 domain containing 2 | up |
| MBL2 | 4153 | mannose binding lectin 2 | up |
| MCL1 | 4170 | myeloid cell leukemia 1 | down |
| MCTP2 | 55784 | multiple C2 domains, transmembrane 2 | down |
| MCTS1 | 28985 | malignant T-cell amplified sequence 1 | up |
| ME1 | 4199 | malic enzyme 1 | up |
| ME2 | 4200 | malic enzyme 2 | down |
| MEOX1 | 4222 | mesenchyme homeobox 1 | down |
| METTL2B | 55798 | methyltransferase like 2B | up |
| MFN2 | 9927 | mitofusin 2 | down |
| MFSD12 | 126321 | major facilitator superfamily domain containing 12 | up |
| MICAL2 | 9645 | microtubule associated monooxygenase, calponin and LIM domain containing 2 | down |
| MIR145 | 406937 | microRNA 145 | down |
| MMP2 | 4313 | matrix metallopeptidase 2 | down |
| MMP24-AS1 | 101410538 | MMP24 antisense RNA 1 | down |
| MPHOSPH9 | 10198 | M-phase phosphoprotein 9 | up |
| MPZ | 4359 | myelin protein zero | down |
| MR1 | 3140 | major histocompatibility complex, class I-related | up |
| MROH1 | 727957 | maestro heat like repeat family member 1 | up |
| MRVI1 | 10335 | murine retrovirus integration site 1 homolog | down |
| MSR1 | 4481 | macrophage scavenger receptor 1 | up |
| MTDH | 92140 | metadherin | up |
| MTFR1 | 9650 | mitochondrial fission regulator 1 | up |
| MTPAP | 55149 | mitochondrial poly(A) polymerase | up |
| MTUS1 | 57509 | microtubule associated tumor suppressor 1 | down |
| MUC1 | 4582 | mucin 1, cell surface associated | down |
| MUC13 | 56667 | mucin 13, cell surface associated | down |
| MVP | 9961 | major vault protein | down |
| MYO7A | 4647 | myosin VIIA | up |
| MZB1 | 51237 | marginal zone B and B1 cell specific protein | down |
| N4BP2L2 | 10443 | NEDD4 binding protein 2-like 2 | up |
| NABP1 | 64859 | nucleic acid binding protein 1 | up |
| NCF2 | 4688 | neutrophil cytosolic factor 2 | up |
| NCKAP5L | 57701 | NCK associated protein 5 like | up |
| NDN | 4692 | necdin, MAGE family member | down |
| NDNF | 79625 | neuron-derived neurotrophic factor | down |
| NEFL | 4747 | neurofilament, light polypeptide | down |
| NEIL1 | 79661 | nei like DNA glycosylase 1 | up |
| NIPAL3 | 57185 | NIPA like domain containing 3 | down |
| NMRAL1 | 57407 | NmrA-like family domain containing 1 | up |
| NMT1 | 4836 | N-myristoyltransferase 1 | up |
| NNMT | 4837 | nicotinamide N-methyltransferase | up |
| NOC3L | 64318 | NOC3 like DNA replication regulator | up |
| NOC4L | 79050 | nucleolar complex associated 4 homolog | up |
| NPC1L1 | 29881 | NPC1 like intracellular cholesterol transporter 1 | up |
| NPY | 4852 | neuropeptide Y | down |
| NRAV | 100506668 | negative regulator of antiviral response (non-protein coding) | up |
| NRXN3 | 9369 | neurexin 3 | down |
| NSG1 | 27065 | neuron specific gene family member 1 | down |
| NTHL1 | 4913 | nth-like DNA glycosylase 1 | up |
| NUDT11 | 55190 | nudix hydrolase 11 | down |
| NUDT14 | 256281 | nudix hydrolase 14 | up |
| OBSCN | 84033 | obscurin, cytoskeletal calmodulin and titin-interacting RhoGEF | up |
| OCA2 | 4948 | OCA2 melanosomal transmembrane protein | up |
| OGDHL | 55753 | oxoglutarate dehydrogenase-like | up |
| OLFML2A | 169611 | olfactomedin like 2A | down |
| OLR1 | 4973 | oxidized low density lipoprotein receptor 1 | up |
| ONECUT2 | 9480 | one cut homeobox 2 | up |
| OR7E14P | 10819 | olfactory receptor family 7 subfamily E member 14 pseudogene | down |
| ORM1 | 5004 | orosomucoid 1 | up |
| P2RX5 | 5026 | purinergic receptor P2X 5 | down |
| P2RY1 | 5028 | purinergic receptor P2Y1 | down |
| P4HA1 | 5033 | prolyl 4-hydroxylase subunit alpha 1 | up |
| PAAF1 | 80227 | proteasomal ATPase associated factor 1 | up |
| PAFAH2 | 5051 | platelet activating factor acetylhydrolase 2 | down |
| PALLD | 23022 | palladin, cytoskeletal associated protein | down |
| PAMR1 | 25891 | peptidase domain containing associated with muscle regeneration 1 | down |
| PANK2 | 80025 | pantothenate kinase 2 | up |
| PANK3 | 79646 | pantothenate kinase 3 | down |
| PARP10 | 84875 | poly(ADP-ribose) polymerase family member 10 | up |
| PARVA | 55742 | parvin alpha | down |
| PATZ1 | 23598 | POZ/BTB and AT hook containing zinc finger 1 | up |
| PBX1 | 5087 | PBX homeobox 1 | down |
| PCDH18 | 54510 | protocadherin 18 | down |
| PCDH20 | 64881 | protocadherin 20 | down |
| PCDH7 | 5099 | protocadherin 7 | down |
| PCSK5 | 5125 | proprotein convertase subtilisin/kexin type 5 | down |
| PCSK7 | 9159 | proprotein convertase subtilisin/kexin type 7 | down |
| PDCD4-AS1 | 282997 | PDCD4 antisense RNA 1 | down |
| PDGFRA | 5156 | platelet derived growth factor receptor alpha | down |
| PDZRN3 | 23024 | PDZ domain containing ring finger 3 | down |
| PECAM1 | 5175 | platelet and endothelial cell adhesion molecule 1 | down |
| PELI2 | 57161 | pellino E3 ubiquitin protein ligase family member 2 | down |
| PEX10 | 5192 | peroxisomal biogenesis factor 10 | up |
| PF4 | 5196 | platelet factor 4 | up |
| PFKFB3 | 5209 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 | up |
| PGBD5 | 79605 | piggyBac transposable element derived 5 | up |
| PGK1 | 5230 | phosphoglycerate kinase 1 | up |
| PGR | 5241 | progesterone receptor | down |
| PHGDH | 26227 | phosphoglycerate dehydrogenase | up |
| PI3 | 5266 | peptidase inhibitor 3 | down |
| PIAS2 | 9063 | protein inhibitor of activated STAT 2 | down |
| PIK3R2 | 5296 | phosphoinositide-3-kinase regulatory subunit 2 | up |
| PIPOX | 51268 | pipecolic acid and sarcosine oxidase | up |
| PKIG | 11142 | protein kinase (cAMP-dependent, catalytic) inhibitor gamma | down |
| PLA2G15 | 23659 | phospholipase A2 group XV | up |
| PLA2G2A | 5320 | phospholipase A2 group IIA | down |
| PLA2G4A | 5321 | phospholipase A2 group IVA | down |
| PLAGL2 | 5326 | PLAG1 like zinc finger 2 | up |
| PLAT | 5327 | plasminogen activator, tissue type | down |
| PLEK2 | 26499 | pleckstrin 2 | up |
| PLEKHH3 | 79990 | pleckstrin homology, MyTH4 and FERM domain containing H3 | up |
| PLEKHO1 | 51177 | pleckstrin homology domain containing O1 | down |
| PLEKHS1 | 79949 | pleckstrin homology domain containing S1 | up |
| PLG | 5340 | plasminogen | up |
| PLGLB2 | 5342 | plasminogen-like B2 | up |
| PLS1 | 5357 | plastin 1 | down |
| PLSCR3 | 57048 | phospholipid scramblase 3 | up |
| PMP22 | 5376 | peripheral myelin protein 22 | down |
| POFUT1 | 23509 | protein O-fucosyltransferase 1 | up |
| POLB | 5423 | polymerase (DNA) beta | up |
| POLR1C | 9533 | polymerase (RNA) I subunit C | up |
