Abstract
Background
Temperature stress is a major environmental factor affecting not only plant growth and development, but also fruit postharvest life and quality. MicroRNAs (miRNAs) are a class of non-coding small RNAs that play important roles in various biological processes. Harvested banana fruit can exhibit distinct symptoms in response to different temperature stresses, but the underlying miRNA-mediated regulatory mechanisms remained unknown.
Results
Here, we profiled temperature-responsive miRNAs in banana, using deep sequencing and computational and molecular analyses. In total 113 known miRNAs and 26 novel banana-specific miRNAs were identified. Of these miRNAs, 42 miRNAs were expressed differentially under cold and heat stresses. Degradome sequencing identified 60 target genes regulated by known miRNAs and half of these targets were regulated by 15 temperature-responsive miRNAs. The correlative expression patterns between several miRNAs and their target genes were further validated via qRT-PCR. Our data showed that miR535 and miR156 families may derive from a common ancestor during evolution and jointly play a role in fine-tuning SPL gene expression in banana. We also identified the miRNA-triggered phased secondary siRNAs in banana and found miR393-TIR1/AFB phasiRNA production displaying cold stress-specific enrichment.
Conclusions
Our results provide a foundation for understanding the miRNA-dependent temperature stress response in banana. The characterized correlations between miRNAs and their response to temperature stress could serve as markers in the breeding programs or tools for improving temperature tolerance of banana.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5395-1) contains supplementary material, which is available to authorized users.
Keywords: Banana, sRNAome, Degradome, miRNA, Temperature stress
Background
Banana is a major tropical crop plant worldwide. By far the most important cultivars belong to the triploid AAA group of Musa acuminata, commonly referred to as Cavendish group bananas. They accounted for the majority of banana exports, and year-round supply makes them critical for global food security. Bananas are usually refrigerated between 13 °C and 15 °C during storage and transport. At lower temperatures < 12 °C, the peel of bananas gradually turns brown and the ripening permanently stalls. On the other hand, bananas fail to develop an even yellow peel and stay green when ripening at temperatures > 25 °C. There have been reports on the gene regulation of temperature stress response in banana, and some cold resistance-related genes, including a few transcription factors have been characterized [1, 2]. Comparatively, genes associated with fruit heat resistance have rarely been reported. Accumulation of soluble sugars in the peel induced by a high temperature has been proposed as a major factor suppressing chlorophyll degradation, causing the stay-green ripening phenomenon [3]. Overall, the essential factors for the distinct symptoms of banana fruit under temperature stress still remain elusive, and the exploration of underlying mechanism may have practical value for improving banana fruit resistance to temperature stress.
Small regulatory RNAs are major components of eukaryotic transcriptomes. Plants produce many distinct types of DCL/AGO-associated regulatory small RNAs, among which miRNAs and phased siRNAs (phasiRNAs) are the two major types [4, 5]. For more than two decades, miRNAs have been widely studied as important regulatory molecules involved in almost all aspects of the plant life cycle. In plants, the target site usually shows near perfect complementarity to the miRNA sequence, so most target mRNAs are cleaved by RISC, although exceptions have been described where the translation of the mRNA is suppressed without a cleavage [6]. As a subset of plant secondary siRNAs, phasiRNAs can be produced from both protein-coding and non-coding genes. They derive from double-stranded precursors whose synthesis typically depends on an initial miRNA or siRNA interaction and subsequent RDR activity [7, 8]. When there is a single, initiating cleavage event, the dsRNAs all begin at the same nucleotide, so the siRNAs are successively processed from that terminus in a sequential pattern, termed ‘phasing’. Some of the phasiRNAs are capable of acting in trans or cis [9, 10], so it is intriguing to identify biologically meaningful phasiRNAs from their generating loci (PHAS loci) via computational survey and experimental validation. So far, four TAS gene families (TAS1–4) with three specialized initial miRNAs (miR173, miR390 and miR828) have been identified and well characterized in Arabidopsis [7, 11, 12]. Recent studies have demonstrated the identification of phasiRNAs in other plants [10, 13, 14].
While initial studies have largely demonstrated the role of miRNAs in morphogenesis and developmental processes in plants, increasing number of studies show that plant miRNAs also target genes involved in stress responses [15]. The first study revealed the up-regulation of miR393, miR397b and miR402 in response to cold, whereas miR319c appeared to be induced by cold but not by dehydration, salinity, or ABA [16]. Recent reports have also demonstrated the role of miRNAs in cold stress response in various plants [17–19]. Also, there are reports on heat-responsive miRNAs in plants [20–22]. Despite these efforts, miRNA investigation in banana has a relatively short history. Although a number of miRNAs have been reported in banana [23–26], so far no banana miRNA sequences have been deposited in the miRNA database - miRBase. Moreover, the role of miRNAs in the temperature stress response of banana fruit remains largely unknown. In this study, we profiled the miRNAs of banana fruit and characterized their expression pattern under cold and heat stress. We comprehensively identified the temperature-responsive miRNAs and their targets, and characterized the miRNA-triggered phasiRNA production in banana. These data provide novel insights into the miRNA-mediated temperature response of banana fruit.
Results
Changes in sRNA population of banana under temperature stress
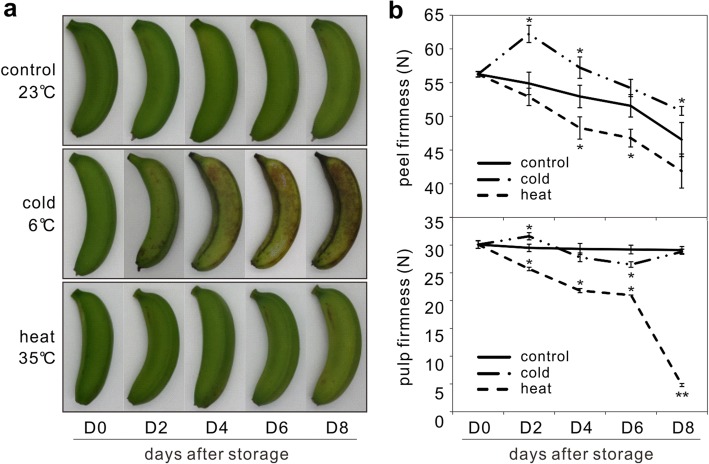
Harvested banana fruit were kept at 23 °C (control), 6 °C (cold-stressed) and 35 °C (heat-stressed), respectively for 8 days. Fruit were photographed every other day to monitor the injury symptoms under different temperature stresses (Fig. 1a). Chilling injury of brown spots could be observed as soon as the 2nd day of 6 °C-stored fruit and symptoms intensified as the time progressed. Comparatively, the control and 35 °C-stored fruit showed no change on the appearance (Fig. 1a). The peel firmness of the heat-stressed fruit decreased much faster than the control, and more strikingly a sharp drop of pulp firmness was detected in the heat-stressed fruit after the 6th day. However, cold stress increased the peel firmness especially on the 2nd day while it caused a fluctuation in the pulp firmness (Fig. 1b). Six sRNA libraries of banana peel samples collected from control, cold-stressed and heat-stressed fruit were made for deep sequencing, which yielded 37.3 million high quality reads. Among 3.7 million unique reads, ranging from 18- to 25-nt, ~ 70% were perfectly matched to at least one locus in the banana genome. These reads were used for further analysis (Additional file 1: Table S1). The 20~24-nt sRNAs constituted over 75% of the identified banana sRNAs, and the 21-nt sRNAs was the most abundant class in all samples. The total 21-nt sRNAs were less abundant in the temperature-stressed fruit than in the control, especially for the heat-stressed fruit. In addition, the total 24-nt sRNAs were less abundant in the cold-stressed fruit (Fig. 2a). On the contrary, the unique 21-nt sRNAs were more abundant in the cold-stressed fruit than in the control, and the expression of the unique 24-nt sRNAs was higher than the 21-nt class, but only for the control fruit (Fig. 2a). In the temperature-stressed fruit, both total and unique 18~20-nt sRNAs became more abundant. The increased percentage of these shorter sRNAs may result from the increased RNA degradation products caused by temperature stress. These dynamic changes in the sRNA population clearly showed that cold and heat stresses have differential effects on the sRNA biosynthesis.
Fig. 1.
