Abstract
Background
Development of autoimmune diseases is the result of a complex interplay between hereditary and environmental factors, with multiple genes contributing to the pathogenesis in human disease and in experimental models for disease. The T-box protein 3 is a transcriptional repressor essential during early embryonic development, in the formation of bone and additional organ systems, and in tumorigenesis.
Methods
With the aim to find novel genes important for autoimmune inflammation, we have performed genetic studies of collagen-induced arthritis (CIA), a mouse experimental model for rheumatoid arthritis.
Results
We showed that a small genetic fragment on mouse chromosome 5, including Tbx3 and three additional protein-coding genes, is linked to severe arthritis and high titers of anti-collagen antibodies. Gene expression studies have revealed differential expression of Tbx3 in B cells, where low expression was accompanied by a higher B cell response upon B cell receptor stimulation in vitro. Furthermore, we showed that serum TBX3 levels rise concomitantly with increasing severity of CIA.
Conclusions
From these results, we suggest that TBX3 is a novel factor important for the regulation of gene transcription in the immune system and that genetic polymorphisms, resulting in lower expression of Tbx3, are contributing to a more severe form of CIA and high titers of autoantibodies. We also propose TBX3 as a putative diagnostic biomarker for rheumatoid arthritis.
Electronic supplementary material
The online version of this article (10.1186/s13075-018-1797-3) contains supplementary material, which is available to authorized users.
Keywords: Collagen-induced arthritis, Transcriptional regulation, TBX3, TBX5, Biomarker, Eae39r
Background
Rheumatoid arthritis (RA), affecting 0.5–1% of the world’s population, is characterized by chronic inflammation, which primarily affects the synovial joints leading to tissue damage and physical disability. Furthermore, the consequences of immunological abnormalities and systemic inflammation may lead to premature mortality. The disease follows a heterogeneous clinical course with a gender bias towards women. The etiology mainly remains unknown due to the multifactorial nature and complexity of the disease. Both genetic and environmental factors are reported to play a role in the predisposition to RA [1]. Genes within the major histocompatibility complex (MHC), also called the human leukocyte antigen (HLA) complex, are the strongest genetic factors for RA [2]. In addition, a large number of non-MHC genes have been shown to contribute to RA pathogenesis [3, 4].
Despite the introduction of new, effective therapies for RA, not all patients respond to the available treatments. Therefore, there is an unmet need for novel drugs and better biomarkers that can improve early detection, determine prognosis and disease activity, stratify patients for various therapeutic options, and monitor the response to therapy [5]. Novel biomarkers should be robust in their association with disease activity and offer predictive value in selection of appropriate therapy.
Experimental animal models, providing a genetically homogeneous and well-controlled experimental environment, are invaluable tools in genetic studies and drug and biomarker discovery in RA, whereby collagen-induced arthritis (CIA) is the most commonly used model [6]. After immunization with bovine collagen type II (CII), the B10.RIII mouse strain develops CIA, whereas the MHC-matched RIIIS/J mouse strain is resistant to arthritis development. The two particular mouse strains are therefore suitable for studies of non-MHC genes involved in the development of CIA [7, 8]. A strategy involving gene-segregating crosses between the two mouse strains, and development of congenic mice, allows for identification of novel non-MHC candidate genes and related disease pathways [9].
The genetic locus Eae39, located on mouse chromosome 5, was previously linked to experimental autoimmune encephalomyelitis (EAE), an experimental model for multiple sclerosis. Subsequently, it was also shown to influence CIA development and severity [8, 10]. Eae39 congenic and sub-congenic lines were bred on the genetic background of the B10.RIII strain [7]. In the present study, the Eae39 sub-locus, Eae39r, was investigated. This locus comprises four protein-coding genes: Mediator complex subunit 13-like (Med13l), T-box transcription factor 3 (Tbx3), T-box transcription factor 5 (Tbx5), and Probable RNA-binding protein 19 (Rbm19). The MED13L protein is part of the cyclin-dependent kinase (CDK)-8 module of the mediator complex and acts as a key regulator of RNA polymerase II-dependent gene transcription [11, 12]. The Rbm19 gene encodes for a protein contributing to ribosomal (r) RNA processing, a key step in ribosome biogenesis [13]. It belongs to a group of RNA binding proteins (RBPs) that have been associated with neurological disorders, cancers and inflammatory diseases [14]. However, the exact function and role of Rbm19 in disease associations have not been well-studied.
The T-box genes Tbx3 and Tbx5 are closely linked and share 98% and 96% homology between mouse and human, respectively [15]. TBX5 acts as a transcription activator [16], whereas TBX3 functions predominantly as a transcriptional repressor. Nevertheless, TBX3 contains both activation and repression domains that are suggested to be modulated in different cellular milieus [17, 18]. Among other functions, both proteins are involved in bone development [18–20] and remodeling [21]. Mutations in the human TBX3 and TBX5 genes cause ulnar-mammary syndrome (UMS, OMIM 181450) and Holt-Oram syndrome (HOS, OMIM 142900), respectively. Both syndromes cause defects in limb development [16, 22], which asserts their link to bone pathways recently recognized to interact with immune pathways [23]. However, the role of the TBX3 and TBX5 proteins in the immune system, their link to RA, and their biomarker potential, remains unexplored.
With the aim to elucidate the role of the Eae39r genetic locus in autoimmune inflammation, we investigated CIA pathogenesis in B10.RIIIS/J-Eae39r congenic and sub-congenic mice, followed by studies of polymorphisms, gene transcription, and protein expression. We report that mice with allelic variants in the upstream and downstream regions of the Tbx3, Tbx5, and Rbm19 genes, develop more severe CIA compared to littermate controls. In addition, these mice develop significantly enhanced anti-CII antibody responses and have altered B cell response upon activation in vitro. In the present study, we identified reduced expression of Tbx3 in the spleens of sub-congenic mice and showed that activation of B cells is concomitant with a changed level of active TBX3 protein. This study gives novel insight into CIA candidate genes and provides the first evidence for TBX3 as an RA candidate gene and putative RA biomarker, thereby furthering our understanding of the disease pathogenesis.
Methods
Mice
BR.RIIIS/J-Eae39r congenic mice were produced by introduction of the Eae39r fragment from the CIA-resistant RIIIS/J donor strain, purchased from Jackson Laboratory (Bar Harbor, ME, USA), to the CIA susceptible B10.RIII background strain, provided by J. Klein (Tübingen, Germany), as previously described [8]. The sub-congenic line BR.RIIIS/J-Eae39r2 was produced by further inter-crossing heterozygous BR.RIIIS/J-Eae39r mice. All mice were kept and bred under standard conditions in the animal facility at the Department of Drug Design and Pharmacology, Faculty of Health and Medical Sciences, University of Copenhagen, Denmark. The Danish Animal Experiment Inspectorate license number is 2010/561–1920 and 2015-15-0201-00794.
Genotyping
Genomic DNA (gDNA) was purified from mouse ear biopsies with High Pure PCR template preparation kit (11796828001; Roche Holding AG, Basel, Switzerland) according to the manufacturer’s protocol. Purified gDNA was used for genotyping by high-resolution melting (HRM) SNP genotyping for rs33583463 (5:118596773 bp; mouse genome assembly GRCm38) and rs29824716 (5:120043597 bp; mouse genome assembly GRCm38), and by PCR-agarose gel electrophoresis method for the homemade microsatellite marker D5tbxhm17 (5:119660373 bp; mouse genome assembly GRCm38). HRM analysis was carried out on the Roche LightCycler 480 using High Resolution Melting Master (04909631001, Roche Holding AG, Basel, Switzerland) as described elsewhere [26]. Microsatellite marker genotyping was determined by analyzing PCR products on a MegaBACE1000 genotyping system (Amersham Biosciences, Little Chalfont, UK) or on a 3% agarose gel as previously described [9]. Primer sequences are provided in the Additional file 1: Table S1. Based on genotype, mice were allocated into homozygous congenic, sub-congenic or wild type control groups for various experiments.
Induction and evaluation of CIA
CIA was induced in 8–10 weeks old male mice by intra-dermal injection of 100 μg bovine CII (CII-7806; Sigma-Aldrich, St Louis, MO, USA) emulsified in incomplete Freund’s adjuvant (IFA) (F5506; Sigma-Aldrich, St Louis, MO, USA) at the base of the tail (day 0) followed by a booster dose of 50 μg CII emulsified in IFA on day 35. Clinical disease was monitored in a blinded manner three times a week, whereby arthritis was evaluated based on inflammation (erythema and swelling) of affected joints. Each mouse received a score according to a scoring system whereby each inflamed toe (first phalanx excluded), all inflamed knuckles and inflamed wrist or ankle were assigned one point giving a maximum of 6 points per paw and a theoretical maximum score of 24 per mouse. According to the Danish Animal Experiment Inspectorate approved humane endpoints, mice receiving scores above 10 were euthanized. The experiment was terminated when no mice (irrespective of group) developed arthritis in previously non-affected paws on three consecutive observations or when the groups were left too small for adequate statistical power.
For comparison of CIA development between congenic, sub-congenic and control groups, arthritis scores for mice belonging to a particular genotype were analyzed together in order to calculate mean score versus days after immunization (disease progression), mean maximum score (disease severity), incidence, ethical survival, mean day of onset, and area under the curve (AUC; the sum of scores during the course of the disease).
A total of 24 male B10.RIII mice were used for the time-course CIA study, whereby 20 mice were immunized and 4 were left unimmunized. CIA was induced on day 0, by subcutaneous immunization with 100 μg CII emulsified in IFA containing 50 μg Mycobacterium tuberculosis H37Ra (DIFCO laboratories, Detroit, MI, USA) and boosted on day 35 with 50 μg CII in IFA. CIA was scored as aforementioned and four mice were sacrificed on different days of the experiment, covering pre-clinical (day 7, 15, and 25), onset/progressive (day 42) and late (day 68) phases of CIA. Blood was collected for serum analysis, paws were snap-frozen in liquid nitrogen for later analysis and spleen was used for isolating CD19+ B cells for DNA activity assay.
Enzyme-linked immunosorbent assay (ELISA)
For anti-CII ELISA, serum was prepared from blood collected on days 0 and 15 (by submandibular bleeding), and on the last day of CIA (by cardiac puncture). The levels of CII-specific IgM, IgG1, IgG2c, and IgG3 antibodies were determined as previously described [8]. Briefly, serum dilutions were applied to 96-well micro-titer plates (Nunc maxisorp, Roskilde, Denmark) coated overnight with CII in PBS (0.5 μg/well) and subsequently blocked with 1% BSA/PBS solution. The antigen-antibody binding signal was revealed by biotinylated secondary antibodies: goat anti-mouse IgM (1020–08), anti-IgG1 (1070–08), anti-IgG2c (1079–08), and anti-IgG3 (1100–08) (SouthernBiotech, Birmingham, AL, USA) and Avidin-HRP (554058, BD Pharmingen, San Jose, CA, USA), followed by detection with 2,2′-Azinobis [3-ethylbenzothiazoline-6-sulfonic acid]-diammonium salt (ABTS) substrate (A1888; Sigma-Aldrich, St Louis, MO, USA). A SpectraMax Microplate Reader (Molecular Devices Corporation, Sunnyvale, CA, USA) was used to read the absorbance at a wavelength of 405 nm with a wavelength correction set to 492 nm. Pooled serum from arthritic mice were used as a standard and the antibody levels were measured as arbitrary concentrations.