| POLR2D | 5433 | polymerase (RNA) II subunit D | up |
| PON1 | 5444 | paraoxonase 1 | up |
| PON3 | 5446 | paraoxonase 3 | up |
| POSTN | 10631 | periostin | up |
| POT1 | 25913 | protection of telomeres 1 | up |
| PP7080 | 25845 | uncharacterized LOC25845 | down |
| PPBP | 5473 | pro-platelet basic protein | up |
| PPM1L | 151742 | protein phosphatase, Mg2+/Mn2+ dependent 1L | down |
| PPP1R14D | 54866 | protein phosphatase 1 regulatory inhibitor subunit 14D | down |
| PPP1R35 | 221908 | protein phosphatase 1 regulatory subunit 35 | up |
| PPP1R9A | 55607 | protein phosphatase 1 regulatory subunit 9A | down |
| PPP2CB | 5516 | protein phosphatase 2 catalytic subunit beta | down |
| PRAP1 | 118471 | proline-rich acidic protein 1 | up |
| PRDM6 | 93166 | PR domain 6 | down |
| PRKG2 | 5593 | protein kinase, cGMP-dependent, type II | down |
| PROC | 5624 | protein C, inactivator of coagulation factors Va and VIIIa | up |
| PROSC | 11212 | proline synthetase co-transcribed homolog (bacterial) | down |
| PROSER2 | 254427 | proline and serine rich 2 | up |
| PRPF40B | 25766 | pre-mRNA processing factor 40 homolog B | up |
| PRUNE2 | 158471 | prune homolog 2 (Drosophila) | down |
| PTBP3 | 9991 | polypyrimidine tract binding protein 3 | up |
| PTGER3 | 5733 | prostaglandin E receptor 3 | down |
| PTK2 | 5747 | protein tyrosine kinase 2 | up |
| PTPMT1 | 114971 | protein tyrosine phosphatase, mitochondrial 1 | up |
| PTPN18 | 26469 | protein tyrosine phosphatase, non-receptor type 18 | down |
| PTPRCAP | 5790 | protein tyrosine phosphatase, receptor type C associated protein | down |
| PYGB | 5834 | phosphorylase, glycogen; brain | down |
| PYGM | 5837 | phosphorylase, glycogen, muscle | down |
| RAB27B | 5874 | RAB27B, member RAS oncogene family | down |
| RAD54L2 | 23132 | RAD54-like 2 (S. cerevisiae) | up |
| RAP1A | 5906 | RAP1A, member of RAS oncogene family | down |
| RASD2 | 23551 | RASD family member 2 | down |
| RASSF10 | 644943 | Ras association domain family member 10 | up |
| RBMS2 | 5939 | RNA binding motif single stranded interacting protein 2 | down |
| RBP4 | 5950 | retinol binding protein 4 | up |
| RCAN1 | 1827 | regulator of calcineurin 1 | down |
| RCSD1 | 92241 | RCSD domain containing 1 | down |
| RDH16 | 8608 | retinol dehydrogenase 16 (all-trans) | up |
| RECK | 8434 | reversion inducing cysteine rich protein with kazal motifs | down |
| REEP2 | 51308 | receptor accessory protein 2 | down |
| RETNLB | 84666 | resistin like beta | down |
| RGS4 | 5999 | regulator of G-protein signaling 4 | up |
| RGS5 | 8490 | regulator of G-protein signaling 5 | down |
| RHBDD1 | 84236 | rhomboid domain containing 1 | up |
| RHNO1 | 83695 | RAD9-HUS1-RAD1 interacting nuclear orphan 1 | up |
| RHPN1 | 114822 | rhophilin, Rho GTPase binding protein 1 | up |
| RNASE6 | 6039 | ribonuclease A family member k6 | up |
| RNF113A | 7737 | ring finger protein 113A | up |
| RNF138 | 51444 | ring finger protein 138 | down |
| RNF144A | 9781 | ring finger protein 144A | down |
| RNF219 | 79596 | ring finger protein 219 | up |
| RPA3 | 6119 | replication protein A3 | up |
| RPAP3 | 79657 | RNA polymerase II associated protein 3 | up |
| RPIA | 22934 | ribose 5-phosphate isomerase A | up |
| RPL13 | 6137 | ribosomal protein L13 | up |
| RPL14 | 9045 | ribosomal protein L14 | up |
| RPL35A | 6165 | ribosomal protein L35a | up |
| RPL36 | 25873 | ribosomal protein L36 | up |
| RPRM | 56475 | reprimo, TP53 dependent G2 arrest mediator candidate | down |
| RPS21 | 6227 | ribosomal protein S21 | up |
| RRP1 | 8568 | ribosomal RNA processing 1 | up |
| RUNX1T1 | 862 | RUNX1 translocation partner 1 | down |
| S1PR1 | 1901 | sphingosine-1-phosphate receptor 1 | down |
| SAA1 | 6288 | serum amyloid A1 | up |
| SAA4 | 6291 | serum amyloid A4, constitutive | up |
| SCARB1 | 949 | scavenger receptor class B member 1 | up |
| SCNN1A | 6337 | sodium channel epithelial 1 alpha subunit | down |
| SCP2 | 6342 | sterol carrier protein 2 | down |
| SCRG1 | 11341 | stimulator of chondrogenesis 1 | down |
| SDCCAG3 | 10807 | serologically defined colon cancer antigen 3 | up |
| SDK1 | 221935 | sidekick cell adhesion molecule 1 | down |
| SDS | 10993 | serine dehydratase | up |
| SEC14L2 | 23541 | SEC14 like lipid binding 2 | up |
| SEC14L4 | 284904 | SEC14 like lipid binding 4 | up |
| SEMA4D | 10507 | semaphorin 4D | up |
| SERPINA1 | 5265 | serpin family A member 1 | up |
| SERPINA10 | 51156 | serpin family A member 10 | up |
| SERPINA3 | 12 | serpin family A member 3 | up |
| SERPINA5 | 5104 | serpin family A member 5 | up |
| SERPINA6 | 866 | serpin family A member 6 | up |
| SERPINC1 | 462 | serpin family C member 1 | up |
| SERPIND1 | 3053 | serpin family D member 1 | up |
| SERPINF2 | 5345 | serpin family F member 2 | up |
| SERTAD4-AS1 | 574036 | SERTAD4 antisense RNA 1 | down |
| SFRP2 | 6423 | secreted frizzled related protein 2 | down |
| SGCD | 6444 | sarcoglycan delta | down |
| SGCE | 8910 | sarcoglycan epsilon | down |
| SGSM1 | 129049 | small G protein signaling modulator 1 | down |
| SHFM1 | 7979 | split hand/foot malformation (ectrodactyly) type 1 | up |
| SIGLEC7 | 27036 | sialic acid binding Ig like lectin 7 | up |
| SLAMF7 | 57823 | SLAM family member 7 | down |
| SLC13A5 | 284111 | solute carrier family 13 member 5 | up |
| SLC16A14 | 151473 | solute carrier family 16 member 14 | down |
| SLC16A4 | 9122 | solute carrier family 16 member 4 | up |
| SLC17A5 | 26503 | solute carrier family 17 member 5 | down |
| SLC22A7 | 10864 | solute carrier family 22 member 7 | up |
| SLC23A2 | 9962 | solute carrier family 23 member 2 | up |
| SLC25A14 | 9016 | solute carrier family 25 member 14 | up |
| SLC25A24 | 29957 | solute carrier family 25 member 24 | down |
| SLC25A29 | 123096 | solute carrier family 25 member 29 | up |
| SLC25A47 | 283600 | solute carrier family 25 member 47 | up |
| SLC27A5 | 10998 | solute carrier family 27 member 5 | up |
| SLC28A2 | 9153 | solute carrier family 28 member 2 | down |
| SLC2A1 | 6513 | solute carrier family 2 member 1 | up |
| SLC2A2 | 6514 | solute carrier family 2 member 2 | up |
| SLC30A4 | 7782 | solute carrier family 30 member 4 | down |
| SLC39A4 | 55630 | solute carrier family 39 member 4 | up |
| SLC3A2 | 6520 | solute carrier family 3 member 2 | up |
| SLC7A6OS | 84138 | solute carrier family 7 member 6 opposite strand | up |
| SLC9A3 | 6550 | solute carrier family 9 member A3 | down |
