The gut microbiota is essential for the survival of many organisms, including ruminants that rely on microorganisms for nutrient acquisition from dietary inputs for the production of products such as milk and meat. While alteration of the adult ruminant microbiota to improve production is possible, changes are often unstable and fail to persist. In contrast, the early-life microbiota may be more amenable to sustained modification. However, few studies have determined the impact of early-life interventions on downstream production. Here, we investigated the impact of agriculturally relevant calf diets, including calf starter and corn silage, on gut microbial communities, growth, and production through the first lactation cycle. Thus, this work serves to further our understanding of early-life microbiota acquisition, as well as informing future practices in livestock management.
KEYWORDS: dairy calf, dairy cow, microbiota, rumen
ABSTRACT
Gastrointestinal tract (GIT) microorganisms play important roles in the health of ruminant livestock and affect the production of agriculturally relevant products, including milk and meat. Despite this link, interventions to alter the adult microbiota to improve production have proven ineffective, as established microbial communities are resilient to change. In contrast, developing communities in young animals may be more easily altered but are less well studied. Here, we measured the GIT-associated microbiota of 45 Holstein dairy cows from 2 weeks to the first lactation cycle, using Illumina amplicon sequencing of bacterial (16S rRNA V4), archaeal (16S rRNA V6 to V8), and fungal (internal transcribed region 1 [ITS1]) communities. Fecal and ruminal microbiota of cows raised on calf starter grains and/or corn silage were correlated to lifetime growth as well as milk production during the first lactation cycle, in order to determine whether early-life diets have long-term impacts. Significant diet-associated differences in total microbial communities and specific taxa were observed by weaning (8 weeks), but all animals reached an adult-like composition between weaning and 1 year. While some calf-diet-driven differences were apparent in the microbiota of adult cows, these dissimilarities did not correlate with animal growth or milk production. This finding suggests that initial microbial community establishment is affected by early-life diet but postweaning factors have a greater influence on adult communities and production outcomes.
IMPORTANCE The gut microbiota is essential for the survival of many organisms, including ruminants that rely on microorganisms for nutrient acquisition from dietary inputs for the production of products such as milk and meat. While alteration of the adult ruminant microbiota to improve production is possible, changes are often unstable and fail to persist. In contrast, the early-life microbiota may be more amenable to sustained modification. However, few studies have determined the impact of early-life interventions on downstream production. Here, we investigated the impact of agriculturally relevant calf diets, including calf starter and corn silage, on gut microbial communities, growth, and production through the first lactation cycle. Thus, this work serves to further our understanding of early-life microbiota acquisition, as well as informing future practices in livestock management.
INTRODUCTION
The mammalian gastrointestinal tract (GIT) houses a diverse microbial community that can significantly affect both the lifetime and evolutionary success of its host (1). These microbial communities contribute to host survival by extracting nutritionally (2) and developmentally (3) relevant compounds from dietary inputs. Microbial colonization of the GIT begins early in life (4, 5) and gut communities, once established, are resistant to change without extreme or sustained interventions (6, 7). Therefore, efforts to alter the GIT microbiota to improve any number of host outcomes may be most effective in early life, before microbial communities have reached an adult-like steady state. However, factors affecting microbial acquisition and long-term maintenance in the GIT are not well understood.
In mammals, there is a dramatic shift toward an adult-like microbiota during weaning, likely as a result of changes in diet (for a review, see reference 8). This indicates that diet is a strong contributing factor in the establishment of the gut microbiota and could serve as a tool to directionally alter the microbiota in early life. Manipulation of the microbiota through diet is of interest in a number of areas, particularly in agriculture, where the use of other therapeutics (such as antibiotics and probiotics) is tightly regulated (9). In the beef and dairy industries, differences in the microbiota have been linked to bovine milk production (10, 11), weight gain (12), and methane emissions (13). Strong associations occur between these traits and the microbiota of the rumen, because this enlarged foregut compartment houses the microbial community responsible for most dietary digestion and fermentation (14). Therefore, it may be possible to utilize diet to manipulate bovine GIT microbial communities to increase animal performance (15).
Although diet has been investigated as a means of altering the adult cow microbiota (16, 17) and production (18, 19), calf feed has not been as fully assessed. Previous dietary work with dairy calves focused on weight gain, as this is positively associated with increased downstream milk production (20). However, studies investigating the calf GIT microbiota failed to follow calves to maturity (21–23). Given that early-life feeding has demonstrated long-term effects on the adult microbiota of other ruminants (24–26) and that microbial communities in preweaned dairy calves are highly variable (27, 28) and amenable to change (6), we posit that early-life dietary interventions in dairy calves can affect microbial colonization, with long-term consequences for the adult cow microbiota and production.
To investigate this, we utilized Illumina amplicon sequencing of bacterial (16S rRNA V4), archaeal (16S rRNA V6 to V8), and fungal (internal transcribed region 1 [ITS1]) communities of dairy cows from 2 weeks after birth to the middle of the first lactation cycle. We compared calves raised on low-fiber, high-protein starter grains (previously investigated in reference 28) to those raised on high-fiber corn silage or a mixture of starter grains and corn silage. We then correlated the fecal and ruminal microbiota with outcomes including weight gain and milk production, to determine the short- and long-term impacts of the calf diet.
RESULTS
Sample collection and sequencing.
In this study, 9 bulls were retained through weaning (8 weeks) and 33 cows were retained through the first lactation cycle (>2 years). Three cows were removed from the study prior to the 2-year sampling due to poor health (cow 5013), reproductive issues (cow 5055), and a broken leg (cow 5025). Feces were sampled from animals at 2, 4, and 8 weeks and 1 and 2 years of age, during their first lactation cycle, while ruminal contents were sampled from bulls sacrificed at weaning and from a subset of cannulated cows (n = 12) at 1 and 2 years. Feces were not sampled from 5 cows at 2 weeks and from 8 cows at 8 weeks. Replacement samples were obtained at 3 weeks but samples were not taken at 9 weeks, because heifers were on a different diet by that time (see Table S1 in the supplemental material). All other fecal samples (n = 183) and all ruminal samples (n = 162) were obtained at the target age (Table S2).