For interleukin-2 (IL-2) cytokine ELISA, supernatants from triplicate CD4+ T lymphocyte cultures were collected at 48 h and analyzed by standard protocol for sandwich ELISA described elsewhere (BD Biosciences, Immune function analysis application handbook).
Sequencing and bioinformatics analysis
The borders of the congenic and sub-congenic fragments were mapped using the aforementioned genotyping markers, spanning the Eae39r fragment, and were fine-mapped by sequencing the genetic regions around those borders by Sanger sequencing (GATC biotech, Germany). The genomic locations of the borders and of single nucleotide polymorphisms (SNPs) used for fine-mapping are shown in Fig. 1. Using the defined borders, genetic elements located in the congenic and sub-congenic fragments, their biotype [27] and expression details [28] were determined, followed by literature mining. PubMed [29] and Google Scholar [30] databases were searched using the keywords collagen-induced arthritis, rheumatoid arthritis, autoimmunity, inflammation, lymphocytes, and cell signaling. The retrieved information was used to prioritize the genes for further studies.
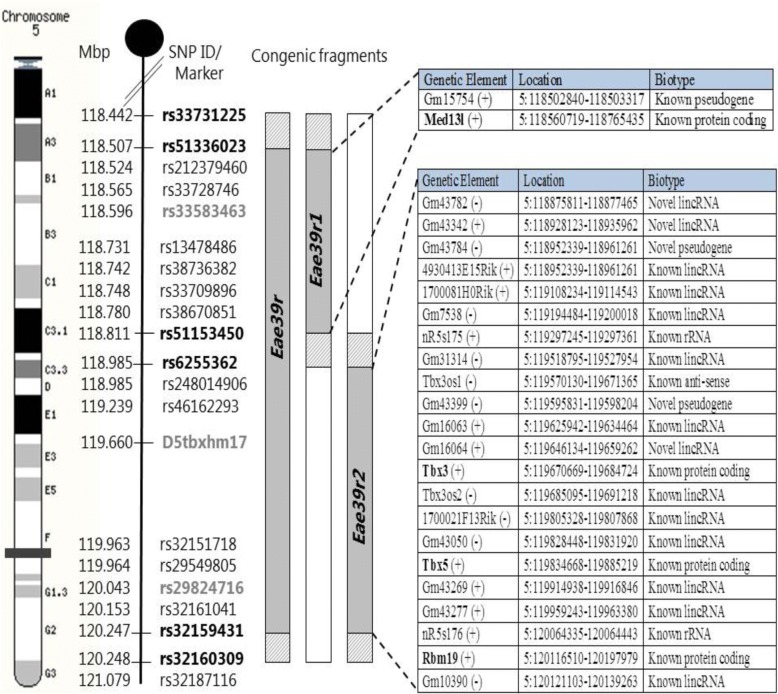
Fig. 1.
Markers defining the location of the Eae39r locus, and the Eae39r1 and Eae39r2 sub-loci on mouse chromosome 5, and the genetic elements included in the Eae39r region. SNP Ids in bold (black) represent the borders of the congenic and sub-congenic fragments. The SNP Ids/microsatellite markers shown in gray were used for genotyping. Solid gray represents RIIIS/J alleles bred on to the B10.RIII background (white), while the diagonally lined gray represents regions of unknown genotype. The genetic elements defining the Eae39r fragment are shown in tabular form along with their location and biotype (according to Ensembl release 74) and protein coding genes are bolded. Mbp, mega base pair; SNP, single nucleotide polymorphism; lincRNA, long intervening noncoding RNA; rRNA, ribosomal RNA. (+) = forward strand, (−) = reverse strand
To identify genetic variations in protein coding genes located in the Eae39r2 sub-congenic fragment, Sanger sequencing (GATC biotech, Germany), followed by computational analysis, was performed. All coding exons and regulatory regions of the Tbx3 and Tbx5 genes, including promoters (2 kb upstream of transcriptional start site), 5’ UTR, and 3’ UTR, in addition to regions containing documented non-synonymous SNPs for the Rbm19 gene were sequenced (primers are listed in Additional file 1: Table S2). The genomic locations of these regions, based on mouse genome build GRCm38, were retrieved from Ensembl [27].
The identified variants were classed based on their relationship to genes in the dbSNP [31] and Ensembl [27] databases. Furthermore, web-based prediction tools were used to check for possible effect of intronic and 5’/3’UTR variants on transcription factor binding [32, 33] and micro RNA (miRNA) binding [34], respectively. For prediction of transcription factor binding site (TFBS) 20 nt region around the confirmed SNPs, between B10.RIII and B10.RIIIS/J-Eae39r2 mice, was checked for difference in binding using two prediction tools, CONSITE [32] and PROMO [33]. The disruption/introduction of TFBS was reported with respect to background strain i.e. B10.RIII.
Isolation of B and T lymphocytes
Mouse spleens were harvested and mashed through a 40-μm cell strainer to prepare single-cell suspensions followed by depletion of red blood cells using BD Lysing Buffer (555899; BD Biosciences, San Jose, CA, USA). CD19+ B lymphocytes for cell proliferation assay, real time PCR, and TBX3 activity assay, were positively selected from splenic cell suspensions by using the EasySep™ CD19 selection kit (18954; Stemcell Technologies, Vancouver, Canada) according to the manufacturer’s recommended protocol. Pan T lymphocytes for real time PCR and CD4+ T lymphocytes for cell proliferation assay were isolated from the CD19− flow-through using EasySep™ T cell isolation kit (19851; Stemcell Technologies, Vancouver, Canada) and EasySep™ CD4+ T cell enrichment kit (19752; Stemcell Technologies, Vancouver, Canada), respectively. Cell purity was > 90% for CD19+ B cells and > 92% for pan T and CD4+ T cells, as determined by flow cytometry.
Cell cultures, stimulation, and proliferation assay
Purified B lymphocytes were plated at 3 × 105 per well in round-bottomed 96-well cell culture plates (Corning, New York, USA). The cells were cultured in complete Dulbecco’s modified eagle’s medium (DMEM) GlutaMAX-I (61965026), 5% heat-inactivated fetal bovine serum (FBS- 10082139), 1 mM HEPES (15630056), 50 μM 2-Mercaptoethanol (31350010), and 1% penicillin/streptomycin (10378016; Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), alone or in the presence of 0–10 μg/ml lipopolysaccharide (LPS) (L2880; Sigma-Aldrich, St Louis, MO, USA), 0–40 μg/ml goat anti-mouse IgM F (ab’)2 (115–006-075; Jackson ImmunoResearch Laboratories, Inc. Baltimore, PA, USA) or a combination of 0–4 μg/ml anti-mouse CD40 antibody, clone 1C10 (14–0401; eBioscience, San Diego, CA, USA) and 10 ng/ml mouse IL-4 recombinant protein (14–8041; eBioscience, San Diego, CA, USA). For CD4+ T cells, 105 purified cells per well were stimulated with plate-bound purified anti-mouse CD3 antibody, 0-3 μg/ml, clone 145-2C11 (550275; BD Biosciences, San Jose, CA, USA) and anti-mouse CD28 antibody, fixed at 3 μg/ml, clone 37.51 (16–0281; eBioscience, San Diego, CA, USA). After culturing for 48 h at 37 °C in 5% CO2, the cells were pulsed for 16–18 h with 1 μCi per well 3H-thymidine (Perkin Elmer, Waltham, MA, USA), and incorporation was measured on a TopCount Scintillation Counter (Perkin Elmer, Waltham, MA, USA) as counts per minute (cpm).
For the time-course CIA, purified B cells from CII immunized mice, at the indicated time points, were cultured for 48 h in complete DMEM 5% FCS at 37 °C, 5% CO2 with or without stimulation with a fixed concentration of anti-mouse IgM F (ab’)2 (10 μg/ml).
Flow cytometric analysis
Single-cell suspensions of spleen tissue were prepared in complete DMEM as described above: 1 × 106 cells were pre-incubated with 1 μg of Fc receptor block (anti-CD16/CD32 antibody clone 93; 14–0161-81, eBioscience, San Diego, CA), prior to staining with 0.1 μg of antibodies for 15 min on ice. Rat anti-mouse CD19 fluorescein isothiocyanate (anti CD19-FITC) antibody clone 1D3 (553785), anti-mouse CD4-phycoerytherin (anti CD4-PE) antibody clone GK1.5 (553730), and anti-mouse CD8-phycoerytherin-cyanine 5 (anti CD8-PE-Cy5) antibody clone RPA-T8 (555368), all purchased from BD Biosciences (San Jose, CA, USA) were used. The stained cells were analyzed using Gallios Flow Cytometer (Beckman Coulter, Brea, CA, USA) and FlowJo software (Tree Star Inc., Ashland, OR, USA).
Reverse transcriptase-polymerase chain reaction (RT-PCR)
For transcript quantification, snap-frozen whole ankle joints (including synovium, adjacent tissues, and bones) were pulverized using a mortar and pestle, followed by homogenization with IKA T-10 Ultra-Turrax homogenizer (IKA-Works, Staufen im Breisgau, Germany). Snap-frozen spleen tissue was also homogenized with IKA T-10 Ultra-Turrax homogenizer. Total RNA was extracted from joint and spleen homogenates using the RNeasy Plus Universal Mini Kit (73404; Qiagen, Hilden, Germany) and from purified splenic B and T lymphocytes using the RNeasy Micro Kit (74034; Qiagen, Hilden, Germany) followed by removal of gDNA remnants by Turbo DNA free kit (AM1907; Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The concentration and purity of total RNA was measured on the NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA), while integrity was verified on a 1% agarose gel. Complementary DNA (cDNA) was synthesized from 1 μg of extracted RNA using Superscript III First Strand Synthesis Kit (11752–250; Invitrogen, Carlsbad, CA, USA) and SYBR green-based real time PCR was performed with PrecisionPlus qPCR Mastermix (Primerdesign Ltd., Eastleigh, UK) and LightCycler480 Real-Time PCR System (Roche, Mannheim, Germany). Relative quantification was performed by calculating 2^(−delta delta CT) (2-ΔΔCT), whereby ΔΔCT was taken as the difference in normalized crossing threshold (CT) values between experimental (sub-congenic) and control samples, and the CT values of both groups were normalized to Ubiquitin C (Ubc) as a housekeeping gene. Primer sequences are listed in Additional file 1: Table S3.
