Abstract
Abnormal expression of long noncoding RNAs (lncRNAs) is closely associated with the pathogenesis of multiple malignancies, and lncRNA small nucleolar RNA host genes (SNHGs) play critical roles in tumor progression. However, the mechanism by which SNHG3 contributes to osteosarcoma (OS) remains elusive. The association between SNHG3 expression and the clinicopathological characteristics in OS patients was analyzed using the TCGA (The Cancer Genome Atlas) dataset. Cell viability and colony number were estimated by MTT and colony formation assays. MiR-196a-5p-specific binding with SNHG3 or HOXC8 was confirmed by the luciferase report assay. As a result, the expression of SNHG3 was dramatically increased in OS tissue as compared with the adjacent normal tissues. High expression of SNHG3 was associated with tumor size and acted as an independent prognostic factor of poor survival in OS patients. Knockdown of SNHG3 inhibited cell viability and colony formation, but its overexpression reversed these effects. SNHG3 was further identified to act as a sponge of miR-196a-5p, which counteracted the tumor-promoting effects caused by SNHG3 in OS cells. The expression of miR-196a-5p had a negative correlation with SNHG3 and the poor survival in OS patients. In conclusion, lncRNA SNHG3 promoted cell growth by sponging miR-196a-5p and indicated a poor prognosis in OS patients.
Keywords: growth, HOXC8, miR-196a-5p, osteosarcoma, SNHG3
Introduction
Osteosarcoma (OS) is one of the most common cancerous bone tumors, and its morbidity is gradually increasing in adolescents and children within 20 years old.1 Despite the great advances in OS treatment, the prognosis of OS patients is still poor owing to its distant metastasis. Noncoding RNAs (ncRNAs) are partially responsible for the tumor progression in OS.2 Therefore, identification of the target ncRNAs may provide the promising markers for OS patients.
Dysregulation of long noncoding RNAs (lncRNAs) is associated with the tumorigenesis of OS.3 Small nucleolar RNAs (snoRNAs) as lncRNAs play a key role in OS. SNHG1 indicates a poor survival and promotes the growth of OS cells by sponging miR-326.4 SNHG6 functions as an oncogenic factor in OS cells by regulating p21.5 SNHG12 contributes to a poor prognosis and facilitates the proliferation and invasion by sponging miR-195,6 but knockdown of SNHG20 inhibits the tumorigenesis of OS by regulating miR-139/RUNX2 axis.7
However, the functional role of SNHG3 in OS is unreported. In this study, we found that upregulation of SNHG3 expression was associated with tumor size and acted as an independent prognostic factor of poor survival in OS patients. Ectopic SNHG3 expression promoted cell growth by sponging miR-196a-5p, and its knockdown reversed this effect. MiR-196a-5p had a negative correlation with SNHG3 expression and the poor survival in OS patients. Our findings indicated that SNHG3 might be a potential marker for poor survival of OS patients.
Materials and methods
Clinical data
Clinical data from 127 cases of OS patients as well as the relative expression levels of SNHG3 and miR-196a-5p were downloaded from The Cancer Genome Atlas (TCGA) database (https://xenabrowser.net/heatmap/). OS patients did not receive any chemotherapy, and the clinicopathological characteristics of these patients are summarized in Supplementary Table S1. The protocols were approved by the Ethics Committee of Shibei Hospital of Jingan District.
Bioinformatic analysis
The miRNAs that may bind with SNHG3 were identified using starBasev2.0 (http://starbase.sysu.edu.cn/starbase2/index.php) according to the high binding potency, and the target genes of miR-196a-5p were identified using the TargetScanHuman7.1 (http://www.targetscan.org/vert_71/) according to cumulative weighted context scores.
Cell lines and cell culture
OS cell lines (MG-63, 143B, HOS, SW-1353, Saos-2, U-2OS) were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) in a humidified atmosphere containing 5% CO2 at 37°C.