Archaeal, bacterial, and fungal amplicons were sequenced for all ruminal samples, and archaeal and bacterial amplicons were sequenced for all fecal samples. Fungal PCR of calf feces failed to yield sufficient DNA; therefore, only fecal samples from 1- and 2-year old cows were sequenced for this amplicon. After filtering in mothur, 540,000 high-quality archaeal sequences (mean ± standard error [SE], 1,500 ± 85 sequences per sample), 13.9 million high-quality bacterial sequences (mean ± SE, 39,000 ± 1,000 sequences per sample), and 5.3 million high-quality fungal sequences (mean ± SE, 23,000 ± 985 sequences per sample) were obtained. Sequence coverage was deemed sufficient, with Good’s coverage values of >96.5%, for all bacterial and fungal communities and most archaeal communities. A total of 16 fecal archaeal communities (2 weeks, 12 communities; 4 weeks, 3 communities; 8 weeks, 1 community) had low Good’s coverage values (0 to 94%) even after repeated sequencing yielded a minimum of 30,000 contigs per sample (Table S2). After normalization, samples retained sufficient Good’s coverage for archaeal (>86%), bacterial (>88%), and fungal (>75%) samples.
Calf diet correlation with microbiota at weaning.
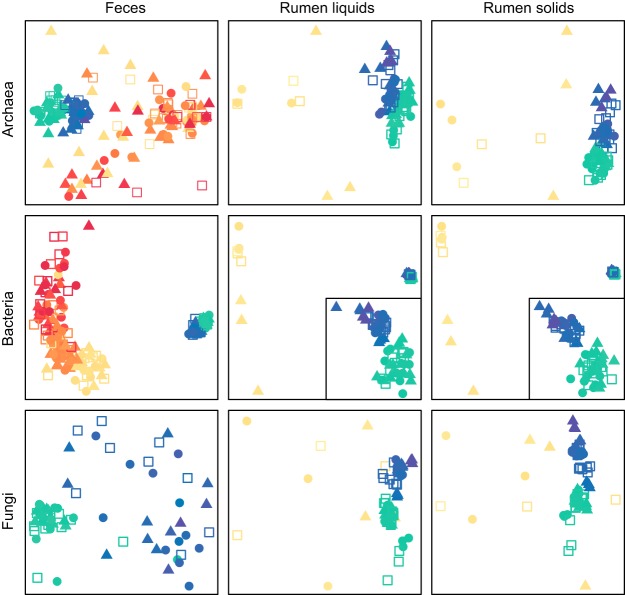
Calves increased consumption of supplemental feeds from birth to weaning (8 weeks) in an exponential manner (Fig. S1), and there were no differences in the log values of supplement consumption between diet groups (P = 0.681, repeated-measures linear regression). Calf diet did not significantly correlate with the overall structure or composition of any ruminal community at weaning (permutational multivariate analysis of variance [PERMANOVA]) (Fig. 1 and Table S3). However, silage-fed calves had lower bacterial diversity with greater archaeal diversity and fungal richness in ruminal liquids; bacterial diversity was also significantly greater in silage-fed ruminal solids (analysis of variance [ANOVA] and Tukey’s honest significant difference [HSD] test) (Fig. 2; also see Fig. S2 and Table S3). Ruminal archaeal communities of silage-fed calves had greater interanimal variation at 8 weeks (permutational analysis of multivariate dispersions [PERMDISP] and Tukey’s HSD test) (Table S3).
FIG 1.
Total fecal and ruminal microbiota in dairy cows raised on different diets. nMDS plots of Bray-Curtis diversity index values for archaeal, bacterial, and fungal communities in feces and ruminal liquids and solids are presented. Points are colored by animal age, i.e., 2 weeks (red), 4 weeks (orange), 8 weeks (yellow), 1 year (green), and 2 years (blue). Calf diet is indicated by circles (calf starter), triangles (corn silage), and open squares (mixed diet). Ruminal bacterial plots have inset plots displaying the 1- and 2-year clusters in more detail.
FIG 2.
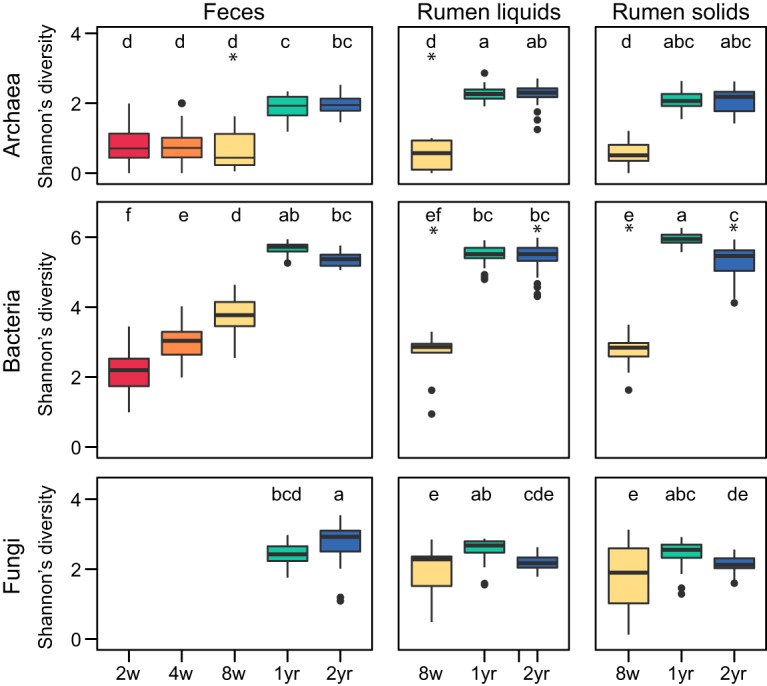
Diversity of fecal and ruminal microbiota in cows raised on different diets. Shannon’s diversity index values for archaeal, bacterial, and fungal communities in feces and ruminal liquids and solids are presented. Boxes are colored by animal age, i.e., 2 weeks (red), 4 weeks (orange), 8 weeks (yellow), 1 year (green), and 2 years (blue). Ages with significantly different diversity index values across amplicons are indicated by different letters. Asterisks denote groups containing significant diet differences (P < 0.05, Tukey’s HSD test) (Table S3).