| SLC9A7 | 84679 | solute carrier family 9 member A7 | up |
| SLCO1B3 | 28234 | solute carrier organic anion transporter family member 1B3 | up |
| SMAD9 | 4093 | SMAD family member 9 | down |
| SMTN | 6525 | smoothelin | down |
| SNHG7 | 84973 | small nucleolar RNA host gene 7 | up |
| SNORA24 | 677809 | small nucleolar RNA, H/ACA box 24 | up |
| SNRPE | 6635 | small nuclear ribonucleoprotein polypeptide E | up |
| SNRPG | 6637 | small nuclear ribonucleoprotein polypeptide G | up |
| SNX32 | 254122 | sorting nexin 32 | up |
| SOS2 | 6655 | SOS Ras/Rho guanine nucleotide exchange factor 2 | down |
| SOSTDC1 | 25928 | sclerostin domain containing 1 | down |
| SOX9-AS1 | 400618 | SOX9 antisense RNA 1 | up |
| SPINK4 | 27290 | serine peptidase inhibitor, Kazal type 4 | down |
| SPTAN1 | 6709 | spectrin alpha, non-erythrocytic 1 | up |
| SRCAP | 10847 | Snf2-related CREBBP activator protein | up |
| STAMBPL1 | 57559 | STAM binding protein like 1 | up |
| STK31 | 56164 | serine/threonine kinase 31 | up |
| STON1 | 11037 | stonin 1 | down |
| STXBP5 | 134957 | syntaxin binding protein 5 | down |
| SULT2A1 | 6822 | sulfotransferase family 2A member 1 | up |
| SULT2B1 | 6820 | sulfotransferase family 2B member 1 | up |
| SUPT3H | 8464 | SPT3 homolog, SAGA and STAGA complex component | up |
| SYK | 6850 | spleen tyrosine kinase | up |
| TAF1A | 9015 | TATA-box binding protein associated factor, RNA polymerase I subunit A | up |
| TBL3 | 10607 | transducin beta like 3 | up |
| TCEAL7 | 56849 | transcription elongation factor A like 7 | down |
| TCF4 | 6925 | transcription factor 4 | down |
| TDO2 | 6999 | tryptophan 2,3-dioxygenase | up |
| TF | 7018 | transferrin | up |
| TFF1 | 7031 | trefoil factor 1 | down |
| TGFB1I1 | 7041 | transforming growth factor beta 1 induced transcript 1 | down |
| THBS1 | 7057 | thrombospondin 1 | down |
| THBS4 | 7060 | thrombospondin 4 | down |
| THSD1 | 55901 | thrombospondin type 1 domain containing 1 | up |
| TIPIN | 54962 | TIMELESS interacting protein | up |
| TJP2 | 9414 | tight junction protein 2 | up |
| TM4SF4 | 7104 | transmembrane 4 L six family member 4 | up |
| TMEM119 | 338773 | transmembrane protein 119 | down |
| TMEM131 | 23505 | transmembrane protein 131 | down |
| TMEM133 | 83935 | transmembrane protein 133 | down |
| TMEM182 | 130827 | transmembrane protein 182 | up |
| TMEM185B | 79134 | transmembrane protein 185B | up |
| TMEM191A | 84222 | transmembrane protein 191A (pseudogene) | up |
| TMEM27 | 57393 | transmembrane protein 27 | up |
| TMEM45A | 55076 | transmembrane protein 45A | up |
| TMEM88 | 92162 | transmembrane protein 88 | down |
| TMEM8B | 51754 | transmembrane protein 8B | down |
| TMEM9B | 56674 | TMEM9 domain family member B | down |
| TMTC1 | 83857 | transmembrane and tetratricopeptide repeat containing 1 | down |
| TOMM20 | 9804 | translocase of outer mitochondrial membrane 20 | up |
| TPM2 | 7169 | tropomyosin 2 (beta) | down |
| TPSAB1 | 7177 | tryptase alpha/beta 1 | down |
| TPSB2 | 64499 | tryptase beta 2 (gene/pseudogene) | down |
| TPSG1 | 25823 | tryptase gamma 1 | down |
| TRDV3 | 28516 | T cell receptor delta variable 3 | down |
| TRIM59 | 286827 | tripartite motif containing 59 | up |
| TRMT10B | 158234 | tRNA methyltransferase 10B | up |
| TRPA1 | 8989 | transient receptor potential cation channel subfamily A member 1 | down |
| TSEN2 | 80746 | tRNA splicing endonuclease subunit 2 | up |
| TSNARE1 | 203062 | t-SNARE domain containing 1 | up |
| TSPAN11 | 441631 | tetraspanin 11 | down |
| TTC39C | 125488 | tetratricopeptide repeat domain 39C | up |
| TTR | 7276 | transthyretin | up |
| TUB | 7275 | tubby bipartite transcription factor | up |
| TULP3 | 7289 | tubby like protein 3 | up |
| TUSC3 | 7991 | tumor suppressor candidate 3 | down |
| TWSG1 | 57045 | twisted gastrulation BMP signaling modulator 1 | down |
| TYMS | 7298 | thymidylate synthetase | down |
| TYROBP | 7305 | TYRO protein tyrosine kinase binding protein | up |
| UBQLN1 | 29979 | ubiquilin 1 | up |
| UGT2B4 | 7363 | UDP glucuronosyltransferase family 2 member B4 | up |
| UGT3A1 | 133688 | UDP glycosyltransferase family 3 member A1 | up |
| UNC93A | 54346 | unc-93 homolog A (C. elegans) | up |
| UPB1 | 51733 | beta-ureidopropionase 1 | up |
| UQCC2 | 84300 | ubiquinol-cytochrome c reductase complex assembly factor 2 | up |
| USP53 | 54532 | ubiquitin specific peptidase 53 | down |
| UTP23 | 84294 | UTP23, small subunit processome component | up |
| VAT1L | 57687 | vesicle amine transport 1-like | down |
| VNN1 | 8876 | vanin 1 | up |
| VTN | 7448 | vitronectin | up |
| VWF | 7450 | von Willebrand factor | down |
| WDR24 | 84219 | WD repeat domain 24 | up |
| WDR72 | 256764 | WD repeat domain 72 | up |
| WDR83 | 84292 | WD repeat domain 83 | up |
| WDYHV1 | 55093 | WDYHV motif containing 1 | up |
| WFDC1 | 58189 | WAP four-disulfide core domain 1 | down |
| WFDC3 | 140686 | WAP four-disulfide core domain 3 | up |
| WIF1 | 11197 | WNT inhibitory factor 1 | up |
| WLS | 79971 | wntless Wnt ligand secretion mediator | down |
| WNT3 | 7473 | Wnt family member 3 | up |
| WNT9A | 7483 | Wnt family member 9A | down |
| XKRX | 402415 | XK related, X-linked | up |
| YEATS4 | 8089 | YEATS domain containing 4 | up |
| ZDHHC14 | 79683 | zinc finger DHHC-type containing 14 | down |
| ZDHHC24 | 254359 | zinc finger DHHC-type containing 24 | up |
| ZEB1 | 6935 | zinc finger E-box binding homeobox 1 | down |
| ZFAND1 | 79752 | zinc finger AN1-type containing 1 | up |
| ZFP36 | 7538 | ZFP36 ring finger protein | down |
| ZNF182 | 7569 | zinc finger protein 182 | up |
| ZNF251 | 90987 | zinc finger protein 251 | up |
| ZNF415 | 55786 | zinc finger protein 415 | down |
| ZNF420 | 147923 | zinc finger protein 420 | up |
| ZNF502 | 91392 | zinc finger protein 502 | up |
| ZNF511 | 118472 | zinc finger protein 511 | up |
| ZNF579 | 163033 | zinc finger protein 579 | up |
| ZNF623 | 9831 | zinc finger protein 623 | up |
| ZNF655 | 79027 | zinc finger protein 655 | down |
| ZNF91 | 7644 | zinc finger protein 91 | down |
| ZNF93 | 81931 | zinc finger protein 93 | up |
| ZNRD1 | 30834 | zinc ribbon domain containing 1 | up |
| ZP3 | 7784 | zona pellucida glycoprotein 3 (sperm receptor) | up |
| ZSCAN18 | 65982 | zinc finger and SCAN domain containing 18 | down |
| ZXDB | 158586 | zinc finger, X-linked, duplicated B | up |
| Table 1 (continued) | |||
S3.
