Despite the considerable loss inflicted on important crops yearly by Pectobacterium and Dickeya diseases, investigations on key virulence and interbacterial competition assets relying on extensive comparative genomics are still surprisingly lacking for these genera. Such approaches become more powerful over time, underpinned by the growing amount of genomic information in public databases. In particular, our findings point to new functional associations among well-known genomic themes enabling alternative means of neutralizing SRE diseases through disruption of pivotal virulence programs. By elucidating novel transcriptional and genomic associations, this study adds valuable information on virulence candidates that could be decisive in molecular applications in the near future. The utilization of 100 genomes of Pectobacterium and Dickeya strains in this study is unprecedented for comparative analyses in these taxa, and it provides novel insights on the biology of economically important plant pathogens.
Keywords: bacterial capsules, bacteriophages, genomics, pectobacterium, transcriptome analysis, type VI secretion systems
ABSTRACT
Soft-rot Enterobacteriaceae (SRE), typified by Pectobacterium and Dickeya genera, are phytopathogenic bacteria inflicting soft-rot disease in crops worldwide. By combining genomic information from 100 SRE with whole-transcriptome data sets, we identified novel genomic and transcriptional associations among key pathogenicity themes in this group. Comparative genomics revealed solid linkage between the type I secretion system (T1SS) and the carotovoricin bacteriophage (Ctv) conserved in 96.7% of Pectobacterium genomes. Moreover, their coactivation during infection indicates a novel functional association involving T1SS and Ctv. Another bacteriophage-borne genomic region, mostly confined to less than 10% of Pectobacterium strains, was found, presumably comprising a novel lineage-specific prophage in the genus. We also detected the transcriptional coregulation of a previously predicted toxin/immunity pair (WHH and SMI1_KNR4 families), along with the type VI secretion system (T6SS), which includes hcp and/or vgrG genes, suggesting a role in disease development as T6SS-dependent effectors. Further, we showed that another predicted T6SS-dependent endonuclease (AHH family) exhibited toxicity in ectopic expression assays, indicating antibacterial activity. Additionally, we report the striking conservation of the group 4 capsule (GFC) cluster in 100 SRE strains which consistently features adjacently conserved serotype-specific gene arrays comprising a previously unknown organization in GFC clusters. Also, extensive sequence variations found in gfcA orthologs suggest a serotype-specific role in the GfcABCD machinery.
IMPORTANCE Despite the considerable loss inflicted on important crops yearly by Pectobacterium and Dickeya diseases, investigations on key virulence and interbacterial competition assets relying on extensive comparative genomics are still surprisingly lacking for these genera. Such approaches become more powerful over time, underpinned by the growing amount of genomic information in public databases. In particular, our findings point to new functional associations among well-known genomic themes enabling alternative means of neutralizing SRE diseases through disruption of pivotal virulence programs. By elucidating novel transcriptional and genomic associations, this study adds valuable information on virulence candidates that could be decisive in molecular applications in the near future. The utilization of 100 genomes of Pectobacterium and Dickeya strains in this study is unprecedented for comparative analyses in these taxa, and it provides novel insights on the biology of economically important plant pathogens.
INTRODUCTION
Conflicts among microbes and against their hosts unfold under intense selective pressure, resulting in an abundant inventory of colonization traits in unicellular organisms from all domains of life (1–6). In prokaryotic genomes specifically, the allocation of gene inventories is recognized by frequent conservation within functionally clustered units. Such a pattern is determined by the strong selection imposed on the organization of prokaryotic genomes (7). The typical simultaneous occurrence of essential processes, such as transcription, translation, and protein localization in the prokaryotic cytoplasm, drives selective pressure on genome organization (8). In this scenario, the occurrence of clustered units constitutes a cohesive solution to facilitate cotranscription of functionally associated genes (9–12). In fact, this feature has been broadly exploited through guilt-by-association methods in several studies, enabling the elucidation of novel gene functions (9, 13, 14).
Bacteria recently reclassified in the Pectobacteriaceae family (15), responsible for causing soft rot-blackleg/wilt disease, are commonly designated, based on the previous taxonomic classification, as soft-rot Enterobacteriaceae (SRE). This group is mainly represented by the Pectobacterium and Dickeya genera (16). These microbes have attracted attention for having great impact on global food security, as they infect a considerable range of plant hosts (16, 17). Hence, roughly 100 SRE genome sequencing projects are publicly available, either completed or in progress (18–22). Underpinned by such a wealth of public data, recent transcriptome-based reports have depicted important facets of SRE biology. For example, the role of small RNAs (sRNAs) in the adaptive response of Pectobacterium atrosepticum exposed to nutrient-deficient environments has been investigated, revealing 68 regulated sRNAs in these conditions and leading to the discovery of nine novel sRNAs (23). In P. atrosepticum, the impact on the transcription of 26% of the genome upon deletion of expI revealed the critical role of quorum-sensing regulation for disease development (24). Similarly, transcriptome analyses demonstrated the impact of 32 isolated stress conditions, mimicking those found during infection, on Dickeya dadantii strain 3937 regulatory patterns, providing a detailed landscape on environmental triggers for gene expression (25). Furthermore, an investigation of the role played by the D. dadantii strain 3937 PecS global regulator during early colonization of leaf tissues uncovered more than 600 genes in its regulon (26). Thus, as a general rule, transcriptome-based approaches are powerful means to explore key strategies utilized by plant pathogens and plants’ critical physiological programs, which can optimize plant genetic enhancement, resulting in positive long-term impacts on food security (27–29).
The deployment of several exoenzyme families specializing in breaking plant cell wall is one of the most conspicuous strategies presented by SRE; hence, it is one of the most exhaustively investigated (30). The activity of plant cell wall-degrading enzymes (PCWDE) causes the release of by-products which can be taken up as nutrients by the bacterial cell (16). Some of these PCWDE include cellulases, pectate lyases (PLs), and pectin lyases (PNLs), among others (16, 31). Another important asset for disease development is the ability to biosynthesize high-molecular-weight polysaccharides, which may either be secreted extracellularly (EPS, or exopolysaccharide) or remain attached to the bacterial cell surface (e.g., lipopolysaccharide [LPS] and capsular polysaccharides [CPS]) (32).
Horizontal gene transfer is a powerful driver in the acquisition of novel functions by bacterial pathogens (33). In this context, genomically integrated bacteriophages (prophages) and toxin/antitoxin systems frequently exported by bacterial secretion systems (e.g., type I, III, and VI secretion systems) typify pathogenically important horizontally acquired islands (HAIs) (34, 35). The expression of HAIs is also a ubiquitous strategy to induce pathogenesis and to succeed in interbacterial competition (36). Specifically, the bacteriophage-like type VI secretion system (T6SS) has been implicated as a crucial ecological asset of several Gram-negative bacteria either as a virulence or bacterial competition agent (37).
In this article, regions of interest were gleaned by analyzing an original transcriptomic data set obtained from Pectobacterium carotovorum subsp. brasiliense strain PBR 1692 (here referred to as Pcb1692) during disease development. The aim was to survey the transcriptional activation of critical genomic regions required for virulence or interbacterial competition in Pcb1692 and assess their conservation in 100 SRE genomes. We report here the in planta coactivation of the carotovoricin homolog (Ctv) prophage in Pcb1692 along with a T1SS module immediately upstream. Comparative analysis added supporting evidence by unveiling an exquisite genomic conservation of the T1SS+Ctv block in Pectobacterium genomes. These results presumably point to a systemic association between these two themes in the Pectobacterium genus in which T1SS may export Ctv elements through the periplasm. Extensive gene neighborhood and protein domain architecture analyses, combined with large-scale sequencing, also shed light on the strong topological and transcriptional association of a previously predicted toxin/immunity pair (WHH- and SMI1_KNR4-containing gene products) with the T6SS machinery. Further evidence uncovered the infection-induced upregulation in Pcb1692 of a highly conserved group 4 capsule (GFC or G4C) biosynthesis cluster in SRE. The analyses also demonstrated that the GFC cluster may be the only capsule production region conserved in Pcb1692. Sequence analyses of gene products encoded by the gfcA locus in a large number of organisms unveiled high sequence variation, suggesting they have a role in GFC machinery as serotype-specific membrane proteins.
RESULTS AND DISCUSSION
Transcriptome sequencing of Pcb1692 during in planta infection.
Pcb1692 has been reported as one of the most aggressive Pectobacterium species known to date (17, 38, 39). Aiming to examine the transcriptional landscape of Pcb1692 during infection in potato tubers, a whole-transcriptome data set was generated, including samples harvested 24 and 72 h postinfection (hpi) along with an in vitro control (see Materials and Methods). The data set comprised 14 to 20 million transcriptome sequencing (RNA-Seq) paired-end reads for each stage, with 97% of the mapped reads in each sample uniquely mapped to the reference genome, implying good overall quality (Table 1). Subsequent analyses identified 1,743 protein-coding genes (43.5% of total annotated in Pcb1692) under infection-induced regulation (log2 fold change of >1 or <−1; false discovery rate [FDR], <0.01) in the wild-type strain in at least one time range (see Table S1.1 in the supplemental material). Importantly, a recent study depicted D. dadantii’s transcriptome during infection on Arabidopsis thaliana, compared at 6 and 24 hpi, as being able to detect 13.5% of its protein-coding genes (575 out of 4,244) under regulation (up or down) (26). We take advantage of this recently published gene expression experiment, featuring a closely related wild-type SRE strain infecting a non-crop host, to comparatively examine up/downregulation in our original gene expression data set obtained from Pcb1692.
TABLE 1.
Mapping summary of RNA-Seq reads on Pcb1692 reference genome
| Samplea | No. of: |
% Uniquely mapped |
Total mapped reads |
No. of raw read inputs |
|||
|---|---|---|---|---|---|---|---|
| Unmapped reads |
Multiple matches |
Uniquely mapped reads |
No. | % | |||
| Cwt | 751,568 | 416,981 | 13,835,241 | 97.07 | 14,252,222 | 95.0 | 15,003,790 |
| Wt-24hpi | 1,856,371 | 555,623 | 18,131,044 | 97.03 | 18,686,667 | 91.0 | 20,543,037 |
| Wt-72hpi | 2,852,242 | 333,569 | 11,124,857 | 97.09 | 11,458,426 | 80.1 | 14,310,667 |
Abbreviations: Cwt, in vitro control wild type; Wt-24hpi, wild type 24 hpi; Wt-72hpi, wild type 72 hpi.
SRE’s most well-recognized strategy to accomplish plant tissue invasion involves disruption of plant cell wall by deploying a variety of pectinase-, cellulase-, and protease-related protein families (16). PLs and PNL are pectinases that participate in this process by breaking α-1-4-linkages of homogalacturonan backbone in the pectin molecule via a transelimination mechanism (40). The data set presented here emphasizes the role of PL in Pcb1692 infection by the significant activation of seven out of eight genes, occurring simultaneously during the first 24 hpi. This encompasses orthologs of well-documented genes (pelABCINZ, PCBA_RS04055, PCBA_RS04060, PCBA_RS04065, PCBA_RS10065, PCBA_RS03200, and PCBA_RS04070) and one unannotated gene (PCBA_RS18630) (Table S1.1). These findings are similar to those detected in D. dadantii, in which all nine PLs (pelABCDEILNZ) were activated while infecting A. thaliana (26). The pectin lyase gene pnl (PCBA_RS19200), which encodes a major PCWDE (24), is massively activated (>5 log2 fold change), ranking among the top 1% most upregulated genes 24 hpi in Pcb1692 (Table S1.2). This finding suggests a specific high transcriptional demand of pnl in Pcb1692 during early disease development. Conversely, in the D. dadantii transcriptome, the pnl ortholog (Dda3937_03551) surprisingly shows no detectable regulation (Table S1.3) (26). Together, these results strengthen the notion of diversity in PCWDE pools transcriptionally activated by pectinolytic pathogens to surpass various obstacles imposed by hosts in distinct plant tissues (41). Whereas for some PCWDE families (e.g., PLs) the upregulation is basal, regardless of host characteristics, other PCWDE families (e.g., PNL) are transcriptionally activated under specific host-imposed cues.