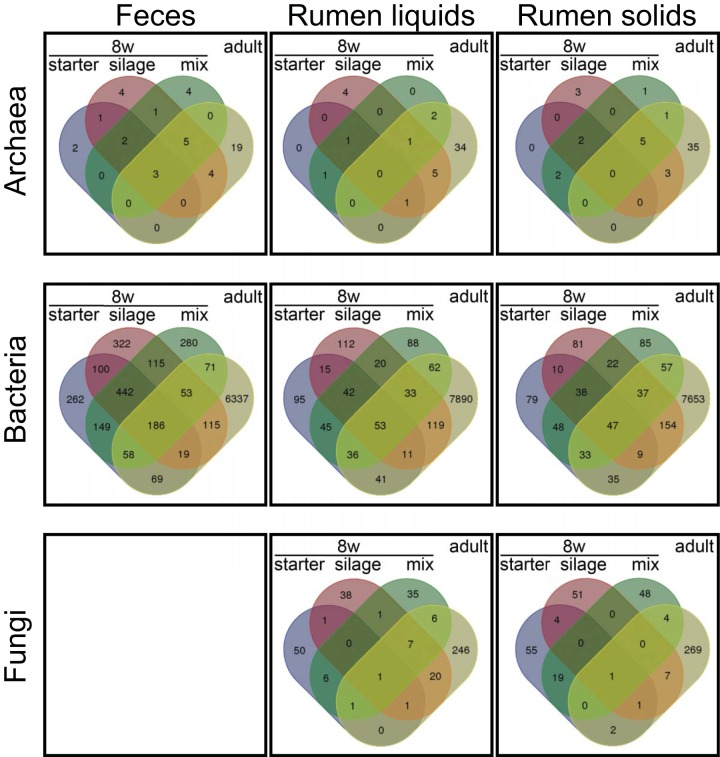
Among the three calf diets, the ruminal microbiota of silage-fed calves at weaning had the most operational taxonomic units (OTUs) in common with adult communities and those of starter-fed calves had the least (Fig. 3). In particular, the ruminal communities of silage-fed animals had greater abundances of a number of adult-associated OTUs that were absent in starter-fed calves (Table S4). Notably, silage-fed animals also had lower abundances (0 to 60%) of a calf-associated Methanobrevibacter OTU (A-OTU1), which dominated starter- and mixed-diet-fed animals to >90% but was absent in all adult animals. In contrast, starter-fed and/or mixed-diet-fed calves had more highly abundant OTUs at weaning that were not present in adults (Table S4).
FIG 3.
Microbial taxa shared by calves and cows. Venn diagrams of OTUs shared by calves at 8 weeks and adult cows are shown. Calf groups are split by diet, and the “adult” category includes all samples at 1 and 2 years.
Short-chain organic acid (SCOA) concentration profiles in ruminal liquids (Table S5) varied with animal age (PERMANOVA) (Table S3) and covaried with the overall archaeal (r = 0.400, P = 4.95E−02) and bacterial (r = 0.556, P = 0.010) but not fungal (Mantel test, P = 0.129) microbiota. Regardless of diet, calves had significantly higher concentrations and molar fractions of butyrate, propionate, and valerate but lower molar fractions of acetate and total branched volatile fatty acid (VFAs) (ANOVA) (Table S3 and Fig. S3). Calves also tended to accumulate higher concentrations of fermentation intermediates, including ethanol, lactate, and succinate, than did cows.
Overall, calves fed starter had the highest concentrations of total VFAs, with both starter-fed and mixed-diet-fed calves reaching total levels similar to those of adult cows (Fig. S3). Concentrations and molar fractions of specific SCOAs varied across calf diet groups, without consistent trends across diets. Unlike adult cows, silage-fed calves had the highest concentrations of formate and succinate. Similar to adult cows, however, silage-fed calves also had the lowest concentrations of ethanol, propionate, and valerate, as well as the lowest molar proportions of propionate. Starter-fed calves were similarly inconsistent, with high lactate concentrations but also high, adult-like, concentrations of acetate and molar proportions of valerate being common among calves (Fig. S3). Calf butyrate concentrations occurred within the same range as for adult cows, with starter-fed calves having the highest concentrations among calves. However, butyrate molar proportions did not differ among diet/age groups (Table S3).
Few specific OTUs correlated with SCOA concentrations, and none correlated with molar fractions (Table S4). Strong correlations were mostly driven by age, with calf-associated OTUs being positively correlated with calf-associated fermentation intermediates. Specifically, calf-dominating Methanobrevibacter (A-OTU1) was positively associated with ethanol and lactate concentrations. Also, an unclassified fungal OTU found only in calves (F-OTU46) was positively associated with ethanol. With the removal of age as a factor, to investigate possible dietary effects, only Methanobrevibacter (A-OTU1) remained strongly correlated with ethanol in calves alone (Table S4).
In contrast to the ruminal microbiota, calf diet significantly correlated with overall fecal archaeal and bacterial communities in calves. At 4 weeks, corn silage and mixed diet archaeal communities differed in structure (PERMANOVA) (Table S3). At weaning (8 weeks), archaeal and bacterial microbiota from corn-silage-fed calves differed in both structure and composition from those of calves fed starter or mixed diet (PERMANOVA) (Table S3). Also, fecal archaeal communities of silage-fed calves had greater interanimal variation at 8 weeks (PERMDISP and Tukey’s HSD test) (Table S3). Similar to trends observed for ruminal samples, more archaeal and fungal OTUs in feces were shared between adults and silage-fed calves, compared to calves fed the other diets (Fig. 3). Specific trends were not apparent for bacterial OTUs, due to the high diversity and interanimal variation among calf fecal samples.
Calf diet influence on long-term ruminal microbiota.
Overall, all calves achieved an adult-like microbiota between weaning and 1 year of age, although additional differences were apparent between 1- and 2-year-old cows (Fig. 1 and 2). Adult cows generally displayed lower beta-diversity than calves (Table S6), and calf diet was significantly correlated with the structure and/or composition of some adult ruminal but not fecal microbial communities (PERMANOVA) (Table S3). Overall, at 1 year, fungi differed in animals raised on corn silage and bacteria differed across all diets. At 2 years, archaea and bacteria differed in animals raised on calf starter and fungi differed across all diets (PERMANOVA) (Table S3). Additionally, 2-year bacterial diversity was higher in the rumens of cows raised on silage, relative to a mixed diet (ANOVA and Tukey’s HSD test), and interanimal variation in ruminal microbial communities differed within calf diet groups (PERMDISP and Tukey’s HSD test) (Table S3). No significant diet effects were observed in the structure, composition, variation, or diversity of communities in feces from 1- or 2-year-old animals.
In general, the ruminal communities of cows raised on different calf diets contained similar taxa but differed in the abundance of OTUs within the taxa. The taxa included the bacterial taxa Prevotella, Succiniclasticum, and unclassified Succinivibrionaceae and the fungal taxa Caeomyces, Piromyces, and Oripinomyces; Methanobrevibacter archaeal OTUs also differed across diets. In contrast, silage-fed cows had more abundant Fibrobacter, while starter-fed and mixed-diet-fed cows had more abundant Cyllamyces.