Physiological changes in banana fruit under different temperature stresses. a Harvested banana fruit were stored at 23 °C (control), 6 °C (cold) and 35 °C (heat), respectively. Photos were taken every other day from D0 (before storage) to D8 (8 days after storage when symptoms were clearly observed for temperature-stressed fruit). The experiment was conducted twice independently with similar results, and typical photos of the fruit were presented. b Changes in fruit firmness under cold and heat treatments. Firmness was measured on unpeeled and peeled fruit fingers for peel and pulp firmness, respectively. Data were presented as the means ± standard errors (n = 9) as denoted by the error bars. Asterisks indicated significant differences in expression between control and temperature-stressed samples (*p < 0.05; **p < 0.01)
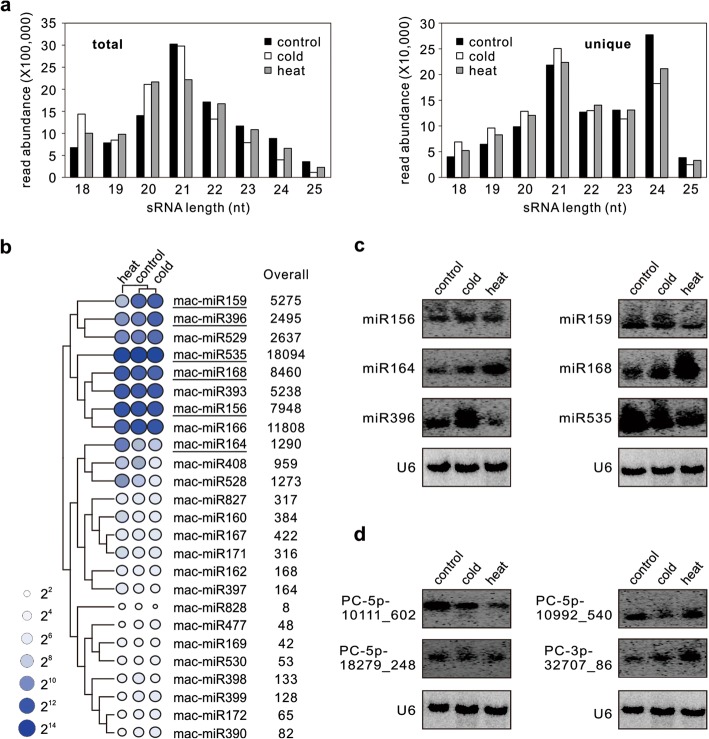
Fig. 2.
sRNA and miRNA expression profiling in banana. a Length distribution of total (left panel) and unique (right panel) sRNA sequences in banana stored at control temperature and subjected to cold and heat treatments. b Heat map and overall read abundance of the known miRNAs in the control, cold- and heat-stressed samples. Read abundance was normalized and expressed as reads per million (RPM) of genome-matched reads in each sample, as denoted by color and illustrated at the top-leaf corner of the panel. Known miRNAs selected for validation were underlined. c, d RNA gel blot analysis of selected known (c) and banana-specific (d) miRNAs in different samples. Total RNA was isolated from control, cold-stressed and heat-stressed banana peel and probed with end labeled antisense oligonucleotides. U6 served as a loading control. Blotting results from probing and reprobing the same filter were grouped together
MiRNA profiling in banana
Sequencing reads (21~22-nt) that perfectly mapped to the genome (22 million) were subjected to further analyses for miRNA identification. We identified 113 miRNAs belonging to 25 miRNA families either widely conserved or previously reported. All these miRNAs were collectively referred to as known miRNAs in this study. Expression levels of these known miRNAs, as reflected by normalized reads (RPM), showed a great variation among families in all samples (Additional file 1: Table S2). Most notably, the highest read abundance (~ 18,000 RPM) was detected for miR535 family; the level exceeded most conserved miRNAs. Other high abundance (~ 8000 RPM in total) was detected for miR156, miR166 and miR168 families, which were about 1.5~12 times more than other relatively abundant miRNA families, including miR159, miR393, miR529, miR396, miR164, miR528 and miR408, whose total abundance ranged from 1000 to 5000 RPM (Fig. 2b; Additional file 1: Table S2). While lower expression (between 200 and 500 RPM) was observed for the miR167, miR160, miR171 and miR827 families, their expression level was about twice higher than any of the remaining ten known miRNA families (Fig. 2b; Additional file 1: Table S2). Many known miRNA families comprised multiple species of different mature sequences and distinct length predominance, and showed differential expression among the samples analyzed (Additional file 1: Tables S2).
After excluding sRNA reads homologous to known miRNAs (≤1 mismatch, miRBase 21) and other non-coding sRNAs (Rfam v12.0), the remaining 21~22-nt sRNAs were subjected to novel miRNA identification based on a series of stringent criteria recognized by the research community [27]. A total of 26 miRNA candidates, which had typical hairpin precursor structure and star sequences (miRNA*) found, met the screening criteria (Additional file 1: Table S3), thus were considered novel or banana-specific miRNAs. Among them, 20 belong to the 21-nt class of miRNAs and only six to the 22-nt class (Additional file 1: Table S3). In general, the banana-specific miRNAs were much less abundant compared to the known miRNAs in all samples examined; the highest total read abundance being only ~ 800 RPM, and most yielded levels below 10 RPM (Additional file 1: Table S3).
To validate the sRNA sequencing data, we performed RNA gel blot analysis for selected miRNAs representing known examples in the control and cold/heat-stressed samples (Fig. 2c). We found that the blotting results for most highly expressed known miRNAs were reflective of the relative abundances of sequenced sRNAs. For example, miR164 and miR168 were more accumulated in the heat-stressed sample. Both miR159 and miR396 were induced by cold but greatly repressed by heat stress (Fig. 2c). However, divergence between the two analyses was also observed. Such contradictions between in vivo RNA levels and sequencing results for miRNAs have been previously reported for Arabidopsis and grapevine [11, 28]. On the other hand, the low level expression of novel miRNAs was confirmed by RNA gel blot analysis as signal was detectable only for those banana-specific miRNAs with total abundance > 100 RPM (Fig. 2d). As reported above for known miRNAs, RPM values for selected banana-specific miRNAs corresponded to relative signal intensity observed in RNA gel blots in some cases, but divergence was observed as well (Fig. 2d).
The impact of temperature stress on miRNA expression
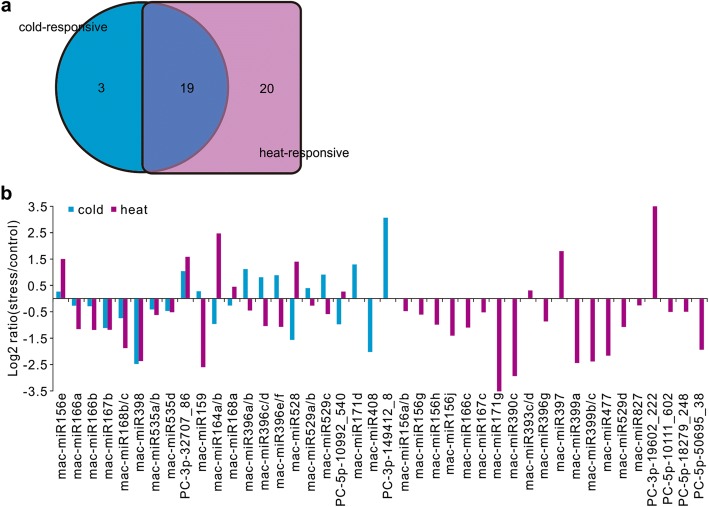
To detect the effect of different temperature stresses on banana miRNAs, the expression of miRNAs in banana fruit with cold stress, heat stress and without stress were examined. By comparing the expression level of miRNAs between the control and temperature-stressed fruit (p < 0.01), we identified 22 cold-responsive and 39 heat-responsive miRNAs, respectively. Some miRNA families responded to both stressors while a few miRNA families showed stress-specific response (Fig. 3a, Additional file 1: Table S4). About equal numbers of miRNAs were up-regulated and down-regulated by cold, while more miRNAs were down-regulated than up-regulated in the heat-stressed fruit. Among the overlapping group of 19 miRNAs responding to both cold and heat stresses, ten of which showed opposite expression change; the other nine with the same trend (Fig. 3b). Two known miRNAs and one banana-specific miRNA were found to respond only to cold stress. Comparatively, more miRNAs showed obviously altered expression just under heat stress, including 20 known miRNAs and four banana-specific miRNAs. Among these heat-responsive miRNAs, most were down-regulated (Additional file 1: Table S4). We further examined the overlapping group of miRNAs responding to both temperature stresses, and found some miRNAs with targets identified by degradome and having opposite expression patterns between cold and heat stress. For example, mac-miR164a/b targeting a NAC gene was down-regulated in the cold-stressed fruit but up-regulated in the heat-stressed fruit. Mac-miR159 targeting a TCP gene was induced by cold but repressed by heat. For the three mac-miR396 members that target growth-regulating factor were all up-regulated by cold but down-regulated by heat (Fig. 3b, Additional file 1: Table S4).
Fig. 3.
Expression and temperature responsiveness of miRNAs in banana. a Venn diagram showing the number of common and specific miRNAs responding to cold or heat stress. The circular and square areas represented cold- and heat-responsive miRNAs, respectively. The overlapping area represented miRNAs responding to both temperature stresses. b Fold change of miRNAs in cold- and heat-treated banana fruit on the 5th day of storage, relative to non-treated control. Fold change was calculated on the basis of normalized reads of stressed vs. unstressed samples. Only differentially expressed (p < 0.01) miRNAs were shown
MiRNA target identification and expression validation of temperature-responsive miRNAs and their targets
To identify targets for the banana miRNAs reported here, we performed degradome sequencing to generate a total of 37.4 million short reads representing 5′ ends of uncapped, polyadenylated RNAs. About 70% of the unique reads can be perfectly aligned to the banana transcriptome (https://banana-genome-hub.southgreen.fr/download). These reads were subsequently screened and analyzed with the Cleaveland 3.0 [29]. Based on the miRNA cleavage features on the transcripts, a total of 60 targets falling into five categories (0 to 4) were identified (p < 0.05), all for 37 of the 13 known miRNAs or families (Table 1 and Additional file 1: Table S5). Among the 60 targets for the known miRNA families, 90% fell into category 0, which represented the most abundant degradome tags corresponding to the cleavage site and matching cognate transcripts. The number of identified gene targets varied for different miRNAs, ranging from one to eights. For example, miR159 targeted multiple genes encoding TCP transcription factors, miR156 targeted SQUAMOSA PROMOTER BINDING-LIKE (SPL) genes, and miR167 targeted AUXIN RESPONSE FACTOR (ARF) genes (Table 1 and Additional file 1: Table S5). Although most of the genes (48 of 60) identified were the conserved targets for these miRNAs across a wide range of plant species, a few of them (12 of 60) had not been reported in other species. For instance, miR156 was also found to target one gene coding for CCR4-NOT transcription factor and miR396 that mainly targets GROOWTH REGULATING FACTOR (GRF) genes was also found to target one gene encoding metallopeptidase (Table 1 and Additional file 1: Table S5).
