The digestive tracts of animals are home to a community of microorganisms, the gut microbiota, which affects the growth, development, and health of the host. Interactions among microbes in this inner ecosystem can influence which species colonize the gut and can lead to changes in host physiology. We investigated a mutually beneficial interaction between two bacterial species from the gut microbiota of fruit flies. By coculturing the bacteria in vitro, we were able to identify a metabolic gene required for the bacteria to grow better together than they do separately. Our data suggest that one species consumes the waste products of the other, leading to greater productivity of the microbial community and modifying the nutrients available to the host. This study provides a starting point for investigating how these and other bacteria mutually benefit by sharing metabolites and for determining the impact of mutualism on host health.
KEYWORDS: Acetobacter, Lactobacillus, gluconeogenesis, mutualism, symbiosis
ABSTRACT
Interactions between species shape the formation and function of microbial communities. In the gut microbiota of animals, cross-feeding of metabolites between microbes can enhance colonization and influence host physiology. We examined a mutually beneficial interaction between two bacteria isolated from the gut microbiota of Drosophila, i.e., Acetobacter fabarum and Lactobacillus brevis. After developing an in vitro coculture assay, we utilized a genetic screen to identify A. fabarum genes required for enhanced growth with L. brevis. The screen, and subsequent genetic analyses, showed that the gene encoding pyruvate phosphate dikinase (ppdK) is required for A. fabarum to benefit fully from coculture. By testing strains with mutations in a range of metabolic genes, we provide evidence that A. fabarum can utilize multiple fermentation products of L. brevis. Mutualism between the bacteria in vivo affects gnotobiotic Drosophila melanogaster; flies associated with A. fabarum and L. brevis showed >1,000-fold increases in bacterial cell density and significantly lower triglyceride storage than monocolonized flies. Mutation of ppdK decreased A. fabarum density in flies cocolonized with L. brevis, consistent with the model in which Acetobacter employs gluconeogenesis to assimilate Lactobacillus fermentation products as a source of carbon in vivo. We propose that cross-feeding between these groups is a common feature of microbiota in Drosophila.
IMPORTANCE The digestive tracts of animals are home to a community of microorganisms, the gut microbiota, which affects the growth, development, and health of the host. Interactions among microbes in this inner ecosystem can influence which species colonize the gut and can lead to changes in host physiology. We investigated a mutually beneficial interaction between two bacterial species from the gut microbiota of fruit flies. By coculturing the bacteria in vitro, we were able to identify a metabolic gene required for the bacteria to grow better together than they do separately. Our data suggest that one species consumes the waste products of the other, leading to greater productivity of the microbial community and modifying the nutrients available to the host. This study provides a starting point for investigating how these and other bacteria mutually benefit by sharing metabolites and for determining the impact of mutualism on host health.
INTRODUCTION
An animal’s health can be profoundly influenced by the community of microorganisms in its gut. This community, the gut microbiota, affects infection resistance, nutrient acquisition, and behavior, among other traits (1–6). The outcome of these effects often depends on the taxonomic composition of the microbiota, which in turn can be influenced by host factors such as diet and genetics (7–12). Interactions among gut microbes also play a central role in shaping the composition and function of these communities (13–15). Thus, investigating the molecular basis for microbe-microbe interactions is a major focus of microbiota research.
Examination of human gut microbes in vitro and in gnotobiotic mice has revealed metabolic cross-feeding as a key force shaping the gut microbiota. Degradation of polysaccharides and monosaccharides by primary fermenters like Bacteroides species produces organic acids and other metabolites, which serve as growth substrates for a range of other microbes, including methanogens, acetogens, and sulfate reducers (14, 15). In gnotobiotic animals with microbiota consisting of only two or a few species, metabolic interactions have been shown to affect colonization and resource utilization. For example, the sulfate-reducing bacterium Desulfovibrio piger cannot colonize the intestine effectively without the primary fermenter Bacteroides thetaiotaomicron being present to provide sulfate via sulfatase activity (16). Another study showed that the metabolism of B. thetaiotaomicron shifted to being focused on fructan catabolism during cocolonization with the methanogen Methanobrevibacter smithii, rather than D. piger; both M. smithii and B. thetaiotaomicron reach higher densities when colonizing together, rather than alone (17). Lastly, among Bacteroidetes, there is an array of cross-feeding interactions in which diverse polysaccharide substrates are catabolized via the collective action of many species, each contributing a different enzymatic repertoire (18–20). Extrapolating the insights gained from these studies, which utilize simplified communities (relative to the thousands of species found in native human gut microbiota), presents a challenge, although recent work suggests that it may be feasible (21, 22).
Drosophila melanogaster is an attractive model animal for microbiota studies because its gut microbiota has lower diversity, typically consisting of just a few species that are cultivable and easily manipulated (23–25). The ease of rearing Drosophila axenically (free of microbes) and gnotobiotically (with defined microbial communities) has enabled large-scale studies, including studies of the association of host genetic variation with host responses to microbiota (26) and metagenome-wide studies of the impact of microbiota genetic variation on host traits (27). Microbiota affect Drosophila immunity (28, 29), development (30, 31), nutrition (32–35), and starvation resistance (36), as well as behavioral responses such as preferences for food, mates, and oviposition sites (37–39). Genetic manipulation of both the host and the microbiota presents an exciting opportunity to investigate the mechanistic bases of these impacts (30, 40–42).
Recent studies examining interactions among members of the Drosophila microbiota suggest that these interactions are varied and consequential. Newell and Douglas tested the impact of single-species, dual-species, and multispecies microbiota on bacterial cell density, as well as host nutrient allocation and development (33). That study found that a positive interaction led Lactobacillus brevis and Acetobacter species to reach higher densities in cocolonized versus monocolonized flies, but that pattern was not seen for other Lactobacillus species paired with Acetobacter species. Flies colonized with both genera were spared from the high triglyceride (TAG) levels that developed in axenic or monocolonized flies, suggesting that interspecies interactions are essential for microbiota function (33). Other studies indicated that, when they are cultured together, microbiota members produce unique metabolites that act as signals sensed by Drosophila. Fischer et al. showed that flies exhibit a strong oviposition preference for Acetobacter-Lactobacillus-Saccharomyces cocultures, due to the unique mixture of volatile esters and aldehydes they produce (38). A separate study by Leitão-Gonçalves et al. found that a combination of Acetobacter pomorum and Lactobacillus, but not either bacterium alone, modulated feeding preferences and increased egg laying under conditions of amino acid deprivation; the authors suggested that an unknown metabolite, unique to this multispecies microbiota, regulated host behaviors (43).
The objective of this study was to investigate interactions between Acetobacter and Lactobacillus in the Drosophila microbiota. We developed an in vitro coculture assay to examine positive interactions between Acetobacter species and L. brevis. The assay was applied in a transposon mutant screen to identify genes important for the growth of Acetobacter fabarum DsW_054 in coculture. We found that mutualism between A. fabarum and L. brevis relies on metabolite exchange and affects Drosophila nutrient storage.
RESULTS
Mutualism between Acetobacter and L. brevis in vitro.