Overall SCOA profiles and levels of specific SCOAs were significantly different in 2-year-old cows raised on different diets (PERMANOVA) (Table S3). Cows raised on the mixed diet had significantly different profiles (PERMANOVA), as well as higher concentrations of acetate and butyrate (ANOVA) (Table S3 and Fig. S3), than cows raised on starter or silage alone. Also, levels of the branched VFAs isobutyrate and isovalerate plus 2-methylbutyrate were higher in 2-year-old cows that were raised on the mixed diet, despite not differing by diet in calves. Molar fractions of VFAs did not differ among cows raised on different diets.
Lack of calf diet correlation with production measures.
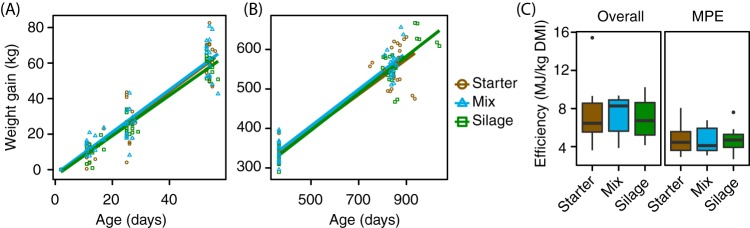
Animals gained weight from 2 to 8 weeks (1.18 kg/day; P = 6.69E−30) and from 8 weeks to lactation (0.456 kg/day; P = 1.35E−30) following linear trends, with faster growth in calves. Calf diet did not affect weight gain in calves (2 to 8 weeks; P = 0.432) (Fig. 4A) or in adults (8 weeks to >2 years; repeated-measures linear regression, P = 0.797) (Fig. 4B). Additionally, neither 2-year milk production efficiency (MPE) (ANOVA, P = 0.965) nor overall efficiency (P = 0.959) varied by calf diet (Fig. 4C). No alpha- or beta-diversity measure of microbial communities significantly correlated with efficiency or MPE (PERMANOVA and ANOVA) (Table S3). However, one unclassified Neocallimastigomycota OTU (F-OTU19) displayed a strong positive correlation with efficiency (Kendall test) (Table S4). SCOA profile structure and composition correlated with MPE (PERMANOVA); however, no individual SCOA levels significantly differed overall or by MPE (ANOVA) (Table S3).
FIG 4.
Calf diet effects on weight gain and milk production. (A and B) Weight gain of animals over time from 2 days to weaning (8 weeks) (A) and from 8 weeks to the first lactation cycle (B). Data were normalized to initial day 2 weights. (C) Milk and overall efficiency of dairy cows raised on different diets. Overall efficiency was determined by total energy expenditure (mastitis, gestation, weight maintenance and gain/loss, and milk production) (Table S8) per kilogram of DMI. MPE was determined as total milk production per kilogram of DMI. Calf starter (brown), corn silage (green), and mixed diet (blue) are indicated.
DISCUSSION
Purposeful alteration of GIT microbial communities has been proposed as a means to improve production of agriculturally relevant products such as milk (10, 11). Early-life interventions are a promising avenue, because the developing microbiota in preweaned animals may be more easily altered than those in adults (6, 27, 28). Here, we compared the GIT microbiota and production traits of dairy cows raised on calf starter grains (investigated in depth previously [28]) to those of cows raised on corn silage or a mixture of starter and silage, in order to determine whether early-life diets had long-term effects on the adult cows.
Similar to calves raised on calf starter (28), GIT microbial communities in calves fed silage or mixed diets became more alike and more diverse with age. Animals had a succession of microorganisms representing similar taxa but different specific OTUs present at different ages. All animals achieved an adult-like microbiota between weaning (8 weeks) and 1 year of age, regardless of diet. Therefore, age is a major driving force in the establishment of microbial communities in developing dairy calves, confirming previous reports (27, 28). In this study, however, diet-driven differences were also apparent, indicating that preweaning feed has a lesser, but still detectable, effect on the developing GIT microbiota.
While all calves progressed toward a similar adult microbiota, silage-fed animals achieved a more adult-like microbiota by weaning (8 weeks), compared to the other diets. Specifically, silage-fed calves at weaning had greater numbers and abundances of OTUs shared with adults, as well as fewer and less abundant OTUs strictly present in calves (Fig. 3). At weaning, there was a striking difference in archaeal communities, with silage-fed animals transitioning away from the calf-dominating Methanobrevibacter (A-OTU1) and away from the calf-associated unclassified fungi (see Table S4 in the supplemental material).
Importantly, under typical modern production conditions, dairy calves receive minimal maternal care and have limited interaction with adult animals (29). Calves are exposed to adult-cow-associated microorganisms present in the environment or through contact with humans who interact with adult animals (28). Our data suggest that a calf diet of corn silage may accelerate ruminal microbiota development into a more adult-like state by continually seeding the rumen with fiber, thus making complementary resources and nutrients readily available whenever exposure to adult-associated microbes occurs. In this way, corn-silage-fed calves are likely able to maintain an adult-like ruminal microbiota well before full GIT development.
In addition, analysis of ruminal SCOAs indicated accelerated microbiota development in corn-silage-fed calves at weaning. Specifically, silage was associated with lower concentrations of VFA intermediates, such as ethanol and lactate. These VFA intermediates generally do not accumulate in adult ruminants, as members of the microbiota convert them to VFAs that are readily absorbed by the host (14). Conversion away from intermediates in silage-fed calves suggests that a more complex, adult-like microbiota was present. Similar trends have been seen in calves reared with the addition of calf starter grains to a milk-only diet (22), and the more pronounced results in this study may be due to the greater similarity of silage to adult feeds, in terms of protein, starch, and fiber contents (Table S1). Silage-fed calves also had low, adult-like concentrations and proportions of propionate, the main substrate of gluconeogenesis in ruminants (30). This is in contrast to starter-fed and mixed-diet-fed calves, which had significantly higher propionate levels that may indicate reduced GIT development and a failure to meet nutritional needs from ingested feeds (30).