Transcriptional profile and conservation of type VI secretion system and associated effectors in SRE.
The T6SS is a bacteriophage-like structure that depends on the expression of 13 core genes (tss cluster) to assemble a complex that spans the cell envelope. In addition to the tss cluster, a hemolysin-coregulated protein (Hcp) forms the T6SS tube structure, which is propelled across the membranes with a piercing structure on its tip, consisting primarily of two proteins (VgrG/PAAR), into the target cell (42, 43). Furthermore, it has also been demonstrated that this piercing structure is able to accommodate independent proteins which function as effectors (34). Our data set underscores the importance of T6SS by showing the overwhelming activation of all 13 genes in the tss cluster (>3-log2 fold change in transcriptional variation during the first 24 hpi). Moreover, a total of four Hcp secretion islands (HSIs), one adjacent to the tss cluster (HSI-1) and another three in different genomic contexts (HSI-2, -3, and -4), were also upregulated for at least one time point. These HSI exhibit an overall activation of two PAAR-coding and four vgrG genes (Table S2.1). In comparison, wild-type D. dadantii displays similar upregulation of most tss and four hcp genes out of six conserved in the genome upon infection on A. thaliana (Table S1.3). However, none of the VgrG- or PAAR-encoding genes (five and three, respectively) are transcriptionally regulated in D. dadantii in the same experiment (26). Notably, the role of VgrG-PAAR as a delivery mechanism for Rhs effectors into target cells is established in D. dadantii facing in vitro competition (44). Thus, although structural components of T6SS sheath and tube are similarly regulated when infecting distant hosts in Pcb1692 and D. dadantii, genes encoding the piercing tip components may be activated under specific cues (26) (Table S1.3).
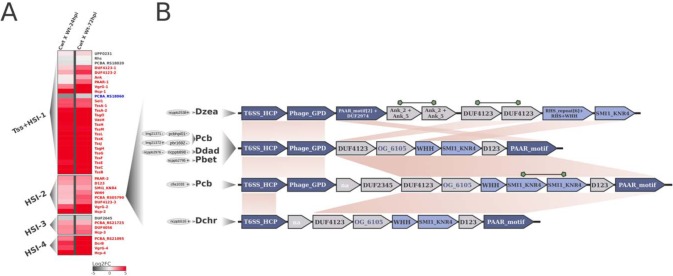
Interestingly, the second major HSI in the Pcb1692 genome (HSI-2) spans ∼6.6 kb, harboring eight genes under positive regulation for at least one time point during infection (Fig. 1A). Of these, six (excluding only hcp and vgrG) are uncharacterized/unannotated (Table S2.1). Aiming to garner deeper functional insight on this region, we combined the analysis of domain architectures conserved in protein sequences with contextual genomic information from 100 SRE organisms. First, conserved domain analysis confirmed VgrG (PCBA_RS05800; Pfam entry PF05954) and PAAR gene products (PCBA_RS05770; Pfam entry PF05488) in this region, implying that the cluster is able to source both the Hcp tube and the piercing tip (VgrG/PAAR) assembly (Fig. 1B). Further, two uncharacterized genes (PCBA_RS05780 and PCBA_RS05785) flanked by the genes encoding VgrG and PAAR caught our attention for containing domain architectures SMI1_KNR4 and WHH (Pfam entry PF14414 and PF09346), respectively, in their products. Interestingly, the association between the superfamilies SUKH and HNH/ENDOVII, which encompass SMI1_KNR4 and WHH families, respectively, has been recognized as a recurrent theme in bacterial genomes (14). This duplet (WHH-SMI1_KNR4) was described as an immunity/toxin pair associated with contact-dependent growth inhibition (CDI) systems (e.g., T5SS and T6SS), although evidence actually linking these genes to the secretion systems remains scarce (14, 45–47). The WHH family was originally identified as a restriction endonuclease derived from the HNH domain (Pfam entry PF01844; CL0263) (48). Our analyses revealed that members of the WHH family are present in only 18 out of 100 SRE strains. In 3 of these 18 strains (Pectobacterium parmentieri strains CFIA1002 and RNS08421a and P. carotovorum subsp. brasiliense CFIA1033), the WHH-containing gene was found to be duplicated (Table S2.2). Out of the 18 genomes conserving WHH-containing genes, tight association to SMI1_KNR4 family members immediately downstream in 13 genomes was found, which corroborates the original report for this duplet (Fig. 1B and Table S2.2) (14). Analysis of the 13 genomes conserving this association (WHH-SMI1_KNR4) in SRE determined that one was not suitable for contextual genomic inspection in this specific region due to incompleteness of genome assembly (namely, P. betavasculorum strain NCPPB 2793). By assessing the remaining suitable structures conserving the WHH-SMI1_KNR4 duplet, we found 83% (10 out of 12) linkage with upstream HCP, and PAAR-encoding genes were mostly downstream (Fig. 1B). A duplication in the immunity gene encoding SMI1_KNR4 was also found in one strain, which corroborates the original report of this family (14) (Fig. 1B). These results imply a strong association of the WHH-SMI1_KNR4 duplet with HSIs in SRE genomes, such as P. carotovorum subsp. brasiliense (five strains, including Pcb1692), D. dadantii (two strains), Dickeya zeae (one strain), Dickeya chrysanthemi (one strain), and P. betavasculorum (one strain) (Fig. 1B and Table S2.2). These observations, taken together with the coordinated upregulation at the transcriptional level of WHH-SMI1_KNR4-encoding genes, along with all other HSI-2 neighboring elements in Pcb1692, suggest the T6SS-dependent secretion of these genes (Fig. 1). In this context, we provide a report of coordinated transcriptional regulation of a SUKH-1/HNH-ENDOVII system along with the surrounding HSI, in which the WHH-containing gene product may work as a T6SS effector recruited during infection.
FIG 1.
Transcriptional variation in planta of T6SS-related genes in Pcb1692 and linkage between the endonuclease WHH and HSI in SRE genomes. (A) Each row on the heatmap represents a gene labeled after its (i) respective encoded protein name, (ii) functional domain, or (iii) locus tag given annotation availability. The two columns of the heatmaps represent pairwise comparisons between samples; thus, each cell shows the variation in transcription found in each comparison, illustrated by a color scale ranging from −5 (dark gray) to 5 (red), representing the log2 fold change in expression. For each gene label, statistical significance of regulation is represented with the following colors: red, upregulation; blue, downregulation; gray, no support for regulation. Column labels: Cwt, control wild type; Wt-24hpi/Wt-72hpi, wild type at 24/72 hpi in potato tubers. Four segments in the heatmap separate T6SS-related clusters, type VI secretion (Tss) and Hcp secretion island (HSI), located in different genomic regions, namely, Tss+HSI-1, HSI-2, HSI-3, and HSI-4. (B) Gene neighborhood of WHH-SMI1_KNR4 duplets in the 10 SRE strains conserved within HSIs. Among the 100 SRE strains analyzed, those exhibiting linkage between WHH-SMI1_KNR4- and Hcp/PAAR-encoding genes are represented. Each gene arrangement is associated with its respective strain(s), represented on the left side by 4-letter abbreviations: Dzea, Dickeya zeae; Pcb, Pectobacterium carotovorum subsp. brasiliense; Ddad, Dickeya dadantii; Pbet, Pectobacterium betavasculorum; Dchr, Dickeya chrysanthemi. For each species, the respective number of strains conserving that particular gene arrangement is represented in gray ellipses on the left. The WHH-SMI1_KNR4 duplets (Pfam PF14414 and PF09346, respectively) are highlighted in light blue. Hcp (T6SS_HCP; Pfam PF05638), VgrG (Phage_GPD; Pfam PF05954), and PAAR (PAAR_motif; Pfam PF05488) genes are highlighted in dark blue. Syntenically conserved blocks across the different gene arrangements are linked by light-brown areas. Duplicated genes in a single arrangement are linked by green shapes. Successfully clustered products in orthologous groups for which no conserved domains were detected are highlighted in dark blue.
Analysis of antibacterial activity of type VI secretion system-related toxins.
Aiming to investigate the potential of HSI-borne genes from Pcb1692 to have antibacterial activity, four relevant genes were cloned into an expression vector and ectopically expressed in Escherichia coli, as described by Koskiniemi et al. in 2013 (44). To this end, the previously described WHH-containing gene product (PCBA_RS05785) was selected alongside three other putative toxins (PCBA_RS05775, PCBA_RS18045, and PCBA_RS22965) for this analysis. PCBA_RS05775 belongs to the cell division cycle protein 123 family (D123; Pfam entry PF07065), which seems not to be accompanied by an adjacent immunity-encoding gene (Table S2.3). The PCBA_RS18045 gene is located in HSI-1, within the Pcb1692 tss gene cluster, and possesses a PAAR domain followed by an alpha/beta hydrolase fold with a GxSxG catalytic lipase motif (Pfam entry PF12697), characteristic of phospholipases (49). This phospholipase family member is followed by a putative immunity gene downstream that carries an Ank domain (Pfam entry PF00023). The PCBA_RS22965 gene has a conserved HNH/EndoVII fold derivative known as AHH (Pfam entry PF14412). Similar to WHH family members, the AHH proteins were predicted to be functionally associated with CDI systems in the original report of the family (14). By scanning 100 SRE genomes, we found that 23 of these conserve AHH-encoding genes, out of which one (Dickeya zeae strain CSLRW192) harbors two copies of the gene (Table S2.3). In most SRE genomes harboring members of the AHH family, no detectable domain in the downstream immunity protein can be observed. In Pcb1692, similar to six other SRE genomes conserving AHH-encoding genes, a contig break occurs up to five genes prior to this AHH member, hindering direct gene neighborhood-based conclusions. Nonetheless, in other strains from P. carotovorum subsp. brasiliense, P. carotovorum subsp. carotovorum, and P. atrosepticum species conserving 70% to 95% of overall genomic synteny with Pcb1692 (Fig. S1), the AHH genes are solidly neighbored by hcp, vgrG, and a DcrB domain (Pfam entry PF08786)-encoding gene, suggesting a role as a T6SS-associated effector (Table S2.3). Such an arrangement precisely matches the previously assessed HSI-4 region in Pcb1692 (Fig. 1A). In addition to the original report of the AHH family, these results suggest that the AHH member plays a role in Pcb1692 infection as a T6SS-associated nuclease.
The results show that no cell death occurred due to expression of three of the four effectors between 60 and 240 min (Fig. 2A). However, expression of the AHH effector caused a reduction in E. coli growth, indicating that it has a toxic effect on E. coli. This growth inhibition could be counteracted by coexpression of the toxin and immunity gene (AHH+i) in E. coli, suggesting that the previously observed reduced growth was due to the expression of the AHH nuclease (Fig. 2B). Thus far, no experimental data exist for the AHH and WHH nucleases, as well as the D123 protein, as T6SS effectors. This report elucidates the antibacterial activity of an AHH-containing protein.
FIG 2.