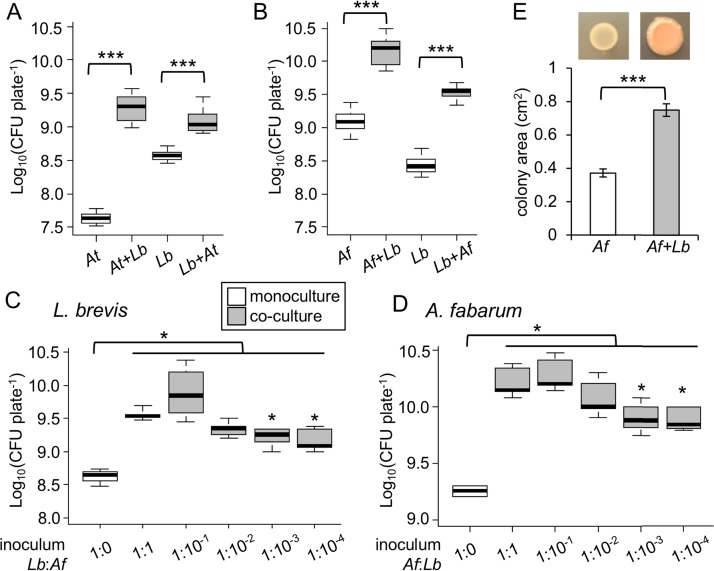
We sought to understand the basis for positive interactions between Acetobacter species and L. brevis found in the gut microbiota of Drosophila. First, we tested whether positive interactions between the bacteria could be observed under in vitro growth conditions. When Acetobacter tropicalis and L. brevis were mixed in equal proportions and cultured on agar plates, each species reached a significantly higher cell density, compared to a monoculture control (Fig. 1A). We obtained comparable results for cocultures of L. brevis and A. fabarum DsW_054, a genetically tractable isolate from wild-caught Drosophila suzukii (41, 44) (Fig. 1B). Next, we varied the ratio of L. brevis to A. fabarum in the inoculum, to observe the impact on the final density of the bacteria. The final density of L. brevis was lower at the 10−4 and 10−3 dilutions of A. fabarum, compared to the 1:1 inoculum (Fig. 1C). However, all of the cocultures resulted in significantly more L. brevis than the monoculture (P < 0.05, Wilcoxon test [n = 3 to 5]), even at the lowest dose of A. fabarum, which consisted of 10 to 100 CFU per plate. Essentially the same pattern was observed when we performed the reverse experiment; even the lowest starting dose of L. brevis significantly increased the A. fabarum density in coculture, compared to monoculture (P < 0.05, Wilcoxon test [n = 4 or 5]) (Fig. 1D). These experiments show that mutualism can occur across a wide range of starting densities for either partner. In addition to reaching a higher cell density in coculture, A. fabarum covered a larger surface area and developed an orange color on coculture plates (Fig. 1E). Collectively, these data demonstrate that L. brevis and some Acetobacter isolates from the gut microbiota of Drosophila can mutually benefit from growing together, consistent with prior studies of interspecies interactions in vivo (33).
FIG 1.
Acetobacter and L. brevis mutualism in vitro. Bacteria were inoculated independently or in a 1:1 mixture on YPD agar plates. (A and B) CFU after 6 days of culture for A. tropicalis in monoculture (At) or coculture (At+Lb), A. fabarum in monoculture (Af) or coculture (Af+Lb), and L. brevis in monoculture (Lb) or coculture (Lb+xx) (n = 9). ***, P < 0.001 in Wilcoxon test comparing the bracketed treatments. In each box plot, the box delineates the first and third quartiles, the dark line is the median, and the whiskers show the range. White bars indicate monocultures and gray bars indicate cocultures. (C and D) CFU of the species indicated after 6 days of coculture initiated with different ratios of L. brevis to A. fabarum (n = 3 to 5). *, P < 0.05 in Wilcoxon test, indicating significantly lower CFU in comparison with a 1:1 ratio inoculum or between the bracketed treatments. (E) Colony areas measured digitally using ImageJ software; bars indicate the mean ± standard deviation (n = 18).
Genetic basis for mutualistic growth of Acetobacter.
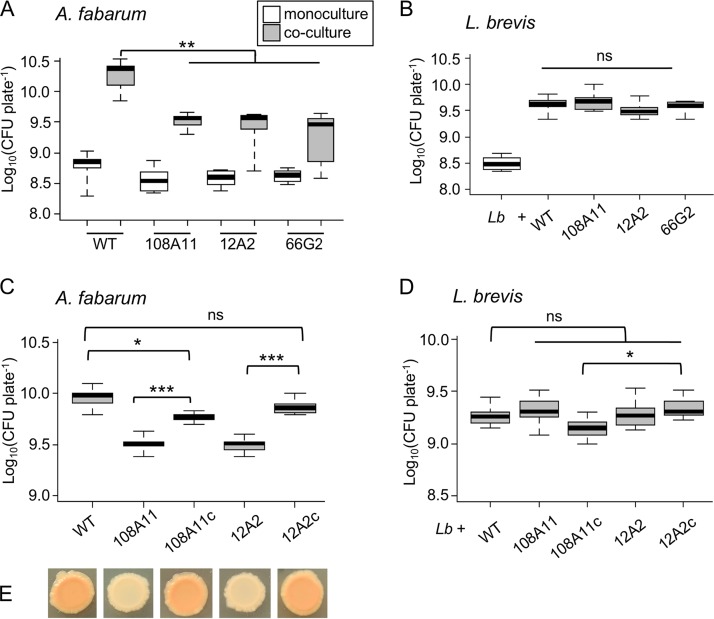
A genetic screen of A. fabarum DsW_054 was performed in order to identify mutations that lead to reductions in mutualistic growth with L. brevis. About 3,000 independent transposon mutants were screened visually for reduced growth on coculture plates and normal growth in monoculture. One mutant with the desired phenotype, 108A11, was isolated. The transposon insertion site was found to be in the gene encoding pyruvate phosphate dikinase (PPDK), the enzyme that catalyzes the conversion of pyruvate to phosphoenolpyruvate. This represents the first step in gluconeogenesis, suggesting that A. fabarum relies on a carbon source other than glucose under coculture conditions. We found a significant reduction in the density of 108A11 in coculture, compared to the wild-type strain (P < 0.001, Wilcoxon test [n = 9]) (Fig. 2A). In monoculture, growth of the mutant was reduced slightly but was not significantly different from that of the wild-type strain (P > 0.05, Wilcoxon test [n = 9]), suggesting that the loss of PPDK does not cause a generalized growth defect on yeast extract-peptone-dextrose (YPD) plates. Interestingly, L. brevis grew to a similar density in coculture with ppdK mutants, compared to coculture with the wild-type strain (Fig. 2B), suggesting that the reduced growth of Acetobacter did not affect L. brevis.
FIG 2.
PPDK gene contribution to A. fabarum growth in coculture. (A and C) CFU of A. fabarum grown in monoculture (white bars) or coculture (gray bars). (B and D) CFU of L. brevis (Lb) grown in monoculture or coculture with the indicated strains of A. fabarum. WT, wild type. The 108A11, 12A2, and 66G2 strains are independent transposon ppdK mutants, and the 108A11c and 12A2c strains have a wild-type copy of ppdK reintroduced for complementation. Pairwise Wilcoxon tests compared the bars indicated with brackets; ns, not significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001, after Bonferroni correction (n = 9 to 12). (E) Representative images of coculture colonies containing L. brevis and the strains of A. fabarum indicated by the labels of the plot in panel C.
To further assess the importance of PPDK for A. fabarum growth with L. brevis, two additional independent transposon mutants were tested (12A2 and 66G2), and similar reductions in coculture growth were seen (Fig. 2A). These additional mutants were isolated and mapped as part of a separate study (41). A wild-type ppdK gene was introduced into mutants 108A11 and 12A2 to determine whether this gene is necessary and sufficient to explain the mutant phenotypes. The complemented strains (108A11c and 12A2c) showed restored mutualism in coculture, reaching higher cell densities than the mutants (P < 0.001, Wilcoxon test [n = 12]) (Fig. 2C). The 12A2c strain reached a density comparable to that of the wild-type strain when grown with L. brevis, while the density of the 108A11c strain was slightly lower (P < 0.05, Wilcoxon test [n = 12]). L. brevis densities did not vary significantly when cocultures with wild-type A. fabarum were compared to cocultures with the mutants or complemented mutants (Fig. 2D). However, a pairwise comparison of L. brevis densities in the cocultures with the 108A11c and 12A2c strains indicated a lower density with 108A11c (P < 0.05, Wilcoxon test [n = 12]). We noted that both complemented mutant strains developed the characteristic color seen for the wild-type strain when grown in coculture (Fig. 2E), while ppdK mutants did not, suggesting that gluconeogenesis is required for the production of pigment(s) or secreted and/or surface molecules (e.g., polysaccharides). Altogether, the data indicate that A. fabarum requires PPDK to fully benefit from growth with L. brevis and therefore is likely to utilize something other than glucose as a source of carbon. Given that glucose is the main substrate provided in the coculture medium, A. fabarum must obtain carbon from a metabolite produced by L. brevis.
Impacts of mutations in metabolic genes.