Table 1.
Target of banana known miRNAs (or families)a
| miRNA | Target | ASb | Categoryc | Reads at the cleavage site (tpb)d | Target annotation |
|---|---|---|---|---|---|
| Conserved targets for known miRNAs | |||||
| miR156 | Achr6T36010 | 3 | 0 | 722.4 | Putative Squamosa promoter-binding-like protein 12 |
| Achr10T23280 | 1 | 0 | 214.0 | Putative Squamosa promoter-binding-like protein 12 | |
| Achr5T21990 | 1 | 0 | 240.8 | Putative Squamosa promoter-binding-like protein 12 | |
| Achr10T02970 | 1 | 0 | 989.9 | Putative Squamosa promoter-binding-like protein 12 | |
| Achr3T11170 | 1.5 | 0 | 124.9 | Putative Squamosa promoter-binding-like protein 17 | |
| Achr5T25090 | 1.5 | 0 | 133.8 | Putative Squamosa promoter-binding-like protein 18 | |
| Achr7T22940 | 1.5 | 0 | 2060.1 | Putative Squamosa promoter-binding-like protein 16 | |
| Achr8T25550 | 1.5 | 0 | 124.9 | Putative Squamosa promoter-binding-like protein 17 | |
| miR159 | Achr2T12990 | 3.5 | 0 | 693.8 | Putative Transcription factor PCF6 |
| Achr4T22960 | 3.5 | 0 | 292.5 | Putative Transcription factor PCF6 | |
| Achr4T31520 | 3.5 | 0 | 194.4 | Putative Transcription factor PCF6 | |
| Achr5T02530 | 3.5 | 0 | 881.1 | Putative Transcription factor PCF6 | |
| Achr7T01160 | 3.5 | 0 | 881.1 | Putative Transcription factor PCF6 | |
| Achr5T06660 | 3 | 0 | 160.5 | Putative Transcription factor TCP4 | |
| Achr8T19990 | 3 | 0 | 454.8 | Putative Transcription factor TCP4 | |
| Achr10T0102 | 3 | 0 | 481.6 | Putative Transcription factor TCP4 | |
| miR160 | Achr6T18900 | 1.5 | 0 | 321.1 | Putative Auxin response factor 22 |
| Achr9T29480 | 1.5 | 0 | 695.6 | Putative Auxin response factor 17 | |
| Achr11T00640 | 2 | 0 | 27,825.2 | Auxin response factor 18 | |
| Achr5T18540 | 2 | 0 | 13,029.7 | Auxin response factor 18 | |
| Achr5T14630 | 2 | 0 | 4053.4 | Auxin response factor 18 | |
| Achr5T03960 | 2 | 0 | 6822.5 | Auxin response factor 18 | |
| Achr4T18240 | 2 | 0 | 4080.1 | Auxin response factor 18 | |
| Achr8T18930 | 2 | 0 | 3852.7 | Auxin response factor 18 | |
| miR164 | Achr3T23360 | 3 | 0 | 936.4 | Putative NAC domain-containing protein |
| Achr9T27530 | 3.5 | 0 | 1337.7 | no apical meristem protein, putative | |
| Achr9T26140 | 2 | 0 | 6207.1 | NAC domain-containing protein | |
| Achr9T10210 | 2 | 0 | 20,347.1 | NAC domain-containing protein | |
| miR167 | AchrUn_randomT06470 | 3.5 | 0 | 5468.7 | Auxin response factor 12 |
| Achr11T25770 | 4 | 0 | 5415.2 | Auxin response factor 6 | |
| Achr3T23290 | 4 | 0 | 5281.4 | Auxin response factor 17 | |
| Achr5T00590 | 4 | 0 | 4535.0 | Auxin response factor 17 | |
| Achr5T26580 | 4 | 0 | 5281.4 | Auxin response factor 6 | |
| Achr5T02450 | 4 | 0 | 4454.7 | Auxin response factor 6 | |
| miR168 | Achr3T27070 | 4 | 0 | 1150.5 | Protein argonaute 1B |
| miR171 | Achr4T07190 | 3 | 0 | 165,720.2 | Putative Scarecrow-like protein 15 |
| Achr3T29970 | 3 | 0 | 8802.4 | Putative Scarecrow-like protein 15 | |
| Achr4T03460 | 3 | 0 | 508.3 | Putative Scarecrow-like protein 15 | |
| Achr5T20860 | 2 | 0 | 1551.8 | Putative Scarecrow-like protein 6 | |
| miR393 | Achr6T20580 | 3 | 0 | 4666.5 | Transport inhibitor response 1-like protein |
| Achr6T16510 | 3 | 0 | 3424.6 | Transport inhibitor response 1-like protein | |
| Achr6T08000 | 3 | 0 | 4882.8 | Transport inhibitor response 1-like protein | |
| Achr9T01140 | 3 | 0 | 4693.3 | Transport inhibitor response 1-like protein | |
| AchrUn_randomT10710 | 2 | 0 | 3917.4 | Transport inhibitor response 1-like protein | |
| Achr10T12000 | 2 | 0 | 3244.0 | Transport inhibitor response 1-like protein | |
| miR396 | Achr3T15140 | 2.5 | 0 | 615.4 | growth regulating factor protein, putative |
| Achr7T27480 | 2.5 | 0 | 615.4 | growth-regulating factor, putative | |
| miR535 | Achr10T02970 | 4 | 0 | 989.9 | Putative Squamosa promoter-binding-like protein 12 |
| Achr4T05720 | 4 | 0 | 695.6 | Putative Squamosa promoter-binding-like protein 12 | |
| Non-conserved targets for known miRNAs | |||||
| miR156 | Achr10T05030 | 3.5 | 2 | 240.8 | CCR4-NOT transcription factor, putative, expressed |
| miR164 | AchrUn_randomT19770 | 3 | 0 | 20,775.2 | Hypothetical protein |
| Achr11T02240 | 4 | 4 | 13.4 | Probable aquaporin TIP1–1 | |
| Achr8T12920 | 4 | 4 | 13.4 | Probable aquaporin TIP1–1 | |
| miR396 | Achr5T20170 | 4 | 1 | 267.5 | Hypothetical protein |
| Achr8T26530 | 2.5 | 0 | 615.4 | metalloendopeptidase/ metallopeptidase/ zinc ion binding protein, putative | |
| miR528 | AchrUn_randomT22730 | 3.5 | 0 | 1172.8 | Putative Polyphenol oxidase A1, chloroplastic |
| AchrUn_randomT25220 | 3.5 | 4 | 8.9 | Putative Polyphenol oxidase A1, chloroplastic | |
| Achr8T34370 | 2.5 | 2 | 615.4 | Polyphenol oxidase, chloroplastic | |
| miR530 | Achr2T17590 | 1 | 0 | 2354.4 | Hypothetical protein |
| miR535 | Achr4T12470 | 4 | 0 | 668.9 | Putative uncharacterized protein |
| miR828 | Achr2T17960 | 3 | 0 | 107.0 | Hypothetical protein |
a A detailed list was included in Additional file 1: Table S5. b The alignment score (AS) threshold was set to 4 for known miRNAs. c Category 0: > 1 raw read at the position, abundance at position is equal to the maximum on the transcript, and there is only one maximum on the transcript. Category 1: > 1 raw read at the position, abundance at position is equal to the maximum on the transcript, and there is more than one maximum position on the transcript. Category 2: > 1 raw read at the position, abundance at position is less than the maximum but higher than the median for the transcript. Category 3: > 1 raw read at the position, abundance at position is equal to or less than the median for the transcript. Category 4: only 1 raw read at the position. d Reads at the cleavage site were normalized to transcripts per billion (tpb)
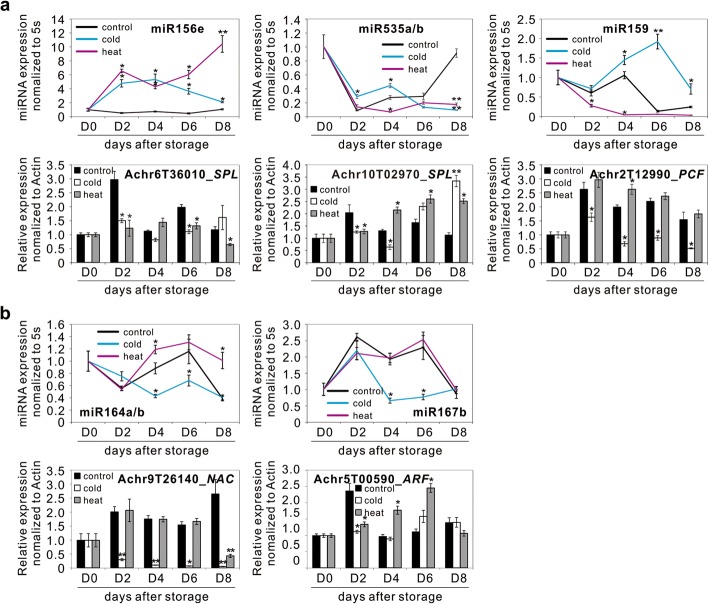
We selected several temperature-responsive miRNAs with their corresponding target genes and validated their differential expression in a time-course manner. Some miRNA-target pairs, including miR156/535-SPL and miR159-TCP modules displayed an inverse correlation (Fig. 4a). However, discrepancies were also observed such as miR164-NAC and miR167-ARF modules (Fig. 4b), suggesting other levels of regulation may also take effect.
