It is well known that the forest litter layer is inhabited by a complex microbial community of bacteria and fungi. However, while the importance of fungi in the turnover of natural biomass is well established, the role of their bacterial counterparts is less extensively studied. We show that Chitinophaga pinensis, a prominent member of an important bacterial genus, is capable of using both plant and fungal biomass as a nutrient source but is particularly effective at deconstructing dead fungal material. The turnover of dead fungus is key in natural elemental cycles in the forest. We show that C. pinensis can perform extensive degradation of this material to support its own growth while also releasing sugars that may serve as nutrients for other microbial species. Our work adds detail to an increasingly complex picture of life among the environmental microbiota.
KEYWORDS: β-glucan polysaccharides, bacteria, Bacteroidetes, biomass recycling, carbohydrate active enzymes, polysaccharide utilization loci
ABSTRACT
The genome and natural habitat of Chitinophaga pinensis suggest it has the ability to degrade a wide variety of carbohydrate-based biomass. Complementing our earlier investigations into the hydrolysis of some plant polysaccharides, we now show that C. pinensis can grow directly on spruce wood and on the fungal fruiting body. Growth was stronger on fungal material, although secreted enzyme activity was high in both cases, and all biomass-induced secretomes showed a predominance of β-glucanase activities. We therefore conducted a screen for growth on and hydrolysis of β-glucans isolated from different sources. Most noncrystalline β-glucans supported good growth, with variable efficiencies of polysaccharide deconstruction and oligosaccharide uptake, depending on the polysaccharide backbone linkage. In all cases, β-glucan was the only type of polysaccharide that was effectively hydrolyzed by secreted enzymes. This contrasts with the secretion of enzymes with a broad range of activities observed during growth on complex heteroglycans. Our findings imply a role for C. pinensis in the turnover of multiple types of biomass and suggest that the species may have two metabolic modes: a “scavenging mode,” where multiple different types of glycan may be degraded, and a more “focused mode” of β-glucan metabolism. The significant accumulation of some types of β-gluco-oligosaccharides in growth media may be due to the lack of an appropriate transport mechanism, and we propose that this is due to the specificity of expressed polysaccharide utilization loci. We present a hypothetical model for β-glucan metabolism by C. pinensis that suggests the potential for nutrient sharing among the microbial litter community.
IMPORTANCE It is well known that the forest litter layer is inhabited by a complex microbial community of bacteria and fungi. However, while the importance of fungi in the turnover of natural biomass is well established, the role of their bacterial counterparts is less extensively studied. We show that Chitinophaga pinensis, a prominent member of an important bacterial genus, is capable of using both plant and fungal biomass as a nutrient source but is particularly effective at deconstructing dead fungal material. The turnover of dead fungus is key in natural elemental cycles in the forest. We show that C. pinensis can perform extensive degradation of this material to support its own growth while also releasing sugars that may serve as nutrients for other microbial species. Our work adds detail to an increasingly complex picture of life among the environmental microbiota.
INTRODUCTION
Bacteria of the Bacteroidetes phylum are prolific degraders of complex carbohydrates (1), and this process has wide-ranging environmental impacts (2). The decomposition of polysaccharides enables soluble sugars to be made available to other organisms and recycles carbon, nitrogen, and water (1), but it can also lead to the disruption of aggregate soil particles that are held together by complex carbohydrates and are important for land cohesion and carbon retention (3). As soils are vital carbon sinks, this can have significant impacts on the climate. A detailed understanding of biomass decomposition is needed to help manage land use in ways that consider the health of the soil microbiota, but relatively little is currently known about the contribution made by bacteria compared to what is known about their fungal counterparts (4).
The environment of the forest soil and litter offers rich nutrient and carbon sources for both primary degraders and opportunistic species (5). Depending on the season and extent of decomposition, forest litter can contain more than 100 mg carbohydrate per gram, mainly derived from dead plant and fungal biomass (6). Fallen plant material provides abundant carbohydrate in the forms of cellulose, pectins, and hemicelluloses (4). In softwood forests, β-mannans dominate, and the abundant glucomannan likely contains few α-d-Galp substitutions and some irregular O-acetylation (7, 8). In the same environment, decomposing mycelium provides a rich variety of glycans, including chitin (9), mannosylated glycoproteins with long galactan chains (10, 11), and various β-glucans (9). β-(1→3)-Glucans dominate (65% to 90% of fungal cell wall glucan), but there may also be β-(1→6)-linked branches, and some β-(1→6)-Glc backbones (11). Soil and litter areas where dead mycelium is actively degraded are “hotspots” of enzyme activity, and the Bacteroidetes genus Chitinophaga is particularly enriched in these microenvironments in forest and agricultural soils (5, 12).
The type strain Chitinophaga pinensis is a Bacteroidetes species isolated from softwood forest litter, and it belongs to a genus that is considered to be strongly chitinolytic but not cellulolytic (12). We previously demonstrated the growth of C. pinensis and the induction of specific enzymes by larch wood arabinogalactan (13), and more recently we performed a proteomic assessment comparing enzyme secretion during growth on glucose and complex plant β-mannans (14, 15). We found that the secretion of a selection of carbohydrate active enzymes (CAZymes) was induced in the presence of β-mannan, likely via the action of polysaccharide utilization loci (PULs). A PUL is a discrete genetic locus inducible by a specific glycan structure that contains genes encoding proteins that can deconstruct that carbohydrate and transport it into the cell. The glycan is recognized by a SusD-like protein, hydrolyzed by a consortium of specifically acting glycoside hydrolases (GHs), and brought into the periplasm by a SusC-like protein (16). In our earlier proteomic experiments, we detected protein components of ten PULs, four of which appear to be induced by β-mannan, showing that these loci are indeed involved in carbohydrate metabolism by C. pinensis (15). However, we also detected a significant subset of enzymes and PULs that were expressed during growth on all carbon sources tested, including some enzymes that were highly expressed during growth on glucose (15). A large proportion of these enzymes had predicted activity on a β-glucan polysaccharide or were found in a PUL together with a β-glucanase.
We now present a comprehensive study of growth on a range of β-glucan polysaccharides derived from both plant and microbial sources of biomass. These are likely to be highly abundant carbon sources in the microhabitat of C. pinensis, where plant matter is bound together in a network-like mat by dead and living mycelium as well as exopolysaccharides secreted by fungi and bacteria. We compare the deconstruction of a range of different β-glucans with that of the complex plant heteroglycans glucomannan and arabinoxylan, which contain multiple sugars and linkages and which we have previously shown support growth by C. pinensis (13).
In this study, we demonstrate that C. pinensis displays different behaviors in terms of growth, enzyme secretion, and oligosaccharide uptake, depending upon the origin and the structure of the polysaccharide provided as the carbon source. We also show that growth of the bacterium on complex plant and fungal biomass is accompanied by high levels of secreted enzyme activities, with a consistent focus on the degradation of β-glucans. Growth on isolated heteroglycans leads to the secretion of enzymes with a wide range of carbohydrate-degrading activities and an efficient uptake of produced oligosaccharides. Conversely, growth on β-glucans from various sources leads to the secretion of enzymes solely with endo-glucanase activities and can be characterized by a significant accumulation of oligosaccharides in the external environment, depending on the backbone linkage of the β-glucan in question. We propose that this is due to the specificity of PULs and non-PUL enzymes produced in the presence of glucose that allows certain β-glucan polysaccharides to be degraded but does not allow the efficient uptake of the released oligosaccharides. We present a hypothetical model for this process of β-glucan metabolism and propose that the release of oligosaccharides might contribute to a nutrient pool for the soil microbiota.
RESULTS AND DISCUSSION
Deconstruction of complex plant and microbial biomass.