| Group | Gene symbol | Gene description | GSE49355_liver metastasis vs. normal | GSE62321_liver metastasis vs. normal | |||||
| FC | P | FDR | FC | P | FDR | ||||
| FC, fold-change; FDR, false discovery rat | |||||||||
| Up-regulated genes | ACSM2A | acyl-CoA synthetase
medium-chain family member 2A |
2.450000 | 0.000614 | 0.003200 | 4.100000 | 0.000531 | 0.005380 | |
| APOB | apolipoprotein B | 5.890000 | 0.000088 | 0.000634 | 2.880000 | 0.001366 | 0.011400 | ||
| APOH | apolipoprotein H | 18.10000 | 0.000004 | 0.000047 | 2.110000 | 0.000966 | 0.008680 | ||
| F5 | coagulation factor V | 5.060000 | 0.000000 | 0.000002 | 2.420000 | 0.000039 | 0.000634 | ||
| FTCD | formimidoyltransferase
cyclodeaminase |
2.510000 | 0.000863 | 0.004250 | 2.445000 | 0.000434 | 0.004525 | ||
| LYRM4 | LYR motif containing 4 | 2.120000 | 0.000000 | 0.000005 | 2.070000 | 0.001437 | 0.011900 | ||
| PLG | plasminogen | 5.080000 | 0.000701 | 0.003570 | 2.770000 | 0.000147 | 0.001900 | ||
| SERPINA1 | serpin family
A member 1 |
7.010000 | 0.000000 | 0.000000 | 3.970000 | 0.000006 | 0.000129 | ||
| UPB1 | beta-ureidopropionase 1 | 2.290000 | 0.006336 | 0.022200 | 2.010000 | 0.001004 | 0.008960 | ||
| Down-regulated genes | CDC42EP3 | CDC42 effector protein 3 | 0.430000 | 0.000327 | 0.001900 | 0.390000 | 0.000004 | 0.000093 | |
| CXCL14 | C-X-C motif
chemokine ligand 14 |
0.069000 | 0.000000 | 0.000000 | 0.027000 | 0.000000 | 0.000000 | ||
| DDR2 | discoidin domain receptor
tyrosine kinase 2 |
0.370000 | 0.000112 | 0.000776 | 0.330000 | 0.000005 | 0.000109 | ||
| DOCK5 | dedicator of
cytokinesis 5 |
0.450000 | 0.000182 | 0.001160 | 0.480000 | 0.000003 | 0.000066 | ||
| EMCN | endomucin | 0.490000 | 0.000613 | 0.003200 | 0.380000 | 0.000106 | 0.001450 | ||
| GIMAP6 | GTPase, IMAP
family member 6 |
0.420000 | 0.000002 | 0.000023 | 0.450000 | 0.000074 | 0.001080 | ||
| GNAI1 | G protein subunit
alpha i1 |
0.430000 | 0.000031 | 0.000265 | 0.400000 | 0.000049 | 0.000775 | ||
| HPSE | heparanase | 0.450000 | 0.000948 | 0.004610 | 0.480000 | 0.000025 | 0.000437 | ||
| IGKC | immunoglobulin
kappa constant |
0.230000 | 0.000241 | 0.001390 | 0.200000 | 0.000027 | 0.000475 | ||
| MZB1 | marginal zone B and B1
cell specific protein |
0.270000 | 0.000007 | 0.000079 | 0.470000 | 0.001146 | 0.009950 | ||
| PARVA | parvin alpha | 0.490000 | 0.007658 | 0.026000 | 0.435000 | 0.001358 | 0.010515 | ||
| RNF138 | ring finger protein 138 | 0.490000 | 0.000000 | 0.000000 | 0.460000 | 0.000003 | 0.000079 | ||
| SGCD | sarcoglycan delta | 0.400000 | 0.000152 | 0.000997 | 0.480000 | 0.000103 | 0.001420 | ||
Significant GOs and pathways
All of the identified genes were used to predict the functional categories with GO annotation. They were involved in different biological processes, molecular functions and cellular components. It was found that the differential expression of the 22 genes mainly participated in 153 significant GOs (Supplementary Table S4). It was concluded that the specific genes were mainly involved in immune response, metabolic process and cell adhesion. Blood coagulation, platelet activation and degranulation, acute-phase responses, negative regulation of endopeptidase activity, and complement activation may take part in liver metastasis of colon carcinoma.
S4.
GO annotation of 22 genes
| Gene_name | GO_name |
| ACSM2A | Medium-chain fatty-acyl-CoA
metabolic process |
| ACSM2A | Triglyceride homeostasis |
| ACSM2A | Fatty acid metabolic process |
| ACSM2A | Glucose homeostasis |
| APOB | Blood coagulation |
| APOB | Small molecule metabolic process |
| APOB | Cellular response to prostaglandin stimulus |
| APOB | Lipoprotein catabolic process |
| APOB | Triglyceride mobilization |
| APOB | Lipoprotein biosynthetic process |
| APOB | Regulation of cholesterol
biosynthetic process |
| APOB | Positive regulation of lipid storage |
| APOB | Positive regulation of cholesterol storage |
| APOB | Response to carbohydrate stimulus |
| APOB | Response to selenium ion |
| APOB | Very-low-density lipoprotein
particle assembly |
| APOB | Low-density lipoprotein particle clearance |
| APOB | Low-density lipoprotein particle remodeling |
| APOB | Positive regulation of macrophage derived foam cell differentiation |
| APOB | Cholesterol transport |
| APOB | Lipoprotein transport |
| APOB | Triglyceride catabolic process |
| APOB | Cholesterol efflux |
| APOB | Artery morphogenesis |
| APOB | Fertilization |
| APOB | Sperm motility |
| APOB | Lipoprotein metabolic process |
| APOB | Cellular response to tumor necrosis factor |
| APOB | Receptor-mediated endocytosis |
| APOB | Retinoid metabolic process |
| APOB | Phototransduction, visible light |
| APOB | Cholesterol homeostasis |
| APOB | Cholesterol metabolic process |
| APOB | Post-embryonic development |
| APOB | Leukocyte migration |
| APOB | Response to lipopolysaccharide |
| APOB | Response to virus |
| APOB | Lipid metabolic process |
| APOB | In utero embryonic development |
| Table S4 (continued) | |
In order to identify the key pathways the specific genes were involved in, we performed pathway analysis. Fifty-six KEGG biological pathways were annotated (Supplementary Table S5). The major regulated biological pathways include complement and coagulation cascades, metabolic pathways, PI3K-protein kinase B (AKT) signaling pathway, pathways in cancer, focal adhesion, Staphylococcus aureus infection, carbon metabolism, chemokine signaling pathway, and biosynthesis of amino acids. The results revealed the genes play an important role in pathways related to cancer cell migration, such as PI3K-AKT signaling pathway, focal adhesion and chemokine signaling pathway.
S5.
Pathway annotation of 22 genes
| Gene_name | Path_name |
| ACSM2A | Metabolic pathways |
| ACSM2A | Butanoate metabolism |
| APOB | Vitamin digestion and absorption |
| APOB | Fat digestion and absorption |
| CXCL14 | Chemokine signaling pathway |
| CXCL14 | Cytokine-cytokine receptor interaction |
| F5 | Complement and coagulation cascades |
| FTCD | Metabolic pathways |
| FTCD | One carbon pool by folate |
| FTCD | Histidine metabolism |
| GNAI1 | Chemokine signaling pathway |
| GNAI1 | Cocaine addiction |
| GNAI1 | Regulation of lipolysis in adipocytes |
| GNAI1 | Long-term depression |
| GNAI1 | Renin secretion |
| GNAI1 | Gastric acid secretion |
| GNAI1 | Pertussis |
| GNAI1 | Progesterone-mediated oocyte maturation |
| GNAI1 | Gap junction |
| GNAI1 | GABAergic synapse |
| GNAI1 | Morphine addiction |
| GNAI1 | Circadian entrainment |
| GNAI1 | Estrogen signaling pathway |
| GNAI1 | Melanogenesis |
| GNAI1 | Retrograde endocannabinoid signaling |
| GNAI1 | Chagas disease (American trypanosomiasis) |
| GNAI1 | Cholinergic synapse |
| GNAI1 | Serotonergic synapse |
| GNAI1 | Glutamatergic synapse |
| GNAI1 | Leukocyte transendothelial migration |
| GNAI1 | Toxoplasmosis |
| GNAI1 | Sphingolipid signaling pathway |
| GNAI1 | Axon guidance |
| GNAI1 | Dopaminergic synapse |
| GNAI1 | Platelet activation |
| GNAI1 | Tight junction |
| GNAI1 | Parkinson’s disease |
| GNAI1 | Adrenergic signaling in cardiomyocytes |
| GNAI1 | Oxytocin signaling pathway |
| GNAI1 | cGMP-PKG signaling pathway |
| GNAI1 | Alcoholism |
| GNAI1 | cAMP signaling pathway |
| Table S5 (continued) | |
Dynamic gene network analysis
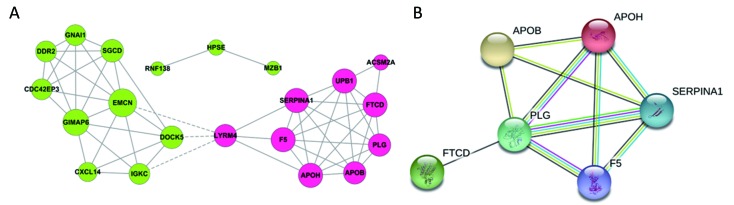
All the screened 22 DEGs were then subjected to a gene co-expression analysis network with k-core algorithm to determine which genes may play a potential role in the colon carcinoma metastasis. The gene-gene interaction network was constructed as shown in Figure 3A. The degree of a node describes the number of links of one gene with others, which had shown in the gene network. The node with larger diameter in the network means more important values. Importantly, six genes (SERPINA1, UPB1, FTCD, F5, EMCN, GIMAP6) belonged to the most significant genes, which involved in acute-phase response, metabolic process, angiogenesis, endothelial cell migration and proliferation, cell adhesion. The SERPINA1, UPB1, FTCD and F5 genes were up-regulated, but EMCN and GIMAP6 genes were down-regulated (Figure 3A). Furthermore, it was obvious that CXCL14, which was associated with cell migration and immune response, was also down-regulated.
3.