qRT-PCR and Western blot analysis
To detect the expression levels of SNHG3 and miR-196a-5p in OS cells, we conducted quantitative real-time polymerase chain reaction (qRT-PCR) analysis. GAPDH gene or U6 was used as an endogenous control. The primers used are listed in Supplementary Table S2. U-2OS and MG-63 cell lines were harvested and extracted using lysis buffer. The primary antibodies against PCNA (ab29, Mouse monoclonal antibody, Abcam, Cambridge, MA, USA) and β-actin (ab16039, Rabbit polyclonal antibody, Abcam, Cambridge, MA, USA) were diluted at a ratio of 1:1000 according to the instructions and incubated overnight at 4°C. The specific details for qRT-PCR and Western blot analysis were described as previously reported.4
Luciferase reporter assay
U-2OS and MG-63 cell lines were seeded into 24-well plates. After 24 h incubation, 6 ng of pmirGLO report vector carrying wild type (WT) or mutated (Mut) SNHG3 and WT or Mut HOXC8 5′UTR was co-transfected with miR-196-5p mimic (100 nM) or inhibitor into the U-2OS and MG-63 cell lines. Luciferase activities were examined with a dual-luciferase Reporter System (Promega, Madison, Wisconsin, USA).
Plasmid and miR-196-5p mimic and inhibitor
Plasmid-mediated SNHG3 or si-SNHG3 vector and miR-196a-5p mimic, inhibitor or miR-NC were purchased from Genepharma (Shanghai, China). U-2OS and MG-63 cells were planted in 6-well plates 24 h prior to SNHG3 or si-SNHG3, and miR-196a-5p inhibitor or mimic transfection with 50%–70% confluence, and then were transfected with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Cell viability and colony formation assays
Cell viability was assessed using the MTT assay and cell colony number was assessed by colony formation assay. The specific details were described as previously reported.4
Statistical analysis
Student’s t test, chi-square test and analysis of variance (ANOVA) were used to analyze the statistical significance for the comparisons of two groups. Pearson’s correlation analysis was used to analyze the correlations of SNHG3 with miR-196a-5p expression in OS tissues. Survival and recurrence curves were plotted using the Kaplan–Meier method and assessed for the statistical significance using a log-rank test. Statistical significance was set at P < 0.05.
Results
Upregulation of SNHG3 expression was associated with poor survival in OS patients
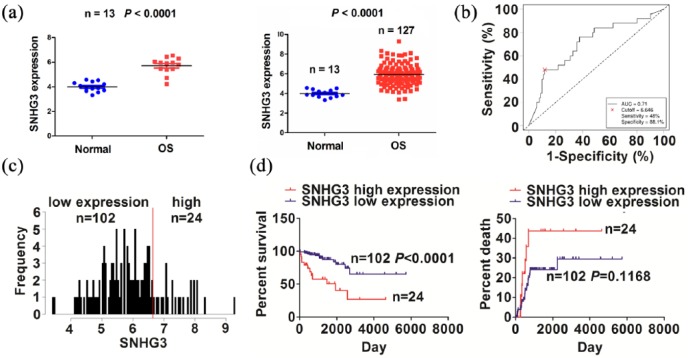
The expression of SNHG3 was examined in OS tissues using the TCGA dataset, indicating that SNHG3 expression level was dramatically increased in paired (n = 13) and unpaired OS tissues (n = 127, Figure 1(a)), as compared with the adjacent normal tissues (n = 13). We then analyzed the association between SNHG3 expression and the clinicopathological characteristics in OS patients. As shown in Figure 1(b), the cutoff value of SNHG3 was obtained in OS patients, and according to its cutoff value, the patients were divided into high or low SNHG3 expression group (Figure 1(c)). Receiver operating characteristic (ROC) curve showed that the sensitivity, specificity, and area under the curve (AUC) value of SNHG3 were 48.0%, 88.1%, and 0.71, respectively.
Figure 1.
The association of SNHG3 expression with the prognosis of OS patients. (a) TCGA analysis of the expression levels of SNHG3 in paired and unpaired OS tissues. (b) ROC curve analysis of the cutoff value of SNHG3 in OS patients. (c) Grouping the OS patients according to the cutoff value of SNGH3. (d) Kaplan–Meier analysis of the association of SNGH3 expression with the prognosis in OS patients.
As shown in Supplementary Table S3, high expression of SNHG3 was positively associated with tumor size (P = 0.004), but had no association with other factors (P > 0.05). Kaplan–Meier analysis revealed that the patients with high SNHG3 expression displayed a poorer survival, but had no difference in tumor recurrence, as compared with the patients with low SNHG3 expression (Figure 1(d)). Univariate and multivariate analyses indicated that high SNHG3 expression was an independent prognostic factor of poor survival in OS patients (Supplementary Table S4).