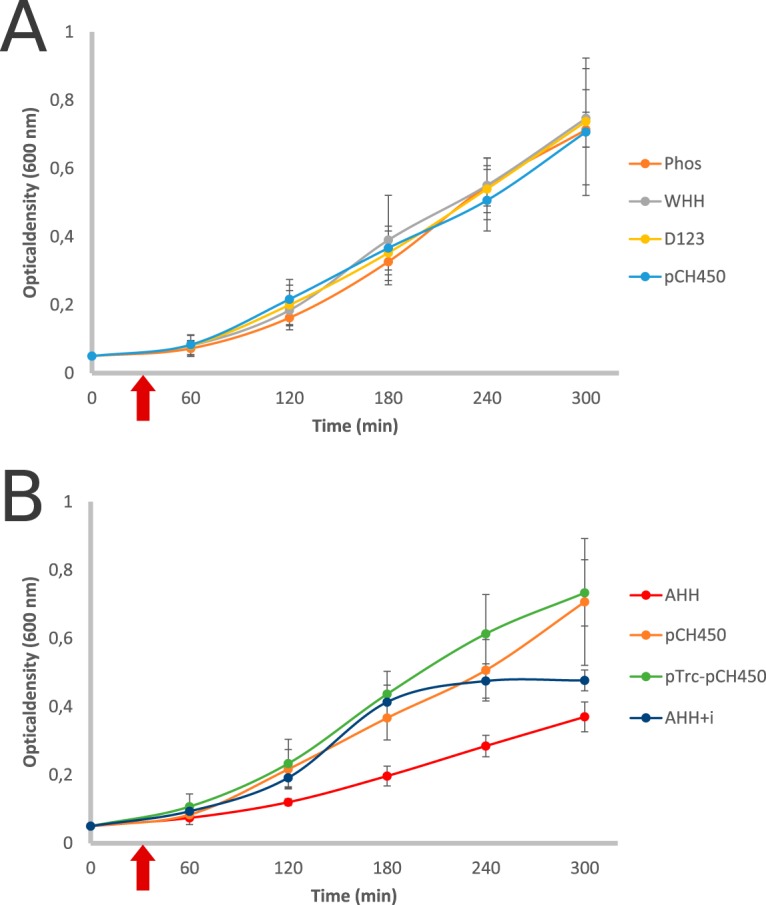
Growth of E. coli DH5α expressing effectors from pCH450 induced with 0.2% l-arabinose after 30 min of growth or effector and immunity protein from pCH450 and pTrc100, respectively. (A) Expression of phospholipase, WHH nuclease, and D123 effectors in E. coli shows growth comparable to that of the empty vector control (p450). Individual expression of these effectors does not contribute to bacterial growth inhibition under the conditions analyzed. (B) Expression of AHH nuclease decreases the growth rate of E. coli. Expression of both AHH and its immunity protein (AHH+i) negates the toxic effect of the effector alone; however, after 240 min a plateau in growth is reached due to protein overexpression.
Several T6SS effectors targeting eukaryotes, bacteria, or both have been identified thus far (25, 50, 51). Here, we have demonstrated that one of the four effectors identified inhibits growth of E. coli cells. It is possible that, for the other effectors tested, the in vitro conditions used were not conducive to growth inhibition. We may also need to consider that our hypothesis that the T6SS of Pcb1692 is mainly antibacterial is not entirely correct. Bernal et al. (52) have noted that, thus far, no plant-targeting effectors have been identified as T6SS dependent. As the D123 protein has no reported function and lacks a downstream immunity protein, it could be a candidate for a plant-targeted effector. Since some effectors have interkingdom activity, it may be necessary to reevaluate whether the effectors we have identified as putative antibacterial effectors have key functions within the eukaryotic host. Moreover, it must also be taken into consideration that T6SS could have functions other than contact-dependent antibacterial competition, which may favor conservation of effectors involved in other biological programs that will enforce successful host colonization.
Genomic conservation and transcriptional regulation of prophages during in planta infection.
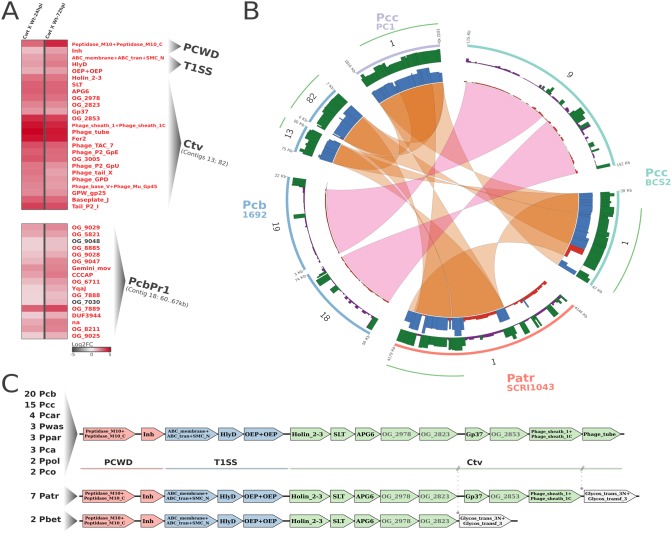
The impact of HAIs originating from prophages in bacterial pathogenic behavior is pervasive for both plant and animal pathogens (53–55). Importantly, selective advantage conferred by prophage-borne genes to plant pathogens has been associated with the activity of bacterial secretion systems, as reported in Pseudomonas syringae and Ralstonia solanacearum (35, 56, 57). In this context, by using a combination of relevant genome-wide approaches (see Materials and Methods), we detected two prominent prophage regions in Pcb1692 exhibiting significant activation at the transcriptional level during infection (Fig. 3A). The first region encompasses 20 genes in the Pcb1692 genome strongly upregulated in the first 24 hpi (Fig. 3A). Moreover, 90% of these genes are syntenic with genomic segments in at least 54/60 other Pectobacterium strains (Fig. 3B and Table S3.1). Interestingly, a genomic segment in P. carotovorum subsp. carotovorum similar to the one found in Pcb1692 was originally described as being of prophage origin, regarded as carotovoricin (Ctv) consisting of 19 genes (58) (Table S3.2). The report of this bacteriocin has been recently corroborated by in silico analyses in the same species, even extending the putative range of the cluster to 22 genes in P. carotovorum subsp. carotovorum (termed PecaPC1-p2) (Table S3.3) (35). In this context, we also detected in P. atrosepticum a segment of 12.7 kb containing 11 gene products highly similar to Ctv sequences (ECA_RS18415-ECA_RS18485). Intriguingly, this segment is located within a previously characterized bacteriophage spanning ∼36 kb, designated ECA41 (59). In addition, ECA41 contribution to virulence toward potato hosts was reported, although the mechanism of putative effectors’ action remains unknown (59). Curiously, those three reports describing Ctv, PecaPC1-p2, and ECA41 occurred without knowledge convergence based on the detectable homology linking the regions in P. atrosepticum and P. carotovorum subsp. carotovorum, which our analyses now reconcile under the same Ctv-homologous root. This preliminary observation, combined with interpretations of the above-mentioned reports, may provide a robust background for future studies on the role of carotovoricin in pathogenesis.
FIG 3.
Transcriptional variation in Pcb1692 and genomic range of Ctv and PcbPr1 insertions in four Pectobacterium strains. (A) The heatmaps show transcriptional variation profiles of T1SS+Ctv block and serralysin/inhibitor genes (top) and from a 7-kb segment of PcbPr1 (bottom). The heatmap is represented as described for Fig. 1. For Ctv and PcbPr1, contig coordinates are represented in parentheses immediately below labels, which corresponds to panel B in this figure. Genes are preferentially represented by the domain architecture or by the respective orthologous group (OG_#). na, none of the previously described labels is applicable. (B) Drastically contrasting degree of conservation between Ctv and PcbPr1 is represented in four selected Pectobacterium strains. Genomic regions from P. carotovorum subsp. carotovorum strain PC1 (light purple) and BCS2 (light green), P. atrosepticum strain SCRI1043 (light orange), and Pcb1692 (light blue) are represented in circular ideograms. The next two consecutive inner radii show the degree of conservation of the regions, represented by bars. The height of the bars in each radius represents the frequencies of BLASTP (protein sequence similarity; purple/green color range) and MCScanX (syntenic conservation; red/blue color range) hits for each gene product in the respective strains (PC1, BCS2, SCRI1043, and Pcb1692) compared pairwise to those of the remaining 99 SRE strains. The color range key used in the bars represents low and high conservation in BLASTP and MCScanX comparisons. Genes with 30 or fewer positive hits (out of 99) are shown in purple and red, respectively, in each radius. Genes with more than 30 positive hits are shown in green and blue. The links binding genomic regions represent the insertion range of prophages for which conserved synteny could be detected (Ctv, orange; PcbPr1, pink). The green outer radius adjacent to the ideograms represents the range of BLASTP similarity to the original Ctv described by Yamada et al. (58) (Table S3.2). (C) Gene neighborhood screening of T1SS+Ctv block. Genes were separated into plant cell wall degradation (PCWD; light red), T1SS (blue), and Ctv (light green) groups or a group unrelated to these classes (gray). The genomic organization found in Pectobacterium genomes is to the right of the names for species conserving arrangements abbreviated as described in Table S8. To the left of each species name is the number of strains harboring the particular arrangement.
Furthermore, we observed that Ctv is neighbored by a complete set of T1SS genes in P. carotovorum subsp. brasiliense (PCBA_RS08610-PCBA_RS08620) immediately upstream (Fig. 3C and Table S3.4). In addition, conserved domain analyses revealed two genes in the Ctv cluster containing SLT (Pfam entry PF01464) and Gp37 (Pfam entry PF09646) architectures, both reportedly conserved in bacterial virulence factors (60, 61). Therefore, the presence of predicted effector functionalities in these genes, along with a known secretion system, prompted us to assess the degree of conservation of the T1SS+Ctv structure. The results support a remarkable conservation of this genomic architecture, displaying a prominent block of at least 13 genes in Pectobacterium genomes. In contrast, the T1SS+Ctv genomic association is consistently absent from all 39 Dickeya genomes analyzed (Table S3.4). The 13 genes in the T1SS+Ctv gene block include (i) two genes encoding a peptidase/inhibitor pair, (ii) three T1SS components, and (iii) eight Ctv genes under solid conservation in 96.7% of Pectobacterium genomes (59 out of 61). Importantly, this high conservation trend seems to occur regardless of the overall synteny between these genomes (Fig. S1). The T1SS is recognized for its relative simplicity, composed of three core proteins which form a tunnel-like structure in the inner membrane, enabling molecule transfer from cytosol to the extracellular space fueled by ATP hydrolysis (62). Although typically regarded as a signal-independent system, there is a known group of products exported in a T1SS-dependent manner that contains N-terminal signals. This group encompasses bacteriocins and microcins (63–65). Since Ctv constitutes a bacteriocin system, an additional layer of support to the T1SS-Ctv functional association could be added by searching for signal peptides within Ctv sequences. Given the generally poor conservation described in T1SS signal peptides, sensitive search setups were carried out (66, 67). The predictions from two different methods corroborate the possible T1SS/Ctv association by the combined detection of N-terminal signal peptides in five proteins encoded by genes in this region. In addition, some genes lacking known conserved domains, for which the protein sequences were clustered in orthologous groups (see Materials and Methods) OG_2978, OG_2823, and OG_2853, are collectively conserved in 96% to 100% of Pectobacterium genomes (Fig. 3C and Table S3.4). Curiously, these highly conserved genes in Pectobacterium encode small products (up to 109 amino acids) and could undertake a role in bacterial competition as small antimicrobial peptides. Here, we unraveled the strikingly dominant theme encompassing T1SS and Ctv clusters in Pectobacterium genomes tied by different sources of evidence. This linkage suggests either that Ctv is an addictive selfish element colonizing these genomes or that, similar to other reported T1SS/bacteriocin associations, this system was recruited in the Pectobacterium lineage to export Ctv-borne products either toward or through the cell membranes.