DNA binding activity assay for TBX3
TBX3 activity was measured in the nuclear lysate of unstimulated and B cell receptor (BCR)-stimulated CD19+ B cells, and joint tissue homogenates using pre-validated TFact™ DNA binding ELISA kit (TFE-7159; Assay Biotechnology, Sunnyvale, CA, USA). The nuclear extraction was performed with few changes while the immunoassay was done according to the manufacturer’s protocol. Briefly, the cells were lysed on ice for 15 min in a hypo-osmotic lysis buffer (10 mM HEPES, 10 mM KCl, 1.5 mM MgCl2, and 0.1 mM DTT) containing protease and phosphatase inhibitors-PPI (05892791001; Roche Diagnostics GmbH, Mannheim, Germany). The nuclear proteins were then extracted in the provided nuclear extraction buffer with PPI by incubating the samples on ice for 40 min and vortexing intermittently. A Pierce BCA Protein Assay (23227; Thermo Fisher Scientific, Waltham, MA, USA) was employed to determine the amount of nuclear proteins in all samples using known concentrations of BSA as standards.
The extracted nuclear samples were applied to the wells pre-coated with oligonucleotides specific for binding TBX3 and incubated overnight at 4 °C. Following incubation with TBX3 primary antibody (for 2 h), and HRP-conjugated secondary antibody (for 1 h), 3, 3′, 5, 5’-tetramethylbenzidine (TMB) substrate was added to allow color development, which was quenched after 20 min by addition of stop solution. Absorbance of each well was determined at a wavelength of 450 nm (correction set to 540 nm) using a SpectraMax Microplate Reader (Molecular Devices Corporation, Sunnyvale, CA, USA). Active TBX3 levels were measured as a relative concentration in comparison to standard curve of nuclear lysate positive control (NLPC) provided in the assay kit.
Serum quantification of TBX3
TBX3 was quantitated in serum samples, from the time-course CIA study, using a pre-validated mouse TBX3 ELISA Kit (MBS2886293; MyBioSource, Inc. San Diego, CA, USA). Standard sandwich ELISA was performed according to the manufacturer’s protocol with few changes in incubation time and temperature to reduce the background signal. Briefly, 100 μl of appropriate dilutions of TBX3 standard (0–2.5 ng/ml) and samples were applied per well to the 96-well microtiter plate pre-coated with TBX3 capture antibody. The plate was incubated overnight at 4 °C followed by addition of the recommended amount of biotin-conjugated detection antibody (detection reagent A). After 2 h incubation at room temperature and washing steps, avidin-HRP (detection reagent B) was added at room temperature for 1 h. TMB substrate was added to allow color development, which was quenched after 20 min by addition of 0.1 M H2SO4 stop solution. Absorbance of each well was determined at a wavelength of 450 nm (correction set to 540 nm) using a SpectraMax spectrophotometer. The sensitivity limit of the assay was 0.091 ng/ml, and the inter-assay and intra-assay coefficient of variation (CV) was ≤ 10%, as reported by the manufacturer.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7.03. All data on CIA (except disease incidence and ethical survival data), ELISA, in vitro proliferation, and TBX3 activity were analyzed using the Mann-Whitney U test. The chi-square test and Fischer’s exact test were applied for analysis of CIA incidence and ethical survival data, respectively. RT-PCR data were analyzed by two-way Student’s t test. P-values less than 0.05 were considered significant in all cases.
Results
The Eae39r2 locus controls arthritis development and anti-CII antibody response
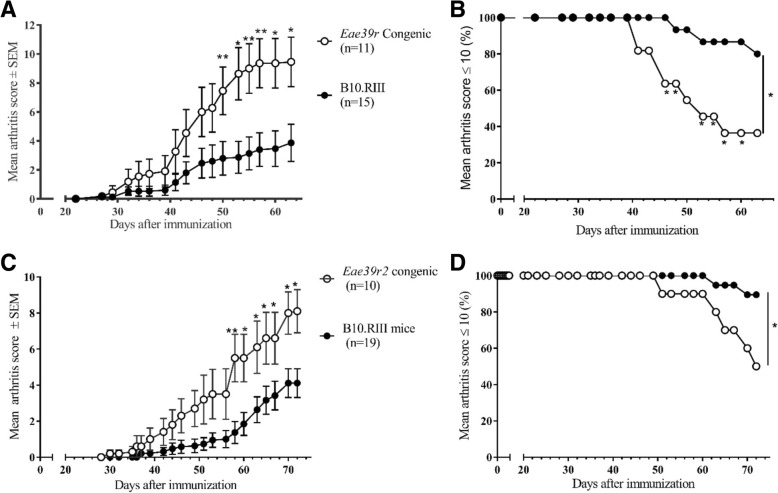
In order to further dissect the genetics of the previously reported non-MHC locus Eae39r [8, 35] and its impact on CIA in mice, we established an B10.RIIIS/J-Eae39r congenic mouse line, carrying the Eae39r fragment from the CIA-resistant RIIIS/J (H-2r) strain on the background genome of the CIA-susceptible B10.RIII (H-2r) strain, and two Eae39r sub-congenic mouse lines: B10.RIIIS/J-Eae39r1 and B10.RIIIS/J-Eae39r2 (Fig. 1). Arthritis development was studied in the B10.RIIIS/J-Eae39r congenic and B10.RIIIS/J-Eae39r2 sub-congenic mice by immunization with bovine CII and the results from the clinical arthritis scoring showed that B10.RIIIS/J-Eae39r congenic and B10.RIIIS/J-Eae39r2 sub-congenic mice developed significantly more severe arthritis compared to their littermate control mice (Fig. 2a-d and Table 1). These results show that the Eae39r2 locus harbors genes that control the development of CIA. Similar results were obtained in a second independent CIA study comparing the B10.RIIIS/J-Eae39r2 sub-congenic and control mice (data not shown).
Fig. 2.
Collagen-induced arthritis (CIA) development in B10.RIIIS/J-Eae39r and B10.RIIIS/J-Eae39r2 congenic and sub-congenic mice in comparison to littermate controls. a CIA progression in B10.RIIIS/J-Eae39r congenic mice represented as mean arthritis score for each group ± standard error of the mean (SEM). b Ethical survival curve for B10.RIIIS/J-Eae39r congenic mice - based on the animal healthcare inspectorate guidelines that mice attaining a total score > 10 should be euthanized. c CIA progression and d ethical survival in B10.RIIIS/J-Eae39r2 sub-congenic mice. Data were analyzed using the Mann-Whitney U test for disease progression and the chi-squared test for ethical survival; *p < 0.05, **p < 0.01
Table 1.
Collagen-induced arthritis (CIA) phenotypes in Eae39r congenic and Eae39r2 sub-congenic mice
| Disease parameter (phenotype) | Experimental group | |||||
|---|---|---|---|---|---|---|
| Eae39r congenic (n = 11) | Littermate controls (n = 15) | p valuea | Eae39r2 sub-congenic (n = 10) | Littermate controls (n = 19) | p valuea | |
| Incidence | 9/11 (82%) | 8/15 (53%) | 0.2167 | 10/10 (100%) | 15/19 (79%) | 0.2680 |
| Mean max scoreb | 9.7 ± 1.6 | 3.9 ± 1.3 | 0.0041** | 8.30 ± 1.14 | 4.4 ± 0.8 | 0.0118* |
| AUCc | 80.8 ± 18 | 29.7 ± 11.2 | 0.0120* | 67.8 ± 17.6 | 25.7 ± 7.2 | 0.0134* |
| Mean day of disease onset | 42.2 ± 2.6 | 36.3 ± 3.4 | 0.1942 | 47.9 ± 3.4 | 53.6 ± 2.9 | 0.2529 |
| Number of mice euthanizedd | 7/11 (64%) | 3/15 (20%) | 0.0426* | 5/10 (50%) | 2/19 (10.5%) | 0.0302* |
aData on the incidence and number of mice euthanized were analyzed using the chi-squared test and the other variables were analyzed using the Mann-Whitney U test; *p < 0.05, **p < 0.01
bMean max score, indicating collagen-induced arthritis severity, is the mean (± SEM) of the maximum score for all affected mice in the group
cArea under curve (AUC) is the mean (± SEM) of the sum of scores for mice of the corresponding genotype (days 22–63 for Eae39r and days 30–72 for Eae39r2)
dMice having a score above 10 were euthanized during the experiment as per recommendations of animal welfare authorities
The disease-enhancing effect from the Eae39r2 genetic fragment was also accompanied by significantly higher titers of anti-CII antibodies, compared with littermate controls, in both early (day 15) and late (day 72) phases of disease (Table 2). This shows that genes in this region, in addition, control antibody responses to CII, which constitute a major part of the CIA pathogenesis [36].
Table 2.
Anti-collagen type II antibody levels in serum of Eae39r2 sub-congenic mice
| Congenic fragment | Days after collagen type II immunization | Antibody isotype | Antibody titers in groups of mice (AU/ml)a | Statisticsb | |
|---|---|---|---|---|---|
| Homozygous Congenic | Littermate controls | p value | |||
| Eae39r2 | 15 (preclinical phase) | IgG1 | 252 ± 46 | 96 ± 26 | 0.0041** |
| IgG2C | 148 ± 11 | 77 ± 10 | < 0.0001**** | ||
| IgG3 | 7 ± 0.5 | 4 ± 0.3 | 0.0067** | ||
| IgM | 55 ± 3 | 37 ± 4 | 0.0002*** | ||
| 72 (clinical phase) | IgG1 | 5044 ± 1363 | 1852 ± 486 | 0.0002*** | |
| IgG2C | 822 ± 113 | 323 ± 45 | < 0.0001**** | ||
| IgG3 | 9 ± 0.5 | 5 ± 0.4 | < 0.0001**** | ||
| IgM | 11 ± 4 | 9 ± 3 | 0.144 | ||
aArbitrary units per ml (AU/ml) are calculated in comparison to a standard curve of pooled serum from arthritic mice in the same experiment
bData were analyzed using the Mann-Whitney U test; *p < 0.05, **p < 0.01, ***p < 0.001
Tbx3 and Tbx5 are candidate genes in the Eae39r2 locus
Sanger sequencing of documented SNPs from mouse genome databases, in and around the Eae39r locus, allowed the determination of borders of the congenic and sub-congenic fragments as closely as possible, thereby narrowing down the list of genes that could be responsible for the CIA-enhancing effect. The Eae39r fragment spanned over 1.8 Mbp, between rs33731225 (5:118442102 bp) and rs32160309 (5:120248221 bp); and Eae39r2 was defined to be 1.4 Mbp, between rs51153450 (5:118811970 bp) and rs32160309 (5:120248221 bp) (Fig. 1). Since Eae39r2 was the smallest fragment controlling CIA development, further focus of this study was restricted to this fragment. Using online resources, all protein-coding and non-protein-coding genetic elements located within the Eae39r2 locus were listed (Table 3).