SNHG3 promoted OS cell proliferation and colony formation
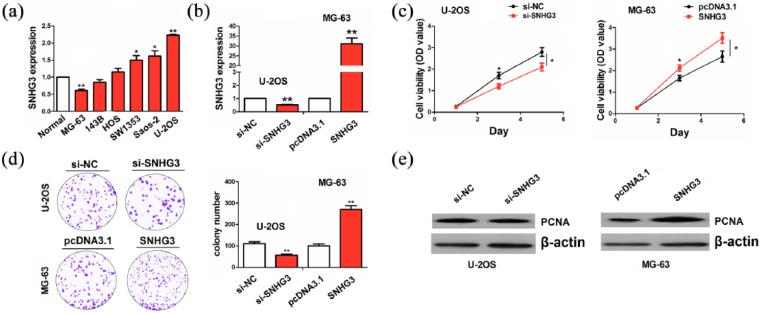
Having verified the positive association of SNHG3 with tumor size in OS tissues, we further evaluated the functional role of SNHG3 in OS cells. The expression level of SNHG3 was examined in different OS cell lines by qRT-PCR analysis, indicating that SNHG3 exhibited a higher expression in U-2OS cells but a lower expression in MG-63 cells, as compared with normal tissues (Figure 2(a)). Then, si-SNHG3 and SNHG3 plasmids were, respectively, transfected intro U-2OS and MG-63 cell lines. After transfection for 48 h, the transfection efficiency of si-SNHG3 or SNHG3 was determined by qRT-PCR analysis (Figure 2(b)). MTT and colony formation assays showed that cell viability and colony number were markedly reduced by the transfection with si-SNHG3 in U-2OS cells, but increased by SNHG3 in MG-63 cells, as compared with the control group (Figure 2(c) and (d)). Western blot analysis indicated that the protein expression of PCNA was downregulated by knockdown of SNHG3 in U-2OS cells, but upregulated by SNGH3 overexpression in MG-63 cells (Figure 2(e)).
Figure 2.
SNGH3 promoted OS cell growth. (a) qRT-PCR analysis of the expression levels of SNGH3 in different OS cell lines. (b) qRT-PCR analysis of the transfection efficiency of SNGH3 or si-SNHG3 in U-2OS or MG-63 cell line. (c) MTT analysis of the effects of SNGH3 or si-SNHG3 on OS cell viability. (d) Colony formation analysis of the effects of SNGH3 or si-SNHG3 on OS cell colony number. (e) Western blot analysis of the effects of SNGH3 or si-SNHG3 on PCNA expression. Data were the means ± SEM of three experiments. *P < 0.05; **P < 0.01.
SNHG3 acted as a sponge of miR-196a-5p in OS cells
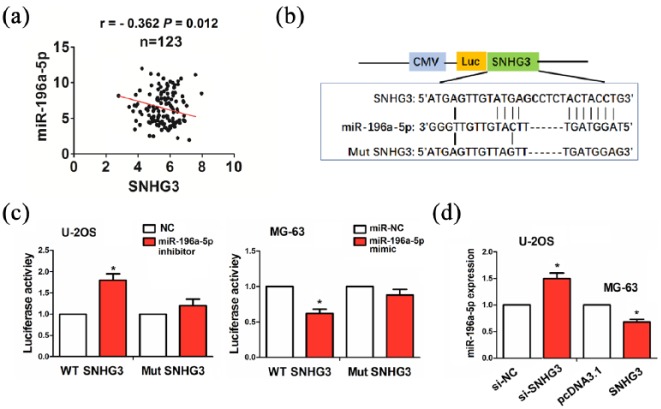
To reveal the molecular mechanism by which SNHG3 contributes to OS, we used the starBasev2.0 to identify three miRNAs that might bind to SNHG3, of which miR-196a-5p had the strongest binding ability with SNHG3 (Supplementary Table S5). Pearson correlation analysis revealed that SNHG3 had a negative correlation with miR-196a-5p expression in OS tissues (Figure 3(a)). The binding sites of miR-196a-5p with WT or Mut SNHG3 were indicated in Figure 3(b). The transfection efficiency of miR-196a-5p inhibitor or mimic in U2-OS or MG-63 cell line was determined by qRT-PCR analysis (Supplementary Figure S1). To confirm whether SNHG3 can bind to miR-196a-5p, we co-transfected U-2OS and MG-63 cells with WT or Mut SNHG3 and miR-196a-5p inhibitor or mimic, indicating that miR-196a-5p inhibitor increased the luciferase activity of WT SNHG3 in U-2OS cells, and its mimic had an opposite effect in MG-63 cells, but miR-196a-5p inhibitor or mimic had no effects on the luciferase activity of Mut SNHG3 as compared with the control group (Figure 3(c)). We further observed the effects of SNHG3 on miR-196a-5p expression and found that knockdown of SNHG3 increased the expression of miR-196a-5p in U-2OS cells, while overexpression of SNHG3 decreased its expression level in MG-63 cells, as compared with the control group (Figure 3(d)).