Some calf-diet-driven differences in the microbiota were still present in adult animals, a phenomenon also observed in goats and sheep (24–26). Similar to age-related changes (28), overall community structures (Bray-Curtis index values) differed, and comparable taxa but different OTUs were present in adult cows raised on different diets. The presence or abundance of adult-associated OTUs at weaning had no correlation with actual adult OTU abundances, and no significant differences in OTU abundances in calves persisted in adults. Therefore, the apparent long-term effects of calf diet on adult microbial communities appear to be weak and indirect. These effects may be mediated by differences in internal development, as supplements have been shown to affect rumen size, weight, and/or papillation (31–34), or by different programming of the immune system by early-life microorganisms (3, 35). Taken together, these findings indicate that early-life feeds aimed at physiological and/or immune system development, as mediated by the microbiota or other factors, may have more impact on later adult microbiota than those targeted at the establishment of specific adult-associated microorganisms.
Although the calf diet correlated with the ruminal microbiota of adult cows, it did not significantly affect weight gain, milk production, or overall efficiency. Additionally, no adult microbial communities differed as a result of the calf diet. This finding indicates that the weaning transition and factors after weaning contribute significantly to the establishment of an adult-like microbiota and may allow underdeveloped animals to “catch up,” such that early-life differences are no longer apparent or have an impact on production. Thus, we propose that efforts to directionally alter the microbiota toward improved production and efficiency should be explored in calves during the weaning transition, prior to the stabilization of host-specific communities observed in adult cows.
Overall, this work concurrently evaluated the effects of preweaning diet on growth, milk production, and associated GIT microbial communities in dairy cows over time. While it does not appear that the preweaning diet can be used to promote specific microorganisms or to improve efficiency in adult cows, there were no significant detriments to feeding calves corn silage rather than commercially prepared calf starter grains. Given that corn silage is often significantly cheaper than calf starter grains and is more readily accessible on dairy farms, feeding corn silage to calves could result in substantial cost savings for dairy producers. Our study also confirms that preweaning calf management appears to be most effective at promoting physiological development and animal growth, as has historically been the case, while the weaning transition may be an opportune time to alter microbial communities for long-term production gains. These results have implications for management and feeding practices across numerous agricultural systems, as more efficient, less environmentally costly food production is needed to meet global demands.
MATERIALS AND METHODS
Animals and diet.
This study was completed under animal use protocol A01501, approved by the Institutional Animal Care and Use Committee of the College of Agriculture and Life Sciences, University of Wisconsin-Madison. Three cohorts of 15 Holstein dairy calves (3 male and 12 female calves each; total n = 45) were raised at the U.S. Dairy Forage Research Center (USDFRC) research farm near Prairie du Sac, Wisconsin. All calves received pasteurized milk with Milk Balancer protein blend (Land O’Lakes, St. Paul, MN) added to 15% total milk solids. From 0 to 5 days of age, calves were fed 6 quarts (∼5.7 liters) of milk per day; from 6 days until weaning (approximately 56 days), they were fed 8 quarts (∼7.6 liters) per day. From birth, each cohort was also offered ad libitum access to one of three randomly assigned supplemental feeds, namely, calf starter (58.25% whole corn, 1.75% molasses, and 40% Future Cow Ampli-Calf Mixer Pellet B150; Purina Animal Nutrition, Shoreview, MN) (diet A) (28), corn silage (USDFRC) (diet B), or a 25:75 (by weight) calf starter/corn silage mixture (approximately 50:50 by dry matter) (diet C). Detailed nutritional analyses of feeds are provided in Table S1 in the supplemental material. Supplement offered to and refused by each calf was weighed daily to determine supplement intake (Table S7).
After weaning, calves were transitioned through a series of standard heifer diets until they were placed, after calving, on the lactating herd total mixed ration (TMR) diet of approximately 58% silage/haylage, 11% grains, 22% corn, 7% soy, and 3% minerals (by dry matter) (Table S1). TMR offered to and refused by each cow was weighed and subsamples were taken daily during the milk sampling period (150 to 158 days in milk [DIM]), to determine nutrient intake using established methods (36). All animals had ad libitum access to water throughout the trial.
Rumen sampling and feces collection.
Samples were collected between June 2012 and July 2015. The 9 male calves (3 per diet cohort) were sacrificed near weaning (56 to 68 days) to obtain ruminal samples. Ruminal liquids were obtained by straining total ruminal contents through four layers of cheesecloth. Solids remaining in the cheesecloth were squeezed to remove free liquid and were transferred to a separate sterile container. Twelve heifers (9.5 to 11 months of age, 4 per diet cohort) were ruminally cannulated (animal use protocol A01307), and ruminal liquids and solids were collected through the cannula before morning feeding on 3 consecutive days at 1 year (365 ± 7 days) and in the middle of the first lactation cycle after 2 years (154 to 156 DIM) (Fig. S4). All samples were immediately transported on wet ice and stored at −80°C prior to DNA extraction.
Fresh feces were obtained by hand from the rectum of animals, using clean nitrile gloves. Samples were taken at 2 weeks of age (14 ± 3 days [n = 40]) or 3 weeks of age (21 ± 2 days [n = 5]), in an effort to avoid sampling during illness. Feces were also collected from all animals at 4 weeks (27 ± 2 days), 8 weeks (54 ± 2 days), and 1 year (365 ± 7 days) of age and in the middle of the first lactation cycle (155 ± 1 DIM) (Fig. S4). Samples were stored at −20°C on site, transported on wet ice, and then stored at −80°C prior to DNA extraction.
Milk collection.
Total milk production was measured for each cow over 9 consecutive days during the middle of the first lactation cycle (151 to 159 DIM), offset 1 day from feed sampling because morning milk production is affected by the previous day’s feed. Milk samples were collected from all three daily milkings on those days. Milk was stored at 4°C and submitted to AgSource Cooperative Services (Verona, WI) for near-infrared spectroscopic prediction to determine composition, including fat, lactose, protein, nonfat solids, and milk urea nitrogen levels and somatic cell counts (SCCs) (37).
Growth and health.
Cow health was assessed by standard daily monitoring for disease, including fecal and attitude scores, as described (28). Calves were treated with antibiotics and electrolytes for scours (diarrhea) and with antibiotics plus fever reducer for respiratory disease, as required. Cows were treated with propylene glycol for ketosis, to restore energy balance, and with antibiotics for mastitis, as necessary. Calf fecal samples taken during treatment for scours (n = 16) or respiratory disease (n = 3) are noted (Table S2). No samples were taken during illness among adult animals.
Calves were weighed at 2 days and near the time of fecal sampling from 2 to 8 weeks. Cows were weighed on 3 consecutive days at 1 year (365 to 367 days) and on 2 sets of 3 days bracketing the milk sampling period during the first lactation cycle (149 to 151 and 159 to 161 DIM).