Acetobacter and Lactobacillus are important components of cocoa fermentations (45, 46), and a study by Adler et al. identified lactate produced by Lactobacillus as the major source of carbon for Acetobacter under these conditions (47). The complete metabolic flux analysis of Acetobacter pasteurianus in the study pinpointed PPDK as the key enzyme for assimilation of carbon from lactate via gluconeogenesis, while ethanol was used predominantly for energy generation via alcohol dehydrogenase (ADH). During growth on these substrates, lactate dehydrogenase (LDH) converts lactate to pyruvate to feed gluconeogenesis, while excess pyruvate is converted to acetoin via α-acetolactate (47). Based on our finding that PPDK is important for the growth of A. fabarum in coculture, a basic model for mutualistic growth of L. brevis and A. fabarum is as follows: the heterofermentative L. brevis converts glucose to lactic acid and ethanol while A. fabarum utilizes these products for carbon and energy, respectively.
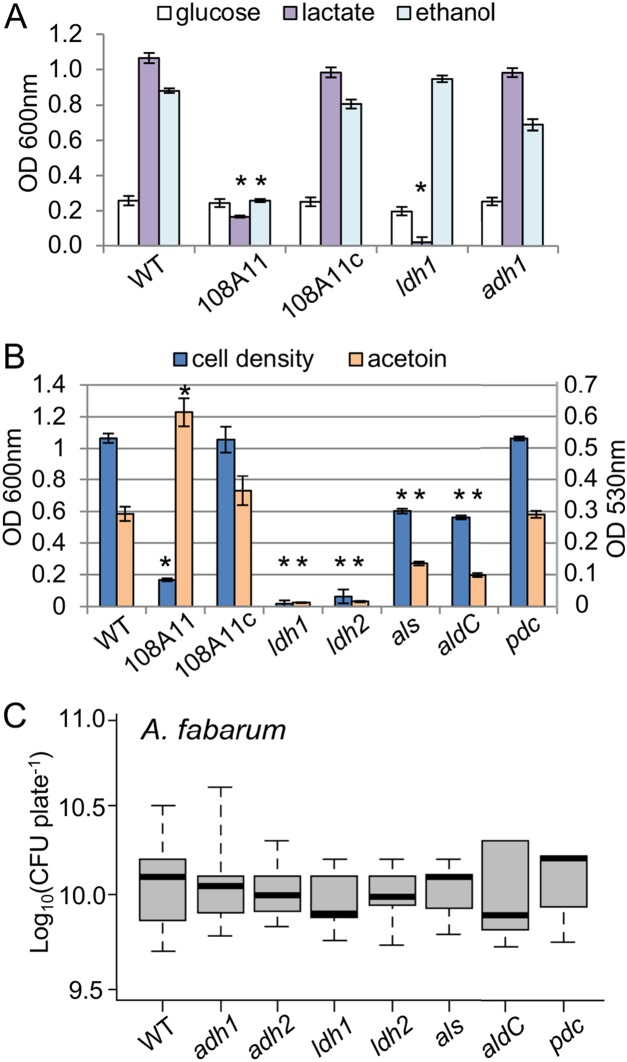
To test this model, we investigated the contributions of particular A. fabarum metabolic genes to growth on different carbon sources, utilizing transposon mutants mapped previously (41). Liquid cultures were incubated statically for 48 h to mimic the structured, diffusion-limited conditions in coculture colonies. A. fabarum density was nearly 5-fold higher in lactate medium and 4-fold higher in ethanol medium than in glucose medium, indicating a preference for those substrates (Fig. 3A). As expected, the ppdK mutant showed reduced growth in lactate and ethanol media but reached a cell density similar to that of the wild-type strain in glucose medium. Reintroduction of ppdK in the complemented strain restored growth in lactate and ethanol media, confirming that gluconeogenesis is required for proliferation in those media (Fig. 3A). Mutation of ldh eliminated growth in lactate but not in glucose or ethanol, while mutation in the pyrroloquinoline quinone-dependent ADH (adh) had no effect on growth in lactate or glucose and caused only a minor reduction in growth in ethanol (Fig. 3A). A kinetic analysis of adh mutant growth found a reduced growth rate in ethanol medium but a final yield similar to that of the wild-type strain (data not shown). Additional ADHs are encoded in the A. fabarum genome (44), so it is not surprising that a mutation in adh did not abolish the ability to utilize ethanol.
FIG 3.
Effects of mutations in metabolic genes on A. fabarum growth and mutualism. (A) Cell density (OD600) of static liquid cultures of A. fabarum in YP medium amended with the indicated carbon source. *, P < 0.05 in Tukey’s HSD test, pairwise with the wild-type strain under the same conditions (n = 6 to 8). WT, wild type; 108A11, ppdK mutant; 108A11c, complemented 108A11; ldh1, ldh mutant 10B7, adh1, adh mutant 21G4. (B) Cell density (blue bars) (OD600) and acetoin production (orange bars) (OD530) of A. fabarum in YP-lactate medium after 48 h (mean ± standard deviation [n = 8]). Strains and statistics are as in panel A plus the following: ldh2, ldh mutant 92G1; als, als mutant; aldC, aldC mutant; pdc, pdc mutant. (C) CFU of A. fabarum in coculture with L. brevis; no significant difference was found in pairwise Wilcoxon tests (n = 9 to 12). Strains are as in panels A and B plus the following: adh2, adh mutant 5F1.
In another set of experiments, we measured cell density and acetoin production of selected mutants in lactate medium. Our objectives were to verify that acetoin is produced as an overflow metabolite of lactate metabolism in A. fabarum and to evaluate the relative contributions of several genes to this pathway. We tested two independent ldh mutants, as well as one each for the genes for acetolactate synthase (ALS) (als), acetolactate decarboxylase (ALDC) (aldC), and pyruvate decarboxylase (pdc), enzymes that may contribute to acetoin production. As anticipated, mutation of ldh abolished growth in lactate, as well as acetoin production (Fig. 3B). The als and aldC mutations reduced but did not eliminate growth and acetoin production in lactate, while mutation of pdc had no effect. Finally, the ppdK mutant showed an increase in acetoin production despite reduced growth in lactate, and wild-type phenotypes were restored by complementation (Fig. 3B). These data fit the model that lactate is assimilated through gluconeogenesis via LDH and PPDK, while excess pyruvate is converted to acetoin by ALS and ALDC. Altogether, the data are consistent with the metabolic flux analysis of A. pasteurianus by Adler et al. (47).
We tested the impact of mutations in metabolic genes on growth in coculture. Our rationale was that, if these mutations interfere with growth on lactate and/or ethanol, they should also affect the mutualism. Surprisingly, all of the mutants reached densities in coculture that were not significantly different from that of the wild-type strain (P > 0.05, Wilcoxon test [n = 9 to 12]) (Fig. 3C). This was unexpected, given the phenotype of the PPDK mutant (Fig. 2A) and the fact that lactate is the major metabolic product of Lactobacillus fermentation. L. brevis reached similar densities in cocultures with all of the A. fabarum strains tested in these experiments (data not shown).
Microbiota mutualism in Drosophila.
Next, we examined mutualism between the bacteria in the context of their host by generating gnotobiotic Drosophila melanogaster. Prior studies have highlighted the importance of interspecies interactions within the microbiota in determining its composition and function. In particular, Acetobacter density can be significantly increased in the presence of L. brevis, and flies colonized by both types of bacteria have significantly lower TAG levels than those colonized by Acetobacter alone (33). Based on these findings, we predicted increased densities of both A. fabarum and L. brevis in hosts colonized by the two species (coculture flies), relative to those colonized by only one species (monoculture). Furthermore, we predicted that mutations that disrupt bacterial mutualism in vitro would reduce the densities in coculture flies and potentially affect host TAG stores.