Figure 3.
SNHG3 acted as a sponge of miR-196a-5p in OS cells. (a) Pearson correlation analysis of the correlation of SNHG3 with miR-196a-5p expression in OS tissues. (b) The binding sites of miR-196a-5p with WT or Mut SNHG3. (c) The luciferase activities of WT or Mut SNHG3 after co-transfection with miR-196a-5p inhibitor or mimic and WT or Mut SNHG3 in OS cells. (d) qRT-PCR analysis of the effects of SNGH3 or si-SNHG3 on miR-196a-5p expression in OS cells. Data were the means ± SEM of three experiments. *P < 0.05.
MiR-196a-5p reverses SNHG3-induced cell proliferation in OS cells
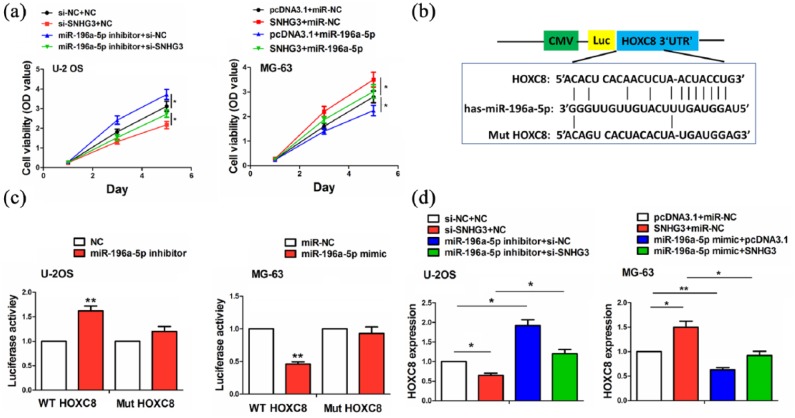
After co-transfection with si-SNHG3 and miR-196a-5p inhibitor in U-2OS cells or SNHG3 and miR-196a-5p mimic in MG-63 cells, we assessed the cell viability using MTT assay and found that miR-196a-5p inhibitor promoted the cell viability and counteracted the anti-proliferation effect caused by SNHG3 knockdown in U-2OS cells, while miR-196a-5p mimic inhibited the cell viability and reversed the proliferation-promoting effect caused by SNHG3 overexpression in MG-63 cells (Figure 4(a)). The binding sites of miR-196a-5p with WT or Mut HOXC8 3′UTR were indicated in Figure 4(b). To confirm whether miR-196a-5p can bind to HOXC8 3′UTR, we co-transfected U-2OS and MG-63 cells with WT or Mut HOXC8 3′UTR and miR-196a-5p inhibitor or mimic, indicating that miR-196a-5p inhibitor increased the luciferase activity of WT HOXC8 3′UTR in U-2OS cells, and miR-196a-5p mimic had an opposite effect in MG-63 cells, but miR-196a-5p inhibitor or mimic had no effects on the luciferase activity of Mut HOXC8 3′UTR as compared with the control group (Figure 4(c)). We further observed the effects of the co-transfection with miR-196a-5p inhibitor and si-SNHG3 or miR-196a-5p mimic and SNHG3 on HOXC8 expression and found that miR-196a-5p inhibitor increased HOXC8 expression in U-2OS cells and knockdown of SNHG3 reversed this effect, while miR-196a-5p mimic decreased HOXC8 expression and overexpression of SNHG3 reversed this effect in MG-63 cells, as compared with the control group (Figure 4(d)).