Fig. 4.
Differential expression of miRNAs and their corresponding targets in control and temperature stressed samples during storage. a Anti-correlation pattern of miR156/535-SPL and miR159-TCP modules. b The miR164-NAC and miR167-ARF modules displaying no obvious correlation. In both (a) and (b), the top panel showed miRNA accumulation and the bottom panel displayed the corresponding target gene expression. The relative expression was normalized to 5 s rRNA for miRNAs and Actin for target genes. Normalized expression in control fruit at day 0 was arbitrarily set to 1. Data were presented as the means ± standard errors (n = 3) as denoted by the error bars. Asterisks indicated significant differences in expression between control and temperature-stressed samples (*p < 0.05; **p < 0.01)
SPL gene family co-targeted by miR156 and miR535 in banana
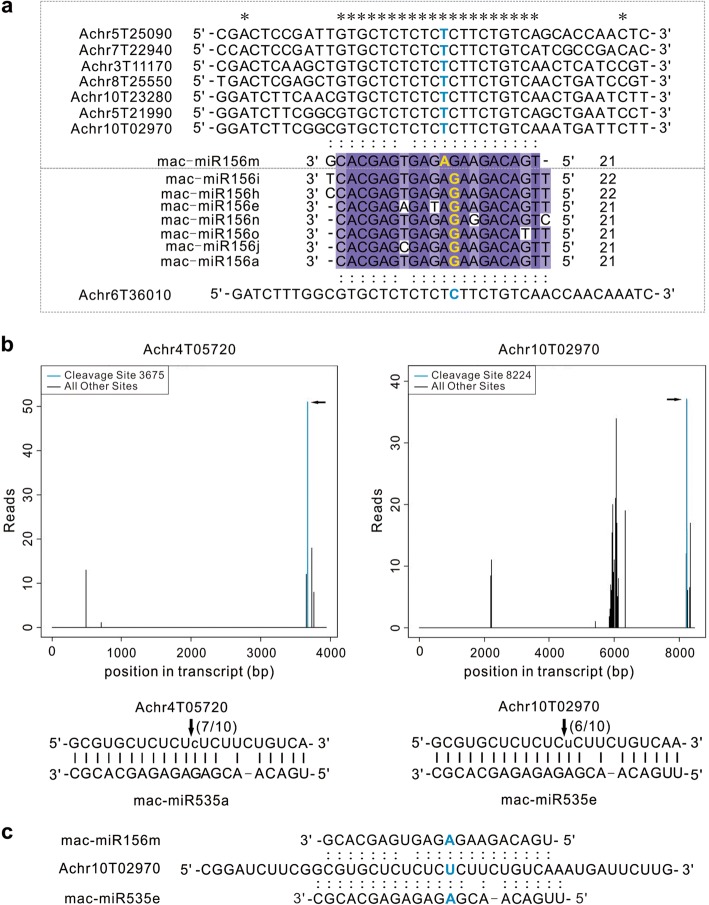
We found that miR156 displayed a highly conserved target relationship in banana, especially for miR156m variant that was identified to target seven various SPL genes, while other miR156 variants collectively targeted one SPL gene (Additional file 1: Table S5). Interestingly, there was one nucleotide offset between the cleavage sites of these two groups of SPL targets; accordingly miR156m was shorter by 1 nt at the 5′ end (Fig. 5a). In addition to miR156 family, four sequence-specific miRNAs belonging to miR535 family were also identified to target three SPL genes, two of which (Achr4T05720, Achr10T02970) was further confirmed by RLM-5’-RACE (Fig. 5b). It was also shown that Achr10T02970 could be cleaved by both miR156m and miR535e at the same location (Fig. 5c). Actually, sequence alignment showed a high similarity among miR156 and miR535 variants and their targets, explaining such a co-targeting phenomenon. While co-targeting the same gene family, different miR156 and miR535 variants exhibited very distinctive expression patterns by temperature stress. For example, mac-miR535a/b was down-regulated by heat stress, whereas mac-miR156e was up-regulated under both stresses, especially heat. Such differential expression patterns were validated by stem-loop qPCR (Fig. 4a). Considering the abundantly and differentially expressed miR535 isoforms, our result suggest that miR535 family may play an equally considerable role, as the well-conserved miR156 family does, in fine tuning SPL gene expression in banana.
Fig. 5.
MiR156 and miR535 families co-targeting SPL gene family in banana. a Sequence alignment of miR156 variants and their targets. The cleavage site detected in the degradome is indicated in blue and yellow letters. b Target plot (t-plot) and alignment of validated targets for mac-miR535a/b and mac-miR535e, respectively. The arrow indicates signatures consistent with miRNA-directed cleavage. Cleavage frequency as determined by gene-specific 5′-rapid amplification of cDNA ends (5’-RACE) at the indicated position is shown in parentheses. Primer sequences used for 5’-RACE are provided in Additional file 1: Table S7. c Achr10T09270 is co-targeted by mac-miR156m and mac-miR535e. The cleavage sites detected in the degradome are highlighted in blue letters
Characterization of PHAS loci and their triggers in banana
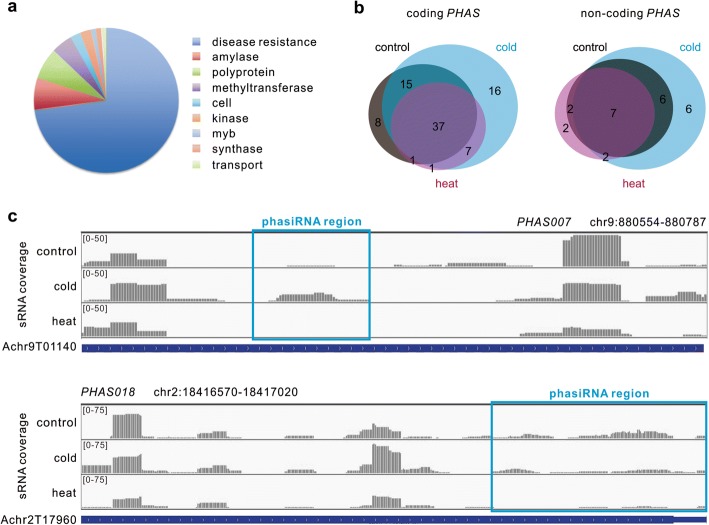
PhasiRNAs are a major class of sRNAs in plants [9]. They are produced from not only diverse protein-coding genes, including large gene families such as NB-LRRs, PPRs, and MYBs, but also non-coding transcripts [10, 30]. To explore whether phasiRNA pathways are conserved and responsive to temperature stresses in banana, a genome-wide identification of PHAS loci was performed. For this analysis, an approach as described in [31] was used. In total, we identified 110 PHAS loci (Additional file 1: Table S6), of which 85 loci shown homology to known protein-coding genes, with the remaining 25 loci with no homology found, and thus considered as non-coding loci. Among the coding loci, the majority showed sequence similarity to NB-LRR genes (62 or 73%), with other proportions mainly represented by genes coding for alpha-amylase, methyltransferase and polyprotein (Fig. 6a). However, the other two large gene families (MYB and PPR genes) for generating the majority of phasiRNAs in most eudicots appeared to be rare in banana, where only one gene for MYB protein and none for PPR were identified (Additional file 1: Table S6). Some other coding genes were also found to generate phasiRNAs as well, including genes encoding kinase, starch synthase and transport inhibitor (Additional file 1: Table S6). All non-coding PHAS loci were relatively short (< 400 bp), compared to coding PHAS loci, suggesting that in banana non-coding PHAS loci mainly generate relatively short transcripts.
Fig. 6.