To assess the metabolism of polysaccharides in a naturalistic setting, C. pinensis was grown on select types of carbohydrate-rich biomass that the species is likely to encounter in its native habitat. The bacterium was grown in minimal medium supplemented with 5 g · liter−1 of either spruce wood or Agaricus bisporus fruiting body. The wood material had been milled to a fine powder and contained cell walls of cellulose, galactoglucomannan, xylan, and lignin, in addition to proteins and extractives (17). For some growth experiments, the milled wood was additionally autoclaved to further open the cell wall structure, but no differences in growth or enzyme profiles were observed (data not shown). The A. bisporus fungal material was highly enriched in cell walls, which comprised chitin, chitosan, β-(1→3)- and β-(1→6)-d-glucans, and some α-(1→3)-d-glucans, as well as lipids and proteins (18). Growth and protein concentrations were monitored over 8 days, and the activities of secreted proteins were repeatedly measured against a broad panel of polysaccharides from different sources. In addition, the culture medium was sampled at these times and analyzed by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) to detect any oligosaccharides. The first samples (zero time points) were taken within 4 h of inoculation on day 0.
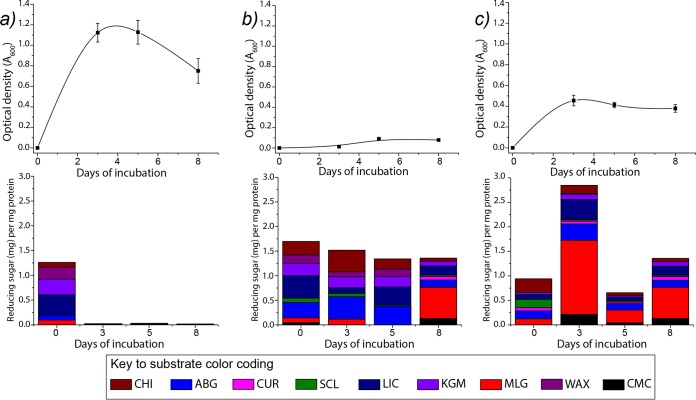
Our data show that, compared to that on glucose, the bacterium is capable of very limited growth on spruce wood, but good growth was achieved on the fungal material (Fig. 1). By visual observation, particulate matter in only the fungus culture became dispersed within 3 to 4 days of growth, suggesting solubilization that did not occur in the spruce cultures. HPAEC-PAD analyses showed no accumulation of any detectable oligosaccharides during growth on either type of biomass. However, our activity assays showed that carbohydrate-active enzymes (CAZymes) were secreted during growth on both materials (Fig. 1). As in a previous study (15), there was no significant CAZyme activity in glucose-grown cultures once the cells had adapted to the medium (i.e., from the second sampling point onwards) (Fig. 1a). Chitinase activity was found at approximately equal levels in the wood- and fungus-grown cultures, which echoes our previous proteomic observation that predicted chitinase enzymes could be detected in C. pinensis secretomes regardless of the carbon source (15). CAZyme activity was measurable throughout the growth period in the spruce-grown cultures, with the strongest activities targeting glucans with β-(1→3)- and/or β-(1→6)-linkages (Fig. 1b). There was some activity on glucomannan (KGM), but it is perhaps surprising that this was not a more dominant activity, as this is the preferred polysaccharide growth substrate of C. pinensis (13) and is abundant in the wood material. There was also some xylanase activity and some hydrolysis of arabinogalactan. In late spruce cultures (day 8), the hydrolysis of plant mixed-linkage β-glucans (MLG) became the dominant activity, perhaps reflecting changes to the growth substrate that rendered cell wall β-glucans more accessible, inducing a change in the profile of secreted enzymes. This may have resulted from an earlier removal of pectins and hemicelluloses by the activities detected in our assays performed on days 3 and 5 (Fig. 1b).
FIG 1.
Growth curves (top) and profiles of secreted glycoside hydrolase activity (bottom) for C. pinensis grown on different types of carbohydrate or complex biomass: glucose (a), spruce wood (b), and fungal fruiting body (c). Three biological replicates were performed for each experiment. Samples were collected for analysis after 3, 5, and 8 days of cultivation. The zero time point samples were collected 4 h after inoculation. CHI, chitin; ABG, linear A. bisporus β-glucan; CUR, curdlan; SCL, scleroglucan; LIC, lichenan; KGM, konjac glucomannan; MLG, barley mixed-linkage β-glucan; WAX, wheat arabinoxylan; CMC; carboxymethylcellulose. Data for the enzyme activity for reducing sugars represent average values from triplicate experiments. Table S3 in the supplemental material shows the enzyme assay data in numerical form, including standard deviation error values.
The strongest levels of activity in any culture were detected in early (day 3) fungus-grown cultures, although these levels dropped somewhat during growth (Fig. 1c). The strongest activities again focused on β-(1→3)- and β-(1→6)-d-glucans, and there was particularly high activity on MLG. This activity may have been induced by the β-glucan polysaccharides present in the fungal material which, due to their noncrystalline structures, would be more accessible to recognition by bacterial proteins than the cellulose in the wood material. In the fungus-grown cultures, the activities on KGM and arabinogalactan were greatly reduced compared to those in the spruce-grown cultures, which indicates that the responsible enzymes are secreted at a higher level when their respective substrates are available in the inducing biomass. In contrast, a high overall level of β-glucanase activity was a common factor regardless of the growth material available in the environment. Intrigued by this observation, we investigated growth on a series of isolated β-glucans that differ in their origins, backbone linkages, and degrees of branching.
Polysaccharide degradation by C. pinensis.
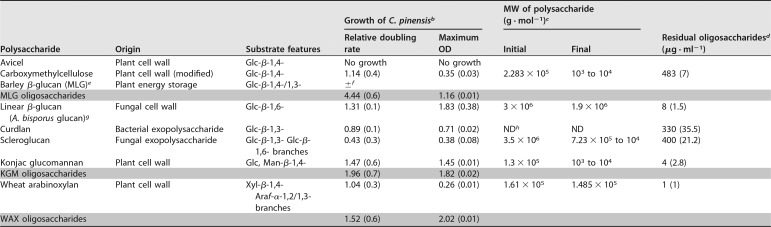
C. pinensis was incubated with a range of β-glucan polysaccharides as the sole carbon source. These were provided at 5 g · liter−1 in M9 minimal growth medium lacking glucose. Growth was monitored by regular measurements of the optical density at 600 nm (OD600) to produce growth curves (see Fig. S1 in the supplemental material). The doubling rates of these cultures, determined over the exponential phase of growth, were normalized to those of cultures grown on glucose (Table 1). Culture samples were collected at the beginning and late exponential stages of growth and analyzed by size-exclusion chromatography with coupled multiangle laser-light scattering (SEC-MALLS) to determine the average molecular weights of carbohydrates in the medium (Table 1 and Fig. S2). At the end of cultivation, samples of the growth medium were analyzed by HPAEC-PAD to quantify the residual identifiable oligosaccharides in solution (Table 1 and Fig. S3). In addition, samples of growth medium were collected during the mid-exponential growth phase and assayed for hydrolytic activity against a select panel of relatively soluble polysaccharides (Fig. 2). For comparative purposes, the same experiments were performed using KGM and wheat arabinoxylan (WAX) as the carbon source; these complex heteroglycans derived from the plant cell wall were previously shown to support growth by C. pinensis (13). The activities detected in all cultures were ascribed to secreted proteins likely utilized in the natural environment for nutrient acquisition. The different carbon sources tested induced very different profiles of enzyme activity, presumably due to changes in gene expression and/or posttranslational regulation. Most striking is the observation that all β-glucans induced only endo-glucanase activities, while KGM and WAX both induced a broad range of different activities. Indeed, very high overall levels of activity were detected in the KGM and WAX cultures compared to that in the glucan-grown cultures, and activities in the KGM and WAX cultures were not exclusively related to the deconstruction of the provided carbohydrate (discussed below).
TABLE 1.
Deconstruction of carbon sourcea
Carbon source was included in the medium at 5 g · liter−1 at the start of cultivation. The shaded rows show data for oligosaccharides produced by preincubating polysaccharides with an appropriate endo-acting glycoside hydrolase. Two biological replicates were performed for each experiment, and standard deviations (SD) are shown in parentheses. Cultures were regularly sampled to assess the deconstruction of polysaccharides. Growth curves for konjac glucomannan and wheat arabinoxylan were presented in reference 13.
bGrowth is given as the doubling rate for growth on a polysaccharide divided by the doubling rate for growth on glucose (i.e., a relative doubling rate of >1 indicates that growth was faster for this carbon source than for glucose).
cApproximate average molecular weight (MW) of polysaccharides was calculated from data obtained by SEC-MALLS analysis (see Fig. S1 in the supplemental material). Changes in average MW show the extent to which the polymers were degraded.
dOligosaccharides remaining in the liquid growth medium following the final stages of growth were analyzed by HPAEC-PAD (see Fig. S2 for the exact time point at which these samples were taken). Structures were quantified based on the peak area of the most closely eluting appropriate linear standard.
eMLG, mixed-linkage β-glucan.
f±, some growth was observed, but it was too slow to be measured.
gA. bisporus glucan, a linear β-glucan from Agaricus bisporus.
hND, not determined due to solubility issue.