SCOA analysis.
Analysis of SCOAs was performed on ruminal liquids following standard methods, using high-performance liquid chromatography (HPLC) (38). In brief, portions of ruminal liquid samples were mixed with calcium hydroxide and cupric sulfate and then analyzed on an Aminex HPX-87H HPLC column (Bio-Rad, Hercules, CA, USA) with a Pro-Star autosampler (Varian, Palo Alto, CA, USA) and a refractive index detector (Knauer, Berlin, Germany). The flow rate of the mobile phase (0.015 N H2SO4, 0.0034 M EDTA) was 0.70 ml/min, at 45°C. Samples were compared to two standard curves, one containing 10 mM concentrations of acetate, butyrate, formate, isobutyrate, isovalerate, 2-methylbutyrate, propionate, succinate, and valerate and one containing 10 mM lactate. Standards were run for every 10 samples, and crotonate (2-butenoate) was added to all samples as an internal standard. Separate analyses confirmed that detector responses for each individual SCOA were linear to at least 300 mM.
Total VFA concentrations were calculated as the sum of acetate, propionate, isobutyrate, butyrate, isovalerate, 2-methylbutyrate, and valerate. Total branched-chain VFAs included isobutyrate, isovalerate, and 2-methylbutyrate. Molar fractions of VFAs were also calculated, compared to total VFA concentrations per sample.
DNA extraction and sequencing.
Total genomic DNA was extracted from samples following a mechanical disruption and hot/cold phenol extraction protocol published previously (39), with feces being processed in a similar manner as ruminal solids. For all samples, 25:24:1 phenol/chloroform/isoamyl alcohol was used in place of phenol/chloroform, and samples required up to six additional washes. All DNA samples were resuspended in water, quantified with a Qubit fluorometer (Invitrogen, San Diego, CA, USA), and stored at −20°C.
For bacteria, PCR was performed using universal primers flanking the V4 region of the bacterial 16S rRNA (40). Conditions, purification, and sequencing were as described previously (28). Briefly, PCR was performed using 50 ng of DNA with HotStart ReadyMix (KAPA Biosystems, Wilmington, MA, USA), and products were purified by gel extraction (Zymo Research, Irvine, CA). Samples were quantified with a Qubit fluorometer, pooled on an equimolar basis, and sequenced with a MiSeq v2 kit (2 by 250 bp; Illumina, San Diego, CA, USA), using custom sequencing primers (40).
For archaea and fungi, a two-step PCR protocol was employed, as described previously (28). In short, the archaeal 16S rRNA V6 to V8 region or the fungal ITS1 region was amplified (41) and Illumina adapters with unique indices were added. PCR products were column purified (PureLink; Invitrogen) after the first PCR and gel extracted (Zymo Research) after the second. Samples were quantified with a Qubit fluorometer, pooled on an equimolar basis, and sequenced with a MiSeq v3 kit (2 by 300 bp; Illumina).
Sequence cleanup.
All sequences were demultiplexed on the Illumina MiSeq system. Further sequence processing was performed using mothur v1.36.1 (42), following a protocol adapted from reference 40. The full cleanup pipeline is detailed in reference 28. Briefly, paired-end sequences were combined into contigs, and poor-quality sequences were removed. Bacterial and archaeal sequences were aligned against the SILVA 16S rRNA gene reference alignment database v119 (43). Fungal sequences were aligned by Needleman-Wunsch pairwise sequence alignment within the data set. For all amplicons, sequences were preclustered, and chimera detection and removal were performed. Bacterial and archaeal sequences were classified using the GreenGenes v13.8 database (44); fungal sequences were classified using the UNITE dynamic ITS v6 database (45). Singletons were removed to facilitate downstream analyses.
All sequences were grouped into 97% OTUs by uncorrected pairwise distances and furthest neighbor clustering. Coverage was assessed by Good’s coverage, as calculated in mothur. Bacterial (9,000 sequences), archaeal (100 sequences), and fungal (250 sequences) communities were normalized to equal sequence counts near their lowest respective samples, and these normalized OTU tables were used in all further analyses.
Animal statistics.
All tests were assessed at a significance of P < 0.05. Differences between diet groups were assessed by repeated-measures linear regression over time and by calf for weight gain and supplement intake, using the lme4 (46) and lmerTest (47) packages in R v3.2.3 (48). Weight gain was normalized to day 2 weights, and supplement intake was log transformed. MPE was calculated based on energy-corrected milk (ECM) production, body weight change and maintenance, gestation, and mastitis infection, as described (Table S8) (49, 50). ECM values from the three daily milkings were summed per day, and dry matter intake (DMI) was calculated as feed offered minus feed refused (in kilograms). Animal weights before and after the milk sampling period were averaged across 3 days each. Daily ECM, DMI, and milk SCC values were averaged across the period for each cow. SCCs in milk were then converted to potential milk production loss (50). Efficiency was defined as total energy output (in megajoules) of the five energetic demands (i.e., ECM, weight maintenance, weight change, gestation, and mastitis) per kilogram of DMI. Efficiencies were compared between diet groups with ANOVA and were correlated to OTUs at a minimum of 0.5% relative abundance in at least one sample using Kendall’s correlations, with microbial data averaged across consecutive sampling days to avoid animal effects. All code is available at https://github.com/kdillmcfarland/GS01.
Microbiota statistics.
Beta-diversity was visualized by nonmetric multidimensional scaling (nMDS) plots of the Bray-Curtis metric calculated with square-root-transformed data in R (vegan package [51]). Differences in the spread of Bray-Curtis values within age and diet groups were calculated by PERMDISP (vegan) and pairwise between groups with Tukey’s HSD test.
All tests were assessed at a significance of P < 0.05, and strong correlations were defined as those with coefficients of ≥0.7 or ≤−0.7. Alpha-diversity was determined with Shannon’s diversity index and Chao’s richness index calculated in mothur. Differences in community diversity and richness were assessed overall by ANOVA and pairwise between groups with significant ANOVA results by Tukey’s HSD test from multiple comparisons in R. All samples were modeled for calf diet, age group, and diet within age groups (diet/age group) (model 1). Two-year samples were modeled for milk and overall efficiency (model 2), with ruminal OTU tables averaged across the 3 consecutive days at 1 or 2 years to avoid animal effects.