The second prophage, which apparently has not been described thus far, has low similarity to other SRE genomes, being mostly confined into P. carotovorum subsp. brasiliense strains (here referred to as PcbPr1) and one P. carotovorum subsp. carotovorum strain (Fig. 3B). BLASTP searches of PcbPr1 sequences resulted in 5 proteins out of 64 (7.8%) returning matches against at least 90 SRE species (out of 99) (Fig. 3B). In contrast, overall 82.3% of Pcb1692 protein-coding sequences (3,376 out of 4,099) successfully match at least 90 SRE (Fig. S2). The low conservation of PcbPr1 is also observable in terms of genomic organization, as 93.7% of the whole PcbPr1 structure (60 out of 64 genes) conserves synteny with less than 10% of all Pectobacterium genomes analyzed (Fig. 3B and Fig. S2). Despite the low degree of conservation, PcbPr1 harbors an internal segment spanning 7 kb, in which 88.2% of the genes (15 out of 17) are cohesively upregulated over the first 72 hpi (Fig. 3A and Table S4). Importantly, 12 out of 17 genes in this region encode proteins for which known conserved domains could not be detected. In addition, most of these gene products were clustered in small orthologous groups with sequences from other P. carotovorum subsp. brasiliense and P. carotovorum subsp. carotovorum isolates, implying lineage-specific origin (Fig. 3A). This observation supports the previous synteny analysis, reinforcing the possibility of PcbPr1 comprising a lineage-specific insertion in P. carotovorum subsp. brasiliense and P. carotovorum subsp. carotovorum. Notably, genomic conservation of functional lineage-specific prophages is observable in both animal and plant pathogens, as typified in studies using E. coli and R. solanacearum (68–70). Interestingly, it has been reported that two HAIs in R. solanacearum, namely, Rasop1 and Rasop2, showed no similarity to other phages previously sequenced in the species (35). In order to assess gene expression patterns of Rasop1 and Rasop2 during infection, we gleaned data from an existing whole-transcriptome experiment (71). Upregulation of 58.6% and 62.5% of Rasop1 (27 out of 46) and Rasop2 (20 out of 32) genes was detected upon infection of tomatoes (Table S5) (71). The evidence presented underscores lineage-specific insertion conservation as an important strategy for some organisms, probably providing a competitive advantage against close species not bearing the same traits. Therefore, PcbPr1 conservation and upregulation during infection might have implications in Pcb1692 success when facing in planta competition against other Pectobacterium spp. As one of the few relatively large lineage-specific genomic regions compared to those of other Pectobacterium spp., we speculate that PcbPr1 could be a key asset for the reported high level of virulence observed in P. carotovorum subsp. brasiliense.
Characterization of dedicated polysaccharide-biosynthetic clusters.
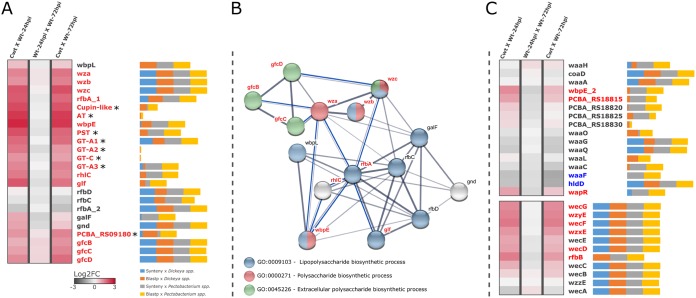
The composition of serotype-specific polysaccharide gene clusters gives rise to biochemical diversity in bacterial cell surfaces (72). The fundamental role of cell surface in virulence has been well established in both animal and plant pathogens (32, 73–75). In SRE, however, aside from the establishment of an E. coli rffG homolog in P. atrosepticum as a probable player in enterobacterial common antigen biosynthesis (76), overall knowledge on this subject remains limited. In this context, our data set enabled detection of a 25-kb region in Pcb1692 functionally associated with polysaccharide biosynthesis, exhibiting significant coactivation of 17 out of 24 genes within the first 24 hpi in potato tubers (Fig. 4A). A homologous region in D. dadantii 3937 exhibits a different pattern while infecting A. thaliana leaves by regulating only 3/24 genes positively and 2/24 negatively (Table S1.3). Through orthology-based annotation of Pcb1692 genes compared with those of model organisms (see Materials and Methods) and subsequent integration with the STRING correlational database (77), most of the gene products were annotated into the LPS biosynthetic pathway (Fig. 4B). Out of the remaining eight unannotated entries, seven were successfully characterized by conserved domain inspection through HMM profiles (78, 79), revealing well-known domains in polysaccharide production, such as an integral membrane polysaccharide-specific transporter (PST; Pfam entry PF14667), an acetyltransferase (AT; Pfam entry PF13302), a cupin-like protein (Pfam entry PF05523), and four glycosyltransferases (GT; PFAM clans CL0110 and CL0111) (Fig. 4A and Table S6.1). The remaining unannotated gene (PCBA_RS09180) is related to gfcA-yjbE, described in E. coli, and will be discussed in detail in the next section. As a general rule, all eight of these entries unsurprisingly display high sequence variation, mostly comprising weakly conserved blocks among SRE (Fig. 4A). These lineage-specific genes presumably comprise a serotype-specific block in SRE polysaccharide biosynthesis.
FIG 4.
In planta transcriptional profile, genomic conservation, and annotation of dedicated polysaccharide clusters in Pcb1692. (A) Heatmap of GFC operon as described in the legend to Fig. 1. Asterisks next to genes indicate those for which no orthology relationship could be assessed; as such, they are not represented in the network. Next to each gene name, horizontal bars (percentage-stacked) display their respective conservation compared with SRE genomes of Dickeya (39 strains) and Pectobacterium (60 strains) genera. These bars include overall frequencies of BLASTP (sequence similarity) and MCScanX (syntenic conservation) searches of Pcb1692 gene products against 99 SRE complete data sets. The bars are divided in segments: syntenic with Dickeya spp. (blue), BLASTP positive against Dickeya spp. (red), syntenic with Pectobacterium spp. (gray), and BLASTP positive against Pectobacterium spp. (yellow). (B) Correlational network denoting the annotated gene association between successfully predicted Pcb1692 orthologs in E. coli and P. aeruginosa according to the STRING database (77) as well as showing respective GO term annotations. Thickness of network links are proportional to the combined scores obtained from the STRING algorithm. Blue lines represent new coregulation evidence found in our transcriptome data set between genes for which other categories of associations are known. Those entries not annotated in the three major GO terms shown in the network are highlighted in gray. (C) Transcriptional variation of waa (top) and wec (bottom) gene clusters represented as described for panel A.
By integrating our annotations with STRING networks, we next observed new coexpression (coordinately upregulated in our data set) associations between Pcb1692 orthologs during the course of infection (Fig. 4B and Table S6.2). The coexpression linking several genes encoding GTs and modification enzymes (e.g., rhlC, rfbA, glf, and wbpE) in our data set were undetected in STRING, indicating an uncommon genomic architecture in Pcb1692, detailed below (Fig. 4B). Some other associations are surprisingly novel, such as those among gfcBCD and wza-wzc genes, all reportedly participants of capsule biosynthesis (80, 81) (Fig. 4B). In this regard, functional studies have elucidated the role of Wza-Wzc as a transmembrane complex which spans the entire periplasm and which is required for capsule assembly (82). As for gfcABCD, although the exact functions remain unknown, these genes are regarded as the typifying theme of group 4 capsule production clusters (32, 81, 83). Therefore, the presence of wza-wzb-wzc and gfc orthologs indicates that Pcb1692 possesses, and significantly activates during infection, the genetic apparatus to produce GFC. The contrast between expression patterns found in experiments involving D. dadantii and Pcb1692 suggests that the transcriptional activation of the region is triggered by specific cues related to the host.
It is noteworthy that a recently proposed model for programmatic large-scale identification of GFC operons based on E. coli did not include internal serotype-specific genes (83). Indeed, a typical GFC operon in E. coli carries the gfc genes immediately upstream of wza-wzb-wzc paralogs (i.e., gfcE, etp, and etk) with no adjacent serotype-specific genes. However, our report supports the presence of lineage-specific genes flanked by wza-wzb-wzc and gfcBCD in Pcb1692, for which transcriptional upregulation is required upon infection, potentially representing a serotype-specific block (Fig. 4A). Moreover, the presence of poorly conserved gene organizations upstream of gfc genes seems to be a general feature in SRE genomes, as depicted by extensive gene neighborhood analysis (Fig. S3 and Table S6.3). These blocks in SRE may vary from 13 to 27 genes harboring diverse family compositions consistently flanked by wza-wzb-wzc upstream and gfcBCD downstream. Although blocks ranging from 16 to 20 serotype-specific genes seem to be preferred across SRE genomes, there is no direct association between any particular species and specific block sizes (Fig. S3 and Table S6.4). This unusual organization of the GFC lineage-specific region in SRE compared to the one in E. coli might explain why some coexpression associations (e.g., gnd and rhlC) in our data set were not present in the STRING database as mentioned above (Fig. 4B). The functional implications for SRE, and especially Pcb1692 harboring this lineage-specific gene array flanked by wza-wzb-wzc and gfcABCD, will be discussed in detail in another section.
Importantly, a recent study predicted that 40% of the bacterial lineages analyzed conserve more than one capsule system (83). This observation prompted us to systematically survey the Pcb1692 genome for other possible capsule region occurrences. Since LPS, EPS, and capsules share many gene families in their functional units, which greatly hinders fully automatic predictions, the regions in the Pcb1692 genome were manually inspected following an initial automatic screening (see Materials and Methods). The analysis allowed additional detection of two conspicuous polysaccharide clusters in Pcb1692, although neither of these regions conserves characteristic domains that typify capsule-related regions (Table S6.1). Thus, these findings indicate that GFC is the only known capsule group produced in Pcb1692. Nonetheless, these two additional regions carry eight and seven waa and wec orthologs, respectively, which are commonly associated with LPS biosynthesis and could be important assets during infection; therefore, they are worth investigating (84, 85). By analyzing their transcriptional patterns, an increased demand for GFC and wec gene transcription within the first 24 hpi was observed. In contrast, in the waa region, only 3 out of 16 genes were upregulated (Fig. 4C). Further, significant downregulation of two genes (waaF and hldD) and three borderline predictions of downregulation (waaG, waaQ, and waaC) (Table S6.1) indicated a general low demand for transcription of the waa region during infection. Curiously, between 24 and 72 hpi a slightly negative transcriptional modulation occurs in most of the genes in the three regions; however, it does not significantly impact the overall trend for upregulated genes (Fig. 4C). Unlike GFC or waa gene clusters, the wec region is remarkably conserved in nearly all SRE, encompassing 6 (out of 11) genes displaying infection-induced upregulation in Pcb1692 (Fig. 4C). These observations elucidate the unequal transcriptional demand for three distinct polysaccharide clusters during infection of potato by Pcb1692. While wec and especially gfc gene clusters seem to be consistently recruited at the transcriptional level, the transcription profile of the waa cluster is mostly flat, suggesting different functional demands during infection for these regions.