FIG 2.
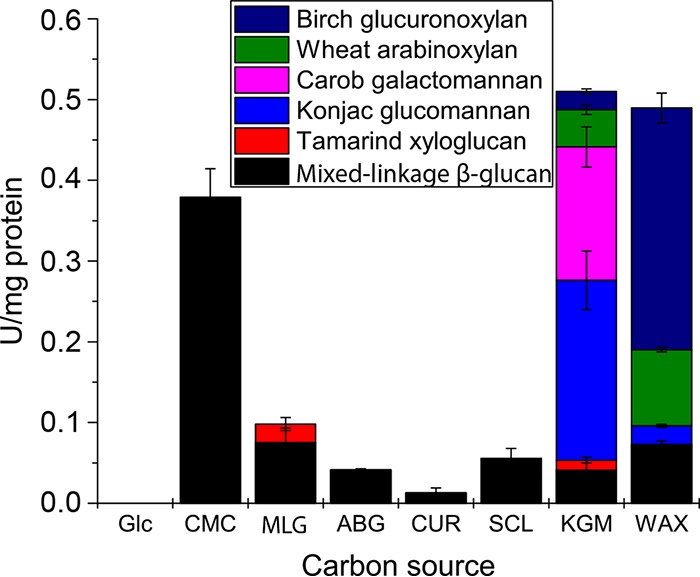
Hydrolytic activity on complex polysaccharide substrates (average values from two duplicate experiments) of proteins collected from growth medium of C. pinensis supplemented with different carbohydrate carbon sources. Glc, glucose; CMC, carboxymethyl-cellulose; MLG, barley mixed-linkage β-glucan (MLG); ABG, linear A. bisporus β-glucan; CUR, curdlan; SCL, scleroglucan; KGM, konjac glucomannan; WAX, wheat arabinoxylan. 1 U = 1 mmol reducing sugar (glucose equivalents) released/min. We previously showed that glucose-grown cultures do not secrete any detectable polysaccharide-degrading enzymes (15). Activity data shown for KGM cultures were also originally presented in reference 15. Table S4 shows these data in numerical form, including standard deviation error values.
Heteroglycans from the plant cell wall.
The konjac glucomannan (KGM) used in our studies is an excellent analogue for an abundant woody glycan likely to be encountered by C. pinensis. We have previously shown that it supported strong growth and induced the secretion of enzymes with a range of activities (15), and we now show that KGM was almost completely depolymerized by the bacterium during cultivation, with only a small amount of short oligosaccharides remaining (Table 1). Mannanase activity was detected in KGM-induced secretomes (Fig. 2 and reference 15), and indeed, the C. pinensis genome encodes two enzymes from the β-mannanase-dominated family GH26 (Cpin_3128 and Cpin_5393), as well as several potential β-mannanases from uncharacterized subfamilies of GH5 (15). By comparison, growth on WAX was very weak unless the polysaccharide was predigested to xylo-oligosaccharides (XOs) by an exogenous enzyme (CjGH10A endo-xylanase from Cellvibrio japonicus [19]) (Table 1). A genome analysis showed one putative β-xylanase from GH10 (Cpin_4240, annotated as a probable gene fragment on www.cazy.org), one GH30 (Cpin_4557, belonging to a subfamily known to contain some xylanases [20]), and one putative β-xylanase from family GH43 (Cpin_2866), some members of which can act very weakly to degrade xylan (21, 22).
The apparent discrepancy between xylanase activity in cultures and in cell-free secretomes (Fig. 2) may relate to a proximity effect, whereby enzymes in the medium during cultivation have limited access to the substrate and concentrated enzymes in the isolated secretome have a longer contact time with the substrate. It may also be that the relevant xylan-degrading enzymes function more effectively after washing in buffer and the removal of inhibitory compounds or other proteins. Regardless, in the natural environment of a softwood forest, the lack of an effective xylan-degrading system might not be a disadvantage, as xylan polysaccharides are not likely to be particularly abundant. However, the strong growth on XOs (Table 1) suggests that C. pinensis may be able to take up and metabolize XOs released by other microorganisms. Indeed, our analysis of the genome predicts that the bacterium can secrete multiple β-xylosidases and α-l-arabinofuranosidases that are able to deconstruct short decorated XOs (see Table S1 for details on these predicted enzymes).
For both the heteroglycans KGM and WAX, the uptake of oligosaccharides was highly efficient. At the end of incubation with KGM, less than 5 μg ml−1 of oligosaccharides remained in the medium (Table 1), and the structures that did remain were short complex gluco-manno-oligosaccharides (GMOs) that may be recalcitrant to enzymatic hydrolysis (23) (Fig. S3). At the end of incubation with WAX poly- and oligosaccharides, there were only trace amounts of detectable XOs. Therefore, for both of these plant heteroglycans, rapid growth correlated with efficient uptake of oligosaccharides.
The reducing sugar activity screens show that both KGM and WAX induced the secretion of a large overall amount of enzymes with hydrolytic activity, including enzymes capable of hydrolyzing different polysaccharides (Fig. 2), including β-glucans, mannans, and xylans. This echoes the broad predicted activity profile of enzymes detected in our previous proteomic analysis of KGM-induced secretomes (15) and suggests that the β-mannan- and β-xylan-degrading machinery is not produced in isolation from CAZymes targeting other polysaccharides. We identified several PULs in the C. pinensis genome that may target one or more structural elements of xylan or mannan (Table S1), but none that obviously encode a complete degradation pathway as has been demonstrated for various glycans in other bacterial species (see references 24–27 for examples). We identified predicted PULs in the C. pinensis genome that appear to target mannan or xylan as well as another type of glycan (Table S1). In fact, many of the CAZymes that we predict to target mannan or xylan are not found in PULs, and the mechanism by which their expression might be induced by specific glycans is unknown. Furthermore, for those enzymes not found in PULs, the protein machinery by which the oligosaccharides they produce are taken into the cell is also unclear. Nonetheless, it is apparent that CAZymes targeting a wide range of polysaccharides are produced in response to growth on KGM and WAX and that the products of enzymatic digestion are efficiently taken up into the cell.
β-Glucans from plant and microbial sources.
The β-glucan polysaccharides utilized in our growth and activity screens were derived from both plant and microbial sources. All are composed exclusively of glucose residues but differ in the linkages of their backbones and in the presence or absence of decorations to the main chain (Table 1). Growth on plant glucans with a predominantly β-(1→4)-linked backbone was poor or nil; although carboxymethylcellulose (CMC) supported fairly rapid growth, the final OD was low. In contrast, growth on microbial glucans with β-(1→3)- or β-(1→6)-linked backbones was quite strong. Furthermore, preincubation of barley mixed-linkage β-glucan (MLG) with a commercial cellulase enzyme (C1184; Sigma-Aldrich), resulting in oligosaccharides somewhat enriched with the β-(1→3) linkage, resulted in a potent growth substrate, although there was no measurable ability to grow on polymeric MLG (Table 1).