PhasiRNA-generating (PHAS) network in banana. a All PHAS loci were grouped into coding and non-coding genes, and coding PHAS loci were further classified based on their annotation, as shown in the pie chart. b Venn diagram for coding and non-coding PHAS loci differentially accumulated in the control and temperature-stressed samples. c Two examples of coding PHAS loci displaying differential variations in phasiRNA production in banana upon cold and heat stress. In both panels, each track represented small RNA abundance based on mapping results in the control, cold- and heat-stressed samples. Changes of the phasiRNA accumulation were highlighted in the blue boxes. The gene models from which the sRNA generated were presented below the panels
Next, we determined which miRNAs served as triggers of phasiRNA production for these PHAS loci. Six miRNA families were found to function as phasiRNA triggers (Table 2), integrating dagradome data, the miRNAs and the PHAS loci. Most miRNA triggers were 22-nt with an initial ‘U’. Several interactions between miRNA triggers and PHAS loci were conserved in many other plants, including miR393-TIR/AFB, miR828-MYB, and miR482/miR2118-NB-LRR; another three miRNAs, including miR162, miR166 and miR319/miR159, were also identified as triggers. Among them, miR162 and one miR319/miR159 member were also able to trigger phasiRNAs from NBS-type genes, but these two were of 21-nt in length, for which the triggering mechanism is not clear. Some miRNA triggers (miR319/miR159, miR393 and miR482/miR2118) could also target non-coding PHAS loci, in addition to coding genes (Table 2). Most identified PHAS loci (92 or 84%) could not find any known or novel miRNA triggers characterized in this study, indicating a complicated mechanism involved in triggering phasiRNA production. We also found great variations of PHAS loci distribution and phasiRNA production in banana fruit upon temperature stress. In general, more PHAS loci were found in the cold-stressed fruit while less loci in the heat-stressed fruit, regardless of coding or non-coding PHAS loci (Fig. 6b). As examples, two PHAS loci exhibited distinct phasiRNAs production in the temperature-stressed samples. The miR393-triggered phasiRNAs were preferentially produced only upon cold stress while the miR828-triggered phasiRNA production was largely abolished upon heat stress (Fig. 6c).
Table 2.
PHAS loci and trigger miRNAs in banana
| Trigger miRNA | Sequence | Length (nt) | PHAS loci annotation |
|---|---|---|---|
| miR162 | UCUAUAAACCUCUGCAUCCGG | 21 | NBS-type resistance protein RGC5(1e-05) |
| miR166 | UCGUACCAGGCUUCAUUUCCC | 21 | non-coding |
| miR319/miR159 | UUGGACUGAAGGGAGCUCCUCU | 22 | non-coding |
| miR319/miR159 | UUGGACUGAAGGGAGCUCCUCU | 22 | non-coding |
| miR319/miR159 | GAGAGCUUCCUUCAGUCCACU | 21 | Resistance protein (Fragment)(3e-41) |
| miR393 | UUCCAAAGGGAUCGCAUUGAUC | 22 | non-coding |
| miR393 | UUCCAAAGGGAUCGCAUUGAUC | 22 | Transport inhibitor response family protein(3e-37) |
| miR482/miR2118 | UCUUGCCGAUUCCUCCCAUCCC | 22 | NBS resistance protein (Fragment)(8e-30) |
| miR482/miR2118 | UCUUGCCGAUUCCUCCCAUCCC | 22 | NBS-LRR disease resistance protein (Fragment)(3e-126) |
| miR482/miR2118 | UCUUGCCGAUUCCUCCCAUCCC | 22 | NBS-LRR disease resistance protein (Fragment)(2e-79) |
| miR482/miR2118 | UCUUGCCGAUUCCUCCCAUCCC | 22 | NBS-LRR disease resistance protein (Fragment)(1e-85) |
| miR482/miR2118 | UUUCCAAUACCUCCCAUGCCAA | 22 | non-coding |
| miR482/miR2118 | UUUCCAAUACCUCCCAUGCCAA | 22 | non-coding |
| miR482/miR2118 | UUUCCAAUACCUCCCAUGCCAA | 22 | Os05g0492600 protein(2e-168) |
| miR482/miR2118 | UUUCCGAUUCCUCCCAUCCCUA | 22 | NBS-type disease resistance protein (Fragment)(4e-84) |
| miR482/miR2118 | UUGCCGAUUCCUCCCAUCCCUA | 22 | NBS-LRR disease resistance protein (Fragment)(6e-121) |
| miR482/miR2118 | UUGCCGAUUCCUCCCAUCCCUA | 22 | NBS-LRR resistance protein (Fragment)(3e-76) |
| miR828 | UCUUGCUCAAGUGAGUAUUCCA | 22 | R2R3 MYB(5e-12) |
Discussion
MiRNA and sRNA expression in banana
The large-scale miRNA identification via deep sequencing with experimental approaches has been performed in diverse plant species, including many monocots. However, until recently, quite few studies have been done on characterizing miRNAs in banana [23, 24], especially in banana fruit subjected to different temperature stress. In this paper, genome-wide analysis of banana miRNAs were performed with high-throughput sequencing and their response to temperature stress was also analyzed, providing useful information to widen our understanding of miRNAs in monocotyledonous plants. Deep sequencing of the banana sRNA libraries revealed 113 known miRNAs as well as 26 banana-specific miRNAs (Additional file 1: Tables S2 and S3). Most of the conserved miRNA families were identified in our data, but all the banana-specific miRNAs exhibited relatively low expression levels (Additional file 1: Table S3), in consistent with the notion that non-conserved miRNAs are often expressed at a lower level than conserved miRNAs. These miRNAs have not been previously reported in other plants, possibly due to their inappreciable or inducible expression pattern. Deep sequencing and whole-genome data mining enable us to discover probably most of the miRNAs in banana under temperature stress. For all the identified miRNAs, only those showing significant temperature stress responses were further analyzed.
Among the conserved miRNA families, miR156 family appeared to be greatly expanded in banana, having 16 members. MiR535 family was also distinguished because it was the most highly expressed miRNA family in banana, with eight members (Additional file 1: Table S2). In addition, miR535 family was found to co-target SPL gene family, together with miR156 family. To date, the origin of most plant MIRNAs has not been fully elucidated, although several evolutionary paths have been proposed [31–34]. It has been shown that the origin of plant MIRNAs is dependent on the occurrence of various duplications, probably followed by chromosomal rearrangements and loss of duplicated genes, paralleling the coevolution with the mRNA targets [35, 36]. Considering the high sequence homology between miR156 and miR535 variants, these two miRNA families may derive from a common ancestor after duplication events during the evolution. And banana apparently has evolved an expanded miR156/miR535 gene family to conform to their target SPL family that comprises a large number of gene members, with the functional importance in banana stress response to be further determined.
PhasiRNAs are a major class of sRNAs in plants. In this study, we identified 110 PHAS loci in banana, triggered by six miRNA families. Among the three major protein-coding PHAS gene families in eudicots, only NB-LRRs yielded abundant phasiRNAs in banana. For other phasiRNA pathways conserved in plants, only miR828-MYB and miR393-TIR1/AFB were identified in banana. On the other hand, some phasiRNA pathways appeared to be newly evolved in banana, although the identification of their trigger miRNAs was unsuccessful (Additional file 1: Table S6). All these results indicate the dynamic feature of PHAS pathways in banana during evolution. In modern plants, auxin plays an important role in the regulation of leaf morphology, lateral root growth, and developmental patterning [37, 38], with ARF and TIR genes being the main components. In banana, profuse ARF and TIR genes were targeted by miR160/miR167, and miR393, respectively (Additional file 1: Table S5). Moreover, miR393-TIR1/AFB phasiRNA production displayed a cold stress-specific enrichment (Fig. 6c). However, the well-conserved miR390-TAS3-ARF pathway appeared to be minimized in banana and the reason for such functional diversification of auxin-related pathways deserves further investigation.
The temperature-responsive miRNAs in banana
MiRNAs were first revealed to be involved in plant cold response by [16]. Based on a recent review [39], 18 studies in various plant species have confirmed the involvement of miRNAs in response to cold stress, using microarray and sequencing platforms. It has been proposed that response of a particular miRNA to the same cold stress differs depending on the plant species, indicating the divergence of miRNA function during plant evolution. Another explanation would be the tissue-specific miRNA expression pattern that leads to the inconsistency. In those reports, the tested tissues include seedlings, leaves, leaf buds, anther and ovary. Compare to cold-responsive miRNAs, there have been fewer reports of heat-responsive miRNAs in plants, although the specificity of miRNA response to heat stress is also demonstrated [40, 41]. As a typical tropical fruit, banana is very sensitive to cold stress, which usually results in peel pitting, discoloration and abnormal fruit ripening. On the other hand, high temperature stress can result in the ‘stay-green’ ripening disorder of banana fruit. These facts indicate that banana might have evolved differential regulatory mechanisms to react to cold and heat stress. However, information on the role of miRNAs is lacking for harvested banana fruit so far. Exploring the temperature-responsive miRNAs in banana can provide useful information for improving the temperature tolerance and avoidance of ripening disorder of banana fruit. In this study, the expression levels of miRNAs in banana with and without temperature stress treatments were separately compared; around 30% of the miRNAs showed significant changes under temperature stress (Additional file 1: Table S4). Some of these miRNAs have been reported as temperature-responsive in other plants. For example, specific members of miR396 and miR171 families, previously reported as up-regulated by cold in Arabidopsis [42], were also found to be differentially induced after cold treatment in banana, indicating that they also conserve in monocots. On the other hand, strong suppression of miR167b, miR398, miR528 and miR408 expression by cold was widely observed in banana. Such down-regulation of miR398 and miR408 partly agreed with previous reports in wheat and grapevine [43], but different from the observation in Arabidopsis [42]. In addition, both miR167b and miR528 have never been reported as cold-responsive in plants. Likewise, some previously identified cold-induced conserved miRNAs, such as miR319, miR393 and miR402 in Arabidopsis [16], were not found in our data, suggesting that the induction levels of certain miRNAs may be too low to be detected as significant, or they were unaffected by cold in banana. Previous studies have put great emphasis on the cold-induced miRNAs, but cold-repressed miRNAs have comparatively received little attention. Our study showed almost equal numbers of up-regulated and down-regulated miRNAs to cold stress, and these cold-responsive miRNAs had relatively equivalent expression levels (Additional file 1: Table S4). These data suggest that both kinds of regulation for miRNAs were involved in the banana cold response.