Analysis of gfcA-related sequences and genomic contexts.
The inability to assess either orthology or domain conservation for PCBA_RS09180, in addition to the current lack of information regarding the function of gfcA, prompted an in-depth comparative investigation into this locus. Based on results presented above, 88% of the SRE genomes analyzed carry orphan (i.e., not clustered by OrthoMCL) genes upstream of gfcBCD (Table S6.3). In the remaining 22% of strains, we detected gene products from small clusters populated with sequences from three (OG_5444; Dickeya solani, D. dadantii, and D. chrysanthemi), two (OG_10199; D. solani and D. chrysanthemi), and one (OG_7594; D. dianthicola) species (Table S6.3). This clustering pattern in groups populated by a small number of species implies lineage-specific expansions. Furthermore, overall only 10% of SRE strains carry the YjbE domain (Pfam entry PF11106) originally described in E. coli. These include the two P. betavasculorum strains analyzed and six out of seven P. atrosepticum strains in the analysis. Hence, the presence of some PCBA_RS09180-related sequences in small clusters, represented by no more than three species (e.g., OG_5444 and OG_7594) due to the high level of sequence variation, constitutes a typical serotype-specific pattern.
In order to test these results more broadly, we next expanded the gene neighborhood analysis beyond the SRE group. Sequence-based searches were performed in order to retrieve a set of PCBA_RS09180-related proteins in bacteria. Unsurprisingly, BLASTP (86) search using PCBA_RS09180 sequence against NCBI (nonredundant protein database) was ineffective. On a preliminary level, this is consistent with the observations found in SRE that showed high sequence variation in PCBA_RS09180-related sequences. We then utilized a more sensitive approach (87) to obtain distantly related PCBA_RS09180 proteins. Within 50 positive matches, 27 were supported by publicly available genome-wide data and were suitable for gene neighborhood screening (Table S6.5). From these, three encode YjbE-containing products (Pfam entry PF11106), corroborating the relationship between PCBA_RS09180 and yjbE and/or gfcA. On the other hand, the remaining 24 gene products display no detectable domain (similar to PCBA_RS09180). The results revealed that even in evolutionarily distant organisms, 91% (22 out of 24) of the genes distantly related to that encoding PCBA_RS09180 are confined upstream of gfc and/or yjb homologous operons (Table S6.6). Thus, although this locus has been termed yjbE and/or gfcA and the respective domain described in E. coli is designated YjbE, this locus is in fact highly variable and the YjbE domain is weakly represented. An additional confirmation was obtained by consulting a large repository for domain architecture annotation (88). The presence of the YjbE domain in bacteria is at least 20-fold less frequent than that of YjbF, YjbH, and Caps_synth_GfcC (Table S6.7). Together, these results uncover the highly variable nature of the gfcA locus in a broad range of prokaryotes, which could be a serotype-specific player in the capsule biosynthesis machinery.
The gfcA-encoded products from 100 SRE strains were then aligned to assess possible conserved residues in their sequences. The results revealed a conspicuously conserved N-terminal segment harboring an MKKTLxxLxxxxAxxxxxxA motif (Fig. S4). Further, we conducted predictions for the secondary structure of PCBA_RS09180 sequence compared to those of GfcA and YjbE using two different methods (89, 90). Together, the analyses cohesively show two possible transmembrane sections in both PCBA_RS09180 and GfcA with apparent cytoplasmic orientation of the N-terminal region. Conversely, secondary-structure predictions for YjbE were inconclusive. Also, the prediction of an N-terminal signal peptide in PCBA_RS09180 and GfcA also strengthens the possibility of shared functionality. In addition, these findings indicate a closer functional relationship between PCBA_RS09180 and GfcA than PCBA_RS09180 and YjbE. Strikingly, further extensive signal peptide predictions made by two different methods in 100 gfcA sequences from SRE also returned 96% overlapping positive results in N-terminal regions. This result matches those for the conserved region found in the previous alignment, reinforcing their functional conservation (Fig. S4). Moreover, secondary-structure predictions in the same set of sequences supported that 80% of gfcA orthologs can conserve between one to three transmembrane regions, which is in accordance with the results from GfcA. Together, these results suggest that PCBA_ RS09180, and presumably SRE orthologs, could be directed to the inner membrane (similar to GfcA) to function as membrane proteins. This corroborates the current hypothesis of GfcABCD performing an auxiliary role in polysaccharide translocation across the membranes (91).
Analysis of GFC elements and wza-wzc system organization in Pcb1692.
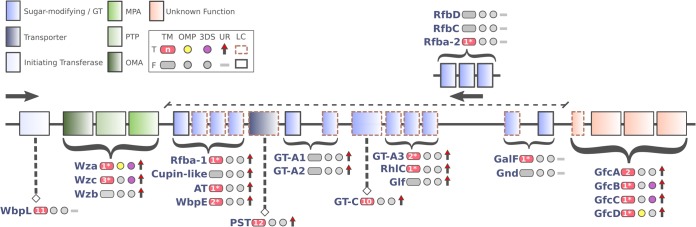
The currently accepted model for polysaccharide biosynthesis via the Wza-Wzc transmembrane conduit involves association of several functional modules, such as (i) a series of peripheral GTs and sugar modification enzymes, (ii) a Wzx-like (PST family) transmembrane flipping protein, (iii) an initiation sugar transferase, and (iv) a Wzy-like polymerase (32). The Wza-Wzc system is often associated with two highly similar systems that produce group 1 and 4 capsules (32). Curiously, the gene arrangement in SRE’s GFC region resembles the one from group 1 capsule described in other species carrying a serotype-specific block downstream of wza-wzb-wzc (Fig. S3) (92). Nonetheless, the genome-wide absence of the group 1 capsule-characteristic wzi gene suggests it could not be produced in the vast majority of SRE species, except for P. betavasculorum and P. carotovorum subsp. actinidiae (data not shown). In this context, the results for Pcb1692 were examined in light of comparative analyses with SRE and then superimposed onto the canonical Wza-Wzc model in order to propose an organization for the GFC machinery in P. carotovorum subsp. brasiliense. A total of eight GTs or modification enzymes, predicted to be soluble or weakly attached to the inner membrane, located in this region exhibit cohesive upregulation within the first 24 hpi in Pcb1692 (Fig. 5). These enzymes exhibit zero to two transmembrane (TM) sections, which is in accordance with the Wza-Wzc functional model (Fig. 5). The conserved PST within the GFC region presents 12 TM helices, which is described as the consensus number of TM sections for Wzx-like flippases (93). The functional domain detected in this sequence is also classified in the same Pfam clan (CL0222), which encompasses the domain found in the canonical Wzx (Pfam entry PF01943). Further, in Pcb1692, the Glycos_transf_4 (Pfam entry PF00953) domain found in the WbpL ortholog (PCBA_RS09080) is also conserved in WbaP/WecA sequences, which are the reported initiation transferase linked to Wzc in other bacteria (Table S6.1) (94, 95). This WbpL ortholog also features 11 TM helices solidly predicted, which matches the exact number found in WecA from E. coli (Fig. 5). As for the Wzy-like activity, however, the direct association to this Pcb1692 region is unclear. The WzyE activity in E. coli is described as being similar to that of members of the GT-B subfamily (96, 97). However, GT-B is the only unrepresented GT subfamily within the described GFC region in Pcb1692. Interestingly, a GT-C member found within the Pcb1692 GFC region conserves 10 TM helices, matching the number found in Pcb1692's direct ortholog of WzyE (PCBA_RS14975). The presence of periplasmic loops could also be predicted in this GT-C member, consistent with previously described Wzy proteins (Fig. 5) (98, 99).
FIG 5.
Schematic organization of GFC region organization in Pcb1692. At the top left, the color assignments of known or unknown protein functionalities are shown using the following abbreviations: MPA, membrane-periplasmic auxiliary protein; PTP, protein tyrosine phosphatase; OMA, outer membrane auxiliary protein; GT, glycosyltransferase. A second box, framed by continuous lines, shows a binary table with graphical representations for the presence (T, true) or absence (F, false) of five parameters analyzed in the protein sequences. Abbreviations: TM, transmembrane segments; OMP, outer membrane protein; 3DS, available three-dimensional structure; UR, significant infection-induced upregulation in Pcb1692; LC, low conservation among SRE (detailed below), as assessed by synteny and sequence similarity. The number of detected TMs is within the pink shape. Numbers with an asterisk indicate the prediction methods diverge, whereas the lack of an asterisk indicates these methods converge in the number of TMs predicted. LC is attributed to sequences exhibiting fewer than 40 and 70 positive hits, respectively, in protein similarity (BLASTP) and synteny-based (MCScanX) pairwise comparisons using 100 SRE strains (see Table S6.1 for details). The arrows indicate the transcriptional orientation (sense or antisense) of the genes within the region, and the curly braces indicate ranges of genes transcribed in a given orientation for which gene names are displayed (top to bottom) according to the genomic order. A dashed line above the region representation highlights the serotype-specific block found between wza-wzb-wzc and gfcABCD in Pcb1692.
By inspecting outer membrane (OM) propensity (see Materials and Methods) in sequences from the GFC region, only GfcD and Wza returned positive results. This opens a question on the localization of GfcB, which is generally regarded as an OM protein but surprisingly could not be predicted as one (Fig. 5) (PDB entry 2IN5). The only structurally resolved encoded product from the gfc operon currently published is GfcC, which is hypothesized to be a periplasmic protein (91). Together, the results presented here show that Pcb1692 conserves canonical elements of the Wza-Wzc system (i.e., wzx-wzy and wecA) separated from the GFC region (Fig. 4), which could support their encoded products as putative participants in the machinery. Additionally, evidence also indicates that PST, GT-4, and WbpL proteins encoded by genes within the serotype-specific block of the GFC region remarkably mimics structural features found in Wzx, Wzy, and WecA, respectively, suggesting they may undertake similar roles in the wza-wzc system (Fig. 5). Combined, these analyses expand the current view of GFC region gene organization by applying extensive contextual comparisons among SRE genomes, domain analysis, and membrane topology predictions. The presence of serotype-specific blocks flanked by wza-wzb-wzc and gfc genes in SRE sheds light on a novel genomic architecture that combines the known pattern from group 1 capsule regions with typical genes involved in group 4 capsule biosynthesis.
Conclusions.
In this article, we presented an integrative approach that takes advantage of extensive public information to produce a reliable background for genome-wide studies in SRE organisms, including 100 strains from Pectobacterium and Dickeya genera. By combining this platform with an original transcriptome data set, we shed light on strong genomic associations among virulence and interbacterial competition determinants in the SRE group, supported by coordinated transcriptional upregulation of these elements in Pcb1692 throughout disease development in potato tubers. Collectively, these findings provide strong evidence for gene associations and/or infection-induced transcriptional demand within important pathogenicity themes, including PCWDE, T6SS, Ctv, and the capsule biosynthesis machinery, which may pave the way for biotechnological applications in the near future.
MATERIALS AND METHODS
Culture media, growth conditions, and total RNA extraction.