In contrast with observations of enzyme secretion during growth on heteroglycans (see above), our experiments revealed that all of the β-glucan growth substrates induced the secretion of enzymes that were solely capable of degrading the β-glucan substrates in our screen. However, C. pinensis is not a cellulolytic species (12). The genome encodes no apparent cellulose-degrading lytic polysaccharide monooxygenases (LPMOs), and in fact, none have yet been discovered in any Bacteroidetes species, nor does it encode any predicted cellobiohydrolases (CBHs). CBH enzymes are classically considered to be required for the efficient deconstruction of crystalline cellulose from the plant cell wall, although it must be noted that Cytophaga hutchinsonii, the only cultured cellulolytic member of the Bacteroidetes to have been studied in detail, possesses no genes encoding identifiable CBHs (28). C. pinensis did not grow on crystalline cellulose (Avicel), indicating that it is not cellulolytic in the same manner as C. hutchinsonii (Table 1). However, the genome does encode several putative endo-glucanases and β-glucosidases, suggesting the potential to deconstruct other β-glucans. An analysis of growth media showed effective depolymerization of all β-glucans during growth (Table 1), with roughly similar levels of hydrolysis of plant MLG by all β-glucan-induced secretomes. An exception to this was the much higher level of MLG hydrolysis detected in CMC-induced secretomes (Fig. 2). CMC is an artificially modified form of cellulose, but it is an ideal substrate for endo-glucanases and is often used to discover or characterize such enzymes. An endo-β-1,4-glucanase would be able to hydrolyze MLG quite effectively. It is striking that the MLG hydrolysis induced by CMC was so much greater than that induced by MLG itself. This suggests that enzymes hydrolyzing β-(1→4)-d-Glc linkages may be induced by long stretches of this linkage, which are more common in CMC than in MLG.
Despite the effective depolymerization of CMC, bacterial growth was weak, with a low final OD. An analysis of the growth medium revealed the accumulation of large amounts of cello-oligosaccharides (COs) (∼500 μg · ml−1) (Table 1), indicating very poor uptake of these degradation products. Preliminary growth trials had indicated very little ability to grow on soluble β-(1→4)-linked cello-oligosaccharides (COs; degree of polymerization [dp], 2 to 6), showing that COs with a dp of up to 6 are not favored growth substrates. We propose that this is due to the lack of a PUL induced by such short COs (discussed below).
The fungal and bacterial β-glucans tested were also degraded quite efficiently, as judged by reductions in molecular weight and the production of oligosaccharides (Table 1). Indeed, five potential β-1,3-glucanases and three potential β-1,6-glucanases were predicted by analysis of the C. pinensis genome; most are predicted to be secreted, indicating a strong capacity for the hydrolysis of these microbial β-glucans in the external environment (see Table S2). Several of these enzymes are found in PULs that are predicted to target microbial polysaccharides (discussed below).
Agaricus bisporus glucan is a linear β-(1→6)-d-glucan extracted from a fungal cell wall: this supported strong and rapid growth, and we observed an approximate 1.5-fold reduction in molecular weight during cultivation (Table 1), indicating effective cleavage of the polysaccharide backbone. The bacterial exopolysaccharide curdlan, which features a linear β-(1→3)-linked backbone that forms a triple helical structure in solution (29), supported moderately strong growth. Although SEC-MALLS analysis of curdlan cultures was not possible due to solubility issues, the production of oligosaccharides again indicates that backbone cleavage occurred. Finally, the fungal exopolysaccharide scleroglucan, which presents a linear β-(1→3)-linked backbone with regularly spaced β-(1→6)-linked Glc decorations, supported slower and weaker growth. At the end of bacterial cultivation, branched scleroglucan was deconstructed in a way that produced a dual population of products: short oligosaccharides with molecular weights (MW) in the range of 103 to 104, and longer glycan chains with an average MW of 7 × 105 (Table 1 and Fig. S2). This contrasts with, for instance, glucomannan, CMC, and A.bisporus glucan which were depolymerized to a single relatively homogenous population of oligosaccharides with a narrow MW range of 103 to 104 (Fig. S1), and likely indicates a limited capacity for backbone deconstruction in heavily decorated regions of scleroglucan.
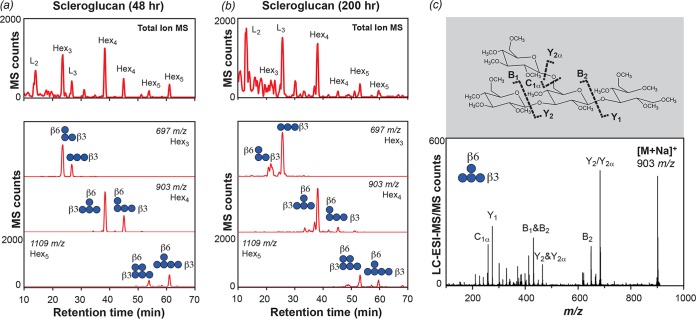
An HPAEC-PAD analysis showed that the concentration of scleroglucan-derived oligosaccharides at the end of cultivation was 400 μg · ml−1 (Table 1), among the highest levels of accumulation we have observed. A total of 150 μg · ml−1 oligosaccharides in the medium did not match any linear β-(1→3)-linked gluco-oligosaccharide (GO) standards. These structures were therefore sequenced by liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) (Fig. 3). In samples taken from scleroglucan cultures in early exponential phase, our MS analyses found high levels of linear laminaribiose (L2) and laminaritriose (L3) and identified the most abundant branched structures as tri-, tetra-, and pentasaccharides with Glc substitutions at or near the nonreducing ends (Fig. 3). By the end of cultivation, oligosaccharides substituted at the nonreducing end were lost or significantly reduced in abundance. Conversely, oligosaccharides substituted next to the nonreducing end, or at two adjacent nonreducing end positions, remained at the end of the growth period. These were not degraded further and were apparently not taken up by the bacterium. Thus, in the extracellular deconstruction of scleroglucan by C. pinensis, branched oligosaccharides with consistent structures accumulate at high levels in the external environment. We predict that only one exo-β-glucosidase (Cpin_6735) was secreted into the medium under these growth conditions. This is found in a PUL together with an endo-β-1,6-glucanase (Cpin_6736) and an endo-β-1,3-glucanase (Cpin_6737) (Table S2), but the enzyme may have a limited capacity to remove these specific branching structures. Such specificity of action echoes that of the GH31 α-xylosidases, exo-acting enzymes that can remove Xyl substitutions from only the terminal backbone units of xyloglucan oligosaccharides (30). The presence of nonterminal β-(1→6)-linked decorations on scleroglucan may be the cause of the lower growth rate compared to that of the curdlan cultures.
FIG 3.
Branched gluco-oligosaccharides accumulate in the medium of scleroglucan cultures. Total ion chromatograms and selected extracted ion chromatograms (Hex3 to Hex6) for samples taken during exponential (a) and late stationary (b) phases of growth (48 and 200 h of incubation, respectively). The most abundant oligosaccharide structure at 200 h was d-Glcp-(1→6)(1→3)-β-d-Glcp-(1→3)-β-d-Glcp; its corresponding fragmentation spectrum is shown (c). Fig. S4 shows the LC-MS profiles of other abundant structures.
Gluco-oligosaccharide uptake by C. pinensis.
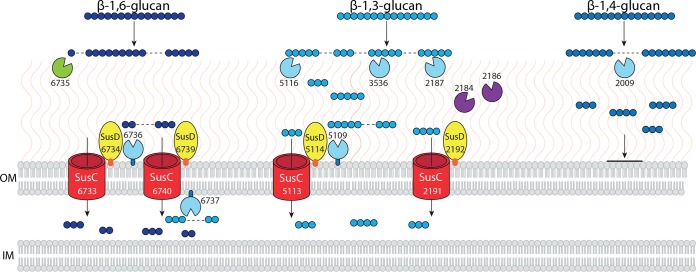
A PUL is an operon that encodes proteins for the sensing, degradation, and uptake of a specific carbohydrate structure (16). From analysis of the C. pinensis genome, we predict that there are six PULs with the capacity to degrade one or more types of β-glucan (summarized in Table S2). Only one PUL (Cpin_6806-6816) encodes a predicted endo-β-1,4-glucanase, and it also encodes predicted exo-acting enzymes targeting α-Man and α-Fuc structures, suggesting that this is not an exclusively glucan-targeting PUL. There are two small PULs that each encode only one enzyme: Cpin_3588-3590 is predicted to target β-1,6-glucans, and Cpin_5109-5117 is predicted to target β-1,3-glucans. The PUL Cpin_2184-2192 encodes enzymes predicted to degrade β-1,3-glucans and chitin and may therefore target mixed component fungal cell wall material. Finally, the PUL Cpin_6730-6742 encodes an endo-β-1,3-glucanase, an endo-β-1,6-glucanase, and an exo-β-glucosidase that may scavenge GOs. Clearly the theoretical capacity for the degradation and uptake of β-glucans with various backbone linkages provided by the genome is extensive. In our previous proteomic analysis of protein secretion by C. pinensis, we detected PULs that were expressed either during growth on all substrates or during growth on all glucose-containing substrates, while others were induced by β-mannan polysaccharides (15). We used these earlier data to produce a model of β-glucan metabolism (Fig. 4), showing only those enzymes and PULs whose expression was confirmed to be active in the presence of Glc and not to be induced by β-mannan (15). This model is supported by our new observations of growth, polysaccharide deconstruction, and oligosaccharide accumulation and incorporates new predictions of cellular localization and enzyme activity (Table S2).