Table 3.
Genetic elements located in the Eae39r2 region, their expression, and literature overview
| Genetic Elementa | Expressionb | Literature summary |
|---|---|---|
| Tbx3 (+) | Expressed in colon, lung, heart, liver, adrenal gland and spleen | Controls cell proliferation and differentiation [37, 60, 62]; enhances tumor invasiveness [18]; regulates gene splicing [68]; modulates number of cell signaling pathways involved in the development of limb, mammary gland, and heart [18]; regulates bone formation [69, 70] and bone resorption [71]; downregulated in the early phase of collagen-induced arthritis (CIA) [21]; linked to rheumatoid arthritis (RA) in a genome-wide association study (GWAS) [72]; and modulated in RA twin lymphoblastoid B cell lines compared with healthy co-twin LCLs [73]. |
| Tbx5 (+) | Expressed in heart, lung, bone, and urinary bladder | Controls cell proliferation, differentiation and migration [74, 75]; involved in cell signaling pathways [76–78]; regulates bone growth and maturation [20, 79] and system development [80, 81]; downregulated in the early phase of CIA [21]; linked to osteochondrosis in GWAS [82] and RA by activation of RA synovial fibroblasts [83]. |
| Rbm19 (+) | Expressed in heart, lung, uterus, and skeletal muscle | Key role in synthesis of the small eukaryotic ribosome subunit [84], pre-implantation development [13], and mammary gland morphology [85]. |
| 1700081H04Rik (+) | Expressed in testis, brain, heart, kidney and liver | No data available |
| n-R5s175 (+) | Expressed in kidney, liver, testis, brain, spleen, skeletal tissue, and lung | No data available |
| Tbx3os1 (−) | Expressed in liver, lung, colon, brain, kidney, skeletal muscle, spleen, and testis | No data available |
| Gm16063 (+) | Expressed in liver, lung, colon, brain, kidney, skeletal muscle, spleen, and testis | No data available |
| Gm16064 (+) | Expressed in colon, kidney, lung, spleen, brain, liver, skeletal muscle, and testis | No data available |
| Tbx3os2 (−) | Expressed in kidney, lung, brain, colon, liver, adrenal gland, and spleen | No data available |
| Gm10390 (−) | Expressed in brain, liver, spleen, testis, heart, skeletal muscle, colon, and lung | No data available |
aBased on Ensembl release 74; protein coding genes are shown in bold; (+) indicates genetic element annotated on forward strand and (−) indicates reverse strand
bExpression documented for C57BL6 mouse strain in atlas by European Bioinformatics Institute [28]; organs of high expression are shown in bold
Among all, Tbx3 and Tbx5 were chosen as potential candidate genes based on their conservation between mouse and human, documented role in transforming growth factor-beta (TGF-β) [37], bone morphogenic protein (BMP) [38, 39], Hedgehog (Hh) [18], and Wnt/β-catenin [40, 41] bone signaling pathways, the link of mutations in these genes to human disorders involving bone phenotype [16, 22], and reported expression in spleen and bone of the C57BL6 mouse strain [28].
Genetic variations in regulatory regions of the Tbx3 and Tbx5 genes
Sequencing of the Tbx3 and Tbx5 genes revealed a number of variations in regulatory and intronic regions, but none in the coding region (Table 4). Computational analysis of these variations predicted that some of these might affect promoter function, transcription factor binding, or microRNA (miRNA) binding (Table 4).
Table 4.
Genetic variations between B10.RIII and Eae39r2 sub-congenic mice
| Genetic variations related to the mouse Tbx3 gene | ||||
| SNP ID | Location in genomea | Allelesb | Functional class | Predicted consequencec |
| rs234834055 | 5:119669859 | C/− | Promoter variant | May affect gene expression level |
| rs48522093 | 5:119670996 | C/G | 5’ UTR variant | Introduction of new TFBS for f (alpha)-f (epsilon) (undocumented role) |
| rs221652067 | 5:119674641 | −/AATT | Intronic variant | No TF binding site in this region |
| rs48057748 | 5:119675826 | G/T | Intronic variant | No TF binding site in this region |
| rs260549092 | 5:119677177 | −/A | Intronic variant | No TF binding site in this region |
| rs260388829 | 5:119680214 | C/T | Intronic variant | Disruption of TFBS for c-Rel (member of NF-ĸB TF family, linked to CIA development, and lymphoid cell growth and development [86]) and Sp1 (involved in cellular processes including growth, differentiation and apoptosis) |
| rs223041783 | 5:119681737 | −/(GA)6 | Intronic variant | No TF binding site in this region |
| No SNP id | 5:119684553 | A/− | 3’ UTR variant | Lies very close to mmu-miR-511 binding site (miR-511negatively regulates growth and metastasis of tumor cells in humans [87]) |
| rs254153782 | 5:119684580 | A/C | 3’ UTR variant | No miRNA binding site in this region |
| Genetic variations related to the mouse Tbx5 gene | ||||
| SNP ID | Location in genomea | Allelesb | Functional class | Predicted consequencec |
| rs247725267 | 5:119834384–119,834,391 | (TTCC)2/− | Promoter variant | May affect expression level of gene |
| rs46426184 | 5:119859957 | A/G | Intron variant | No TF binding site in this region |
| rs222502452 | 5:119860635 | C/T | Intron variant | No TF binding site in this region |
| rs32145767 | 5:119860705 | G/A | Intron variant | No TF binding site in this region |
| rs242983646 | 5:119860887 | A/C | Intron variant | Disruption of c-fos binding site (involved in cell proliferation, growth and survival pathways, including bone and immune cells [88]) |
| rs252594029 | 5:119861058 | T/C | Intron variant | No TF binding site in this region |
| rs246838998 | 5:119862700 | A/G | Intron variant | No TF binding site in this region |
| rs213393759 | 5:119862716 | G/A | Intron variant | No TF binding site in this region |
| rs32145773 | 5:119863349 | T/C | Intron variant | Disruption of Sox5 binding site (linked to Th17 cell differentiation [89]) |
| rs32146450 | 5:119867116 | C/T | Intron variant | No TF binding site in this region |
| rs50869445 | 5:119868930 | T/C | Intron variant | No TF binding site in this region |
| rs48190630 | 5:119868943 | T/C | Intron variant | Introduction of new TFBS for C/EBPbeta (regulating the expression of genes involved in immune and inflammatory responses [90]) |
| rs32147927 | 5:119868969 | C/T | Intron variant | No TF binding site in this region |
| rs228127170 | 5:119868977 | G/A | Intron variant | No TF binding site in this region |
| rs49608882 | 5:119869004 | C/T | Intron variant | No TF binding site in this region |
| rs45644322 | 5:119869140 | G/A | Intron variant | No TF binding site in this region |
| rs48943542 | 5:119869144 | G/A | Intron variant | No TF binding site in this region |
| rs6373056 | 5:119871760 | G/A | Intron variant | No TF binding site in this region |
| rs48054187 | 5:119878634 | C/T | Intron variant | No TF binding site in this region |
| rs265584798 | 5:119883860 | C/T | 3’ UTR variant | No miRNA binding site in this region |
| rs255866543 | 5:119884274 | A/G | 3’ UTR variant | Lies very close to mmu-miR-6918-5p binding site (undocumented role) |
| rs232783988 | 5:119885135 | −/G | 3’ UTR variant | No miRNA binding site in this region |
| rs237950985 | 5: 119885225 | C/T | Downstream variant | NA |
| rs237459586 | 5:119885320–119,885,324 | TCTTT/− | Downstream variant | NA |
| rs258422458 | 5:119885423 | C/T | Downstream variant | NA |
Reduced expression of Tbx3 in B lymphocytes
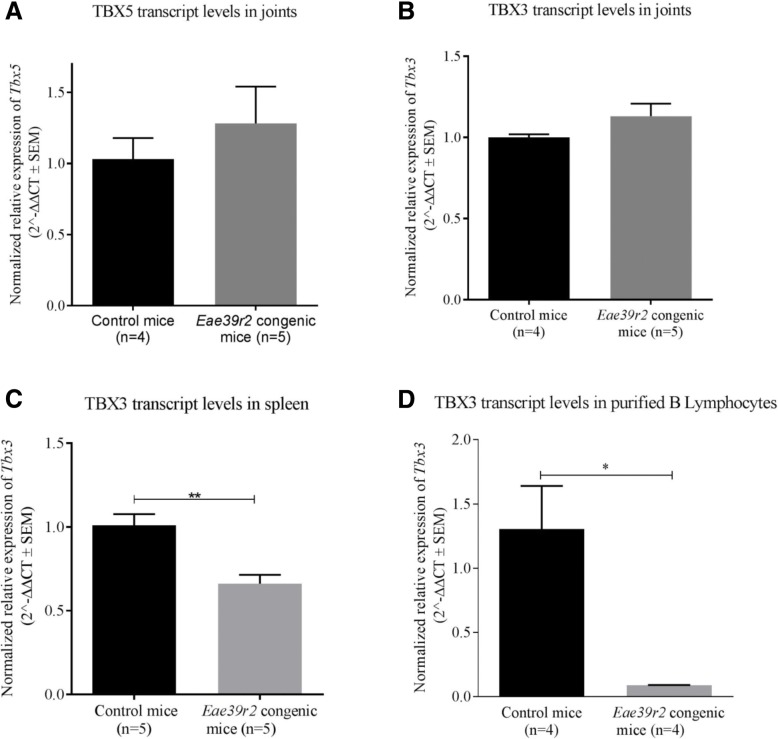
Since variations in the regulatory regions of the genes may affect DNA transcription, mRNA levels of Tbx3 and Tbx5 were measured in the spleens and joints of naïve B10.RIIIS/J-Eae39r2 sub-congenic and littermate control mice. Both Tbx3 and Tbx5 were equally expressed in the joints of naïve sub-congenic and control mice (Fig. 3a, b). On the contrary, the sub-congenic mice had significantly less Tbx3 mRNA in the spleen as compared to the control mice (p = 0.0036, Fig. 3c), while Tbx5 was not detectable in the spleen.
Fig. 3.
Effect of the Eae39r2 sub-congenic fragment on Tbx5 (a) and Tbx3 (b) mRNA levels in the joints, Tbx3 mRNA levels in spleen (c) and purified splenic B lymphocytes (d). Values are presented as mean of 2^-ΔΔCt and bars represent standard error of the mean (SEM). Statistical analyses were performed using two-way Student’s t test; *p < 0.05, **p < 0.01
In order to determine which cell type mainly contributed to the expression of Tbx3 in the spleen, quantitative RT-PCR was carried out on purified B and pan T lymphocytes. We found that Tbx3 expression was only detectable in B lymphocytes, and B10.RIIIS/J-Eae39r2 sub-congenic mice had significantly less Tbx3 transcripts in B lymphocytes as compared to the control mice (p = 0.0286, Fig. 3d).