Total structure (relative abundance with Bray-Curtis measure) and composition (presence/absence with Jaccard index) of OTUs and SCOA concentrations were evaluated for changes using PERMANOVA (vegan), with the same models as for ANOVA (models 1 and 2). Also, fecal OTUs at 2 and 4 weeks were modeled by scours versus healthy, since some of the samples were taken during illness (model 3). Pairwise comparisons between groups with significant PERMANOVA results were completed using PERMANOVA with the Benjamini-Hochberg correction for multiple comparisons. OTUs contributing to differences between groups in PERMANOVA were identified by analysis of similarity percentages (SIMPER) (vegan). OTUs contributing at least 1% of the SIMPER variation were assessed further using the Kruskal-Wallis test with the Benjamini-Hochberg correction for multiple comparisons.
Covariation of the microbiota with SCOAs in ruminal liquids was tested using Mantel tests for dissimilarity matrices. Differences between SCOA concentrations and molar fractions within age groups and diets were assessed overall by ANOVAs with the Benjamini-Hochberg correction for multiple comparisons and pairwise between groups with Tukey’s HSD test. OTUs with at least 0.5% relative abundance in one or more samples were associated with SCOAs by Kendall’s correlations with 3-day averaged ruminal OTU tables. All code is available at https://github.com/kdillmcfarland/GS01.
Accession number(s).
All DNA sequences were deposited in the NCBI Sequence Read Archive under accession no. PRJNA319127.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff at the USDFRC for daily animal care and, in particular, Ron Skoyen for his assistance. We also thank all members of the Suen laboratory for their support and careful reading of the manuscript. Finally, we thank the Wisconsin Energy Institute information technology group for access to computational resources.
This work was supported by U.S. Department of Agriculture, National Institute of Food and Agriculture, HATCH grant WIS01729 and foundational grant 2015-67015-23246 (to G.S.) and USDA Agricultural Research Service CRIS project 5090-21000-024-00D (to P.J.W.).
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02141-18.
REFERENCES
- 1.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergman EN. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev 70:567–590. doi: 10.1152/physrev.1990.70.2.567. [DOI] [PubMed] [Google Scholar]
- 3.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. 2016. How colonization by microbiota in early life shapes the immune system. Science 352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fonty G, Gouet P, Jouany J-P, Senaud J. 1987. Establishment of the microflora and anaerobic fungi in the rumen of lambs. Microbiology 133:1835–1843. doi: 10.1099/00221287-133-7-1835. [DOI] [Google Scholar]
- 5.Klein-Jöbstl D, Schornsteiner E, Mann E, Wagner M, Drillich M, Schmitz-Esser S. 2014. Pyrosequencing reveals diverse fecal microbiota in Simmental calves during early development. Front Microbiol 5:622. doi: 10.3389/fmicb.2014.00622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanez-Ruiz DR, Abecia L, Newbold CJ. 2015. Manipulating rumen microbiome and fermentation through interventions during early life: a review. Front Microbiol 6:1133. doi: 10.3389/fmicb.2015.01133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weimer PJ, Stevenson DM, Mantovani HC, Man SLC. 2010. Host specificity of the ruminal bacterial community in the dairy cow following near-total exchange of ruminal contents. J Dairy Sci 93:5902–5912. doi: 10.3168/jds.2010-3500. [DOI] [PubMed] [Google Scholar]
- 8.Lallès JP. 2012. Long term effects of pre- and early postnatal nutrition and environment on the gut. J Anim Sci 90(Suppl 4):421–429. doi: 10.2527/jas.53904. [DOI] [PubMed] [Google Scholar]
- 9.Bajagai YS, Klieve AV, Dart PJ, Bryden WL. 2016. Probiotics in animal nutrition: production, impact and regulation. No. 179 Food and Agricultural Organization of the United Nations, Rome, Italy. [Google Scholar]
- 10.Jami E, White BA, Mizrahi I. 2014. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS One 9:e85423. doi: 10.1371/journal.pone.0085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jewell KA, McCormick CA, Odt CL, Weimer PJ, Suen G. 2015. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Appl Environ Microbiol 81:4697–4710. doi: 10.1128/AEM.00720-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carberry CA, Kenny DA, Han S, McCabe MS, Waters SM. 2012. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl Environ Microbiol 78:4949–4958. doi: 10.1128/AEM.07759-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace RJ, Rooke JA, McKain N, Duthie C-A, Hyslop JJ, Ross DW, Waterhouse A, Watson M, Roehe R. 2015. The rumen microbial metagenome associated with high methane production in cattle. BMC Genomics 16:839. doi: 10.1186/s12864-015-2032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flint HJ, Bayer EA, Rincon MT, Lamed R, White BA. 2008. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol 6:121–131. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 15.Malmuthuge N, Griebel PJ, Guan LL. 2015. The gut microbiome and its potential role in the development and function of newborn calf gastrointestinal tract. Front Vet Sci 2:36. doi: 10.3389/fvets.2015.00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Menezes AB, Lewis E, O’Donovan M, O’Neill BF, Clipson N, Doyle EM. 2011. Microbiome analysis of dairy cows fed pasture or total mixed ration diets. FEMS Microbiol Ecol 78:256–265. doi: 10.1111/j.1574-6941.2011.01151.x. [DOI] [PubMed] [Google Scholar]
- 17.Patra AK, Yu Z. 2012. Effects of essential oils on methane production and fermentation by, and abundance and diversity of, rumen microbial populations. Appl Environ Microbiol 78:4271–4280. doi: 10.1128/AEM.00309-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torok VA, Percy NJ, Moate PJ, Ophel-Keller K. 2014. Influence of dietary docosahexaenoic acid supplementation on the overall rumen microbiota of dairy cows and linkages with production parameters. Can J Microbiol 60:267–275. doi: 10.1139/cjm-2013-0805. [DOI] [PubMed] [Google Scholar]