In banana, more miRNAs differentially respond to heat stress than to cold stress (Additional file 1: Table S4), suggesting that heat stress might have imposed a wider range of effect on miRNA expression. Moreover, compare to the relatively equal number of down- and up-regulated miRNAs responding to cold, the number of down-regulated miRNAs was almost three times that of up-regulated ones among the identified heat-responsive miRNAs in banana, indicating that overall heat stress tended to inhibit miRNA expression. Several heat-responsive miRNAs identified from banana were also previously reported in other plants. For example, it has been reported that miR156 integrates the response to recurring heat stress with development in Arabidopsis through SPL TFs [44]. We found that in banana, multiple miR156 variants showed differential response upon heat stress, where miR156e was significantly induced while others got suppressed. Our result indicated that the miR156-SPL module might also mediate the heat stress response in banana. Other significantly heat-induced miRNAs included miR164a/b, miR528 and miR397. A similar up-regulation pattern of miR164 has been previously reported in heat-stressed switchgrass [40], but the case was opposite in wheat [41]. On the contrary, a large number of banana miRNAs, including miR159, miR166c, miR171g, miR390c, miR396a-f, miR398, miR399a/b/c and miR477, all showed down-regulation in response to heat stress (Additional file 1: Table S4). Similarly, down-regulation of miR159 was observed under heat in wheat [41] but an opposite trend was found in switchgrass [40]. All the discrepancies may result from the species-specific miRNA levels, or that miRNAs respond to heat stress only in specific tissues or at specific developmental stages.
MiRNA-mediated pathways involved in banana temperature stress response
Based on miRNA expression profiling and dagradome combined with qRT-PCR validation, we found that several miRNA-target pairs might have played important roles in the regulation of banana temperature stress response. SPL transcription factors, which are unique to plants, are involved in many important biological events [45]. Most members of the SPL family are targets of miR156 and recent evidence showed that miR156 can down-regulate SPL expression by either mRNA cleavage or translational repression [46]. In banana, SPL genes can also be targeted by miR535 and our data confirmed the anti-correlation between miR156/miR535 and their corresponding target SPL genes (Fig. 4a). Moreover, an inverse expression pattern of the two tested SPL genes was observed at the later stage of storage, suggesting that SPL transcript accumulation may be balanced via the co-regulation of miR156 and miR535.
Another well anti-correlated module in banana was miR159 targeting PCF/TCP transcription factor. Upon cold treatment, miR159 gradually accumulated and remained at a higher level compared to the control. Accordingly, its PCF target gradually became suppressed as the injury of cold stress aggravated. Such pattern was opposite in the heat-treated sample (Fig. 4a). In rice, two miR319-targeted PCF genes (OsPCF5 and OsPCF8) have been characterized as negative regulators of cold tolerance [47], which seemed to be different from our observation. Moreover, an earlier study [48] proposed that in Arabidopsis, TCPs are normally not targeted by miR159. However, our data showed that a banana PCF gene was targeted by miR159. In addition, we did not identify any miR319 variants in banana. These results implicated that banana may possess a functional specialization of miR159/miR319 different from Arabidopsis. In banana, without miR319, miR159 may have evolved to exert its regulation on PCF.
Besides the above mentioned miRNAs, we also found other miRNAs having differential responses to temperature stress. For example, miR397 showed a significant induction after heat stress treatment. Previous studies have shown that miR397 targets laccase (LAC) in rice, Arabidopsis, Populus and litchi [49, 50]. In banana, at least three annotated laccase genes were predicted as miR397 targets. We suggest that the downregulation of LAC was mediated by the induction of miR397, which might be involved in the thermotolerance through protection from oxidative damage. However, there has been no direct evidence for the relationship between oxidative damage and ripening disorder upon heat stress in banana. On the contrary, both miR398 and miR399 showed downregulation in response to heat. Target prediction identified their targets encoding putative cytochrome c oxidase subunit and ubiquitin-related modifier 1 homolog, respectively. MiR398 and its target sites on Cu/Zn superoxide dismutase genes (CSDs) mRNA are conserved in dicots and monocots [49, 51], with an important role in the oxidative stress tolerance in plants [51]. MiR399 has been reported to be responsive to phosphate starvation in Arabidopsis via targeting genes coding for putative ubiquitin-conjugating enzymes [52]. Under heat stress, banana fruit would consume more energy, so repressing of these two miRNAs would accumulate their target genes to adapt to the nutrient deficiency.
Conclusions
Altogether, our work delivered new insights into the role of miRNAs in the temperature stress response of banana fruit. Most targets of temperature-responsive miRNAs in banana were transcription factors including a large group of target genes involved in auxin signaling, and other functional genes associated with redox/nutrient homeostasis. Emerging studies have demonstrated the utility of miRNAs for genetic improvement in crops [53, 54], so the characterized candidate miRNAs in our study may serve as markers in breeding programs or tools for biotechnological approaches for improving temperature stress tolerance of banana.
Methods
Plant materials and sample collection
Mature-green ‘Brazilian’ banana fruit (Musa spp. AAA group, cv. Cavendish) were harvested from a plantation located in Gaozhou County, Guandong province. Fruits were harvested at 80% of maturity and selected for uniformity and free of defects. All fruits were soaked in 0.2% (w/v) Sporgon solution (Bayer, Germany) for 5 min to eliminate potential microbes and air dried. Based on our previous trials, three storage temperatures were set in this study: control at 23 °C, cold stress temperature at 6 °C and heat stress temperature at 35 °C. For each temperature treatment, 120 fruit fingers were selected, packed in 0.025-mm-thick polyethylene bags, and divided into three groups of forty fingers each as biological replicates. Peel samples from control, cold- and heat-stressed fruit were collected 0, 2, 4, 6 and 8 d after treatment. All the samples were immediately frozen in liquid nitrogen and stored at − 80 °C until use.
Observation of fruit appearance and firmness measurement under temperature stress
At each sampling time, photos were taken for fruit stored at different temperatures, which could represent a typical state. For each treatment, fruit firmness was measured at three equidistant points around the middle position on three unpeeled (peel firmness) and three peeled (pulp firmness) fruit fingers, respectively.
Small RNA library construction and sequencing
Peel samples from control, cold- and heat-stressed fruits were collected on the 5th day of storage, when the typical injury symptoms appeared. Two replicate total RNA from different samples was extracted using Trizol reagent (Invitrogen, CA, USA) following the manufacturer’s procedure. The total RNA quantity and purity were evaluated by Agilent Bioanalyzer 2100 and RNA 6000 Nano LabChip Kit (Agilent, CA, USA) with RIN number > 7.0. Approximately 1 μg of total RNA was used to prepare small RNA library according to protocol of TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA). Then the single-end sequencing (36 bp) was performed on an Illumina Hiseq2000 at the LC-BIO (Hangzhou, China) following the vendor’s recommended protocol.
Small RNA data analysis
Briefly, the raw reads were subjected to the Illumina pipeline filter (Solexa 0.3), and then the dataset was further processed with an in-house program, ACGT101-miR (LC Sciences, Houston, Texas, USA) to remove adaptor, low complexity, common RNA families (rRNA, tRNA, snRNA, snoRNA) and repeats. Subsequently, unique sequences with length of 18~25-nt were mapped to specific species precursors in miRBase 21 by BLAST search to identify known and novel 5p- and 3p-derived miRNAs. Length variation at both 3′ and 5′ ends and one mismatch inside of the sequence were allowed in the alignment. Unique sequences mapping to specific species mature miRNAs in hairpin arms were identified as known miRNAs, while unique sequences mapping to the other arm of known specific species precursor hairpin opposite to the annotated mature miRNA-containing arm were considered to be novel 5p- or 3p-derived miRNA candidates and the mapped pre-miRNAs were further compared against the specific species genomes using BLAST to determine their genomic locations.