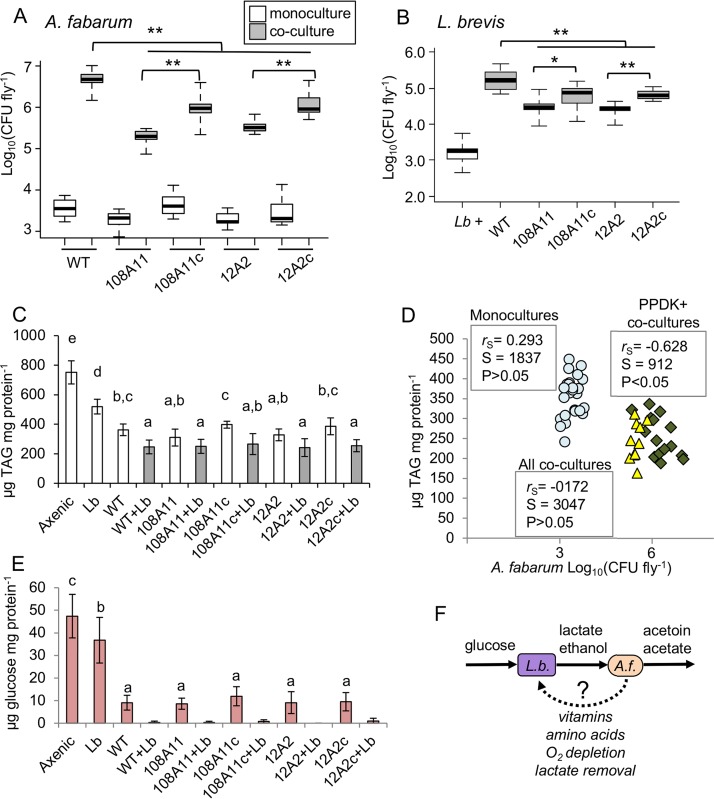
Drosophila were raised gnotobiotically from the embryo stage under monoculture and coculture conditions. Bacterial cell density in whole flies was determined for females on the fifth day after eclosion. We found the density of A. fabarum to be increased for all cocultures, compared to monocultures, by at least 2 orders of magnitude (Fig. 4A). In the monoculture flies, there was not a significant difference in A. fabarum density when the wild-type strain was compared to ppdK mutants or complemented mutants (P > 0.01, Wilcoxon test [n = 18]). However, both ppdK mutants reached significantly lower densities than the wild-type strain in coculture flies with L. brevis (P < 0.01, Wilcoxon test [n = 18]). Reintroduction of ppdK in the complemented mutants significantly increased the A. fabarum density in coculture flies (P < 0.01, Wilcoxon test [n = 18]) but not to a level on par with that of the wild-type strain (Fig. 4A). L. brevis density was also increased by about 2 orders of magnitude in coculture flies, compared to monoculture flies (Fig. 4B). In contrast to our in vitro results, there were significant differences in L. brevis density in vivo depending on the A. fabarum genotype. L. brevis reached significantly lower densities in cocultures with ppdK mutants, compared to the complemented mutants or the wild-type strain (P < 0.05, Wilcoxon test [n = 18]). Mirroring the A. fabarum densities in same samples (Fig. 4A), L. brevis did not reach as high a density in cocultures with the complemented mutants, compared to the wild-type strain (P < 0.01, Wilcoxon test [n = 18]) (Fig. 4B). Altogether, these results are consistent with our prediction that A. fabarum and L. brevis engage in mutualism when colonizing Drosophila, and they suggest that gluconeogenesis via PPDK in A. fabarum contributes to that mutualism.
FIG 4.
Mutualism in vivo and its impact on Drosophila nutrition. (A and B) Cell densities of A. fabarum (A) and L. brevis (Lb) (B) in pools of three female gnotobiotic flies at 5 days post eclosion. White bars indicate single-species associations, and gray bars indicate coculture associations including L. brevis and the indicated strains of A. fabarum. WT, wild type; 108A11 and 12A2, transposon ppdK mutants; 108A11c and 12A2c, complemented ppdK mutants. Pairwise Wilcoxon tests compared the bars indicated with brackets; *, P < 0.05; **, P < 0.01, after Bonferroni correction (n = 15 to 18). (C) TAG contents of fly homogenates from the indicated treatment groups, normalized to the protein concentrations determined for the same samples (mean ± standard deviation [n = 18]). Treatments that do not share a letter above the bars are significantly different by Tukey’s HSD test, P < 0.05. (D) Correlation between A. fabarum abundance and TAG contents in fly homogenates. Values are matched on the basis of vial, and correlation statistics are from Spearman’s rank order test. Circles, monocultures; diamonds, cocultures with A. fabarum with ppdK; triangles, cocultures with A. fabarum ppdK mutants. (E) Glucose contents of Drosophila diet after gnotobiotic rearing with the indicated bacteria, normalized to soluble protein concentrations (mean ± standard deviation [n = 12]). Treatments that do not share a letter above the bars are significantly different by Tukey’s HSD test, P < 0.05. Coculture treatments were not included in the statistics because ≥50% of the samples did not have detectable glucose. (F) Model for metabolic mutualism between L. brevis (L.b.) and A. fabarum (A.f.). Solid arrows indicate connections with experimental support, while the dotted arrow represents hypothesized benefits to L. brevis.
To avoid unwanted effects on L. brevis or Drosophila, neither our in vitro assays nor the gnotobiotic fly experiments utilized antibiotics to select for the maintenance of transposon or complementation constructs in A. fabarum. We assessed the stability of these genetic elements over a 15-day incubation period by plating a subset of the samples from flies monocolonized with A. fabarum strains on selective media. For transposon mutants, comparable numbers of CFU were obtained on YPD kanamycin plates, which selected for the transposon, and nonselective plates (94.4 ± 15.1% [n = 12]). In contrast, samples from flies monocolonized with complemented mutants (108A11c and 12A2c) had fewer CFU on YPD tetracycline plates, compared to nonselective plates (36.7 ± 8.7% [n = 15]), suggesting that a significant fraction of the A. fabarum cells had lost the complementation construct by the time flies were harvested. This provides a possible explanation for why the complemented mutants did not reach as high a density as the wild-type strain in coculture flies (Fig. 4A).
Impact of microbiota mutualism on Drosophila.
To ascertain how mutualism between microbiota members may affect the host, we assayed TAG concentrations in gnotobiotic flies. TAG levels are significantly elevated in axenic versus gnotobiotic flies, and flies monocolonized with Acetobacter have significantly lower TAG levels than those colonized only with Lactobacillus (33, 48, 49) (Fig. 4C). Consistent with prior work on other Acetobacter species, we found that flies colonized with both A. fabarum and L. brevis had significantly less TAG than those colonized with A. fabarum alone; this was true regardless of A. fabarum genotype (Fig. 4C). Interestingly, the presence or absence of PPDK in A. fabarum did not affect TAG levels, as coculture flies with ppdK mutants had TAG levels comparable to those of coculture flies with either wild-type A. fabarum or the complemented mutants. This was unexpected, because mutations in ppdK did significantly decrease the cell density of A. fabarum in coculture flies (Fig. 4A), and Newell and Douglas showed a significant negative correlation between Acetobacter density and host TAG levels (33).
To explore this further, we examined the correlation of A. fabarum density and TAG concentrations in gnotobiotic fly homogenates, grouped on the basis of vial. Consistent with the findings of Newell and Douglas, we found that there was not a significant correlation between A. fabarum density and TAG levels in monoculture flies (P > 0.05, Spearman’s test [n = 25]) (Fig. 4D, circles). However, unlike in the prior study, there was not a significant correlation between A. fabarum density and TAG levels in coculture flies (P > 0.05, Spearman’s test [n = 20]). This discrepancy appears to be driven by the samples from flies colonized by ppdK mutants, which had lower TAG levels than expected based on their bacterial density (Fig. 4D, triangles). When only the coculture flies with wild-type A. fabarum or complemented ppdK mutants were considered, there was a significant negative correlation between Acetobacter density and TAG levels (P < 0.05, Spearman’s test [n = 15]) (Fig. 4D, diamonds).