Functional associations between screened genes. (A) From the total differential genes, 22 specific genes about liver metastasis of colon carcinoma were constructed a gene co-expression network with k-core algorithm. Red cycle nodes represent up-regulated genes, blue cycle nodes represent down-regulated genes; (B) Protein-protein interaction (PPI) network of screened genes. Each node represents one gene; edges indicate the interaction relationship.
A PPI network of genes
To further define the interaction between the screened 22 DEGs, we used STRING database to construct the PPI network. The PPI network consisted of 6 nodes interacting by 29 edges, the remaining 16 DEGs failed to form the PPI pairs. It was concluded that FTCD, APOB, APOH, PLG, F5, SERPINA1 were closely linked (Figure 3B).
Prognostic values of highlighted DEGs
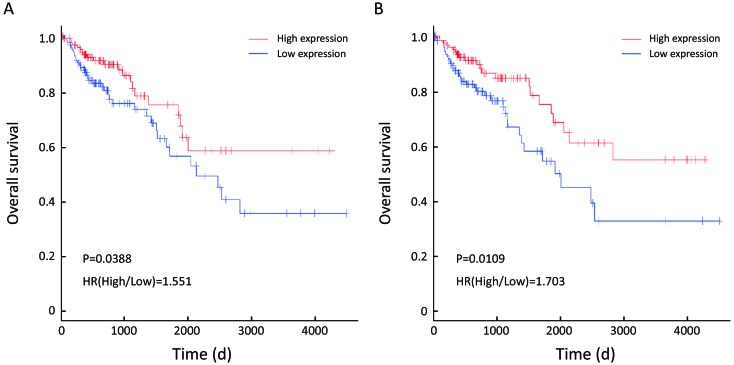
To evaluate the prognostic values of the 22 DEGs, we further investigated the associations of the DEGs with OS of patients by Kaplan-Meier and log-rank analysis. Because neither ACSM2A nor FTCD’s positive expression rate, the percentage of sample numbers with gene expression accounting for all sample numbers, was less than 50% in TCGA database, survival analysis was used to estimate the prognosis value of the other 20 genes. We found that patients with lower CXCL14, SERPINA1 expression demonstrated poorer survival than patients with higher expression (P=0.0388; P=0.0109; Figure 4). However, it was contradictory that SERPINA1 expression up-regulated in liver metastasis tissues indicated benefit prognosis. We therefore further researched the gene CXCL14 which were specifically involved in anti-liver metastasis process of colon carcinoma and predicted beneficial prognosis.
4.
Association of expression of C-X-C motif chemokine ligand 14 (CXCL14) and SERPINA1 with overall survival (OS) of 250 patients from The Cancer Genome Atlas (TCGA) data. Kaplan-Meier survival analysis of OS based on expression status provided associations of differential expression genes (DEGs) with OS of 250 patients from TCGA data. Cut-off values for genes were the median respectively. (A) CXCL14 [hazard rate (HR)=1.551; P=0.0388]; (B) SERPINA1 (HR=1.703; P=0.0109). With x-axis from left to right, the expression of CXCL14 was from high to low.
GSEA analysis of CXCL14
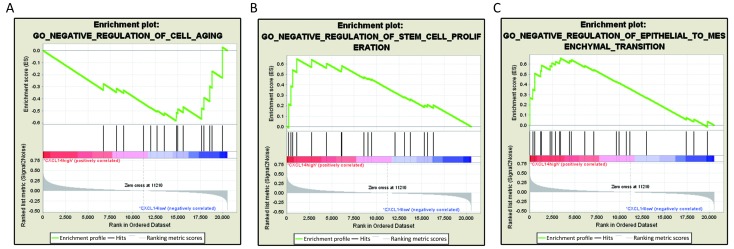
Based on above results, we have found that CXCL14 play a key role in liver metastasis of colon carcinoma. Then, it was quite necessary to predict biological functions of this gene. Analysis of GSEA, a powerful tool to infer the biological function, was performed. The results showed that genes associated with cell aging, negative regulation of stem cell proliferation and epithelial to mesenchymal transition (EMT), which were closely related to cancer metastasis (22-24) were significantly enriched in CXCL14-high samples of colon carcinoma (Figure 5). These observations suggested that CXCL14 may be a predicted indicator of patients with colon carcinoma liver metastasis.
5.
Gene set enrichment analysis (GSEA) analysis of C-X-C motif chemokine ligand 14 (CXCL14). GSEA showed that CXCL14 was associated with (A) Cell aging; (B) Stem cell proliferation; and (C) Epithelial to mesenchymal transition (EMT).
Validation of CXCL14 expression and its clinical relevance with clinical samples
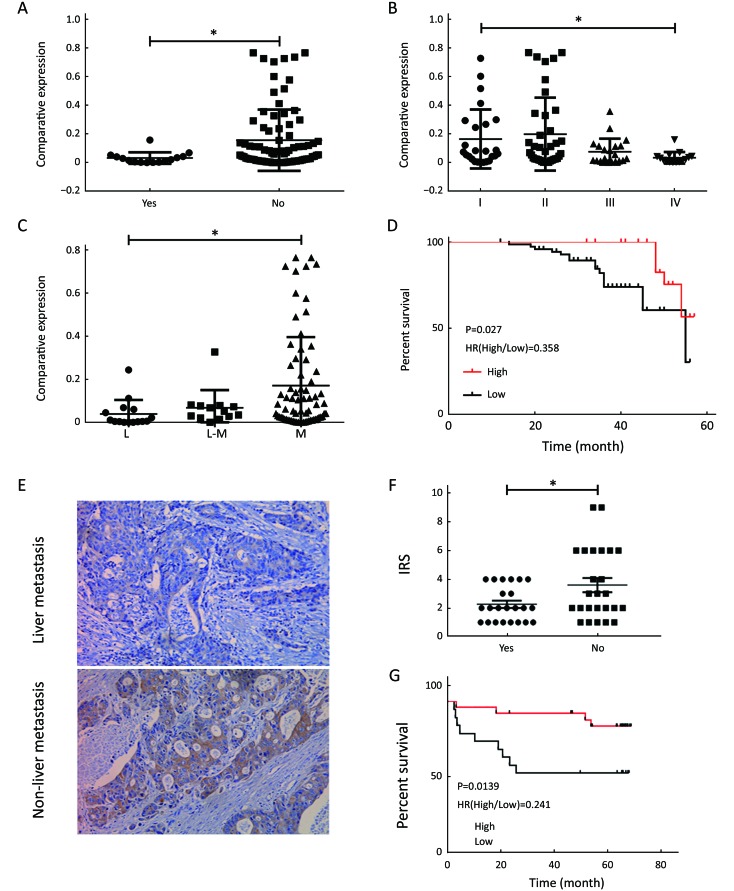
To further demonstrate the clinical significance of CXCL14 expression in patients with colon carcinoma, the association between CXCL14 expression and various clinicopathological variables was investigated by real-time quantitative PCR in 103 colon carcinoma patients. The clinicopathological data of the patients are detailed in Table 1. CXCL14 expression showed a high level in colon carcinoma patients with early stage, non-liver metastasis, middle histological differentiation (Figure 6A, B, C). At the protein level, the results also showed that CXCL14 expression was lower in patients with liver metastasis (Figure 6D, E). Then 103 colon carcinoma samples were stratified into “high” and “low” according to the median 0.045 127 of CXCL14 level. We found that low expression of CXCL14 was strongly correlated with advanced liver metastasis (P=0.01), overall stage (P=0.0001), abnormal CA72-4 value (P=0.0001), tumor size (P=0.001) and site of lesion (P=0.006) (Table 2).
6.
Association of C-X-C motif chemokine ligand 14 (CXCL14) expression with clinical characteristics and overall survival (OS) of patients with colon carcinoma. The mRNA expression level of CXCL14 in different groups of (A) Colon carcinoma patients with liver metastasis (Yes) and without liver metastasis (No); (B) TNM stage; and (C) Histological differentiation. L, low differentiation; M, moderate differentiation (n=103). (D) Kaplan-Meier curves show the association between mRNA expression level of CXCL14 and OS (n=103); (E) Immunohistochemical staining results of tumor tissue in colon cancer with liver metastasis, and without liver metastasis (×200); (F) Immune responsive score (IRS) of CXCL14 in colon cancer with liver metastasis (Yes) and without liver metastasis (No) (n=45); (G) Kaplan-Meier curves show the association between expression of CXCL14 and OS according to the immunohistochemical results (n=45) (*, P<0.05).
2.