Wild-type Pcb1692 was grown on a nutrient plate at 37°C for 16 h. Overnight cultures were prepared by inoculating a single colony into 10 ml Luria-Bertani (LB) broth at 37°C for 16 h with constant shaking at 200 rpm. To obtain potato inoculated with Pcb1692, healthy potato tubers (Solanum tuberosum cv. Mondial) were inoculated with the Pcb1692 (optical density at 600 nm [OD600] of 1) wild-type strain as previously described (117). The experiments were performed using three biological replicates, with three potato tubers per replicate. Macerated potato tissue was scooped out at 24 and 72 hpi and homogenized in double-distilled water. Bacterial cells were recovered by grinding the scooped, macerated potato tissues in 20 ml of double-distilled water using an autoclaved pestle and mortar. Starch material was removed by centrifuging ground tissue at 10,000 rpm for 1 min. Potato-inoculated and in vitro-cultured bacterial cells were stabilized using RNAprotect reagent (Qiagen, USA) according to the manufacturer’s instructions. Total RNA from in vitro-grown and potato-inoculated bacteria was extracted as previously described using the RNeasy minikit (Qiagen, Hilden, Germany).
Total RNA quality and cDNA library construction.
The concentration and purity of each extracted total RNA sample was evaluated using spectrophotometric analysis (NanoDrop ND-1000; NanoDrop Technologies, Wilmington, DE) at a ratio of 230 to 260 nm. Using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.), total RNA sample concentration, RNA integrity number (RIN), and 28S/18S ratio were determined. Aliquots (200 ng) of total RNA from Pcb1692 extracted from in vitro-grown cells (16 h) and infected potato tubers at 24 and 72 hpi was used to prepare cDNA libraries. The Illumina sequencing service was provided by BGI Co., Ltd. (China). TruSeq RNA sample prep kit v2 (Illumina, USA) was used to construct the cDNA libraries by following the manufacturer’s protocol. In summary, the mRNA was cleaved into small fragments, followed by the synthesis of first-strand cDNA with random hexamer-primed reverse transcription. RNase H and DNA polymerase I were used to synthesize second-strand cDNA. The double-stranded cDNA was subjected to terminal modification by addition of adenosine and ligated with adapters. Adaptor-ligated fragments with suitable sizes were selected and enriched by PCR using the PureLink PCR purification kit (Invitrogen, USA) to create the cDNA libraries for sequencing. Paired-end sequencing (PE91) was performed on an Illumina HiSeq 2000 sequencing platform.
Read mapping, gene expression, and statistical analysis.
Quality assessment of the raw data were performed by fastqc software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc), and then low-quality regions were trimmed by Trimmomatic v 0.36 (100). The trimmed RNA-Seq reads were mapped to the Pcb1692 reference genome (GCF_000173135.1) using hisat2 v 2.1.0 (101). Raw read counts were performed in the R environment (https://www.r-project.org/) by the featureCounts package (102), and subsequent statistical analysis of differential expression was performed by the edgeR package (103). The threshold parameters used to assign differential expression were an FDR of <0.01 (104), absolute log2 fold change of >1 (upregulated), or log2 fold change of <−1 (downregulated). Gene transcriptional profiles were graphically rendered by Gitools (105).
Orthology analysis, domain architecture detection, and gene neighborhood screenings.
Orthology relationships between protein sequences were assessed using the OrthoMCL (inflation, 1.5) pipeline (106), which takes tabular results from a BLASTP search under specific parameters (options: -seg yes -outfmt 6 -num_threads 3 -num_alignments 100000 -evalue 1e-05) as input. The sequences are clustered in orthologous groups labeled with OG numeric tags (e.g., OG_1 and OG_2). In parallel, all sequences were characterized using HMMER3 (107) supported by the Pfam database in order to generate in-house predictions of conserved domain architectures through hidden Markov model (HMM) profiles (108). Orthology and domain architecture information are then combined with genomic coordinates from each genome by custom Perl scripts in order to generate annotated gene neighborhood screenings.
Ectopic expression of predicted T6SS-dependent toxins.
Removal of the rat FABP1 gene from pTrc99A (AddGene) removed some cut sites from the multiple-cloning site (MCS). KnpI and BamHI cut sites were reintroduced into the plasmid by incorporation of the cut sites into the 5′ region of the PCR primers (H5907_F/R) (Table 2). PCR amplification of PCBA_RS05790 (encoding a hypothetical protein upstream of WHH-containing nuclease) incorporated XbaI, KpnI, and BamHI upstream of the open reading frame (ORF) and BamHI and SalI downstream of the ORF. The PCR product was subcloned into pJET1.2/blunt and excised with XbaI/SalI restriction digestion. pTrc99A (without rat FABP1) was digested with XbaI/SalI and ligated with the excised PCR product. The gene was removed using BamHI digest and the linear plasmid religated. In this manner, BamHI and KnpI were reintroduced into the MCS to facilitate cloning of effector genes. The resulting plasmid is referred to as pTrc100, as its MCS differs slightly from that of pTrc99A, but the rest of the plasmid remains unchanged.
TABLE 2.
Primers used in this study
| Primer name and purpose | Sequencea (5′–3′) |
|---|---|
| Modification of pTrc99A to pTrc100 | |
| H_5907_F | TCTAGAGGTACCGGATCCATGTGGCAGTTTAATGACCTGCA |
| H_5907_R | GTCGACGGATCCTTAATCCAGACGGATAAATG |
| Open reading frame of effectors | |
| WHH_F | GAGCTCGGTACCATGGCGAATATCAGAAAGCAAAGCG |
| WHH_R | CTGCAGGTCGACTTAATAATCCTCGCTACGCCACATCG |
| Phos_F | GAGCTCGGTACCATGCCAGGTGCTGCACGTTTAG |
| Phos_R | CTGCAGGTCGACTTATATTCCCCTGAATGTATTGG |
| AHH_F | GAGCTCGGTACCATGGACTGGGTTGATCCGTTTGGGTTGGC |
| AHH_R | CTGCAGGGTACCTCAATATTTAAACGTCCCTGAAACAAGTG |
| D123_F | GTCGACGGTACCATGTATTCTGAACATAAGGCGAC |
| D123_R | CTGCAGAAGCTTCTATAATAAATAGGGTCTTCTTTG |
| Immunity gene for AHH nuclease | |
| AHH-I_F | GAGCTCGGTACCATGAACATTTACACGCTAGAAGC |
| AHH-I_R | CTGCAGGTCGACCTAATCTGATACTCCAACC |
Cut sites are underlined.
Effector genes (PCBA_RS05785, WHH-containing nuclease, SacI/PstI; PCBA_RS05775, D123-containing protein, KpnI/PstI digest; PCBA_RS18045, phospholipase, SacI/PstI digest; and PCBA_RS22965, AHH-containing nuclease, SacI/PstI digest) were ligated into arabinose-inducible expression plasmid pCH450. Immunity genes (PCBA_RS08690, AHHi, KpnI/PstI digest) were ligated into isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible pTrc100 and transformed into E. coli DH5α. Overnight cultures were grown in 0.1% glucose to repress expression from the arabinose-inducible promoter. Cultures were washed in 10 mM MgSO4 adjusted to an OD600 of 0.05 in fresh LB broth supplemented with 5 μg/ml tetracycline, 50 μg/ml ampicillin, and 1 mM IPTG as required. After 30 min, expression from pCH450 was induced with 0.2% l-arabinose. Where necessary, expression from pTrc100 with IPTG was induced immediately. Optical density (at 600 nm) was measured hourly.
Collinearity analysis, prophage origin predictions, and capsule region inspection.
In order to predict colinear conservation in SRE genomes, we first obtained genomic annotation and protein sequence data from all Pectobacterium and Dickeya strains available in the RefSeq database (https://www.ncbi.nlm.nih.gov/refseq) (see Table S7 in the supplemental material) until December 2017, totaling 100 strains, including Pcb1692. We then performed the synteny analysis using MCScanX (109), allowing a minimum match size of two genes to call syntenic blocks (option -s 2). MCScanX takes the tabular results of a nonstringent BLASTP search (86) as the input (option -evalue 1). The total number of positive matches of gene products (from one organism) against all other SRE protein data sets resulting from both BLASTP and MCScanX were parsed by in-house Perl scripts (https://www.perl.org/). Graphical ideograms were scripted in Circos (110), compiling syntenic regions in the genomes as ribbon links between chromosomes, as well as BLASTP and MCScanX hit counts from pairwise comparisons previously described. Prophage origin predictions were conducted by using PhiSpy software (111) in parallel with BLASTP search support (options -qcov_hsp_perc 40 -evalue 1e-05) against Phast (112) and NCBI bacteriophage sequence databases. In parallel, we performed genome-wide synteny predictions on Pcb1692 and P. carotovorum subsp. carotovorum strain BCS2 using recently predicted prophage regions published previously (35). The capsule regions were initially surveyed using HMM-profile search (113) based on the curated database of capsule-related domains made public by Rendueles et al. (83). The Pcb1692 genome next was programmatically scanned for segments harboring at least three consecutive matches carrying the above-mentioned capsule-related domains with two nonmatches gap, allowing for greater sensitivity. These segments were then manually inspected to identify detectable functionality in the adjacent genes that could support the existence of a capsule region.
Sequence search, alignment, and membrane topology predictions of GfcA-related entries.
PCBA_RS09180-related sequences were obtained using the HHblits online tool with default parameters (87). HHblits positive hits were filtered by at least 40 matched amino acid columns in the HMM-HMM alignment, representing ∼40% of the query (PCBA_RS09180) sequence (Table S6.5). All above-threshold entries were retrieved. Out of 50 above-threshold entries, 27 were supported by publicly available genome-wide data, making them suitable for gene neighborhood screening. Genomic data for these 27 structures (complete genome/scaffold/contig) were then obtained from the RefSeq database for gene neighborhood screening. The CDART database was inspected to obtain extensive representativeness of YjbE, YjbF, Caps_synth_GfcC, and YjbH (Pfam entries PF11106, PF11102, PF06251, and PF06082). (88). Sequence alignments were performed using Clustal Omega (114). Prediction of transmembrane segments was conducted by using TMpred (89) and TMHMM (90) online servers in parallel. Signal peptides were predicted by SignalP 4.1 server (66) and Signal-BLAST (67). Detection of putative outer membrane proteins was conducted by using HHomp (115).
STRING network integration.
In order to integrate Pcb1692 with the STRING database (77), orthology annotations with Escherichia coli strain K-12 and Pseudomonas aeruginosa strain PAO1 were used. The Pcb1692 entries conserving orthology with the model organisms were supplied to the STRING correlational database. This provided association between annotated orthologs with the respective Gene Ontology (GO) terms (116) for each sequence.
Accession number(s).
Data determined in the course of this work have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through GEO accession number GSE102557.
Supplementary Material
ACKNOWLEDGMENTS
This research study was funded by the National Research Foundation (NRF), South Africa, through Competitive Funding for Rated Researchers (CFRR 98993), NRF Bioinformatics and Functional Genomics (BFG 93685), and the NRF Research Technology and Transfer Fund (RTF 98654). D.Y.S. received an NRF BFG postdoctoral fellowship. D.B.-R. received a University of Pretoria postdoctoral fellowship. N.M. was funded by NRF Freestanding, PSA, and University of Pretoria Postgraduate bursaries. C.K.T. was funded by a University of Pretoria bursary. Any opinion, findings, conclusions, or recommendations expressed in this material are those of the author(s), and the NRF does not accept any liability in this regard.
Author responsibilities were as follows: conception or design of the study, D.B.-R., C.K.T., and L.N.M.; acquisition, analysis, or interpretation of the data, D.B.-R., C.K.T., N.M., D.Y.S., S.K., and L.N.M.; writing, D.B.-R., N.M., and L.N.M.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02050-18.