Microbiota can decrease Drosophila TAG storage through diet modification; bacteria limit the amount of glucose available to their host by consuming the sugar (27, 48). Based on the fact that coculture flies have lower TAG levels than monoculture flies, we hypothesized that mutualism between A. fabarum and L. brevis reduces host TAG levels by depleting glucose in the diet. To test this hypothesis, we measured soluble glucose and protein contents of the diet on the same day flies were harvested for other analyses. Microbiota did not significantly affect the protein concentration in the food (F11,144 = 1.643; P > 0.05, ANOVA); therefore, the glucose values were normalized based on the concentration of soluble protein. Microbiota did significantly affect the glucose content (F6,84 = 87.99; P < 10−15, ANOVA). All diet samples containing bacteria had significantly lower glucose levels than those from axenic vials (P < 0.001, Tukey’s honestly significant difference [HSD] test) (Fig. 4E). All A. fabarum treatments resulted in significantly lower glucose levels than L. brevis (P < 10−7, Tukey’s HSD test). We were unable to reliably compare glucose contents in the diet from coculture treatments because ≥50% of the samples for each of the treatments did not contain detectable amounts of glucose (limit of detection, 0.1 μg glucose/ml). Nevertheless, diet from coculture flies had at most one-fifth the amount of glucose as diet from flies with A. fabarum alone. This finding is consistent with the hypothesis that coculture microbiota significantly decrease the host TAG content via consumption of dietary glucose.
DISCUSSION
We investigated how two gut bacteria, A. fabarum and L. brevis, engage in mutualism. Our ability to quantify the benefits of coculture in vitro provided an opportunity to apply genetic tools in A. fabarum and gain insight into the mechanisms behind this interaction. We conclude that the mutualism has a metabolic basis, i.e., A. fabarum requires gluconeogenesis to benefit fully from growth with L. brevis and therefore must rely on one or more metabolites produced by L. brevis as a carbon source. While the full extent of metabolic cross-feeding between them remains to be elucidated, our results provide some clues and directions for future inquiry. More broadly, this work highlights the importance of interspecies interactions in shaping the composition and function of microbiota. There is increasing confidence that mechanistic dissection of pairwise interactions may enable accurate modeling and modulation of dynamics in more complex communities (21, 22). Such advances will open new avenues for treatment of a range of human aliments that are caused or potentiated by gut microbes (50, 51).
For Drosophila melanogaster, microbiota composition affects TAG storage, among other phenotypes (29). Here we show that mutualism between A. fabarum and L. brevis significantly lowers TAG levels in coculture gnotobiotic flies, relative to those with only Acetobacter. Our data are consistent with a model in which this two-species microbiota consumes more glucose, thereby reducing the concentration available to the host. Reductions in TAG levels caused by the microbiota are known to decrease starvation resistance in Drosophila (36) and may affect survival during infection.
Interestingly, mutation of ppdK in A. fabarum decreased the density of both microbes in flies but did not significantly affect TAG levels. It is possible that bacterial density above a certain threshold is sufficient to deplete dietary glucose and to depress host TAG levels and the cocultures with ppdK mutants were able to reach that threshold. It should be noted that the loss of PPDK reduced but did not eliminate mutualism, as both partners still showed densities at least 1 order of magnitude higher in coculture flies without PPDK than in monoculture flies (Fig. 4A and B). Another possibility is that, despite reaching a lower density, microbiota without PPDK use the same amount of glucose as those with PPDK. The A. fabarum ppdK mutant may compete with L. brevis for glucose while the wild-type A. fabarum does not. Additional experiments (e.g., tracking bacterial densities and the consumption of isotopically labeled metabolites over time) may distinguish between these hypotheses. Metabolomics were successfully applied in a recent study of the honey bee gut microbiota, in which some cross-feeding was shown to occur (52).
The in vitro coculture data suggest that A. fabarum can utilize multiple metabolites produced by L. brevis. Based on the literature, it is predicted that lactate is the primary source of carbon for A. fabarum under these conditions; LDH converts lactate to pyruvate, which is assimilated through gluconeogenesis via PPDK (47). However, ldh mutants reached a density in coculture similar to that of the wild-type strain, despite not being able to grow on lactate alone (Fig. 3). Therefore, something else must serve as a source of carbon for A. fabarum, and ethanol is the most plausible candidate. A metabolic flux analysis of A. pasteurianus grown on lactate and ethanol gave robust evidence that Acetobacter exhibited split metabolism under those conditions, obtaining nearly all carbon from lactate while utilizing ethanol for energy via ADH (47). Our experiments with ldh mutants suggest that A. fabarum can efficiently reroute its metabolism in order utilize ethanol as a sole carbon source. This could explain why there is no apparent defect in mutualism for ldh mutants. Alternatively, other metabolites that have yet to be identified may serve as connections between the bacteria.
The benefits of coculture to L. brevis remain to be elucidated (summarized in Fig. 4F). Removal of oxygen and/or lactate by A. fabarum may enhance L. brevis growth. This would be analogous to the consumption of H2 by secondary fermenters in the human gut microbiota, removing a waste product of saccharolytic primary fermenters and improving their growth (15). If lactate removal were the only benefit, however, we might expect every lactobacillus to exhibit mutualism with Acetobacter. Instead, L. brevis was unique among the three lactobacilli tested by Newell and Douglas in reaching a higher density in gnotobiotic flies with Acetobacter than in monocolonized flies (33). The fact that the L. brevis density in coculture is not affected by mutation of ldh in A. fabarum also argues against lactate removal as the major benefit of the mutualism. The genome of L. brevis DmCS_003 lacks many biosynthetic pathways, including those for most amino acids, B vitamins, purines, and pyrimidines (49). Therefore, it is possible that L. brevis benefits from A. fabarum provisioning these factors.
Notably, L. brevis was not affected by a 10-fold reduction in A. fabarum cell density in vitro when cocultured with the ppdK mutant versus the wild-type strain, but L. brevis did show a lower density in vivo when colonized with the ppdK mutant (Fig. 4B). This indicates an important difference in the mutualism between the two conditions and a role for the host in modulating interactions between the bacteria. The host likely adds a spatiotemporal dimension, by mixing diet and microbes throughout development and by providing a number of distinct environments in the digestive tract (53–55). Digestive physiology and/or interactions with the immune system also could account for the difference (3). Investigating the influences of these factors on A. fabarum and L. brevis mutualism is a focus for future research.
Finally, it is informative to consider how Acetobacter-Lactobacillus mutualism fits into the broader ecological context of interactions between D. melanogaster and its microbiota. Larvae acquire microbiota from the chorion but also by consuming the fermenting substrate in which they were laid (56, 57). Female flies assess a range of cues when choosing oviposition sites, seeking to optimize the success of their offspring, which must develop with ephemeral food sources (58, 59). These cues include metabolites produced through metabolic interactions among microbes. Fischer et al. showed that flies are attracted to volatile compounds produced in Acetobacter-Lactobacillus-Saccharomyces cocultures, including breakdown products of acetoin and acetaldehyde (38). Those authors proposed that a particular blend of volatiles may signal to the host that an oviposition site contains a microbial community that is beneficial for larval development and is protected from parasites or competitors. We also expect cocultures of L. brevis and Acetobacter to produce these metabolites. In either case, the volatiles would signal that a potential substrate for oviposition is lower in sugar and higher in protein than one lacking interspecies metabolic exchange, i.e., a nutrient profile more conducive to rapid larval development (32). Food preferences of both larvae and adults are influenced by microbes in the food source, as well as the identity of the microbes with which the animals were raised (39, 43, 60). These findings suggest a dynamic in which cooperation between microbes could be reinforced because the host is conditioned to seek out microbiota with a familiar metabolic profile. The role of host behavior in microbiota assembly and the relative contributions of cooperation and competition among microbes in the formation of these communities are exciting areas of investigation (13, 14). The Drosophila microbiota will be a valuable experimental system for this research.
Based on our data and the studies discussed here, we propose that a typical microbiota for Drosophila is one with at least two complementary metabolic types, namely, primary saccharolytic fermenters (yeasts and/or lactic acid bacteria) and secondary acetogenic oxidizers that consume the products of the first group (Acetobacter and/or other acetic acid bacteria). A similar dynamic has been observed in a range of traditional fermentations, including those of cocoa (45, 61) and vinegar (62, 63), and is likely widespread in nature. This could also be just one of several common configurations for Drosophila microbiota. More research into microbial interactions in these communities will yield additional insights into how microbiota assemble and function.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Unless otherwise noted, all cultures were grown at 30°C in a YPD broth containing 10 g/liter peptone, 10 g/liter yeast extract, and 8 g/liter dextrose. For solid medium, 18 g/liter of agar was added to the YPD broth. Acetobacter liquid cultures were incubated with shaking at 250 rpm, while L. brevis cultures were incubated statically.