TBX3 is associated with B cell proliferation but not development of B cells
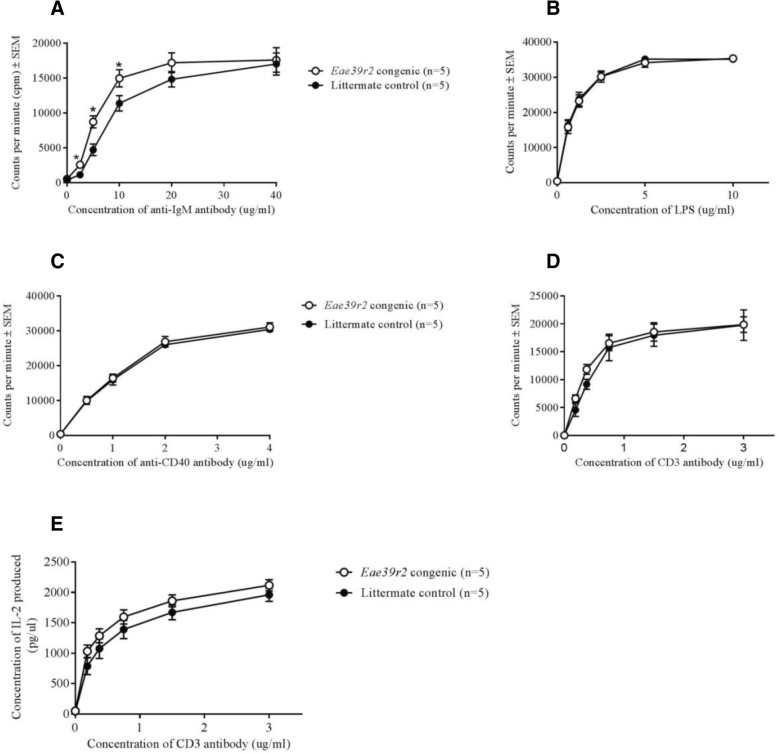
To investigate whether the observed differential expression of Tbx3 in the Eae39r2 sub-congenic mouse spleen was accompanied by altered immune cell activation, an in vitro proliferation assay was performed with purified CD19+ B cells from naïve B10.RIIIS/J-Eae39r2 sub-congenic and control mice. B cell proliferation was significantly increased in the sub-congenic mice in response to anti-IgM stimulation (p = 0.0317, Fig. 4a). However, no difference in proliferation of B cells upon stimulation with either LPS or anti-CD40 antibody/IL-4 was observed (Fig. 4b-c). Furthermore, no difference in proliferation or IL-2 production by purified CD4+ T cells, following stimulation with anti-CD3/CD28 antibodies, was observed when comparing sub-congenic mice with littermate controls (Fig. 4d-e).
Fig. 4.
In vitro proliferative response of CD19+ B cells and CD4+ T cells from B10.RIIIS/J-Eae39r2 sub-congenic mice. 3H-thymidine incorporation, represented as cpm ± SEM, of CD19+ B cells following in vitro stimulation with titrated concentrations (0–40 μg/ml) of anti-IgM antibody (a); titrated concentrations (0–10 μg/ml) of lipopolysaccharide LPS (b); combination of fixed concentration (10 ng/ml) of IL-4 and titrated concentrations (0–4 μg/ml) of anti-CD40 antibody (c). d 3H-thymidine incorporation of CD4+ T cells following in vitro stimulation with combination of titrated concentrations (0–3 μg/ml) of anti-CD3 antibody and fixed concentration of anti-CD28 antibody (3 μg/ml). e IL-2 production (mean ± SEM) in the supernatant of CD4+ T cells stimulated with anti-CD3/CD28 antibodies. Statistical analyses were performed using the Mann-Whitney U test; *p < 0.05
To determine whether the reduced expression of Tbx3 had an effect at the B cell developmental level, the total and relative numbers of splenic B cells in naïve B10.RIIIS/J-Eae39r2 sub-congenic and control mice were determined by automated cell counter and flow cytometry, respectively. No difference in the absolute number of splenic B cells (mean ± SEM, B10.RIIIS/J-Eae39r2 sub-congenic 38.4 × 106 ± 2.8 × 106 (n = 5), control 36.9 × 106 ± 3.0 × 106 (n = 7)) and relative number of splenic B cells (mean ± SEM: B10.RIIIS/J-Eae39r2 sub-congenic 59.9% ± 0.8 (n = 10), control 58.7% ± 0.5 (n = 7)) was found between the two groups of mice, indicating that TBX3 is associated with proliferation of B cells, but not the number.
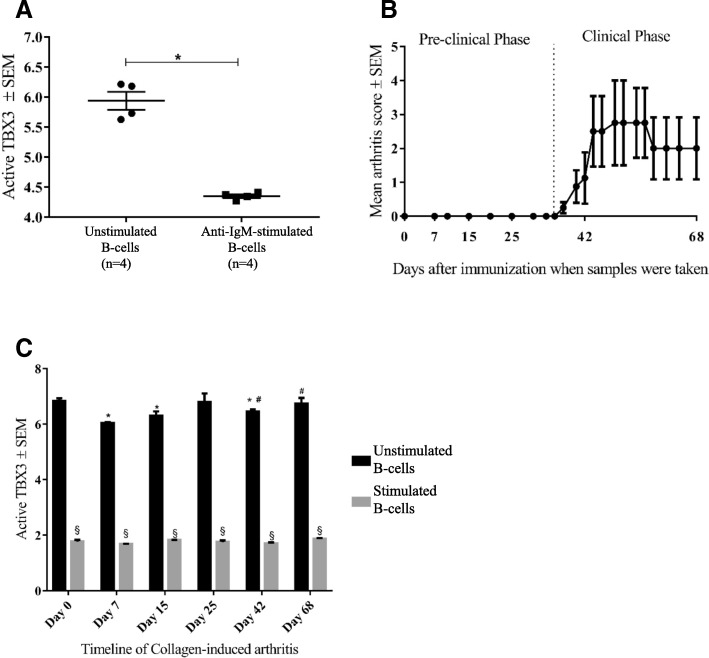
We further analyzed changes in intra-nuclear TBX3 levels in relation to B cell activation and proliferation by comparing the quantity of active TBX3 in unstimulated B cells and in B cells activated through the BCR by anti-IgM antibody. We found that TBX3 activity in B cells was strongly downregulated upon in vitro stimulation with anti-IgM antibody (p = 0.0286; Fig. 5a). These data suggested that TBX3 is involved in BCR downstream pathways and might contribute to humoral immune responses.
Fig. 5.
Quantitative analysis of active TBX3 in nuclear lysates of CD19+ B cells from B10.RIII mice. a Comparison of active TBX3 levels in CD19+ B cells isolated from naïve B10.RIII mice (n = 4), with and without in vitro stimulation with a fixed concentration (10 μg/ml) of anti-IgM antibody for 48 h. b, c Collagen-induced arthritis (CIA) development and corresponding TBX3 activity in B10.RIII mice whereby four mice were killed at indicated time points after collagen type II (CII) immunization and levels of active TBX3 were determined with and without ex vivo re-stimulation with fixed concentration (10 μg/ml) of anti-IgM antibody for 48 h. Statistical analyses were performed using the Mann-Whitney U test; *significant (p < 0.05) compared to day 0 unstimulated samples; #significant compared to day 7 unstimulated samples; §significant compared to respective unstimulated samples
TBX3 activity varies in splenic B cells during the course of CIA
Based on our findings, we sought to examine the activity of TBX3 in B cells during the course of CIA. In that time-course study, mice started to show signs of arthritis on day 37 post immunization, which peaked around day 50 and slightly declined in the late phase, as shown in Fig. 5b. The overall incidence of disease was 75%, which is in accordance with previous reports on the B10.RIII mouse strain [42]. By comparing the intra-nuclear TBX3 activity in purified B cells, we found that the amount of active TBX3 falls significantly in the early pre-clinical phase (days 7 and 15) and early clinical phase (day 42) after receiving the first and booster dose of collagen, respectively (Fig. 5c). Furthermore, a significant decrease in the level of TBX3 was observed upon ex vivo re-stimulation of the B cells with anti-IgM antibody, at all timepoints of sampling in CIA (Fig. 5c). On the contrary, no significant variation in TBX3 activity was found in joint tissue homogenates from the same CIA time-points as for splenic B cells (data not shown).
TBX3 - a potential biomarker of arthritis
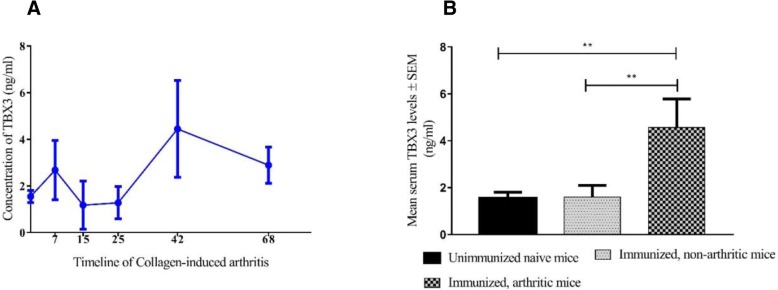
By studying the kinetics of TBX3 protein expression in serum, we made two key observations: first, TBX3 can be measured in mouse serum and second, it is dynamic along the timeline of CIA, with a considerable rise in the clinical phases (days 42 and 68) of disease in comparison to naïve and pre-clinical levels (Fig. 6a). Moreover, we showed that arthritic mice (sick mice from days 42 and 68 samples; n = 6) had significantly higher TBX3 levels in serum than immunized, non-arthritic mice (days 7, 15, 25, and non-arthritic mice from days 42 and 68; n = 14) (p = 0.0087) and naïve control mice (p = 0.0095) Fig. 6b. Spearman’s correlation coefficient was calculated to determine the relationship between the CIA clinical score and TBX3 serum levels (rs = 0,62, n = 24, p = 0,0012). These data suggest that severity of arthritis in the CIA model is associated with an increased serum concentration of TBX3.
Fig. 6.