FIG 4.
Hypothetical model of the protein machinery involved in focused β-glucan metabolism by C. pinensis, including PULs and individual enzymes. See Table S3 for a summary of all predictions made regarding each enzyme and protein shown. Endo-acting enzymes are colored light blue; these are endo-β-glucanases that have specificity for the indicated linkages. Cpin_2184 and Cpin_2186 are putative chitinases shown in purple; these are coexpressed in a PUL with an endo-β-1,3-glucanase, Cpin_2187. The sole exo-acting β-glucosidase is colored green. Protein localization is based on predictions of signal peptides and lipo tags using LipoP (www.cbs.dtu.dk/services/LipoP/), as well as predictions of localization made using PSORTb (http://www.psort.org/psortb/). Additionally, the presence of a C-terminal domain for targeting to the type 9 secretion system (T9SS) was used to identify secreted proteins. OM, outer membrane; IM, inner membrane. Protein molecules are not drawn to scale, and their physical proximity within the membrane is purely hypothetical. Proteins with a lipo tag are shown tethered to the OM and either face toward to the external environment or into the periplasmic space depending upon the localization prediction made by PSORTb. Glucose monomers are shown as small blue circles, with different light or dark shading depending on the backbone linkage type, as indicated. A dashed line within a glucan structure indicates a predicted site of enzymatic bond cleavage.
Despite the fact that the genome encodes enzymes and PULs predicted to target all linkages of β-glucan, our data show that there are differences in the extent to which different types of GO are taken up by C. pinensis (Table 1). Despite a significant reduction in the molecular weight of CMC, growth was very poor and there was an accumulation of almost 500 μg · ml−1 of COs in the medium, suggesting a failure to transport these oligosaccharides into the cell. The predicted endo-β-1,4-glucanase encoded by PUL Cpin_6806-6816 should hydrolyze CMC, COs of which would in theory be transported into the cell by the SusC-like protein Cpin_6808. However, none of the proteins in this PUL were detected in our previous proteomic work, suggesting that this PUL is not expressed in the presence of glucose. We did detect the endo-β-1,4-glucanase Cpin_2009, which we expect to be involved in CMC deconstruction and is therefore included in our model (Fig. 4). However, the gene encoding this enzyme is not located within a PUL, and so we are unable to identify any means by which the resulting COs would be taken up into the cell. If this enzyme is not associated with a concerted protein machinery for uptake of the oligosaccharides it produces, this would account for the significant accumulation of COs we observed in the growth medium (Table 1). The PUL Cpin_6806-6816 may be specifically induced by a very long CO structure, explaining both its absence in our previous proteomic study and the inability to grow on COs up to a dp of 6. We propose that Cpin_2009 is responsible for the MLG hydrolysis in secretomes induced by CMC, where it is found at a much higher level than in cultures induced by MLG or other glucans (Fig. 2), suggesting that a long unbroken stretch of β-(1→4)-linkages is necessary to induce the expression of that gene.
Our earlier proteomic experiments confirmed that a PUL targeting β-(1→3)-d-glucans and chitin (Cpin_2184-2192), a PUL targeting β-(1→3)-d-glucans (Cpin_5109-5117), and a PUL targeting both β-(1→3)- and β-(1→6)-d-glucans (Cpin_6730-6742) are all detected during growth on glucose, indicating either constitutive or glucose-dependent expression (15). These PULs are therefore expected to be expressed during growth on all of the plant and microbial β-glucans tested here, giving C. pinensis the capacity to effectively produce and take up GOs with β-(1→3) or β-(1→6) linkages. The chitinases coexpressed by the PUL Cpin_2184-2192 along with the endo-β-1,3-glucanase Cpin_2187 account for the chitin-degrading activity detected in biomass-grown cultures (Fig. 1).
Less than 10 μg · ml−1 of residual oligosaccharides were detected in growth medium after cultivation on the β-(1→6)-linked A.bisporus glucan, which we predict to induce the aforementioned PUL Cpin_6730-6742, which putatively targets both β-(1→3)- and β-(1→6)-d-glucans (Table S2). However, large amounts of some specific structures were residual after growth on the β-(1→3)-linked curdlan and scleroglucan. At the end of incubation, curdlan cultures contained around 330 μg · ml−1 oligosaccharides, mostly L2. Approximately 50% of the residual oligosaccharides in scleroglucan cultures were also L2, in addition to the specific branched structures discussed above. This strongly indicates import specificity for longer linear structures through a β-(1→3)-d-glucan-specific SusC-like protein.
Our model in Fig. 4 shows that some GOs are taken up for further metabolism while others remain in the environment. As discussed above, we propose that the accumulation of large amounts of oligosaccharides in the external environment during growth has two main causes. For β-(1→4)-linked COs, there is a failure to induce the relevant PUL (Cpin_6808-6816), perhaps due to the unnatural structural features of CMC. Meanwhile, for β-(1→3)-d-glucans, only longer and unbranched structures can be successfully transported into the cell via SusC-like proteins in the PULs Cpin_2184-2192, Cpin_5109-5117, and Cpin_6730-6742. There may be a biological rationale for this external accumulation of oligosaccharides, but this is currently unclear. For example, metabolizable sugars released by C. pinensis-mediated deconstruction of polysaccharides may enter into an environmental pool that is utilized for nutrition by other microbial species. A similar phenomenon has been described among bacterial symbionts of the human digestive system (31), another environment rich in complex biomass and diverse microbial life; Rakoff-Nahoum et al. have described syntrophic interactions among Bacteroidales mediated by glycoside hydrolases secreted by one species permitting the growth of another by deconstructing polysaccharides (31). Our data showing growth on arabinoxylan-derived oligosaccharides but not polysaccharide may indicate that C. pinensis can in turn take nutrition from such a common pool of partly degraded polysaccharides. This remains a speculative hypothesis based on our observations to date.
Conclusions and outlook.
Taken together, the data from our integrated growth studies, biochemical assays, and carbohydrate characterization show that C. pinensis deconstructs a range of complex glycans derived from both plant and fungal biomass. The species is therefore likely to contribute to the decomposition of a wide variety of materials in its natural environment, including elements of plant and fungal cell walls. Our experiments using complex biomass show that growth is stronger on fungal material than on a type of wood that is common to the native habitat of C. pinensis. These complex growth materials induced a range of CAZyme activities, but with a general focus on endo-glucanase activity in both cases. Growth on isolated β-glucans from plant and microbial sources induced the secretion of solely endo-glucanase enzymes, whereas growth on complex plant cell wall heteroglycans led to the secretion of enzymes with a variety of activities. This may indicate that C. pinensis has two major modes of metabolism: a “focused” mode of extracting glucose from β-glucans, and a “scavenging” mode that enables survival on complex plant glycans while hunting for other carbon sources.