TABLE 1.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Relevant characteristics or sequencea | Reference |
|---|---|---|
| Strains | ||
| Acetobacter fabarum | ||
| DsW_054 | From D. suzukii gut | 44 |
| 108A11 | Tn5::ppdK | This study |
| 108A11c | 108A11 plus pMQ97-Tc-ppdK | This study |
| 12A2 | Tn5::ppdK | 41 |
| 12A2c | 12A2 plus pMQ97-Tc-ppdK | This study |
| 66G2 | Tn5::ppdK | 41 |
| ldh1 (10B7) | Tn5::ldh | 41 |
| ldh2 (92G1) | Tn5::ldh | 41 |
| adh1 (21G4) | Tn5::adh | 41 |
| adh2 (5F1) | Tn5::adh | 41 |
| als (36G1) | Tn5::als | 41 |
| aldC (26G11) | Tn5::aldC | 41 |
| pdc (63F5) | Tn5::pdc | 41 |
| Acetobacter tropicalis DmCS_007 | From D. melanogaster gut | 49 |
| Lactobacillus brevis DmCS_003 | From D. melanogaster gut | 49 |
| Escherichia coli S17-1 (λ-pir) | Conjugation donor | 67 |
| Saccharomyces cerevisiae InvSc1 | URA− | 66 |
| Plasmids | ||
| pRL27 | Tn5 delivery vector; Kmr | 64 |
| pKO3 | Tcr | 65 |
| pMQ97 | RK2 ori, URA3, 2 μm | 66 |
| pMQ97-Tc | Tcr | This study |
| pMQ97-Tc-ppdK | ppdK with native promoter | This study |
| Primers | ||
| Extsx | GACAACAAGCCAGGGATG | 68 |
| Intsx | CGCACTGAGAAGCCCTTAGAGC | 68 |
| Arb1 | GGCCACGCGTCGACTAGTACN10GATAT | 65 |
| Arb2 | GGCCACGCGTCGACTAGTAC | 65 |
| Arb6 | GGCCACGCGTCGACTAGTACN10ACGCC | 65 |
| Tc_Fwd | GTGAATCCGTTAGCGAGGTGC | This study |
| Tc_Rev | CCGATCTCGGCTTGAACGAATTG | This study |
| ppdK_YC_Fwd | CGTAACAAACGGATAGAACCAC | This study |
| ppdK_YC_Rev | CGGAAACGAGGTGGAGAGTAAG | This study |
Kmr, kanamycin resistance; Tcr, tetracycline resistance.
Transposon mutant library of Acetobacter fabarum DLS_54.
Transposon mutants of A. fabarum were generated via conjugal transfer of the plasmid vector pRL27 (64) from Escherichia coli S17, using the method of reference 41. Briefly, donor and recipient strains were grown overnight in lysogeny broth and YPD broth, respectively. Cells were harvested by centrifugation and washed twice in fresh medium before the two species were mixed in a 1:1 ratio. After a 4-hour coincubation, cells were plated on YPD plates with 50 μg/ml kanamycin and 25 μg/ml chloramphenicol to select for Acetobacter transconjugates. A total of 3,008 individual colonies were arrayed in sterile, 96-well plates containing 100 μl/well of YPD medium with 50 μg/ml kanamycin, with wild-type and sterile controls in each plate. After 48 h of incubation at 30°C, cultures were mixed with 50 μl of sterile 75% glycerol for storage at −80°C.
Screen of A. fabarum mutant library.
Cultures of each library plate were grown by inoculating a fresh 96-well plate of YPD agar containing 50 μg/ml kanamycin, using a sterile, 48-pin, multiwell transfer device. After a 24-hour incubation at 30°C, with gentle agitation, cultures were spotted onto YPD agar, with or without L. brevis, using the transfer device. L. brevis was introduced prior to spotting by spread plating of 50 μl of overnight liquid culture. Agar plates were incubated for 1 week prior to visual screening for colonies showing reduced growth on YPD agar with L. brevis, compared to the wild-type strain, but unaffected growth on YPD agar alone.
Sequencing and complementation of ppdK mutant.
Primers and plasmids used in this study are listed in Table 1. All enzymes were from New England Biolabs and were used according to the manufacturer’s recommendations. The transposon insertion site of 108A11 was mapped by Sanger sequencing of arbitrarily primed PCR products, as described (65). Briefly, PCR was performed in two rounds with OneTaq polymerase using purified genomic DNA from 108A11. The first round used primers Arb1, Arb6, and Extsx, and the second round used primers Arb2 and Intsx. Products were purified with the Thermo GeneJet PCR purification kit and were sequenced with primer Intsx.
The Saccharomyces cerevisiae-bacteria shuttle vector pMQ97 was modified by adding the tetracycline resistance gene tetA from pKO3. The plasmid was linearized by digestion with HindIII and blunted with DNA polymerase I Klenow fragment. The tetA gene was amplified with Phusion polymerase and primers Tc_Fwd and Tc_Rev and then was ligated to pMQ97 with T4 DNA ligase. E. coli was transformed with the ligation product, and construction of the resultant plasmid, pMQ97-Tc, was confirmed by restriction digestion and Sanger sequencing. For complementation, the ppdK gene of A. fabarum was PCR amplified with primers ppdK_YC_Fwd and ppdK_YC_Rev. The product and SmaI-linearized pMQ97-Tc were used to transform S. cerevisiae, enabling recombination cloning as described (66). Plasmid construction was confirmed by restriction digestion and Sanger sequencing. The plasmid was introduced into mutant A. fabarum via conjugation with E. coli S17, and selection was performed on YPD plates with 50 μg/ml kanamycin and 20 μg/ml tetracycline.
Coculture assay.
Overnight liquid cultures of Acetobacter and Lactobacillus were normalized via resuspension in phosphate-buffered saline (PBS) to an optical density at 600 nm (OD600) of 0.2. Cells were mixed together in equal volumes. Two 5-μl spots of mixed cultures were pipetted onto YPD agar plates containing 10 g/liter peptone, 10 g/liter yeast extract, 20 g/liter dextrose, and 15 g/liter agar. The plates were incubated for 6 days at 30°C. Following incubation, cells were collected in sterile PBS. Tenfold serial dilutions in PBS were performed for each sample, and six dilutions were spotted onto YPD agar in triplicate 5-μl aliquots. Plates were incubated for 3 days prior to counting colonies. Colony counts of Acetobacter and Lactobacillus were recorded for the dilution containing between 5 and 50 individual colonies. The species were distinguished by differences in colony color and morphology. The median count of three replicate spots was used to determine the CFU.
YP medium growth experiments.
Aliquots of the overnight cultures were transferred to corresponding yeast extract-peptone (YP) medium containing 1% yeast extract and 1% peptone. Added carbon sources included 1% glucose, 1.25% ethanol, or 1% lactic acid. Components were sterilized separately before being added to sterile water to make YP medium. The inoculated YP medium was cultured statically at 30°C. Samples of the bacterial cultures were collected after 48 h. The cell density of each sample was determined by measuring the OD600. The cells were removed from the samples via centrifugation (1 min at 16,000 × g), and the supernatants were tested via the Voges-Proskauer assay (Gibson Bioscience). Acetoin levels were then quantified by measuring the OD530 in a microplate reader. All measurements were blanked with the respective sterile medium.
Coculture with gnotobiotic Drosophila.
Drosophila melanogaster Canton S (without Wolbachia) was used in all experiments. Gnotobiotic Drosophila were generated using the method described by Newell and Douglas (33). Briefly, embryos were collected in deionized water, sterilized by two washes in 0.6% hypochlorite and one in sterile water, and then aseptically transferred to sterile Drosophila diet in a biological safety cabinet. The diet contained 100 g/liter yeast, 100 g/liter glucose, and 12 g/liter agar, and embryos were added at a density of 20 to 40 embryos per vial. Overnight cultures of the bacteria used were pelleted by centrifugation, washed once in PBS, and resuspended in PBS at an OD600 of 0.2. For monocultures, 50 μl of a single strain was added to each vial; cocultures received 50 μl of a 1:1 mixture of the two strains. At 5 days post eclosion, flies were anesthetized with CO2, and three females were pooled and homogenized in 100 μl of sterile PBS, with 1.4-mm ceramic beads, for each measurement. CFU were determined by 10-fold serial dilutions in a 96-well plate and spot plating on YPD agar, as described above.