Association between CXCL14 expression and clinicopathological features of patients with colon carcinoma (N=99)
| Variables | Total | CXCL14 | χ2 | P | |
| High | Low | ||||
| CXCL14, C-X-C motif chemokine ligand 14; CEA, carcinoembryonic antigen; CA, carbohydrate antigen. | |||||
| Gender | 0.020 | 0.887 | |||
| Male | 66 | 33 | 33 | ||
| Female | 33 | 17 | 16 | ||
| Age (year) | 0.250 | 0.617 | |||
| <60 | 51 | 27 | 24 | ||
| ≥60 | 48 | 23 | 25 | ||
| Site of lesion | 7.492 | 0.006 | |||
| Colon | 41 | 14 | 27 | ||
| Rectum | 58 | 36 | 22 | ||
| Pathology | 0.497 | 0.481 | |||
| Poor | 27 | 11 | 16 | ||
| Well | 72 | 39 | 33 | ||
| Tumor size (cm) | 10.924 | 0.001 | |||
| <4 | 62 | 34 | 28 | ||
| ≥4 | 37 | 16 | 21 | ||
| Pathological type | 1.611 | 0.204 | |||
| Adenocarcinoma | 87 | 46 | 41 | ||
| Others | 12 | 4 | 8 | ||
| Lymph node metastasis | 3.119 | 0.077 | |||
| No | 55 | 37 | 28 | ||
| Yes | 34 | 13 | 21 | ||
| Liver metastasis | 6.581 | 0.010 | |||
| No | 84 | 47 | 37 | ||
| Yes | 15 | 3 | 12 | ||
| Stage number | 14.696 | 0.0001 | |||
| I/II | 53 | 37 | 26 | ||
| III/IV | 36 | 13 | 23 | ||
| CEA | 0.863 | 0.353 | |||
| Normal | 67 | 36 | 31 | ||
| High | 32 | 14 | 18 | ||
| CA 19-9 | 0.304 | 0.581 | |||
| Normal | 79 | 41 | 38 | ||
| High | 20 | 9 | 11 | ||
| CA 72-4 | 13.142 | 0.0001 | |||
| Normal | 76 | 46 | 30 | ||
| High | 23 | 4 | 19 | ||
To examine the potential of CXCL14 to predict liver metastasis, logistic regression analysis was used. Univariate analyses revealed that low CXCL14 level [odds ratio (OR)=2.13; P=0.04], high CA 72-4 level (OR=6.9; P=0.01), and advanced overall stage (OR=6.0; P=0.02) were associated with liver metastasis. In multivariate analyses, CXCL14 (OR=1.24; P=0.03) and CA 72-4 levels (OR=2.35; P=0.04) were independent predictor of liver metastasis (Table 3).
3.
Logistic regression model analysis of liver metastasis predictors in patients with colon carcinoma
| Characteristics | Univariate | Multivariate | |||||
| OR | 95% CI | P | OR | 95% CI | P | ||
| CEA, carcinoembryonic antigen; CA, carbohydrate antigen; CXCL14, C-X-C motif chemokine ligand 14; OR, odds ratio; 95% CI, 95% confidence interval. | |||||||
| Sex (Female vs. Male) | 1.06 | 0.46−2.45 | 0.89 | 1.04 | 0.35−2.99 | 0.94 | |
| Age (<60vs. ≥60) (year) | 1.13 | 0.51−2.49 | 0.76 | 1.24 | 0.43−3.53 | 0.69 | |
| Pathology (Poor to Well) | 0.34 | 0.10−1.18 | 0.09 | 2.45 | 0.51−12.56 | 0.32 | |
| Tumor size (≥4 vs. <4) (cm) | 1.50 | 0.66−3.40 | 0.34 | 3.10 | 0.49−19.63 | 0.52 | |
| Lymph node metastasis (No vs. Yes) | 2.01 | 0.86−4.68 | 0.11 | 1.00 | 0.31−32.76 | 0.99 | |
| Stage number (III/IV vs. I/II) | 6.00 | 1.34−26.81 | 0.02 | 0.11 | 0.04−0.30 | 0.94 | |
| CEA (Normal vs. High) | 1.41 | 0.60−3.28 | 0.43 | 0.32 | 0.08−3.02 | 0.78 | |
| CA 19-9 (Normal vs. High) | 1.25 | 0.47−3.36 | 0.65 | 0.14 | 0.03−0.72 | 0.02 | |
| CA 72-4 (Normal vs. High) | 6.90 | 2.14−22.24 | 0.01 | 2.35 | 1.54−5.97 | 0.04 | |
| CXCL14 (Low vs. High) (Negative) | 2.13 | 1.51−3.49 | 0.04 | 1.24 | 0.43−3.53 | 0.03 | |
Moreover, Kaplan-Meier survival analysis revealed that higher CXCL14 expression was significantly associated with better survival in patients with colon carcinoma (Figure 6D, G). Simultaneously, as shown in Table 4, univariate analysis revealed that low CXCL14 level [hazard ratio (HR)=0.348; P=0.001] and liver metastasis (HR=2.742; P=0.037) were significantly associated with poor prognosis. Importantly, a multivariate Cox’s regression analysis revealed that CXCL14 level (HR=0.388; P=0.0001) and liver metastasis (HR=1.174; P=0.045) were independent prognostic factors for the OS of patients with colon carcinoma (Table 4). Collectively, these results suggest that CXCL14 expression status plays an important role in predicting prognosis and liver metastasis in patients with colon carcinoma.
4.
Cox’s proportional hazard model analysis of prognostic factors in patients with colon carcinoma
| Variables | Univariate | Multivariate | |||||
| HR | 95% CI | P | HR | 95% CI | P | ||
| CEA, carcinoembryonic antigen; CA, carbohydrate antigen; CXCL14, C-X-C motif chemokine ligand 14; HR, hazard ratio; 95% CI, 95% confidence interval. | |||||||
| Sex (Female vs. Male) | 1.343 | 0.548−3.294 | 0.533 | 1.138 | 0.641−2.021 | 0.658 | |
| Age (<60vs. ≥60) (year) | 0.944 | 0.400−2.226 | 0.893 | 1.083 | 0.634−1.851 | 0.769 | |
| Site of lesion (Rectum vs. Colon) | 0.810 | 0.342−1.921 | 0.632 | 0.867 | 0.473−1.587 | 0.643 | |
| Pathology (Poor to Well) | 1.687 | 0.520−5.480 | 0.287 | 0.463 | 0.352−1.632 | 0.341 | |
| Tumor size (≥4 vs. <4) (cm) | 1.259 | 0.520−3.049 | 0.594 | 1.162 | 0.656−2.060 | 0.607 | |
| Pathological type
(Adenocarcinoma vs. Others) |
1.870 | 0.405−8.632 | 0.527 | 0.696 | 0.303−1.597 | 0.392 | |
| Lymph node metastasis (No vs. Yes) | 1.630 | 0.625−2.369 | 0.265 | 0.497 | 0.092−2.674 | 0.415 | |
| Stage number (III/IV vs. I/II) | 1.875 | 0.788−5.407 | 0.140 | 3.342 | 0.522−21.382 | 0.203 | |
| CEA (Normal vs. High) | 0.774 | 0.310−1.933 | 0.560 | 0.940 | 0.493−1.795 | 0.852 | |
| CA 19-9 (Normal vs. High) | 1.231 | 0.427−3.565 | 0.698 | 1.223 | 0.490−3.057 | 0.666 | |
| CA 72-4 (Normal vs. High) | 1.490 | 0.499−4.447 | 0.475 | 3.132 | 0.906−10.821 | 0.071 | |
| CXCL14 (High vs. Low) | 0.348 | 0.181−0.668 | 0.001 | 0.388 | 0.245−0.617 | 0.0001 | |
| Liver metastasis (Yes vs. No) | 2.742 | 0.669−11.250 | 0.037 | 1.174 | 0.594−2.322 | 0.045 | |
Discussion
In relation to disease relapse, liver metastasis is the most major recurrent mode of colon carcinoma. When patients were firstly diagnosed, some of them were found to have distant metastasis, which might result in unfavorable prognosis. Thus, it is critical to identify an effective indicator that predicts the liver metastasis of colon carcinoma to provide new methods for therapy. It is notable that RNA-sequencing data and microarray-based expression profiling data provide a more comprehensive and accurate understanding of carcinogenesis and cancer progression at the molecular level. In this study, mRNA profiling by microarray from GEO was used to identify a number of novel genes related to colon carcinoma liver metastasis.
To identify new predictors regulating liver metastasis in colon carcinoma, we compared mRNA expression levels in M vs. N and T vs. N. Based on the fact that the GSE49355 and GSE62321 were from the same panel of patients but different platform, we took the intersection analysis rather than the union analysis for obtaining more accurate genes. The results showed that 22 genes were specifically related to liver metastasis. To further demonstrate their function and signaling pathway, we performed annotation analysis and verified that the genes were strongly associated with: 1) cell migration, adhesion, proliferation (cell adhesion/focal adhesion/ chemokine signaling pathway/PI3K-AKT signaling pathway/APOH/F5/CXCL14); and 2) immune response (innate immune response/complement activation/acute-phase response/SERPINA1/CXCL14).