REFERENCES
- 1.Alouf JE. 2003. Molecular features of the cytolytic pore-forming bacterial protein toxins. Folia Microbiol (Praha) 48:5–16. doi: 10.1007/BF02931271. [DOI] [PubMed] [Google Scholar]
- 2.Aznar A, Patrit O, Berger A, Dellagi A. 2015. Alterations of iron distribution in Arabidopsis tissues infected by Dickeya dadantii. Mol Plant Pathol 16:521–528. doi: 10.1111/mpp.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg HC. 1975. Chemotaxis in bacteria. Annu Rev Biophys Bioeng 4:119–136. doi: 10.1146/annurev.bb.04.060175.001003. [DOI] [PubMed] [Google Scholar]
- 4.Costerton JW, Stewart PS, Greenberg EP. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Gossani C, Bellieny-Rabelo D, Venancio TM. 2014. Evolutionary analysis of multidrug resistance genes in fungi–impact of gene duplication and family conservation. FEBS J 281:4967–4977. doi: 10.1111/febs.13046. [DOI] [PubMed] [Google Scholar]
- 6.Kubheka GC, Coutinho TA, Moleleki N, Moleleki LN. 2013. Colonization patterns of an mCherry-tagged Pectobacterium carotovorum subsp. brasiliense strain in potato plants. Phytopathology 103:1268–1279. doi: 10.1094/PHYTO-02-13-0049-R. [DOI] [PubMed] [Google Scholar]
- 7.Touchon M, Rocha EP. 2016. Coevolution of the organization and structure of prokaryotic genomes. Cold Spring Harb Perspect Biol 8:a018168. doi: 10.1101/cshperspect.a018168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Esnault E, Valens M, Espeli O, Boccard F. 2007. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet 3:e226. doi: 10.1371/journal.pgen.0030226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. 1999. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A 96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sabatti C, Rohlin L, Oh MK, Liao JC. 2002. Co-expression pattern from DNA microarray experiments as a tool for operon prediction. Nucleic Acids Res 30:2886–2893. doi: 10.1093/nar/gkf388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamames J, Casari G, Ouzounis C, Valencia A. 1997. Conserved clusters of functionally related genes in two bacterial genomes. J Mol Evol 44:66–73. doi: 10.1007/PL00006122. [DOI] [PubMed] [Google Scholar]
- 12.Yin Y, Zhang H, Olman V, Xu Y. 2010. Genomic arrangement of bacterial operons is constrained by biological pathways encoded in the genome. Proc Natl Acad Sci U S A 107:6310–6315. doi: 10.1073/pnas.0911237107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anantharaman V, Aravind L. 2003. Application of comparative genomics in the identification and analysis of novel families of membrane-associated receptors in bacteria. BMC Genomics 4:34. doi: 10.1186/1471-2164-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang D, Iyer LM, Aravind L. 2011. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res 39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adeolu M, Alnajar S, Naushad S, Gupta RS. 2016. Genome-based phylogeny and taxonomy of the “Enterobacteriales”: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int J Syst Evol Microbiol 66:5575–5599. [DOI] [PubMed] [Google Scholar]
- 16.Pérombelon MCM. 2002. Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol 51:1–12. [Google Scholar]
- 17.Marquez-Villavicencio MP, Groves RL, Charkowski AO. 2011. Soft rot disease severity is affected by potato physiology and Pectobacterium taxa. Plant Dis 95:232–241. doi: 10.1094/PDIS-07-10-0526. [DOI] [PubMed] [Google Scholar]
- 18.Alic S, Naglic T, Llop P, Toplak N, Koren S, Ravnikar M, Dreo T. 2015. Draft genome sequences of Dickeya sp. isolates B16 (NIB Z 2098) and S1 (NIB Z 2099) causing soft rot of phalaenopsis orchids. Genome Announc 3:e00973-15. doi: 10.1128/genomeA.00973-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li XS, Yuan KX, Cullis J, Levesque CA, Chen W, Lewis CT, De Boer SH. 2015. Draft genome sequences for Canadian isolates of Pectobacterium carotovorum subsp. brasiliense with weak virulence on potato. Genome Announc 3:e00240-15. doi: 10.1128/genomeA.00240-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onkendi EM, Ramesh AM, Kwenda S, Naidoo S, Moleleki L. 2016. Draft genome sequence of a virulent Pectobacterium carotovorum subsp. brasiliense isolate causing soft rot of cucumber. Genome Announc 4:e01530-15. doi: 10.1128/genomeA.01530-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panda P, Lu A, Armstrong KF, Pitman AR. 2015. Draft genome sequence for ICMP 5702, the type strain of Pectobacterium carotovorum subsp. carotovorum that causes soft rot disease on potato. Genome Announc 3:e00875-15. doi: 10.1128/genomeA.00875-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raoul Des Essarts Y, Mondy S, Helias V, Faure D. 2015. Genome sequence of the potato plant pathogen Dickeya dianthicola strain RNS04.9. Genome Announc 3:e00581-15. doi: 10.1128/genomeA.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwenda S, Gorshkov V, Ramesh AM, Naidoo S, Rubagotti E, Birch PR, Moleleki LN. 2016. Discovery and profiling of small RNAs responsive to stress conditions in the plant pathogen Pectobacterium atrosepticum. BMC Genomics 17:47. doi: 10.1186/s12864-016-2376-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Coulthurst SJ, Pritchard L, Hedley PE, Ravensdale M, Humphris S, Burr T, Takle G, Brurberg MB, Birch PR, Salmond GP, Toth IK. 2008. Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog 4:e1000093. doi: 10.1371/journal.ppat.1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang F, Wang X, Wang B, Chen L, Zhao Z, Waterfield NR, Yang G, Jin Q. 2016. The Pseudomonas aeruginosa type VI secretion PGAP1-like effector induces host autophagy by activating endoplasmic reticulum stress. Cell Rep 16:1502–1509. doi: 10.1016/j.celrep.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Pedron J, Chapelle E, Alunni B, Van Gijsegem F. 2017. Transcriptome analysis of the Dickeya dadantii PecS regulon during the early stages of interaction with Arabidopsis thaliana. Mol Plant Pathol 19:647–663. doi: 10.1111/mpp.12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellieny-Rabelo D, de Oliveira EAG, da Silva Ribeiro E, Costa EP, Oliveira AEA, Venancio TM. 2016. Transcriptome analysis uncovers key regulatory and metabolic aspects of soybean embryonic axes during germination. Sci Rep 6:36009. doi: 10.1038/srep36009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao L, Tu ZJ, Millett BP, Bradeen JM. 2013. Insights into organ-specific pathogen defense responses in plants: RNA-seq analysis of potato tuber-Phytophthora infestans interactions. BMC Genomics 14:340. doi: 10.1186/1471-2164-14-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanui CK, Shyntum DY, Priem SL, Theron J, Moleleki LN. 2017. Influence of the ferric uptake regulator (Fur) protein on pathogenicity in Pectobacterium carotovorum subsp. brasiliense. PLoS One 12:e0177647. doi: 10.1371/journal.pone.0177647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toth IK, Birch PR. 2005. Rotting softly and stealthily. Curr Opin Plant Biol 8:424–429. doi: 10.1016/j.pbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Allen C, Reverchon S, Robert-Baudouy J. 1989. Nucleotide sequence of the Erwinia chrysanthemi gene encoding 2-keto-3-deoxygluconate permease. Gene 83:233–241. doi: 10.1016/0378-1119(89)90109-1. [DOI] [PubMed] [Google Scholar]
- 32.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 33.Pallen MJ, Wren BW. 2007. Bacterial pathogenomics. Nature 449:835–842. doi: 10.1038/nature06248. [DOI] [PubMed] [Google Scholar]
- 34.Durand E, Cambillau C, Cascales E, Journet L. 2014. VgrG, Tae, Tle, and beyond: the versatile arsenal of type VI secretion effectors. Trends Microbiol 22:498–507. doi: 10.1016/j.tim.2014.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Varani AM, Monteiro-Vitorello CB, Nakaya HI, Van Sluys MA. 2013. The role of prophage in plant-pathogenic bacteria. Annu Rev Phytopathol 51:429–451. doi: 10.1146/annurev-phyto-081211-173010. [DOI] [PubMed] [Google Scholar]
- 36.Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 37.Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103:1528–1533. doi: 10.1073/pnas.0510322103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duarte V, de Boer SH, Ward LJ, de Oliveira AM. 2004. Characterization of atypical Erwinia carotovora strains causing blackleg of potato in Brazil. J Appl Microbiol 96:535–545. doi: 10.1111/j.1365-2672.2004.02173.x. [DOI] [PubMed] [Google Scholar]
- 39.Durrant A. 2016. Antimicrobial production by Pectobacterium carotovorum subspecies brasiliensis and its role in competitive fitness of the potato pathogen. Lincoln University, Lincoln, Nebraska. [Google Scholar]
- 40.Carpita NC, Gibeaut DM. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3:1–30. doi: 10.1111/j.1365-313X.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- 41.Toth IK, Bell KS, Holeva MC, Birch PR. 2003. Soft rot erwiniae: from genes to genomes. Mol Plant Pathol 4:17–30. doi: 10.1046/j.1364-3703.2003.00149.x. [DOI] [PubMed] [Google Scholar]
- 42.Basler M. 2015. Type VI secretion system: secretion by a contractile nanomachine. Philos Trans R Soc Lond B Biol Sci 370:20150021. doi: 10.1098/rstb.2015.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shneider MM, Buth SA, Ho BT, Basler M, Mekalanos JJ, Leiman PG. 2013. PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500:350–353. doi: 10.1038/nature12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. 2013. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110:7032–7037. doi: 10.1073/pnas.1300627110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aoki SK, Diner EJ, de Roodenbeke CT, Burgess BR, Poole SJ, Braaten BA, Jones AM, Webb JS, Hayes CS, Cotter PA, Low DA. 2010. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayes CS, Aoki SK, Low DA. 2010. Bacterial contact-dependent delivery systems. Annu Rev Genet 44:71–90. doi: 10.1146/annurev.genet.42.110807.091449. [DOI] [PubMed] [Google Scholar]
- 47.Ruhe ZC, Low DA, Hayes CS. 2013. Bacterial contact-dependent growth inhibition. Trends Microbiol 21:230–237. doi: 10.1016/j.tim.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shub DA, Goodrich-Blair H, Eddy SR. 1994. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem Sci 19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 49.Alcoforado Diniz J, Liu YC, Coulthurst SJ. 2015. Molecular weaponry: diverse effectors delivered by the type VI secretion system. Cell Microbiol 17:1742–1751. doi: 10.1111/cmi.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347. doi: 10.1038/nature10244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernal P, Llamas MA, Filloux A. 2018. Type VI secretion systems in plant‐associated bacteria. Environ Microbiol 20:1–15. doi: 10.1111/1462-2920.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Addy HS, Askora A, Kawasaki T, Fujie M, Yamada T. 2012. The filamentous phage φRSS1 enhances virulence of phytopathogenic Ralstonia solanacearum on tomato. Phytopathology 102:244–251. doi: 10.1094/PHYTO-10-11-0277. [DOI] [PubMed] [Google Scholar]
- 54.Reeve JN, Shaw JE. 1979. Lambda encodes an outer membrane protein: the lom gene. Mol Gen Genet 172:243–248. doi: 10.1007/BF00271723. [DOI] [PubMed] [Google Scholar]
- 55.Vaca-Pacheco S, Paniagua-Contreras GL, García-González O, de la Garza M. 1999. The clinically isolated FIZ15 bacteriophage causes lysogenic conversion in Pseudomonas aeruginosa PAO1. Curr Microbiol 38:239–243. doi: 10.1007/PL00006794. [DOI] [PubMed] [Google Scholar]
- 56.Genin S, Denny TP. 2012. Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol 50:67–89. doi: 10.1146/annurev-phyto-081211-173000. [DOI] [PubMed] [Google Scholar]