Determination of Drosophila TAG concentrations and diet contents.
Five days post eclosion, female flies were pooled in groups of three and homogenized as described above but in 100 μl of TET buffer (10 mM Tris [pH 8], 1 mM EDTA, 0.1% Triton X-100). Debris was pelleted by centrifugation (1 min at 16,000 × g). One aliquot of supernatant was frozen at −20°C for subsequent protein determination, while another was heated at 72°C for 20 min and then frozen for subsequent TAG measurement. Protein contents were determined by using the Bio-Rad DC protein assay kit, following the manufacturer’s instructions. TAG contents were determined using the Cayman Chemical TAG colorimetric assay kit, as directed by the manufacturer’s instructions.
After removal of 5-day-old flies, 50-mg aliquots of used diet from the top 5 mm of the vials were transferred from the vials to microcentrifuge tubes. The diet was thoroughly mixed with 500 μl of 100 mM Tris-HCl (pH 7.5) by vortex-mixing. Soluble protein contents were determined using the Bio-Rad DC protein assay kit. Glucose contents were determined using the Invitrogen Amplex Red glucose/glucose oxidase assay kit, according to the instructions.
Statistical analyses.
All statistic analyses were performed in R (version 2.15.3 or later), and all P values were adjusted for multiple comparisons with the p.adjust function, using the Bonferroni method. Pairwise Wilcoxon rank sum tests were performed with the wilcox.test function. For the in vitro growth experiments shown in Fig. 3A and B, all comparisons were made pairwise between the wild-type strain and mutants under the same conditions, because not all mutants were assayed at the same time. For fly TAG and food glucose data, a linear mixed effects model was implemented using the multcomp and lme4 packages, with experiment as a random effect; this accounted for any “block” variations among independent experiments. Pairwise comparisons were made via Tukey’s HSD test (ghlt function in multcomp, with correction of P values for multiple comparisons by the single-step method). Correlations were tested by Spearman’s method, using the cor.test function.
ACKNOWLEDGMENTS
We thank Alec Walter, Ghymizu Espinoza, and Luiza Nawrot for technical assistance, the students of Bio 310 in the autumn of 2016 for help with the genetic screen, John Chaston of Brigham Young University for sharing transposon mutants and for critical reading of the manuscript, Robert Shanks of the University of Pittsburgh for sharing plasmids, and William Metcalf of the University of Illinois at Urbana-Champaign for sharing strains.
This work was supported by a grant for scholarly and creative activity to P.D.N. from the Office of the Provost of the State University of New York at Oswego and by an internship to A.J.S. via NSF STEP grant 1161127 to the Chemistry Department of the State University of New York at Oswego.
REFERENCES
- 1.Kostic AD, Howitt MR, Garrett WS. 2013. Exploring host-microbiota interactions in animal models and humans. Genes Dev 27:701–718. doi: 10.1101/gad.212522.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 3.Buchon N, Broderick NA, Lemaitre B. 2013. Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat Rev Microbiol 11:615–626. doi: 10.1038/nrmicro3074. [DOI] [PubMed] [Google Scholar]
- 4.Cryan JF, Dinan TG. 2012. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci 13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 5.Stecher B, Hardt WD. 2011. Mechanisms controlling pathogen colonization of the gut. Curr Opin Microbiol 14:82–91. doi: 10.1016/j.mib.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Leulier F, MacNeil LT, Lee WJ, Rawls JF, Cani PD, Schwarzer M, Zhao L, Simpson SJ. 2017. Integrative physiology: at the crossroads of nutrition, microbiota, animal physiology, and human health. Cell Metab 25:522–534. doi: 10.1016/j.cmet.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, Spector TD, Clark AG, Ley RE. 2014. Human genetics shape the gut microbiome. Cell 159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaston JM, Dobson AJ, Newell PD, Douglas AE. 2016. Host genetic control of the microbiota mediates the Drosophila nutritional phenotype. Appl Environ Microbiol 82:671–679. doi: 10.1128/AEM.03301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. 2013. The long-term stability of the human gut microbiota. Science 341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108:4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carmody RN, Gerber GK, Luevano JM Jr, Gatti DM, Somes L, Svenson KL, Turnbaugh PJ. 2015. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adair KL, Douglas AE. 2017. Making a microbiome: the many determinants of host-associated microbial community composition. Curr Opin Microbiol 35:23–29. doi: 10.1016/j.mib.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Abreu NA, Taga ME. 2016. Decoding molecular interactions in microbial communities. FEMS Microbiol Rev 40:648–663. doi: 10.1093/femsre/fuw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischbach MA, Sonnenburg JL. 2011. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell Host Microbe 10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. 2013. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci U S A 110:13582–13587. doi: 10.1073/pnas.1312524110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samuel BS, Gordon JI. 2006. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A 103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN, Sonnenburg JL. 2010. Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141:1241–1252. doi: 10.1016/j.cell.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rakoff-Nahoum S, Foster KR, Comstock LE. 2016. The evolution of cooperation within the gut microbiota. Nature 533:255–259. doi: 10.1038/nature17626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venturelli OS, Carr AC, Fisher G, Hsu RH, Lau R, Bowen BP, Hromada S, Northen T, Arkin AP. 2018. Deciphering microbial interactions in synthetic human gut microbiome communities. Mol Syst Biol 14:e8157. doi: 10.15252/msb.20178157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman J, Higgins LM, Gore J. 2017. Community structure follows simple assembly rules in microbial microcosms. Nat Ecol Evol 1:109. doi: 10.1038/s41559-017-0109. [DOI] [PubMed] [Google Scholar]
- 23.Broderick NA, Lemaitre B. 2012. Gut-associated microbes of Drosophila melanogaster. Gut Microbes 3:307–321. doi: 10.4161/gmic.19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandler JA, Lang JM, Bhatnagar S, Eisen JA, Kopp A. 2011. Bacterial communities of diverse Drosophila species: ecological context of a host-microbe model system. PLoS Genet 7:e1002272. doi: 10.1371/journal.pgen.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong CN, Ng P, Douglas AE. 2011. Low-diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ Microbiol 13:1889–1900. doi: 10.1111/j.1462-2920.2011.02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dobson AJ, Chaston JM, Newell PD, Donahue L, Hermann SL, Sannino DR, Westmiller S, Wong AC, Clark AG, Lazzaro BP, Douglas AE. 2015. Host genetic determinants of microbiota-dependent nutrition revealed by genome-wide analysis of Drosophila melanogaster. Nat Commun 6:6312. doi: 10.1038/ncomms7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaston JM, Newell PD, Douglas AE. 2014. Metagenome-wide association of microbial determinants of host phenotype in Drosophila melanogaster. mBio 5:e01631-14. doi: 10.1128/mBio.01631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. 2009. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev 23:2333–2344. doi: 10.1101/gad.1827009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martino ME, Ma D, Leulier F. 2017. Microbial influence on Drosophila biology. Curr Opin Microbiol 38:165–170. doi: 10.1016/j.mib.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 30.Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, Yoon JH, Ryu JH, Lee WJ. 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 31.Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab 14:403–414. doi: 10.1016/j.cmet.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 32.Wong AC, Dobson AJ, Douglas AE. 2014. Gut microbiota dictates the metabolic response of Drosophila to diet. J Exp Biol 217:1894–1901. doi: 10.1242/jeb.101725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newell PD, Douglas AE. 2014. Interspecies interactions determine the impact of the gut microbiota on nutrient allocation in Drosophila melanogaster. Appl Environ Microbiol 80:788–796. doi: 10.1128/AEM.