It is well known that metastasis is closely related to colon cancer patients’ survival, and almost 80% of metastases occurred in liver. Therefore, we analyzed the prognosis value of the screened 22 specific liver metastasis genes though TCGA database. Because neither ACSM2A nor FTCD’s positive expression rate was less than 50%, survival analysis was used to focus on the other 20 specific genes. The results suggest that CXCL14 and SERPINA1 may be favorable prediction factors for colon carcinoma patients’ survival. However, our present study found that SERPINA1 expressed higher level in metastatic liver tissues when comparing with normal tissues. Recent studies have been reported that SERPINA1, a protease inhibitor that can act on a variety of targets such as serine proteases, has been proposed as a poor prognosis biomarker for various diseases, including papillary thyroid carcinoma (25), lung cancer (26) and breast carcinoma (27). As for our inconsistent results of SERPINA1, we decided to focus on the CXCL14 in proceeding research.
CXCL14, is an orphan member of the CXC chemokine subfamily. CXCL14 mRNA and protein are ubiquitously expressed in normal tissues, but are absent in tumor cell lines and in primary tumors (28,29). CXCL14 level in colon carcinoma tissues with lymphoid metastasis was significantly lower than that in tumor tissues without lymphoid metastasis (30). However, the effect of CXCL14 on colon carcinoma liver metastasis remains unclear. In our study, we provided evidences to show that the expression of CXCL14 was down-regulated in M vs. N and it was closely correlated with a beneficial survival outcome. Combined with the similar results that CXCL14 mediated suppression of tumor metastasis in lung cancer and Ewing sarcoma (31), we could conclude that it may play an important role in regulating colon carcinoma metastasis. On the contrary, Liu and colleagues previously described that CXCL14 induced metastasis (32). CXCL14-positive cancer associated fibroblast involved in ovarian cancer metastatic progression. This inconsistency may be caused by the intrinsic characteristic differences of different subtypes of human cancer.
Here, we firstly reported the role of CXCL14 in colon carcinoma liver metastasis. However, its underlying mechanism remains to be elucidated. Combined with the evidence that: 1) absence of CXCL14 expression in many malignant tissues is in agreement with the deficiency of effective antitumor immune responses in cancer patients. CXCL14 may act as chemo-attractant for monocytes, dendritic cells (DC) and (natural killer) NK cells; 2) CXCL14 may also influ¬ence the proliferation, invasion and migration of tumor cells via auto/paracrine pathways; 3) CXCL14 may suppress tumor vasculature by inhibiting the chemotaxis of vascular smooth muscle cells and the formation of microvas¬cular systems (33,34), and therefore suppresses the metabolism and growth of a tumor. Thus, it might indicate that CXCL14 plays an important role in regulating liver metastasis of colon carcinoma through suppression of cancer cells and migration of leukocytes.
Our data suggest that modulating CXCL14 expression could exert tumor suppression effects on colon carcinoma. CXCL14 expression is suppressed by epidermal growth factor (EGF) and can be restored by treatment with an EGF receptor (EGFR) tyrosine kinase inhibitor in head and neck squamous cell carcinoma (HNSCC) cells (35). Reversing the promoter hypermethylation of CXCL14 could be a feasible approach to restore anti-tumor immune responses to treat oral cancers (36). In summary, up-regulating the CXCL14 level may be a valuable adjuvant treatment to improve the outcomes of patients.
Conclusions
Taken together, our data firstly indicated that the CXCL14 expression level was down-regulated in metastatic liver tissues compared to non-tumor tissues. The absence of CXCL14 contributed to the cancer metastasis that then causes poor outcomes of patients. This is the first report that CXCL14 exerts an anti-metastasis effect on colon carcinoma via the screening of bioinformatics and the further validation of clinical samples. The expression level of CXCL14 may be a valuable adjuvant parameter to predict the liver metastasis and prognosis of patients with colon carcinoma and provides a potential future therapeutic strategy.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No.8177061284).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Weitz J, Koch M, Debus J, et al Colorectal cancer. Lancet. 2005;365:153–65. doi: 10.1016/S0140-6736(05)17706-X. [Weitz J, Koch M, Debus J, et al. Colorectal cancer. Lancet 2005;365:153-65] [DOI] [PubMed] [Google Scholar]
- 2.Pawlik TM, Choti MA Surgical therapy for colorectal metastases to the liver. J Gastrointest Surg. 2007;11:1057–77. doi: 10.1007/s11605-006-0061-3. [Pawlik TM, Choti MA. Surgical therapy for colorectal metastases to the liver. J Gastrointest Surg 2007;11:1057-77] [DOI] [PubMed] [Google Scholar]
- 3.Gao J, Li N, Dong Y, et al miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34:4142–52. doi: 10.1038/onc.2014.348. [Gao J, Li N, Dong Y, et al. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene 2015;34:4142-52] [DOI] [PubMed] [Google Scholar]
- 4.Li H, Rokavec M, Jiang L, et al Antagonistic effects of p53 and HIF1A on microRNA-34a regulation of PPP1R11 and STAT3 and hypoxia-induced epithelial to mesenchymal transition in colorectal cancer cells. Gastroenterology. 2017;153:505–20. doi: 10.1053/j.gastro.2017.04.017. [Li H, Rokavec M, Jiang L, et al. Antagonistic effects of p53 and HIF1A on microRNA-34a regulation of PPP1R11 and STAT3 and hypoxia-induced epithelial to mesenchymal transition in colorectal cancer cells. Gastroenterology 2017;153:505-20] [DOI] [PubMed] [Google Scholar]
- 5.Cohen SA, Yu M, Baker K, et al The CpG island methylator phenotype is concordant between primary colorectal carcinoma and matched distant metastases. Clin Epigenetics. 2017;9:46. doi: 10.1186/s13148-017-0347-1. [Cohen SA, Yu M, Baker K, et al. The CpG island methylator phenotype is concordant between primary colorectal carcinoma and matched distant metastases. Clin Epigenetics 2017;9:46] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galbán S, Apfelbaum AA, Espinoza C, et al A bifunctional MAPK/PI3K antagonist for inhibition of tumor growth and metastasis. Mol Cancer Ther. 2017;16:2340–50. doi: 10.1158/1535-7163.MCT-17-0207. [Galbán S, Apfelbaum AA, Espinoza C, et al. A bifunctional MAPK/PI3K antagonist for inhibition of tumor growth and metastasis. Mol Cancer Ther 2017;16:2340-50] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nannini M, Pantaleo MA, Maleddu A, et al Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev. 2009;35:201–9. doi: 10.1016/j.ctrv.2008.10.006. [Nannini M, Pantaleo MA, Maleddu A, et al. Gene expression profiling in colorectal cancer using microarray technologies: results and perspectives. Cancer Treat Rev 2009;35:201-9] [DOI] [PubMed] [Google Scholar]
- 8.Kleivi K, Lind GE, Diep CB, et al Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses. Mol Cancer. 2007;6:2. doi: 10.1186/1476-4598-6-2. [Kleivi K, Lind GE, Diep CB, et al. Gene expression profiles of primary colorectal carcinomas, liver metastases, and carcinomatoses. Mol Cancer 2007;6:2] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamasaki M, Takemasa I, Komori T, et al The gene expression profile represents the molecular nature of liver metastasis in colorectal cancer. Int J Oncol. 2007;30:129–38. [Yamasaki M, Takemasa I, Komori T, et al. The gene expression profile represents the molecular nature of liver metastasis in colorectal cancer. Int J Oncol 2007;30:129-38] [PubMed] [Google Scholar]
- 10.Koh KH, Rhee H, Kang HJ, et al Differential gene expression profiles of metastases in paired primary and metastatic colorectal carcinomas. Oncology. 2008;75:92–101. doi: 10.1159/000155211. [Koh KH, Rhee H, Kang HJ, et al. Differential gene expression profiles of metastases in paired primary and metastatic colorectal carcinomas. Oncology 2008;75:92-101] [DOI] [PubMed] [Google Scholar]
- 11.Koehler A, Bataille F, Schmid C, et al Gene expression profiling of colorectal cancer and metastases divides tumours according to their clinicopathological stage. J Pathol. 2004;204:65–74. doi: 10.1002/path.1606. [Koehler A, Bataille F, Schmid C, et al. Gene expression profiling of colorectal cancer and metastases divides tumours according to their clinicopathological stage. J Pathol 2004;204:65-74] [DOI] [PubMed] [Google Scholar]