Figure 4.
miR-196a-5p reversed the tumor-promoting effects of SNHG3 in OS cells. (a) MTT analysis of the cell viability after co-transfection with miR-196a-5p inhibitor and si-SNHG3 or miR-196a-5p mimic and SNHG3 in OS cells. (b) The binding sites of miR-196a-5p with WT or Mut 3′UTR of HOXC8. (c) The luciferase activities of WT or Mut 3′UTR of HOXC8 after co-transfection with miR-196a-5p inhibitor or mimic and WT or Mut 3′UTR of HOXC8 in OS cells. (d) qRT-PCR analysis of the effects of co-transfection with miR-196a-5p inhibitor and si-SNHG3 or miR-196a-5p mimic and SNHG3 in OS cells. Data were the means ± SEM of three experiments. *P < 0.05; **P < 0.01.
High expression of miR-196a-5p was associated with favorable survival in OS patients
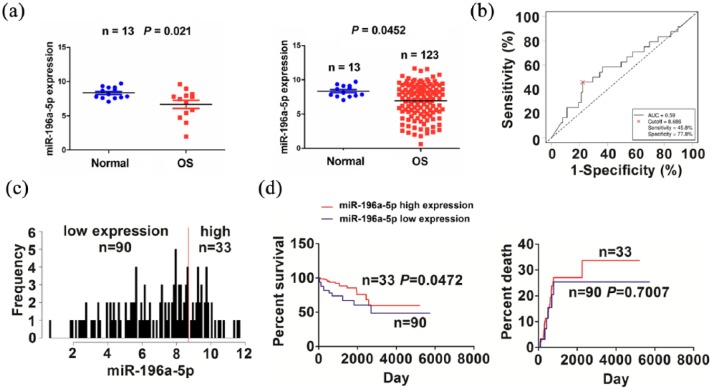
TCGA cohort indicated that miR-196a-5p expression level was downregulated in paired (n = 13) and unpaired OS tissues (n = 123, Figure 5(a)) as compared with the adjacent normal tissues. We then analyzed the association of miR-196a-5p expression with the clinicopathological characteristics in OS patients. As shown in Figure 5(b), the cutoff value of miR-196a-5p was obtained in OS, and according to its cutoff value, the patients were divided into high or low miR-196a-5p expression group (Figure 5(c)). As shown in supplementary Table S6, high expression of miR-196a-5p had no association with the clinical factors (P > 0.05). Kaplan–Meier analysis demonstrated that the patients with high miR-196a-5p expression exhibited a favorable survival, but had no difference in tumor recurrence, as compared with the patients with low miR-196a-5p expression (Figure 5(d)). Univariate and multivariate analyses revealed that miR-196a-5p expression was not an independent prognostic factor of overall survival in OS patients (supplementary Table S7).
Figure 5.
The association of miR-196a-5p expression with the prognosis of OS patients. (a) TCGA analysis of the expression levels of miR-196a-5p in paired and unpaired OS tissues. (b) ROC curve analysis of the cutoff value of miR-196a-5p in OS patients. (c) Grouping the OS patients according to the cutoff value of miR-196a-5p. (d) Kaplan–Meier analysis of the association of miR-196a-5p expression with the prognosis in OS patients.
Discussion
SNHG6/20 acts as oncogenic factors by sponging miRNAs.5,7 However, the functional role of SNHG3 in OS is still undocumented. We found that the increased expression of SNHG3 was associated with tumor size and poor survival, acting as an independent prognostic factor in OS patients. SNHG3 is also associated with the lymph node metastasis and poor prognosis in multiple cancer patients,8 suggesting that SNHG3 might be a prognostic marker in cancer.
SNHG3 promotes the proliferation and invasion and regulates energy metabolism in ovarian cancer.8,9 Our results showed that SNHG3 promoted the proliferation and colony formation of OS cells, but knockdown of SNHG3 reversed these effects. PCNA as a proliferation marker is involved in regulating the cell growth. We found that SNHG3 upregulated the protein expression of PCNA, but knockdown of SNHG3 downregulated its expression. These results indicated that SNHG3 might act as an oncogenic factor in cancer.
Small nucleolar RNA host genes (SNHGs) can act as competing endogenous RNAs (ceRNAs) to regulate OS cells by sponging miRNA. SNHG12 sponges miR-195 to promote the growth and invasion of OS cells.6 We also identified miR-196a-5p-specific binding with SNHG3 and validated it as a ceRNA of miR-196a-5p, indicating that SNHG3 might act as a sponge of miR-196a-5p in OS cells.