In scavenging mode, such as when complex heteroglycans are more abundant in the local microenvironment, a variety of different enzymes are produced to maximize the sugar collected, and the uptake of oligosaccharides from these substrates is highly efficient. Conversely, when microbial material is more locally abundant and β-glucans are the primary available glycans, the PUL system allows C. pinensis to switch to a more “focused” β-glucan-degrading behavior. The bacterium secretes enzyme cocktails that can very effectively deconstruct complex high-molecular-weight β-glucans, but uptake of the oligosaccharides produced is affected by the linkage of the polysaccharide backbone, the length of the oligosaccharides produced, and the presence of branching. This specificity of uptake is mediated via SusC-like proteins encoded in glucan-degrading PULs and via the selectivity of PULs to be induced by defined glucan structures. From a biological perspective, this behavior may be due to the relatively high metabolic value of glucose compared to that of the monosaccharides available from the more heterogeneous glycans analyzed here, or it may reflect some mutuality in the microbial community. Other organisms may utilize the GOs produced by C. pinensis, which in turn relies on other species to depolymerize glycans such as arabinoxylan. Additional study of C. pinensis in a broader microbial context is needed to explore this further, to determine whether cross-feeding of GOs to other relevant species from the soil microbiota is possible, and to understand whether C. pinensis can indeed metabolize XOs produced by other bacterial or fungal members of the community.
MATERIALS AND METHODS
Carbohydrates utilized.
Avicel cellulose was purchased from Fluka Biochemika. Birch wood xylan, chitin, and chitosan were purchased from Sigma-Aldrich. Xyloglucan (tamarind seed), wheat arabinoxylan, barley mixed-linkage β-glucan, larch arabinogalactan, konjac glucomannan, carob galactomannan, sugar beet arabinan, lichenan, and carboxymethylcellulose-4M were purchased from Megazyme. Curdlan was obtained from Waco Chemicals (Richmond, VA, USA). Scleroglucan (CS11) was obtained from Cargill. Monosaccharide standards were obtained from Sigma-Aldrich. Oligosaccharide standards were purchased from Megazyme.
Extraction and purification of mushroom polysaccharides.
Fungal polysaccharide was isolated from the basidiomycete species Agaricus bisporus. The fruiting bodies of A. bisporus were extracted and fractionated as previously described to obtain the linear (1→6)-linked β-d-glucan (32). The structure of the polysaccharide was confirmed by monosaccharide composition and linkage analyses (see Table S2 in the supplemental material).
(i) Monosaccharide composition analysis of mushroom polysaccharides by GC-MS.
TFA hydrolysis was performed with 1 mg of freeze-dried material using 2 M trifluoroacetic acid (TFA) at 120°C for 3 h. The hydrolyzed samples were dried until complete removal of the TFA, reduced with sodium borohydride (NaBH4) at 60°C for 1 h, and acetylated with pyridine and acetic anhydride (1:1 [vol/vol], 200 μl) at 100°C for 30 min. The alditol acetates were extracted with ethyl acetate and analyzed by gas chromatography with coupled mass spectrometry (GC-MS). The monosaccharide composition was quantified by external calibration using neutral sugar standards by the comparison of the retention times and electron impact profiles.
(ii) Glycosidic linkage analysis of mushroom polysaccharides by GC-MS.
Per-O-methylation of A. bisporus polysaccharide fractions was carried out using NaOH-dimethyl sulfoxide (Me2SO)-methyl iodide (MeI) as described by Pettolino et al. (33), with some modifications. The samples (1 mg) were dissolved in dimethyl sulfoxide (400 μl) and powdered NaOH (20 mg) was added. Methyl iodide was added with 10-min intervals (5 × 50 μl) under continuous stirring and an inert (Ar) atmosphere. The methylated polysaccharides were recovered in the organic phase after partition (3×) with H2O and dichloromethane (DCM; 2:1 [vol/vol]). The samples were hydrolyzed by the addition of 1 ml 2 M TFA at 121°C for 3 h and were dried, reduced with sodium borodeuteride (NaBD4) at 60°C for 1 h, and acetylated with pyridine and acetic anhydride (1:1 [vol/vol], 200 μl) at 100°C for 30 min. The per-O-methylated alditol acetates were extracted with ethyl acetate, analyzed by GC-MS, and identified from the fragmentation of their positive ions by comparison with polysaccharide standards. The results were expressed as a relative percentage of each component.
Preparation of complex biomass carbon sources.
Spruce wood chips were subjected to ball milling and ground to a fine powder, using a method described by Giummarella et al. (17). For some experiments, the spruce wood powder was additionally pretreated by autoclaving for 2 h at 121°C. A fruiting body of A. bisporus was lyophilized and ground to a fine powder.
Strain growth.
All reagents used in bacterial growth were purchased from Sigma-Aldrich, unless otherwise stated, and were of microbiological grade. A lyophilized pellet of cultured C. pinensis strain UQM 2034T was purchased from DSMZ (designated DSM-2588) and propagated on Casitone-yeast extract (CY) and LB media and plates with kanamycin at 50 μg · ml−1, to which the bacterium has innate immunity. In common with previous observations (34), a yellow pigment was produced during growth on these media, due to the production of a flexirubin pigment (35, 36).
To screen for growth on complex forms of biomass, 50-μl samples from 10-ml starter cultures grown in LB were inoculated in 10 ml M9 minimal medium (37). The standard glucose was absent from this medium. Cultures were supplemented with 0.5% of either glucose, spruce wood, or A. bisporus fruiting body, prepared as described above. Samples of 500 μl were taken after 3, 5, and 8 days of incubation, and absorbance readings (A600) were taken to use as an indirect measure of cell density. Samples were allowed to stand and settle for 5 min before OD readings were taken. Samples were retained for later analysis of carbohydrate structures by HPAEC-PAD (see below). In addition, secretomes were assayed for enzymatic activity (see “Reducing sugar assays of enzyme activity” below).
In a first screen for growth on a range of isolated polysaccharides, 50-μl samples from 10-ml starter cultures grown in LB were inoculated in M9 minimal medium (10 ml). The standard glucose was again absent from this medium, which was instead supplemented with a specific carbon source at 0.5%. The carbon sources tested were glucose, carboxymethylcellulose, barley β-glucan, a linear β-1,6-glucan extracted from A. bisporus, curdlan, scleroglucan, konjac glucomannan, and wheat arabinoxylan. Minimal medium not supplemented with carbon source served as a control and supported no growth. Cultures were incubated at 25°C with rotary shaking at 180 rpm. Secretomes were harvested from these cultures by centrifugation to pellet cells (4,000 × g for 15 min at 4°C) when cultures were observed to have reached cell densities indicating maximum growth (between 5 and 25 days). Secretomes were assayed for enzymatic activity (see “Reducing sugar assays of enzyme activity” below).
To generate detailed growth curves, cultures of 15 ml in M9 medium (lacking glucose but supplemented with another carbon source at 0.5%, as described above) were inoculated with 50 μl from a 10-ml LB starter culture (three biological replicates). Samples of 500 μl were taken at regular intervals, and OD readings (OD600) were used as an indirect measure of cell density. Samples were allowed to stand and settle for 5 min before OD readings were taken. Samples were retained for later analysis of carbohydrate structures by HPAEC-PAD (see below). During growth on all carbohydrates, the pH remained stable at around 7.0 throughout incubation.
Carbohydrate analysis of growing cultures.
Samples (500 μl) were taken from growing cultures of C. pinensis and analyzed by SEC-MALLS, HPAEC-PAD, and LC-ESI-MS/MS to assess the degradation of polysaccharides and the release of oligosaccharides.
Size-exclusion chromatography with coupled multiangle light scattering.
Samples were analyzed by size exclusion chromatography (SEC; SECcurity 1260; PSS, Mainz, Germany) coupled to a multiple-angle laser light scattering detector (MALLS; BIC-MwA7000; Brookhaven Instrument Corp., Holtsville, NY) and a refractive index detector (SECcurity 1260; PSS, Mainz, Germany) to determine the MW distribution of the polysaccharide substrates in the medium after growth of C. pinensis. Samples of 100 μl were injected directly into a combined column setup with a SUPREMA precolumn, a SUPREMA 1,000 Å column, and two SUPREMA 3,000 Å analytical columns (PSS, Mainz, Germany) eluting with 1 ml · min−1 of 100 mM NaNO3 plus 1 mM NaN3 (Milli-Q) as the mobile phase at 40°C. Calibration of the detectors was performed by the injection of pullulan standards (PSS, Mainz, Germany) with molar masses ranging from 342 × 103 to 708 × 103 Da (PSS, Germany). The Mark-Houwink parameters for pullulan in aqueous solutions at 40°C were K = 1.0176 × 10−3 dl · g−1 and a = 0.525 (38), and the dn/dc was 0.149 ml · g−1 (39). Data collection and analysis were performed using WinGPC software (PSS, Mainz, Germany).