Kinetics of the expression of the TBX3 protein during development of collagen-induced arthritis (CIA) in the B10.RIII mouse strain. Levels of TBX3 in serum from CIA-immunized mice (a); comparison of serum-TBX3 levels between naïve mice (n = 4), immunized, non-arthritic mice (n = 14) and arthritic mice (n = 6) (b). Statistical analyses were performed using the Mann-Whitney U test; **p < 0.01
Discussion
In this study, we have shown that a 1.4-Mbp locus located on mouse chromosome 5, comprising three protein coding genes and a number of non-protein coding genetic elements (Fig. 1), controls the severity of clinical arthritis in CIA along with anti-CII antibody titers. Based on a literature survey and the link to other bone diseases in humans, we selected Tbx3 and Tbx5, as potential candidate genes for CIA. We demonstrated variations in the regulatory part of the Tbx3 gene, which may affect the observed difference in expression level of Tbx3 in splenic B lymphocytes, in addition to the increased in vitro B cell activation and proliferation upon stimulation through cell-bound IgM. Moreover, our functional data showed that TBX3 activity is inversely related to B cell activation both in vitro and in the CIA model. This suggests that TBX3 is a player in CIA pathogenesis by modulating B cell proliferative responses, but that it does not affect the development of B lymphocytes.
Identification of quantitative trait loci (QTLs), genetic fine-mapping by using congenic and sub-congenic mice, and the discovery of novel candidate genes, have led to the identification of molecular interactions that underlie complex diseases [43–45]. In this study, we fine-mapped a previously reported QTL, Eae39, linked to CIA [8, 10]. By breeding of sub-congenic lines and by using the same immunization protocol, we have shown that the Eae39 region contains a smaller QTL (Eae39r), which encompasses 1.8 Mbp and has an enhancing effect on CIA severity. The genes within the fragment are Med13l, Tbx3, Tbx5, and Rbm19, coding for the mediator complex subunit 13-like protein, the T-box protein 3, T-box protein 5, and RNA binding motif protein 19, respectively. CIA experiments with sub-congenic mice provide an attractive opportunity to study the individual (or combined) contribution of these genes towards arthritis development. We have previously shown that B10.RIIIS/J-Eae39r1 sub-congenic mice (with only Med13l in the sub-congenic fragment) develop arthritis to the same extent as the wild-type B10.RIII mice [46]. In the current study, we showed that B10.RIIIS/J-Eae39r2 sub-congenic mice (with Tbx3, Tbx5 and Rbm19 in the sub-congenic fragment) develop more severe arthritis compared to the control mice. Taken together, it is reasonable to conclude that the CIA-enhancing effect seen in the B10.RIIIS/J-Eae39r mice is due to the genes located in the Eae39r2 fragment and that MED13L has minimal or no effect on CIA development. Additionally, the Eae39r2 region controls the anti-CII antibody response.
Within the Eae39r2 locus, two evolutionarily related genes, Tbx3 and Tbx5, were chosen as potential candidate genes controlling CIA severity based on the consensus criterion recommended by the Complex Trait Consortium [47]. We cannot, however, disregard the involvement of non-coding genes within Eae39r2. In this study we have reported a number of polymorphisms in regulatory regions of the Tbx3 and Tbx5 genes when comparing the B10.RIII and RIIIS/J parental mouse strains. Some of the polymorphisms are predicted to affect the transcription and protein expression levels by modulating promoter activity, transcription factor binding, or microRNA binding. We propose that one or more of these variants are responsible for the allele-specific differences in Tbx3 transcript levels observed in total spleen and splenic B lymphocytes, but not in joints. This can be explained by tissue-specific regulation of Tbx3 that has been previously documented in mice [18] and humans [18, 25].
It has recently been acknowledged that B lymphocytes and autoantibodies are key in aspects of RA pathogenesis [48, 49] and the clinical success of B cell targeted therapies further strengthens this [6, 50]. Although not directly demonstrated, we believe that decreased expression of Tbx3 in B lymphocytes from mice with a 1.4-Mbp fragment derived from a genetically different mouse strain, in addition to the increased in vitro proliferation of these cells in response to anti-IgM stimulation, is a finding that can explain the enhanced antibody response and more severe disease in these mice. We suggest that the increased proliferation of B cells from the B10.RIIIS/J-Eae39r2 sub-congenic mice in response to anti-IgM stimulation, but not to LPS or anti-CD40/IL-4 treatment, might be the result of TBX3 interacting with specific components of the intracellular pathways activated. Anti-IgM antibodies stimulate B cells through the B cell receptor (BCR) thereby activating further downstream signaling cascades like the phosphatidylinositol 3-kinase (PI3K), protein kinase B (AKT) and mitogen-activated protein kinase (MAPK) pathways [51–53]. TBX3 has previously been shown to directly interact with these signaling cascades in other cell types to regulate cell survival and proliferation [54–57] and a similar mechanism might operate in B cells.
The importance of TBX3 in relation to proliferation is underpinned by studies of its role in oncogenesis [24, 25, 58, 59]. TBX3 functions primarily as a transcriptional repressor and initial reports suggested that it can promote proliferation by repressing the cell cycle regulators p14/p19ARF and p21 [60–62]. More recently, it was recognized that although TBX3 promotes migration and invasiveness of cancer cells, it negatively regulates cell proliferation [37, 63]. In line with these reports of an anti-proliferative role for TBX3, our study suggests that it might be controlling cell cycle regulators in B lymphocytes under normal conditions. Lack of, or low expression of Tbx3, may thus lead to increased activation of B lymphocytes, manifested by proliferation and increased autoantibody production, which subsequently would cause severe arthritis in the CIA model. To this end, we demonstrated that the level of functionally active TBX3 in B cells falls after in vitro activation through the BCR. Furthermore, our in vivo results from the time-course CIA study show that TBX3 activity falls significantly when the immune system is challenged through heterologous CII immunization. The lowest TBX3 activity was observed a week after the first injection of immunogen, which corresponds to the time point reported for the highest B cell activation and proliferation of CII-specific IgM-positive B cells in the CIA model [64]. This, again, suggests that TBX3 interacts with signaling pathways downstream of the IgM-BCR, thereby being modulated by, or modulating, the activation signal, resulting in consequences for the outcome of the B cell response.
To our knowledge, this is the first study to suggest the involvement of TBX3 in B cell proliferation and development of CIA, and adds mechanistic links to three independent genome-wide association studies (GWAS) that linked TBX3 to RA [65–67]. Whether the TBX3 genetic variant(s) linked to RA in these studies represents a true susceptibility variant(s) is still to be investigated. Furthermore, our preliminary data show that TBX3 is a putative RA biomarker and that an increase in TBX3 serum levels in untreated arthritis reflects dysregulation of currently unidentified disease pathways. Although the current biomarker study has limitations in terms of group size and that the studies are performed in an experimental model, the conservation of this protein and disease pathways between man and mouse might overcome the discovery step, which is often a bottleneck in the biomarker field. Future studies in human cohorts are needed to prove the biomarker potential of the multifaceted protein, TBX3.
Conclusion
We have fine-mapped the CIA enhancing Eae39r locus and suggest that the therein located Tbx3 gene is a disease candidate gene for experimental arthritis in the mouse. From studies of gene transcription and B cell proliferation, we propose that TBX3 impacts CIA development and autoantibody production by regulating B lymphocyte activation. Taken together, these results indicate that TBX3 is involved in RA pathogenesis and possibly also in other autoimmune diseases dependent on B cell activation and autoantibody production. By investigating intracellular TBX3 protein activity in B cells upon stimulation through the B cell receptor, we have shown that activation of B cells is concomitant with low levels of active TBX3 in vitro. Moreover, we observed that serum levels of TBX3 follow arthritis severity. We therefore propose TBX3 as a candidate biomarker that can potentially aid in diagnosis and assessment of RA disease severity.
Additional file
Table S1. List of primers used for genotyping of mice. Table S2. List of primers for Tbx3 and Tbx5 sequencing. Table S3. List of primers used for real time PCR. (DOCX 22 kb)
Acknowledgements
We thank Durita Poulsen and other members of the Immunopharmacology group for technical assistance, and Mathilde Caldara, Heidi Thousgaard Jensen, and Mette Lynggaard Rådbjerg for taking care of the animals.
Funding
This work was supported by The Danish Rheumatism Association, The AP Møller Research grant for Medical Science and The Oticon foundation. The funding bodies did not contribute in the design of the study, experimental work, data analysis and interpretation, or writing the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- BCR
B cell receptor
- bp
Base pairs
- BSA
Bovine serum albumin
- CIA
Collagen-induced arthritis
- CII
Collagen type II
- DMEM
Dulbecco’s modified Eagle’s medium
- ELISA
Enzyme-linked immunosorbent assay
- FBS
Fetal bovine serum
- FCS
Fetal calf serum
- GWAS
Genome-wide association studies
- IFA
Incomplete Freund’s adjuvant
- IL
Interleukin
- LPS
Lipopolysaccharide
- Mbp
Mega base pair
- MHC
Major histocompatibility complex
- PBS
Phosphate-buffered saline
- QTL
Quantitative trait locus
- RA
Rheumatoid Arthritis
- SNP
Single nucleotide polymorphism
- TBX3
T-box protein 3
- TFBS
Transcription factor binding site
- UTR
Untranslated region
Authors’ contributions
Study concept and design: SS, ÅA. Experimentation and data acquisition: SS, AK, DV, ERM, ÅA. Data analysis: SS, DV, EK, RG. Data interpretation: SS, EK, RG, ÅA. Manuscript writing: SS, ÅA. Manuscript review: SS, AK, DV, ERM, EK RG, ÅA. All authors read and approved the final manuscript.
Ethics approval
All animal experiments included in this study were approved by local ethical authorities (Danish Animal Experiment Inspectorate license number: 2010/561–1920 and 2015-15-0201-00794) and were performed in accordance with standard guidelines on the three Rs; replacement, reduction, and refinement.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Samra Sardar, Email: samrasardar@gmail.com.
Alish Kerr, Email: kerral@tcd.ie.
Daniëlle Vaartjes, Email: daniellevaartjes@hotmail.com.
Emilie Riis Moltved, Email: emoltved@gmail.com.
Edita Karosiene, Email: edita.karosiene@gmail.com.
Ramneek Gupta, Email: rg@bioinformatics.dtu.dk.
Åsa Andersson, Email: asa.andersson@hh.se.