Upregulation of miR-196a boosts the tumor proliferation and invasion10,11 and enhances the radio-resistance.12 We found that miR-196a-5p expression was decreased in OS tissues and associated with favorable survival. Restoration of miR-196a-5p expression repressed cell proliferation and reversed SNHG3-induced tumor-promoting effects. We further identified HOXC8 as a target of miR-196a-5p, which downregulated the expression of HOXC8 and reversed SNHG3-caused HOXC8 upregulation. These results suggested that SNHG3 contributed to OS growth by sponging miR-196a-5p/HOXC8 axis.
In conclusion, our findings indicated that SNHG3 promoted the growth of OS cells by sponging miR-196a-5p/HOXC8 axis and provided the potential marker for OS patients.
Supplemental Material
Supplemental material, Supplementary_data_(2) for LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma by Jun Chen, Zhouyi Wu and Yong Zhang in International Journal of Immunopathology and Pharmacology
Supplemental Material
Supplemental material, Supplementary_Figure_S1_(1) for LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma by Jun Chen, Zhouyi Wu and Yong Zhang in International Journal of Immunopathology and Pharmacology
Footnotes
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval and consent to participate: This study was approved by the Hospital’s Protection of Human Subjects Committee.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Supplemental material: Supplemental material for this article is available online.
References
- 1. Harrison DJ, Schwartz CL. (2017) Osteogenic sarcoma: Systemic chemotherapy options for localized disease. Current Treatment Options in Oncology 18(4): 24. [DOI] [PubMed] [Google Scholar]
- 2. Wang C, Jing J, Cheng L. (2018) Emerging roles of non-coding RNAs in the pathogenesis, diagnosis and prognosis of osteosarcoma. Investigational New Drugs 36: 1116–1132. [DOI] [PubMed] [Google Scholar]
- 3. Wang Y, Zeng X, Wang N, et al. (2018) Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Molecular Cancer 17(1): 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang J, Cao L, Wu J, et al. (2018) Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. International Journal of Oncology 52(1): 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ruan J, Zheng L, Hu N, et al. (2018) Long noncoding RNA SNHG6 promotes osteosarcoma cell proliferation through regulating p21 and KLF2. Archives of Biochemistry and Biophysics 646: 128–136. [DOI] [PubMed] [Google Scholar]
- 6. Zhou S, Yu L, Xiong M, et al. (2018) LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochemical and Biophysical Research Communications 495(2): 1822–1832. [DOI] [PubMed] [Google Scholar]
- 7. Wang W, Luo P, Guo W, et al. (2018) LncRNA SNHG20 knockdown suppresses the osteosarcoma tumorigenesis through the mitochondrial apoptosis pathway by miR-139/RUNX2 axis. Biochemical and Biophysical Research Communications 503: 1927–1933. [DOI] [PubMed] [Google Scholar]
- 8. Hong L, Chen W, Wu D, et al. (2018) Upregulation of SNHG3 expression associated with poor prognosis and enhances malignant progression of ovarian cancer. Cancer Biomarkers 22(3): 367–374. [DOI] [PubMed] [Google Scholar]
- 9. Li N, Zhan X, Zhan X. (2018) The lncRNA SNHG3 regulates energy metabolism of ovarian cancer by an analysis of mitochondrial proteomes. Gynecologic Oncology 150(2): 343–354. [DOI] [PubMed] [Google Scholar]
- 10. Hou T, Ou J, Zhao X, et al. (2014) MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. British Journal of Cancer 110(5): 1260–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lu YC, Chang JT, Liao CT, et al. (2014) OncomiR-196 promotes an invasive phenotype in oral cancer through the NME4-JNK-TIMP1-MMP signaling pathway. Molecular Cancer 13: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suh YE, Raulf N, Gäken J, et al. (2015) MicroRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. International Journal of Cancer 137(5): 1021–1034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, Supplementary_data_(2) for LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma by Jun Chen, Zhouyi Wu and Yong Zhang in International Journal of Immunopathology and Pharmacology
Supplemental material, Supplementary_Figure_S1_(1) for LncRNA SNHG3 promotes cell growth by sponging miR-196a-5p and indicates the poor survival in osteosarcoma by Jun Chen, Zhouyi Wu and Yong Zhang in International Journal of Immunopathology and Pharmacology