High-performance anion-exchange chromatography with pulsed amperometric detection.
Oligo- and monosaccharides were analyzed on a Dionex ICS-3000 high-performance liquid chromatography (HPLC) system operated by Chromeleon software version 6.80 (Dionex) using a Dionex CarboPac PA200 column. Solvent A was water, solvent B was 1 M sodium hydroxide, and solvent C was 1 M sodium acetate. Depending on the analytes, different gradients were employed. For the detection of monosaccharides or xylooligosaccharides on a Dionex CarboPac PA1 column the gradients were as follows: prewash and column calibration, −14 to −7 min 60% B, 40% C (1 ml · min−1); −6 to 0 min 100% A (1 ml · min−1); sample injection, 0 to 5 min 100% A (0.5 ml · min−1); gradient elution, 5 to 20 min 0% to 30% B (0.5 ml · min−1). For the detection of manno- or gluco-oligosaccharides on a Dionex CarboPac PA200 column, the gradients were as follows: prewash and column calibration, −5 to 0 min 15% B (0.5 ml · min−1); sample injection, 0 to 16 min 15% B (0.5 ml · min−1); gradient elution, 16 to 30 min 33% B (0.5 ml · min−1), 30 to 31 min 33% B and 50% D (0.5 ml · min−1); column wash and final elution, 31 to 35 min 15% B (0.5 ml · min−1). Carbohydrates were identified and quantified by comparing the peak retention time and area to those of standards of known concentration. The concentration of unidentifiable oligosaccharides was estimated by comparison to the peak area of the most closely eluting standard.
Liquid chromatography-electrospray ionization-tandem mass spectrometry.
Oligosaccharides for which no standards were available were sequenced via derivatization and LC-ESI-MS/MS. Samples were reduced in 2% borohydride for 30 min. The reaction was quenched with acidic methanol and dried under inert gas flow. Samples were then resuspended in dimethyl sulfoxide, and methylation was performed as previously described (40). After partition in dichloromethane-water, the organic phase was recovered, dried, and resuspended in 50% acetonitrile. Derivatized oligosaccharides were separated through an SB-C18 column (250 mm by 4.6 mm; Agilent Technologies) in a Capillary LC (Micromass, UK) at a flow rate of 10 μl · min−1 with a gradient of increasing acetonitrile content (30% to 60%). The LC was coupled to a Q-TOF2 (Micromass, UK) for MS and MS/MS analysis. The Q-TOF was operated at 3.3 kV and 60 V in the capillary and the cone, respectively. Argon was used as collision gas for MS/MS analysis of selected ions, at a voltage of 35 to 90 V. Glycosidic bond fragments indicate the presence of branching points by a difference of 14 m/z in the Y ion. The presence of a 1→6 linkage at the nonreducing end was indicated by the corresponding 3,5A2 fragment (m/z 329).
Reducing sugar assays of enzyme activity.
For the activity screen, the substrate at 1 mg · ml−1 was incubated with ∼50 μg · ml−1 protein for 16 h at 25°C in 50 mM sodium phosphate buffer (pH 7). Hydrolysis of polysaccharide substrates was measured by an increase in reducing sugars, using the 3,5-dinitrosalicylic acid reducing sugar assay (DNSA) (41, 42) versus a standard curve of Glc (1 to 6 mM). Refer to the “Carbohydrates utilized” for a full list of the polysaccharides that were assayed.
Reaction samples were added to an equal volume of DNSA reagent (1% [wt/vol] DNSA, 0.2% [vol/vol] phenol, 1% [wt/vol] NaOH, 0.005% [wt/vol] glucose, and 0.05% [wt/vol] NaSO3) to terminate the reaction, and the color was developed by boiling for 20 min and cooling on ice for 5 min, prior to measuring A575 on a Cary 50 spectrophotometer.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from the Knut and Alice Wallenberg foundation via the Wallenberg Wood Science Centre (to L.S.M.) and the Swedish Research Council Vetenskapsrådet (project 621-2014-5295 to F.V. and project 2017-04906 to L.S.M.).
Spruce wood was a kind donation from Martin Lawoko of KTH Royal Institute of Technology, Stockholm, Sweden.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02231-18.
REFERENCES
- 1.Thomas F, Hehemann JH, Rebuffet E, Czjzek M, Michel G. 2011. Environmental and gut bacteroidetes: the food connection. Front Microbiol 2:93. doi: 10.3389/fmicb.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griffiths E, Gupta RS. 2001. The use of signature sequences in different proteins to determine the relative branching order of bacterial divisions: evidence that Fibrobacter diverged at a similar time to Chlamydia and the Cytophaga-Flavobacterium-Bacteroides division. Microbiology 147:2611–2622. doi: 10.1099/00221287-147-9-2611. [DOI] [PubMed] [Google Scholar]
- 3.Schimel JP, Schaeffer SM. 2012. Microbial control over carbon cycling in soil. Front Microbiol 3:348. doi: 10.3389/fmicb.2012.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.López-Mondéjar R, Zühlke D, Becher D, Riedel K, Baldrian P. 2016. Cellulose and hemicellulose decomposition by forest soil bacteria proceeds by the action of structurally variable enzymatic systems. Sci Rep 6:25279. doi: 10.1038/srep25279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brabcova V, Novakova M, Davidova A, Baldrian P. 2016. Dead fungal mycelium in forest soil represents a decomposition hotspot and a habitat for a specific microbial community. New Phytol 210:1369–1381. doi: 10.1111/nph.13849. [DOI] [PubMed] [Google Scholar]
- 6.Gholz HL, Wedin DA, Smitherman SM, Harmon ME, Parton WJ. 2000. Long-term dynamics of pine and hardwood litter in contrasting environments: toward a global model of decomposition. Glob Chang Biol 6:751–765. doi: 10.1046/j.1365-2486.2000.00349.x. [DOI] [Google Scholar]
- 7.Lundqvist J, Teleman A, Junel L, Zacchi G, Dahlman O, Tjerneld F, Stålbrand H. 2002. Isolation and characterization of galactoglucomannan from spruce (Picea abies). Carbohydr Polym 48:29–39. doi: 10.1016/S0144-8617(01)00210-7. [DOI] [Google Scholar]
- 8.Willför S, Sjöholm R, Laine C, Roslund M, Hemming J, Holmbom B. 2003. Characterisation of water-soluble galactoglucomannans from Norway spruce wood and thermomechanical pulp. Carbohydr Polym 52:175–187. doi: 10.1016/S0144-8617(02)00288-6. [DOI] [Google Scholar]
- 9.Bowman SM, Free SJ. 2006. The structure and synthesis of the fungal cell wall. Bioessays 28:799–808. doi: 10.1002/bies.20441. [DOI] [PubMed] [Google Scholar]
- 10.Ruthes AC, Rattmann YD, Malquevicz-Paiva SM, Carbonero ER, Cordova MM, Baggio CH, Santos AR, Gorin PA, Iacomini M. 2013. Agaricus bisporus fucogalactan: structural characterization and pharmacological approaches. Carbohydr Polym 92:184–191. doi: 10.1016/j.carbpol.2012.08.071. [DOI] [PubMed] [Google Scholar]
- 11.Smiderle FR, Ruthes AC, van Arkel J, Chanput W, Iacomini M, Wichers HJ, Van Griensven LJ. 2011. Polysaccharides from Agaricus bisporus and Agaricus brasiliensis show similarities in their structures and their immunomodulatory effects on human monocytic THP-1 cells. BMC Complement Altern Med 11:58. doi: 10.1186/1472-6882-11-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sangkhobol V, Skerman VBD. 1981. Chitinophaga, a new genus of chitinolytic myxobacteria. Int J Syst Evol Bacteriol 31:285–293. doi: 10.1099/00207713-31-3-285. [DOI] [Google Scholar]