- 19.Jami E, Shterzer N, Yosef E, Nikbachat M, Miron J, Mizrahi I. 2014. Effects of including NaOH-treated corn straw as a substitute for wheat hay in the ration of lactating cows on performance, digestibility, and rumen microbial profile. J Dairy Sci 97:1623–1633. doi: 10.3168/jds.2013-7192. [DOI] [PubMed] [Google Scholar]
- 20.Soberon F, Raffrenato E, Everett RW, Van Amburgh ME. 2012. Preweaning milk replacer intake and effects on long-term productivity of dairy calves. J Dairy Sci 95:783–793. doi: 10.3168/jds.2011-4391. [DOI] [PubMed] [Google Scholar]
- 21.Cannon SJ, Fahey GC Jr, Pope LL, Bauer LL, Wallace RL, Miller BL, Drackley JK. 2010. Inclusion of psyllium in milk replacer for neonatal calves. 2. Effects on volatile fatty acid concentrations, microbial populations, and gastrointestinal tract size. J Dairy Sci 93:4744–4758. doi: 10.3168/jds.2010-3077. [DOI] [PubMed] [Google Scholar]
- 22.Malmuthuge N, Li M, Goonewardene LA, Oba M, Guan LL. 2013. Effect of calf starter feeding on gut microbial diversity and expression of genes involved in host immune responses and tight junctions in dairy calves during weaning transition. J Dairy Sci 96:3189–3200. doi: 10.3168/jds.2012-6200. [DOI] [PubMed] [Google Scholar]
- 23.Jatkauskas J, Vrotniakiene V. 2010. Effects of probiotic dietary supplementation on diarrhoea patterns, faecal microbiota and performance of early weaned calves. Vet Med (Praha) 55:494–503. doi: 10.17221/2939-VETMED. [DOI] [Google Scholar]
- 24.Abecia L, Waddams KE, Martínez-Fernandez G, Martín-García AI, Ramos-Morales E, Newbold CJ, Yáñez-Ruiz DR. 2014. An antimethanogenic nutritional intervention in early life of ruminants modifies ruminal colonization by archaea. Archaea 2014:841463. doi: 10.1155/2014/841463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abecia L, Martín-García AI, Martínez G, Newbold CJ, Yáñez-Ruiz DR. 2013. Nutritional intervention in early life to manipulate rumen microbial colonization and methane output by kid goats postweaning. J Anim Sci 91:4832–4840. doi: 10.2527/jas.2012-6142. [DOI] [PubMed] [Google Scholar]
- 26.Yáñez-Ruiz DR, Macías B, Pinloche E, Newbold CJ. 2010. The persistence of bacterial and methanogenic archaeal communities residing in the rumen of young lambs. FEMS Microbiol Ecol 72:272–278. doi: 10.1111/j.1574-6941.2010.00852.x. [DOI] [PubMed] [Google Scholar]
- 27.Jami E, Israel A, Kotser A, Mizrahi I. 2013. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J 7:1069–1079. doi: 10.1038/ismej.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dill-McFarland KA, Breaker JD, Suen G. 2017. Microbial succession in the gastrointestinal tract of dairy cows from 2 weeks to first lactation. Sci Rep 7:40864. doi: 10.1038/srep40864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan MA, Bach A, Weary DM, von Keyserlingk MAG. 2016. Transitioning from milk to solid feed in dairy heifers. J Dairy Sci 99:885–902. doi: 10.3168/jds.2015-9975. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Q, Koser SL, Bequette BJ, Donkin SS. 2017. Effect of propionate on mRNA expression of key genes for gluconeogenesis in liver of dairy cattle. J Dairy Sci 98:8698–8709. doi: 10.3168/jds.2015-9590. [DOI] [PubMed] [Google Scholar]
- 31.Suarez-Mena FX, Hill TM, Heinrichs AJ, Bateman HG, Aldrich JM, Schlotterbeck RL. 2011. Effects of including corn distillers dried grains with solubles in dairy calf feeds. J Dairy Sci 94:3037–3044. doi: 10.3168/jds.2010-3845. [DOI] [PubMed] [Google Scholar]
- 32.Suárez BJ, Van Reenen CG, Stockhofe N, Dijkstra J, Gerrits WJJ. 2007. Effect of roughage source and roughage to concentrate ratio on animal performance and rumen development in veal calves. J Dairy Sci 90:2390–2403. doi: 10.3168/jds.2006-524. [DOI] [PubMed] [Google Scholar]
- 33.Warner RG, Flatt WP, Loosli JK. 1956. Dietary factors influencing the development of the ruminant stomach. J Agric Food Chem 4:788–792. doi: 10.1021/jf60067a003. [DOI] [Google Scholar]
- 34.Tamate H, McGilliard AD, Jacobson NL, Getty R. 1962. Effect of various dietaries on the anatomical development of the stomach in the calf. J Dairy Sci 45:408–420. doi: 10.3168/jds.S0022-0302(62)89406-5. [DOI] [Google Scholar]
- 35.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weimer PJ, Waghorn GC, Odt CL, Mertens DR. 1999. Effect of diet on populations of three species of ruminal cellulolytic bacteria in lactating dairy cows. J Dairy Sci 82:122–134. doi: 10.3168/jds.S0022-0302(99)75216-1. [DOI] [PubMed] [Google Scholar]
- 37.Tsenkova R, Atanassova S, Toyoda K, Ozaki Y, Itoh K, Fearn T. 1999. Near-infrared spectroscopy for dairy management: measurement of unhomogenized milk composition. J Dairy Sci 82:2344–2351. doi: 10.3168/jds.S0022-0302(99)75484-6. [DOI] [PubMed] [Google Scholar]
- 38.Weimer P, Shi Y, Odt C. 1991. A segmented gas/liquid delivery system for continuous culture of microorganisms on insoluble substrates and its use for growth of Ruminococcus flavefaciens on cellulose. Appl Microbiol Biotechnol 36:178–183. doi: 10.1007/BF00164416. [DOI] [Google Scholar]
- 39.Stevenson D, Weimer P. 2007. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl Microbiol Biotechnol 75:165–174. doi: 10.1007/s00253-009-2033-5. [DOI] [PubMed] [Google Scholar]
- 40.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. 2013. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH. 2013. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS One 8:e47879. doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiss M, Larsson K-H. 2013. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22:5271–5277. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 46.Bates D, Maechler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- 47.Kuznetsova A, Brockhoff PB, Christensen RHB. 2017. lmerTest package: tests in linear mixed effects models. J Stat Softw 82:1–26. doi: 10.18637/jss.v082.i13. [DOI] [Google Scholar]
- 48.R Core Team. 2017. R: a language and environment for statistical computing, version 3.4.3. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- 49.National Research Council. 2001. Nutrient requirements of dairy cattle, 7th rev ed National Academy Press, Washington, DC. [Google Scholar]
- 50.Dairy Records Management Systems. 2013. The DHI glossary. Dairy Records Management Systems, Raleigh, NC. [Google Scholar]
- 51.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH. 2015. vegan: community ecology package 2.2-1. https://CRAN.R-project.org/package=vegan.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.