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ridley EV, Wong AC, Westmiller S, Douglas AE. 2012. Impact of the resident microbiota on the nutritional phenotype of Drosophila melanogaster. PLoS One 7:e36765. doi: 10.1371/journal.pone.0036765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sannino DR, Dobson AJ, Edwards K, Angert ER, Buchon N. 2018. The Drosophila melanogaster gut microbiota provisions thiamine to its host. mBio 9:e00155-18. doi: 10.1128/mBio.00155-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Judd AM, Matthews MK, Hughes R, Veloz M, Sexton CE, Chaston JM. 2018. Bacterial methionine metabolism genes influence Drosophila melanogaster starvation resistance. Appl Environ Microbiol 84:e00662-18. doi: 10.1128/AEM.00662-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. 2010. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc Natl Acad Sci U S A 107:20051–20056. doi: 10.1073/pnas.1009906107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer CN, Trautman EP, Crawford JM, Stabb EV, Handelsman J, Broderick NA. 2017. Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. eLife 6:e18855. doi: 10.7554/eLife.18855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wong AC, Wang QP, Morimoto J, Senior AM, Lihoreau M, Neely GG, Simpson SJ, Ponton F. 2017. Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in Drosophila. Curr Biol 27:2397–2404.e4. doi: 10.1016/j.cub.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 40.Matos RC, Schwarzer M, Gervais H, Courtin P, Joncour P, Gillet B, Ma D, Bulteau AL, Martino ME, Hughes S, Chapot-Chartier MP, Leulier F. 2017. D-Alanylation of teichoic acids contributes to Lactobacillus plantarum-mediated Drosophila growth during chronic undernutrition. Nat Microbiol 2:1635–1647. doi: 10.1038/s41564-017-0038-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White KM, Matthews MK, Hughes RC, Sommer AJ, Griffitts JS, Newell PD, Chaston JM. 2018. A metagenome-wide association study and arrayed mutant library confirm Acetobacter lipopolysaccharide genes are necessary for association with Drosophila melanogaster. G3 (Bethesda) 8:1119–1127. doi: 10.1534/g3.117.300530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Storelli G, Strigini M, Grenier T, Bozonnet L, Schwarzer M, Daniel C, Matos R, Leulier F. 2018. Drosophila perpetuates nutritional mutualism by promoting the fitness of its intestinal symbiont Lactobacillus plantarum. Cell Metab 27:362–377.E8. doi: 10.1016/j.cmet.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leitão-Goncalves R, Carvalho-Santos Z, Francisco AP, Fioreze GT, Anjos M, Baltazar C, Elias AP, Itskov PM, Piper MDW, Ribeiro C. 2017. Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol 15:e2000862. doi: 10.1371/journal.pbio.2000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winans NJ, Walter A, Chouaia B, Chaston JM, Douglas AE, Newell PD. 2017. A genomic investigation of ecological differentiation between free-living and Drosophila-associated bacteria. Mol Ecol 26:4536–4550. doi: 10.1111/mec.14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moens F, Lefeber T, De Vuyst L. 2014. Oxidation of metabolites highlights the microbial interactions and role of Acetobacter pasteurianus during cocoa bean fermentation. Appl Environ Microbiol 80:1848–1857. doi: 10.1128/AEM.03344-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adler P, Bolten CJ, Dohnt K, Hansen CE, Wittmann C. 2013. Core fluxome and metafluxome of lactic acid bacteria under simulated cocoa pulp fermentation conditions. Appl Environ Microbiol 79:5670–5681. doi: 10.1128/AEM.01483-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adler P, Frey LJ, Berger A, Bolten CJ, Hansen CE, Wittmann C. 2014. The key to acetate: metabolic fluxes of acetic acid bacteria under cocoa pulp fermentation-simulating conditions. Appl Environ Microbiol 80:4702–4716. doi: 10.1128/AEM.01048-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang JH, Douglas AE. 2015. Consumption of dietary sugar by gut bacteria determines Drosophila lipid content. Biol Lett 11:20150469. doi: 10.1098/rsbl.2015.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newell PD, Chaston JM, Wang Y, Winans NJ, Sannino DR, Wong AC, Dobson AJ, Kagle J, Douglas AE. 2014. In vivo function and comparative genomic analyses of the Drosophila gut microbiota identify candidate symbiosis factors. Front Microbiol 5:576. doi: 10.3389/fmicb.2014.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shreiner AB, Kao JY, Young VB. 2015. The gut microbiome in health and in disease. Curr Opin Gastroenterol 31:69–75. doi: 10.1097/MOG.0000000000000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, Quraishi MN, Kinross J, Smidt H, Tuohy KM, Thomas LV, Zoetendal EG, Hart A. 2016. The gut microbiota and host health: a new clinical frontier. Gut 65:330–339. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kesnerova L, Mars RAT, Ellegaard KM, Troilo M, Sauer U, Engel P. 2017. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol 15:e2003467. doi: 10.1371/journal.pbio.2003467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inamine H, Ellner SP, Newell PD, Luo Y, Buchon N, Douglas AE. 2018. Spatiotemporally heterogeneous population dynamics of gut bacteria inferred from fecal time series data. mBio 9:e01453-17. doi: 10.1128/mBio.01453-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong AC, Luo Y, Jing X, Franzenburg S, Bost A, Douglas AE. 2015. The host as the driver of the microbiota in the gut and external environment of Drosophila melanogaster. Appl Environ Microbiol 81:6232–6240. doi: 10.1128/AEM.01442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buchon N, Osman D, David FP, Yu Fang H, Boquete JP, Deplancke B, Lemaitre B. 2013. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Rep 3:1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 56.Bakula M. 1969. The persistence of a microbial flora during postembryogenesis of Drosophila melanogaster. J Invertebr Pathol 14:365–374. doi: 10.1016/0022-2011(69)90163-3. [DOI] [PubMed] [Google Scholar]
- 57.Erkosar B, Storelli G, Defaye A, Leulier F. 2013. Host-intestinal microbiota mutualism: “learning on the fly.” Cell Host Microbe 13:8–14. doi: 10.1016/j.chom.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Kim G, Huang JH, McMullen JG II, Newell PD, Douglas AE. 2018. Physiological responses of insects to microbial fermentation products: insights from the interactions between Drosophila and acetic acid. J Insect Physiol 106:13–19. doi: 10.1016/j.jinsphys.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reaume CJ, Sokolowski MB. 2006. The nature of Drosophila melanogaster. Curr Biol 16:R623–R628. doi: 10.1016/j.cub.2006.07.042. [DOI] [PubMed] [Google Scholar]
- 60.Farine JP, Habbachi W, Cortot J, Roche S, Ferveur JF. 2017. Maternally-transmitted microbiota affects odor emission and preference in Drosophila larva. Sci Rep 7:6062. doi: 10.1038/s41598-017-04922-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Camu N, De Winter T, Verbrugghe K, Cleenwerck I, Vandamme P, Takrama JS, Vancanneyt M, De Vuyst L. 2007. Dynamics and biodiversity of populations of lactic acid bacteria and acetic acid bacteria involved in spontaneous heap fermentation of cocoa beans in Ghana. Appl Environ Microbiol 73:1809–1824. doi: 10.1128/AEM.02189-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pothakos V, Illegems K, Laureys D, Spitaels F, Vandamme P, De Vuyst L. 2016. Acetic acid bacteria in fermented food and beverage ecosystems, p 73–99. In Matsushita K, Toyama H, Tonouchi N, Okamoto-Kainuma A (ed), Acetic acid bacteria. Springer, Tokyo, Japan. [Google Scholar]
- 63.Wolfe BE, Dutton RJ. 2015. Fermented foods as experimentally tractable microbial ecosystems. Cell 161:49–55. doi: 10.1016/j.cell.2015.02.034. [DOI] [PubMed] [Google Scholar]
- 64.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 65.Monds RD, Newell PD, Schwartzman JA, O'Toole GA. 2006. Conservation of the Pho regulon in Pseudomonas fluorescens Pf0-1. Appl Environ Microbiol 72:1910–1924. doi: 10.1128/AEM.72.3.1910-1924.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simon R, Priefer U, Puhler A. 1983. A broad host range system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 68.Lauro FM, Tran K, Vezzi A, Vitulo N, Valle G, Bartlett DH. 2008. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J Bacteriol 190:1699–1709. doi: 10.1128/JB.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]