- 13.McKee LS, Brumer H. 2015. Growth of Chitinophaga pinensis on plant cell wall glycans and characterisation of a glycoside hydrolase family 27 beta-l-arabinopyranosidase implicated in arabinogalactan utilisation. PLoS One 10:e0139932. doi: 10.1371/journal.pone.0139932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsbrink J, Tuveng TR, Pope PB, Bulone V, Eijsink VG, Brumer H, McKee LS. 2017. Proteomic data on enzyme secretion and activity in the bacterium Chitinophaga pinensis. Data Brief 11:484–490. doi: 10.1016/j.dib.2017.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsbrink J, Tuveng TR, Pope PB, Bulone V, Eijsink VG, Brumer H, McKee LS. 2017. Proteomic insights into mannan degradation and protein secretion by the forest floor bacterium Chitinophaga pinensis. J Proteomics 156:63–74. doi: 10.1016/j.jprot.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Grondin JM, Tamura K, Déjean G, Abbott DW, Brumer H. 2017. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol 199:e00860-16. doi: 10.1128/JB.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giummarella N, Zhang L, Henriksson G, Lawoko M. 2016. Structural features of mildly fractionated lignin carbohydrate complexes (LCC) from spruce. RSC Adv 6:42120–42131. doi: 10.1039/C6RA02399A. [DOI] [Google Scholar]
- 18.Novaes-Ledieu M, Garcia Mendoza C. 1981. The cell walls of Agaricus bisporus and Agaricus campestris fruiting body hyphae. Can J Microbiol 27:779–787. doi: 10.1139/m81-121. [DOI] [PubMed] [Google Scholar]
- 19.Harris GW, Jenkins JA, Connerton I, Cummings N, Leggio LL, Scott M, Hazlewood GP, Laurie JI, Gilbert HJ, Pickersgill RW. 1994. Structure of the catalytic core of the family F xylanase from Pseudomonas fluorescens and identification of the xylopentaose-binding sites. Structure 2:1107–1116. doi: 10.1016/S0969-2126(94)00112-X. [DOI] [PubMed] [Google Scholar]
- 20.St John FJ, Gonzalez JM, Pozharski E. 2010. Consolidation of glycosyl hydrolase family 30: a dual domain 4/7 hydrolase family consisting of two structurally distinct groups. FEBS Lett 584:4435–4441. doi: 10.1016/j.febslet.2010.09.051. [DOI] [PubMed] [Google Scholar]
- 21.Mewis K, Lenfant N, Lombard V, Henrissat B. 2016. Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization. Appl Environ Microbiol 82:1686–1692. doi: 10.1128/AEM.03453-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cartmell A, McKee LS, Peña MJ, Larsbrink J, Brumer H, Kaneko S, Ichinose H, Lewis RJ, Viksø-Nielsen A, Gilbert HJ, Marles-Wright J. 2011. The structure and function of an arabinan-specific α-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases. J Biol Chem 286:15483–15495. doi: 10.1074/jbc.M110.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKee LS, Sunner H, Anasontzis GE, Toriz G, Gatenholm P, Bulone V, Vilaplana F, Olsson L. 2016. A GH115 alpha-glucuronidase from Schizophyllum commune contributes to the synergistic enzymatic deconstruction of softwood glucuronoarabinoxylan. Biotechnol Biofuels 9:2. doi: 10.1186/s13068-015-0417-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, Creagh AL, Haynes CA, Kelly AG, Cederholm SN, Davies GJ, Martens EC, Brumer H. 2014. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506:498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, Rogowski A, Hamilton BS, Chen R, Tolbert TJ, Piens K, Bracke D, Vervecken W, Hakki Z, Speciale G, Munōz-Munōz JL, Day A, Peña MJ, McLean R, Suits MD, Boraston AB, Atherly T, Ziemer CJ, Williams SJ, Davies GJ, Abbott DW, Martens EC, Gilbert HJ. 2015. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luis AS, Briggs J, Zhang X, Farnell B, Ndeh D, Labourel A, Baslé A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schückel J, Venditto I, Ralet M-C, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. 2018. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol 3:210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pluvinage B, Grondin JM, Amundsen C, Klassen L, Moote PE, Xiao Y, Thomas D, Pudlo NA, Anele A, Martens EC, Inglis GD, Uwiera RER, Boraston AB, Abbott DW. 2018. Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont. Nat Commun 9:1043. doi: 10.1038/s41467-018-03366-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu Y, Han L, Hefferon KL, Silvaggi NR, Wilson DB, McBride MJ. 2016. Periplasmic Cytophaga hutchinsonii endoglucanases are required for use of crystalline cellulose as sole carbon and energy source. Appl Environ Microbiol doi: 10.1128/aem.01298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishinari K, Zhang H, Funami T. 2009. 20 - Curdlan, p 567–591. In Philips GO, Williams PA (ed), Handbook of hydrocolloids, 2nd ed Woodhead Publishing, Cambridge, UK. [Google Scholar]
- 30.Larsbrink J, Izumi A, Ibatullin FM, Nakhai A, Gilbert HJ, Davies GJ, Brumer H. 2011. Structural and enzymatic characterization of a glycoside hydrolase family 31 alpha-xylosidase from Cellvibrio japonicus involved in xyloglucan saccharification. Biochem J 436:567–580. doi: 10.1042/BJ20110299. [DOI] [PubMed] [Google Scholar]
- 31.Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smiderle FR, Alquini G, Tadra-Sfeir MZ, Iacomini M, Wichers HJ, Van Griensven LJLD. 2013. Agaricus bisporus and Agaricus brasiliensis (1→6)-β-d-glucans show immunostimulatory activity on human THP-1 derived macrophages. Carbohydr Polym 94:91–99. doi: 10.1016/j.carbpol.2012.12.073. [DOI] [PubMed] [Google Scholar]
- 33.Pettolino FA, Walsh C, Fincher GB, Bacic A. 2012. Determining the polysaccharide composition of plant cell walls. Nat Protoc 7:1590–1606. doi: 10.1038/nprot.2012.081. [DOI] [PubMed] [Google Scholar]
- 34.Glavina del Rio T, Abt B, Spring S, Lapidus A, Nolan M, Tice H, Copeland A, Cheng JF, Chen F, Bruce D, Goodwin L, Pitluck S, Ivanova N, Mavromatis K, Mikhailova N, Pati A, Chen A, Palaniappan K, Land M, Hauser L, Chang YJ, Jeffries CD, Chain P, Saunders E, Detter JC, Brettin T, Rohde M, Goker M, Bristow J, Eisen JA, Markowitz V, Hugenholtz P, Kyrpides NC, Klenk HP, Lucas S. 2010. Complete genome sequence of Chitinophaga pinensis type strain (UQM 2034). Stand Genomic Sci 2:87–95. doi: 10.4056/sigs.661199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoner TA, Fuchs SW, Schonau C, Bode HB. 2014. Initiation of the flexirubin biosynthesis in Chitinophaga pinensis. Microb Biotechnol 7:232–241. doi: 10.1111/1751-7915.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reichenbach H, Kohl W, Böttger–Vetter A, Achenbach H. 1980. Flexirubin-type pigments in Flavobacterium. Arch Microbiol 126:291–293. doi: 10.1007/BF00409934. [DOI] [Google Scholar]
- 37.Miller JH. 1975. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 38.Sullivan MA, Powell PO, Witt T, Vilaplana F, Roura E, Gilbert RG. 2014. Improving size-exclusion chromatography separation for glycogen. J Chromatogr A 1332:21–29. doi: 10.1016/j.chroma.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 39.Ciric J, Oostland J, de Vries JW, Woortman AJJ, Loos K. 2012. Size exclusion chromatography with multi detection in combination with matrix-assisted laser desorption ionization-time-of-flight mass spectrometry as a tool for unraveling the mechanism of the enzymatic polymerization of polysaccharides. Anal Chem 84:10463–10470. doi: 10.1021/ac302704q. [DOI] [PubMed] [Google Scholar]
- 40.Ciucanu I, Kerek F. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res 131:209–217. doi: 10.1016/0008-6215(84)85242-8. [DOI] [Google Scholar]
- 41.Miller GL. 1959. The use of dinitrosalicylic acid for the determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 42.McKee LS. 2017. Measuring enzyme kinetics of glycoside hydrolases using the 3,5-dinitrosalicylic acid assay In Abbot DW, Lammerts van Bueren A (ed), Protein-carbohydrate interactions: methods and protocols. Springer, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.