References
- 1.Klareskog L, Padyukov L, Lorentzen J, Alfredsson L. Mechanisms of disease: genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nat Clin Pract Rheumatol. 2006;2(8):425–433. doi: 10.1038/ncprheum0249. [DOI] [PubMed] [Google Scholar]
- 2.Newton JL, Harney SMJ, Wordsworth BP, Brown MA. A review of the MHC genetics of rheumatoid arthritis. Genes Immun. 2004;5(3):151–157. doi: 10.1038/sj.gene.6364045. [DOI] [PubMed] [Google Scholar]
- 3.Kochi Y, Suzuki A, Yamamoto K. Genetic basis of rheumatoid arthritis: a current review. Biochem Biophys Res Commun. 2014;452(2):254–262. doi: 10.1016/j.bbrc.2014.07.085. [DOI] [PubMed] [Google Scholar]
- 4.Besenyei T, Kadar A, Tryniszewska B, Kurko J, Rauch TA, Glant TT, et al. Non-MHC risk alleles in rheumatoid arthritis and in the syntenic chromosome regions of corresponding animal models. Clin Dev Immunol. 2012;2012:284751. doi: 10.1155/2012/284751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor PC, Moore A, Vasilescu R, Alvir J, Tarallo M. A structured literature review of the burden of illness and unmet needs in patients with rheumatoid arthritis: a current perspective. Rheumatol Int. 2016;36(5):685–695. doi: 10.1007/s00296-015-3415-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sardar S, Andersson Å. Old and new therapeutics for rheumatoid arthritis: in vivo models and drug development. Immunopharmacol Immunotoxicol. 2016;38(1):2–13. doi: 10.3109/08923973.2015.1125917. [DOI] [PubMed] [Google Scholar]
- 7.Jirholt J, Cook A, Emahazion T, Sundvall M, Jansson L, Nordquist N, et al. Genetic linkage analysis of collagen-induced arthritis in the mouse. Eur J Immunol. 1998;28:3321–3328. doi: 10.1002/(SICI)1521-4141(199810)28:10<3321::AID-IMMU3321>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 8.Lindvall T, Karlsson J, Holmdahl R, Andersson Å. Dissection of a locus on mouse chromosome 5 reveals arthritis promoting and inhibitory genes. Arthritis Res Ther. 2009;11(1):R10. doi: 10.1186/ar2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beutler B, Du X, Xia Y. Precis on forward genetics in mice. Nat Immunol. 2007;8(7):659–664. doi: 10.1038/ni0707-659. [DOI] [PubMed] [Google Scholar]
- 10.Karlsson J, Zhao X, Lonskaya I, Neptin M, Holmdahl R, Andersson Å. Novel quantitative trait loci controlling development of experimental autoimmune encephalomyelitis and proportion of lymphocyte subpopulations. J Immunol. 2003;170(2):1019–1026. doi: 10.4049/jimmunol.170.2.1019. [DOI] [PubMed] [Google Scholar]
- 11.Davis MA, Larimore EA, Fissel BM, Swanger J, Taatjes DJ, Clurman BE. The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Genes Dev. 2013;27(2):151–156. doi: 10.1101/gad.207720.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adegbola A, Musante L, Callewaert B, Maciel P, Hu H, Isidor B, et al. Redefining the MED13L syndrome. Eur J Hum Genet. 2015;23(10):1308–1317. doi: 10.1038/ejhg.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Tomasini AJ, Mayer AN. RBM19 is essential for preimplantation development in the mouse. BMC Dev Biol. 2008;8:115. doi: 10.1186/1471-213X-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neelamraju Y, Hashemikhabir S, Janga SC. The human RBPome: from genes and proteins to human disease. J Proteome. 2015;2015:61–70. doi: 10.1016/j.jprot.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 15.Papaioannou VE. The T-box gene family: emerging roles in development, stem cells and cancer. Development. 2014;141(20):3819–3833. doi: 10.1242/dev.104471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh TK, Packham E, Bonser J, Robinson TE, Cross SJ, Brook JD. Characterization of the TBX5 binding site and analysis of mutations that cause Holt-Oram syndrome. Hum Mol Genet. 2001;10(18):1983–1994. doi: 10.1093/hmg/10.18.1983. [DOI] [PubMed] [Google Scholar]
- 17.Carlson H, Ota S, Campbell CE, Hurlin PJ. A dominant repression domain in Tbx3 mediates transcriptional repression and cell immortalization: relevance to mutations in Tbx3 that cause ulnar-mammary syndrome. Hum Mol Genet. 2001;10(21):2403–2413. doi: 10.1093/hmg/10.21.2403. [DOI] [PubMed] [Google Scholar]
- 18.Washkowitz AJ, Gavrilov S, Begum S, Papaioannou VE. Diverse functional networks of Tbx3 in development and disease. Wiley Interdiscip Rev Syst Biol Med. 2012;4(3):273–283. doi: 10.1002/wsbm.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson-Brown JJ, et al. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Dev Dyn. 1996;206(4):379–390. doi: 10.1002/(SICI)1097-0177(199608)206:4<379::AID-AJA4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 20.Bickley SRB, Logan MPO. Regulatory modulation of the T-box gene Tbx5 links development, evolution, and adaptation of the sternum. Proc Natl Acad Sci U S A. 2014;111(50):17917–17922. doi: 10.1073/pnas.1409913111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denninger KCM, Litman T, Marstrand T, Moller K, Svensson L, Labuda T, et al. Kinetics of gene expression and bone remodelling in the clinical phase of collagen-induced arthritis. Arthritis Res Ther. 2015;17:43. doi: 10.1186/s13075-015-0531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank DU, Emechebe U, Thomas KR, Moon AM. Mouse Tbx3 mutants suggest novel molecular mechanisms for ulnar-mammary syndrome. PLoS One. 2013;8(7):1–7. doi: 10.1371/journal.pone.0067841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Amelio P, Sassi F. Osteoimmunology: from mice to humans. Bonekey Rep. 2016;5:1–6. doi: 10.1038/bonekey.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lomnytska M, Dubrovska A, Hellman U, Volodko N, Souchelnytskyi S. Increased expression of cSHMT, Tbx3 and utrophin in plasma of ovarian and breast cancer patients. Int J Cancer. 2006;118(2):412–421. doi: 10.1002/ijc.21332. [DOI] [PubMed] [Google Scholar]
- 25.Willmer T, Cooper A, Sims D, Govender D, Prince S. The T-box transcription factor 3 is a promising biomarker and a key regulator of the oncogenic phenotype of a diverse range of sarcoma subtypes. Oncogene. 2016;5(2):e199. doi: 10.1038/oncsis.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thomsen N, Ali RG, Ahmed JN, Arkell RM. High Resolution Melt Analysis (HRMA); a Viable Alternative to Agarose Gel Electrophoresis for Mouse Genotyping. PLoS One. 2012;7(9):e45252. doi: 10.1371/journal.pone.0045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ensembl, the vertebrate genome browser. http://www.ensembl.org/index.html. Accessed 10 Feb. 2018.
- 28.The European Bioinformatics Institute database. https://www.ebi.ac.uk/gxa/home. Accessed 10 Feb 2018.
- 29.PubMed database. Available from: https://www.ncbi.nlm.nih.gov/pubmed/. Accessed 18 Nov 2018.
- 30.Google scholar. Available from: https://scholar.google.com/. Accessed 18 Nov 2018.
- 31.NCBI SNP database. Available from: https://www.ncbi.nlm.nih.gov/SNP/. Accessed 2 June 2018.
- 32.Consite prediction tool. http://consite.genereg.net/cgi-bin/consite. Accessed 10 Jan 2018.
- 33.PROMO predcition tool. http://alggen.lsi.upc.es/. Accessed 10 Jan 2018.
- 34.MicroRNA database. Available from: http://mirdb.org/. Accessed 10 Feb 2018.
- 35.The Mouse Microsatellite markers Database. http://www.informatics.jax.org/marker. Accessed 20 April 2018.
- 36.Dahdah A, Habir K, Nandakumar KS, Saxena A, Xu B, Holmdahl R, et al. Germinal center B cells are essential for collagen-induced arthritis. Arthritis Rheumatol. 2018;70(2):193–203. doi: 10.1002/art.40354. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Weinberg MS, Zerbini L, Prince S. The oncogenic TBX3 is a downstream target and mediator of the TGF-β1 signaling pathway. Mol Biol Cell. 2013;24:3569–3576. doi: 10.1091/mbc.E13-05-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tümpel S, Sanz-Ezquerro JJ, Isaac A, Eblaghie MC, Dobson J, Tickle C. Regulation of Tbx3 expression by anteroposterior signalling in vertebrate limb development. Dev Biol. 2002;250(2):251–262. [PubMed] [Google Scholar]
- 39.Lee HS, Cho HH, Kim HK, Bae YC, Baik HS, Jung JS. Tbx3, a transcriptional factor, involves in proliferation and osteogenic differentiation of human adipose stromal cells. Mol Cell Biochem. 2007;296(1–2):129–136. doi: 10.1007/s11010-006-9306-4. [DOI] [PubMed] [Google Scholar]
- 40.Renard CA, Labalette C, Armengol C, Cougot D, Wei Y, Cairo S, et al. Tbx3 is a downstream target of the Wnt/β-catenin pathway and a critical mediator of β-catenin survival functions in liver cancer. Cancer Res. 2007;67(3):901–910. doi: 10.1158/0008-5472.CAN-06-2344. [DOI] [PubMed] [Google Scholar]
- 41.Lüdtke TH, Rudat C, Wojahn I, Weiss AC, Kleppa MJ, Kurz J, et al. Tbx2 and Tbx3 Act downstream of Shh to maintain canonical Wnt signaling during branching morphogenesis of the murine lung. Dev Cell. 2016;39(2):239–253. doi: 10.1016/j.devcel.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 42.Brand DD, Kang AH, Rosloniec EF. The mouse model of collagen-induced arthritis. Methods Mol Med. 2004;102(1):295–312. doi: 10.1385/1-59259-805-6:295. [DOI] [PubMed] [Google Scholar]
- 43.Bonnet N, Somm E, Rosen CJ. Diet and gene interactions influence the skeletal response to polyunsaturated fatty acids. Bone. 2014;68:100–107. doi: 10.1016/j.bone.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomes-Solecki M, Santecchia I, Werts C. Animal models of leptospirosis: of mice and hamsters. Front Immunol. 2017;8:58. doi: 10.3389/fimmu.2017.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma Y, Bramwell KK, Lochhead RB, Paquette JK, Zachary JF, Weis JH, Teuscher C, Weis JJ. Borrelia burgdorferi arthritis associated locus Bbaa1 regulates Lyme arthritis and K/B×N serum transfer arthritis through intrinsic control of type I IFN production. J Immunol. 2014;193(12):6050–6060. doi: 10.4049/jimmunol.1401746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sardar S, Kanne K, Andersson Å. Analysis of polymorphisms in the Mediator complex subunit 13-like (Med13L) gene in the context of immune function and development of experimental arthritis. Arch Immunol Ther Exp. 2018;66(5):365–377. doi: 10.1007/s00005-018-0516-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, et al. The nature and identification of quantitative trait loci: a community’s view. Nat Rev Genet. 2003;4(11):911–916. doi: 10.1038/nrg1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panayi GS. B cells: a fundamental role in the pathogenesis of rheumatoid arthritis? Rheumatology (Oxford) 2005;44(Suppl 2):ii3–ii7. doi: 10.1093/rheumatology/keh616. [DOI] [PubMed] [Google Scholar]