- 57.Guidot A, Prior P, Schoenfeld J, Carrere S, Genin S, Boucher C. 2007. Genomic structure and phylogeny of the plant pathogen Ralstonia solanacearum inferred from gene distribution analysis. J Bacteriol 189:377–387. doi: 10.1128/JB.00999-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamada K, Hirota M, Niimi Y, Nguyen HA, Takahara Y, Kamio Y, Kaneko J. 2006. Nucleotide sequences and organization of the genes for carotovoricin (Ctv) from Erwinia carotovora indicate that Ctv evolved from the same ancestor as Salmonella typhi prophage. Biosci Biotechnol Biochem 70:2236–2247. doi: 10.1271/bbb.60177. [DOI] [PubMed] [Google Scholar]
- 59.Evans TJ, Coulthurst SJ, Komitopoulou E, Salmond GP. 2010. Two mobile Pectobacterium atrosepticum prophages modulate virulence. FEMS Microbiol Lett 304:195–202. doi: 10.1111/j.1574-6968.2010.01901.x. [DOI] [PubMed] [Google Scholar]
- 60.Mushegian AR, Fullner KJ, Koonin EV, Nester EW. 1996. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci U S A 93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Summer EJ, Gonzalez CF, Carlisle T, Mebane LM, Cass AM, Savva CG, LiPuma J, Young R. 2004. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J Mol Biol 340:49–65. doi: 10.1016/j.jmb.2004.04.053. [DOI] [PubMed] [Google Scholar]
- 62.Holland IB, Schmitt L, Young J. 2005. Type 1 protein secretion in bacteria, the ABC-transporter dependent pathway (review). Mol Membr Biol 22:29–39. doi: 10.1080/09687860500042013. [DOI] [PubMed] [Google Scholar]
- 63.Duquesne S, Destoumieux-Garzón D, Peduzzi J, Rebuffat S. 2007. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat Prod Rep 24:708–734. doi: 10.1039/b516237h. [DOI] [PubMed] [Google Scholar]
- 64.Duquesne S, Petit V, Peduzzi J, Rebuffat S. 2007. Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. J Mol Microbiol Biotechnol 13:200–209. doi: 10.1159/000104748. [DOI] [PubMed] [Google Scholar]
- 65.Kanonenberg K, Schwarz CK, Schmitt L. 2013. Type I secretion systems–a story of appendices. Res Microbiol 164:596–604. doi: 10.1016/j.resmic.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 66.Emanuelsson O, Brunak S, Von Heijne G, Nielsen H. 2007. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 67.Frank K, Sippl MJ. 2008. High-performance signal peptide prediction based on sequence alignment techniques. Bioinformatics 24:2172–2176. doi: 10.1093/bioinformatics/btn422. [DOI] [PubMed] [Google Scholar]
- 68.Gabriel DW, Allen C, Schell M, Denny TP, Greenberg JT, Duan YP, Flores-Cruz Z, Huang Q, Clifford JM, Presting G, González ET, Reddy J, Elphinstone J, Swanson J, Yao J, Mulholland V, Liu L, Farmerie W, Patnaikuni M, Balogh B, Norman D, Alvarez A, Castillo JA, Jones J, Saddler G, Walunas T, Zhukov A, Mikhailova N. 2006. Identification of open reading frames unique to a select agent: Ralstonia solanacearum race 3 biovar 2. Mol Plant Microbe Interact 19:69–79. doi: 10.1094/MPMI-19-0069. [DOI] [PubMed] [Google Scholar]
- 69.Guidot A, Coupat B, Fall S, Prior P, Bertolla F. 2009. Horizontal gene transfer between Ralstonia solanacearum strains detected by comparative genomic hybridization on microarrays. ISME J 3:549. doi: 10.1038/ismej.2009.14. [DOI] [PubMed] [Google Scholar]
- 70.Perna NT, Plunkett G III, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 71.Jacobs JM, Babujee L, Meng F, Milling A, Allen C. 2012. The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. mBio 3:e00114-12. doi: 10.1128/mBio.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmid J, Sieber V, Rehm B. 2015. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front Microbiol 6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barras F, van Gijsegem F, Chatterjee AK. 1994. Extracellular enzymes and pathogenesis of soft-rot Erwinia. Annu Rev Phytopathol 32:201–234. doi: 10.1146/annurev.py.32.090194.001221. [DOI] [Google Scholar]
- 74.Kao CC, Sequeira L. 1991. A gene cluster required for coordinated biosynthesis of lipopolysaccharide and extracellular polysaccharide also affects virulence of Pseudomonas solanacearum. J Bacteriol 173:7841–7847. doi: 10.1128/jb.173.24.7841-7847.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rocchetta HL, Burrows LL, Lam JS. 1999. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 63:523–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toth I, Thorpe C, Bentley S, Mulholland V, Hyman L, Perombelon M, Salmond G. 1999. Mutation in a gene required for lipopolysaccharide and enterobacterial common antigen biosynthesis affects virulence in the plant pathogen Erwinia carotovora subsp. atroseptica. Mol Plant Microbe Interact 12:499–507. doi: 10.1094/MPMI.1999.12.6.499. [DOI] [PubMed] [Google Scholar]
- 77.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. 2005. STRING: known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res 33:D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform 23:205–211. [PubMed] [Google Scholar]
- 79.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. 2010. The Pfam protein families database. Nucleic Acids Res 38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Drummelsmith J, Whitfield C. 1999. Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30). Mol Microbiol 31:1321–1332. doi: 10.1046/j.1365-2958.1999.01277.x. [DOI] [PubMed] [Google Scholar]
- 81.Peleg A, Shifrin Y, Ilan O, Nadler-Yona C, Nov S, Koby S, Baruch K, Altuvia S, Elgrably-Weiss M, Abe CM, Knutton S, Saper MA, Rosenshine I. 2005. Identification of an Escherichia coli operon required for formation of the O-antigen capsule. J Bacteriol 187:5259–5266. doi: 10.1128/JB.187.15.5259-5266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Collins RF, Beis K, Dong C, Botting CH, McDonnell C, Ford RC, Clarke BR, Whitfield C, Naismith JH. 2007. The 3D structure of a periplasm-spanning platform required for assembly of group 1 capsular polysaccharides in Escherichia coli. Proc Natl Acad Sci U S A 104:2390–2395. doi: 10.1073/pnas.0607763104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rendueles O, Garcia-Garcera M, Neron B, Touchon M, Rocha EPC. 2017. Abundance and co-occurrence of extracellular capsules increase environmental breadth: Implications for the emergence of pathogens. PLoS Pathog 13:e1006525. doi: 10.1371/journal.ppat.1006525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. 2007. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J Bacteriol 189:2618–2628. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Regue M, Climent N, Abitiu N, Coderch N, Merino S, Izquierdo L, Altarriba M, Tomas JM. 2001. Genetic characterization of the Klebsiella pneumoniae waa gene cluster, involved in core lipopolysaccharide biosynthesis. J Bacteriol 183:3564–3573. doi: 10.1128/JB.183.12.3564-3573.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic Local Alignment Search Tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 87.Remmert M, Biegert A, Hauser A, Soding J. 2011. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods 9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 88.Geer LY, Domrachev M, Lipman DJ, Bryant SH. 2002. CDART: protein homology by domain architecture. Genome Res 12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hofmann K. 1993. TMbase–a database of membrane spanning proteins segments. Biol Chem Hoppe-Seyler 374:166. [Google Scholar]
- 90.Moller S, Croning MD, Apweiler R. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 91.Sathiyamoorthy K, Mills E, Franzmann TM, Rosenshine I, Saper MA. 2011. The crystal structure of Escherichia coli group 4 capsule protein GfcC reveals a domain organization resembling that of Wza. Biochemistry 50:5465–5476. doi: 10.1021/bi101869h. [DOI] [PubMed] [Google Scholar]
- 92.Rahn A, Whitfield C. 2003. Transcriptional organization and regulation of the Escherichia coli K30 group 1 capsule biosynthesis (cps) gene cluster. Mol Microbiol 47:1045–1060. doi: 10.1046/j.1365-2958.2003.03354.x. [DOI] [PubMed] [Google Scholar]
- 93.Islam ST, Lam JS. 2013. Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environ Microbiol 15:1001–1015. doi: 10.1111/j.1462-2920.2012.02890.x. [DOI] [PubMed] [Google Scholar]
- 94.Valvano MA. 2003. Export of O-specific lipopolysaccharide. Front Biosci 8:s452–s471. doi: 10.2741/1079. [DOI] [PubMed] [Google Scholar]
- 95.Wang L, Liu D, Reeves PR. 1996. C-terminal half of Salmonella enterica WbaP (RfbP) is the galactosyl-1-phosphate transferase domain catalyzing the first step of O-antigen synthesis. J Bacteriol 178:2598–2604. doi: 10.1128/jb.178.9.2598-2604.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Islam ST, Lam JS. 2014. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol 60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- 97.Zhao G, Wu B, Li L, Wang PG. 2014. O-antigen polymerase adopts a distributive mechanism for lipopolysaccharide biosynthesis. Appl Microbiol Biotechnol 98:4075–4081. doi: 10.1007/s00253-014-5552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Daniels C, Vindurampulle C, Morona R. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol Microbiol 28:1211–1222. doi: 10.1046/j.1365-2958.1998.00884.x. [DOI] [PubMed] [Google Scholar]
- 99.Kim TH, Sebastian S, Pinkham JT, Ross RA, Blalock LT, Kasper DL. 2010. Characterization of the O-antigen polymerase (Wzy) of Francisella tularensis. J Biol Chem 285:27839–27849. doi: 10.1074/jbc.M110.143859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liao Y, Smyth GK, Shi W. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 103.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Benjamini Y, Yekutieli D. 2005. False discovery rate–adjusted multiple confidence intervals for selected parameters. J Am Stat Assoc 100:71–81. doi: 10.1198/016214504000001907. [DOI] [Google Scholar]
- 105.Perez-Llamas C, Lopez-Bigas N. 2011. Gitools: analysis and visualisation of genomic data using interactive heat-maps. PLoS One 6:e19541. doi: 10.1371/journal.pone.0019541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Baum LE, Petrie T. 1966. Statistical inference for probabilistic functions of finite state Markov chains. Ann Math Statist 37:1554–1563. doi: 10.1214/aoms/1177699147. [DOI] [Google Scholar]
- 109.Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Lee TH, Jin H, Marler B, Guo H, Kissinger JC, Paterson AH. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Akhter S, Aziz RK, Edwards RA. 2012. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res 40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–W352. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Eddy SR. 1998. Profile hidden Markov models. Bioinformatics 14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 114.Sievers F, Higgins DG. 2018. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci 27:135–145. doi: 10.1002/pro.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Remmert M, Linke D, Lupas AN, Soding J. 2009. HHomp–prediction and classification of outer membrane proteins. Nucleic Acids Res 37:W446–W451. doi: 10.1093/nar/gkp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S, Ami GOH, Web Presence Working Group . 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Moleleki LN, Pretorius RG, Tanui CK, Mosina G, Theron J. 2016. A quorum sensing-defective mutant of Pectobacterium carotovorum ssp. brasiliense 1692 is attenuated in virulence and unable to occlude xylem tissue of susceptible potato plant stems. Mol Plant Pathol 18:32–44. doi: 10.1111/mpp.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.