The remaining unmapped sequences were compared against the banana genome (Musa acuminata DH-Pahang v1, banana-genome.cirad.fr) using BLAST to identify novel banana-specific miRNAs. The hairpin RNA structures containing sequences were predicated from the flanking 120-nt sequences using RNAfold software (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) [55]. The criteria for secondary hairpin structure prediction were: (1) number of nucleotides in one bulge in stem (≤12). (2) number of base pairs in the stem region of the predicted hairpin (≥16). (3) cutoff of free energy (dG < − 35 kcal/mol). (4) length of hairpin (up and down stems + terminal loop ≥50). (5) length of hairpin loop (≤200). (6) number of nucleotides in one bulge in mature region (≤4). (7) number of biased errors in one bulge in mature region (≤2). (8) number of biased bulges in mature region (≤2). (9) number of errors in mature region (≤4). (10) number of base pairs in the mature region of the predicted hairpin (≥12). (11) percent of mature in stem (≥80). Based on the criteria, only 21~22-nt sRNAs with a good hairpin structure and a miRNA/miRNA* duplex accounting for more than 75% reads matching to the precursor locus were considered as potential banana-specific miRNAs.
Degradome library construction, sequencing and data analysis
Mixed total RNAs were used, with equal amounts from all banana fruit at different stages of storage and also under temperature stress. Approximately 20 μg of total RNA was used to prepare a degradome library with following steps: (1) Biotinylated Random Primers (BRPs) were used to incubate with ~ 150 ng of poly (A)+ RNA (2) RNA fragments annealed with BRPs were captured by Strapavidin. (3) 5′ adaptor ligation to only those RNAs containing 5′ monophosphates. (4) Reverse transcription and PCR. (5) Libraries were sequenced using the 5′ adaptor only, resulting in the sequencing of the first 36 nucleotides of the inserts that represented the 5′ ends of the original RNAs. Then the single-end sequencing (36 bp) was performed on an Illumina Hiseq2500 at the LC-BIO (Hangzhou, China) following the vendor’s recommended protocol. Raw sequencing reads were obtained using Illumina’s Pipeline v1.5 software following sequencing image analysis by Firecrest Module and base-calling by Bustard Module. The extracted sequencing reads were used in the standard data analysis. A public software package, CleaveLand 3.0 was used for analyzing sequencing data generated, with alignment score (AS) threshold set to 4. Reads at the cleavage site were normalized to transcripts per billion (tpb).
Analysis of differential expressed miRNAs under temperature stress
Total number of reads perfectly matching the banana genome in a given library was used for the normalization of read abundance, which was denoted as RPM (reads per million genome-matched reads). The miRNA differential expression based on normalized deep-sequencing counts was analyzed by Student t test based on the experimental design. The significance threshold was set to be p < 0.01 with fold changes (stress reads/control reads) > 1.2-fold for identifying differentially expressed miRNAs upon cold or heat stress. TBtools software was used for the construction of the heat map [56].
PHAS loci identification
PhasiRNAs are produced with the ‘phasing’ pattern and this pattern can be used to identify phasiRNA-generating loci (PHAS loci), based on reference-aligned sRNA-seq data. After mapping sRNA reads to the reference genome, unique sRNAs were denoted with their matching coordinates. Two-nucleotide offset was added for sRNA matching to the antisense strand due to the 2-nt overhang at the 3′ end of the sRNA duplex. A genome-wide search was performed using a nine-cycle sliding window with each shift of three cycles, and a window was reported when ≥10 distinct sRNA reads fell into it; more than half of the mapped distinct sRNAs were 21-nt in length, and with ≥3 distinct reads in a certain register. All reported windows with overlapping region were then combined into a single longer window and a P-value was calculated for each window [31, 57]. In addition to a P-value threshold of 0.001, additional criteria were applied to ensure that only highly confident PHAS loci were identified: (1) the length of phasiRNA-producing region ≥100 bp. (2) over 30% of the siRNAs from a given PHAS locus were produced in phase. (3) the sRNA genomic matches were ≤ 10. The PHAS loci identification was done separately for each sample, and results were merged to a non-redundant list based on the genomic coordinates. Next, the potential miRNA triggers of PHAS loci were predicted using ‘reverse computation’ method as described in [31].
RNA gel blot
Total RNA was extracted from peel samples of control, cold- and heat-stressed fruit with Plant RNA Isolation Reagent® (Invitrogen). For gel blot, 20 μg of total RNA from banana fruit samples was separated on a 15% denaturing polyacrylamide gel and transferred to Amersham Hybond™-NX membranes (GE Healthcare, Waukesha, WI, USA). RNA was cross linked using EDC (N-(3-dimethylaminopropyl)-N′-ethyl-carbodiimide hydrochloride (Sigma, St Louis, MO, USA). The probes of 21~22-nt DNA oligonucleotides (Additional file 1: Table S7) reverse complementary to banana miRNA candidates were labeled with γ-[32p]-ATP by T4 polynucleotide kinase (NEB, Ipswich, MA, USA). Non-incorporated nucleotides were removed using microspin G-25 column (GE Healthcare, Buckinghamshire, UK). Blots were also probed with a DNA probe complementary to U6 to confirm uniform loading. The prepared membrane filters were hybridized at 42 °C overnight, then washed twice at 55 °C with washing buffer containing 2 × SSC and 2% SDS. Membranes were then exposed to phosphorscreens and scanned with a Typhoon TRIO Variable Mode Imager (GE Healthcare). Membrane exposure time was adjusted, dependent on signal intensity.
RLM-5’-race
Five micrograms of total RNA isolated from banana peel was used for ligating 5’ RNA adaptors at 37 °C and then reverse transcription at 42 °C for nested PCR for 5′ RLM-RACE, following the manufacturer’s instructions for FirstChoice RLM-RACE Kit (Ambion, Austin, TX, USA). Gene-specific primers (Additional file 1: Table S7) were designed to conduct nested PCRs, and PCR products were gel purified, cloned into the pMD18-T vector (TaKaRa) and sequenced.
Quantitative real-time PCR validation of differentially expressed miRNAs and their target genes
Quantitative real-time PCR was carried out using the same RNA samples used for gel blot analysis. Total RNA (1 μg) was treated with DNase I and reverse transcribed with PrimeScript™ RT Kit (Takara), according to the manufacturer’s instructions but using specific stem-loop RT primers for miRNAs and oligo-dT primer for target mRNAs (Additional file 1: Table S7). Real-time PCR analysis was carried out using SYBR® Premix Ex Taq™ II (TaKaRa) on an ABI 7500 PCR System (Applied Biosystems), according to the standard protocol. The analysis was performed using three independent cDNA preparations and triplicate PCR reactions. The relative expression was calculated using 2-(∆∆Ct) method with 5 s ribosomal RNA (5 s rRNA) and Actin as references for miRNAs and target genes, respectively.
Additional file
Table S1. Statistics of sRNA sequences from banana. Table S2. Summary of known miRNAs in banana. Table S3. Summary of species-specific miRNAs in banana. Table S4. Summary of differentially expressed miRNAs in respond to temperature stress in banana. Table S5. Target genes of known miRNAs in banana. Table S6. PHAS loci in banana. Table S7. Probes for RNA gel blot and primers for qPCR and RLM-5’-RACE. (XLSX 83 kb)
Acknowledgements
We would like to thank Prof. Rui Xia (South China Agricultural University, China) for the help of PHAS loci analysis and valuable suggestion on the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (31772371 and 31301822), and Natural Science Foundation of Guangdong Province (2015A030313686). The work was also supported by Guangdong Provincial Key Laboratory of Applied Botany, Key Laboratory of Plant Resource Conservation and Sustainable Utilization, CAS, and Key Laboratory of Post-Harvest Handling of Fruits, Ministry of Agriculture.
Availability of data and materials
All raw sequencing data were deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE77590.