- 49.Bugatti S, Vitolo B, Caporali R. B cells in rheumatoid arthritis: from pathogenic players to disease biomarkers. Biomed Res. 2014;2014:681678. doi: 10.1155/2014/681678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burmester GR, Feist E, Dörner T. Emerging cell and cytokine targets in rheumatoid arthritis. Nat Rev Rheumatol. 2014;10(2):77–88. doi: 10.1038/nrrheum.2013.168. [DOI] [PubMed] [Google Scholar]
- 51.Scharenberg AM, Humphries LA, Rawlings DJ. Calcium signalling and cell-fate choice in B cells. Nat Rev Immunol. 2007;7:778. doi: 10.1038/nri2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kurosaki T. Regulation of B-cell signal transduction by adaptor proteins. Nat Rev Immunol. 2002;2:354. doi: 10.1038/nri801. [DOI] [PubMed] [Google Scholar]
- 53.Satpathy S, Wagner SA, Beli P, Gupta R, Kristiansen TA, Malinova D, et al. Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol Syst Biol. 2015;11(6):810. doi: 10.15252/msb.20145880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang F, Lee JT, Navolanic PM, Steelman LS, Shelton JG, Blalock WL, et al. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia. 2003;17(3):590–603. doi: 10.1038/sj.leu.2402824. [DOI] [PubMed] [Google Scholar]
- 55.Woyach JA, Johnson AJ, Byrd JC. The B-cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120(6):1175–1185. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peres J, Mowla S, Prince S. The T-box transcription factor, TBX3, is a key substrate of AKT3 in melanomagenesis. Oncotarget. 2014;6(3):1821–1833. doi: 10.18632/oncotarget.2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460(7251):118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- 58.Ito A, Asamoto M, Hokaiwado N, Takahashi S, Shirai T. Tbx3 expression is related to apoptosis and cell proliferation in rat bladder both hyperplastic epithelial cells and carcinoma cells. Cancer Lett. 2005;219(1):105–112. doi: 10.1016/j.canlet.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 59.Fan W, Huang X, Chen C, Gray J, Huang T. TBX3 and its isoform TBX3 + 2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast CANCER CELL LInes. Cancer Res. 2004;64(15):5132–5139. doi: 10.1158/0008-5472.CAN-04-0615. [DOI] [PubMed] [Google Scholar]
- 60.Carlson H, Ota S, Song Y, Chen Y, Hurlin PJ. Tbx3 impinges on the p53 pathway to suppress apoptosis, facilitate cell transformation and block myogenic differentiation. Oncogene. 2002;21(24):3827–3835. doi: 10.1038/sj.onc.1205476. [DOI] [PubMed] [Google Scholar]
- 61.Brummelkamp TR, Kortlever RM, Lingbeek M, Trettel F, MacDonald ME, Van Lohuizen M, et al. TBX-3, the gene mutated in ulnar-mammary syndrome, is a negative regulator of p19ARF and inhibits senescence. J Biol Chem. 2002;277(8):6567–6572. doi: 10.1074/jbc.M110492200. [DOI] [PubMed] [Google Scholar]
- 62.Wensing LA, Campos AH. TBX3, a downstream target of TGF-beta1, inhibits mesangial cell apoptosis. Exp Cell Res. 2014;328(2):340–350. doi: 10.1016/j.yexcr.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 63.Li J, Ballim D, Rodriguez M, Cui R, Goding CR, Teng H, et al. The anti-proliferative function of the tgf-β1 signaling pathway involves the repression of the oncogenic tbx2 by its homologue tbx3. J Biol Chem. 2014;289(51):35633–35643. doi: 10.1074/jbc.M114.596411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palm A-KE, Friedrich HC, Mezger A, Salomonsson M, Myers LK, Kleinau S. Function and regulation of self-reactive marginal zone B cells in autoimmune arthritis. Cell Mol Immunol. 2015;12(4):493–504. doi: 10.1038/cmi.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robert P, Seielstad M, Lee T, D P, Remmers EF, Ding B, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis — a genomewide study. N Engl J Med 2007;357(12):1199–1209. [DOI] [PMC free article] [PubMed]
- 66.Freudenberg J, Lee HS, Han BG, Shin H, Do KYM, Sung YK, et al. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011;63(4):884–893. doi: 10.1002/art.30235. [DOI] [PubMed] [Google Scholar]
- 67.Julia A, Ballina J, Caete JD, Balsa A, Tornero-Molina J, Naranjo A, et al. Genome-wide association study of rheumatoid arthritis in the Spanish population: KLF12 as a risk locus for rheumatoid arthritis susceptibility. Arthritis Rheum. 2008;58(8):2275–2286. doi: 10.1002/art.23623. [DOI] [PubMed] [Google Scholar]
- 68.Kumar P, Franklin S, Emechebe U, Hu H, Moore B, Lehman C, et al. TBX3 regulates splicing in vivo: a novel molecular mechanism for ulnar-mammary syndrome. PLoS Genet. 2014;10(3):e1004247. doi: 10.1371/journal.pgen.1004247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Govoni KE, Lee SK, Chadwick RB, Yu H, Kasukawa Y, Baylink DJ, et al. Whole genome microarray analysis of growth hormone-induced gene expression in bone: T-box3, a novel transcription factor, regulates osteoblast proliferation. Am J Physiol Endocrinol Metab. 2006;291(1):E128–E136. doi: 10.1152/ajpendo.00592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Govoni KE, Linares GR, Chen ST, Pourteymoor S, Mohan S. T-box 3 negatively regulates osteoblast differentiation by inhibiting expression of osterix and runx2. J Cell Biochem. 2009;106(3):482–490. doi: 10.1002/jcb.22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yao C, Yao GQ, Sun BH, Zhang C, Tommasini SM, Insogna K. The transcription factor T-box 3 regulates colony-stimulating factor 1-dependent Jun dimerization protein 2 expression and plays an important role in osteoclastogenesis. J Biol Chem. 2014;289(10):6775–6790. doi: 10.1074/jbc.M113.499210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freudenberg J, Lee H-S, Han B-G, Do SH, Kang YM, Sung Y-K, et al. Genome-wide association study of rheumatoid arthritis in Koreans: population-specific loci as well as overlap with European susceptibility loci. Arthritis Rheum. 2011;63(4):884–893. doi: 10.1002/art.30235. [DOI] [PubMed] [Google Scholar]
- 73.Haas CS, Creighton CJ, Pi X, Maine I, Koch AE, Haines GK, et al. Identification of genes modulated in rheumatoid arthritis using complementary DNA microarray analysis of lymphoblastoid B cell lines from disease-discordant monozygotic twins. Arthritis Rheum. 2006;54(7):2047–2060. doi: 10.1002/art.21953. [DOI] [PubMed] [Google Scholar]
- 74.Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9(1):204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Georges R, Nemer G, Morin M, Lefebvre C, Nemer M. Distinct expression and function of alternatively spliced Tbx5 isoforms in cell growth and differentiation. Mol Cell Biol. 2008;28(12):4052–4067. doi: 10.1128/MCB.02100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, et al. Foxf genes integrate Tbx5 and Hedgehog pathways in the second heart field for cardiac septation. PLoS Genet. 2014;10(10):e1004604. doi: 10.1371/journal.pgen.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nishimoto S, Wilde SM, Wood S, Logan MPO. RA Acts in a coherent feed-forward mechanism with Tbx5 to control limb bud induction and initiation. Cell Rep. 2015;12(5):879–891. doi: 10.1016/j.celrep.2015.06.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang KK, Xiang M, Zhou L, Liu J, Curry N, Heine Suñer D, et al. Gene network and familial analyses uncover a gene network involving Tbx5/Osr1/Pcsk6 interaction in the second heart field for atrial septation. Hum Mol Genet. 2015;25(6):1140–1151. doi: 10.1093/hmg/ddv636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pizard A, Burgon PG, Paul DL, Bruneau BG, Seidman CE, Seidman JG. Connexin 40, a target of transcription factor Tbx5, patterns wrist, digits, and sternum. Mol Cell Biol. 2005;25(12):5073–5083. doi: 10.1128/MCB.25.12.5073-5083.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, et al. A murine model of Holt-Oram syndrome defines roles of the T-Box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106(6):709–721. doi: 10.1016/s0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 81.Moskowitz IPG, Pizard A, Patel VV, Bruneau BG, Kim JB, Kupershmidt S, et al. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131(16):4107–16. doi: 10.1242/dev.01265. [DOI] [PubMed] [Google Scholar]
- 82.Rangkasenee N, Murani E, Brunner RM, Schellander K, Cinar MU, Luther H, et al. Genome-wide association identifies TBX5 as candidate gene for osteochondrosis providing a functional link to cartilage perfusion as initial factor. Front Genet. 2013;4:78. doi: 10.3389/fgene.2013.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Karouzakis E, Trenkmann M, Gay RE, Michel BA, Gay S, Neidhart M. Epigenome analysis reveals TBX5 as a novel transcription factor involved in the activation of rheumatoid arthritis synovial fibroblasts. J Immunol. 2014;193(10):4945–4951. doi: 10.4049/jimmunol.1400066. [DOI] [PubMed] [Google Scholar]
- 84.Kallberg Y, Segerstolpe Å, Lackmann F, Persson B, Wieslander L. Evolutionary conservation of the ribosomal biogenesis factor Rbm19/Mrd1: implications for function. PLoS One. 2012;7(9):e43786. doi: 10.1371/journal.pone.0043786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pausch H, Emmerling R, Schwarzenbacher H, Fries R. A multi-trait meta-analysis with imputed sequence variants reveals twelve QTL for mammary gland morphology in Fleckvieh cattle. Genet Sel Evol. 2016;48(1):14. doi: 10.1186/s12711-016-0190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Campbell IK, Gerondakis S, O’Donnell K, Wicks IP. Distinct roles for the NF-kappaB1 (p50) and c-Rel transcription factors in inflammatory arthritis. J Clin Invest. 2000;105(12):1799–1806. doi: 10.1172/JCI8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao G, Dong W, Meng X, Liu H, Liao H, Liu S. MiR-511 inhibits growth and metastasis of human hepatocellular carcinoma cells by targeting PIK3R3. Tumour Biol. 2015;36(6):4453–4459. doi: 10.1007/s13277-015-3085-z. [DOI] [PubMed] [Google Scholar]
- 88.Wagner EF, Robert E. Fos/AP-1 proteins in bone and the immune system. Immunol Rev. 2005;208:26–140. doi: 10.1111/j.0105-2896.2005.00332.x. [DOI] [PubMed] [Google Scholar]
- 89.Suto A, Tanaka S, Nakajima H. Sox5 and Th17 cell differentiation. Oncotarget. 2015;6(24):19952–19953. doi: 10.18632/oncotarget.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365(Pt 3):561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of primers used for genotyping of mice. Table S2. List of primers for Tbx3 and Tbx5 sequencing. Table S3. List of primers used for real time PCR. (DOCX 22 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.