- 12.Del Rio M, Mollevi C, Vezzio-Vie N, et al Specific extracellular matrix remodeling signature of colon hepatic metastases. PLoS One. 2013;8:e74599. doi: 10.1371/journal.pone.0074599. [Del Rio M, Mollevi C, Vezzio-Vie N, et al. Specific extracellular matrix remodeling signature of colon hepatic metastases. PLoS One 2013;8:e74599] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin AY, Chua MS, Choi YL, et al Comparative profiling of primary colorectal carcinomas and liver metastases identifies LEF1 as a prognostic biomarker. PLoS One. 2011;6:e16636. doi: 10.1371/journal.pone.0016636. [Lin AY, Chua MS, Choi YL, et al. Comparative profiling of primary colorectal carcinomas and liver metastases identifies LEF1 as a prognostic biomarker. PLoS One 2011;6:e16636] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright GW, Simon RM A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19:2448–55. doi: 10.1093/bioinformatics/btg345. [Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics 2003;19:2448-55] [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Crawford N, Lukes L, et al Metastasis predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis. 2005;22:593–603. doi: 10.1007/s10585-005-6244-6. [Yang H, Crawford N, Lukes L, et al. Metastasis predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis 2005;22:593-603] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gene Ontology Consortium The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–6. doi: 10.1093/nar/gkj021. [Gene Ontology Consortium. The Gene Ontology (GO) project in 2006. Nucleic Acids Res 2006;34:D322-6] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, et al Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Morris JH, Cook H, et al The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-D368] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer B, Sandmann T, Horn T, et al A map of directional genetic interactions in a metazoan cell. Elife. 2015;6:4. doi: 10.7554/eLife.05464. [Fischer B, Sandmann T, Horn T, et al. A map of directional genetic interactions in a metazoan cell. Elife 2015;6:4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu JY, Li F, Wang LP, et al CTL- vs Treg lymphocyte-attracting chemokines, CCL4 and CCL20, are strong reciprocal predictive markers for survival of patients with oesophageal squamous cell carcinoma . Br J Cancer. 2015;113:747–55. doi: 10.1038/bjc.2015.290. [Liu JY, Li F, Wang LP, et al. CTL- vs Treg lymphocyte-attracting chemokines, CCL4 and CCL20, are strong reciprocal predictive markers for survival of patients with oesophageal squamous cell carcinoma. Br J Cancer 2015;113:747-55 ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo C, Hou J, Ao S, et al HOXC10 up-regulation promotes gastric cancer cell proliferation and metastasis through MAPK pathway. Chin J Cancer Res. 2017;29:572–80. doi: 10.21147/j.issn.1000-9604.2017.06.12. [Guo C, Hou J, Ao S, et al. HOXC10 up-regulation promotes gastric cancer cell proliferation and metastasis through MAPK pathway. Chin J Cancer Res 2017;29:572-80] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reno TA, Kim JY, Raz DJ Triptolide inhibits lung cancer cell migration, invasion, and metastasis. Ann Thorac Surg. 2015;100:1817–24. doi: 10.1016/j.athoracsur.2015.05.074. [Reno TA, Kim JY, Raz DJ. Triptolide inhibits lung cancer cell migration, invasion, and metastasis. Ann Thorac Surg 2015;100:1817-24] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li S, Li Q Cancer stem cells and tumor metastasis (Review) Int J Oncol. 2014;44:1806–12. doi: 10.3892/ijo.2014.2362. [Li S, Li Q. Cancer stem cells and tumor metastasis (Review). Int J Oncol 2014;44:1806-12] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai LM, Lyu XM, Luo WR, et al EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN . Oncogene. 2015;34:2156–66. doi: 10.1038/onc.2014.341. [Cai LM, Lyu XM, Luo WR, et al. EBV-miR-BART7-3p promotes the EMT and metastasis of nasopharyngeal carcinoma cells by suppressing the tumor suppressor PTEN. Oncogene 2015;34:2156-66 ] [DOI] [PubMed] [Google Scholar]
- 25.Vierlinger K, Mansfeld MH, Koperek O, et al Identification of SERPINA1 as single marker for papillary thyroid carcinoma through microarray meta analysis and quantification of its discriminatory power in independent validation. BMC Med Genomics. 2011;4:30. doi: 10.1186/1755-8794-4-30. [Vierlinger K, Mansfeld MH, Koperek O, et al. Identification of SERPINA1 as single marker for papillary thyroid carcinoma through microarray meta analysis and quantification of its discriminatory power in independent validation. BMC Med Genomics 2011;4:30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Topic A, Ljujic M, Nikolic A, et al Alpha-1-antitrypsin phenotypes and neutrophil elastase gene promoter polymorphisms in lung cancer. Pathol Oncol Res. 2011;17:75–80. doi: 10.1007/s12253-010-9283-5. [Topic A, Ljujic M, Nikolic A, et al. Alpha-1-antitrypsin phenotypes and neutrophil elastase gene promoter polymorphisms in lung cancer. Pathol Oncol Res 2011;17:75-80] [DOI] [PubMed] [Google Scholar]
- 27.López-árias E, Aguilar-Lemarroy A, Felipe Jave-Suárez L, et al Alpha 1-antitrypsin: a novel tumor-associated antigen identified in patients with early-stage breast cancer. Electrophoresis. 2012;33:2130–7. doi: 10.1002/elps.201100491. [López-árias E, Aguilar-Lemarroy A, Felipe Jave-Suárez L, et al. Alpha 1-antitrypsin: a novel tumor-associated antigen identified in patients with early-stage breast cancer. Electrophoresis 2012;33:2130-7] [DOI] [PubMed] [Google Scholar]
- 28.Frederick MJ, Henderson Y, Xu X, et al In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue . Am J Pathol. 2000;156:1937–50. doi: 10.1016/S0002-9440(10)65067-5. [Frederick MJ, Henderson Y, Xu X, et al. In vivo expression of the novel CXC chemokine BRAK in normal and cancerous human tissue. Am J Pathol 2000;156:1937-50 ] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meuter S, Moser B Constitutive expression of CXCL14 in healthy human and murine epithelial tissues. Cytokine. 2008;44:248–55. doi: 10.1016/j.cyto.2008.08.009. [Meuter S, Moser B. Constitutive expression of CXCL14 in healthy human and murine epithelial tissues. Cytokine 2008;44:248-55] [DOI] [PubMed] [Google Scholar]
- 30.Lin K, Zou R, Lin F, et al Expression and effect of CXCL14 in colorectal carcinoma. Mol Med Rep. 2014;10:1561–8. doi: 10.3892/mmr.2014.2343. [Lin K, Zou R, Lin F, et al. Expression and effect of CXCL14 in colorectal carcinoma. Mol Med Rep 2014;10:1561-8] [DOI] [PubMed] [Google Scholar]
- 31.Sand LG, Scotlandi K, Berghuis D, et al CXCL14, CXCR7 expression and CXCR4 splice variant ratio associate with survival and metastases in Ewing sarcoma patients. Eur J Cancer. 2015;51:2624–33. doi: 10.1016/j.ejca.2015.08.020. [Sand LG, Scotlandi K, Berghuis D, et al. CXCL14, CXCR7 expression and CXCR4 splice variant ratio associate with survival and metastases in Ewing sarcoma patients. Eur J Cancer 2015;51:2624-33] [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Zhang J, Sun X, et al Down-regulation of miR-29b in carcinoma associated fibroblasts promotes cell growth and metastasis of breast cancer. Oncotarget. 2017;8:39559–70. doi: 10.18632/oncotarget.17136. [Liu Y, Zhang J, Sun X, et al. Down-regulation of miR-29b in carcinoma associated fibroblasts promotes cell growth and metastasis of breast cancer. Oncotarget 2017;8:39559-70] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hara T, Nakayama Y CXCL14 and insulin action. Vitam Horm. 2009;80:107–23. doi: 10.1016/S0083-6729(08)00605-5. [Hara T, Nakayama Y. CXCL14 and insulin action. Vitam Horm 2009;80:107-23] [DOI] [PubMed] [Google Scholar]
- 34.Ozawa S, Kato Y, Ito S, et al Restoration of BRAK/CXCL14 gene expression by gefitinib is associated with antitumor efficacy of the drug in head and neck squamous cell carcinoma. Cancer Sci. 2009;100:2202–9. doi: 10.1111/j.1349-7006.2009.01281.x. [Ozawa S, Kato Y, Ito S, et al. Restoration of BRAK/CXCL14 gene expression by gefitinib is associated with antitumor efficacy of the drug in head and neck squamous cell carcinoma. Cancer Sci 2009;100:2202-9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ang KK, Berkey BA, Tu X, et al Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–6. [Ang KK, Berkey BA, Tu X, et al. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res 2002;62:7350-6] [PubMed] [Google Scholar]
- 36.Konda JD, Olivero M, Musiani D, et al Heat-shock protein 27(HSP27, HSPB1) is synthetic lethal to cells with oncogenic activation of MET, EGFR and BRAF. Mol Oncol. 2017;11:599–611. doi: 10.1002/1878-0261.12042. [Konda JD, Olivero M, Musiani D, et al. Heat-shock protein 27(HSP27, HSPB1) is synthetic lethal to cells with oncogenic activation of MET, EGFR and BRAF. Mol Oncol 2017;11:599-611] [DOI] [PMC free article] [PubMed] [Google Scholar]