Abbreviations
- AGO
Argonaute
- AS
Alignment score
- DCL
Dicer-like
- miRNAs
MicroRNAs
- nt
Nucleotide
- PHAS loci
Phased siRNA-generating loci
- phasiRNAs
Phased siRNAs
- qRT-PCR
Quantitative reverse transcription real-time PCR
- RISC
RNA-induced silencing complex
- RLM-5’-RACE
RNA ligase-mediated rapid amplification of cDNA 5′ end
- RPM
Reads per million genome-matched reads
- sRNAs
Small RNAs
- tpb
Transcripts per billion
Authors’ contributions
HZ designed the research, analyzed data and wrote the manuscript draft. YZ collected plant materials, measured fruit firmness and performed RT-qPCR. RT performed RNA gel blot. HQ and XD contributed the reagents and analytical tools. YJ edited and revised the manuscript. All of the authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sreedharan S, Shekhawat UK, Ganapathi TR. MusaSAP1, a A20/AN1 zinc finger gene from banana functions as a positive regulator in different stress responses. Plant Mol Biol. 2012;80(4–5):503–517. doi: 10.1007/s11103-012-9964-4. [DOI] [PubMed] [Google Scholar]
- 2.Zhao ML, Wang JN, Shan W, Fan JG, Kuang JF, Wu KQ, Li XP, Chen WX, He FY, Chen JY, et al. Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 2013;36(1):30–51. doi: 10.1111/j.1365-3040.2012.02551.x. [DOI] [PubMed] [Google Scholar]
- 3.Yang X, Pang X, Xu L, Fang R, Huang X, Guan P, Lu W, Zhang Z. Accumulation of soluble sugars in peel at high temperature leads to stay-green ripe banana fruit. J Exp Bot. 2009;60(14):4051–4062. doi: 10.1093/jxb/erp238. [DOI] [PubMed] [Google Scholar]
- 4.Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 5.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc Natl Acad Sci U S A. 2004;101(34):12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320(5880):1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 7.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121(2):207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127(3):565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 9.Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25(7):2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, Gonzalez AJ, Yan Z, Kitto SL, Grusak MA, et al. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25(23):2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20(24):3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19(18):2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arikit S, Xia R, Kakrana A, Huang K, Zhai J, Yan Z, Valdes-Lopez O, Prince S, Musket TA, Nguyen HT, et al. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell. 2014;26(12):4584–4601. doi: 10.1105/tpc.114.131847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu H, Xia R, Zhao B, An YQ, Dardick CD, Callahan AM, Liu Z. Unique expression, processing regulation, and regulatory network of peach (Prunus persica) miRNAs. BMC Plant Biol. 2012;12:149. doi: 10.1186/1471-2229-12-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sunkar R, Li YF, Jagadeeswaran G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012;17(4):196–203. doi: 10.1016/j.tplants.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Sunkar R, Zhu JK. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell. 2004;16(8):2001–2019. doi: 10.1105/tpc.104.022830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Zhang Y, Ren Y, Xu J, Zhang Z, Wang Y. Genome-wide identification of cold-responsive and new microRNAs in Populus tomentosa by high-throughput sequencing. Biochem Biophys Res Commun. 2012;417(2):892–896. doi: 10.1016/j.bbrc.2011.12.070. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Xu Y, Huan Q, Chong K. Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics. 2009;10:449. doi: 10.1186/1471-2164-10-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhu X, Chen X, Song C, Zou Z, Wang Y, Wang M, Fang W, Li X. Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 2014;14:271. doi: 10.1186/s12870-014-0271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruszka K, Pacak A, Swida-Barteczka A, Nuc P, Alaba S, Wroblewska Z, Karlowski W, Jarmolowski A, Szweykowska-Kulinska Z. Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J Exp Bot. 2014;65(20):6123–6135. doi: 10.1093/jxb/eru353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu F, Wang W, Sun X, Liang Z, Wang F. Conserved and novel heat stress-responsive microRNAs were identified by deep sequencing in Saccharina japonica (Laminariales, Phaeophyta) Plant Cell Environ. 2015;38(7):1357–1367. doi: 10.1111/pce.12484. [DOI] [PubMed] [Google Scholar]
- 22.Yu X, Wang H, Lu Y, de Ruiter M, Cariaso M, Prins M, van Tunen A, He Y. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J Exp Bot. 2012;63(2):1025–1038. doi: 10.1093/jxb/err337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bi F, Meng X, Ma C, Yi G. Identification of miRNAs involved in fruit ripening in Cavendish bananas by deep sequencing. BMC Genomics. 2015;16:776. doi: 10.1186/s12864-015-1995-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chai J, Feng R, Shi H, Ren M, Zhang Y, Wang J. Bioinformatic identification and expression analysis of banana microRNAs and their targets. PLoS One. 2015;10(4):e0123083. doi: 10.1371/journal.pone.0123083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Hont A, Denoeud F, Aury JM, Baurens FC, Carreel F, Garsmeur O, Noel B, Bocs S, Droc G, Rouard M, et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature. 2012;488(7410):213–217. doi: 10.1038/nature11241. [DOI] [PubMed] [Google Scholar]
- 26.Wen JZ, Liao JY, Zheng LL, Xu H, Yang JH, Guan DG, Zhang SM, Zhou H, Qu LH. A contig-based strategy for the genome-wide discovery of microRNAs without complete genome resources. PLoS One. 2014;9(2):e88179. doi: 10.1371/journal.pone.0088179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, et al. Criteria for annotation of plant MicroRNAs. Plant Cell. 2008;20(12):3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pantaleo V, Szittya G, Moxon S, Miozzi L, Moulton V, Dalmay T, Burgyan J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010;62(6):960–976. doi: 10.1111/j.0960-7412.2010.04208.x. [DOI] [PubMed] [Google Scholar]
- 29.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25(1):130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia R, Zhu H, An YQ, Beers EP, Liu Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 2012;13(6):R47. doi: 10.1186/gb-2012-13-6-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia R, Meyers BC, Liu Z, Beers EP, Ye S. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell. 2013;25(5):1555–1572. doi: 10.1105/tpc.113.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36(12):1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 33.Felippes FF, Schneeberger K, Dezulian T, Huson DH, Weigel D. Evolution of Arabidopsis thaliana microRNAs from random sequences. RNA. 2008;14(12):2455–2459. doi: 10.1261/rna.1149408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piriyapongsa J, Jordan IK. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA. 2008;14(5):814–821. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maher C, Stein L, Ware D. Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 2006;16(4):510–519. doi: 10.1101/gr.4680506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 37.Quint M, Gray WM. Auxin signaling. Curr Opin Plant Biol. 2006;9(5):448–453. doi: 10.1016/j.pbi.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang R, Estelle M. Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr Opin Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Megha S, Basu U, Kav NNV. Regulation of low temperature stress in plants by microRNAs. Plant Cell Environ. 2018;41(1):1–15. doi: 10.1111/pce.12956. [DOI] [PubMed] [Google Scholar]
- 40.Hivrale V, Zheng Y, Puli COR, Jagadeeswaran G, Gowdu K, Kakani VG, Barakat A, Sunkar R. Characterization of drought- and heat-responsive microRNAs in switchgrass. Plant Sci. 2016;242:214–223. doi: 10.1016/j.plantsci.2015.07.018. [DOI] [PubMed] [Google Scholar]
- 41.Kumar RR, Pathak H, Sharma SK, Kala YK, Nirjal MK, Singh GP, Goswami S, Rai RD. Novel and conserved heat-responsive microRNAs in wheat (Triticum aestivum L.) Funct Integr Genomics. 2015;15(3):323–348. doi: 10.1007/s10142-014-0421-0. [DOI] [PubMed] [Google Scholar]
- 42.Liu HH, Tian X, Li YJ, Wu CA, Zheng CC. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14(5):836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Sun YF, Song N, Wei JP, Wang XJ, Feng H, Yin ZY, Kang ZS. MicroRNAs involving in cold, wounding and salt stresses in Triticum aestivum L. Plant Physiol Biochem. 2014;80:90–96. doi: 10.1016/j.plaphy.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 44.Stief A, Altmann S, Hoffmann K, Pant BD, Scheible WR, Baurle I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell. 2014;26(4):1792–1807. doi: 10.1105/tpc.114.123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birkenbihl RP, Jach G, Saedler H, Huijser P. Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol. 2005;352(3):585–596. doi: 10.1016/j.jmb.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Gandikota M, Birkenbihl RP, Hohmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3’ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49(4):683–693. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- 47.Yang C, Li D, Mao D, Liu X, Ji C, Li X, Zhao X, Cheng Z, Chen C, Zhu L. Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.) Plant Cell Environ. 2013;36(12):2207–2218. doi: 10.1111/pce.12130. [DOI] [PubMed] [Google Scholar]
- 48.Palatnik JF, Wollmann H, Schommer C, Schwab R, Boisbouvier J, Rodriguez R, Warthmann N, Allen E, Dezulian T, Huson D, et al. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell. 2007;13(1):115–125. doi: 10.1016/j.devcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 49.Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell. 2004;14(6):787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 50.Yao F, Zhu H, Yi C, Qu H, Jiang Y. MicroRNAs and targets in senescent litchi fruit during ambient storage and post-cold storage shelf life. BMC Plant Biol. 2015;15:181. doi: 10.1186/s12870-015-0509-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18(8):2051–2065. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fujii H, Chiou TJ, Lin SI, Aung K, Zhu JK. A miRNA involved in phosphate-starvation response in Arabidopsis. Curr Biol. 2005;15(22):2038–2043. doi: 10.1016/j.cub.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 53.Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, Arora R, Bragg JN, Vogel JP, Singh S, et al. Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci U S A. 2011;108(42):17550–17555. doi: 10.1073/pnas.1113971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang YC, Yu Y, Wang CY, Li ZY, Liu Q, Xu J, Liao JY, Wang XJ, Qu LH, Chen F, et al. Overexpression of microRNA OsmiR397 improves rice yield by increasing grain size and promoting panicle branching. Nat Biotechnol. 2013;31(9):848–852. doi: 10.1038/nbt.2646. [DOI] [PubMed] [Google Scholar]
- 55.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31(13):3429–31. [DOI] [PMC free article] [PubMed]
- 56.Chen C, Xia R, Chen H, He Y. TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. 2018:289660. 10.1101/289660.
- 57.Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci U S A. 2007;104(9):3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Statistics of sRNA sequences from banana. Table S2. Summary of known miRNAs in banana. Table S3. Summary of species-specific miRNAs in banana. Table S4. Summary of differentially expressed miRNAs in respond to temperature stress in banana. Table S5. Target genes of known miRNAs in banana. Table S6. PHAS loci in banana. Table S7. Probes for RNA gel blot and primers for qPCR and RLM-5’-RACE. (XLSX 83 kb)
Data Availability Statement
All raw sequencing data were deposited in the NCBI Gene Expression Omnibus (GEO) under accession number GSE77590.